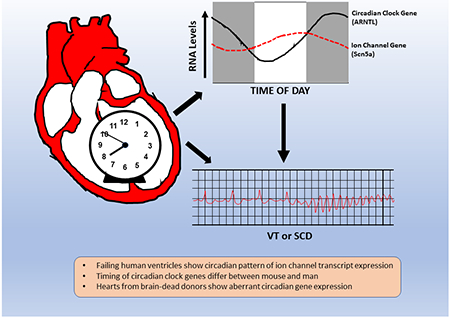
Abstract
Background -
Ventricular tachyarrhythmias (VT) and sudden cardiac death (SCD) show a circadian pattern of occurrence in heart failure patients. In the rodent ventricle, a significant portion of genes, including some ion channels, shows a circadian pattern of expression. However genes that define electrophysiologic properties in failing human heart ventricles have not been examined for a circadian expression pattern.
Methods -
Ventricular tissue samples were collected from patients at the time of cardiac transplantation. Two sets of samples (n=37 and 46, one set with a greater arrhythmic history) were selected to generate pseudo-time series according to their collection time. A third set (n=27) of samples was acquired from the non-failing ventricles of brain-dead donors. The expression of 5 known circadian clock genes and 19 additional ion channel genes plausibly important to electrophysiologic properties were analyzed by RT-PCR, and then analyzed for the percentage of expression variation attributed to a 24 hour circadian pattern.
Results -
The 5 known circadian clock gene transcripts showed a strong circadian expression pattern. Compared to rodent hearts, the human circadian clock gene transcripts showed a similar temporal order of acrophases but with a ~ 7.6 hours phase shift. Five of the ion channel genes also showed strong circadian expression. Comparable studies of circadian clock gene expression in samples recovered from non-heart failure brain-dead donors showed acrophase shifts, or weak or complete loss of circadian rhythmicity, suggesting alterations in circadian gene expression.
Conclusions -
Ventricular tissue from failing human hearts display a circadian pattern of circadian clock gene expression, but phase-shifted relative to rodent hearts. At least 5 ion channels show a circadian expression pattern in the ventricles of failing human hearts, which may underlie a circadian pattern of VT/SCD. Non-failing hearts from brain-dead donors show marked differences in circadian clock gene expression patterns, suggesting fundamental deviations from circadian expression.
Keywords: left ventricle, heart failure, ion channel, circadian rhythm, human, Basic Science Research, Arrhythmias
Graphical Abstract
Introduction
Arrhythmias (VT or Ventricular Tachyarrhythmias, SCD or Sudden Cardiac Death) are a major source of mortality in patients with heart failure of both ischemic and non-ischemic origin. In the United States ~ 5.7 million patients have heart failure, leading to ~ 300,000 deaths in 2017.1 It is estimated that 20-50% of patients with congestive heart failure (CHF) succumb to lethal arrhythmias.2 A circadian pattern of sudden cardiac death and ventricular tachyarrhythmias has been observed, with major peaks reported in the morning after awakening, afternoon/evening, or both, although some reports have failed to observe this pattern.3–9 However, even normal hearts display a circadian pattern of ECG parameters.10
The mechanisms of arrhythmogenesis in CHF involve multiple dynamic processes. Among these, proarrhythmic changes in ion currents are observed in cardiomyocytes isolated from patients and animal models with CHF.11–15 Although the expression of RNA transcripts encoding ion channels that define specific currents are reported to be altered in failing relative to non-failing human hearts, there is no consensus as to which transcripts change, or their directional changes.16 While several factors could generate such disparities (age/sex, disease etiology, anatomic site), studies typically do not consider the time of human specimen acquisition on the expression of genes relevant arrhythmias. Thus the question arises; does a circadian expression pattern of genes relevant to electrophysiologic properties and arrhythmias exist in the human heart?
Gene expression analyses shows that 2-13% of all rodent cardiac gene transcripts display circadian expression.17 A core set of circadian clock genes (CLOCK, ARNTL, PER1-3, CRY, and NRD1D1) define the molecular clock in most tissues18. However most circadian-expressed transcripts are not part of the circadian clock mechanism, but likely underlie circadian variation in heart rate, contractile function, responses to adrenergic stimulation, workload, and metabolic substrate utilization. Indeed, rodent hearts show a circadian expression patterns for RNAs encoding sodium channels, potassium channel and subunits, and calcium channels/exchangers, and studies have linked circadian changes in electrophysiologic parameters and/or arrhythmogenic vulnerability to gene expression changes.19–25
While the expression of several circadian clock gene transcripts in human cardiac ventricular and atrial tissues has been reported26, 27 no studies have specifically examined the circadian expression of genes encoding proteins that define electrophysiologic properties in human ventricles. As rodents and humans display differential contributions of ion channels to electrophysiologic properties, analysis of human tissues is essential to understanding human pathophysiology.28 In this report we utilized LV free wall ventricular tissue recovered from end-stage heart failure patients at the time of cardiac transplantation. With collection times dispersed throughout one 24 hour interval, we measured the expression of 5 transcripts reported to have a robust circadian expression pattern in the myocardium of other species, and 19 ion channel gene transcripts, so as to generate a pseudo-time series of their expression in human LV. Our results confirmed the expression of circadian clock genes expressed in the human heart, compared their expression to the reported circadian parameters of homologous genes in rodent hearts, and identified at least 5 ion channel gene transcripts that show a circadian expression.
Methods
The data, methods of analysis, and materials that support the findings of this study, are available from the corresponding author upon reasonable request.
Patients/tissues
Full-thickness transmural LV tissue was collected at the time of cardiac transplantation from two sets of end-stage heart failure patients (F1 and F2), who were consented under the University of Pittsburgh Institutional Review Board approved protocol 0404033. Patient medical records were reviewed for history of arrhythmias (ventricular tachycardia or fibrillation), and atrial or ventricular pacing. Set F1 included 15 patient cases with a history of arrhythmic burden (sustained ventricular tachycardia or fibrillation, or pacing frequencies >5%). Set F2 arose from patients with no history of arrhythmic burden. Similar LV samples were obtained from brain-dead donors who did not have heart failure (set NF), and whose hearts were not suitable as donors for cardiac transplantation. Samples from brain-dead patients were collected under protocols approved by Institutional review boards at the University of Pennsylvania (Gift of Life Donor Program) and the Cleveland Clinic Foundation (IRB 2378). Patient population characteristics (mean+/−SD) are presented in Table 1.
Table 1.
Demographics of Failing and Non-Failing Patient Populations
| SET | (N) | AGE | SEX | RACE | NICM/ISCH | VAD | ARRHYTHMIC HX |
|---|---|---|---|---|---|---|---|
| F1 | 37 | 49.2+/12.3 | 23M/14F | 25W/6B/1H/5ND | 35/2 | 5 | 15 AP or VP >5% or sustained VT/VF |
| F2 | 45 | 45.1+/14.3 | 29M/17F | 34W/7B/1H/1A/3ND | 44/2 | 17 | 11 Non-sustained VT |
| SET | (N) | AGE | SEX | RACE | LVEF (n=15) | ISCHEMIC TIME (min) | BRAIN DEATH to X-CLAMP (hrs) (n=19) |
| NF | 27 | 53.2+/−14.5 | 8M/19F | 14W/11B/2H | 59.3+/−12.2 | 63.9+/−22.4 | 22.8+/−16.9 Range; 4-74 Median 18 |
F1, Heart Failure Set 1; F2, Heart Failure Set 2; NF1, Non- failing set. M; male. F; female. W; White. B; Black. H; Hispanic. A; Asian. ND; No Data. VAD; ventricular assist device supported. HX; history. AP; atrial pacing. VP; ventricular pacing. VT; ventricular tachyarrhythmia. VF; ventricular fibrillation. NICM; non-ischemic cardiomyopathy. ISCH; Ischemic cardiomyopathy. LVEF; LV ejection fraction. X-clamp; aortic cross-clamping
RNA, RT-PCR
RNA was isolated (RNEasy, Qiagen, Germantown MD), and integrity confirmed by agarose gel electrophoresis. RNA was copied to cDNA (Applied Biosystem High Capacity, Foster City, CA), and used in fluorescence-based kinetic real-time PCR (Applied Biosystems model 7000). PCR primer sequences/sources are listed (Table s1). Reactions were run in duplicate, corrected for amplification efficiency, and normalized to the expression of the reference transcript GAPDH using the δδCt method and calculated relative to the mean each group (F1, F2, or NF).
Data Analysis
Correction sample time collection from time of day to Hours After Sunrise
For each sample we calculated a time of collection relative to time of sunrise for the date of collection. Acrophase (time of peak expression) data is reported as hours after sunrise (HASR) with time of sunrise =0. See Online methods for expanded details.
Rayleigh and Rao Spacing tests
HASR time of collection was converted from a 24 hour clock time to degree on a 360° circle, with HASR time 0/24 equal to degrees 0/360. The Rayleigh test, used to test the null hypothesis that the time of collection follows a uniform distribution (versus the alternative hypothesis of a unimodal distribution), revealed a uniform distribution of sample collection times in all groups (F1, F2, or NF). The Rao spacing test statistic, used to test the null hypothesis that sample collection times were evenly distributed around the clock, showed that sample groups had a uniform temporal distribution (see Table s2).
Determination of Circadian Rhythmicity
To smooth individual sample variation of expression data that could obscure detection of circadian rhythms, the mean value from a moving window of gene expression data (using 9 temporally-consecutive samples for F1, 11 for F2, and 7 for NF) was calculated, generating 37 (F1), 45 (F2), or 27 (NF1) moving window mean value data-points per transcript for each sample set. This data was then analyzed by Chronos-Fit v1.06 software29, which utilizes a partial Fourier-analysis and a stepwise regression technique, and calculates the F-fit statistic (significance of fit to a circadian expression pattern with a period length of 24 hours), acrophase (time of peak expression), MESOR (Midline Estimating Statistic of Rhythm, a rhythm adjusted mean), amplitude ((fitted peak expression value-fitted minimum expression value)/2), and percent rhythm (percentage of the data variance accounted for by the fitted cosine function).
Comparison of Human and Mouse Acrophases
The expression acrophase of known circadian genes in mouse hearts30 and human brains31 were compared to circadian gene acrophases determined in this report by calculation of circular correlation coefficients.32
Results
Temporal Expression of Circadian Clock Genes in Two Sets of Failing Human Heart Tissues
We assessed the expression of core circadian clock genes (ARNTL, DBP, NFIL3, NR1D1, PER2) in a two groups of LV samples (F1; n=37; F2; n=45) collected from patients at the time of orthotopic heart transplantation. Table 1 shows patient demographics; Figure 1 shows temporal distribution of sample collection time as hours after sunrise (HASR). All patients had end-stage heart failure, predominantly of non-ischemic origin. Set F1 (with a greater arrhythmic history) and set F2 (with no significant arrhythmic history), were separately analyzed using two different sets of gene-specific RT-PCR primers (Table s1). Figure 2 shows averaged expression for each gene (ARNTL, DBP, NFIL3, NR1D1, PER2) independently determined from the two sets of samples (2A, set F1; 2B, set F2). (Note NFIL3 was not examined in the F1 set). Table 2 shows the circadian expression parameters for these core circadian clock genes in the two sample sets, yielding nearly identical acrophases for ARNTL, DBP, NR1D1, and PER2.
Figure 1.
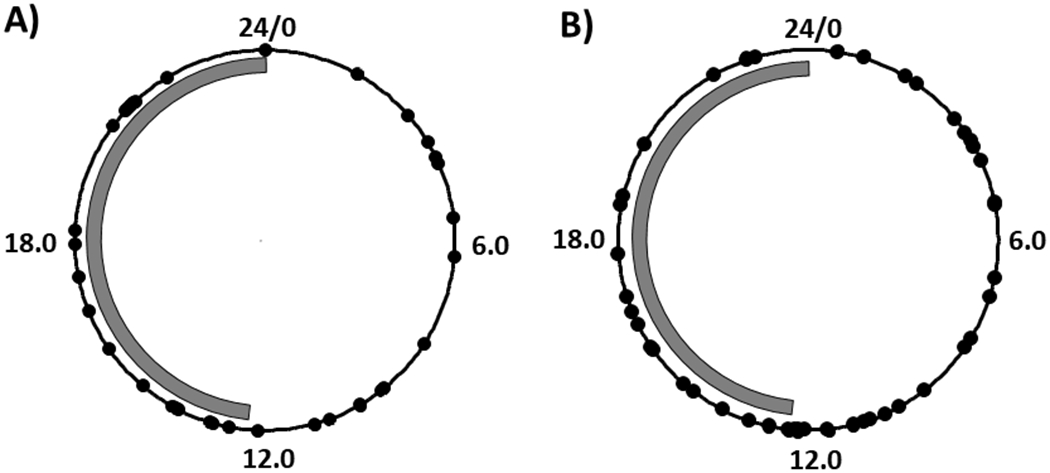
Time of collection of failing human heart samples. A) Set 1; n=37. B) Set 2; n=45. Data in hours after sunrise (HASR). Each sample represented by one dot. Gray bar represents approximate yearly average length of night in Pittsburgh PA (11.75 hours).
Figure 2.
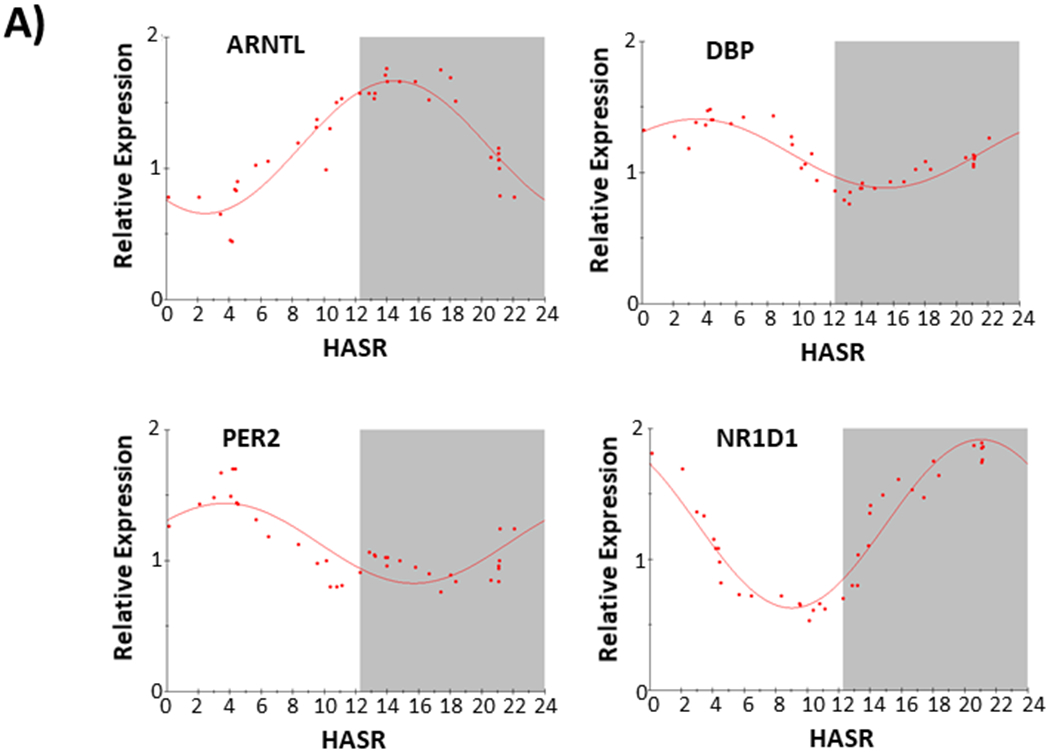
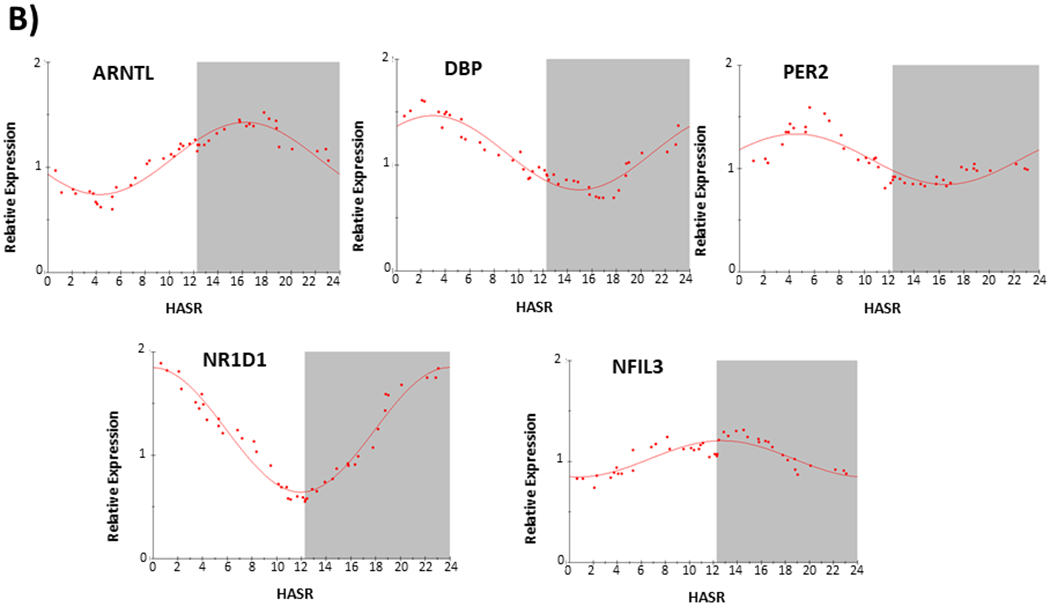
Circadian expression of core circadian clock gene (ARNTL, DBP, NFIL3, NR1D1, and PER2) transcripts in failing human hearts. A) Data from F set 1, n=37. B) Data from F set 2, n=45. HASR; hours after sunrise. Gene transcript in upper left corner of each plot. Each data point (organized according to collection time in HASR) is calculated from (A, F set 1) the mean of 9 consecutive samples, with 37 datapoints calculated from 37 samples, or (B, F set 2) the mean of 11 consecutive samples, with 45 datapoints calculated from 45 samples. The best fit from partial Fourier-analysis (calculated using Chronosfit 1.06) is represented by the solid line. Approximate light/dark interval (based upon approximate yearly average length of night in Pittsburgh PA) is represented with dark-phase in gray.
Table 2.
Circadian Expression Parameters of Core Circadian Clock Transcripts in Human Failing Hearts. Circadian Expression Parameters of NR1D1, ARNTL, DBP, PER2, NFIL3 Transcript Expression in Two Sets of Failing Human Hearts. Acrophase reported in fractional Hours after Sunrise. % Rhythm; percentage of variation ascribed to a circadian expression with one 24 hour period. MAX-MIN; difference between the curve fitted peak and trough values. MESOR is the mean of the curve fitted values. NFIL3 analysis was not performed on set 1.
| F HEART SET | GENE | ACROPHASE | % RHYTHYM | MAX-MIN | MESOR |
|---|---|---|---|---|---|
| 1 | NR1D1 | 23.92 | 95.6 | 1.21 | 1.24 |
| 2 | NR1D1 | 21.01 | 91.9 | 1.29 | 1.27 |
| 1 | ARNTL | 16.25 | 91.8 | 0.69 | 1.08 |
| 2 | ARNTL | 14.47 | 87.0 | 1.01 | 1.16 |
| 1 | DBP | 2.95 | 89.9 | 0.7 | 1.11 |
| 2 | DBP | 3.47 | 81.8 | 0.53 | 1.14 |
| 1 | PER2 | 4.51 | 72.6 | 0.49 | 1.09 |
| 2 | PER2 | 3.66 | 68.4 | 0.61 | 1.13 |
| 1 | NFIL3 | 12.68 | 72.3 | 0.36 | 1.02 |
| 2 | NFIL3 | ND |
In addition the acrophase for ARNTL (14.47 to 16.25 HASR, or an average Time of Day (TOD) of 20.77 to 22.55) and PER2 (3.63 to 4.51 HASR, average TOD 9.93-10.8) transcript expression closely approximated those local TOD acrophases previously reported for ARNTL (21.31 TOD) and PER2 (9.48 TOD) in human heart LV samples26 (see Supplement for comment on relating HASR to TOD).
The Cardiac Circadian Clock is Phase Advanced in Human Hearts Relative to Rodent Hearts
Using data generated from the F2 sample set, the acrophases from the five core clock gene transcripts were also compared to that reported for rodent studies in Table 1 of Yan et al30. Wheel and spoke plots and circadian correlation coefficients (Fig 3a) showed a similar temporal order of core circadian clock genes expression between failing human LV and normal mouse hearts (Circular Correlation Coefficient = 0.677), with human heart core circadian clock acrophases being advanced by ~7.6 hours relative to that of homologous transcripts in mouse hearts. Interestingly a similar temporal offset (human advanced by ~6.5 hrs) was observed between a set of core circadian clock genes examined in human prefrontal cortex and mouse whole brains.31 When compared to data reported from human post-mortem brain samples (Fig 3b), human heart core circadian clock acrophases appeared ~ 1.56 hours advanced (but with a weaker correlation; Circular Correlation Coefficient = 0.227).
Figure 3.
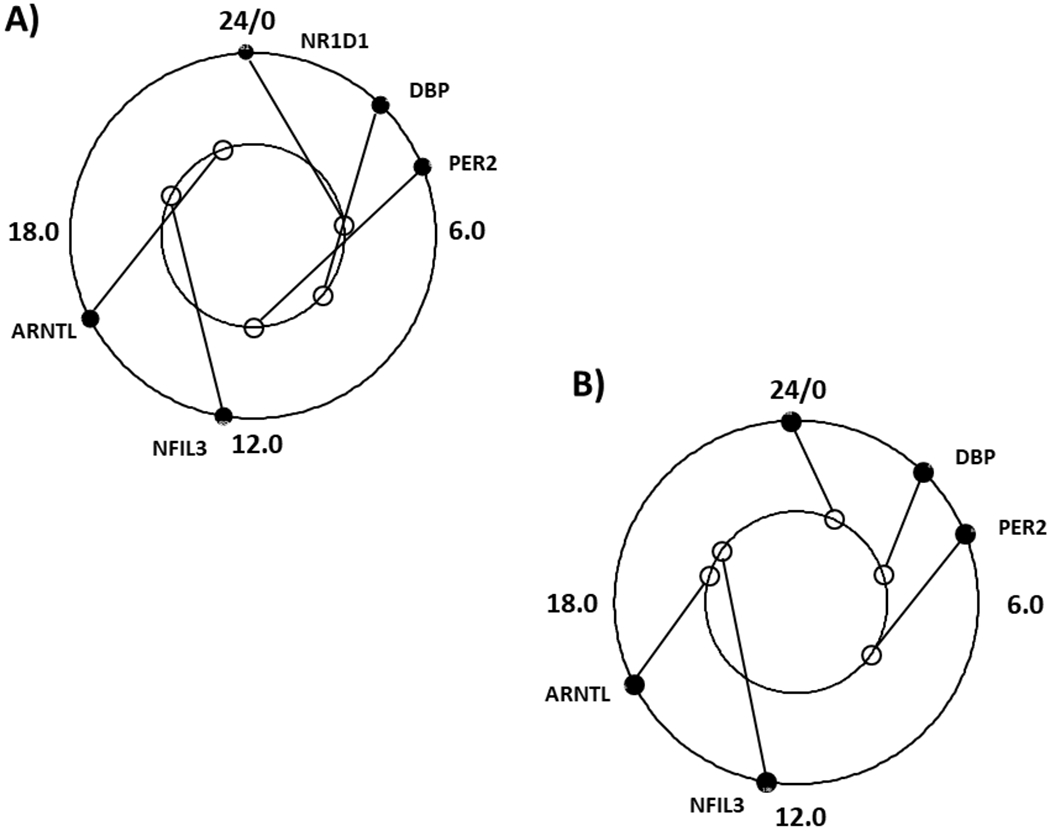
Comparison of core circadian clock transcript expression in human heart to mouse hearts or human brain. A) Outer circle; Human heart (data this report), inner circle Mouse heart (data from Table 1, Yan et al 30). B) Comparison of core circadian clock gene transcript expression in human heart (data this report) to human brains (average of all brain regions from Figure 5S in Li et al31. Outer circle human heart; inner circle human brain. Circadian clock transcripts labelled ARNTL, DBP, PER2, NFIL3, NR1D1. Reference hours after sunrise (HASR) are numbered.
Circadian Expression of Ion Channel Gene Transcripts in Failing Human hearts
We also examined the expression of 19 different gene transcripts arising from ion channel genes or genes plausibly related to cardiac arrhythmias in the F2 sample set. Five gene transcripts (SCN5A, KCNA5, KCNB1, SLC8A1, KCNA4) showed strong evidence of a circadian expression pattern (~70% or more expression variance ascribed to a 24 hour circadian pattern; Figure 4).
Figure 4.
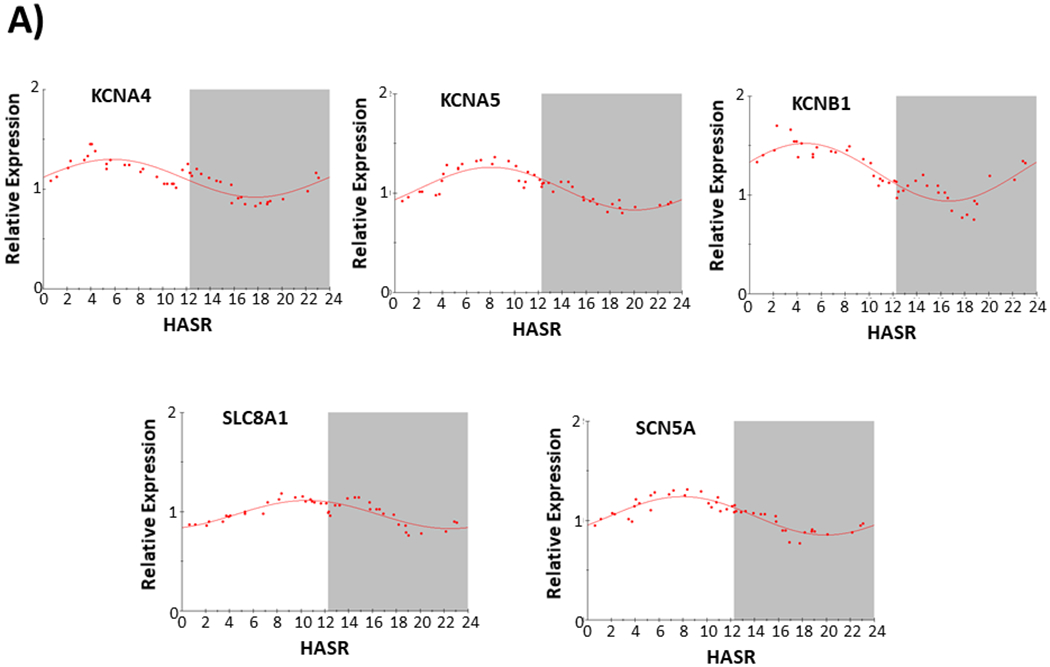
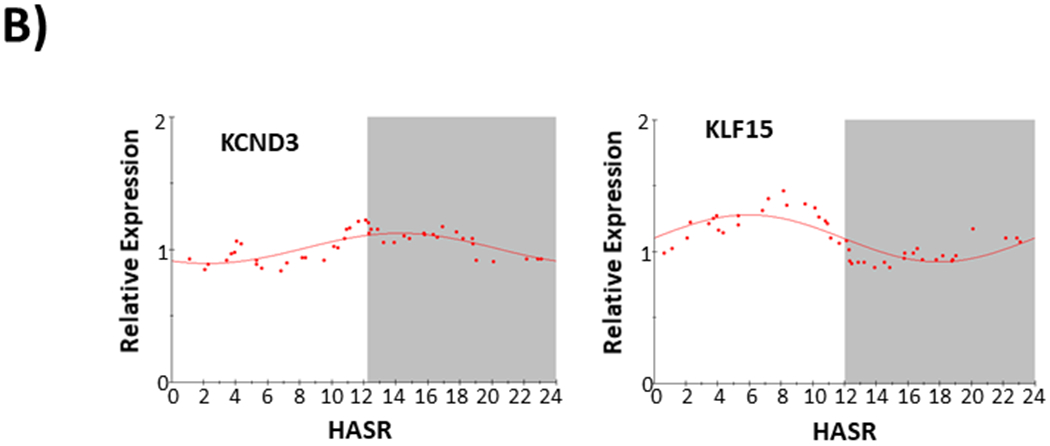
Circadian Expression of ion channel and related transcripts (KCNA5, KCNA4, KCNB1, SCN5A, SLC8A1, KCND3, KLF15) in failing human hearts. Abbreviations and data analysis are as described in Figure 2 legend.
Transcripts from an additional two genes show a circadian pattern of expression (KFL15 and KCND3) with ~ 60% expression variance ascribed to a 24 hour circadian pattern. All other gene transcripts (summarized Table 3) showed a lesser (56.9 to 16.8%) expression variance ascribable to a circadian pattern. In failing human hearts, we could not detect a significant circadian pattern of expression for KCNH2, reported to have a strong circadian expression pattern in normal rodent hearts.21
Table 3. Circadian Expression Parameters of Ion Channel Genes or Genes Plausibly Relevant to Cardiac Arrhythmias.
Circadian Expression Parameters of Transcripts Relevant to Arrhythmia in Failing Human Hearts. Circadian parameters as described in Table 2.
| GENE | ACROPHASE | % RHYTHYM | MAX-MIN | MESOR |
|---|---|---|---|---|
| SCN5A | 7.9 | 83.7 | 0.39 | 1.04 |
| KCNA5 | 8.05 | 82.9 | 0.43 | 1.04 |
| KCNB1 | 4.62 | 81.3 | 0.58 | 1.23 |
| SLC8A1 | 10.5 | 73.3 | 0.29 | 0.97 |
| KCNA4 | 5.78 | 69.6 | 0.38 | 1.11 |
| KLF15 | 5.9 | 62.4 | 0.36 | 1.1 |
| KCND3 | 14.2 | 59.4 | 0.23 | 1.01 |
| KCNJ8 | 5.99 | 56.9 | 0.44 | 1.08 |
| ATPA2A | 6.2 | 55.1 | 0.26 | 1.04 |
| SIRT1 | 2.94 | 54.9 | 0.36 | 1.13 |
| CACNA1C | 6.3 | 54.1 | 0.13 | 1.01 |
| KCNJ2 | 23.49 | 46.5 | 0.22 | 1.03 |
| KCNC4 | 10.2 | 44.2 | 0.3 | 1.15 |
| KCNQ1 | 9.86 | 43.5 | 0.23 | 1.06 |
| SCN1B | 2.11 | 38.8 | 0.16 | 1.05 |
| KCNH2 | 8.89 | 33.7 | 0.1 | 1.02 |
| KCNS3 | 9.2 | 32.7 | 0.2 | 1.09 |
| KCNK1 | 5.73 | 29.2 | 0.23 | 1.08 |
| KCNA7 | 23.12 | 16.8 | 0.2 | 1.19 |
Attenuation and Loss of Circadian Clock Rhythmicity in Non-failing Hearts from Brain-dead Donors
We performed similar analysis on a set (NF) of 27 non-failing hearts that were not suitable for use in cardiac transplantation. Table 1 describes demographics of patients (and Fig s1 shows time of sample acquisition). Relative to the F2 set, the NF set showed marked acrophase shifts (ARNTL, NR1D1), or complete loss of circadian rhythmicity (NFIL3, PER2), in 4 of the 5 core circadian clock gene transcripts (Figure 5). To test whether the smaller sample size of the NF set (n=27) precluded the detection of circadian patterns of gene expression observed in the larger F2 set (n=45), we analyzed a subset (n=27) of F2 samples (F2/27) selected to closely match the collection time of the NF sample set (Fig s2). While the smaller subset of failing samples attenuated the strength of calculated rhythmicity, a circadian expression of circadian clock gene transcripts was still detectable in the F2/27 subset resembling that observed in the larger F2 (n=46) sample set, suggesting that the smaller set of NF samples did not necessarily preclude detection of circadian patterns of gene expression (Table 4).
Figure 5.
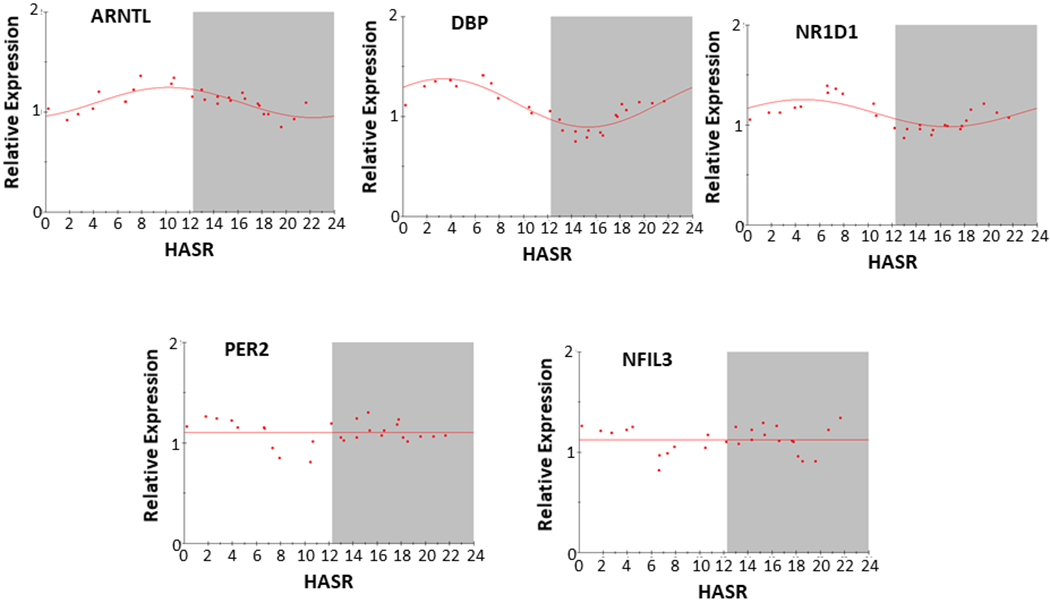
Circadian expression of core circadian clock gene (ARNTL, DBP, NFIL3, NR1D1, and PER2) transcripts in non-failing human hearts. Abbreviations and data analysis are similar to that described in Figure 1 legend. Here, each data point is calculated from the mean of 7 consecutive samples, with 27 datapoints calculated from 27 samples.
Table 4.
Circadian Expression Parameters of ARNTL, DBP, NFIL3, NR1D1, PER2 Transcript Expression in Non-failing Human Hearts (NF, N=27) and a Sample-size Matched Sub-set of Failing Human Hearts (F2/27) Derived from the F2 Set (where N=45). Circadian parameters as described in Table 2.
| GENE | F2 ACROPHASE | % RHYTHM | F2/27 ACROPHASE | % RHYTHM | NF ACROPHASE | % RHYTHM |
|---|---|---|---|---|---|---|
| NR1D1 | 23.92 | 95.6 | 23.14 | 87.7 | 4.61 | 53.9 |
| ARNTL | 16.25 | 91.8 | 14.56 | 72.83 | 10.2 | 62.2 |
| DBP | 2.95 | 89.9 | 2.76 | 85.71 | 3.4 | 80.8 |
| PER2 | 4.51 | 72.6 | 2.16 | 60.81 | none | 0 |
| NFIL3 | 12.68 | 72.3 | 12.13 | 44.32 | none | 0 |
Discussion
Circadian biology influences multiple aspects of human cardiac physiology, with circadian variations in heart rate, blood pressure, sympathetic and parasympathetic regulation, and cardiac function.33 The incidence of myocardial infarction, arrhythmias, and SCD are often, though not always, reported to show a circadian pattern of occurrence.3–9, 34 Circadian perturbations that occur in shift workers and after conversion to ‘summer’ or daylight savings times are also associated with increased incidences of myocardial infarctions and atrial fibrillation.35, 36
This study sought to test the hypothesis that circadian patterns of ion channel transcript expression occur in failing human hearts, potentially providing mechanistic insight into circadian patterns of arrhythmias. To perform these analyses, we examined transcript levels using a pseudo-time series of samples generated from failing human hearts collected at the time of excision for cardiac transplantation. We compared results on core circadian clock gene expression to that reported for mouse hearts and human brains. We also sought to compare circadian gene expression patterns between failing and non-failing human hearts. These studies generated 4 primary results; 1) we confirmed the TOD circadian pattern of ARNTL and PER2 transcript acrophases previously reported for human cardiac ventricles, and extended these findings to additional core circadian clock gene transcripts (DBP, NR1D1, NFIL3); 2) relative to mouse hearts, human cardiac ventricles show a ~ 7.6 hours acrophase advance in a set of core circadian clock gene transcripts, suggesting a fundamental circadian clock phase shift between these nocturnal and diurnal species; 3) failing human cardiac ventricles show strong evidence for a circadian pattern of ion channel transcript expression (KCNA5, KCNA4, KCNB1, SCN5A, SLC8A1), and 4) left ventricles from brain-dead organ donors show phase-shifted, weaker, or absent circadian rhythmicity for core circadian clock gene transcripts, suggesting a fundamental alteration in their circadian gene expression pattern.
Leibdetseder et al first reported the expression of core circadian clock genes (ARNTL, PER1, PER2) in human cardiac ventricles, with temporal data reported as local time of day.26 Recently the circadian expression pattern of thousands of genes in multiple post-mortem ‘normal’ human tissues (including human atria but not ventricles) was reported, referencing them to the ARNTL2 acrophase.27 In general, our results for the acrophase of circadian clock gene transcripts in human ventricles, reported as HASR (similar to rodent studies that reference hours after light onset), agree with those reports, once corrected for the temporal reference utilized.
We also compared the acrophase for these circadian clock gene transcripts in human failing left ventricle to that reported for these transcripts in normal mouse ventricles30, where we observed that human heart circadian clock gene transcripts showed a composite phase advance of ~ 7.6 hours. Intriguingly, comparison of the same circadian clock gene expression acrophases between mouse and various human brain regions (obtained from recently deceased individuals) showed a similar ~6.5 hour phase advance of human brain circadian clock gene transcripts.31 We also observed the expression of core circadian clock genes in human heart to be phase advanced compared to human brain, similar to that observed between mouse heart and mouse brain (data not shown30, 31). While differences between diurnal (human) and nocturnal (mouse) species in the phase of circadian clock gene expression may not be unexpected, the magnitude of phase shift (~7.6 hours) is perhaps unexpected considering a 12 hour difference in activity onset relative to light onset.
The circadian patterns of normal cardiac electrical function and pathologic arrhythmias has led to the examination of circadian patterns of ion channel expression in rodent species. Various ion channel transcripts (including Kcna5, Kcnd2, Scna5, Slc8a1, Kcnh2, Kcnip2, and Kcnk3) show a circadian expression pattern in rodent hearts, with differences in technical or statistical/analytic methods proposed to account for variances in observed patterns.19–25 Regardless, circadian variation in ion channel expression is hypothesized to contribute to circadian differences in electrical activity and propensity for cardiac arrhythmias. Recognizing that the ion channels that influence murine arrhythmias may not accurately reflect those involved in human cardiac arrhythmias28, we examined a panel of 19 different gene transcripts plausibly related to electrical properties and arrhythmia in failing human hearts. We observed a circadian expression pattern of transcripts for KCNA5, SCNA5, SLC8A1, and KCND3, as well as KLF15, a transcription factor that regulates murine cardiac Kcnip2 expression. We also observed circadian expression patterns for KCNA4, and KCNB1, (not previously reported for rodents), but could not detect circadian expression patterns for KCNH221 in failing human hearts. In addition, while Kcnip2 is reported to have circadian expression in normal mouse hearts22, we could only detect very low KCNIP2 transcript levels (data not shown), inadequate to determine circadian properties, but consistent with reports that KCNIP2 transcript levels are markedly reduced in failing human cardiac ventricles.37 Whether this reflects differences in failing/non-failing hearts, or species differences between humans and mice, cannot be ascertained with the current dataset.
Importantly most transcriptional profiling studies of human myocardium do not consider the time of tissue acquisition, critical if 10% or more of all genes show a circadian expression pattern.17, 27 For example, a prior report observed differences in whole transcriptome expression patterns between tissue recovered from failing hearts at the time of ventricular assist device (VAD) placement and cardiac transplantation, suggesting differences in the underlying disease processes leading to these two therapeutic endpoints.38 However, based upon the experience at our institution, most VAD core tissues are acquired mid-morning (~ 10 AM) during scheduled surgeries, whereas cardiac transplantations occur at all hours, with notable bias towards late evening hours (~ 10 pm), suggesting a potential temporal confounder when comparing tissues acquired at VAD implantation to cardiac transplantation.
Indeed, temporal differences in cardiac gene expression are associated with differential susceptibility to surgery-associated cardiac ischemia-reperfusion injury. Relative to morning surgery (~9 AM), afternoon surgery (~3 PM) was associated with a significantly lower incidence of major adverse events, and lower perioperative release of cardiac troponin.39 Transcriptomic analyses of human atrial samples acquired in morning or afternoon surgeries revealed that NR1D1, PER2, and DBP were expressed at lower levels in the morning, while NFIL3 and ARNTL2 were expressed at higher levels in the afternoon. While that data was not acquired with a 24 hour ‘time series’, if one compares those observations from atrial tissues to the 3 HASR (~ 9 AM) and 9 HASR (~ 3 PM) data in this report from ventricular tissues (see Fig2A), a congruent trend is observed (NR1D1, PER2, DBP higher in the morning; ARNTL, NFIL3 higher in the afternoon).
When compared to data in failing human hearts (this manuscript) and reports regarding circadian clock gene expression in normal and diseased rodent hearts, hearts from brain-dead patients maintained as organ donors have significant alterations in the temporal expression patterns of at least some core circadian clock genes. While the use of cardiac tissue from brain-dead donors for control ‘non-failing’ samples in transcriptome studies represent the ‘best available’ reference, caution should be considered. Most organ donors have suffered traumatic brain injury/herniation as the ultimate cause of death. Organ donors, which may be maintained for over 60 hours after declaration of brain death, have alterations in their adrenal-pituitary axis function, receive pharmaceutical/hormonal support to maintain thermoregulation, kidney function, blood glucose levels, and blood pressure, and are exposed to altered environmental cues of light and feeding/nutritional support, all interventions which could modulate circadian biology.40, 41 While alterations in circadian gene expression and physiologic processes are observed in animals and patients with severe brain injury,42, 43 this appears to be the first report of alterations in cardiac circadian clock gene expression in brain-dead donors.
Limitations
We acknowledge several limitations of this study. This is an observational study limited to specimens collected at the time of surgery and not subjected to experimental manipulations. We did not differentiate cases according to more specific etiologies of heart failure, assess the effects of sex or age on human cardiac circadian rhythms or ion channel expression, or refine the anatomic (epi/mid/endomyocardium) location of the LV samples which may influence ion channel expression levels.37 In addition, the functional importance of variation in transcript levels depend upon post-translational processes not examined here.44 We could not compare our analyses of circadian parameters of these transcripts in failing hearts to non-failing samples as we made the novel observation that non-failing hearts procured from brain-dead patients appear to have significant alterations in their circadian mechanisms. A smaller number of non-failing heart samples weakens, but does not completely obscure, the ability to detect circadian expression patterns of known circadian clock genes.
Conclusions
This study reports the circadian expression pattern of key circadian clock gene and ion channel gene transcripts in the left ventricle of failing human hearts. Relative to mouse hearts, circadian clock gene transcript acrophases are phase advanced (~7.6 hours) in the failing human hearts, revealing a fundamental temporal difference between these diurnal and nocturnal species. A circadian pattern of transcript expression for several ion channel genes exists, revealing a possible mechanistic link to a circadian pattern of arrhythmias in humans with endstage heart failure of non-ischemic origin. Cardiac tissue from ‘non-failing’ brain-dead donors display weaker and/or altered temporal pattern of circadian clock gene transcripts, suggesting alterations of circadian function in human tissues typically utilized as ‘normal’ controls. Such results are important in aligning findings between human and rodent model tissues and reveal limitations in utilizing non-failing cardiac tissue from brain-dead donors as ‘controls’ for failing heart tissues. Studies that refine the stated limitations, examine mechanisms that effect circadian ion channel expression and activity, and expand the investigation of small molecule modulators of the circadian clock mechanism, may reveal novel approaches to modulate circadian arrhythmias.
Supplementary Material
What Is Known?
In rodent hearts, both circadian clock genes and some ion channel genes relevant to arrhythmias display circadian expression.
Failing human heart ventricles, reported to show a circadian pattern of ventricular tachyarrhythmias (VT) and sudden cardiac death (SCD), have not been examined for a circadian expression of genes whose proteins define electrophysiologic properties.
What the Study Adds?
Ventricular tissue from failing human hearts displays a circadian pattern of circadian clock gene expression, but phase-shifted relative to rodent hearts.
At least 5 ion channel genes show a circadian expression pattern in the ventricles of failing human hearts, plausibly relating to a circadian pattern of VT/SCD.
Non-failing hearts from brain-dead donors show marked differences in circadian clock gene expression patterns, suggesting fundamental deviations from circadian expression after brain death.
Acknowledgments:
We thank Andrew Althouse PhD for assistance with statistical analyses, the Heart and Vascular Institute (HVI) nurse coordinators for acquiring patient consents, and the cardiothoracic surgical team for assisting in sample acquisition.
Sources of Funding: Research was performed with institutional support from the HVI, University of Pittsburgh, and National Institutes of Health Grants UL1 RR024153, UL1TR000005 and R01HL105993.
Nonstandard Abbreviations and Acronyms
- VT
Ventricular tachyarrhythmias
- SCD
sudden cardiac death
- NF
non-failing
- HASR
hours after sunrise
- TOD
Time of Day
Footnotes
Disclosures: None
References:
- 1.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, et al. for the; American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart Disease and Stroke Statistics-2017 Update: A Report from the American Heart Association. Circulation. 2017;135:e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ørn S, Dickstein K. How do heart failure patients die? Eur Heart J Suppl. 2002;4:D59–D65. [Google Scholar]
- 3.Willich SN, Levy D, Rocco MB, Tofler GH, Stone PH, Muller JE. (Circadian variation in the incidence of sudden cardiac death in the Framingham Heart Study population. Am J Cardiol. 1987;60:801–806. [DOI] [PubMed] [Google Scholar]
- 4.Englund A, Behrens S, Wegscheider K, Rowland E. Circadian variation of malignant ventricular arrhythmias in patients with ischemic and nonischemic heart disease after cardioverter defibrillator implantation. J Am Coll Cardiol. 1999;34:1560–1568. [DOI] [PubMed] [Google Scholar]
- 5.Tofler GH, Gebara OCE, Mittleman MA, Taylor P, Siegel W, Venditti FJ Jr, Rasmussen CA, Muller JE. Morning peak in ventricular tachyarrhythmias detected by time of implantable cardioverter/defibrillator therapy. Circulation. 1995;92:1203–1208. [DOI] [PubMed] [Google Scholar]
- 6.Wood MA, Simpson PM, London WB, Stambler BS, Herre JM, Bernstein RC, Ellenbogen KA. Circadian pattern of ventricular tachyarrhythmias in patients with implantable cardioverter-defibrillators. J Am Coll Cardiol. 1995;25:901–7. [DOI] [PubMed] [Google Scholar]
- 7.Shusterman V, Aysin B, Gottipaty V, Weiss R, Brode S, Schwartzman D, Anderson KP; for the ESVEM Investigators. Autonomic nervous system activity and the spontaneous initiation of ventricular tachycardia. J Am Coll Cardiol. 1998;32:1891–9. [DOI] [PubMed] [Google Scholar]
- 8.Patton KK, Hellkamp AS, Lee KL, Mark DB, Johnson GW, Anderson J, Bardy GH, Poole JE; SCD-He FT Investigators. Unexpected deviation in circadian variation of ventricular arrhythmias: the SCD-HeFT (Sudden Cardiac Death in Heart Failure Trial). J Am Coll Cardiol. 2014;63:2702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni YM, Rusinaru C, Reinier K, Uy-Evanado A, Chugh H, Stecker EC, Jui J, Chugh SS. Unexpected shift in circadian and septadian variation of sudden cardiac arrest: the Oregon Sudden Unexpected Death Study. Heart Rhythm. 2019;16:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnemeier H, Wiegand UK, Braasch W, Brandes A, Richardt G, Potratz J. Circadian profile of QT interval and QT interval variability in 172 healthy volunteers. Pacing Clin Electrophysiol. 2003;26:377–382. [DOI] [PubMed] [Google Scholar]
- 11.El-Sherif N, Chinushi M, Caref EB, and Restivo M. Electrophysiological mechanism of the characteristic electrocardiographic morphology of torsade de pointes tachyarrhythmias in the long-QT syndrome. Circulation. 1997;96:4392–4399. [DOI] [PubMed] [Google Scholar]
- 12.Beuckelmann DJ, Nabauer M, and Erdmann E. Alterations of K+ currents in isolated human ventricular myocytes from patients with terminal heart failure. Circ Res. 1993;73:379–385. [DOI] [PubMed] [Google Scholar]
- 13.Kaab S, Nuss HB, Chiamvimonvat N, O’Rourke B, Pak PH, Kass DA, Marban E, Tomaselli GF. Ionic mechanism of action potential prolongation in ventricular myocytes from dogs with pacing-induced heart failure. Circ Res. 1996;78:262–273. [DOI] [PubMed] [Google Scholar]
- 14.Nabauer M, Beuckelmann DJ, and Erdmann E. Characteristics of transient outward current in human ventricular myocytes from patients with terminal heart failure. Circ Res. 1993;73:386–394. [DOI] [PubMed] [Google Scholar]
- 15.Beuckelmann DJ, Nabauer M, and Erdmann E. Intracellular calcium handling in isolated ventricular myocytes from patients with terminal heart failure. Circulation. 1992;85:1046–1055. [DOI] [PubMed] [Google Scholar]
- 16.Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiol Rev. 2007;87:425–456. [DOI] [PubMed] [Google Scholar]
- 17.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. [DOI] [PubMed] [Google Scholar]
- 18.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP. The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol. 2013;304:C954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita T, Sekiguchi A, Iwasaki YK, Sagara K, Iinuma H, Hatano S, Fu LT, Watanabe H. Circadian variation of cardiac K+ channel gene expression. Circulation. 2003;107:1917–22. [DOI] [PubMed] [Google Scholar]
- 21.Schroder EA, Burgess DE, Zhang X, Lefta M, Smith JL, Patwardhan A, Bartos DC, Elayi CS, Esser KA, Delisle BP. The cardiomyocyte molecular clock regulates the circadian expression of Kcnh2 and contributes to ventricular repolarization. Heart Rhythm. 2015;12:1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong M, Watanabe E, Yamamoto N, Nagahata-Ishiguro M, Maemura K, Takeda N, Nagai R, Ozaki Y. Circadian expressions of cardiac ion channel genes in mouse might be associated with the central clock in the SCN but not the peripheral clock in the heart. Biol Rhythm Res. 2013;44:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins HE, Rodrigo GC. Inotropic response of cardiac ventricular myocytes to beta-adrenergic stimulation with isoproterenol exhibits diurnal variation: involvement of nitric oxide. Circ Res. 2010;106:1244–52. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Zhu D, Yuan J, Han Z, Wang Y, Qian Z, Hou X, Wu T, Zou J. CLOCK-BMAL1 regulate the cardiac L-type calcium channel subunit CACNA1C through PI3K-Akt signaling pathway. Can J Physiol Pharmacol. 2016;94:1023–32. [DOI] [PubMed] [Google Scholar]
- 26.Leibetseder V, Humpeler S, Svoboda M, Schmid D, Thalhammer T, Zuckermann A, Marktl W, Ekmekcioglu C. Clock genes display rhythmic expression in human hearts. Chronobiol Int. 2009;26:621–36. [DOI] [PubMed] [Google Scholar]
- 27.Ruben MD, Wu G, Smith DF, Schmidt RE, Francey LJ, Lee YY, Anafi RC, Hogenesch JB. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci Transl Med. 2018;10 pii:eaat8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.London B Cardiac arrhythmias: from (transgenic) mice to men. J Cardiovasc Electrophysiol. 2001;12:1089–91. [DOI] [PubMed] [Google Scholar]
- 29.Zuther P and Lemmer B, Chronos-Fit 1.06. 2009; URL: http://www.ma.uni-heidelberg.de/inst/phar/lehre/chrono.html.
- 30.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4:e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci USA. 2013;110:9950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jammalamadaka S, Sarma Y. A correlation coefficient for angular variables; Statistical Theory and Data Analysis 2; North Holland: New York: 1988. [Google Scholar]
- 33.Scheer FA, Hu K, Evoniuk H, Kelly EE, Malhotra A, Hilton MF, Shea SA. Impact of the human circadian system, exercise, and their interaction on cardiovascular function. Proc Natl Acad Sci. 2010;107:20541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T, et al. , for the MILIS Study Group. Circadian variation in the frequency of onset of acute myocardial infarction. New Engl J Med. 1985;313:1315–1322. [DOI] [PubMed] [Google Scholar]
- 35.Janszky I, Ljung R. Shifts to and from daylight saving time and incidence of myocardial infarction. N. Engl. J. Med. 2008;359:1966–1968. [DOI] [PubMed] [Google Scholar]
- 36.Chudow JJ, Dreyfus I, Zaremski L, Mazori AY, Fisher JD, Di Biase L, Romero J, Ferrick KJ, Krumerman A. Changes in atrial fibrillation admissions following daylight saving time transitions. Sleep Med. 2020;69:155–158. [DOI] [PubMed] [Google Scholar]
- 37.Ambrosi CM, Yamada KA, Nerbonne JM, Efimov IR. Gender differences in electrophysiological gene expression in failing and non-failing human hearts. PLoS One. 2013;8:e54635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kittleson MM, Minhas KM, Irizarry RA, Ye SQ, Edness G, Breton E, Conte JV, Tomaselli G, Garcia JG, Hare JM. Gene expression analysis of ischemic and nonischemic cardiomyopathy: shared and distinct genes in the development of heart failure. Physiol Genomics. 2005;21:299–307. [DOI] [PubMed] [Google Scholar]
- 39.Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, Ninni S, Klein C, Ortmans S, Seunes C, et al. Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbα antagonism: a single-centre propensity-matched cohort study and a randomised study. Lancet. 2018;391:59–69. [DOI] [PubMed] [Google Scholar]
- 40.Inaba K, Branco BC, Lam L, Salim A, Talving P, Plurad D, Green DJ, Demetriades D. Organ donation and time to procurement: late is not too late. J Trauma. 2010;68:1362–6. [DOI] [PubMed] [Google Scholar]
- 41.Kotloff RM, Blosser S, Fulda GJ, Malinoski D, Ahya VN, Angel L, Byrnes MC, DeVita MA, Grissom TE, Halpern SD, et al. Management of the Potential Organ Donor in the ICU: Society of Critical Care Medicine/American College of Chest Physicians/Association of Organ Procurement Organizations Consensus Statement. Crit Care Med. 2015;43:1291–325. [DOI] [PubMed] [Google Scholar]
- 42.Boone DR, Sell SL, Micci MA, Crookshanks JM, Parsley M, Uchida T, Prough DS, DeWitt DS, Hellmich HL. Traumatic brain injury-induced dysregulation of the circadian clock. PLoS One. 2012;7:e46204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul T, Lemmer B. Disturbance of circadian rhythms in analgosedated intensive care unit patients with and without craniocerebral injury. Chronobiol Int. 2007;24:45–61. [DOI] [PubMed] [Google Scholar]
- 44.Kojima S, Green CB. Circadian genomics reveal a role for post-transcriptional regulation in mammals. Biochemistry. 2015;54:124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


