Abstract
Background
In about half of all patients with a suspected monogenic disease, genomic investigations fail to identify the diagnosis. A contributing factor is the difficulty with repetitive regions of the genome, such as those generated by segmental duplications. The ATAD3 locus is one such region, in which recessive deletions and dominant duplications have recently been reported to cause lethal perinatal mitochondrial diseases characterized by pontocerebellar hypoplasia or cardiomyopathy, respectively.
Methods
Whole exome, whole genome and long-read DNA sequencing techniques combined with studies of RNA and quantitative proteomics were used to investigate 17 subjects from 16 unrelated families with suspected mitochondrial disease.
Findings
We report six different de novo duplications in the ATAD3 gene locus causing a distinctive presentation including lethal perinatal cardiomyopathy, persistent hyperlactacidemia, and frequently corneal clouding or cataracts and encephalopathy. The recurrent 68 Kb ATAD3 duplications are identifiable from genome and exome sequencing but usually missed by microarrays. The ATAD3 duplications result in the formation of identical chimeric ATAD3A/ATAD3C proteins, altered ATAD3 complexes and a striking reduction in mitochondrial oxidative phosphorylation complex I and its activity in heart tissue.
Conclusions
ATAD3 duplications appear to act in a dominant-negative manner and the de novo inheritance infers a low recurrence risk for families, unlike most pediatric mitochondrial diseases. More than 350 genes underlie mitochondrial diseases. In our experience the ATAD3 locus is now one of the five most common causes of nuclear-encoded pediatric mitochondrial disease but the repetitive nature of the locus means ATAD3 diagnoses may be frequently missed by current genomic strategies.
Funding
Australian NHMRC, US Department of Defense, Japanese AMED and JSPS agencies, Australian Genomics Health Alliance and Australian Mito Foundation.
Keywords: cardiomyopathy, ATAD3, segmental duplication, mitochondrial disease, genomics, quantitative proteomics
Introduction
Mitochondrial energy generation diseases are the most common group of inherited metabolic diseases, affecting at least 20 per 100,000 births1, but comprise more than 350 monogenic disorders2. Most childhood-onset mitochondrial diseases are thought to have autosomal recessive inheritance, with mitochondrial DNA (mtDNA) mutations responsible for about a quarter of cases and other inheritance mechanisms apparently less common1. However, as with many other genetic disorders, up to half of all patients with suspected disease remain without a molecular diagnosis after genomic investigations3. This is partly due to technical limitations and the clinical challenges of interpretation, but another contributor is the genomic architecture of repetitive regions of the genome.
Segmental duplications, also known as low-copy repeats, are highly homologous duplicated sequences that exist in one or more copies within a genome4,5. The human genome is estimated to consist of ~50% repeat sequences with segmental duplications thought to account for ~5% of these5–7. Intrachromosomal segmental duplications can make regions structurally unstable and prone to DNA rearrangements via nonallelic homologous recombination (NAHR)8. These rearrangements can result in structural or copy number variation (CNV), such as duplications and deletions, and have been demonstrated to underlie numerous syndromes8,9 with over 40 “genomic disorders” identified8,10.
Perhaps the most complex genomic site linked to mitochondrial disease, the ATAD3 segmental duplication contains three highly homologous tandemly arrayed genes: ATAD3C, ATAD3B, and ATAD3A. This arrangement is exclusive to hominids, with a single copy of ATAD3 found in other multicellular organisms11. In mice, D. melanogaster and C. elegans, loss of ATAD3 (ATPase family AAA+ domain-containing member 3) causes early embryonic or larval lethality12–14. In 2016, pathogenic variants in ATAD3 were linked to Harel-Yoon Syndrome (HAYOS; MIM: 617183), characterized by global developmental delay and hypotonia, along with other features including axonal neuropathy, optic atrophy or cataracts, cerebellar atrophy and hypertrophic cardiomyopathy15. A variety of genetic mechanisms have now been reported in this locus including de novo dominant single nucleotide variants (SNVs)15, inherited dominant SNVs16, inherited recessive SNVs15,17,18, and ATAD3 gene conversions19. More severe presentations characterized by fatal congenital pontocerebellar hypoplasia and sometimes cardiomyopathy, often with cataracts or corneal clouding, have been linked to ATAD3 variation in multiple patients, the majority with inherited recessive deletions affecting ATAD3A and ATAD3B (MIM: 618810)15,17,19. Likewise, dominant de novo duplications involving ATAD3C, ATAD3B and ATAD3A have recently been reported in five patients with fatal cardiomyopathy and corneal clouding/cataracts (MIM: 618815)20. These ATAD3 CNVs are thought to arise via NAHR-mediated events within the repetitive ATAD3 locus15, resulting in chimeric ATAD3 genes15,19,20 (Figure 1A).
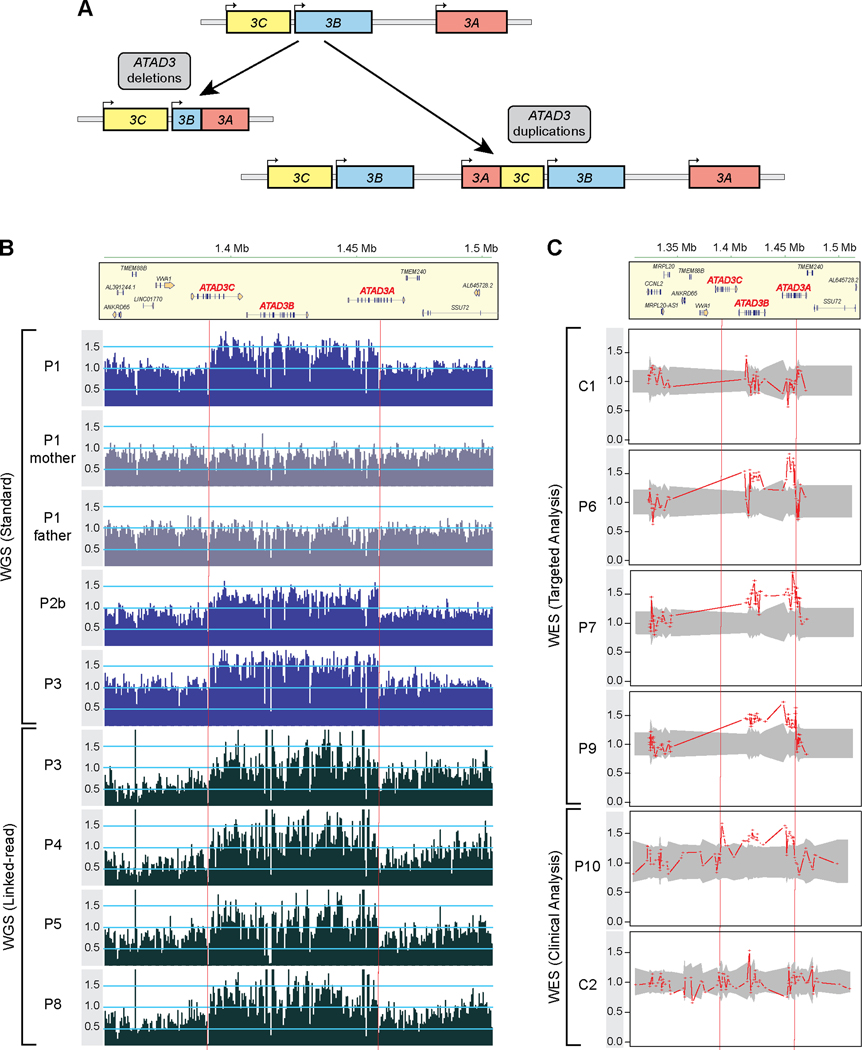
Figure 1. Detection of ATAD3 duplications by genomic investigations.
A) The ATAD3 locus is a site of genetic instability with recurrent pathogenic biallelic deletions and de novo duplications, resulting in chimeric ATAD3B/ATAD3A genes (deletions; expressed under the ATAD3B promoter) or ATAD3A/ATAD3C genes (duplications; expressed under the ATAD3A promoter). B) Read coverage across the chr1 region spanning the ATAD3 locus and flanking genes as determined by either standard WGS in patients P1 and parents, P2b and P3 (upper panels with dark blue (patients) or grey (parents) bars) or 10x Genomics linked-read WGS in P3-P5 and P8 (lower panels with black bars). The number of uniquely mapping reads was determined for each 500 nt non-overlapping window. Read depths per bin were normalized by dividing by the median read count of all chr1 bins with non-zero read counts. Dashed red vertical lines indicate approximate breakpoints of the duplicated region. C) Samples were subjected to WES with either a targeted analysis of mitochondrial genes (control C1, patients P6, P7, and P9) or clinical analysis (control C2 and P10). ExomeDepth analysis across the ATAD3 locus plotted as the ratio of the observed versus expected read depth. Shaded grey areas indicate 95% confidence interval across the matched reference set. See also Figure S1.
ATAD3 localizes to the mitochondrial inner membrane with its N-terminus in the intermembrane space, potentially contacting the mitochondrial outer membrane 12,21. It is found predominantly at the boundary between the inner and outer membranes, rather than in the cristae22, with enrichment also at mitochondria/endoplasmic reticulum contact sites in humans and mice23–25. The precise molecular functions of ATAD3 are unresolved and it has been linked to diverse mitochondrial roles21. Two major roles appear to be in (i) hormone induced steroidogenesis12,23, possibly via organization of cholesterol transfer between the endoplasmic reticulum and mitochondria12,26,27; and (ii) mtDNA organization and segregation, as ATAD3 can be purified as an mtDNA nucleoid component28 and can influence the co-fractionation of mtDNA and cholesterol22. Other functions ascribed to ATAD3 include roles in mitochondrial translation27, adipogenesis and lipid metabolism13,29, iron and heme homeostasis30 and maintenance of mitochondrial networks12,16,31,32, with ATAD3 reported to interact with mitochondrial fission mediator DRP133. Expression of mutant ATAD3 is reported to increase autophagy and mitophagy15,16, with ATAD3 proposed to suppress mitophagy by mediating mitochondrial import and degradation of PINK1 in mice34.
Here, we show that the ATAD3 locus is a hotspot for pathogenic genomic variation leading to mitochondrial disease by describing 17 patients from 16 families with recurrent 68 Kb duplications in the ATAD3 locus (ATAD3dup). These duplications generate an extra copy of the ATAD3B gene and an in-frame ATAD3A/ATAD3C fusion gene that generates a stable chimeric ATAD3A/ATAD3C protein that disrupts ATAD3 oligomerization.
Results
Identification of recurrent de novo ATAD3 duplications
A heterozygous de novo duplication within the ATAD3 locus (GRCh37/hg19; chr1:g.1,385,069 – 1,470,067) was initially identified in a patient (P1) from trio whole genome sequencing (WGS) (Figure 1B). Similar to previously reported patients with biallelic ATAD3 deletions (ATAD3del)15,17,19, her clinical phenotype included hyperlactacidemia, cataracts, white matter abnormalities and perinatal death (Table 1 and individual case reports in Table S1). Strikingly, hypertrophic cardiomyopathy was also observed in this patient, but not commonly noted in ATAD3del patients15,17,19. Likewise, compared to the severe pontocerebellar hypoplasia characteristically observed in ATAD3del patients, P1 had only mild pontocerebellar defects.
Table 1.
Clinical Summary of ATAD3 duplication patientsa.
| Patient | Sex Ancestry | Age at Onset | Birth History | Age at Death | Cardiology | Ophthalmology | Brain imaging / neuropathology | Other Features | Biochemical investigationsb |
|---|---|---|---|---|---|---|---|---|---|
| P1 | F European (Dutch) |
At birth 37/40 | Caesarean 37/40 | 38 days | HCM | Bilateral cataract | MRI: diffuse hyperintense and swollen cerebral white matter on T2, hyperintense basal ganglia on T1, bilateral subependymal pseudocysts, mild pontocerebellar hypoplasia, lactate peak on MRS | Ascites, seizures, burst-suppression pattern (EEG) | Blood: lactate 1.7–2.9 mmol/L, abnormal liver function tests CSF: lactate 8.7 mmol/L Urine: increased TG, PG, IBG, IVG, 3MCG, alanine |
| P2a | M European (Australian) |
At birth 37/40 | Caesarean 37/40 | 6 days | HCM | Dense corneal clouding | Histopathology: widespread gliosis and microglial activation with some neuronal loss but no other apparent structural abnormalities | Hypotonia, seizures, burst-suppression pattern (EEG) | Blood: lactate > 3.5 mmol/L (persistent metabolic acidosis) Urine: slight increase in MMA, MC |
| P2b | M European (Australian) |
At birth 37/40 | Caesarean 37/40 | 5 days | HCM | Dense corneal clouding | Histopathology: microglial activation, gliosis with some neuronal loss and occasional necrotic neurons in the brainstem, cerebellar dentate nucleus, basal ganglia, thalamus, and subiculum. Cerebral white matter was relatively spared | Hypotonia, contractures, seizures | Blood: lactate > 3.5 mmol/L (persistent metabolic acidosis) Urine: slight increase in MMA, MC |
| P3 | M European (Australian) |
In utero 38/40 | Caesarean 38/40 | 9 days | HCM | Bilateral corneal opacification | MRI: mild swelling and diffuse cerebral white matter hyperintensities on T2, bilateral cysts in the temporal lobes (particularly right side), lactate peak on MRS | IUGR, hypotonia, abnormal posturing, burst-suppression pattern (EEG) | Blood: lactate 3–7 mmol/L (persistent metabolic acidosis) CSF: lactate 15.8 mmol/L (1.5 hours postmortem) Urine: slight increase in 3MGA, lactate |
| P4 | F European (Australian) |
In utero 3rd trimester | Caesarean 39/40 | 4 days | Borderline contractility | MRI: increased cerebral white matter signal, high lactate peak and low NAA on MRS Cranial US: Echogenicity of the basal ganglia and left lenticular striate artery | IUGR, hip flexion contractures, seizures, hepatic hemosiderosis and steatosis | Blood: increasing lactates (persistent metabolic acidosis) 7mmol/L (at birth) to 21 mmol/L (50 hours >delivery) | |
| P5 | F Mauritian |
In utero 36/40 | Caesarean 36/40 | 3 days | HCM | Probable cataracts | Not performed | IUGR, hypotonia | Blood: lactate 9.1 mmol/L (persistent metabolic acidosis) |
| P6 | M European (New Zealander) |
In utero 38/40 | Caesarean 38/40 | 3 days (64 hours) | HCM endocardial fibroelastosis | Corneal opacities, microphthalmia, possible cataracts | MRI: severely abnormal cerebral and cerebellar white matter signal and cerebral swelling, subtle basal ganglia signal abnormality | Hydrops fetalis, seizures, burst-suppression pattern (EEG), hepatomegaly with macrovesicular steatosis | Blood: lactate peak 13 mmol/L (persistent metabolic acidosis) Urine: grossly elevated lactate and βOHB with TCA cycle intermediates including moderate increases in fumarate and 3MGA |
| P7 | M European (Australian) |
At birth 37/40 | Caesarean 37/40 | 3 days | HCM endocardial fibrosis | Corneal opacities, periorbital conjunctival edema | Neuropathology: cerebral edema, poor demarcation between cortex and white matter, few areas of apparent softening in the subcortical white matter (right caudate nucleus and right fronto-parietal region) | IUGR, edema, seizures, brother died at 6 days following fetal distress/birth asphyxia | Blood: lactate 4 mmol/L, pyruvate 74 uMol/L, total carnitine 18 μM, free carnitine 12 μM CSF: lactate 4.5 mmol/L Urine: traces of fumaric, 3-methylglutaric and 3MGA |
| P8 | M European (Australian) |
In utero 38/40 | Breech 38/40 | 8 days | HCM endocardial fibroelastosis | Cranial US: intraventricular hemorrhages, patchy hyperechogenicity in the basal ganglia consistent with calcium, right caudate cysts | Edema, hypotonia, hepatomegaly and steatosis | Blood: lactate peak 23.2 mmol/L, minimum pH 7.14 | |
| P9 | F European (Australian)/ East Asian (Japanese) |
In utero IUGR 26/40, reduced fetal movement 34/40 | Caesarean 35/40 | 6 days | Dilated phase of HCM | Cranial US: sub-ependymal cysts and lenticular striate artery echogenicity | IUGR, oligohydramnios, hepatic hemosiderosis, renal failure | Blood: lactate 8.6–25 mmol/L (persistent metabolic acidosis) CSF: lactate 13.5 mmol/L | |
| P10 | M East Asian (Japanese) |
At birth | 40/40 | 2 months | HCM | Corneal clouding | MRI: cerebral white matter hyperintense lesions on T2 | Hypotonia, neonatal asphyxia, bilateral hearing loss | Blood: lactate 9.9 mmol/L, pyruvate 0.36 mmol/L CSF: lactate 9.9 mmol/L, pyruvate 0.34 mmol/L |
| P11 | M East Asian (Japanese) |
At birth | 37/40 | 1 month | HCM | Cranial US: bilateral bright thalami | Neonatal asphyxia, cardiac tamponade | Blood: lactate 12.3 mmol/L, pyruvate 0.46 mmol/L | |
| P12 | F East Asian (Japanese) |
In utero 36/40 | Caesarean 36/40 | 1 month | Heart failure, mild hypertrophy | Corneal clouding, microphthalmia, abnormal iris | Brain CT: mild calcification in lenticular nucleus | IUGR, arthrogryposis neonatal asphyxia | Blood: lactate 12.6 mmol/L, pyruvate 0.35 mmol/L |
| P13 | F East Asian (Japanese) |
At birth | 40/40 | 10 days | HCM | Brain CT: diffuse cerebral white matter hypodensity and swelling plus low density in the basal ganglia and thalamus | Neonatal asphyxia | Blood: lactate12.3 mmol/L, pyruvate 0.25 mmol/L | |
| P14 | M European (Australian) |
In utero 37/40 | Induced 37/40 | 3 days | DCM | Bilateral corneal opacity, bilateral cataracts | Cranial US: bilateral subependymal hemorrhages Histopathology: widespread gliosis and microglial activation with some neuronal loss | Hydrops, seizures, burst-suppression pattern (EEG), congenital alveolar dysplasia, hepatic siderosis and microvesicular steatosis | Blood: lactate 2.3 – 15 mmol/L (persistent metabolic acidosis) Urine: grossly elevated lactate, slight fumarate and 3MGA |
| P15 | M European (Australian) |
In utero 31/40 | Fetal death in utero 36+6/40 | HCM endocardial fibroelastosis | Histopathology: gliosis and microglial activation with some neuronal loss in the brainstem, cerebellum, thalamus and basal ganglia | IUGR, hydrops fetalis | No investigations performed (fetal death in utero) | ||
| P16 | M European (Australian) |
At birth | 41.5/40 | 7 days | HCM | Corneal clouding | Brain MRI: mild signal abnormality and swelling of cerebral white matter, severe generalised encephalopathy, increased lactate on MRS | Seizures, renal failure | Blood: lactate 6.7 – 50 mmol/L (persistent metabolic acidosis) Urine: increased 3MGA, 3methylglutarate, fumarate and lactate |
Abbreviations: ADC, apparent diffusion coefficient; β-OHB, β-hydroxybutyrate; CMV, cytomegalovirus; CSF, cerebral spinal fluid; CT, computed tomography; DCM, dilated cardiomyopathy; EEG, electroencephalogram; HCM, hypertrophic cardiomyopathy; IBC, isobutyrylglycine; IUGR, intrauterine growth restriction; IVG, isovalerylglycine; MC, methylcitrate; MMA, methylmalonic acid; MRI, magnetic resonance imaging; MRS, magnetic resonance spectroscopy; NAA, N-acetylaspartate; PG, propionylglycine; TG, tiglylglycine; US, ultrasound; 3MCG, 3-methylcrotonylglycine; 3MGA, 3-methylglutaconic acid.
See also Table S1.
Reference ranges for blood and CSF lactate and pyruvate levels varied between different centers and we regarded blood and CSF lactate levels as elevated if >2.2 mmol/L and blood and CSF pyruvate as elevated if >0.1 mmol/L.
Re-inspection of available data from WGS and/or whole exome sequencing (WES) of unsolved patients with suspected perinatal or infantile lethal mitochondrial disease revealed 13 additional patients (P2a & b – P13, including monozygotic twins) from 12 families with remarkably similar ATAD3 duplications, detected using a variety of CNV callers including CNVnator35, ExomeDepth36, and XHMM37 (Figures 1B, 1C, S1A, S1B; See methods for individual details). The duplications detected by WES were supported by read depth data from linked-read Chromium WGS (10x Genomics) across the ATAD3 locus for four of the patients (Figure 1B).
An additional ATAD3dup patient (P15) was identified from an in-house clinical SNP microarray database (VCGS; consisting of ~90,000 previously tested patients). The microarray was performed as part of prenatal testing (post-amniocentesis) after an ultrasound scan at 31 weeks gestation indicated the presence of cardiac hypertrophy (Figure S1C). Parental microarray screening indicated the ATAD3 CNV was de novo. We note that microarrays can show poor sensitivity for CNV detection within segmental duplications38 and the probe coverage across the ATAD3 locus is limited on many array platforms, making identification of ATAD3 CNVs challenging15,39. The ATAD3 duplication was only detected in only one of five other confirmed ATAD3dup patients screened on the same array platform (Figure S1C). Therefore, it was not surprising that no other samples with similar ATAD3 duplications were identified within our clinical database.
Duplication CNVs most commonly occur in tandem40, so using the ATAD3A/ATAD3C breakpoints predicted from the sequencing and array technologies, genomic DNA PCR screens were developed to confirm the CNV structure and evaluate inheritance. In all patients, a PCR product spanning the ATAD3A/ATAD3C breakpoint was amplified that enabled molecular determination of the ATAD3 duplication breakpoints by Sanger sequencing (Figure 2A and S2A). The breakpoint PCR confirmed that the ATAD3 CNV arose via a de novo duplication event in 13 of the 13 families with available parental samples for comparison (Figure S2A). We detected no evidence of parental mosaicism for ATAD3dup in our cohort, although one patient (P7) had an older brother who died at six days of age with a suspicious clinical course (DNA unavailable for investigation).
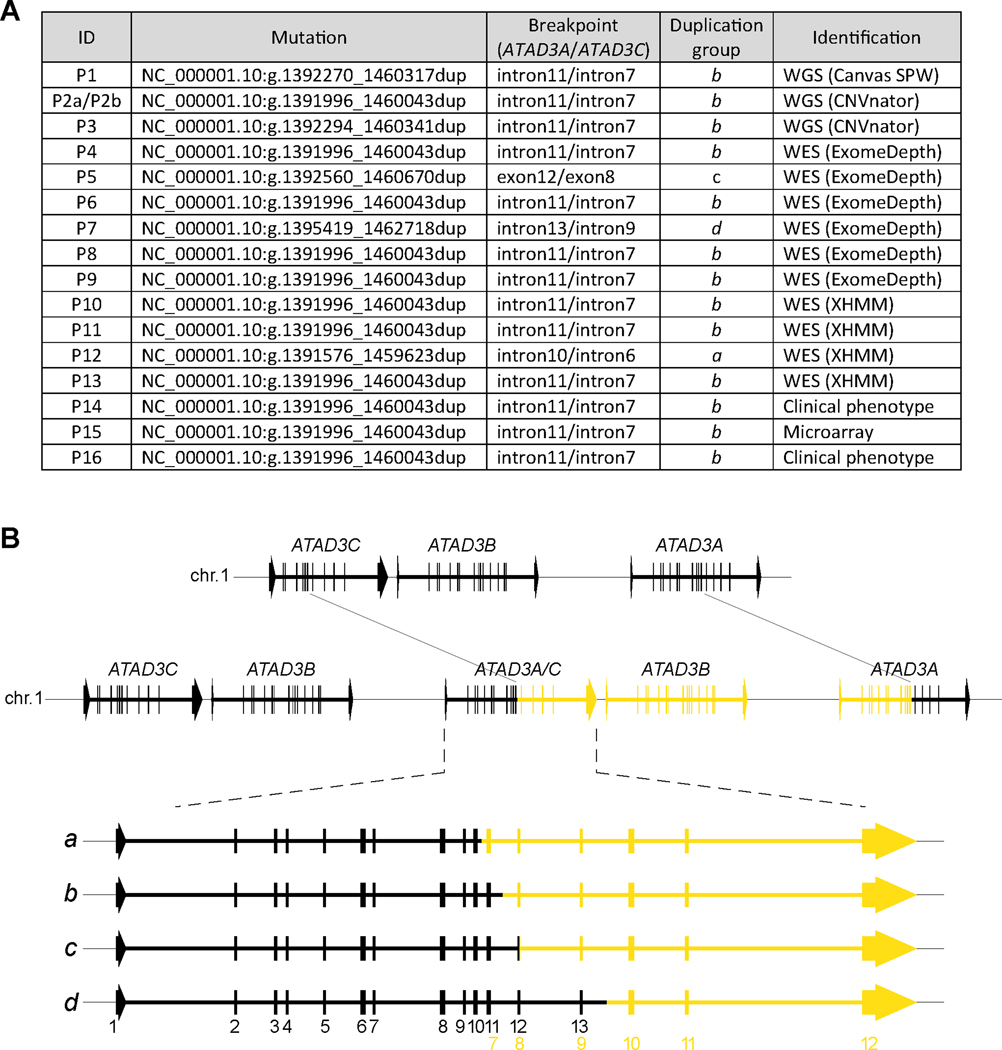
Figure 2. Analysis of ATAD3 duplication breakpoints.
A) Sanger sequencing was used to determine genomic coordinates (GRCh37/hg19) for the ATAD3 duplication in all patients, with the corresponding position of the breakpoint in the ATAD3A and ATAD3C genes indicated. The duplications were assigned to a group according to the location of the breakpoint within the genes. As well, the initial method and variant caller that led to the duplication identification is indicated. Duplications NC_000001.10:g.1392270_1460317dup and NC_000001.10:g.1391996_1460043dup have been previously reported20. B) Schematic of the most common ATAD3 tandem duplication within the cohort (top, in yellow) validated by Sanger sequencing of gDNA breakpoints. The duplicated region (yellow) encompasses part of ATAD3C, all of ATAD3B and part of ATAD3A, resulting in a chimeric ATAD3A/ATAD3C gene. An expanded view of the chimeric ATAD3A/ATAD3C gene (bottom) indicates the different duplication groups (listed in A) and their impact on the gene structure. Corresponding exons are numbered at the bottom. black, ATAD3A; yellow, ATAD3C. See also Figure S2 and Table S2.
Our extensive cohort supports a striking similarity between clinical presentations for ATAD3dup patients (Tables 1 and S1), with hypertrophic cardiomyopathy, hyperlactacidemia and perinatal death being nearly invariant features. Corneal clouding or cataracts, encephalopathy and white matter abnormalities were also common. Compared to the severe pontocerebellar hypoplasia observed in all ATAD3del patients, neurological findings in ATAD3dup patients appear milder, ranging from features consistent with hypoxic ischemia to mild pontocerebellar hypoplasia (P1). Two additional patients (P14 and P16) with consistent clinical features were subsequently confirmed to carry ATAD3 duplications by breakpoint PCR. In total, we identified 17 patients from 16 families carrying recurrent ATAD3 duplications of ~68 Kb with similar breakpoints spanning ATAD3C, ATAD3B and ATAD3A.
Sanger sequencing of the breakpoint PCR products spanning the ATAD3A/ATAD3C duplication junction identified six separate duplication breakpoints within this group of patients (Figure 2A and Table S2) that can be classified into four distinct duplication groups (Figure 2A, groups a - d). In keeping with the role of NAHR in rearrangements of the ATAD3 locus15 and those of other segmental duplications8, the duplications occur in regions with extended stretches of significant homology between ATAD3C and ATAD3A (Table S2), with the homology in regions upstream from the breakpoints ranging from 93.5 – 99.3% identity over >950 bp in nearly all patients. The exception was P5, who had only 92.1% identity over 215 bp. However, due to the ethnic background of this patient (Mauritian), the reference genome and population databases may not accurately reflect all variation within this region and the stretch of homology may in reality extend further.
Expression of a chimeric ATAD3A/ATAD3C gene
The genomic architecture of the ATAD3 locus for the four duplication groups predicts the generation of a chimeric ATAD3A/ATAD3C gene, as well as an extra copy of ATAD3B, in all cases (Figure 2B). This predicted architecture was further supported by Oxford Nanopore Technologies long read WGS from P3, which captured reads spanning the chimeric ATAD3A/ATAD3C gene (Figure S2B). The chimeric ATAD3A/ATAD3C genes are predicted to contain ATAD3A exons 1 through to 10–13 fused to ATAD3C exons 7–10 through to exon 12 (depending on the duplication group) (Figure 2B and Table S2). Using RNA isolated from patient fibroblast lines and heart samples, a stable chimeric ATAD3A/ATAD3C transcript (NM_001170535.1/NM_001039211.2) was detected in all patients studied that was absent from controls (Figure 3A, bottom panel). Stable ATAD3A and ATAD3B transcripts were expressed similarly to controls. The duplication was not predicted to alter the wild-type ATAD3A gene on the affected allele (Figure 2B). This was confirmed by Sanger sequencing of the ATAD3A transcript (Figure 3A, top panel), where heterozygous SNPs in the 3’ untranslated region of ATAD3A in all patients confirmed expression of both alleles.
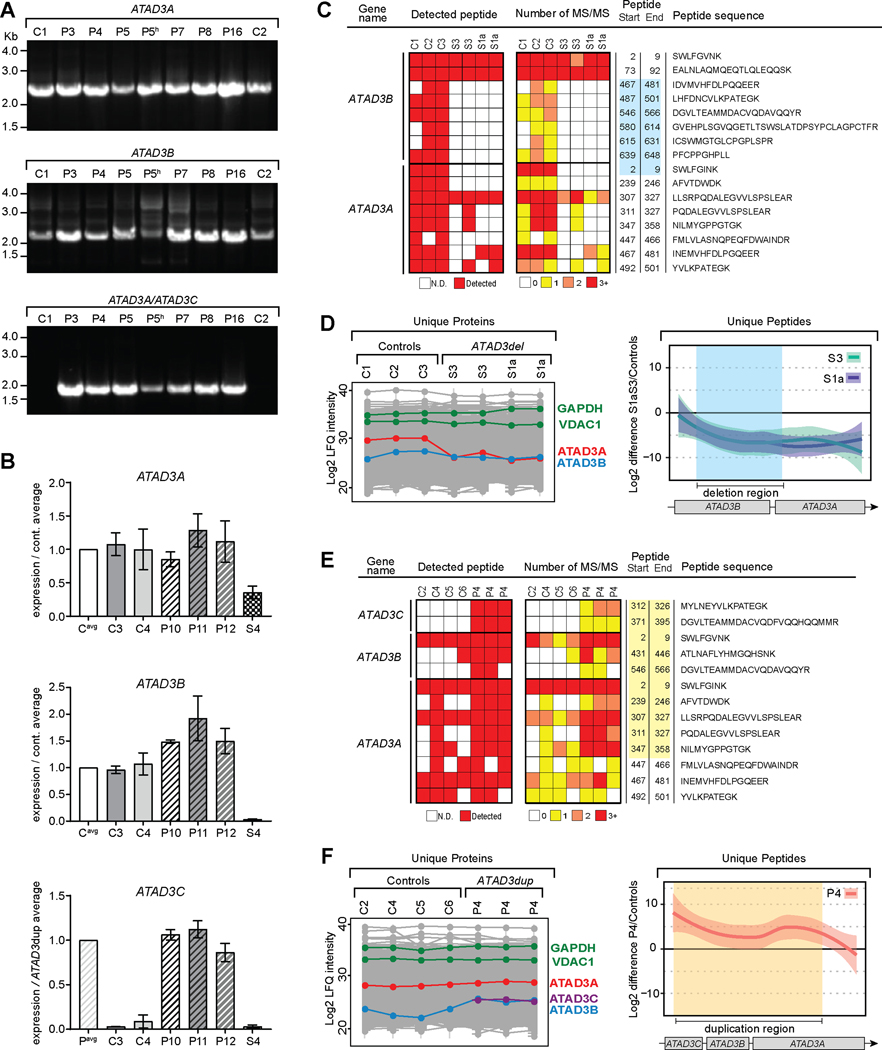
Figure 3. A chimeric ATAD3A/ATAD3C gene generated by the duplication is expressed.
A) cDNA generated from patient and control skin fibroblasts, and P5 heart (indicated by “h”) was used for PCR amplification of transcripts from ATAD3A, ATAD3B and chimeric ATAD3A/ATAD3C genes. Transcripts were amplified using a primer targeting both ATAD3A and ATAD3B, and specific reverse primers for ATAD3A, ATAD3B, and ATAD3C. B) qRT-PCR was performed using cDNA from control and patient skin fibroblasts for quantitative analysis of transcripts using regions corresponding to ATAD3A and ATAD3B exons 15/16 and ATAD3C exons 11/12, which are the last exons of the genes. Values expressed relative to control average (ATAD3A and ATAD3B) or ATAD3dup patient average (ATAD3C). Shaded boxes, controls; dashed boxes, ATAD3dup patients; checked box, ATAD3del patient (S4). n = 3, error = SD. C) Label-free quantitative (LFQ) proteomics was performed on skin fibroblasts from controls (C; n=3) and ATAD3del patients19 S3 and S1a (in duplicate). Unique ATAD3A and ATAD3B peptides were plotted according to their presence, as well as the number of MS/MS spectra observed. The blue box indicates the peptides encoded within the deleted ATAD3 region. ATAD3C peptides were not detected in these samples. N.D., not detected. D) Using the LFQ proteomics data for all unique peptides from C, the relative levels of ATAD3A and ATAD3B proteins in each sample were compared (left panel), alongside the levels of cytosolic (GAPDH) and mitochondrial (VDAC1) marker proteins. The mean difference of Log2 LFQ intensities between S3 and S1a to controls for individual peptides were plotted across the ATAD3 locus with the deleted region indicated (blue shading) (right panel). Missing values for individual peptides were imputed as described in the methods and the loess smoothed curve plotted along with the 95% confidence interval (right panel). E) LFQ proteomics was performed on skin fibroblasts for controls (n=4) and ATAD3dup patient P4 (in triplicate), with unique peptides for ATAD3A, ATAD3B and ATAD3C plotted as in C. The yellow box indicates the peptides encoded within the duplicated ATAD3 region. F) Using the LFQ proteomics data from E, the relative levels of ATAD3A, ATAD3B and ATAD3C proteins were compared across all samples (left panel). The mean difference of Log2 LFQ intensities between P4 to controls were plotted across the ATAD3 locus as in D, with the duplication region indicated (yellow shading) (right panel). See also Figures S3 and S4.
As further confirmation of the predicted impact of the ATAD3 duplication on transcripts, gene-specific qRT-PCR targeting the final exon boundaries was used to compare expression levels of the different ATAD3 transcripts in patients versus controls (Figure 3B). ATAD3A levels were comparable between controls and ATAD3dup patients, consistent with the presence of two full-length copies of ATAD3A. ATAD3dup patients had a ~40–50% increase in ATAD3B transcript levels compared to controls, consistent with expression of a third, duplicated copy of ATAD3B. For validation, ATAD3 transcript levels were also assessed in a previously reported ATAD3del patient (S4)19, with a chimeric ATAD3B/ATAD3A gene expressed under the weaker ATAD3B promoter (Figure 1A). As expected, ATAD3A was ~60% reduced and ATAD3B levels were undetectable in the ATAD3del patient19. ATAD3C, which is normally expressed at low or negligible levels across all tissues, was virtually undetectable in cell lines from controls and ATAD3del patient S4 (Figure 3B and S3). However, in all ATAD3dup patients, transcripts corresponding to ATAD3C could be robustly detected at equivalent levels, confirming expression of the ATAD3A/ATAD3C chimeric gene under the ATAD3A promoter.
Robust confirmation that a stable protein is translated from the chimeric ATAD3A/ATAD3C gene is complicated by the lack of antibodies specific for each ATAD3 protein and the similarity of predicted molecular weights between ATAD3A (~66.2 kDa) and the chimeric ATAD3A/ATAD3C (~66.3 kDa) protein (Figure S4A-E). Therefore, label-free quantitative (LFQ) proteomics was performed on patient fibroblast lines to detect specific ATAD3 peptides and compare expression levels between patients and controls (Figure 3C-F). We initially analyzed two separate ATAD3del patients (S1a and S3; previously reported19), in whom the biallelic deletions were expected to result in expression of a chimeric protein 99.7% identical to ATAD3A, but expressed at a level similar to ATAD3B (Figure 1A). In support of this, the level of ATAD3A in the ATAD3del patients was ~25% of control levels (Figure 3D, left panel), equivalent to that of ATAD3B, with no differences in cytosolic GAPDH or mitochondrial VDAC1. In further confirmation of chimeric ATAD3B/ATAD3A protein expression, individual ATAD3B and ATAD3A specific peptides were mapped against the deleted region (Figure 3C). ATAD3B and ATAD3A specific peptides corresponding to the deleted region within the ATAD3 locus were not detected in ATAD3del patients (Figure 3C), while all other ATAD3A specific peptides were detected at reduced frequency compared to controls (Figure 3D, right panel).
We next analyzed fibroblasts from three ATAD3dup patients (P3, P4 and P5; from duplication groups b and c). Supporting the qRT-PCR data (Figure 3B), ATAD3C specific peptides corresponding to the duplicated region of the gene were robustly detected in all ATAD3dup patients (P3, P4 and P5), but were undetectable in controls as well as the ATAD3del patients (Figure 3E-F and S4F). In all ATAD3dup patients, ATAD3C was detected at lower levels than ATAD3A, consistent with the expression of both the predicted chimeric ATAD3A/ATAD3C protein and two full length copies of ATAD3A (Figure 3F, left panel). Likewise, an increase was also observed for P4 in the abundance of ATAD3B, consistent with the extra copy found in ATAD3dup patients (Figure 3F, left panel), as well as increases in peptides specific to the duplicated ATAD3A region (Figure 3F, right panel). Similar analyses for P3 and P5 were constrained due to pooling of samples prior to analysis preventing evaluation of sample and control variability (Figure S4F). Altogether, the cDNA and proteomic data confirm the presence and stability of the chimeric ATAD3A/ATAD3C protein.
The effect of the ATAD3 duplication on oligomerization and mitochondrial function
Despite the different genomic coordinates of the duplication groups within the chimeric ATAD3A/ATAD3C gene architecture (Figure 2B), all were predicted to create an identical ATAD3A/ATAD3C chimeric protein since ATAD3A exons 11–13 and ATAD3C exons 7–9 have virtually identical reference sequences (Figure 4A and Table S2). The ATAD3A/ATAD3C chimera is 95.1% identical and 96.4% similar to ATAD3A. However, several of the canonical AAA-ATPase residues conserved within the ATAD3A ATPase domain13,28 are altered in ATAD3C, including critical residues in the Sensor-I motif and the “arginine finger” involved in ATP hydrolysis41.
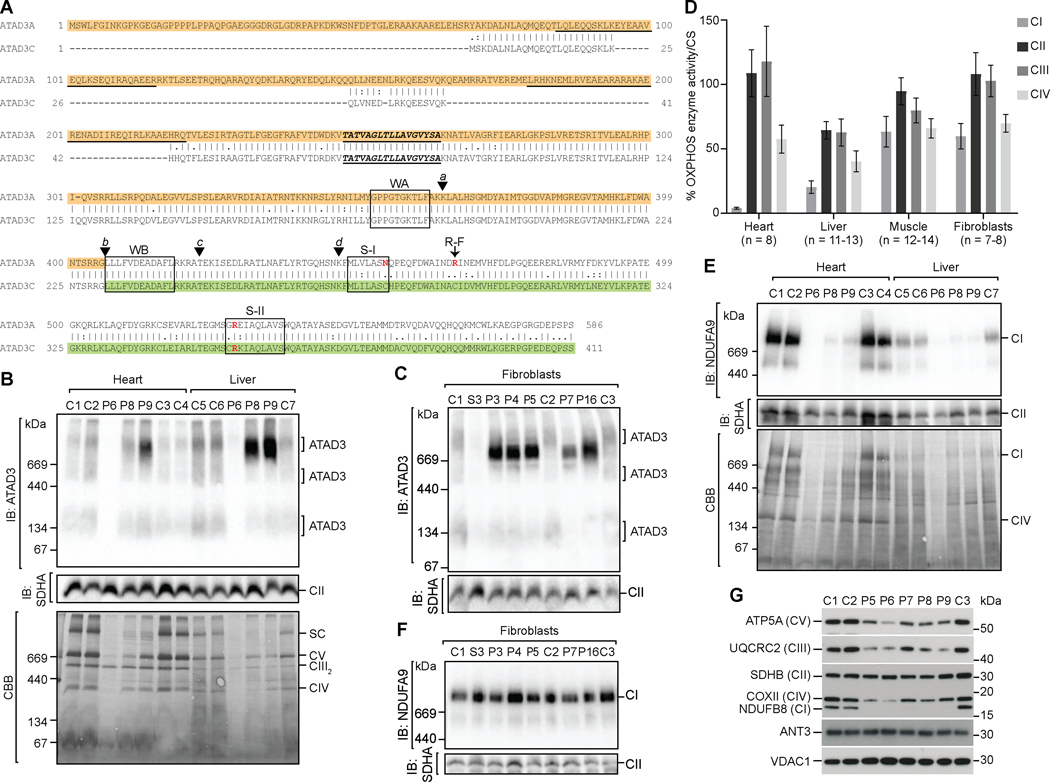
Figure 4. ATAD3 duplication leads to OXPHOS complex I deficiency and ATAD3 oligomerization defects.
A) An alignment of ATAD3A (NP_001164006) with ATAD3C (NP_001034300) showing predicted protein domains and key residues12,28,65: coiled coil regions (underlined); transmembrane domain (underlined/ bold italic text); Walker A and Walker B motifs (WA and WB, boxed); Sensor I and Sensor II motifs (S-I and S-II, key residue in red); and arginine finger residue (R-F, red). The location of the breakpoints corresponding to the deletion groups (a-d) are indicated by arrowheads. The predicted chimeric ATAD3A/ATAD3C protein generated by the most common duplication group group (b) corresponds to the ATAD3A region highlighted in orange, and the ATAD3C region highlighted in green. B) Mitochondria isolated from control and patient heart and liver biopsies were solubilized in 1% digitonin and subjected to blue native (BN)-PAGE and analysis of ATAD3 complexes by immunoblotting (anti-ATAD3), in comparison to OXPHOS complex II (anti-SDHA) (upper panels). Coomassie brilliant blue (CBB) staining of the membrane is shown to compare loading and sample integrity (lower panels). SC, supercomplex. C) Fibroblast mitochondria isolated from control, ATAD3dup patients and ATAD3del patient S3 cell lines analyzed by BN-PAGE as in B. D) OXPHOS enzyme activity normalized to citrate synthase (CS) activity and expressed as the percent of control mean was determined in patient tissue samples. Error = SEM. E) Mitochondria isolated from control and patient heart and liver biopsies were solubilized in 1% Triton X-100 and subjected to BN-PAGE and analysis by immunoblotting for OXPHOS complex I (anti-NDUFA9) and complex II (anti-SDHA) (upper panels), with CBB staining shown for comparison. F) Fibroblast mitochondria isolated from control, ATAD3dup and ATAD3del patient S3 cell lines analyzed by BN-PAGE as in E. G) Total heart tissue lysates from controls and patients (2 μg) analyzed by SDS-PAGE and western blotting with antibodies against an antibody cocktail targeting individual OXPHOS complex subunits, and controls VDAC1 (porin) and ANT3. See also Figure S5 and Table S3.
AAA-ATPases typically function as homo- or heterohexameric oligomers41, with both murine and human ATAD3 observed predominantly in a ~900 kDa complex, with a range of smaller complexes from ~250–900 kDa possibly corresponding to oligomeric ATAD3 structures14,18,42. To determine whether the expression of the chimeric ATAD3A/ATAD3C protein impeded ATAD3 complex assembly, we analyzed isolated mitochondria from control and ATAD3dup tissues and fibroblasts by blue native (BN)-PAGE, which enables separation of detergent solubilized intact protein complexes43. In mitochondria from control heart, liver and fibroblasts, we observed a similar distribution of ATAD3 complexes (~200–900 kDa) to those previously reported (Figure 4B and 4C). In most ATAD3dup mitochondrial samples, a prominent ~900 kDa complex was present that migrated at a slightly faster rate than the control ATAD3 complex. A similar alteration in the distribution of the ATAD3 complexes was observed in ATAD3dup patient heart mitochondria, despite inconsistencies in sample loading (Figure 4B). Unfortunately, there were apparent sample integrity issues with heart and liver biopsies from P6, possibly related to sub-optimal tissue collection (recorded as up to 24hrs postmortem), handling or storage. In contrast to the ATAD3dup patient samples, the ATAD3 complexes were nearly undetectable in mitochondria isolated from ATAD3del patient fibroblasts19 (S3) (Figure 4C).
Examination of the stained BN-PAGE gel of mitochondria solubilized with the detergent digitonin (Figure 4B, bottom panel) showed an obvious decrease in the amount of OXPHOS supercomplexes in ATAD3dup patient heart and liver mitochondria compared to controls. However, no striking differences in OXPHOS complexes were seen in fibroblasts (Figure S5A, left panel), consistent with OXPHOS enzymology studies (Figure 4D). In all ATAD3dup patients with available heart biopsies, complex I (CI; NADH: ubiquinone oxidoreductase) activity was decreased to ~5% of normal control activity (Figure 4D and Table S3). Complex I activity was also decreased in ATAD3dup patient liver biopsies (~20% of control activity, Figure 4D) although this reduction was variable (Table S3). Complex IV (CIV; cytochrome c oxidase) activity was also somewhat decreased in liver and heart (~40% or ~58% of control activity, respectively). Decreased complex I and IV activity was also observed in some ATAD3dup patient muscle and fibroblast samples, however these tended to be less marked and were not considered diagnostically significant. Isolated complex I deficiency, or combined complex I and IV defects, are often observed in disorders of mtDNA replication and expression44. However, few of the patients had overt mtDNA depletion, nor were any found to have mtDNA deletions. The frequent increases in citrate synthase activity and mtDNA/nuclear DNA (nDNA) ratios observed are suggestive of mitochondrial proliferation (Tables S1 and S3).
Due to the structural alterations in OXPHOS supercomplex formation in isolated heart and liver mitochondria from the ATAD3dup patients (Figure 4B, bottom panel), as well as mitochondrial OXPHOS enzymology results, complex I assembly was analyzed. In mitochondria solubilized with Triton X-100 to separate the individual OXPHOS complexes, a strong reduction in assembled complex I was observed in heart and liver from all ATAD3dup patients (Figure 4E). Further analysis of the steady state levels of subunits from individual OXPHOS complexes in total heart extracts showed no detectable complex I subunit NDUFB8 in all ATAD3dup patients, and more modest reductions in complex III, IV and V subunits UQCRC2, MT-COX2, and ATP5A, respectively (Figure 4G). The decreased steady state protein levels correlated with OXPHOS complexes that contain mtDNA-encoded subunits, as no changes were observed in complex II subunit SDHB, nor mitochondrial proteins VDAC1/porin (outer mitochondrial membrane) and ANT3 (inner mitochondrial membrane). Additional analysis of complex III assembly in heart and liver mitochondria indicated a subtle increase in the dimeric complex in patients compared to controls (Figure S5B). In contrast, no complex I defect was detected in ATAD3dup patient fibroblasts (Figure 4F and S5A, right panel), nor was complex III or IV assembly affected (Figure S5C). This was consistent with quantitative proteomic analysis of fibroblasts from patient P4 (Figure S5D).
Discussion
ATAD3, a hotspot for genomic variation and pathogenic CNVs
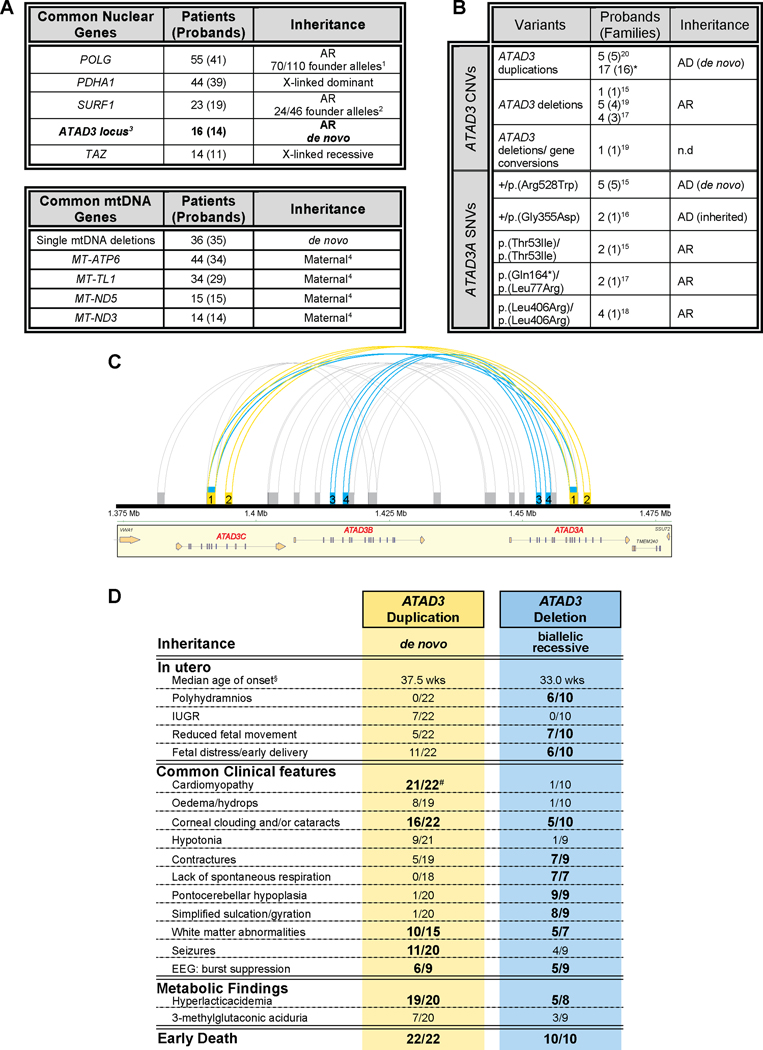
Pediatric mitochondrial disease, including lethal perinatal presentations, is most often thought to have autosomal recessive inheritance1. The de novo nature of ATAD3 duplications markedly alters the reproductive landscape for families, with the recurrence risk estimated to be 1 – 4% for gonadal mosaicism45 rather than 25%. The identification of these recurrent pathogenic duplications and other ATAD3 variants is also shifting the genomic landscape of mitochondrial diagnostics. In our experience of providing genomic diagnoses of mitochondrial disease to more than 500 children from Australia and New Zealand, the ATAD3 locus is now among the five most common nuclear gene causes (Figure 5A). Given all our ATAD3 patients died in the perinatal period, ATAD3 now appears to be the most common gene locus underlying lethal perinatal mitochondrial disease. The four other common genes, all identified ~20–30 years ago46, are prominent within our patient cohort due to common recessive European founder variants (POLG and SURF1)47,48 or due to the vagaries of X chromosomal inheritance (PDHA1 and TAZ). The absence of common European POLG founder mutations in populations such as Japan means that POLG disorders are less common there49. Significantly, the ATAD3 region is the only known locus underlying mitochondrial disease in which segmental duplications lead to recurrent NAHR events. During the preparation of this manuscript, Gunning et al20 reported five additional ATAD3dup patients carrying two of the six de novo breakpoints we describe here (“a” and “b” type duplications). Therefore, 48 ATAD3 patients (39 families) have now been reported since 2016, including 37 with recurrent SNV or CNV mutations (Figure 5B).
Figure 5. ATAD3 regions prone to recurrent rearrangements causing overlapping clinical symptoms.
A) Summary of the five most common nuclear or mtDNA loci in which mutations cause pediatric mitochondrial disease, from our experience with molecular diagnosis of >500 pediatric-onset cases from Australia & New Zealand, including mutations in a total of 74 nuclear genes and 12 mtDNA genes plus mtDNA deletions. aPOLG founder mutations of ancient European origin47 comprise 40 p.(Ala467Thr) alleles, 15 p.(Trp748Ser) alleles and 15 p.(Gly848Ser) alleles; bSURF1 c.311_312insATdel10 founder mutation appears to be of European origin48; cATAD3 defects encompass pathogenic duplication and deletion rearrangements; dUp to a quarter of mtDNA mutations found in pediatric cases appear to be de novo mutations66. AR, autosomal recessive. B) Numerous pathogenic CNVs in the ATAD3 locus and SNVs in ATAD3A have been identified. ATAD3A variants correspond to transcript NM_001170535. *, this study; AD, autosomal dominant; n.d., no data. C) Graphical illustration of tandem repeats in the ATAD3 locus generated using Miropeats67 with a similarity threshold of 900 bp. Regions of similarity associated with pathological ATAD3 rearrangements are indicated for biallelic deletions (blue) and de novo duplications (yellow). All other regions of intragenic similarity in grey. ATAD3 duplication groups a-c fall within region of similarity “1” and duplication group d within similarity region “2”. ATAD3C/ATAD3A deletion breakpoints fall within similarity region “1” and ATAD3B/ATAD3A deletion breakpoints within regions “3” and “4”. Region “1” spans across ATAD3C introns 4–7 and ATAD3A introns 8–11; region “2” across ATAD3C introns 8–9 and ATAD3A introns 12–13; region “3” across ATAD3A and ATAD3B introns 2–4; region “4” across ATAD3A and ATAD3B introns 4–5. D) Comparison of clinical features across all reported patients with ATAD3 duplications and biallelic deletions15,17,19,20 leading to severe presentations. Clinical features associated with more than 50% of ATAD3dup or ATAD3del patients in bold. §, median age of onset in weeks from conception; ATAD3dup patients n = 22 (average age of onset ± SD, 37.0 ± 3.4 weeks) or ATAD3del patients n = 10 (average age of onset ± SD, 32.7 ± 3.4). #, P4 with “borderline contractility” was not considered to have cardiomyopathy; 16/21 patients with cardiomyopathy were diagnosed with hypertrophic cardiomyopathy (HCM). EEG, electroencephalogram; MRI, magnetic resonance imaging.
Many of the patients in this study underwent multiple rounds of genetic and genomic investigations and reanalysis over many years before the ATAD3 duplication was identified. In this cohort, analysis of coverage depth from WGS and WES data proved highly reliable, indicating retrospective analysis of existing sequencing data would readily identify affected individuals. However, it is important to note that microarrays are inefficient at detecting ATAD3 duplications (two of six patients by high-density SNP-array, Figure S1C; two of three patients by custom array-CGH20) and are thus not suited for molecular diagnosis. The software for CNV detection relies on both LogR detection of probe intensity as well as an indication of allelic imbalance from detection of several contiguous B allele values. This can be complicated for duplications, particularly those within segmental duplications, and the detection can be affected by probe sensitivity, allele frequency and DNA quality38. Therefore, it is not surprising that only one ATAD3dup patient was identified in the in-house (VCGS) microarray database over the last 13 years. Many of the analyses were performed on prenatal samples or products of conception (~18,000 since 2014), so ATAD3 duplications may underlie additional unsolved deaths in utero.
Reciprocal ATAD3 deletions and duplications
The ATAD3 locus resembles other chromosomal “hot spots” with genomic instability due to repetitive sequences50,51, many of which are now linked to genomic disorders8,9. Analysis of repeat regions across the ATAD3 locus indicates multiple regions of ≥900 bp of high identity, a subset of which correspond to all reported pathogenic ATAD3 deletion and duplication breakpoints (Figure 5C). The location of these breakpoints is likely explained by the length and identity of homologous low-copy repeat pairs, the GC content and the density of the PRDM6 recombination hot spot motif8,10,52. Indeed, it has been noted that low-copy repeat pairs flanking frequent recurrent de novo deletions share >98% DNA identity10. Since the reported pathogenic ATAD3 duplications and deletions, as well as other CNVs, all arise from the same process (Figure 1A) they would be expected within the population if viable. However, more research is needed to fully define pathogenic versus non-pathogenic ATAD3 variation, as this is complicated by the genomic complexity of the region which makes it difficult to uniquely map reads and clarify breakpoints.
The clinical presentations of all reported ATAD3dup patients are remarkably consistent (Figure 5D). While cardiomyopathy and severe pontocerebellar hypoplasia largely distinguish the ATAD3dup and ATAD3del cohorts, many other clinical features are shared. Nonetheless, the mechanisms behind the genotype-phenotype correlations are not yet clear and some patients with ATAD3A SNVs have both cardiomyopathy and pontocerebellar abnormalities15,18,19. Importantly, the striking clinical consistency of the ATAD3dup phenotype enabled us to readily identify candidate patients from our mitochondrial disease cohorts (Figure 2A, P14 and P16) by searching for similar clinical presentations, with the caveat that patients with divergent presentations would have been overlooked.
Unsurprisingly, the ATAD3dup is absent from gnomAD-SV53 and was identified in only a single patient in DECIPHER54, corresponding to ATAD3dup subject 520. The ATAD3 locus itself is encompassed within the region associated with chromosome 1p36 deletion syndrome (MIM: 607872). One of the most common terminal deletion syndromes, monosomy 1p36 is phenotypically variable, often associated with neurodevelopmental delay and a variety of congenital abnormalities55,56. Numerous genes have been implicated in the variable phenotypes55, the majority of which appear highly intolerant of haploinsufficiency57. It is unlikely that monoallelic loss of ATAD3 would contribute to this syndrome, with no constraint against ATAD3A, ATAD3B and ATAD3C loss of function variants, and ATAD3del carriers appearing normal19,57.
In contrast, only a few patients with exclusive 1p36.3 duplications/triplications have been reported, with milder, variable features usually including global developmental delay or intellectual disability58. While the overall contribution of increased ATAD3 gene dosage to these phenotypes is unclear, an increase of ATAD3B and/or ATAD3C alone would likely play a limited role, with Gunning et al. noting numerous apparently benign ATAD3 specific duplications within DECIPHER that do not involve ATAD3A20. Overall, this supports the idea that increased ATAD3B gene dosage does not contribute to the severe pathogenicity of the ATAD3dup. Instead it appears more likely that the pathogenicity is related to expression of the chimeric ATAD3A/ATAD3C protein. Furthermore, increased ATAD3 levels alone do not specifically correlate with cardiac abnormalities, with atrial septal defects or left ventricular hypertrophy only observed in three of the eight 1p36.3 duplication patients58, and one of the 23 duplications completely overlapping ATAD3 reported in the DECIPHER database.
Complex I deficiency, mtDNA homeostasis and ATAD3
The most common biochemical defect detected in pediatric mitochondrial disease is isolated complex I deficiency, accounting for about one third of all cases59. Severe complex I deficiency was observed in all eight ATAD3dup patients with available heart tissue (Table S3). In fact, six of these ATAD3dup patients were investigated as part of a cohort of 103 complex I deficient mitochondrial disease patients60. The identification of the ATAD3 duplication provides a genetic diagnosis for a further 6% of the patients within that cohort. At the time (2010), patients within the cohort with cardiac associated presentations had a poor molecular diagnostic rate (3/17, or 18%60,61). The ATAD3 duplication now accounts for a further 29% of molecular diagnoses within this group of cardiac patients with isolated complex I deficiency (5/17).
Complex I deficiency was also observed in muscle-specific Atad3 knockout mice42. The impact on OXPHOS function could potentially be explained by issues with mtDNA stability or expression, and in the mouse model, progressive impairments in mtDNA replication and mtDNA depletion were observed42. ATAD3 has been connected with both the maintenance and membrane association of mtDNA nucleoids22,27,28, and abnormalities in mtDNA nucleoids were observed in ATAD3del and ATAD3dup patient fibroblasts19,20. Since ATAD3dup patients also showed milder changes in OXPHOS complexes IV and V, disturbances in mtDNA homeostasis or expression could be causative of these defects. Isolated complex I deficiency is often observed in disorders affecting mtDNA replication or expression62, most likely due to the large number of complex I subunits encoded by mtDNA44. Nevertheless, there is a lack of consistent evidence for mtDNA depletion in this group of ATAD3dup patients across tissues (Table S3), suggesting that the decreased levels of mtDNA observed in some patients might be secondary. In addition, the OXPHOS complexes, through their interaction in supercomplexes, show interdependence for their stability and therefore effects on one complex may secondarily affect others63.
An alternative explanation for the impact on complex I and other OXPHOS complexes could instead be changes in mitochondrial membrane architecture or homeostasis. In mice, the complete loss of ATAD3 is embryonic lethal at a point when embryonic development involves rapid biogenesis of mitochondria to support increasing energy demands14. Reduced mitochondrial mass was observed in muscle fibers and bone marrow cells from the muscle-specific mouse Atad3 knockout42, as well as in C. elegans following Atad-3 knockdown13. A similar loss of mitochondria was observed in flies expressing the dominant ATAD3A p.(Arg528Trp) variant and in the hematopoietic-specific mouse Atad3 knockout, with both involving a concomitant increase in autophagy15,34. Evidence for increased autophagy has been observed in fibroblasts from several ATAD3A patients15,16, as well as ATAD3dup patient P16 (Table S1). This increase could be a secondary response to cellular stress induced by ATAD3-mediated mitochondrial changes, and no real evidence of mitochondrial mass reductions was observed in tissues from ATAD3dup patients, based on mtDNA, enzymatic and proteomic measurements.
Many of the changes in mitochondrial biogenesis and structure may relate to the correlation between ATAD3 and lipid homeostasis. ATAD3A co-purifies with proteins involved in lipid metabolism, has been implicated in both steroid and lipid biosynthesis, and has been shown to facilitate mitochondria/endoplasmic reticulum contacts to support lipid transfer between the organelles23,25–27. This may involve channeling of cholesterol, a key building block in steroidogenesis, into mitochondria where it forms membrane domains that facilitate mtDNA nucleoid replication12,22. Defects in cholesterol homeostasis and mtDNA nucleoid architecture have been observed in both ATAD3dup and ATAD3del patient fibroblasts and may contribute to their clinical presentation19,20.
Mechanisms of ATAD3dup pathogenicity
AAA ATPases are involved in many cellular processes, including DNA replication and protein disassembly, disaggregation and proteolysis41. However, unlike many other AAA ATPases involved in mitochondrial quality control at the inner membrane (e.g. AFG3L2 and SPG7), ATAD3 does not have a proteolytic domain21,64. AAA ATPases typically form hexamers that perform coordinated ATP hydrolysis, with catalysis in one subunit often triggering conformational changes in the next subunit such that progressive ATP hydrolysis occurs within the oligomer48. ATAD3A has conserved AAA ATPase domains, including the Walker A and Walker B motifs required for ATP binding and hydrolysis, as well as Sensor I and II, and arginine finger motifs required for cooperative ATP hydrolysis65 (Figure 4A). The dominant ATAD3A p.(Gly355Asp) Walker A mutation causing spastic paraplegia inhibits ATPase activity16 (Figure 5B). ATAD3C, encoded by a proposed pseudogene15, lacks key Sensor I and arginine finger residues (Figure 4A and 20) and has yet to be confirmed as integral to mitochondrial membranes. Both the Sensor I and arginine finger domains of AAA ATPases are highly intolerant of substitution, with the arginine finger residue interacting in trans with the γ phosphate of ATP bound to the neighboring subunit65.
While AAA ATPases can form homo- or heterocomplexes, the composition of ATAD3 oligomers is unclear due to non-specificity of ATAD3 antibodies, and could theoretically include a combination of ATAD3C, ATAD3B, or ATAD3A molecules. In ATAD3dup patients, it seems that expression of chimeric ATAD3A/ATAD3C interferes with normal ATAD3 oligomerization, likely inhibiting ATPase activity due to the changes in critical residues of the AAA ATPase domain of the protein (Figure 4). Intriguingly, dimerization of ATAD3A reportedly relies on the N-terminal coiled-coil domain11,12,33, which is not altered in the chimeric ATAD3A/ATAD3C protein. Therefore, the observed changes in ATAD3 oligomerization in ATAD3dup patients likely result from inhibited ATP hydrolysis. Nonetheless, it is unclear why severe OXPHOS changes were observed primarily in heart despite similar changes to ATAD3 oligomerization observed in all tissues studied. Relative amounts of ATAD3A to ATAD3B may play a role, with ATAD3B shown to both co-purify with ATAD3A and proposed to negatively regulate ATAD3A interaction with mtDNA nucleoids11. Predictions of ATAD3 complex composition in ATAD3dup patients estimate that ~91% of hexamers would contain at least one chimeric ATAD3A/ATAD3C molecule20. However, the calculations do not take ATAD3B into account and tissue specific differences in ATAD3 ratios could potentially contribute to differences in tissue specificity and clinical presentation between the ATAD3dup and ATAD3del patients. Notably, while ATAD3A expression is relatively consistent across brain regions, ATAD3B expression is markedly higher in cerebellum (Figure S3A and S3B). ATAD3del patients express an ATAD3A-like protein under the ATAD3B promoter (Figure 1A) and this alteration in ratios could contribute to the severe pontocerebellar hypoplasia observed in these patients but not in most ATAD3dup patients.
Summary
We have identified recurrent de novo duplications in the ATAD3 locus that lead to expression of a chimeric ATAD3A/ATAD3C protein, resulting in a characteristic phenotype that includes perinatal lethal mitochondrial hypertrophic cardiomyopathy with tissue-specific isolated complex I deficiency. The remarkable clinical consistency and the ability to discern the duplications on WES and WGS means patients can be readily identified, even from retrospective data, and families advised as to the low recurrence risk. Furthermore, ATAD3dup patients present with intriguing changes to ATAD3 oligomerization that point to a role for ATAD3 interaction and its ATPase function in the disease pathomechanism.
Limitations of Study
Most of the patients with ATAD3 duplications we identified were from retrospective cohorts. The distinctive clinical phenotype led to ascertainment of additional cases based on testing of patients with similar symptoms. Hence the range of clinical phenotypes caused by ATAD3 duplications may be broader than we identified, as exemplified by the single patient within our ATAD3dup cohort that did not have obvious cardiomyopathy. As well, the data imply that the ATAD3A/ATAD3C fusion protein is the primary, dominant cause of mitochondrial dysfunction rather than the extra copy of ATAD3B. Given the tissue specificity of the clinical and biochemical defect, more definitive proof would need carefully titrated expression studies in a cardiomyocyte cell line. These observations underscore the need for further clarification of the molecular mechanisms of ATAD3 in mitochondrial biogenesis, pathogenic versus non-pathogenic ATAD3 variation, and tissue specific determination of the individual contribution and interactions between ATAD3C, ATAD3B, and ATAD3A.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Thorburn (david.thorburn@mcri.edu.au).
Materials Availability
There are restrictions to the availability of the patient cell lines/tissues/genetic material described in this manuscript due to ethics and patient/family consent. Requests directed to the Lead Contact will be considered on an individual basis.
Data and Code Availability
Original/source data for Figures 2, 3 and 4, including gels, western blots, qRT-PCR data and associated supplemental data, is available through the Lead Contact. The published article includes proteomics datasets generated and analyzed during this study.
There are restrictions to the availability of genomics and microarray datasets used in this publication due to patient privacy and ethical restrictions. Any requests should be directed to the Lead Contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Subjects
Patient samples were obtained with familial informed consent as part of diagnostic investigations, and associated studies were approved by the respective institutional human research ethics boards. The overall study was approved by the Royal Children’s Hospital Institutional Review Board (HREC reference numbers 34228, 36291, and 36378). Brief descriptions of the 17 affected individuals (from 16 families) are provided in Table 1, with more detailed clinical information available in Table S1.
Cell culture
Patient fibroblasts generated from skin biopsies were grown at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen).
METHOD DETAILS
Genomic sequencing and CNV detection
DNA from individuals was extracted from blood, fibroblasts and tissues and investigated through whole exome sequencing (WES) with clinical or targeted analysis, whole genome sequencing (WGS), 10x Genomics Chromium linked-read WGS, Oxford Nanopore Technologies long read WGS and microarrays.
Trio WGS was performed on individual P1 and her parents by IIlumina Cambridge Ltd on an Illumina HiSeq X Ten with 2 × 150 bp reads and minimum mean coverage depth of 37x. For patients P2b and P3, singleton WGS was performed at the Kinghorn Centre for Clinical Genomics, also on Illumina HiSeq X Ten sequencers with 2 × 150 bp reads and >30x mean nuclear coverage. For these patients, sequences were aligned to the b37d5 human reference genome using BWA-mem68. Single nucleotide variants (SNVs) and small insertions/deletions (indels) were called using GATK HaplotypeCaller (v3.7.2)69 while copy number variants (CNVs) were called for P1 using canvas SPW70 and variant annotation was performed using SnpEff v4.3m371 and custom scripts were used for variant filtration and prioritization. For P2b and P3, variants were annotated using ENSEMBL’s VEP (v74)72 and converted to an SQLite database using Gemini (v0.11.0 and v0.17.2, respectively)73. Gemini databases were imported into Seave74 for variant filtering and prioritization. Mitochondrial (mt)DNA SNV and indel analysis was performed using Mity75, while structural variants (both nuclear and mtDNA) were identified using ClinSV (Minoche et al, manuscript in preparation), which combines Lumpy76 and CNVnator35 to look for discordantly mapping read pairs, split-mapping reads and depth of coverage changes.
Six individuals, P4-P9, formed part of a previously assembled cohort of complex I deficiency patients (DT24, DT98, DT86, DT106, DT93 and DT101, respectively60). Their DNA underwent singleton WES with targeted analysis of 2,273 mitochondrial and differential diagnosis genes at the Broad Institute of MIT and Harvard as published (per patient 1 in 77). Analysis of mtDNA for SNVs, indels and large deletions was performed as described previously78. Sequencing data for this targeted gene set were also analyzed for large CNVs using ExomeDepth (v.1.1.6)36, with a reference set compiled using sequence data from 92 individuals generated by identical laboratory and computational procedures.
Individuals P10–13 and P14 underwent clinical WES at Juntendo University, Saitama Medical University and the Victorian Clinical Genetics Services, respectively. For P10–13, exomes were captured using SureSelect V5 (for P10) or TruSeq (for P11–13) exome enrichment kits (Agilent Technologies) according to the manufacturers’ protocols. Sequencing was performed using 2 × 150 bp reads on an Illumina HiSeq2500 (for P10) or GAIIx (for P11–13). Whole-exome sequencing was performed using methods and a bioinformatic filtering pipeline as previously published79. The CNV analysis was performed on whole exome sequencing data using XHMM37 (eXome‐Hidden Markov Model) and ExomeDepth36 with default parameters, and the detected CNVs were visualized using R software. For P14, the library was prepared using an Agilent SureSelect QXT CREv1 kit and sequenced on an Illumina HiSeq 4000 sequencer with a targeted mean coverage of 100x and a minimum of 90% of bases sequenced to at least 15x. Data was processed using Cpipe80 with analysis of SNVs and indels in nuclear and mtDNA-encoded genes as well as large mtDNA deletions performed as per81, and for CNVs within the nuclear genome using Ximmer82, which combines CNV callers XHMM37, ExomeDepth36 and CODEX83.
Genomic DNA was extracted from fibroblasts using Nucleobond CB20 for P3, P4, P5 and P8 for 10x Genomics Chromium linked-read WGS (performed at the Victorian Clinical Genetics Services), and was assessed by Qubit (ds DNA High Sensitivity kit) and Agilent TapeStation genomic DNA kits prior to library preparation using the Chromium™ Genome Chip Kit v2 as per manufacturer’s genome protocol (10x Genomics, Pleasanton, CA, USA) and barcoded using the Chromium Controller (10x Genomics). DNA samples were partitioned into gel bead-in-emulsion (GEMs), with the introduction of 16 bp 10x barcodes (bead barcode), 10 bp unique molecular identifiers (UMI), and sample-specific 8 bp barcodes onto the DNA fragments and sequencing library preparation occurring in parallel. Library yield and fragment sizes were determined using an Agilent TapeStation D1000 kit (Agilent, Santa Clara, CA, USA). The barcoded libraries were sequenced on an Illumina NovaSeq 6000 system (paired-end 2× 150 bp). The resulting BCL files were demultiplexed and converted to FASTQ files using bcl2fastq2 (v2.20.0.422). Linked-reads were aligned to the GRCh37/hg19 reference genome using the LongRanger (v2.2.2) pipeline84 to allow structural variant (SV) discovery.
Long-read Whole Genome Sequencing (WGS)
Long-read WGS using Oxford Nanopore Technologies (ONT) was performed to further validate the configuration of the breakpoint in P3. The gDNA isolated using Nucleobond CB20 was prepared using the 1D ligation library prep kit (SQK-LSK109) as per manufacturer’s instructions, using 1500ng input DNA for DNA repair and end-prep and 1000ng end-prep DNA for adaptor ligation. 600ng of adaptor ligated DNA library were sequenced on a PromethION flow cell (FLO-PRO002). Real time data acquisition and base calling were performed using MinKNOW ID 19.06.8 and Guppy v3.0.04. To visualize reads mapping across the duplicated region of the ATAD3 locus, an artificial version of the hg19 chr1 reference sequence was generated by duplicating the sequence in region chr1:1392271–1460342. ONT sequence reads marked as “pass” by Guppy were mapped to the artificial chr1 reference sequence using minimap2 (version 2.17)85 using settings “-ax map-ont -t 4 -MD”. R (version 3.5.0) and the Bioconductor Gviz package (version 1.24.0)86 were used to visualize aligned reads with a mapping quality larger than 0 and read length no less than 2 Kb.
SNP microarray
Chromosomal SNP microarrays were performed on P3, P4, P5, P7, P8 and P15 by the Victorian Clinical Genetics Services (VCGS) using Illumina Infinium Global Screening Array-24 (GSA) v2.0 BeadChips according to manufacturer’s instructions (Illumina, San Diego, CA, USA) and analyzed by Illumina KaryoStudio v1.4.3.0 Build 37. The Infinium GSA-24 array has 16 probes across the region chr1:g.1,378,485–1,470,321 (GRCh37/hg19; six in ATAD3A, four each in ATAD3B and ATAD3C, and 2 intergenic).
Evaluation of CNV boundaries and breakpoints
Intragenic duplications were investigated by long-range PCR amplification using Expand long range dNTPack kit (Roche); primers used and their binding sites within the ATAD3 region are shown in Table S4. Sanger sequencing of the PCR products was performed as described previously60 to define the breakpoints.
Gene-specific RNA studies
To study the effect of the ATAD3 duplication on mRNA expression and splicing, total RNA was extracted from cultured fibroblasts grown with or without cycloheximide treatment (to stabilize transcripts normally degraded via nonsense mediated decay) and cDNA was synthesized as described previously60. cDNA transcripts were amplified by long range PCR as described previously using indicated primers (Table S4). For qRT-PCR assays, RNA was isolated from three different culture plates for each fibroblast line using TRIzol RNA Isolation Reagents (Invitrogen) and then reverse-transcribed to cDNA using SuperScript IV First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. Synthesized cDNA was used as a template in qRT-PCR performed in a Roche LightCycler 480 using Power SYBR Green PCR Master Mix (Applied Biosystems) with primer combinations specific for ATAD3A, ATAD3B or ATAD3C (Table S4), and analysis performed by 2-ΔΔCT method87.
Genomic DNA and protein sequence analyses
Pairwise alignments and sequence comparisons of DNA and protein sequences were performed using EMBOSS Needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/)88. The Miropeats program (v2.02) was employed to compare and graphically display the homology between the three human ATAD3 paralogs67. A threshold of 900 was set for intrachromosomal comparison of human paralogs.
Respiratory chain enzyme and mtDNA analyses
Spectrophotometric enzyme assays assessing mitochondrial oxidative phosphorylation (OXPHOS) and citrate synthase activities were performed in duplicate for samples from individuals P2a, P3–9, P14 and P16 in Melbourne and for P10–13 in Chiba, as described previously89. For heart, skeletal muscle and liver samples, post-nuclear supernatants were prepared at ~4°C by homogenizing 10 – 100 mg (wet weight) of tissue in 9 volumes of Tissue buffer (5 mM HEPES, 1 mM EGTA, 210 mM mannitol, 70 mM sucrose, pH 7.2) with 10 strokes in a glass–glass homogenizer followed by centrifugation at 600 RCF for 10 min. Supernatants were frozen and thawed three times and aliquots for citrate synthase and complex I assays were given 5 x 6 pulses at setting 3, 30% duty cycle on a Branson microtip sonifier. For cultured fibroblasts (four confluent 175 sq.cm. flasks), mitochondria were prepared at ~4°C by resuspending cell pellets in MegaFb buffer (250 mM sucrose, 2 mM HEPES, 0.1 mM EGTA, pH 7.4), followed by two rounds of Teflon/glass homogenization and centrifugation at 600 RCF for 10 min at 4°C. Supernatants from the two rounds were pooled and centrifuged for 10 min at 14,400 RCF and resuspended in 400 μl of MegaFb buffer. An aliquot was retained for assay of complex III and remaining sample was centrifuged for 10 min at 14,400 RCF and resuspended in 1ml of Hypotonic buffer (25 mM potassium phosphate, pH 7.2, 5 mM MgCl2). After centrifugation at 14,400 RCF as before, pellets were resuspended in 300 μl of Hypotonic buffer and frozen and thawed three times.
Enzyme assays measured the change of absorbance in a 1 mL cuvette at 30 °C over 3 min in 50 mM potassium phosphate buffer, pH 7.4 using a Cary 300 Bio Spectrophotometer (Agilent Technologies Australia). Protein levels were measured using bicinchoninic acid (BCA) and enzyme activities were expressed as initial rates (nmol/min/mg) except for complexes III and IV, which were expressed as rate constants (/min/mg). Enzyme activities were expressed as % of the mean of at least 6 matrix-matched control samples.
Rotenone-sensitive complex I (NADH-CoQ1 oxidoreductase) activity was assayed with 50 μM NADH, 1 mM KCN, 10 μM antimycin A, 0.1% (w/v) fatty acid free BSA, 50 μM coenzyme Q1 (CoQ1) with and without 2.5 μM rotenone (Absorbance 340 nm, mM extinction coefficient 6.81).
Complex II (succinate: CoQ1 oxidoreductase) activity was assayed after 10 min pre-incubation with 10 mM sodium succinate, 1 mM KCN, 10 μM antimycin A, 2.5 μM rotenone following addition of 50 μM CoQ1 (Absorbance 280 nm, mM extinction coefficient 12).
Complex III (decylbenzylquinol: cytochrome c oxidoreductase) activity was assayed with 1 mM n-dodecylmaltoside, 1 mM KCN, 2.5 μM rotenone, 0.1% (w/v) fatty acid free BSA, 100 μM decylbenzylquinol and 15 μM cytochrome c. The assay was terminated by addition of a few grains of L-ascorbic acid to the cuvette to fully reduce cytochrome c, prior to a final absorbance reading that was subtracted from each data point before creating a log plot against time to calculate a first-order rate constant from the slope (Absorbance 550 nm, mM extinction coefficient 18.7). A 10mM decylbenzylquinol stock solution was freshly prepared in ethanol each day by reducing 300 μl 10 mM decylbenzylquinone with ~5 mg of KBH4 and 10 μl 0.1 M HCl. After vortexing and 3 min at room temperature, 12 μl 3 M HCl was added to stabilize the decylbenzylquinol and excess KBH4 crystals were removed by brief centrifugation.
Complex IV (cytochrome c oxidase) activity was assayed with 15 μM reduced cytochrome c and the assay was terminated by addition of K3Fe(CN)6 to 1 mM to complete oxidation of cytochrome c prior to a final absorbance reading and calculation of a first-order rate constant (Absorbance 550 nm, mM extinction coefficient 18.7). Reduced cytochrome c was prepared weekly by adding a few grains of ascorbic acid to 0.5 ml 2 mM cytochrome c, with removal of excess ascorbic acid using a PD-10 Sephadex G-25 gel filtration column (Sigma). Prior to use each day, the absorbance at 550 and 565 nm was measured and the solution was discarded if the A550 / A565 was <6.
Citrate synthase activity was assayed with 0.1 mM 5,5’ -dithio-bis-(2-nitrobenzoic acid) (DTNB), 0.1 mM acetyl CoA with and without 0.1 mM oxaloacetic acid (pH adjusted to 7.4) (Absorbance 412 nm, mM extinction coefficient 13.6).
The relative abundance of mtDNA vs nDNA was tested in liver, cardiac or skeletal muscle biopsies from several patient vs control tissues, as described previously90. This quantitative PCR method compares the relative levels of mtDNA to nDNA using TaqMan probes directed against the MT-ND1 gene to represent mtDNA and a single copy nuclear reference gene, CFTR. Results are calculated by the 2-ΔΔCT method87 and are the means of at least two independent runs of triplicate measurements. % mtDNA/nDNA ratios are calculated relative to the mean of at least 6 matrix-matched control samples.
SDS- and Blue Native (BN)-PAGE
For preparation of protein extracts, heart biopsy sections were solubilized using Extraction buffer (1.5% n-Dodecyl-β-d-maltopyranoside, 25 mM HEPES and 100 mM NaCl) for 20 min on ice and then centrifuged at 16,000 rpm for 20 min at 4°C. Protein concentrations of the supernatants were determined by BCA analysis91, and 2–5 μg of each sample was further solubilized in sodium dodecyl sulfate (SDS)/glycerol solubilization buffer (125mM Tris pH8.8, 40% glycerol, 4% SDS, 100mM DTT, 0.01% Bromophenol blue, protease inhibitor cocktail) and analyzed by SDS-PAGE as described previously61.
Mitochondria were isolated from 20–50 mg heart or liver biopsies, or fibroblasts (~1.5 X 106 cells) using differential centrifugation as described92, and protein concentration determined by BCA analysis. BN-PAGE was performed as described previously43,92, using 30 μg of mitochondria isolated from tissue or cells and solubilized in 1% Triton X-100 or 1% Digitonin.
Immunoblotting
SDS-PAGE or BN-PAGE gels were transferred to polyvinylidene difluoride (PVDF) membrane and probed with primary antibodies against ATAD328, ANT3 (Abcam anti-SLC25A6, ab154007), SDHA/complex II 70 kDa subunit (for SDS-PAGE: Molecular Probes, A-11142; for BN-PAGE: Abcam, ab14715), Porin/VDAC1 (Abcam ab14734), Total OXPHOS Human WB Antibody Cocktail (Abcam, ab110411), complex I subunit NDUFA9 raised in-house93, ATAD3A (Abnova, H00055210-D01), ATAD3B (Abnova, H00083858-B01P), Tim23 (BD Bioscience, 611223), V5 (Invitrogen, R960–25), Core1 (ThermoFisher, 459140) and Cox4 (Abcam, ab110261). Blots were incubated with anti-mouse or anti-rabbit IgG secondary antibodies (GE Healthcare) and developed with Clarity Western ECL Substrate (Bio-Rad Laboratories).
Protein expression and immunoprecipitation
ATAD3A (NM_001170535.3), ATAD3C (NM_001039211.3) and chimeric ATAD3A/ATAD3C (NM_001170535.3 exons1-exon11 + NM_001039211.3 exons8–12) were cloned into the CS-CA-MCS vector [obtained from Riken BRC DNA Bank, RDB05963 (deposited by Dr Hiroyuki Miyoshi)]. For co-immunoprecipitation studies, HEK293FT cells seeded at 2×106 cells in 6-cm plates were transfected with the indicated plasmids using Lipofectamine2000 (ThermoFisher Scientific) according to the manufacturer’s protocols. Five days post-transfection, HEK293FT cells overexpressing ATAD3 were lysed for 10 min on ice with 0.15 ml of RIPA buffer (Nacalai Tesque, Inc), and the insoluble fraction was removed by centrifugation. The soluble fraction containing 100 μg protein was incubated with Protein G-Sepharose (Sigma) coated with anti-V5 antibody (Invitrogen). Immunoprecipitations were performed according to the manufacturer’s guidelines.
Quantitative mass spectrophotometry
Mass spectrometry on primary fibroblast material was performed label-free, using sample preparation methodology previously described77 with modifications. Fibroblasts from three to four different control subjects and two to three subcultures of patient cells were solubilized in 1% w/v sodium deoxycholate, 100 mM Tris-HCl (pH 8.1) and incubated at 99°C for 10 min with shaking. Samples were then incubated for 10 min at 60°C in a sonicator waterbath, followed by the addition of 5 mM Tris(2-carboxyethyl)phosphine, 20 mM chloroacetamide and incubation for 5 min at 99°C with shaking. Denatured and alkylated proteins were digested at 37°C overnight using MS grade trypsin (Thermo Scientific). Detergent was removed by extraction into ethyl acetate in the presence of 2% formic acid (FA) on SDB-RPS (styrenedivinylbenzene-reverse phase sulfonate; Supelco Analytical) stage-tips made in-house as described94,95. Peptides reconstituted in 0.5% FA were loaded onto small cation exchange (Empore Cation Exchange-SR, Supelco Analytical) stage-tips and the tips washed with 20% acetonitrile (ACN), 0.5% FA. Peptides were eluted over five fractions of increasing amounts (45–300 mM) of freshly prepared ammonium acetate in 20% ACN, 0.5% FA, followed by a final elution step using 80% ACN, 5% ammonium hydroxide. Fractions were concentrated by vacuum centrifugation and desalted on SDB-XC poly(styrene-divinyl-benzene; Supelco Analytical) stage-tips or Pierce C18 Spin Tips (Thermo Scientific) as per manufacturer instructions.
Peptides were reconstituted in 2% ACN, 0.1% trifluoroacetic acid and analyzed by online nano-HPLC/electrospray ionization-MS/MS on a Q Exactive Plus or Orbitrap Fusion Lumos connected to an Ultimate 3000 HPLC (Thermo-Fisher Scientific). For measurements of peptides derived from S1a, S3 and P4, analysis was performed on a Q Exactive Plus with chromatography conditions and instrument parameters as described previously77. Peptides were loaded onto a trap column (Acclaim PepMap C18 nano-trap column 75 μm x 2 cm, 3 μm particle size, 100 Å pore size; Thermo Fisher Scientific) at 5 μL/min in 2% (v/v) ACN containing 0.1% (v/v) FA for 3 min before switching the pre-column in line with the analytical column (Acclaim PepMap RSLC C18 analytical column 75 μm x 50 cm, 2 μm particle size, 100 Å pore size; Thermo Fisher Scientific). Separation of peptides was performed at 300 nL/min using a 120 min non-linear ACN gradient of buffer A [0.1% (v/v) FA, 2% (v/v) ACN, 5% DMSO] and buffer B [0.1% (v/v) FA in ACN, 5% DMSO]. The mass spectrometer was operated in positive-ionization mode with spray voltage set at 1.9 kV and source temperature at 275°C. Data were collected using the Data Dependent Acquisition (DDA) using m/z 350–1550 at 120000 resolution with AGC target of 5e5. The “top speed” acquisition method mode (3 sec cycle time) on the most intense precursor was used whereby peptide ions with charge states ≥2–5 were isolated with isolation window of 1.6 m/z and fragmented with high energy collision (HCD) mode with stepped collision energy of 30 ±5%. Fragment ion spectra were acquired in Orbitrap at 15000 resolution. Dynamic exclusion was activated for 30s. For P3 and P5, measurements were performed on an Orbitrap Fusion Lumos. Liquid chromatography (LC) coupled MS/MS was carried out on an Orbitrap Lumos mass spectrometer (ThermoFisher Scientific) with a nanoESI interface in conjunction with an Ultimate 3000 RSLC nanoHPLC (Dionex Ultimate 3000). The LC system was equipped with a trap column (Acclaim PepMap C18 nano-trap column 75 μm x 2 cm, 3 μm particle size, 100 Å pore size; Thermo Fisher Scientific) and an analytical column (Acclaim PepMap RSLC C18 analytical column 75 μm x 50 cm, 2 μm particle size, 100 Å pore size; Thermo Fisher Scientific). The tryptic peptides were injected into the trap column at an isocratic flow of 5 μL/min of 2% (v/v) CH3CN containing 0.1% (v/v) FA for 5 min applied before the trap column was switched in-line with the analytical column. The eluents were 5% DMSO in 0.1% v/v FA (solvent A) and 5% DMSO in 100% v/v CH3CN and 0.1% v/v FA (solvent B). The flow gradient was (i) 0–6min at 3% B, (ii) 6–95 min, 3–22% B (iii) 95–105min 22–40% B (iv) 105–110min, 40–80% B (v) 110–115min, 80–80% B (vi) 115–117min, 80–3% and equilibrated at 3% B for 10 minutes before the next sample injection.
The mass spectrometer was operated in positive-ionization mode with spray voltage set at 1.9 kV and source temperature at 275°C. The mass spectrometer was operated in the data-dependent acquisition mode MS spectra scanning from m/z 300–1600 at 120000 resolution with AGC target of 4e5. The “top speed” acquisition method mode (3 sec cycle time, 125 min run) on the most intense precursor was used whereby peptide ions with charge states ≥2–5 were isolated with isolation window of 0.7 m/z and fragmented with high energy collision (HCD) mode with stepped collision energy of 35 ±5%. Fragment ion spectra were acquired in Orbitrap at 50000 resolution. Dynamic exclusion was activated for 30s.
Raw files were analyzed using the MaxQuant platform (96; v.1.6.5.0) searching against the Uniprot human database containing reviewed canonical and isoform variants in FASTA format (June 2019) and a database containing common contaminants. Default search parameters for a label-free (LFQ) experiment were used with the following modifications “Deamidation (NQ)” was added as a variable modification, “Label-free quantification” option was set to “LFQ”, “LFQ min. ratio count” set to 1, “Label min. ratio count” set to 1, Match type “No matching” and “Peptides for quantification” set to “Unique”. Statistical analyses were performed using the Perseus platform (97; v.1.6.2.2). For the peptide heatmaps, peptides.txt files were loaded, and spurious identifications removed as above. “Normalized LFQ intensity” values were log2 transformed and entries containing “NaN” (indicating missing data) were replaced with zero. Heatmaps were generated using the built-in hierarchical clustering function in Perseus platform as above and only peptides that were detected in at least two samples shown. For the protein profile plots, columns and rows from the MaxQuant output file proteinGroups.txt were loaded and identifications annotated as “Only identified by site”, “Reverse”, and “Potential contaminant” were removed from the data. Following this, “Normalized LFQ intensity” values were log2 transformed and profile plots were generated using the built-in profile plot function. For the ATAD3 smoothed peptide plots, LFQ intensity values for missing peptides (NaN) were imputed with the lowest detected log2 value (16.2979 for S3; S1a; 16.7114 for P4) in each experiment, which represents the limit of detection for the instrumentation. The difference of the means was plotted using the geom_smooth function of ggplot2 library in RStudio (v. 3. 6. 2). The curve shows a loess smoothed curve with 95% confidence interval. Matrices for protein and peptide data are available in Tables S5 and S6, respectively.
QUANTIFICATION AND STATISTICAL ANALYSIS
Details of statistical details of experiments for qRT-PCR (Figure 3B), quantitative proteomics (Figure 3D and F) and respiratory chain enzymology (Figure 4D) can be found in the figures and/or figure legends. Analyses of qRT-PCR and respiratory chain enzymology data were performed using GraphPad Prism (v7.04). Analyses of quantitative proteomics data are detailed in the Methods section for “Quantitative mass spectrophotometry”.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-ATAD3 | He et al., 2007 | N/A |
| Rabbit anti-SLC25A6 (ANT3) | Abcam | Cat# ab154007; RRID: AB_2619664 |
| Mouse anti-SDHA (complex II 70 kDa subunit) | Molecular Probes | Cat# A-11142; RRID: AB_221579 |
| Mouse anti-SDHA (complex II 70 kDa subunit) | Abcam | Cat# ab14715; RRID: AB_301433 |
| Mouse anti-VDAC1 (Porin) | Abcam | Cat# ab14734; RRID: AB_443084 |
| Mouse Total OXPHOS Human WB Antibody Cocktail | Abcam | Cat# ab110411; RRID: AB_2756818 |
| Rabbit anti-NDUFA9 | Stroud et al., 2013 | N/A |
| Rabbit anti-ATAD3A | Abnova | Cat# H00055210-D01; RRID: AB_10718149 |
| Mouse anti-ATAD3B | Abnova | Cat# H00083858-B01P; RRID: AB_1571499 |
| Mouse anti-TIM23 | BD Bioscience | Cat# 611223; RRID: AB_398755 |
| Mouse Anti-V5 tag | Invitrogen | Cat# R960-25; RRID: AB_2556564 |
| Mouse anti-UQCRC1 (core 1) 16D10AD9AH5 | Thermo Fisher Scientific | Cat# 459140; RRID: AB_2532227 |
| Mouse anti-COX4 | Abcam | Cat# ab110261; RRID: AB_10862101 |
| Sheep anti-mouse IgG- horseradish peroxidase conjugated antibody | GE Healthcare | Cat# NA931; RRID: AB_772210 |
| Donkey anti-rabbit IgG- horseradish peroxidase conjugated antibody | GE Healthcare | Cat# NA934; RRID: AB_772206 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Human control and ATAD3dup patient tissue biopsies | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Triton X-100 | Sigma Aldrich | Cat# T9284 |
| Digitonin- High purity | Merck | Cat# 300410 |
| Lipofectamine2000 | ThermoFisher Scientific | Cat# 11668030 |
| RIPA buffer | Nacalai Tesque | Cat# 16488-34 |
| Protein G-Sepharose | Sigma | Cat# P3296 |
| MS grade trypsin | Thermo Scientific | Cat# LTS90057 |
| Critical Commercial Assays | ||
| Clarity Western ECL Substrate | Bio-Rad | Cat# 170-5060 |
| SureSelect Human all exome V5 | Agilent Technologies | Cat# 5190-6209 |
| TruSeq exome enrichment kit | Agilent Technologies | Cat# FC-121-1008 |
| SureSelect QXT Reagent Kit CRE V1 | Agilent Technologies | Cat# G9681B |
| SureSelect XT CRE V2 | Agilent Technologies | Cat# 5190-9493 |
| Nucleobond CB20 | Macherey-Nagel | Cat# 740507 |
| Qubit (dsDNA High Sensitivity kit) | Thermo Fisher Scientific | Cat# Q32851 |
| TapeStation genomic DNA ScreenTape | Agilent Technologies | Cat# 5067-5365 |
| TapeStation genomic DNA Reagents | Agilent Technologies | Cat# 5067-5366 |
| Chromium™ Genome Chip Kit v2 | 10x Genomics | Cat# 120257 |
| TapeStation High Sensitivity D1000 ScreenTape | Agilent Technologies | Cat# 5067-5584 |
| TapeStation High Sensitivity D1000 Reagents | Agilent Technologies | Cat# 5067-5585 |
| 1D ligation library prep kit | Oxford Nanopore Technologies | Cat# SQK-LSK109 |
| PromethION flow cell | Oxford Nanopore Technologies | Cat# FLO-PRO002 |
| Infinium Global Screening Array-24 (GSA) v3.0 kit | Illumina | Cat# 20030770 |
| Expand long range dNTPack kit | Roche | Cat# 04 829 034 001 |
| TRIzol RNA isolation reagent | Invitrogen | Cat# 15596018 |
| SuperScript IV First-Strand Synthesis System | Invitrogen | Cat#18091200 |
| Power SYBR Green PCR Master Mix | Applied Biosystems | Cat# 4368577 |
| Lipofectamine2000 | ThermoFisher Scientific | Cat# 11668030 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Human control and ATAD3dup patient skin fibroblast cell lines | This paper | N/A |
| HEK293FT | ThermoFisher Scientific | Cat# R70007 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| See Supplemental Table 2 for complete list of primers | This paper | N/A |
| Recombinant DNA | ||
| CS-CA-MCS plasmid | Riken BRC DNA Bank | Cat# RDB05963 |
| Software and Algorithms | ||
| BWA-mem | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/; RRID: SCR_010910 |
| GATK HaplotypeCaller | Poplin et al., 2017 | https://gatk.broadinstitute.org/hc/en-us; RRID: SCR_001876 |
| canvas SPW | Ivakhno et al., 2018 | https://github.com/Illumina/canvas |
| SnpEff | Cingolani et al., 2012 | http://snpeff.sourceforge.net/; RRID: SCR_005191 |
| ENSEMBL’s VEP | McLaren et al., 2016 | http://www.ensembl.org/info/docs/tools/vep/index.html; RRID: SCR_007931 |
| Gemini | Paila et al., 2013 | https://gemini.readthedocs.io/en/latest/; RRID: SCR_014819 |
| Seave | Gayevskiy et al., 2019 | https://github.com/KCCG/seave |
| Mity | Puttick et al., 2019 | https://github.com/KCCG/mity |
| ClinSV | Minoche et al, manuscript in preparation | https://github.com/KCCG/ClinSV |
| ExomeDepth | Plagnol et al., 2012 | https://cran.r-project.org/web/packages/ExomeDepth/; RRID: SCR_002663 |
| XHMM | Fromer et al., 2012 | https://statgen.bitbucket.io/xhmm/ |
| Cpipe | Sadedin et al., 2015 | https://melbournegenomics.github.io/ |
| Ximmer | Sadedin et al., 2018 | http://ssadedin.github.io/ximmer/; RRID: SCR_016427 |
| bcl2fastq2 | Ilumina | https://sapac.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html; RRID:SCR_015058 |
| LongRanger | 10x Genomics | https://support.10xgenomics.com/genome-exome/software/pipelines/latest/installation |
| MinKNOW | Oxford Nanopore Technologies | https://nanoporetech.com/community |
| Guppy | Oxford Nanopore Technologies | https://nanoporetech.com/community |
| minimap2 | Li, 2018 | https://lh3.github.io/minimap2/ |
| R | N/A | https://www.r-project.org/; RRID: SCR_001905 |
| RStudio | RStudio | https://rstudio.com/; RRID: SCR_000432 |
| ggplot2 | N/A | https://cran.r-project.org/web/packages/ggplot2/index.html; RRID: SCR_014601 |
| Bioconductor Gviz package | Hahne and Ivanek, 2016 | https://bioconductor.org/packages/release/bioc/html/Gviz.html |
| KaryoStudio | Illumina | https://sapac.support.illumina.com/array/array_software/karyostudio.html |
| Miropeats | Parsons, 1995 | http://www.littlest.co.uk/software/bioinf/old_packages/miropeats/ |
| MaxQuant platform | Tyanova, Temu and Cox, 2016 | https://www.maxquant.org/; RRID: SCR_014485 |
| Perseus platform | Tyanova, Temu, Sinitcyn, et al., 2016 | https://www.maxquant.org/perseus/; RRID: SCR_015753 |
| Other | ||
| Empore™ SDB-RPS (styrenedivinylbenzene-reverse phase sulfonate) solid phase extraction disk | Supelco Analytical | Cat# 66886-U |
| Empore™ Cation Exchange-SR solid phase extraction disk | Supelco Analytical | Cat# 66889-U |
| Empore™ SDB-XC poly(styrene-divinyl-benzene) solid phase extraction disk | Supelco Analytical | Cat# 66884-U |
| Pierce C18 Spin Tips | Thermo Scientific | Cat# LTS84850 |
Acknowledgements
We thank Meredith Wilson, Stefanie Eggers, Natalie Tan, Katherine Neas, Tegan Stait, Sabrina Kaminsky and Wendy Fagan for technical assistance, contributions to data collection, and/or clinical discussions. We appreciate the kind gifts of antibodies from Ian Holt and Antonella Spinazzola (anti-ATAD3) and Diana Stojanovski (anti-ANT3). We acknowledge the Melbourne Mass Spectrometry and Proteomics Facility (MMSPF) for provision of instrumentation, training, and technical support.
This research was supported by grants and fellowships from the Australian National Health and Medical Research Council (NHMRC) [1164479 (DRT, JC, DAS, AGC); 1155244 (DRT); 1068278 (CS); 1140851 (DAS)], the US Department of Defense Congressionally Directed Medical Research Programs PR170396 (DRT, JC, MTR, DAS), the New South Wales Office of Health and Medical Research Council Sydney Genomics Collaborative grant (JC), the Victorian Government’s Operational Infrastructure Support Program (DRT, JC), the Australian Mito Foundation (AEF, AGC, CS, DRT, JC, DAS, DHH, MTR), the National Council on Science and Technology (CONACYT) (RR), the Vincent Chiodo Charitable Trust (DRT), the Angela Wright Bennett Foundation and the McCusker Charitable Foundation (GB), the US National Institutes of Health R35GM122455 (VKM), and the Howard Hughes Medical Institute Investigator Program (VKM). Diagnosis of two patients was supported by the Australian Genomics Health Alliance (JC, DRT, CS, GB), which is funded by the NHMRC (1113531) and the Medical Research Future Fund. This work was supported in part by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development, AMED (19ek0109273, 18ek0109177) to KM, AO and YO, the Program for an Integrated Database of Clinical and Genomic Information from AMED (19kk0205014, 18kk0205002) to YO, MEXT-Supported Program for the Private University Research Branding Project, the Japan Society for the Promotion of Science (JSPS) KAKENHI 19H03624 (Grant-in-Aid for Scientific Research (B)) to YO and YK. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Footnotes
Declaration of Interests
RJT is an employee of Illumina, Inc.
References
- 1.Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, Suomalainen A, Thorburn DR, Zeviani M, and Turnbull DM (2016). Mitochondrial diseases. Nat Rev Dis Primers 2, 16080. [DOI] [PubMed] [Google Scholar]
- 2.Rahman J, and Rahman S. (2018). Mitochondrial medicine in the omics era. Lancet 391, 2560–2574. [DOI] [PubMed] [Google Scholar]
- 3.Stenton SL, and Prokisch H. (2018). Advancing genomic approaches to the molecular diagnosis of mitochondrial disease. Essays Biochem 62, 399–408. [DOI] [PubMed] [Google Scholar]
- 4.Stankiewicz P, and Lupski JR (2002). Genome architecture, rearrangements and genomic disorders. Trends Genet 18, 74–82. [DOI] [PubMed] [Google Scholar]
- 5.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, and Eichler EE (2002). Recent segmental duplications in the human genome. Science 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 6.Pu L, Lin Y, and Pevzner PA (2018). Detection and analysis of ancient segmental duplications in mammalian genomes. Genome Res 28, 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho CM, and Lupski JR (2016). Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet 17, 224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mefford HC, and Eichler EE (2009). Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev 19, 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittwald P, Gambin T, Szafranski P, Li J, Amato S, Divon MY, Rodriguez Rojas LX, Elton LE, Scott DA, Schaaf CP, et al. (2013). NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res 23, 1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merle N, Féraud O, Gilquin B, Hubstenberger A, Kieffer-Jacquinot S, Assard N, Bennaceur-Griscelli A, Honnorat J, and Baudier J. (2012). ATAD3B is a human embryonic stem cell specific mitochondrial protein, re-expressed in cancer cells, that functions as dominant negative for the ubiquitous ATAD3A. Mitochondrion 12, 441–448. [DOI] [PubMed] [Google Scholar]
- 12.Gilquin B, Taillebourg E, Cherradi N, Hubstenberger A, Gay O, Merle N, Assard N, Fauvarque MO, Tomohiro S, Kuge O, et al. (2010). The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol. Cell. Biol 30, 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M, Bellance N, Rossignol R, Koopman WJ, Willems PH, Mayatepek E, Bossinger O, and Distelmaier F. (2009). C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS One 4, e7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goller T, Seibold UK, Kremmer E, Voos W, and Kolanus W. (2013). Atad3 function is essential for early post-implantation development in the mouse. PLoS One 8, e54799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harel T, Yoon WH, Garone C, Gu S, Coban-Akdemir Z, Eldomery MK, Posey JE, Jhangiani SN, Rosenfeld JA, Cho MT, et al. (2016). Recurrent De Novo and Biallelic Variation of ATAD3A, Encoding a Mitochondrial Membrane Protein, Results in Distinct Neurological Syndromes. Am J Hum Genet 99, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper HM, Yang Y, Ylikallio E, Khairullin R, Woldegebriel R, Lin KL, Euro L, Palin E, Wolf A, Trokovic R, et al. (2017). ATPase-deficient mitochondrial inner membrane protein ATAD3A disturbs mitochondrial dynamics in dominant hereditary spastic paraplegia. Hum Mol Genet 26, 1432–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peeters-Scholte CMPCD, Adama van Scheltema PN, Klumper F, Everwijn SMP, Koopmans M, Hoffer MJV, Koopmann TT, Ruivenkamp CAL, Steggerda SJ, van der Knaap MS, et al. (2017). Genotype-phenotype correlation in ATAD3A deletions: not just of scientific relevance. Brain 140, e66. [DOI] [PubMed] [Google Scholar]
- 18.Peralta S, González-Quintana A, Ybarra M, Delmiro A, Pérez-Pérez R, Docampo J, Arenas J, Blázquez A, Ugalde C, and Martín MA (2019). Novel ATAD3A recessive mutation associated to fatal cerebellar hypoplasia with multiorgan involvement and mitochondrial structural abnormalities. Mol Genet Metab 128, 452–462. [DOI] [PubMed] [Google Scholar]
- 19.Desai R, Frazier AE, Durigon R, Patel H, Jones AW, Dalla Rosa I, Lake NJ, Compton AG, Mountford HS, Tucker EJ, et al. (2017). ATAD3 gene cluster deletions cause cerebellar dysfunction associated with altered mitochondrial DNA and cholesterol metabolism. Brain 140, 1595–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunning AC, Strucinska K, Munoz Oreja M, Parrish A, Caswell R, Stals KL, Durigon R, Durlacher-Betzer K, Cunningham MH, Grochowski CM, et al. (2020). Recurrent De Novo NAHR Reciprocal Duplications in the ATAD3 Gene Cluster Cause a Neurogenetic Trait with Perturbed Cholesterol and Mitochondrial Metabolism. Am J Hum Genet 106, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baudier J. (2018). ATAD3 proteins: brokers of a mitochondria-endoplasmic reticulum connection in mammalian cells. Biol Rev Camb Philos Soc 93, 827–844. [DOI] [PubMed] [Google Scholar]
- 22.Gerhold JM, Cansiz-Arda S, Lõhmus M, Engberg O, Reyes A, van Rennes H, Sanz A, Holt IJ, Cooper HM, and Spelbrink JN (2015). Human Mitochondrial DNA-Protein Complexes Attach to a Cholesterol-Rich Membrane Structure. Sci Rep 5, 15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Issop L, Fan J, Lee S, Rone MB, Basu K, Mui J, and Papadopoulos V. (2015). Mitochondria-associated membrane formation in hormone-stimulated Leydig cell steroidogenesis: role of ATAD3. Endocrinology 156, 334–345. [DOI] [PubMed] [Google Scholar]
- 24.Poston CN, Krishnan SC, and Bazemore-Walker CR (2013). In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J Proteomics 79, 219–230. [DOI] [PubMed] [Google Scholar]
- 25.Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, and Ting AY (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6, 10.7554/eLife.24463.24001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rone MB, Midzak AS, Issop L, Rammouz G, Jagannathan S, Fan J, Ye X, Blonder J, Veenstra T, and Papadopoulos V. (2012). Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol Endocrinol 26, 1868–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, et al. (2012). Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res 40, 6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. (2007). The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol 176, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Bouzar C, Cottet-Rousselle C, Zagotta I, Lamarche F, Wabitsch M, Tokarska-Schlattner M, Fischer-Posovszky P, Schlattner U, and Rousseau D. (2016). Resveratrol inhibits lipogenesis of 3T3-L1 and SGBS cells by inhibition of insulin signaling and mitochondrial mass increase. Biochim Biophys Acta 1857, 643–652. [DOI] [PubMed] [Google Scholar]
- 30.van den Ecker D, Hoffmann M, Muting G, Maglioni S, Herebian D, Mayatepek E, Ventura N, and Distelmaier F. (2015). Caenorhabditis elegans ATAD-3 modulates mitochondrial iron and heme homeostasis. Biochem Biophys Res Commun 467, 389–394. [DOI] [PubMed] [Google Scholar]
- 31.Harel T, and Lupski JR (2018). Genomic disorders 20 years on-mechanisms for clinical manifestations. Clin Genet 93, 439–449. [DOI] [PubMed] [Google Scholar]
- 32.Fang HY, Chang CL, Hsu SH, Huang CY, Chiang SF, Chiou SH, Huang CH, Hsiao YT, Lin TY, Chiang IP, et al. (2010). ATPase family AAA domain-containing 3A is a novel anti-apoptotic factor in lung adenocarcinoma cells. J Cell Sci 123, 1171–1180. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Sun X, Hu D, Prosdocimo DA, Hoppel C, Jain MK, Ramachandran R, and Qi X. (2019). ATAD3A oligomerization causes neurodegeneration by coupling mitochondrial fragmentation and bioenergetics defects. Nat Commun 10, 1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin G, Xu C, Zhang X, Long J, Rezaeian AH, Liu C, Furth ME, Kridel S, Pasche B, Bian XW, et al. (2018). Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat Immunol 19, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abyzov A, Urban AE, Snyder M, and Gerstein M. (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21, 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, Wood NW, Hambleton S, Burns SO, Thrasher AJ, et al. (2012). A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 28, 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O’Donovan MC, Owen MJ, et al. (2012). Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 91, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alkan C, Coe BP, and Eichler EE (2011). Genome structural variation discovery and genotyping. Nat Rev Genet 12, 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frazier AE, Holt IJ, Spinazzola A, and Thorburn DR (2017). Reply: Genotype-phenotype correlation in ATAD3A deletions: not just of scientific relevance. Brain 140, e67. [DOI] [PubMed] [Google Scholar]
- 40.Newman S, Hermetz KE, Weckselblatt B, and Rudd MK (2015). Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints. Am J Hum Genet 96, 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gates SN, and Martin A. (2020). Stairway to translocation: AAA+ motor structures reveal the mechanisms of ATP-dependent substrate translocation. Protein Sci 29, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peralta S, Goffart S, Williams SL, Diaz F, Garcia S, Nissanka N, Area-Gomez E, Pohjoismaki J, and Moraes CT (2018). ATAD3 controls mitochondrial cristae structure in mouse muscle, influencing mtDNA replication and cholesterol levels. J Cell Sci 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wittig I, Braun HP, and Schägger H. (2006). Blue native PAGE. Nat Protoc 1, 418–428. [DOI] [PubMed] [Google Scholar]
- 44.Fassone E, and Rahman S. (2012). Complex I deficiency: clinical features, biochemistry and molecular genetics. J Med Genet 49, 578–590. [DOI] [PubMed] [Google Scholar]
- 45.Wilkie AOM, and Goriely A. (2017). Gonadal mosaicism and non-invasive prenatal diagnosis for ‘reassurance’ in sporadic paternal age effect (PAE) disorders. Prenat Diagn 37, 946–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frazier AE, Thorburn DR, and Compton AG (2019). Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology. J Biol Chem 294, 5386–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakonen AH, Davidzon G, Salemi R, Bindoff LA, Van Goethem G, Dimauro S, Thorburn DR, and Suomalainen A. (2007). Abundance of the POLG disease mutations in Europe, Australia, New Zealand, and the United States explained by single ancient European founders. Eur J Hum Genet 15, 779–783. [DOI] [PubMed] [Google Scholar]
- 48.Wedatilake Y, Brown RM, McFarland R, Yaplito-Lee J, Morris AA, Champion M, Jardine PE, Clarke A, Thorburn DR, Taylor RW, et al. (2013). SURF1 deficiency: a multi-centre natural history study. Orphanet J Rare Dis 8, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamazaki T, Murayama K, Compton AG, Sugiana C, Harashima H, Amemiya S, Ajima M, Tsuruoka T, Fujinami A, Kawachi E, et al. (2014). Molecular diagnosis of mitochondrial respiratory chain disorders in Japan: focusing on mitochondrial DNA depletion syndrome. Pediatr Int 56, 180–187. [DOI] [PubMed] [Google Scholar]
- 50.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. (2006). Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 38, 1038–1042. [DOI] [PubMed] [Google Scholar]
- 51.Liu P, Carvalho CM, Hastings PJ, and Lupski JR (2012). Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev 22, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paigen K, and Petkov PM (2018). PRDM9 and Its Role in Genetic Recombination. Trends Genet 34, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Khera AV, Francioli LC, Gauthier LD, Wang H, Watts NA, et al. (2019). An open resource of structural variation for medical and population genetics. bioRxiv 578674, 10.1101/578674. [DOI] [Google Scholar]
- 54.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, and Carter NP (2009). DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet 84, 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jordan VK, Zaveri HP, and Scott DA (2015). 1p36 deletion syndrome: an update. Appl Clin Genet 8, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heilstedt HA, Ballif BC, Howard LA, Kashork CD, and Shaffer LG (2003). Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin Genet 64, 310–316. [DOI] [PubMed] [Google Scholar]
- 57.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marquet V, Bourthoumieu S, Dobrescu A, Laroche-Raynaud C, and Yardin C. (2017). Familial 1p36.3 microduplication resulting from a 1p-9q non-reciprocal translocation. Eur J Med Genet 60, 583–588. [DOI] [PubMed] [Google Scholar]
- 59.Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, and Thorburn DR (1999). Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology 52, 1255–1264. [DOI] [PubMed] [Google Scholar]
- 60.Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, et al. (2010). High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet 42, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, Laskowski A, Garone C, Liu S, Jaffe DB, et al. (2012). Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med 4, 118ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamalainen RH, Manninen T, Koivumaki H, Kislin M, Otonkoski T, and Suomalainen A. (2013). Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc Natl Acad Sci U S A 110, E3622–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Protasoni M, Pérez-Pérez R, Lobo-Jarne T, Harbour ME, Ding S, Peñas A, Diaz F, Moraes CT, Fearnley IM, Zeviani M, et al. (2020). Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV. EMBO J 39, e102817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerdes F, Tatsuta T, and Langer T. (2012). Mitochondrial AAA proteases--towards a molecular understanding of membrane-bound proteolytic machines. Biochim Biophys Acta 1823, 49–55. [DOI] [PubMed] [Google Scholar]
- 65.Wendler P, Ciniawsky S, Kock M, and Kube S. (2012). Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta 1823, 2–14. [DOI] [PubMed] [Google Scholar]
- 66.Sallevelt SC, de Die-Smulders CE, Hendrickx AT, Hellebrekers DM, de Coo IF, Alston CL, Knowles C, Taylor RW, McFarland R, and Smeets HJ (2017). De novo mtDNA point mutations are common and have a low recurrence risk. J Med Genet 54, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parsons JD (1995). Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci 11, 615–619. [DOI] [PubMed] [Google Scholar]
- 68.Li H, and Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. (2017). Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178, 10.1101/201178. [DOI] [Google Scholar]
- 70.Ivakhno S, Roller E, Colombo C, Tedder P, and Cox AJ (2018). Canvas SPW: calling de novo copy number variants in pedigrees. Bioinformatics 34, 516–518. [DOI] [PubMed] [Google Scholar]
- 71.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, and Ruden DM (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, and Cunningham F. (2016). The Ensembl Variant Effect Predictor. Genome Biol 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paila U, Chapman BA, Kirchner R, and Quinlan AR (2013). GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol 9, e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gayevskiy V, Roscioli T, Dinger ME, and Cowley MJ (2019). Seave: a comprehensive web platform for storing and interrogating human genomic variation. Bioinformatics 35, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Puttick C, Kumar KR, Davis RL, Pinese M, Thomas DM, Dinger ME, Sue CM, and Cowley MJ (2019). mity: A highly sensitive mitochondrial variant analysis pipeline for whole genome sequencing data. bioRxiv 852210, 10.1101/852210. [DOI] [Google Scholar]
- 76.Layer RM, Chiang C, Quinlan AR, and Hall IM (2014). LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15, R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lake NJ, Webb BD, Stroud DA, Richman TR, Ruzzenente B, Compton AG, Mountford HS, Pulman J, Zangarelli C, Rio M, et al. (2017). Biallelic Mutations in MRPS34 Lead to Instability of the Small Mitoribosomal Subunit and Leigh Syndrome. Am J Hum Genet 101, 239–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lieber DS, Calvo SE, Shanahan K, Slate NG, Liu S, Hershman SG, Gold NB, Chapman BA, Thorburn DR, Berry GT, et al. (2013). Targeted exome sequencing of suspected mitochondrial disorders. Neurology 80, 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohda M, Tokuzawa Y, Kishita Y, Nyuzuki H, Moriyama Y, Mizuno Y, Hirata T, Yatsuka Y, Yamashita-Sugahara Y, Nakachi Y, et al. (2016). A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet 12, e1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sadedin SP, Dashnow H, James PA, Bahlo M, Bauer DC, Lonie A, Lunke S, Macciocca I, Ross JP, Siemering KR, et al. (2015). Cpipe: a shared variant detection pipeline designed for diagnostic settings. Genome Med 7, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akesson LS, Eggers S, Love CJ, Chong B, Krzesinski EI, Brown NJ, Tan TY, Richmond CM, Thorburn DR, Christodoulou J, et al. (2019). Early diagnosis of Pearson syndrome in neonatal intensive care following rapid mitochondrial genome sequencing in tandem with exome sequencing. Eur J Hum Genet 27, 1821–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sadedin SP, Ellis JA, Masters SL, and Oshlack A. (2018). Ximmer: a system for improving accuracy and consistency of CNV calling from exome data. Gigascience 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang Y, Oldridge DA, Diskin SJ, and Zhang NR (2015). CODEX: a normalization and copy number variation detection method for whole exome sequencing. Nucleic Acids Res 43, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, Kyriazopoulou-Panagiotopoulou S, Masquelier DA, Merrill L, Terry JM, et al. (2016). Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hahne F, and Ivanek R. (2016). Visualizing Genomic Data Using Gviz and Bioconductor In Statistical Genomics. Methods in Molecular Biology vol 1418 Mathé E, and Davis S, eds. (New York, NY: Humana Press; ), pp. 335–351. [DOI] [PubMed] [Google Scholar]
- 87.Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 88.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frazier AE, and Thorburn DR (2012). Biochemical analyses of the electron transport chain complexes by spectrophotometry. Methods Mol Biol 837, 49–62. [DOI] [PubMed] [Google Scholar]
- 90.Pagnamenta AT, Taanman JW, Wilson CJ, Anderson NE, Marotta R, Duncan AJ, Bitner-Glindzicz M, Taylor RW, Laskowski A, Thorburn DR, et al. (2006). Dominant inheritance of premature ovarian failure associated with mutant mitochondrial DNA polymerase gamma. Hum Reprod 21, 2467–2473. [DOI] [PubMed] [Google Scholar]
- 91.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, and Klenk DC (1985). Measurement of protein using bicinchoninic acid. Anal Biochem 150, 76–85. [DOI] [PubMed] [Google Scholar]
- 92.Johnston AJ, Hoogenraad J, Dougan DA, Truscott KN, Yano M, Mori M, Hoogenraad NJ, and Ryan MT (2002). Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J Biol Chem 277, 42197–42204. [DOI] [PubMed] [Google Scholar]
- 93.Stroud DA, Formosa LE, Wijeyeratne XW, Nguyen TN, and Ryan MT (2013). Gene knockout using transcription activator-like effector nucleases (TALENs) reveals that human NDUFA9 protein is essential for stabilizing the junction between membrane and matrix arms of complex I. J Biol Chem 288, 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, et al. (2016). Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538, 123–126. [DOI] [PubMed] [Google Scholar]
- 95.Kulak NA, Pichler G, Paron I, Nagaraj N, and Mann M. (2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11, 319–324. [DOI] [PubMed] [Google Scholar]
- 96.Tyanova S, Temu T, and Cox J. (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319. [DOI] [PubMed] [Google Scholar]
- 97.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original/source data for Figures 2, 3 and 4, including gels, western blots, qRT-PCR data and associated supplemental data, is available through the Lead Contact. The published article includes proteomics datasets generated and analyzed during this study.
There are restrictions to the availability of genomics and microarray datasets used in this publication due to patient privacy and ethical restrictions. Any requests should be directed to the Lead Contact.