Abstract
The mechanism by which the cholesterol-dependent cytolysins (CDCs) assemble their giant β-barrel pore in cholesterol-rich membranes has been the subject of intense study in the past two decades. A combination of structural, biophysical, and biochemical analyses has revealed deep insights into the series of complex and highly choreographed secondary and tertiary structural transitions that the CDCs undergo to assemble their β-barrel pore in eukaryotic membranes. Our knowledge of the molecular details of these dramatic structural changes in CDCs has transformed our understanding of how giant pore complexes are assembled and has been critical to our understanding of the mechanisms of other important classes of pore-forming toxins and proteins across the kingdoms of life. Finally, there are tantalizing hints that the CDC pore-forming mechanism is more sophisticated than previously imagined and that some CDCs are employed in pore-independent processes.
Keywords: perfringolysin O, streptolysin O, sterol, CD59, intermedilysin, β-barrel
INTRODUCTION
The establishment of the first life forms required the cell membrane, which separates the systems necessary for life and reproduction from the extracellular milieu. The presence of an unregulated pore in the cell membrane can seriously impact cell viability and/or function. Therefore, the cell membrane was likely one of the first targets in the competition among early life forms for nutrients and ecological niches, which may explain why the pore-forming toxins/proteins are one of the most ubiquitous classes of toxins/proteins and present in species from all kingdoms. The cholesterol-dependent cytolysins (CDCs) make up one of the largest families of bacterial pore-forming toxins. Paul Ehrlich (11) initially described the pore-forming activity of CDCs in 1898, when he reported that Clostridium tetanus had two toxins: a lethal neurotoxin and a hemolysin that lysed rabbit and horse erythrocytes. Today we know that this hemolysin was a CDC. Since then, CDC genes have been identified in nearly 50 gram-positive bacterial species, and now a few have been identified in gram-negative species, as well (26).
The CDCs assemble a giant β-barrel pore in the membrane and are generally considered pathogenesis factors in the gram-positive bacteria (reviewed in 48, 83, 93), whereas the few CDCs from gram-negative organisms are primarily produced by species present only in anaerobic soils and sediments (26). Those gram-positive species that produce a CDC also spend the vast majority of their existence as harmless commensal or soil organisms; thus, the evolution of the CDCs might be due largely to reasons that are unrelated to disease and have more to do with establishing a niche in the host or in one of the many terrestrial environments.
The emphasis of this article is the molecular assembly of the CDC pore, which is one of the most complex mechanisms known for a pore-forming toxin. There have been several thorough reviews on the contribution of the CDCs to bacterial pathogenesis in recent years (22, 43, 48), which will not be covered here. We apologize in advance if we cannot include all structure-function studies because space is limited.
An Overview of the Cholesterol-Dependent Cytolysin Pore-Forming Mechanism
The general scheme by which the CDCs form a pore (Figure 1d) is summarized briefly here to orient readers. In-depth descriptions of each stage of the pore assembly, as we currently understand it, follow. Initially, CDC monomers bind to the membrane via their receptor (cholesterol or CD59) (14, 20), which activates them to interact with each other to form a stable dimer and then assemble into an oligomeric prepore structure (86). Bound monomers do not act as nucleation sites for unbound monomers: Each monomer must bind the membrane where they are activated to interact with each other to form a prepore structure. The prepore structure is defined as the oligomerized complex that is formed by the interaction of the membrane-bound monomers prior to the assembly and membrane insertion of the β-barrel pore.
Figure 1.
Overview of the molecular structure and pore-forming mechanism of the cholesterol-dependent cytolysins (CDCs). (a) Ribbon representations of soluble perfringolysin O (PFO) (77) and (b) a magnified view of D3. The various structures discussed in the text are illustrated. Abbreviations: αHB1 and αHB2, α-helical bundle 1 and 2; CRM, cholesterol recognition motif; D, domain; TMH1 and TMH2, transmembrane hairpin 1 and 2; UDP, undecapeptide. (c) (Top) Overlay of the PFO (blue) and anthrolysin O (ALO; salmon) or (bottom) D4-only α-carbon structures illustrates the differences in the bend angles between D1–3 and D4 and UDP structures. (d) Overview of the CDC pore-forming mechanism. Membrane-bound monomers form unstable dimers, which are infrequently converted to stable dimers when thermal fluctuations displace β5 from β4. Upon stable dimer formation, conformational information is transmitted to the second monomer to induce the dissociation of β5, thereby removing the rate-limiting step to the addition of monomers to stable dimers and higher-order oligomers. After prepore formation, the αHBs unfurl and refold into TMHs. The intermediate structure of the TMHs prior to formation of the β-barrel pore is unknown (brackets). Colors correlate with panels a and b. (e) The folding (95) and rotation (73) models proposed for the vertical collapse of the prepore. All structures were rendered with UCSF Chimera (64).
Once the prepore is completed, it is converted to the pore complex in which each CDC monomer contributes two amphipathic β-hairpins (transmembrane β-hairpins or TMHs), which are derived from the two α-helical bundles (αHBs) in D3 (Figure 1a,b). In a cooperative manner, these hairpins assemble into and insert the membrane-spanning β-barrel pore, which is composed of 35–45 monomers (dependent on the CDC) with a diameter of 250–300 Å. To insert the β-barrel pore, the CDC prepore structure undergoes a downward collapse of approximately 40 Å (8).
THE CHOLESTEROL-DEPENDENT CYTOLYSIN STRUCTURE
Primary Structure
The first DNA-derived primary structures of the CDCs (38, 51, 96, 98) revealed that they exhibited 40–60% identity. These initial studies identified a highly conserved signature motif for the CDCs, an 11-residue peptide (ECTGLAWEWWR) near the CDCs’ C termini that was variously termed the tryptophan-rich loop or the undecapeptide loop (UDP). Few CDCs exhibit any significant variation in this sequence, and as described below, studies have only recently provided insight into its critical function in the CDC pore-forming mechanism.
Nearly all of the CDCs have a canonical amino-terminal signal peptide, which directs their secretion via the general secretory pathway. Streptococcus pneumoniae pneumolysin (PLY) is notable because it lacks a signal peptide and is found in the cytoplasm of the bacterial cell (33). Cell lysis was thought to mediate release, but subsequent studies (1, 3, 66, 67) have provided evidence that an intact peptidoglycan layer largely retains PLY before its release by an as yet unknown mechanism. PLY must be released from the cell, however, because it clearly has extracellular effects (2).
Cholesterol-Dependent Cytolysin Crystal Structure
Rossjohn et al. (78) solved the Clostridium perfringens perfringolysin O (PFO) crystal structure (Figure 1a) in 1997, and it has proven to be a well-behaved protein. Since then, five additional CDC monomer structures have been reported: intermedilysin (ILY) from Streptococcus intermedius, anthrolysin O (ALO) from Bacillus anthracis, suilysin from Streptococcus suis, listeriolysin O (LLO) from Listeria monocytogenes, and streptolysin O (SLO) from Streptococcus pyogenes (4, 15, 39, 65, 101). The CDC crystal structures exhibit a high degree of similarity to that of PFO (Figure 1), and they are largely composed of a β-sheet secondary structure with two prominent α-HBs (αHB1 and αHB2) in domain 3 (D3) that flank its core β-sheet. Two obvious differences in their structures are (a) the bend between D1–3 (large lobe) and D4 (small lobe) and (b) the structure of the conserved UDP when the structures are overlaid with each other, as is shown for PFO and ALO (Figure 1c). The lack of a conserved UDP three-dimensional (3D) structure was surprising considering its conserved primary structure. These differences do not appear to be due to crystal contacts, at least in most cases: It appears that residues within the UDP form different intramolecular contacts with residues outside of this region. Another important feature of the PFO monomer structure is the relatively unstable nature of the interface that D3 (mainly αHB1) forms with D1 and D2 (Figure 1a), which is noncomplementary and largely hydrophilic (78). Initially, these features were unexplained curiosities; however, as described below, they are critical features of the pore-forming mechanism.
THE MEMBRANE-SPANNING DOMAIN OF PERFRINGOLYSIN O
We start our description of the CDC mechanism at the end of the pore-forming process, with the identification of the membrane-spanning structure, for two important reasons: First, the crystal structure of the soluble monomer provided the starting point of the assembly process, and second, the identification of the membrane-spanning structure was solved shortly thereafter and provided us insight into the final structure of the pore. This information enabled us and others to investigate the intervening steps of the pore-forming process.
Each monomer within the large oligomeric pore complex contributes two membrane-spanning TMHs to the formation of the β-barrel pore (84, 85); these TMHs are derived from the two D3 αHBs (αHB1 and αHB2; Figure 1a,b). The large radius of curvature of the CDC pore makes it necessary to employ two hairpins per monomer to allow the TMHs to form intermolecular backbone hydrogen bonds so that a contiguous β-barrel structure is formed in the membrane.
The Cholesterol-Dependent Cytolysin Domain 3 Protein Fold Is Widespread in Nature
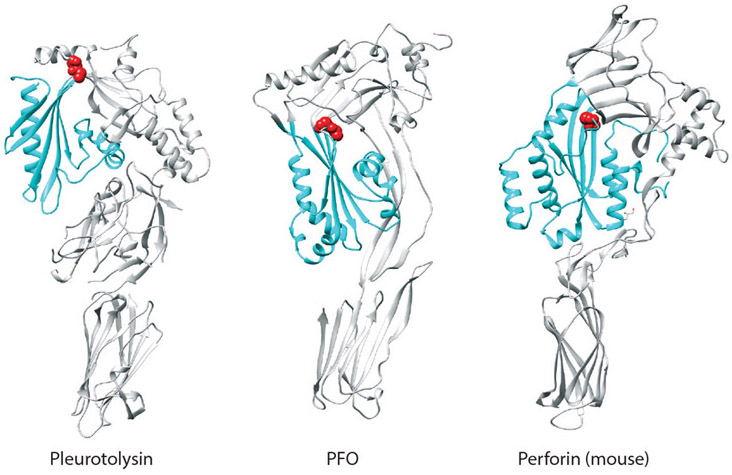
The D3 protein fold and the transition from αHBs to β-strand TMHs were unique to the CDCs, until recently, when the crystal structures of membrane attack complex/perforin (MACPF) family protein members revealed a protein fold similar to the CDC D3 (21, 76, 89) (Figure 2). The CDCs and MACPF proteins do not share any significant similarity in their primary structures, but a double glycine motif is conserved at the junction of β4 and β5 (Figures 1b and 2), which appears to be a hinge region necessary to expose the edge of β4, which then has edge-on interactions with β1 of another monomer (described below) (70). It has been shown for perforin (41) and for a MACPF from the edible oyster mushroom (pleurotolysin) (44) that, like the CDCs, their twin αHBs are converted into two TMHs.
Figure 2.
The cholesterol-dependent cytolysin (CDC) D3-like domains of membrane attack complex/perforin (MACPF) family proteins. Shown are the ribbon representations of the α-carbon backbone of the crystal structures of pleurotolysin from the edible oyster mushroom, Pleurotus ostreatus (44); perfringolysin O (PFO) (77); and mouse perforin (76). The conserved D3 structures are shown in cyan, and the conserved double glycine motif located at the top of β-strand 4 is shown in red.
MEMBRANE RECOGNITION AND BINDING
A defining feature of the CDCs is the absolute dependence of their pore-forming mechanism on the presence of membrane cholesterol, which acts as a receptor or coreceptor for all CDCs (7, 29, 68). The lipid environment of the cholesterol also exerts a significant influence on the recognition and binding of cholesterol by the CDCs, which has made the cholesterol-dependent binding interaction with the cell membrane a much more complex and intriguing subject of investigation in recent years.
Most CDCs appear to bind directly to cholesterol, whereas three of them (the archetype of which is the CDC from S. intermedius, ILY) bind human CD59 (18, 20, 100). Two other CDCs, from Streptococcus tigurinus and Streptococcus pseudopneumoniae, are predicted to bind CD59 based on a conserved Tyr-X-Tyr-X14-Ser-Arg motif present in D4 of CD59-binding CDCs (100). There are hints that receptors or coreceptors other than cholesterol or CD59 may be used by some CDCs with pore-forming activity that exhibits some species selectivity (35). Curiously, the pore-forming activity of the CD59-binding CDCs remained cholesterol dependent (14, 19), which prompted the investigation into the basis of this dependence. These studies revealed the true nature of the cholesterol recognition/binding motif (CRM) and how it is used by the cholesterol-specific CDCs, as well as the CD59-specific CDCs.
Cholesterol-Specific Cholesterol-Dependent Cytolysins
Until 2007, the UDP was tacitly accepted as the CDC CRM. There were good reasons for this, as it is highly conserved, except in the CD59-binding CDCs (18, 20) and had been shown to enter the membrane (24, 57). Subsequent studies (71, 90, 91) showed that three short hydrophobic loops (L1–L3) at the base of D4 (Figure 1a), near the UDP, also entered the membrane and that the CRM function resided within the structure of one or more of these loops. The membrane insertion of loops L1–L3 was also necessary for the pore-forming function of the CD59-binding ILY (90, 91), which suggested a common feature between those CDCs that used cholesterol or CD59 as their receptor.
In 2010, Farrand et al. (14) identified a motif in loop L1, a Thr-Leu pair (Figure 1a), which is conserved in all known CDCs except for the predicted CD59-binding CDC from S. tigurinus, which has a Thr-Ile pair. This motif mediates binding to cholesterol and is now recognized as the CRM of the CDCs. Our current understanding of the binding interaction is that the CRM recognizes the cholesterol 3-β-hydroxyl headgroup and possibly parts of the A-ring. This interaction is likely weak due to the limited interface between the small headgroup of cholesterol and the CDC CRM. After the CRM recognizes the cholesterol, the side chains of amino acids in L2 and L3 insert into the bilayer and stabilize the interaction of the monomer with the membrane (69, 91). The UDP also inserts into the membrane (24, 57, 81) and helps to anchor the monomer to the surface, but as described below, its role is more complex than its service as another membrane anchor point. A recent hydrogen/deuterium (H/D) exchange study (36) with PFO (36) showed that H/D exchange is minimal (compared to that in the soluble monomer) in L1 upon membrane binding, which is consistent with its role as the CRM. The H/D exchange in L3 and the UDP was also decreased, but to a lesser extent, whereas it was not stabilized in L2, even though L2 has been shown to face the bilayer (71).
One of the complicating factors when measuring the affinity of the interaction of CDCs with the bilayer is that they begin to rapidly oligomerize, which quickly increases the avidity of this interaction. Therefore, mutations that affect the rate of oligomerization of the membrane-bound monomers can also have a major impact on the apparent affinity of the binding interaction.
CD59-Specific Cholesterol-Dependent Cytolysins
Nagamune et al. (56) described the human cell–specific nature of the S. intermedius CDC ILY, which was clearly inconsistent with cholesterol serving as its receptor. Giddings et al. (20) subsequently identified the human form of CD59 as the receptor for ILY. CD59 is a glycophosphatidylinositol (GPI)-anchored protein found on most cell types, and it is an inhibitor of the terminal membrane attack complex (MAC) of complement. When MAC is activated, the membrane-bound complement proteins C8α and C9 are bound by CD59, which blocks the assembly of the MAC pore on our cells. My colleagues and I were able to identify this receptor rather quickly owing to a coincidence 14 years earlier in which I was a member of the doctoral committee of Scott Rollins (who was in the lab of Peter Sims), who first purified and characterized human CD59 (75). They showed that CD59’s function was homologously restricted (i.e., human CD59 inhibited human MAC but not MAC from other species). This characteristic was consistent with the species-specific nature of ILY.
The binding site on CD59 for ILY (100) exhibits a remarkable similarity to the CD59 binding site for complement proteins C8α and C9 (30), yet the binding sites for CD59 on each protein bear no obvious similarity. Recently, the cocrystal structure of soluble CD59 (no GPI anchor) and ILY was solved (34), which confirmed most of the contacts for CD59 on ILY mapped by Wickham et al. (100) and suggested that CD59 makes contact with two monomers. Two aromatic residues, Y434 and Y436, identified by Wickham et al. (100) to significantly impact binding, were not present at the cocrystal interface. Their location suggests that they may interact with the GPI anchor of CD59, which was not present in the cocrystal structure. Aromatics residues are necessary for the interaction of the Clostridium septicum α-toxin with the GPI anchor of specific proteins (50) and are frequently found to contact galactose, which is present in the GPI anchor of CD59 (58).
Although ILY binds human CD59, its pore-forming activity remained cholesterol dependent (20, 90). The combined studies of LaChapelle et al. (subsequently Wickham) (40) and Farrand et al. (14) showed that ILY must disengage from CD59 to convert to the pore; thus, it was necessary that ILY form a second cholesterol-dependent interaction identical to that formed by the CRM and loops L2 and L3 in the cholesterol-binding CDCs. The CRM-mediated interaction maintains the interaction of ILY with the membrane as ILY disengages from CD59 (14, 90), but apparently ILY cannot interact with cholesterol prior to binding CD59.
The Lipid Environment of Cholesterol Impacts Membrane Binding
Early studies (23) showed that quantitative binding of PFO to liposomes required 55 mol% of cholesterol to 45 mol% POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine). A sharp transition in PFO liposome binding occurred between 45 and 50 mol% cholesterol in which binding went from 0% to 100%, thereby showing that only a fraction of the membrane cholesterol functioned as a CDC receptor. More recently, Nelson et al. (60) and Flanagan et al. (16) showed that the lipid environment of cholesterol has a dramatic impact on the binding of PFO to membranes. Generally, lipids that pack less well with cholesterol appear to increase binding of PFO to cholesterol, presumably by exposing the small headgroup of cholesterol to the CDC CRM.
The structure of loops L2 and L3 can also impact membrane binding, as specific mutations within loops L2 and L3 have been shown to increase or decrease binding of PFO (14, 32). We recently demonstrated that a single change in an L3 residue of PFO, Asp-434, to a lysine increased the binding affinity approximately 6-fold and the total bound monomers approximately 4-fold (13), although all substitutions for Asp-434 that we tested increased binding to some extent. Because aspartate exhibits the highest free energy of solvation and is therefore the most difficult amino acid to partition into the bilayer (unless it is protonated), it was suggested that the change in the binding parameters was due to replacement of aspartate with amino acids that have a lower free energy of partitioning side chains into the bilayer. Interestingly, the insertion of the β-barrel pore was 50% less efficient in the lysine mutant, although its prepore assembly was equally efficient compared with wild-type PFO. This suggested that the lipid environment surrounding a substantial fraction of the oligomers assembled with the mutant PFO was not compatible with the insertion of the β-barrel. These data support the idea that the CDCs can vary their binding parameters by manipulating the structures of L2–L3, thereby affecting the overall free energy of partitioning them into a specific lipid environment. When one considers that at least 5 residues of L2 and L3 insert into the membrane, the CDCs have a staggering potential to vary the residues that compose this interface (~3 × 106 possible combinations!). Hence, the CDCs may exhibit a high degree of binding specificity on the basis of the structures of the L2 and L3 loops and the lipid environment of the receptor.
Lewis X Glycans as Receptors for Some Cholesterol-Dependent Cytolysins?
Shewell et al. (87) recently suggested that the blood antigen sialyl LewisX (sLeX) is a receptor for PLY and that lacto-N-neotetraosyl ceramide is a receptor for SLO. An intriguing finding, it suggested that some cholesterol-binding CDCs may have a receptor, just as the CD59-binding CDCs do (18, 20, 100). Although Shewell et al. showed PLY bound to sLeX and LeX by surface plasmon resonance with an apparent Kd in the 20 μM range, a 1–10 million molar excess of purified sLeX to PLY was necessary to observe competitive inhibition of binding and hemolytic activity by PLY using human erythrocytes, whereas LeX exhibited no inhibition. Similarly, SLO was also found to bind lacto-N-neotetraose, but as in the studies with PLY, it was necessary to preincubate the SLO with nearly a 3 million–fold excess of the glycan to inhibit binding. Because such excessive molar ratios of glycan were necessary to compete with cell binding of SLO and PLY, it remains unclear whether these glycans are authentic receptors for these or other CDCs.
Allosteric Activation of the Monomers to Oligomerize
Palmer et al. (62) originally proposed that membrane binding allosterically activated the monomers to oligomerize. In support of this model, Heuck et al. (23) found that mutations in αHB1 at the D3–D1,2 interface, which affected the rate of membrane insertion of the TMHs, also affected the rate of membrane insertion of the UDP. Because membrane insertion of the UDP occurs prior to membrane insertion of the β-barrel pore, it was clear that these two events were conformationally coupled. Dowd et al. (9) provided additional support for this allosteric pathway by showing that a specific mutation within the UDP appeared to uncouple the pathway, which prevented the interaction of the monomers at the membrane surface. It also decreased binding, likely owing to the inability to oligomerize, which as indicated above, increases the avidity of the binding interaction. These studies suggested that the UDP is an intimate part of the allosteric pathway that couples membrane binding to activation of the monomers by perturbation of the D3 structure and/or its interface with D1 and D2, as is also suggested by different crystal forms of PFO (79). This role might also explain why CDC pore-forming activity is so sensitive to changes to its structure (31, 65, 81): Even though its 3D structure in the soluble monomer of the CDCs appears to vary considerably, the conserved nature of its primary structure might be necessary to adopt a specific final structure to properly function in this allosteric pathway.
The UDP typically contains the only cysteine in the CDC structure, although this residue is absent in the UDP of CD59-binding CDCs and a few other CDCs. The chemical modification of this cysteine with sulfhydryl-specific reagents significantly reduces pore-forming activity, which is restored if the modification is reversible (31). Its modification was reported to also induce a structural change in the soluble monomer, which was reversible upon restoring the sulfhydryl to its reduced state (31). This phenomenon bears further investigation, as it may provide critical insights into the nature of the structural change(s) induced by the UDP-mediated allosteric pathway. Why most CDCs maintain a cysteine at such a sensitive site also remains unclear, because its oxidation causes such a dramatic loss in pore-forming activity (31). There is limited evidence that it could be an environmental sensor for the CDC listeriolysin (LLO), produced by the intracellular pathogen L. monocytogenes (88). The LLO cysteine sulfhydryl was apparently oxidized and only reduced inside the phagosome by γ-interferon-inducible lysosomal thiol reductase.
Interestingly, ILY binds well to cholesterol-rich liposomes (but not cells) via its CRM, but it remains inert and cannot oligomerize into a functional complex (9). This suggests that the allosteric pathway that couples cholesterol binding to activation of the monomers has been uncoupled from the CRM in ILY and has been transferred to the CD59-binding site. The ILY UDP does not exhibit the UDP consensus sequence, which may have been necessary to uncouple the allosteric pathway from the cholesterol-binding site so that it could be rerouted to the CD59-binding site. Whether the ILY UDP is involved in coupling CD59 binding to monomer activation remains unknown.
ASSEMBLY OF THE PREPORE COMPLEX
The obligatory prepore intermediate is an oligomerized complex that has not assembled and inserted its β-barrel pore. All characterized β-barrel pore-forming toxins (βPFTs) undergo this intermediate step (8, 12, 17, 25, 28, 37, 52, 63, 74, 82, 86, 95). The purpose of the prepore remains unexplained, but it is likely necessary to assemble the β-barrel pore in the bilayer.
The Nonstochastic Assembly of the Prepore Oligomer
Shepard et al. (86) observed that the only sodium dodecyl sulfate (SDS)-resistant oligomeric species of PFO that were detected by gel analysis were the full-sized oligomers, even when its assembly was slowed at 4°C (which also prevents insertion of the β-barrel). Although it was not appreciated at the time, these results suggested that the assembly of the prepore structure was not stochastic.
A critical event in the formation of the prepore complex is the disengagement of β5 from β4 (Figure 1b) (25, 70). This action exposes the edge of β4 in the core β-sheet in one monomer so that it can interact and form backbone hydrogen bonds with the edge of β1 in the core β-sheet of a second monomer. It also allows an intermolecular π-stacking interaction to form between Y181 in β1 and an F318 in β4 (Figure 1b) in PFO (described in detail in the prepore assembly section below). It may also allow the D1 of the monomers to form additional noncovalent intermolecular interactions. If this transition is blocked, the prepore oligomers are sensitive to dissociation by SDS (25, 70).
The twin glycine residues (G324-G325 in PFO; Figure 1b) at the β-turn between the β4 and β5 strands appear to be a flexible hinge region, as side chain residues substituted for either residue prevent disengagement of β5 from β4 (70). This motif is present in all known CDCs and is one of the few CDC motifs that is conserved in the same location within the structures of most of the MACPF proteins (reviewed in 10).
Hotze et al. (27) performed studies that suggested an explanation for the apparent nonstochastic prepore assembly. Their studies suggested that membrane-bound PFO monomers interact in a reversible fashion and that the formation of a stable dimer was the rate-limiting step to pore formation (Figure 1d). On the basis of their studies, they suggested that normal thermal fluctuations within the structure of D3 would occasionally displace or partially displace β5 from β4. If a second monomer was in contact when this occurred, then β1 of the second monomer could form stabilizing intermolecular interactions with β4, thereby prevent-ing the reassociation of β5 with β4 and stabilizing the dimer. Their studies further suggested that conformational information was transmitted to D3 of the second monomer of the stable dimer, which induced the disengagement of its β5 from β4. Hence, formation of the stable dimer opened up the edge of β4 of the second monomer so that β1 of other membrane-bound monomers could readily form stabilizing interactions with a stable dimer or a higher-order oligomer without waiting for thermal fluctuations to initiate the displacement of β5. Hence, the addition of free membrane-bound monomers to existing oligomers would dominate over the initiation of new oligomers, thereby increasing the probability of completing functional pores.
It is likely that D1 also plays an important role in the assembly of the prepore complex, although its role is only now being explored. A recent report of the crystal structure of LLO identified D1–D1 interactions necessary for oligomerization (39), and Kacprzyk-Stokowiec et al. (36) showed that significant changes in the H/D exchange rates occurred in specific regions of D1 of PFO upon formation of the pore complex.
FORMATION OF THE PORE COMPLEX
Once the prepore is completed, a large β-barrel is assembled and inserted into the membrane bilayer to form the pore. The number of monomers per pore complex varies between different CDCs, but for PFO and PLY, typically 35–38 monomers compose a pore complex—unless, as suggested below, incomplete complexes can insert. Less is understood about the prepore-to-pore transition; it is difficult to trap intermediate stages of assembly between the stable prepore and the pore because this transition occurs rapidly once a prepore oligomer is completed (28).
The π-Stacking Interaction
Disengagement of the β5 from β4 exposes residue F318 in β4 (Figure 1b) (70), thereby allowing it to stack with Y181 in β1. The timing of and role of this interaction in pore formation is not fully understood. Also, the mutation of either Y181 or F318 does not yield the same phenotype in PFO: Alanine substitution of Y181 results in an SDS-sensitive prepore, whereas mutation of F318 yields an SDS-resistant prepore. This suggests that Y181 is involved in displacing β5 to allow the assembly of the SDS-resistant prepore. The SDS resistance of the F318 mutant suggests that the stacking interaction is not required for an SDS-resistant prepore. These data suggest that the stacking interaction is necessary to convert from the prepore to the pore and perhaps to lock in the register of the intermolecular backbone hydrogen bonds that subsequently form between TMH1 and TMH2 (70). In view of recent results from our lab (97), our current opinion is that the stacking interaction occurs upon prepore completion and may play a role in tilting the monomers, which, as described below, is necessary to bring two critical charged residues near enough to form a strong electrostatic interaction that triggers the prepore-to-pore transition (80, 97).
Electrostatic Trigger for Prepore-to-Pore Conversion
Prepore-to-pore conversion requires the disruption of the interface that D3, specifically αHB1, forms with domains 1 and 2 (Figure 1a). As indicated above, the crystal structure showed this interface to be metastable (78). The disruption of this interface is necessary so that αHB1 can refold into TMH1, a process that irrevocably commits the prepore to the formation of the pore (28).
Two charged residues of PFO, E183 in β1 and K336 in β5 (Figure 1b), form an intermolecular electrostatic interaction in the prepore that supplies the free energy to disrupt this interface (80). The loss of this interaction traps PFO in a stable prepore state: These two residues are within 4 Å of each other in the prepore complex, and molecular dynamics (MD) simulations suggest that they spontaneously form a salt bridge (80). Because the glutamate is present in β1, which is contiguous with αHB1, we proposed that its interaction with the lysine in β5 stresses the D3–D1,2 interface, which results in its disruption. This scenario is supported by the fact that we could restore pore-forming activity in electrostatic mutants by the introduction of a single destabilizing mutation at the interface between αHB1 and D1,2. To juxtapose these two residues in the prepore, it appears that D3 must tilt approximately 30°: We suspect the stacking interaction of Y181 and F318 contributes to structural changes that bring the charged residues sufficiently close to form their electrostatic interaction, although this remains unproven.
The Vertical Collapse of the Prepore
The perpendicular orientation of the PFO monomers to the membrane (8, 69, 71) suspends D3 40 Å above the bilayer surface; thus, the extended TMHs are only long enough to reach the membrane surface (Figure 1d), yet they do cross the bilayer (84, 85). Czajkowsky et al. (8), using atomic force microscopy (AFM), revealed that the prepore complex underwent a 40 Å vertical collapse upon the formation of the pore complex. This observation was subsequently confirmed by fluorescence-based distance measurements with PFO (71) and cryo-electron microscopy (cryoEM)-based 3D reconstructions of the related PLY prepore and pore (95). The vertical collapse of the prepore to insert the β-barrel pore remains unique to the CDC pore-forming mechanism.
How this collapse is achieved remains unclear, but D2 likely plays a pivotal role because its structure connects D4 with D1–3, and it forms the major fraction of the interface with αHB1 (Figure 1a,c). We (8) and others (95) proposed that once D3 disengaged from its interface with D2, its stabilizing influence on D2 would be lost and D2 would fold to bring D1 and D3 closer to the membrane (8) (D2 folding; Figure 1e). This is consistent with the presence of a bulge on the outer ring surface between D4 and D2 of the PLY-derived cryoEM pore structure (95). Reboul et al. (73) proposed an alternate model in which D2 retains its elongated structure and simply rotates D1 and D3 toward the surface (D2 rotation; Figure 1e), which is also supported by the recent cryoEM study of the CDC from S. suis (suilysin) (42). Neither cryoEM study, however, has provided unambiguous answers to the structural details of the collapse.
The related MACPFs (described above) examined thus far do not undergo vertical collapse upon pore formation (8, 71, 95). The MACPF β-strands are sufficiently long to extend across the membrane surface without a change in height of the oligomeric prepore. Why the CDCs and MACPFs exhibit this difference in the prepore-to-pore conversion remains unclear, but it may be revealed once we more fully understand how and when the β-barrel forms in these two classes of pore-formers.
Assembly and Membrane Insertion of the β-Barrel
Whether the TMHs interact to form a β-barrel prior to membrane insertion, in concert with insertion, or after membrane insertion of the TMHs is unknown. These transitions may be intimately linked and so it may be difficult to experimentally dissect discrete stages of this process. Nonetheless, there are specific thermodynamic aspects of this process and experimental data that provide some insights into β-barrel assembly.
Because D3 is suspended 40 Å above the bilayer, it possibly allows room for the αHBs to unfold and refold into β-hairpins prior to their insertion (Figure 1d, brackets). The D3 core β-strands would act as a template to zipper down the backbone hydrogen bonds to help refold the αHBs into the TMHs, which would form a contiguous pre-β-barrel that plunges into the bilayer. A pre-β-barrel would solve the significant energetic cost of inserting non-hydrogen-bonded peptide bonds into the membrane (reviewed in 99) and is consistent with the highly cooperative all-ornone insertion of the PFO TMHs (25). However, further rigorous experimental analyses will be required to reveal the assembly mechanism.
Sato et al. (80) recently examined the structure of D3 in the low-temperature-trapped prepore complex (86) and the inserted PFO β-barrel pore. In the prepore structure, the αHBs appeared mobile but were not extended into TMHs. These data, along with our recent studies on the electrostatic trigger (97), show that low temperature traps PFO in the prepore state by stabilizing the interaction of αHB1 with D1,2. Reboul et al. (72) predicted that, upon membrane insertion of the β-barrel pore, the β-strands would exhibit a shear (S) = 2/n (see 54, 55), where n is the number of β-strands in the β-barrel. Sato et al. (80) confirmed this experimentally and showed that the TMHs adopted a 20° tilt with a right-handed twist in the β-barrel pore.
Nelson et al. (59) showed that the β-barrel pore of PFO preferentially inserts at the boundary of liquid-ordered microdomains in liposomes, whereas the prepore assembly does not exhibit a similar propensity. These data imply that prepores can form in a variety of lipid environments, but the β-barrel inserts only when the correct lipid environment is encountered during diffusion of the prepore in the membrane.
Can Incomplete Cholesterol-Dependent Cytolysin Oligomers Insert into the Membrane?
Early electron microscopy (EM) images of CDCs showed arc and ring structures, and recent studies using cryoEM, AFM, or electrophysiological approaches have demonstrated or suggested membrane-inserted, incomplete CDC arcs (42, 47, 92, 94). Their importance and origin remain unclear: Are they a phenomenon of synthetic membrane systems that make it thermodynamically possible to insert an incomplete β-barrel? Are they broken rings or an intended end product for some CDCs? Do they represent a significant fraction of the total pore structures in nature? Unam-biguous answers to these questions remain elusive, but whatever the answer(s) it is unlikely to significantly impact our current understanding of the fundamental features of the CDC mechanism.
THE CURIOUS NATURE OF STREPTOLYSIN O–MEDIATED PROTEIN TRANSLOCATION
Studies by Caparon and colleagues (45, 49) have shown that S. pyogenes SLO specifically translocates an NAD+-glycohydrolase (SPN) into the eukaryotic cell at the bacterial–eukaryotic cell synapse, where each is secreted by the general secretory system. Once inside the eukaryotic cell, SPN cleaves NAD+ into ADP-ribose and nicotinamide, and it generates cADP-ribose (5, 6), which is ultimately toxic to the cell. PFO cannot replace SLO in this translocation system, even though it forms pores more efficiently than does SLO when expressed by S. pyogenes.
Remarkably, this translocation process occurs independently of the normal pore-forming pathway for the CDCs: Neither cholesterol binding nor pore formation is required (46, 53, 61), which suggests another component(s) is involved. Because neither the binding nor pore-forming activi-ties of SLO are required for SPN translocation, its contribution to this system remains a mystery.
PERSPECTIVES
The study of the mechanism of pore formation by the CDCs has revealed processes not observed previously in other systems, thereby establishing many new paradigms. These findings have, in turn, also informed the study of other systems, which is illustrated by the insights CDC pore formation provided into the mechanism of the MACPF family of pore-forming proteins. Furthermore, many of the CDC mutants generated in these studies have been used in a wide variety of studies to determine if CDC-mediated cellular effects are pore-dependent or -independent, or to generate more effective toxoids for vaccine purposes. Thus, the impact of the structure-function studies into the CDC pore-forming mechanism spans many fields of study.
Although significant strides have been made in understanding the mechanism of pore formation by the CDCs, these studies have revealed that the CDC mechanism is far more complex than originally conceived. Besides revealing fundamental features of a complex mechanism, a better understanding of these features may impact our view of their role in disease and/or maintenance of the bacteria in their environmental niche. Some of the recent studies described herein have provided tantalizing glimpses into various aspects of CDC function that promise to be fascinating when they are revealed. Finally, the study of the CDC and MACPF protein families suggests that the CDC D3-like motif is widespread in nature and is likely employed in myriad ways when the formation of a pore contributes to the survival of a species.
ACKNOWLEDGMENTS
This work was supported by grant 1R01 AI037657 from the NIH NIAID to R.K.T.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this article.
LITERATURE CITED
- 1.Balachandran P, Hollingshead SK, Paton JC, Briles DE. 2001. The autolytic enzyme LytA of Streptococcus pneumoniae is not responsible for releasing pneumolysin. J. Bacteriol 183:3108–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benton KA, Everson MP, Briles DE. 1995. A pneumolysin-negative mutant of Streptococcus pneumoniae causes chronic bacteremia rather than acute sepsis in mice. Infect. Immun 63:448–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benton KA, Paton JC, Briles DE. 1997. Differences in virulence for mice among Streptococcuspneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect. Immun 65:1237–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourdeau RW, Malito E, Chenal A, Bishop BL, Musch MW, et al. 2009. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J. Biol. Chem 284:14645–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker AL, Carey VJ, Wessels MR. 2005. Role of NADase in virulence in experimental invasive group A streptococcal infection. Infect. Immun 73:6562–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. 2002. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol. Microbiol 44:257–69 [DOI] [PubMed] [Google Scholar]
- 7.Cohen B, Shwachman H, Perkins ME. 1937. Inactivation of pneumococcal hemolysin by certain sterols. Exp. Biol. Med 35:586–91 [Google Scholar]
- 8.Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. 2004. Vertical collapse of a cytolysin prepore moves its transmembrane β-hairpins to the membrane. EMBO J. 23:3206–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowd KJ, Farrand AJ, Tweten RK. 2012. The cholesterol-dependent cytolysin signature motif: a critical element in the allosteric pathway that couples membrane binding to pore assembly. PLOS Pathog. 8:e1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunstone MA, Tweten RK. 2012. Packing a punch: the mechanism of pore formation by cholesterol dependent cytolysins and membrane attack complex/perforin-like proteins. Curr. Opin. Struct. Biol 22:342–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich P 1898. Diskussionsbemerkungen (Crotin und Tetanolysin). Berlin Klin. Wochenschr 36:273–74 [Google Scholar]
- 12.Fang Y, Cheley S, Bayley H, Yang J. 1997. The heptameric prepore of a staphylococcal alpha-hemolysin mutant in lipid bilayers imaged by atomic force microscopy. Biochemistry 36:9518–22 [DOI] [PubMed] [Google Scholar]
- 13.Farrand AJ, Hotze EM, Sato TK, Wade KR, Wimley WC, et al. 2015. The cholesterol-dependent cytolysin membrane-binding interface discriminates lipid environments of cholesterol to support β-barrel pore insertion. J. Biol. Chem 290:17733–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. 2010. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. PNAS 107:4341–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil SC, Ascher DB, Kuiper MJ, Tweten RK, Parker MW. 2014. Structural studies of Streptococcus pyogenes streptolysin O provide insights into the early steps of membrane penetration. J. Mol. Biol 426:785–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan JJ, Tweten RK, Johnson AE, Heuck AP. 2009. Cholesterol exposure at the membrane surface is necessary and sufficient to trigger perfringolysin O binding. Biochemistry 48:3977–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garland WJ, Buckley JT. 1988. The cytolytic toxin aerolysin must aggregate to disrupt erythrocytes, and aggregation is stimulated by human glycophorin. Infect. Immun 56:1249–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelber SE, Aguilar JL, Lewis KL, Ratner AJ. 2008. Functional and phylogenetic characterization of Vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J. Bacteriol 190:3896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giddings KS, Johnson AE, Tweten RK. 2003. Redefining cholesterol’s role in the mechanism of the cholesterol-dependent cytolysins. PNAS 100:11315–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giddings KS, Zhao J, Sims PJ, Tweten RK. 2004. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat. Struct. Mol. Biol 11:1173–78 [DOI] [PubMed] [Google Scholar]
- 21.Hadders MA, Beringer DX, Gros P. 2007. Structure of C8α-MACPF reveals mechanism of membrane attack in complement immune defense. Science 317:1552–54 [DOI] [PubMed] [Google Scholar]
- 22.Hamon MA, Ribet D, Stavru F, Cossart P. 2012. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 20:360–68 [DOI] [PubMed] [Google Scholar]
- 23.Heuck AP, Hotze E, Tweten RK, Johnson AE. 2000. Mechanism of membrane insertion of a multimeric β-barrel protein: Perfringolysin O creates a pore using ordered and coupled conformational changes. Mol. Cell 6:1233–42 [DOI] [PubMed] [Google Scholar]
- 24.Heuck AP, Tweten RK, Johnson AE. 2003. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J. Biol. Chem 278:31218–25 [DOI] [PubMed] [Google Scholar]
- 25.Hotze EM, Heuck AP, Czajkowsky DM, Shao Z, Johnson AE, Tweten RK. 2002. Monomer-monomer interactions drive the prepore to pore conversion of a β-barrel-forming cholesterol-dependent cytolysin. J. Biol. Chem 277:11597–605 [DOI] [PubMed] [Google Scholar]
- 26.Hotze EM, Le HM, Sieber JR, Bruxvoort C, McInerney MJ, Tweten RK. 2013. Identification and characterization of the first cholesterol-dependent cytolysins from gram-negative bacteria. Infect. Immun 81:216–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hotze EM, Wilson-Kubalek E, Farrand AJ, Bentsen L, Parker MW, et al. 2012. Monomer-monomer interactions propagate structural transitions necessary for pore formation by the cholesterol-dependent cytolysins. J. Biol. Chem 287:24534–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotze EM, Wilson-Kubalek EM, Rossjohn J, Parker MW, Johnson AE, Tweten RK. 2001. Arresting pore formation of a cholesterol-dependent cytolysin by disulfide trapping synchronizes the insertion of the transmembrane beta-sheet from a prepore intermediate. J. Biol. Chem 276:8261–68 [DOI] [PubMed] [Google Scholar]
- 29.Howard JG, Wallace KR, Wright GP. 1953. The inhibitory effects of cholesterol and related sterols on haemolysis by streptolysin O. Br. J. Exp. Pathol 34:174–80 [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Qiao F, Abagyan R, Hazard S, Tomlinson S. 2006. Defining the CD59-C9 binding interaction. J. Biol. Chem 281:27398–404 [DOI] [PubMed] [Google Scholar]
- 31.Iwamoto M, Ohno-Iwashita Y, Ando S. 1987. Role of the essential thiol group in the thiol-activated cytolysin from Clostridium perfringens. Eur. J. Biochem 167:425–30 [DOI] [PubMed] [Google Scholar]
- 32.Johnson BB, Moe PC, Wang D, Rossi K, Trigatti BL, Heuck AP. 2012. Modifications in perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry 51:3373–82 [DOI] [PubMed] [Google Scholar]
- 33.Johnson MK. 1977. Cellular location of pneumolysin. FEMS Microbiol. Lett 2:243–45 [Google Scholar]
- 34.Johnson S, Brooks NJ, Smith RA, Lea SM, Bubeck D. 2013. Structural basis for recognition of the pore-forming toxin intermedilysin by human complement receptor CD59. Cell Rep. 3:1369–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jost BH, Lucas EA, Billington SJ, Ratner AJ, McGee DJ. 2011. Arcanolysin is a cholesterol-dependent cytolysin of the human pathogen Arcanobacterium haemolyticum. BMC Microbiol. 11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kacprzyk-Stokowiec A, Kulma M, Traczyk G, Kwiatkowska K, Sobota A, Dadlez M. 2014. Crucial role of perfringolysin O D1 domain in orchestrating structural transitions leading to membrane-perforating pores: a hydrogen-deuterium exchange study. J. Biol. Chem 289:28738–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawate T, Gouaux E. 2003. Arresting and releasing Staphylococcal α-hemolysin at intermediate stages of pore formation by engineered disulfide bonds. Protein Sci. 12:997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kehoe MA, Miller L, Walker JA, Boulnois GJ. 1987. Nucleotide sequence of the streptolysin O (SLO) gene: structural homologies between SLO and other membrane-damaging, thiol-activated toxins. Infect. Immun 55:3228–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koster S, van Pee K, Hudel M, Leustik M, Rhinow D, et al. 2014. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat. Commun 5:3690. [DOI] [PubMed] [Google Scholar]
- 40.LaChapelle S, Tweten RK, Hotze EM. 2009. Intermedilysin-receptor interactions during assembly of the pore complex: assembly intermediates increase host cell susceptibility to complement-mediated lysis. J. Biol. Chem 284:12719–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law RH, Lukoyanova N, Voskoboinik I, Caradoc-Davies TT, Baran K,et al. 2010. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 468:447–51 [DOI] [PubMed] [Google Scholar]
- 42.Leung C, Dudkina NV, Lukoyanova N, Hodel AW, Farabella I, et al. 2014. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. eLife 3:e04247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Los FC, Randis TM, Aroian RV, Ratner AJ. 2013. Role of pore-forming toxins in bacterial infectious diseases. Microbiol. Mol. Biol. Rev 77:173–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukoyanova N, Kondos SC, Farabella I, Law RHP, Reboul CF, et al. 2015. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLOS Biol. 13(2):e1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madden JC, Ruiz N, Caparon M. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell 104:143–52 [DOI] [PubMed] [Google Scholar]
- 46.Magassa N, Chandrasekaran S, Caparon MG. 2010. Streptococcus pyogenes cytolysin-mediated translocation does not require pore formation by streptolysin O. EMBO Rep. 5:400–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchioretto M, Podobnik M, Dalla Serra M, Anderluh G. 2013. What planar lipid membranes tell us about the pore-forming activity of cholesterol-dependent cytolysins. Biophys. Chem 182:64–70 [DOI] [PubMed] [Google Scholar]
- 48.Marriott HM, Mitchell TJ, Dockrell DH. 2008. Pneumolysin: a double-edged sword during the hostpathogen interaction. Curr. Mol. Med 8:497–509 [DOI] [PubMed] [Google Scholar]
- 49.Meehl MA, Caparon MG. 2004. Specificity of streptolysin O in cytolysin-mediated translocation. Mol. Microbiol 52:1665–76 [DOI] [PubMed] [Google Scholar]
- 50.Melton-Witt JA, Bentsen LM, Tweten RK. 2006. Identification of functional domains of Clostridium septicum alpha toxin. Biochemistry 45:14347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mengaud J, Vicente MF, Chenevert J, Pereira JM, Geoffroy C, et al. 1988. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of Listeria monocytogenes. Infect. Immun 56:766–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller CJ, Elliot JL, Collier RL. 1999. Anthrax protective antigen: prepore-to-pore conversion. Biochemistry 38:10432–41 [DOI] [PubMed] [Google Scholar]
- 53.Mozola CC, Magassa N, Caparon MG. 2014. A novel cholesterol-insensitive mode of membrane binding promotes cytolysin-mediated translocation by streptolysin O. Mol. Microbiol 94:675–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murzin AG, Lesk AM, Chothia C. 1994. Principles determining the structure of β-sheet barrels in proteins. I. A theoretical analysis. J. Mol. Biol 236:1369–81 [DOI] [PubMed] [Google Scholar]
- 55.Murzin AG, Lesk AM, Chothia C. 1994. Principles determining the structure of β-sheet barrels in proteins. II. The observed structures. J. Mol. Biol 236:1382–400 [DOI] [PubMed] [Google Scholar]
- 56.Nagamune H, Ohnishi C, Katsuura A, Fushitani K, Whiley RA, et al. 1996. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect. Immun 64:3093–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura M, Sekino N, Iwamoto M, Ohno-Iwashita Y. 1995. Interaction of θ-toxin (perfringolysin O), a cholesterol-binding cytolysin, with liposomal membranes: change in the aromatic side chains upon binding and insertion. Biochemistry 34:6513–20 [DOI] [PubMed] [Google Scholar]
- 58.Nakano Y, Noda K, Endo T, Kobata A, Tomita M. 1994. Structural study on the glycosylphosphatidylinositol anchor and the asparagine-linked sugar chain of a soluble form of CD59 in human urine. Arch. Biochem. Biophys 311:117–26 [DOI] [PubMed] [Google Scholar]
- 59.Nelson LD, Chiantia S, London E. 2010. Perfringolysin O association with ordered lipid domains: implications for transmembrane protein raft affinity. Biophys. J 99:3255–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nelson LD, Johnson AE, London E. 2008. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem 283:4632–42 [DOI] [PubMed] [Google Scholar]
- 61.O’Seaghdha M, Wessels MR. 2013. Streptolysin O and its co-toxin NAD-glycohydrolase protect group A Streptococcus from xenophagic killing. PLOS Pathog. 9:e1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palmer M, Vulicevic I, Saweljew P, Valeva A, Kehoe M, Bhakdi S. 1998. Streptolysin O: a proposed model of allosteric interaction between a pore-forming protein and its target lipid bilayer. Biochemistry 37:2378–83 [DOI] [PubMed] [Google Scholar]
- 63.Panchal RG, Bayley H. 1995. Interactions between residues in staphylococcal α-hemolysin revealed by reversion mutagenesis. J. Biol. Chem 270:23072–76 [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25:1605–12 [DOI] [PubMed] [Google Scholar]
- 65.Polekhina G, Giddings KS, Tweten RK, Parker MW. 2005. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. PNAS 102:600–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price KE, Camilli A. 2009. Pneumolysin localizes to the cell wall of Streptococcus pneumoniae. J. Bacteriol 191:2163–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price KE, Greene NG, Camilli A. 2012. Export requirements of pneumolysin in Streptococcus pneumoniae. J. Bacteriol 194:3651–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prigent D, Alouf JE. 1976. Interaction of streptolysin-O with sterols. Biochim. Biophys. Acta 443:288–300 [DOI] [PubMed] [Google Scholar]
- 69.Ramachandran R, Heuck AP, Tweten RK, Johnson AE. 2002. Structural insights into the membraneanchoring mechanism of a cholesterol-dependent cytolysin. Nat. Struct. Biol 9:823–27 [DOI] [PubMed] [Google Scholar]
- 70.Ramachandran R, Tweten RK, Johnson AE. 2004. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit β-strand alignment. Nat. Struct. Mol. Biol 11:697–705 [DOI] [PubMed] [Google Scholar]
- 71.Ramachandran R, Tweten RK, Johnson AE. 2005. The domains of a cholesterol-dependent cytolysin undergo a major FRET-detected rearrangement during pore formation. PNAS 102:7139–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reboul CF, Mahmood K, Whisstock JC, Dunstone MA. 2012. Predicting giant transmembrane β-barrel architecture. Bioinformatics 28:1299–302 [DOI] [PubMed] [Google Scholar]
- 73.Reboul CF, Whisstock JC, Dunstone MA. 2014. A new model for pore formation by cholesterol-dependent cytolysins. PLOS Comput. Biol 10:e1003791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson SL, Li J, Uzal FA, McClane BA. 2011. Evidence for a prepore stage in the action of Clostridium perfringens epsilon toxin. PLOS ONE 6:e22053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rollins SA, Sims PJ. 1990. The complement-inhibitory activity of CD59 resides in its capacity to block incorporation of C9 into membrane C5b-9. J. Immunol 144:3478–83 [PubMed] [Google Scholar]
- 76.Rosado CJ, Buckle AM, Law RH, Butcher RE, Kan WT, et al. 2007. A common fold mediates vertebrate defense and bacterial attack. Science 317:1548–51 [DOI] [PubMed] [Google Scholar]
- 77.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, et al. 2008. The MACPF/CDC family of pore-forming toxins. Cell. Microbiol 10:1765–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. 1997. Structure of a cholesterol-binding thiol-activated cytolysin and a model of its membrane form. Cell 89:685–92 [DOI] [PubMed] [Google Scholar]
- 79.Rossjohn J, Polekhina G, Feil SC, Morton CJ, Tweten RK, Parker MW. 2007. Structures of perfringolysin O suggest a pathway for activation of cholesterol-dependent cytolysins. J. Mol. Biol 367:1227–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sato TK, Tweten RK, Johnson AE. 2013. Disulfide-bond scanning reveals assembly state and β-strand tilt angle of the PFO β-barrel. Nat. Chem. Biol 9:383–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sekino-Suzuki N, Nakamura M, Mitsui KI, Ohno-Iwashita Y. 1996. Contribution of individual tryptophan residues to the structure and activity of θ-toxin (perfringolysin O), a cholesterol-binding cytolysin. Eur. J. Biochem 241:941–47 [DOI] [PubMed] [Google Scholar]
- 82.Sellman BR, Kagan BL, Tweten RK. 1997. Generation of a membrane-bound, oligomerized pre-pore complex is necessary for pore formation by Clostridium septicum alpha toxin. Mol. Microbiol 23:551–58 [DOI] [PubMed] [Google Scholar]
- 83.Seveau S 2014. Multifaceted activity of listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Subcell. Biochem 80:161–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shatursky O, Heuck AP, Shepard LA, Rossjohn J, Parker MW, et al. 1999. The mechanism of membrane insertion for a cholesterol dependent cytolysin: a novel paradigm for pore-forming toxins. Cell 99:293–99 [DOI] [PubMed] [Google Scholar]
- 85.Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, et al. 1998. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry 37:14563–74 [DOI] [PubMed] [Google Scholar]
- 86.Shepard LA, Shatursky O, Johnson AE, Tweten RK. 2000. The mechanism of assembly and insertion of the membrane complex of the cholesterol-dependent cytolysin perfringolysin O: Formation of a large prepore complex. Biochemistry 39:10284–93 [DOI] [PubMed] [Google Scholar]
- 87.Shewell LK, Harvey RM, Higgins MA, Day CJ, Hartley-Tassell LE, et al. 2014. The cholesterol-dependent cytolysins pneumolysin and streptolysin O require binding to red blood cell glycans for hemolytic activity. PNAS 111(49):E5312–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh R, Jamieson A, Cresswell P. 2008. GILT is a critical host factor for Listeria monocytogenes infection. Nature 455:1244–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Slade DJ, Lovelace LL, Chruszcz M, Minor W, Lebioda L, Sodetz JM. 2008. Crystal structure of the MACPF domain of human complement protein C8α in complex with the C8γ subunit. J. Mol. Biol 379:331–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soltani CE, Hotze EM, Johnson AE, Tweten RK. 2007. Specific protein-membrane contacts are required for prepore and pore assembly by a cholesterol-dependent cytolysin. J. Biol. Chem 282:15709–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Soltani CE, Hotze EM, Johnson AE, Tweten RK. 2007. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. PNAS 104:20226–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sonnen AF, Plitzko JM, Gilbert RJ. 2014. Incomplete pneumolysin oligomers form membrane pores. Open Biol. 4:140044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stevens DL, Bryant AE. 1993. Role of theta-toxin, a sulfhydryl-activated cytolysin, in the pathogenesis of clostridial gas gangrene. Clin. Infect. Dis 16:S195–99 [DOI] [PubMed] [Google Scholar]
- 94.Stewart SE, D’Angelo ME, Piantavigna S, Tabor RF, Martin LL, Bird PI. 2014. Assembly of streptolysin O pores assessed by quartz crystal microbalance and atomic force microscopy provides evidence for the formation of anchored but incomplete oligomers. Biochim. Biophys. Acta 1848:115–26 [DOI] [PubMed] [Google Scholar]
- 95.Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. 2005. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 121:247–56 [DOI] [PubMed] [Google Scholar]
- 96.Tweten RK. 1988. Nucleotide sequence of the gene for perfringolysin O (theta-toxin) from Clostridium perfringens: significant homology with the genes for streptolysin O and pneumolysin. Infect. Immun 56:3235–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wade KR, Hotze EM, Kuiper MJ, Morton CJ, Parker MW, Tweten RK. 2015. An intermolecular electrostatic interaction controls the prepore to pore transition in a cholesterol-dependent cytolysin. PNAS 112(7):2204–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walker JA, Allen RL, Falmagne P, Johnson MK, Boulnois GJ. 1987. Molecular cloning, characterization, and complete nucleotide sequence of the gene for pneumolysin, the sulfhydryl-activated toxin of Streptococcus pneumoniae. Infect. Immun 55:1184–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.White SH, Wimley WC. 1999. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct 28:319–65 [DOI] [PubMed] [Google Scholar]
- 100.Wickham SE, Hotze EM, Farrand AJ, Polekhina G, Nero TL, et al. 2011. Mapping the intermedilysinhuman CD59 receptor interface reveals a deep correspondence with the binding site on CD59 for complement binding proteins C8α and C9. J. Biol. Chem 286:20952–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu L, Huang B, Du H, Zhang XC, Xu J, et al. 2010. Crystal structure of cytotoxin protein suilysin from Streptococcus suis. Protein Cell 1:96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]