Abstract
Campylobacter jejuni and Campylobacter coli are globally recognized as a major cause of bacterial foodborne gastroenteritis. A cross-sectional study was conducted from October 2015 to May 2016 in Mekelle city to isolate, identify, and estimate the prevalence of C. jejuni and C. coli in raw meat samples and to determine their antibiotic susceptibility pattern. A total of 384 raw meat samples were randomly collected from bovine (n = 210), goat (n = 108), and chicken (n = 66), and isolation and identification of Campylobacter spp. were performed using standard bacteriological techniques and PCR. Antibiotic susceptibility test was performed using disc diffusion method. Of the total 384 raw meat samples, 64 (16.67%) were found positive for Campylobacter spp. The highest prevalence of Campylobacter spp. was found in chicken meat (43.93%) followed by bovine meat (11.90%) and goat meat (9.25%). The most prevalent Campylobacter spp. isolated from meat samples was C. jejuni (81.25%). The overall prevalence of Campylobacter in restaurants, butcher shops, and abattoir was 43.93%, 18.30%, and 9.30%, respectively. 96.8%, 81.25%, 75%, and 71% of the Campylobacter spp. isolates were sensitive to norfloxacin, erythromycin, chloramphenicol, and sulphamethoxazole-trimethoprim, respectively. However, 96.9%, 85.9%, and 50% of the isolates were resistant to ampicillin, amoxicillin, and streptomycin, respectively. Strains that developed multi-drug resistant were 68.7%. The result of this study revealed the occurrence of Campylobacter in bovine, goat, and chicken meats. Hence, there is a chance of acquiring infection via consumption of raw or undercooked meat. Thus, implementation of hygienic practices from a slaughterhouse to the retailers, proper handling and cooking of foods of meat are very important in preventing Campylobacter infection.
Introduction
Foodborne diseases occur as a result of the consumption of contaminated foodstuffs especially from animal products such as meat from infected animals or carcasses contaminated with pathogenic bacteria [1, 2]. Food-producing animals are the major reservoirs for many foodborne pathogens such as Campylobacter species, non-Typhi serotypes of Salmonella enterica, Shiga toxin-producing strains of Escherichia coli, and Listeria monocytogenes. Foodborne pathogens cause millions of cases of sporadic illness and chronic complications, as well as large and challenging outbreaks in many countries and between countries [3].
Worldwide, pathogenic Campylobacter species are the leading cause of bacterial-derived foodborne disease and are responsible for the cause of over 400–500 million infections cases each year [4–6]. Campylobacter species are normally carried in the intestinal tracts of many domestic livestock such as poultry, cattle, sheep, goat, pigs, as well as wild animals and birds [7–10]. Fecal matter is a major source of contamination and could reach carcasses through direct deposition [11]. Animal food products can become contaminated by this pathogen during slaughtering and carcass dressing [12]. Humans are infected by ingestion of undercooked or decontaminated meat, or handling of raw products or cross-contamination of raw to cooked foods, swimming in natural waters, direct contact with contaminated animals or animal carcasses, and traveling [13–15].
Pathogenic Campylobacter spp. known to be implicated in human infections include C. jejuni, C. concisus, C. rectus, C. hyointestinalis, C. insulaenigrae, C. sputorum, C. helveticus, C. lari, C. fetus, C. mucosalis, C. coli, C. upsaliensis, and C. ureolyticus [6]. Of these,C.jejuniand C.coli are considered the most commonly reported zoonosis in humans and recognized as the most common causative agents of bacterial gastroenteritis in the world [16–19].
Moreover, Campylobacter with resistance to antimicrobial agents has also been implicated worldwide [4, 20, 21]. The use of antimicrobial agents in food animals has resulted in the emergence and dissemination of antimicrobial-resistant bacteria including antimicrobial-resistant Campylobacter, which has a potentially serious impact on food safety in both animal and human health. The situation seems to deteriorate more rapidly in developing countries where there is widespread and uncontrolled use of antibiotics [22].
In Ethiopia, few studies were conducted on the prevalence and antimicrobial susceptibility of enteric Campylobacteriosis of human beings [23–25] and food of animal origins [9, 20, 26–28]. The absence of a national surveillance program, limited routine culture availability for the isolation of Campylobacter spp. in clinical and research settings, and the need for selective media and unique growth atmosphere make it difficult to give an accurate picture of the burden of the disease in Ethiopia. This fact indicates that Campylobacter as a zoonosis is not given appropriate weight and consideration, particularly in the current study area. As a result, the objectives of this study were to isolate and identify C. jejuni and C. coli from the meat of cattle, goat, and chicken collected from an abattoir, butcher shops, and restaurants in Mekelle City, estimate their prevalence and determine the antibiotic susceptibility pattern of C.jejuniand C. coli isolates.
Material and methods
Ethics approval
This study was reviewed and approved by the Research Ethics Committee of the College of Veterinary Sciences, Mekelle University.
Study area
The study was conducted from October 2015 to May 2016 at an abattoir, butcher shops, and restaurants of Mekelle City. Mekelle is the capital city of Tigray National Regional State of Ethiopia where thousands of cattle and goats are accessible from different districts of the region and the neighboring regions of the country for slaughter. Mekelle is found at 39°29’ East and 13°30’ North of the equator which is 783 kilometers away from Addis Ababa, which is the capital city of Ethiopia. The altitude of the area ranges from 2000–2200 meters above sea level. The mean annual rainfall of the area is 628.8 mm and an annual average temperature of 21°C. The city has seven sub-cities and a total population of 215,546 [29], 308 cafeterias, 292 restaurants, 258 supermarkets, and an active urban-rural exchange of goods which has 30000 micro-and small enterprises [30].
Study design
A cross-sectional study was employed from October 2015 to May 2016 to isolate, identify, and estimate the prevalence and antibiotic susceptibility patterns of C.jejuni and C. coli from bovine, goat, and chicken meat samples collected from the abattoir, butcher shops, and restaurants.
Sample size and sample collection
A total of 384 raw ready-to-eat meat samples comprising of cattle (n = 210), goat (n = 108),and chicken (n = 66) meats were collected from the abattoir (n = 258), butcher shops (n = 60), and restaurants (n = 66) of the study area. All samples were placed in polyethylene plastic bags to prevent spilling and cross-contamination and immediately transported to the Molecular Biology Laboratory of the College of Veterinary Sciences, Mekelle University using an icebox with ice packs.
Bacteriological isolation and identification of Campylobacter species
Approximately 10 grams of raw meat sample was aseptically collected using sterile forceps and scissor and placed into 90ml of buffered peptone water in a sterile plastic bag and homogenized for 1 minute using a stomacher (Lab Blender 400, Seward Medical, London, England)and incubated at 37°C for 48h in the microaerophilic atmosphere (gas mix of 5% O2, 10% CO2, and 85% N2). Then a 0.1ml of the enriched sample was streaked onto Karmali Campylobacter Agar Base (HiMedia Laboratories, Mumbai, India)(Blood free Campylobacter selective agar base medium containing Campylobacter selective supplement comprising cefoperazone, amphotericin B (CCDA selective supplement SR0155E)) [31] and kept in a gas jar containing Campylobacter gas pack systems to maintain the microaerophilic condition and was incubated at a temperature of 37°C for 48h. The colonies were provisionally identified based on staining reaction with Gram’s stain, cellular morphology [32], catalase test, and oxidase test [33], and growth appearance on 5% sheep blood agar (Oxoid Ltd., Basingstoke, Hampshire, England) at 37°C after 24 h [34].
All the thermophilic Campylobacters isolates were tested for Hippurate hydrolysis, H2S production, and susceptibility to Nalidixic acid and Cephalothin as proposed by [34]. Susceptibility tests to Nalidixic acid (30 μg) and Cephalothin (30 μg) were performed using the standard agar disc diffusion method as recommended by Clinical and Laboratory Standards Institutions (CLSI) and isolates were categorized as sensitive and/or resistant according to the interpretation table of the [35]. The presumed Campylobacter isolates were preserved in brain heart infusion broth supplemented with15% glycerol in Eppendorf tubes at -20°C for further analysis. Bacterial strains that were used as quality control organisms in this study were standard strains of S. aureus, S. agalactiae, and E. coli obtained from the National Veterinary Institute (NVI), Debre-Zeit.
Polymerase chain reaction for detection of mapA and ceuE genes
The genomic DNA of the pheno typically resembled isolates of Campylobacter was extracted using the Phenol Chloroform method (Phenol: Chloroform: Isoamyl alcohol mixture (24:25:1)) according to [36]. Then 20μl of each extracted genomic DNA sample was run in an agarose gel electrophoresis and visualized under UV-light gel doc. Then after, a genome-based polymerase chain reaction (PCR) was done as described by [37] using the following species-specific primers: F-5’CTA TTT TAT TTT TGA GTG CTT GTG3’ and R-5’GCT TTA TTT GCC ATTT GTT TTA TTA3’ was used to amplify the mapA gene of C. jejuni(589 bp), and F-5’ATT TGA AAA TTG CTC CAA CTA TG3’ and R-5’TGA TTT TAT TAT TTG TAG CAG CG3’ were used to amplify the ceuE gene of C. coli(462 bp). Each PCR reaction mixture was performed in a 50μl total volume containing 10μl of template DNA,5μl of 5X PC buffer, 5μl of MgCl2, 1μl of each of the primers, 0.75μl of 10 mM of each dNTPs, 0.15μl of Taq DNA polymerase, and27.1μl nuclease-free distilled water. Amplification was carried out with thermal cycling conditions of an initial denaturation at 95°C for 5 min followed by 45 cycles of denaturation at 94°C for 35s, annealing at 54°C for 35s and extension at 72°C for 35s, and with a final extension at 72°C for 6 min. Finally, the PCR products were separated by running on a 1.5% (w/v) agarose gel containing 0.3mg/ml ethidium bromide. Electrophoresis was conducted in a horizontal equipment system for 120 min at 90 V using 1X TAE buffer (40 mM Tris, 1 mM EDTA, and 20 mM glacial acetic acid, pH 8.0). The amplicons were visualized under UV-light gel doc and their molecular weights were estimated by comparing with100bp DNA molecular weight marker(Solis BioDyne, Tartu, Estonia).
Antimicrobial susceptibility testing
The Campylobacter spp. isolates were screened for in vitro antimicrobial susceptibility using the standard agar disc diffusion method as recommended by Clinical and Laboratory Standards Institutions (CLSI) on Mueller-Hinton agar supplemented with 5% sheep blood (Oxoid Ltd., Basingstoke, Hampshire, England). The following nine different antibiotic discs, with their concentrations given in parentheses, were used in the antibiogram testing: Amoxicillin (AML)(10μg), Ampicillin (AMP)(10μg), Chloramphenicol (C)(30μg), Erythromycin (E)(15μg), Gentamycin (CN)(10μg), Norfloxacin (NOR)(10μg), Streptomycin (S)(10μg), Tetracycline (TE)(30μg), and Sulfamethoxazole-trimethoprim (SXT)(25μg) (Oxoid Company, Hampshire, England). After 48h of microaerophilic incubation at 37°C, the clear zones (inhibition zones of bacterial growth around the antibiotic discs(including the discs)diameter for individual antimicrobial agents were measured and then translated into Sensitive (S), Intermediate (I), and Resistant (R) categories according to the interpretation table of the Clinical and Laboratory Standard Institute [35].
Data storage and statistical analysis
All collected data were entered into Microsoft Excel Sheet (Microsoft Corp., Redmond, WA, USA) and analyzed using SPSS version 20 statistical computer software program. Chi-square (χ2) test and Logistic regression were applied to assess the associations. For all tests, a p-value of less than 0.05 was considered statistically significant.
Results
The overall prevalence of Campylobacter species
Out of the total of 384 collected meat samples, 64(16.67%) were positive for the two Campylobacter spp. The highest (43.93%) and lowest (9.25%) prevalence of Campylobacter spp. were recorded in samples taken from chicken (found to be 7.68 times more likely to have Campylobacter contamination compared to other sample types) and goat, respectively. Whereas, the highest (43.93%) and lowest (9.25%) prevalence were recorded in meat samples collected from restaurants (found to be four times more likely to have Campylobacter contamination compared to other sample sources) and abattoir, respectively. Both sample types and sources had significant differences (p = 0.00; χ2 = 43.04or OR = 7.68, CI = 3.40–17.30,and p = 0.00; χ2 = 45.53 or OR = 7.64, CI = 4.01–14.52, respectively) in the prevalence of the two Campylobacter spp. as it is shown in Tables 1 and 2 below.
Table 1. Prevalence of Campylobacter spp. among different sample types and sources.
| Risk Factors | No. of Examined | No. of Positive (%) | χ2 | P-value |
|---|---|---|---|---|
| Type of meat | ||||
| Goat meat | 108 | 10(9.25) | 43.04 | 0.000 |
| Cattle meat | 210 | 25(11.90) | ||
| Chicken meat | 66 | 29(43.93) | ||
| Total | 384 | 64(16.67) | ||
| Sources of meat | ||||
| Abattoir | 258 | 24(9.30) | 45.53 | 0.000 |
| Butcher | 60 | 11(18.33) | ||
| Restaurants | 66 | 29(43.93) | ||
| Total | 384 | 64(16.67) |
Table 2. Logistic regression analysis results of sample types and sample sources.
| Types of sample | No of examined | No of positive (%) | OR(95%CI) | P-value |
|---|---|---|---|---|
| Goat meat* | 108 | 10(9.25) | 1 | |
| Cattle meat | 210 | 25(11.90) | 1.32(0.61–2.86) | 0.476 |
| Chicken meat | 66 | 29(43.93) | 7.68(3.40–17.30) | 0.000 |
| Total | 384 | 64(16.67) | ||
| Abattoir* | 258 | 24(9.30) | 1 | |
| Butcher | 60 | 11(18.33) | 2.18(1.00–4.76) | 0.048 |
| Restaurants | 66 | 29(43.93) | 7.64(4.01–14.52) | 0.000 |
| Total | 384 | 64(16.67) |
OR = Odd ratio; CI = Confidence interval.
Contamination rate of C.jejuni and C. coli in the different sample types
Of the two Campylobacter spp. isolated and identified from cattle, goat, and chicken meat samples C. jejuni and C. coli accounted for 81.25% and 18.75%, respectively. The prevalence of C.jejuni and C.coli in cattle, goat, and chicken meat samples were found to be 76% and 24%, 80% and 20%, and 86.21%and 13.79%, respectively (Table 3).
Table 3. The contamination rate of C. jejuni and C. coli among different sample types.
| Sample Type Prevalence | Campylobacter spp. | |
|---|---|---|
| C.jejuni | C.coli | |
| Cattle meat (n = 25) | 19(76%) | 6(24%) |
| Goat meat (n = 10) | 8(80%) | 2(20%) |
| Chicken meat (n = 29) | 25(86.21%) | 4(13.79%) |
| Total (n = 64) | 52(81.25%) | 12(18.75%) |
PCR amplification results of Campylobacter spp. isolates
Besides the phenotypic characterization, PCR amplification of the 64 samples revealed that 52(81.25%) of the isolates were C.jejuni(having a molecular weight of 589 bp) and the remaining 12(18.75%) isolates were C.coli (having a molecular weight of462 bp) to the targeted genes.
Antimicrobial susceptibility pattern of Campylobacter spp. isolates
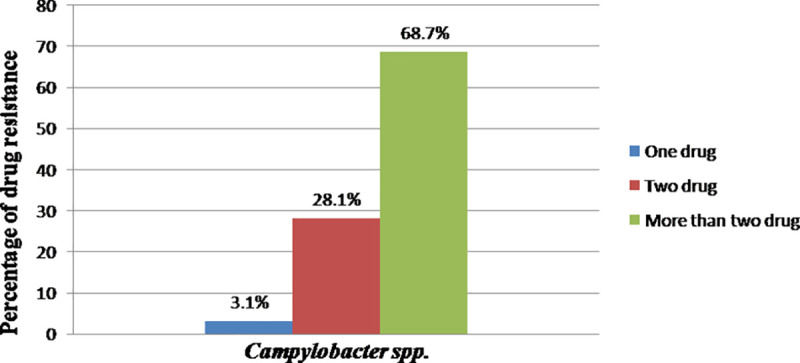
Campylobacter spp. isolated from the different sample types and sources were susceptible to Norfloxacin (96.8%), Erythromycin (81.25%), Chloramphenicol (75%), and Gentamycin (75%). However, the isolates had shown resistance to Ampicillin (96.9%) and Amoxicillin (85.9%) (Table 4). Moreover, 96.8% of the isolates developed resistance for two or more than two drugs as it is shown in Fig 1.
Table 4. In vitro antimicrobial sensitivity pattern of Campylobacter spp. isolates.
| Type of antibiotics | Interpretations | ||
|---|---|---|---|
| Susceptible (%) | Intermediate (%) | Resistant (%) | |
| Ampicillin | 0(0) | 2(3.1) | 62(96.9) |
| Amoxicillin | 3(4.7) | 6(9.4) | 55(85.9) |
| Chloramphenicol | 48(75) | 5(7.8) | 11(17.2) |
| Erythromycin | 52(81.25) | 1(1.6) | 11(17.2) |
| Gentamycin | 48(75) | 8(12.5) | 8(12.5) |
| Norfloxacin | 62(96.8) | 1(1.6) | 1(1.6) |
| Streptomycin | 25(39.06) | 7(10.9) | 32(50) |
| Tetracycline | 42(65.6) | 6(9.4) | 16(25) |
| Sulfamethoxazole-Trimethoprim | 46(71.8) | 4(6.25) | 14(21.9) |
Fig 1. Percentage of drug resistance of Campylobacter spp.
Species based antibiogram result of the isolates revealed that the highest level of sensitivity of both C. jejuni (98.1%) and C. coli (91.7%)was observed against Norfloxacin. Whereas both C.jejuni(96.2%) and C. coli(100%) were showed the highest level of resistance against Ampicillin as it is shown in Table 5.
Table 5. In vitro antimicrobial sensitivity pattern of C. jejuni and C. coli isolates.
| Antibiotic Discs | Interpretations | |||||
|---|---|---|---|---|---|---|
| C. jejuni (N = 52) | C. coli (N = 12) | |||||
| S (%) | I (%) | R (%) | S (%) | I (%) | R (%) | |
| Ampicillin | - | 2(3.8) | 50(96.2) | - | - | 12(100) |
| Amoxicillin | 3(5.8) | 4(7.7) | 45(86.5) | - | 2(16.7) | 10(83.3) |
| Chloramphenicol | 40(76.9) | 4(7.7) | 8(15.4) | 8(66.7) | 1(8.3) | 3(25) |
| Erythromycin | 43(82.7) | - | 9(17.3) | 9(75) | 1(8.3) | 2(16.7) |
| Gentamycin | 41(78.8) | 6(11.5) | 5(9.6) | 7(58.3) | 2(16.7) | 3(25) |
| Norfloxacin | 51(98.1) | - | 1(1.9) | 11(91.7) | 1(8.3) | - |
| Streptomycin | 22(42.5) | 7(13.5) | 23(44.2) | 3(33.3) | - | 9(66.7) |
| Tetracycline | 38(73.1) | 6(11.5) | 8(15.4) | 4(41.7) | - | 8(58.3) |
| Sulfamethoxazole- Trimethoprim | 39(75.5) | 3(5.8) | 10(19.2) | 7(58.3) | 1(8.3) | 4(33.3) |
S = Susceptible, I = Intermediate, R = Resistant.
Discussion
In the current study, the overall prevalence of Campylobacter spp. was found to be 16.67%. The highest prevalence was found in chicken meat samples (43.93%). Chicken meats were found to be 7.68 times more likely to have Campylobacter when compared to goat and cattle meat. The difference in the prevalence of Campylobacter between different types of meat samples was found to be statistically significant (p<0.05) (OR = 7.68, CI = 3.40–17.30). The prevalence of Campylobacter spp. in chicken meat samples was 43.93% which was comparable with those reported by [38], 44%, [39], 56.1%,and [40], 48.02% in Iran. This was higher than the report of [41] who reported a prevalence of 1.93%. However, the present finding was lower than studies conducted by [42–44], and [45] who reported the prevalence of 61.7% and 70.7% and 65% and 81.3% of Campylobacter spp. in Ahvaz, Iran; Washington; Northern Ireland, and Northern Italy, respectively. It is a well-known fact that poultry appeared to be a significant source of Campylobacter and chicken was found to be heavily intestinal carriers of Campylobacter when compared with other food animals [8]. Wide variation (0–90%) in the prevalence of Campylobacter in fresh poultry meat had been reported in different countries [46–48]. These variations in Campylobacter spp. prevalence might be due to differences in hygienic conditions, cross-contamination that may occur during de-feathering, eviscerating, and some other environmental factor such as the temperature of water in the scalding tank.
In this study, the prevalence of Campylobacter spp. in bovine meat was 11.90%. This was comparable to the finding reported from a previous study done by [10] (12.9%) in Nigeria, and [49] (10%) in Iran. However, it was higher than the findings reported by [9] (6.2%) in Ethiopia [50]; (5.6%) in Morogoro, Tanzania; and [51] (0.8%)in Australia. Food of animal origin has been incriminated for being the main source of Campylobacter infection in humans [52]. Since raw meat from beef is widely consumed in the country; the occurrence of Campylobacter in meat increases the likelihood of the pathogen transmission to humans. The present finding was lower than studies conducted by [10] and [53] who reported a prevalence of 69.1% and 22%, respectively. One of the most likely hypotheses to explain the discrepancies is the differences in protocols used for the detection of thermophilic Campylobacter, and especially the absence of an enrichment step for the isolation of thermophilic Campylobacter in [54] work. In general, these variations might be due to approaches of sample collection, the difference in isolation and identification techniques, and differences in sample size.
The prevalence of Campylobacter spp. in goat meat was found to be 9.25%. This finding was in agreement with the reports of [9] and [55], who reported 7.6% and 6.4%, respectively. But it slightly higher than the report of [42] who reported 4.4%. However, the present study was lower than the findings of [5] and [26] who reported 41.2% and 27.5%, respectively.
The meat samples collected from restaurants had the highest prevalence (43.93%). The meat samples collected from restaurants were found to be four times more likely to have Campylobacter compared to meat collected from Butcher and eight times more likely to have Campylobacter compared to meat collected from the abattoir. The difference in the prevalence of Campylobacter between sources of meat samples was found to be statistically significant (P<0.05) (OR = 7.64, CI = 4.01–14.52). This might be due to an extra chance of acquiring contamination from individuals who are working in restaurants during handling or cross-contamination among different carcasses.
In the current study, the bacteriological and PCR characterization of Campylobacter isolates revealed that the prevalence of C.jejuni was higher than C.coli. C. jejuni has been reported to be the most frequent species recovered from the food of animal origin especially chicken meat [48, 56–58]. The prevalence of C. jejuni and C. coli in bovine, goat, and chicken meat were found to be 76% and 24%; 80% and 20%; and 86.3% and 13.7%, respectively. These findings were in agreement with the findings of [9] who reported 78% C.jejuni and 18% C.coli [28]; who reported 78% C. jejuni and 22% C.coli [27]; who reported 93.3% C. jejuni and 6.7% C. coli [26]; who reported 72.5% C.jejuni and 27.5% C. coli in Ethiopia. The prevalence of C.jejuni in raw meat was in agreement with the reports from other countries [48, 57, 59, 60].
Antibiotic resistance in Campylobacter is emerging globally and has already been described by several authors and recognized by the WHO, as a problem of public health importance [61–63]. Campylobacter spp. resistance to antibiotics (C.jejuni and C.coli) can be transferred from different sources to humans. This situation, alarmingly, announces the need to perform an antimicrobial sensitivity test for Campylobacter. Macrolides and Fluoroquinolones are usually considered the drugs of choice for the treatment of foodborne Campylobacteriosis [64–66]. Antibiotic susceptibility patterns have been determined in previous studies conducted in Ethiopia where the 80%-100% of isolates from food animals were sensitive to these antimicrobial agents [9, 20]. However, there are pieces of evidence from different parts of the world that antimicrobial resistance in food animals and human isolates is increasing.
In the current study, 52 C.jejuni and 12 C.coli isolates were investigated for their antimicrobial susceptibility pattern. The percentage of ampicillin and amoxicillin resistant Campylobacter isolates were 96.9% and 85.9%, respectively. This was in agreement with the report of [28], who reported 97.2% and 83.3% for ampicillin and amoxicillin, respectively. Moreover, [67] reported a resistance level of 100% for C. coli. On the other hand, C. coli isolates are generally more resistant than C. jejuni strains [68]. In general, several studies have reported resistance to beta-lactam antibiotics is high in food animals [69–71]. The resistance rate of Campylobacter isolates (25%) to tetracycline in the present study was comparable with the findings of [27] (20.8%)but higher than that of [9] (10%) and [72] (6%). However, it was lower than the report of [73] (77.94%). The resistance level to streptomycin in the current study was 50%, which was higher than reports from Ireland and Thailand by [73] and [74].
Multi-drug resistance isolates always remained susceptible to norfloxacin and erythromycin and chloramphenicol. In the present study, multi-drug resistance to more than two antimicrobial agents was 68.7% which was comparable to the findings of [69] in Belgium, 60% [75]; in Estonia, 60% [49]; in Iran, 75%;and [76] in Korea, 93.4%. However, the current multidrug resistance finding was higher than the report from Addis Ababa and Debre Zeit, Ethiopia, by [9] (20%). Despite global commitments to reduce antimicrobial resistance and protect the effectiveness of antimicrobials, most countries have not yet started implementing government policies to reduce their overuse and misuse of antimicrobials [77]. Hence, the current multidrug resistance finding might be since antibiotics can be bought for human or animal use without a prescription, and similarly, in countries like Ethiopia without standard regulation and treatment guidelines, antibiotics are often over-prescribed by health workers and veterinarians and over-used by the public. Moreover, new resistance mechanisms are emerging and spreading globally. Hence, antibiotic resistance is rising to dangerously high levels in all parts of the world.
Conclusion and recommendations
The present study revealed the occurrence of Campylobacter in bovine, goat, and chicken raw meat samples collected from different sites of the study area. Hence, they can serve as a potential vehicle for transmitting Campylobacter spp. and risk of infection to humans through the consumption of raw or undercooked meat. Therefore, retailers can act as a major source of cross-contamination. Moreover, the current study was revealed the development of antimicrobial resistance by the isolated Campylobacter spp. for certain drugs which is an alert for the concerned bodies. Hence, coordinated actions are needed to reduce or eliminate the risks posed by these pathogens at various stages in the food chain. Moreover, controlled and careful use of antibiotics, both in veterinary and human treatment regimes should be practiced. Finally, further nationwide molecular epidemiology and phenotypic and molecular characterization of the disease should be undertaken.
Acknowledgments
The authors acknowledged owners, managers, and workers of the different abattoir, butcher shops, and restaurants of the study site for their keen interest and cooperation during the collection of the meat samples. We would like to extend our acknowledgement to the College of Veterinary Science Staff members’ who were directly or indirectly helping us during the research period.
Data Availability
All relevant data are within the paper.
Funding Statement
This research work was funded by College of Veterinary Sciences, Mekelle University.
References
- 1.Rosef O, Johnsen G, Stølan A, Klæboe H. Similarity of Campylobacter lariamong human, animal, and water isolates in Norway. Foodborne Pathog Dis.2008;5:33–93. 10.1089/fpd.2007.0027 [DOI] [PubMed] [Google Scholar]
- 2.Wong JSJ, Anderson TP, Chambers ST, On SL, Murdoch DR. Campylobacter fetus-associated epidural abscess and bacterimia. J Clin Microbiol.2009;47:857–8. 10.1128/JCM.00725-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA-ECDC European food safety authority and European Centre for disease prevention and control. The European Union summary report on trends and sourcesof zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016;14. [DOI] [PMC free article] [PubMed]
- 4.Nawal AH. Antimicrobial resistant C.jejuniisolated from humans and animals in Egypt. Glob Vet.2011;6:195–200. [Google Scholar]
- 5.Mpalang RK, Boreux R, Pierrette M, Ni Bitiang K, Daube G, Mol PD. Prevalence of Campylobacter among goats in Congo. J Infect Dev Ctries.2014;8:168–75. 10.3855/jidc.3199 [DOI] [PubMed] [Google Scholar]
- 6.Heredia N, García S. Animals as sources of food-borne pathogens: A review. Animal Nutr. 2018; 4 (2018); 250e255 10.1016/j.aninu.2018.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ertaş HB, Cetinkaya B, Muz A., Ongor H. Genotyping of broiler-originated C. jejuniand C. coli isolates using flatyping and random amplified polymorphic DNA methods. Int J Food Microbial. 2004;94:203–9. [DOI] [PubMed] [Google Scholar]
- 8.Humphery T, O’brien S, Madsen M. Campylobacters as zoonotic pathogens: A food production perspective.Int J of Food Microbiol. 2007;117:237–57. [DOI] [PubMed] [Google Scholar]
- 9.Dadi L, Asrat D. Prevalence and antimicrobial susceptibility of thermotolerant Campylobacter strains in retail raw meat products in Ethiopia. Ethiop J Health Dev.2008;22:195–6. [Google Scholar]
- 10.Salihu MD, Junaidu AU, Oboegbulem SI, Egwu GO. Prevalence and biotypes of Campylobacter spp. isolated from sheep in Sokoto state, Nigeria. Int J Anim VetAd.2009;1:6–9. [Google Scholar]
- 11.Abdalla M, Siham A, Suliman E, YYH, Alian A. Microbial Contamination of Sheep Carcasses at El Kadero Slaughterhouse Khartoum State. Sudan. JVet Sci AnimHusb.2009;48:1–2. [Google Scholar]
- 12.Caprioli A, Morabito S, Brugère H, Oswald E. Enterohemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Res.2005;36:289–311. 10.1051/vetres:2005002 [DOI] [PubMed] [Google Scholar]
- 13.Wingstrand A, Neimann J, Engber J, Nielsen EM, Gerner-Smidt P, Wegener HC, et al. Fresh chicken as main risk factor for Campylobacteriosis, Denmark. Emerg Infect Dis. 2006;12: 280–5. 10.3201/eid1202.050936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuvelink AE, Van heerwaarden C, Zwartkruis-Nahuis A, Tilburg JJ, Bos MH, Heilmann FG, et al. Two outbreaks of Campylobacteriosis associated with the consumption of raw cows’ milk. Int J of Food Microbio.2009;134:70–4. 10.1016/j.ijfoodmicro.2008.12.026 [DOI] [PubMed] [Google Scholar]
- 15.Ricotta EE, Palmer A, Wymore K, Clogher P, Oosmanally N, Robinson T, et al. Epidemiology and antimicrobial resistance of international travel-associated Campylobacter infections in the United States, 2005–2011. Ame J of Public Health. 2014;104:108–14. 10.2105/AJPH.2013.301867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. C.jejunian emerging foodborne pathogen. Emerg Infect Dis.1999;5:28–35. 10.3201/eid0501.990104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rautelin H, Hanninen ML. Campylobacters: the most common bacterial enteropathogens in the Nordic countries. Ann of Medi.2000;32:440–5. 10.3109/07853890009002018 [DOI] [PubMed] [Google Scholar]
- 18.Wesley I. Public health impact of foodborne illness: impetus for the international food safety effort. In: Heredia N., Wesley I., Garcia S.(Eds.), Microbiologically Safe FoodsWiley, John Wiley & Sons, Inc., USA; 2009,pp. 3–13. [Google Scholar]
- 19.EFSA (European Food Safety Authority). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Foodborne Outbreaks in 2013. EFSA J. 2015;13(1):3991. [Google Scholar]
- 20.Kassa T, Gebre-selassie S, Asrat D. The prevalence of thermotolerant Campylobacter species in food animals in Jimma Zone, southwest Ethiopia. Ethiop J Health Dev. 2005;9:225–9. [Google Scholar]
- 21.Ekin IH, Gurturk JK, Boynukara B. Prevalence and characteristic of Campylobacter spp isolated from gallbladder of slaughter sheep in Van, (Eastern) (Turkey). Acta Vet Brno. 2006;75:145–9. [Google Scholar]
- 22.Ebrahim R. Occurrence and resistance to antibiotics of Campylobacter spp. in retail raw sheep and goat meat in ShahreKord, Iran. GlobVet. 2010;.4:504–9. [Google Scholar]
- 23.Gedlu E, Aseffa A. Campylobacter enteritis among children in northwest Ethiopia: A 1-year prospective study. Ann Trop Paediatr.1996;16:207–12. 10.1080/02724936.1996.11747828 [DOI] [PubMed] [Google Scholar]
- 24.Asrat D, Hathaway A, Ekwall E. Studies on enteric Campylobacteriosis in TikurAnbessa and Ethio-Swedish children’s Hospital, Addis Ababa. Ethiopia. Ethiop Med J.1999;37:71–84. [PubMed] [Google Scholar]
- 25.Tafa B, Sawunet T, Tassew H, Asrat D. Isolation and Antimicrobial Susceptibility Patterns of Campylobacterspp among Diarrheic Children at Jimma, Ethiopia. IntJ Bacteriol. 2014; 19:16:3–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tefera W, Daniel A, Girma Z.Prevalence of Thermophilic Campylobacter species in carcasses from sheep and goats in an abattoir in Debre Zeit area, Ethiopia. Ethiop J Health Dev.2009;23(3):229–33. [Google Scholar]
- 27.Yeshimebet C, Daniel A, Patamapom A, Winya L. Prevalence and Antimicrobial susceptibility of thermophilic Campylobacter isolated from sheep at Debre Birhan, North-Shoa, Ethiopia. Kasetsart J (NatSci). 2013;47:551–60. [Google Scholar]
- 28.Faris G. Identification of Campylobacter spp. and their Antibiotic resistance pattern from raw bovine meat in Addis Ababa, Ethiopia. IJMIR. 2015;4(1):001–5. [Google Scholar]
- 29.CSA (Central statistic Authority CSA). Federal Democratic Republic of Ethiopia Population Census Commission. Summary and Statistical Report of population and Housing. 2007.
- 30.Bryant C. Investment opportunities in Mekelle, Tigray state, Ethiopia [online]. 2016. Available from: https://www.ciaonet.org/attachments/15494.Accessed on 27th May 2016.
- 31.Bolton FJ, Hutchinson DN, Coates D. Blood-free selective medium for isolation of C. jejuni from feces. J Clin Microbiol.1984;19:169–71. 10.1128/JCM.19.2.169-171.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nachamkin I. Campylobacter and Arcobacter. In: Murray P.R. and Baron E.G.O., Manual of Clinical Microbiology 7th Eds ASM press; Washington DC: America. Society of Microbiology; 1999,pp. 716–26. [Google Scholar]
- 33.Elmer WK, Stephen DA, Janda PhD, William M, Paul CS, Winn, MDWC Jr. Color Atlas and Text book of Diagnosis Microbiology.5th Edition. 1998, pp.322-6.
- 34.On SLW. Identification methods for Campylobacters, Helicobacters, and related organisms. Clin Microbiol Rev. 1996;9(3):405–22. 10.1128/CMR.9.3.405-422.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CLSI (Clinical and Laboratory Standard Institute) Performance standard for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24.CLSI, Wayne, PA; 2014.
- 36.Sambrook J, Russel WD. Molecular cloning a laboratory manual 3rd ed Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press; 2007. [Google Scholar]
- 37.Denis M, Soumet C, Rivoal K, Ermel G, Blivet D, Salvat G, et al. Development of a m-PCR for simultaneous identification of C jejuniand C. coli. Lett Appl Microbio.1999;29:406–10. [DOI] [PubMed] [Google Scholar]
- 38.Hossein D, Shadi A, Hossein G, Maryam N, Manouchehr AH, Seyed C. Prevalence and Antibiotic susceptibility of Campylobacter spp. isolated from chicken and beef meat. Int Enteric Pathog. 2014;2(2):e17087. [Google Scholar]
- 39.Rahimi E,Tajbakhsh E. Prevalence of Campylobacter spp in poultry meat in the Esfahan city, Iran. BulJ Vet Med. 2008;11:257–62. [Google Scholar]
- 40.Habib I, Sampers I, Utyyendaele M, Berkvens D, De Zutte, L. Base line Data from a Belgium-Wide Survey of Campylobacter Spp. Contamination in Chicken Meat preparation and Consideration for a Reliable Monitoring Program. Appl Environ Microbiol. 2008;72(17):5483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marinou I, Bersimis S, Ioannidis A, Nicolaou C, Mitroussia-Ziouva A, Legakis NJ, et al. Identification and antimicrobial resistance of Campylobacter species isolated from animal sources. Front Microbiol. 2012;3:58 10.3389/fmicb.2012.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahimi E, Kazemeini HR, Safaei S, Allahbakhshi K, Momeni M, Riahi M. Detection and identification of Campylobacter spp from retail raw chicken, turkey, sheep and goat meat in Ahvaz, Iran. Afr J Microbiol Res.2010;4(15):1620–3. [Google Scholar]
- 43.Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, et al. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, D.C., area. Appl Environ Microbiol. 2001;67:431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orla MC, Geraldine D, Sheridan JJ, Blair DA, McDowell DA. A survey on the incidence of Campylobacter spp. and the development of a surface adhesion polymerase chain reaction (SA-PCR) assay for the detection of C. jejuni in retail product. Food Microbiol. 2001;18:287–8. [Google Scholar]
- 45.Pezzotti G, Serafin A, Luzzi I, Mioni R, Milan M, Perin R. Occurrence and resistance to antibiotics of C.jejuniand C.coliin animals and meat in northeastern Italy.Int J Food Microbiol.2003;82:281–7. 10.1016/s0168-1605(02)00314-8 [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen F, Bailey R, Williams S, Henderson P, Wareing DRA, Bolton FJ, et al. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int J Food Microbiol.2002;76:151–64. 10.1016/s0168-1605(02)00027-2 [DOI] [PubMed] [Google Scholar]
- 47.Mayrhofer S, Paulsen P, Smulders FJM, Hilbert H. Antimicrobial resistance profile of five major foodborne pathogens isolated from beef, pork and poultry. Int J Food Microbiol.2004;97:23–9. 10.1016/j.ijfoodmicro.2004.04.006 [DOI] [PubMed] [Google Scholar]
- 48.Hussain I, Mahmood MS, Akhtar M, Khan A. Prevalence of Campylobacter spp. in meat, milk and other food commodities in Pakistan. Food Microbiol. 2007;24:219–22. 10.1016/j.fm.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 49.Taremi M, Soltan-Dallal MM, Gachkar L, MoezArdalan S, Zolfagharian K, Zali MR. Prevalence and antimicrobial resistance of Campylobacter isolated from retail raw chicken and beef meat, Tehran, Iran. Int J Food Microbiol. 2006;108:401–3. 10.1016/j.ijfoodmicro.2005.12.010 [DOI] [PubMed] [Google Scholar]
- 50.Nonga HE, Sells P, Karimuribo ED. Occurrences of thermophilic Campylobacter in cattle slaughtered at Morogoro municipal abattoir, Tanzania. Tropi Animal Heal Productio.2010;42:73–8. 10.1007/s11250-009-9387-7 [DOI] [PubMed] [Google Scholar]
- 51.Vanderlinde PB, Shay B, Murray J. Microbiological quality of Australian beef carcass meat and frozen bulk packed beef. J Food Prot.1998;61:437–43. 10.4315/0362-028x-61.4.437 [DOI] [PubMed] [Google Scholar]
- 52.Oberhelman RA, Taylor DN.Campylobacter infections in developing countries. In: Campylobacter, 2nd ed.; Nachamkin I., Blaser M.J., Eds.; Washington DC: American Society of Microbiology (SAM) Press; 2000, pp. 139–53. 10.1016/s0360-3016(00)00774-4 [DOI] [Google Scholar]
- 53.Thépault A, Poezevara T1, Quesne S, Rose V, Chemaly M, Rivoal K.Prevalence of Thermophilic Campylobacter in Cattle Production at Slaughterhouse Level in France and Link Between C. jejuniBovine Strains and Campylobacteriosis. Front Microbiol. 2018;9:471 10.3389/fmicb.2018.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Châtre P, Haenni M, Meunier D, Botrel MA, Calavas D, Madec JY. Prevalence and antimicrobial resistance of Campylobacter jejuniand Campylobacter coli isolated from cattle between 2002 and 2006 in France. J Food Prot.2010;73:825–31. 10.4315/0362-028x-73.5.825 [DOI] [PubMed] [Google Scholar]
- 55.Rahimi E. Occurrence and Resistance to Antibiotics of Campylobacter Species in Retail Raw Sheep and Goat Meat in Shahr-e Kord, Iran. GlobVet.2010;4(5):504–9. [Google Scholar]
- 56.Zanetti F, Varoli O, Stampi S. Prevalence of thermophilic Campylobacter and Arcobacter butzleri in food of animal origin. Int J Food Microbiol.1996;33:315–21. 10.1016/0168-1605(96)01166-x [DOI] [PubMed] [Google Scholar]
- 57.Ghafir Y, China B, Dierick K, DeZutter L, Daube G. A seven year survey of Campylobacter contamination in meat at different production stages in Belgium. Int J Food Microbiol.2007;116:111–20. 10.1016/j.ijfoodmicro.2006.12.012 [DOI] [PubMed] [Google Scholar]
- 58.Son I, Englen MD, Berrang ME, Fedorka-Cray PJ, Harrison MA. Prevalence of Arcobacter and Campylobacter on broiler carcasses during processing. Int J Food Microbiol.2007;113:16–22. 10.1016/j.ijfoodmicro.2006.06.033 [DOI] [PubMed] [Google Scholar]
- 59.Whyte P, McGill K, Cowley D, Madden RH, Moran L, Scates P, et al. Occurrence of Campylobacter in retail foods in Ireland. Int J Food Microbiol. 2004;95:111–8. 10.1016/j.ijfoodmicro.2003.10.018 [DOI] [PubMed] [Google Scholar]
- 60.Suzuki H, Yamamoto S. Campylobacter contamination in retail poultry meats and by-products in Japan: A literature Survey.J Vet Med Sci. 2009;71(3):255–61. 10.1292/jvms.71.255 [DOI] [PubMed] [Google Scholar]
- 61.WHO. World Health Organization / Department of Communicable Disease Surveillance and Response. Increasing incidence of human Campylobacteriosis. Report and Proceeding of a WHO Consultation of Experts. 2001.
- 62.Elhelberg S, Simonsen J, Gerner-Smid P, Olsen KE, Mølbak K. Spatial distribution and registry-based case-control analysis of Campylobacter infections in Denmark, 1991–2001. Ame J Epidemiol. 2005;162:1008–15. [DOI] [PubMed] [Google Scholar]
- 63.CDC (Centre for Disease Control). Preliminary food net data on the incidence of infection with pathogens transmitted commonly through food-10states, 2009. Morb Mortal Wkl Rep.2010;59:418–22. [PubMed] [Google Scholar]
- 64.Blaser M., Taylor DN, Feldman RA. Feldman, Epidemiology of C. jejuni infections. Epidemiol Rev. 1983;5:157–76. [DOI] [PubMed] [Google Scholar]
- 65.Allos BM. C. jejuniinfections: update on emerging issues and trends. Clin Infect Dis.2001;32:1201–6. 10.1086/319760 [DOI] [PubMed] [Google Scholar]
- 66.Butzler JP. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect.2004;10(10):868–76. 10.1111/j.1469-0691.2004.00983.x [DOI] [PubMed] [Google Scholar]
- 67.Toledo Z, Simaluiza RJ, Fernández H. Occurrence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from domestic animals from Southern Ecuador. Cienc Rural. 2018;48(11):e20180003. [Google Scholar]
- 68.Gallay A, Prouzet-Mauleon V, Kempf I, Lehours P, Labadi L, Camou C, et al. Campylobacter antimicrobial drug resistance among humans, broiler chickens, and pigs, France. Emerging Infect Dis.2007;13:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Looveren M, Daube G, Zutter L, Dumont JM, Lammens C, Wijdooghe M, et al. Antimicrobial Susceptibilities of Campylobacter strains isolated from food animals in Belgium. J Antimicrob Chemother. 2001;48:235–40. 10.1093/jac/48.2.235 [DOI] [PubMed] [Google Scholar]
- 70.Tajada P, Gomez-Garces JL, Alos JI, Balas D, Cogollos R. Antimicrobial susceptibilities of C. jejuniand C. coli to 12 beta-lactam agents and combinations with beta-lactamase inhibitors. Antimicrob Agents Chemother.1996;40:1924–5. 10.1128/AAC.40.8.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nachamkin I, Engberg J, Aarestrup FM. Diagnosis and antimicrobial susceptibility of Campylobacter Spp. in: Campylobacter. 2nd ed., Nachamkin I. and Blaser M.J. (Eds). Washington, DC: ASM Press; 2000,pp. 45–66. [Google Scholar]
- 72.Tan YF, Haresh KK, Chai LC, Son R. Antibiotic susceptibility and genotyping by RAPD of C. jejuni isolated from retailed ready to eat sushi. Int J Food Res.2009;16:31–8. [Google Scholar]
- 73.Sukhapesna J, Amavisit P, Wajjwalku W, Thamchaipenet A, Ukpuaram T. Antimicrobial resistance of C. jejuniisolated from chicken in Nakhon Pathom province Thailand. Kasetsart J (Nat Sci). 2005;39(2):240–6. [Google Scholar]
- 74.Fallon R, Sullivan NO, Mahe M, Carroll C. Antimicrobial resistance of C. jejuniand C. coli isolates from broiler chickens isolated at an Irish poultry processing plant. Lett Appl Microbiol.2003;36:277–81. 10.1046/j.1472-765x.2003.01308.x [DOI] [PubMed] [Google Scholar]
- 75.Praakle-Amin K, Roasto M, Korkeala H, Hänninen ML. PFGE genotyping and antimicrobial susceptibility of Campylobacter in retail poultry meat in Estonia. Int J Food Microbiol. 2007;114:105–12. 10.1016/j.ijfoodmicro.2006.10.034 [DOI] [PubMed] [Google Scholar]
- 76.Hong J, Kim JM, Jung WK, Kim SH, Bae W, Koo HC, et al. Prevalence and antibiotic resistance of Campylobacter sppisolated from chicken meat, pork, and beef in Korea, from 2001 to 2006. J Food Prot.2007;70:860–6. 10.4315/0362-028x-70.4.860 [DOI] [PubMed] [Google Scholar]
- 77.Rogers Van Katwyk S, Grimshaw JM, Nkangu M, Nagi R, Mendelson M, Taljaard M, et al. Government policy interventions to reduce human antimicrobial use: A systematic review and evidence map. PLoS Med. 2019;16(6): e1002819 10.1371/journal.pmed.1002819 [DOI] [PMC free article] [PubMed] [Google Scholar]


