Abstract
The genus Cinnamomum consists of about 250 species spread globally. Out of these, C. verum (C. zeylanicum), also known as true cinnamon or Ceylon cinnamon, has gained worldwide attention due to its culinary uses and medicinal values. Sri Lanka is the largest true cinnamon producer in the world and accounts for about 80–90% of global production. Other than the cultivated species, Sri Lankan natural vegetation is home to seven endemic wild species of the genus Cinnamomum. While these are underutilized, proper identification and characterization are essential steps in any sustainable conservation and utilization strategies. Currently, species identification is purely based on morphological traits, and intraspecific diversity has made it more challenging. In this study, all the eight Cinnamomum species found in Sri Lanka, C. capparu-coronde, C. citriodorum C. dubium, C. litseifolium, C. ovalifolium, C. rivulorum, C. sinharajaense, and C. verum were collected in triplicates and identified using typical morphological traits. DNA extracted with the same collection was assessed with universal barcoding regions, rbcL, matK, and trnH-psbA. While no intraspecific sequence differences were observed in C. citriodorum, C. rivulorum, and C. verum, the others had polymorphic sites in one, two, or all regions assessed. Interestingly, two individuals of C. sinharajaense had identical barcodes to the cultivated species C. verum, while the other one had one variable cite in matK region and three cites in trnH-psbA reigon. Further, one C. dubium and one C. capparu-coronde accession each had identical, rbcL, and trnH-psbA sequences while those had only a single nucleotide variation observed in matK region. Overall, the phylogeny of Cinnamomum species found in Sri Lanka could not be completely resolved with DNA barcoding regions studied.
Introduction
The genus Cinnamomum consists of approximately 250 species [1]. Originally, the genus Cinnamomum was considered a purely Asiatic genus recorded only in the eastern hemisphere, specifically in the Asia Pacific region [2]. Later, several South American species previously included in Phoebe were transferred to Cinnamomum [3]. However, it was shown that these New World species are closer to the Neotropical genus Aiouea than to the Asian species [4]. Nevertheless, some Cinnamomum species have been used for various purposes since ancient times. For example, cinnamon was used to preserve meat and to retard the growth of bacteria. In ancient Rome, it had been used medicinally for cold and flu, as well as to treat diseases associated with the digestive system [5].
Cinnamomum verum J. Presl and C. aromaticum Nees are traded in local and world markets as cinnamon of commerce [6]. Of these, C. verum, also known as Ceylon cinnamon, is considered as the true cinnamon. The species is also known by its synonym C. zeylanicum, which was published only a few months later than C. verum. In addition to being used as a spice and a confectionary flavoring agent, C. verum has been used as an anti-inflammatory, anti-termitic, nematicidal, anti-mosquito, larvicidal, insecticidal, anti-mitotic, and anti-cancer agent. Cinnamon has also been traditionally used as tooth powder and to treat toothaches [7–12].
Cinnamomum verum is endemic to Sri Lanka and is originally found in upcountry rainforests. Portuguese traders brought it under cultivation and later planted it in the coastal areas. Cinnamomum verum was introduced to India in the 19th century by taking seeds from Sri Lanka and planting them in the Western Ghats in Kerala and Tamil Nadu states [2]. Nevertheless, Sri Lanka is the largest true cinnamon producer in the world, accounting for 80–90% of global cinnamon production, earning US$ 190 million foreign exchange by exporting 17,500 metric tons in 2018. This is a 13% increment when compared to exports made in 2017, as of the Department of Export Agriculture, Sri Lanka.
Other than C. verum, there are seven endemic wild species of Cinnamomum in Sri Lanka. They are, C. capparu-coronde Blume, C. citriodorum Thwaites, C. dubium Nees, C. litseifolium Thwaites, C. ovalifolium Wight, C. rivulorum Kosterm. and C. sinharajaense Kosterm [13, 14]. Some of them are restricted to unique environmental conditions whereas others are distributed in a wide range of environmental conditions. For example, C. ovalifolium is only found in upcountry rainforests such as Horton plains and Sri Pada mountain, whereas C. litseifolium is widely found in Kandy, Badulla, Rathnapura, and Nuwara Eliya districts [15, 16]. While some of these species are utilized in Ayurvedic and traditional medicine [17], others are not in use so far. Species such as C. citriodorum, C. dubium, C. litseifolium, and C. rivulorum have unique chemical profiles while species such as C. sinharajaense, C. capparu-coronde have similar chemical constituents to C. verum [18, 19].
Currently accepted species classification and identification of Cinnamomum are purely based on morphological traits such as leaf arrangement, flush color, leaf shape, apex and venation [20, 21]. While C. litseifolium, C. citriodorum, C. sinharajaense, and C. capparu-coronde are easily distinguishable using leaf morphological traits, identification of others, such as C. rivulorum and C. dubium are not that straightforward [22]. Intraspecific diversity of morphological traits makes the identification even more difficult [22].
DNA barcoding, on the other hand, is a universally accepted methodology for molecular level identification of species [23–25]. Most botanists have used the chloroplast coding regions rbcL and matK together with the trnH-psbA intergenic region as in DNA barcoding [26]. Molecular identification has also been implemented to assess both intra and interspecific genetic diversity of Cinnamomum to a certain extent. For example, Abeysinghe and colleagues utilized the chloroplast regions, the trnL intron, the trnT-trnL, trnL-trnF, and trnH-psbA intergenic spacers, as well as the internal transcribed spacer (ITS) of nuclear ribosomal DNA (rDNA) for identification of several wild Cinnamomum species, C. citriodorum, C. capparu-coronde, C. dubium, C. litseifolium, C. rivulorum, C. sinharajaense, C. ovalifolium, an unknown Cinnamomum species, and C. verum [27]. Accordingly, the chloroplast regions alone could not resolve some of the phylogenetic relationships among these species. The sequences of the ITS region turned out to be more useful for the identification of species. Since there was low variation among chloroplast regions studied, Abeysinghe and colleagues have used random amplified polymorphic DNA (RAPD) and sequence-related amplified polymorphic markers (SRAP) to study genetic diversity among selected species in their next study [28]. However, no follow-up studies were done including all the species. In elsewhere, Cinnamomum species used in traditional medicine have been assessed with ITS regions [29]. A recent study used chloroplast regions of trnH-psbA spacer, trnK intron including matK gene, trnL intron, trnL-trnF spacer, and trnQ-rps16 spacer to confirm the identity of plants cultivated under the name Cinnamomum porrectum in Munchen-Nymphenburg Botanical Garden [30]. Further, Yang and colleagues used PCR—Restriction fragment length polymorphism (PCR-RFLP) for rapid identification of the indigenous medicinal crop C. osmophloeum from an adulterant, C. burmannii [31]. The PCR-RFLP can be combined with the morphology-based method known as the deep convolutional neural network (CNN) developed for the same purpose [32].
While there are many advantages of using plant barcoding for species identification and resolving phylogenies, it has failed in the identification of some closely related species in some cases, for example Curcuma (Zingiberaceae) [33], Calligonum [34], Bromeliaceae [35], Picea [36], Berberis [37], and Lauraceae [38]. Therefore, the objective of this study was to assess the possibility of utilizing universally accepted DNA barcoding regions for molecular level identification of Cinnamomum species in Sri Lanka. Both intraspecific and interspecific diversity were assessed with universal barcoding regions, rbcL, matK, and trnH-psbA.
Materials and methods
Sample collection
Permissions for sample collection were obtained from the Research Committee of the Department of Wildlife Conservation, Sri Lanka, and the Department of Forestry, Sri Lanka.
Leaf samples of seven wild Cinnamomum species were collected from the rainforests, the germplasm collections at the National Cinnamon Research and Training Center, Thihagoda, Palolpitiya and Dalpitiya Mid Country Research Station, Department of Export Agriculture (DEA) (S1 Table). Samples of the cultivated C. verum variety ‘Sri Gemunu’ were collected from a vegetative propagated plantation at Nillambe, Sub Research Station, DEA, and seed propagated materials at the National Cinnamon Research and Training Center, Thihagoda, Palolpitiya. The variety Sri Gemunu is one of the two Cinnamomum varieties released by the DEA, Sri Lanka. Three individual plants from each species were considered as biological replicates to assess the intraspecific genetic diversity and named as accessions 001, 002, and 003. Altogether, a total of twenty-four (24) samples were included for the study.
The identity of the collected samples was verified using typical morphological characters. Thirty mature leaves with no symptoms of pest or diseases were collected randomly from the first secondary branch of each studied tree, and leaf morphological characters were recorded following the cinnamon descriptors [20, 22]. Thirteen characters were recorded including four quantitative characters (leaf length, leaf width, petiole length, leaf weight—mean weight of ten mature leaves), and nine qualitative characters (leaf shape, apex, base, texture, margin, arrangement, and color, bark fragrant, and bark taste).
In addition to qualitative characters, the average leaf length, leaf width, leaf weight, and petiole length were calculated and recorded for proper identification purposes (S1 Table). The within-species diversity of the quantitative morphological traits were assessed using Minitab statistical software (18th version). The one-way ANOVA procedure was used for the determination of the significant difference (P<0.05) of means for considered morphological characters.
The collected specimens were further verified with the voucher specimens at the National Herbarium, Sri Lanka. The standard herbarium specimens were prepared by mounting on herbarium sheets and deposited at the National Herbarium, Sri Lanka (S1 Table).
DNA extraction and sequencing
Total genomic DNA from all twenty-four (24) leaf samples was extracted using the cetyltrimethylammonium bromide (CTAB) method [39] with minor modifications [40, 41]. Leaf samples were ground to a fine powder using liquid nitrogen and 200 mg of each sample was used for the extraction. After adding the lysis buffer, C. sinharajaense and C. dubium sample were incubated at 65 oC while shaking for overnight for better lysis without which pipetting was not possible due to jelly like lysate, and the other samples were incubated for one hour while inverting the tubes every 10 minutes. The chloroform: isoamyl alcohol (24:1) extraction was repeated twice to improve the quality of extracted DNA. The extracted DNA samples were re-suspended in 30–50 μL nuclease-free water and stored at 4 oC. DNA quality and quantity were assessed with NanoDrop (NanoDrop 2000 spectrophotometer, Thermo scientific) and running on 0.8% agarose gels with a voltage of 5 Vcm-1 for 30 minutes.
The polymerase chain reaction (PCR) was performed using standard universal plant DNA barcoding primers (Table 1) with a previously optimized reaction mixture [40, 42] containing lx PCR buffer, 1.5 mM MgCl2, 200 μM dNTP (Promega, USA), 0.2 μM of each primer (Integrated DNA Technologies, Singapore), 100 ng of DNA, 0.8 μM spermidine and 1 Unit Go Taq Flexi DNA polymerase (Promega, USA). The PCR cycle consisted of initial denaturation at 94°C for 2 minutes, followed by 35 cycles of 94°C for 1 minute, annealing at 50 oC to 55°C for 30 seconds (Table 1) and 72°C for 30 seconds, and a final extension at 72°C for 3 minutes. Products were separated by electrophoresis (5 Vcm-1) on 1.5% agarose gels and stained with Ethidium Bromide. All the PCR products were sent to Macrogen Inc (Seoul, South Korea-http://dna.macrogen.com) for bi-directional Sanger sequencing using the same primers used for PCR.
Table 1. Primers used for DNA barcoding studies.
| Region | Primer | Sequence 5’-3’ | Annealing temperature | Reference |
|---|---|---|---|---|
| rbcL | rbcLaf-M13_F | 5’ TGT AAA ACG ACG GCC AGT ATG TCA CCA CAA ACA GAG ACT AAA GC 3’ | 55 | [43] |
| rbcLar-M13_R | 5’ CAG GAA ACA GCT ATG ACG TAA AAT CAA GTC CAC CRC G 3’ | 55 | [44] | |
| MatK | MatK_F(390F) | 5’ CGA TCT ATT CAT TCA ATA TTT C 3’ | 55 | [45] |
| MatK_R(1326R) | 5’ TCT AGC CAC GAA AGT CGA AGT 3’ | 55 | [45] | |
| trnH- psbA | psbAF | GTTATGCATGAACGTAATGCTC | 50 | [46] |
| trnHR | CGCGCATGGTGGATTCACAATC | 50 | [46] |
Additional sequences used for the analysis
The trnH-psbA intergenic regions of the accession 001 of all the species were extracted from high-throughput DNA sequencing data stored in a local database. For that, cleaned Sanger sequence data of accession 002 and 003 of each species were used as a query sequence to search (Blastn) high throughput sequence reads of the first accession of respective species using Geneious Prime Software (2020). The same approach was used to assess the quality of Sanger sequencing data of rbcL and matK regions of the accession 001 of all the species. In addition, published sequences of rbcL, matK, and trnH-psbA of C. verum were extracted from the chloroplast sequences deposited in the GenBank (NC_035236.1).
Molecular data analysis
PCR amplification success rates of each region were calculated following the previously described method by Kress [47]. Accordingly, the success rate of PCR amplification refers to the percentage of successful individuals (of the total 24) in two PCR attempts. Both forward and reverse sequencing chromatograms were visually inspected using Geneious Prime software for sequencing errors. The 5’ and 3’ noisy sequences of about 30 bp were removed and cleaned bi-directional sequences of 470 bp and 796 bp were obtained for rbcL and matK regions respectively. However, the reverse sequencing reaction continuously failed in trnH-psbA due to homopolymer runs after 320 bp. Therefore, even though the PCR product size was around 500 bp, the successful sequencing data were obtained only for 308 bp. The consensus sequences for all the samples have been submitted to GenBank (S2 Table).
The alignments were done separately for the three barcode regions, ends were trimed and then only three regions were joined including 470 bp, 796 bp, and 308 bp regions from rbcL, matK, and trnH-psbA respectively. The same regions were included from C. verum, NC_035236.1. In each analysis, the multiple sequence alignment was carried out with Geneious Prime Software using Geneious alignment. The alignment was then exported to Molecular Evolutionary Genetics Analysis (MEGA-X) software for phylogenetic analysis. The maximum likelihood trees were constructed for matK, and trnH-psbA data separately and the combined data set of rbcL, matK, and trnH-psbA with the Tamura–Nei genetic distance model with 100 bootstrap replicates for node supports. A discrete Gamma distribution was used to model evolutionary rate differences among different sites [48, 49]. The intra and interspecific diversities were calculated for each region using the Tamura-Nei model of MEGA–X. All positions with less than 95% site coverage were eliminated and all three codon positions and the noncoding regions were included. Inter and intraspecific sequence divergences for combined barcode regions were also calculated using MEGA–X.
Further, Automatic Barcode Gap Discovery (ABGD) method described by Puillandre et al (2012) delaminate species based on the divergence among organisms [50]. For that, multiple sequence alignment files of matk, and trnH-psbA were separately uploaded to AGBD web (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html). The JC69 Jukes-Cantor measure was set at prior minimum (Pmin) and prior maximum (Pmax) divergence of intraspecific diversity at 0.001 and 0.1 respectively.
Results
We used the standard morphological traits for proper identification of Cinnamomum species and to confirm the identity of collected specimens. The leaf traits such as leaf shape, apex, base, and venation were different among C. litseifolium, C. citriodorum, C. sinharajaense, C. ovalifolium, and C. capparu-coronde. C. sinharajaense has a unique elliptic shape with a narrowly-acuminate apex. Cinnamomum citriodorum shows prominent pinnate venation pattern while all the other species have shown a three-vein structure (S1 Table). Of them, C. ovalifolium, and C. litseifolium have short basal veins while others have long basal veins reaching the leaf apex (S1 Table). The entire leaf margin is prominent in all species. The flat-leaf blade is also prominent in all species except C. litseifolium where it is slightly twisted. However, C. rivulorum, and C. dubium could not easily distinguish each other using leaf morphological characters. Line diagrams of leaf shape, apex, base, and venation of each accession are given in S1 Table. As such, a considerable intraspecific diversity was reported in leaf shape, leaf apex, and leaf base in all the species. The quantitative traits such as leaf length, leaf width, petiole length, and leaf weight also varied considerably within and among species. Nevertheless, the intraspecific diversity was significant only in C. sinharajaense, C. dubium, and C. citriodorum (S1 Table). For example, C. sinharajaense grown in Sinharaja rainforest had significantly longer leaf petioles and longer, wider and heavier leaves compared to the same species grown in Matara (18 cm). However, all three accessions had typical qualitative characteristics to be considered as C. sinharajaense. Cinnamomum dubium collected from Sinharaja rainforest had significantly longer leaf petioles and leaves compared to the accession collected from Matara.
Leaf morphology of the collected samples were further confirmed with herbarium specimens at the National Herbarium. All specimens used in the current study had typical morphological characteristics described previously for species identification purposes.
Good quality DNA could be extracted from all the samples with the modified DNA extraction protocol [40, 41]. PCR success is an important factor considered in selecting barcoding regions [23]. The PCR success rates calculated as the percentage of samples resulting expected band size in two attempts were 100% for rbcL and matK regions and 75% for trnH-psbA region (Table 2). All the samples resulted good quality forward and reverse sequencing data for rbcL and matK regions. However, the reverse reaction of all trnH-psbA sequencing attempts were failed after 320 bp due to homopolymer stretch of T, and therefore, only 308 bp with successful forward and reverse regions were included in the analysis. The alignments done for each region identified polymorphic sites (Table 2). The variability in the rbcL was limited to only one site, at the 56th position, found only in C. ovalifolium accession 001. The trnH-psbA region consists of the highest number of variable sites (11) while matK region had four variable sites. Altogether, 16 variable sites were included in the analysis (Tables 2 and 3). The intraspecific and interspecific diversity indices were calculated for each region (Table 2). Only for rbcL, intraspecific diversity was higher than interspecific diversity.
Table 2. Molecular features of Cinnamomum species barcode.
| Region | PCR amplification success rate % | aAverage sequence length (bp) | bSequence alignment length (bp) | No. of variable sites % | Interspecific diversity % | Intraspecific diversity % |
|---|---|---|---|---|---|---|
| rbcL | 100 | 470 | 470 | 0.21 | 7.7E-06 | 0.00018 |
| ±0.000007 | ±0.000174 | |||||
| MatK | 100 | 796 | 796 | 0.5 | 0.00093 | 0.00053 |
| ±0.000740 | ±0.000320 | |||||
| trnH psbA | 75 | 308 | 308 | 3.57 | 0.011709± | 0.00034 |
| 0.0032 | ±0.001100 |
a length of sequencing data after trimming the noisy 5’ and 3’ ends
b length after multiple alignments.
Table 3. Sequence variation of matK, rbcL, and trnH-psbA regions of Cinnamomum species.
| Region | rbcL | MatK | trnH psbA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | 56 | 52 | 353 | 357 | 795 | 30 | 82 | 83 | 85 | 86 | 89 | 118 | 149 | 154 | 181 | 380 |
| C. cap 001 | C | T | T | T | C | T | A | A | A | G | A | C | C | T | C | T |
| C. cap 002 | C | C | C | T | C | T | A | A | A | G | A | C | A | A | A | C |
| C. cap 003 | C | T | T | T | C | T | A | A | A | G | A | C | C | T | C | T |
| C. cit 001 | C | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. cit 002 | C | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. cit 003 | C | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. dub 001 | C | C | C | T | T | T | G | T | C | T | T | T | A | A | A | C |
| C. dub 002 | C | C | C | T | T | T | A | A | A | G | A | C | A | A | A | C |
| C. dub 003 | C | C | C | T | T | T | G | T | C | T | T | T | A | A | A | C |
| C. lit 001 | C | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. lit 002 | C | C | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. lit 003 | C | T | C | G | C | T | A | A | A | G | A | C | C | A | C | T |
| C. ova 001 | T | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. ova 002 | C | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. ova 003 | C | T | C | T | C | T | A | A | A | G | A | C | C | A | C | T |
| C. riv 001 | C | C | C | T | T | G | A | A | A | G | A | C | A | A | A | C |
| C. riv 002 | C | C | C | T | T | G | A | A | A | G | A | C | A | A | A | C |
| C. riv 003 | C | C | C | T | T | G | A | A | A | G | A | C | A | A | A | C |
| C. sin 001 | C | T | C | T | C | T | G | T | C | T | T | T | C | A | C | T |
| C. sin 002 | C | C | C | T | C | T | G | T | C | T | T | T | A | A | A | C |
| C. sin 003 | C | C | C | T | C | T | G | T | C | T | T | T | A | A | A | C |
| C. ver 001 | C | C | C | T | C | T | G | T | C | T | T | T | A | A | A | C |
| C. ver 002 | C | C | C | T | C | T | G | T | C | T | T | T | A | A | A | C |
| C. ver 003 | C | C | C | T | C | T | G | T | C | T | T | T | A | A | A | C |
| C. ver (NCBI) | C | C | C | T | C | T | G | T | C | T | T | T | A | A | A | C |
C. cap 001, C. capparu-coronde 001- KGG.BS-2018-8-CC-M-1; C. cap 002, C. capparu-coronde 002- KGG.BS-2018-8-CC-M-2; C. cap 003, C. capparu-coronde 003- RAAK.BS-2018-9-CC-D-1; C. cit 001, C. citriodorum 001- KGG.BS-2018-8-C-M-1; C. cit 002, C. citriodorum 002- BS-2019-5-C-N-1; C. cit 003, C. citriodorum 003- BS-2019-5-C-N-2; C. dub 001, C. dubium 001-RHG.BS-2018-11-D-S-1; C. dub 002, C. dubium 002- KGG.BS-2018-8-D-M-1; C. dub 003, C. dubium 003- RHG.BS-2018-11-D-S-2; C. lit 001, C. litseifolium 001- DSA.PCG.BS-2018-5-L-H-1; C. lit 002, C. litseifolium 002- KGG.BS-2018-8-L-M-1; C. lit 003, C. litseifolium 003- RAAK.BS-2018-9-L-D-1; C. ova 001, C. ovalifolium 001- DSA.PCG.BS-2018-5-O-H-1; C. ova 002, C. ovalifolium 002-DSA.PCG.BS-2018-5-O-HP-1; C. ova 003, C. ovalifolium 003-DSA.PCG.BS-2018-5-O-HP-1; C. riv 001, C. rivulorum 001- KGG.BS-2018-8-R-M-1; C. riv 002, C. rivulorum 002- KGG.BS-2018-8-R-M-2; C. riv 3, C. rivulorum 003- KGG.BS-2018-8-R-M-3; C. sin 1, C. sinharajaense 001- RHG.BS-2018-11-S-S-1; C. sin 002, C. sinharajaense 002- KGG.BS-2018-8-S-M-1; C. sin 003, C. sinharajaense 003- KGG.BS-2018-8-S-M-4; C. ver 001, C. verum 001- NL.BS.CHWMRB-2018-6-V-N; C. ver 2, C. ver 002- KGG.BS-2018-8-V-M-1; C. ver 003, C. ver 003- KGG.BS-2018-8-V-M-2; C. ver, C. verum (NC_035236.1).
Interestingly, both C. dubium and C. rivulorum have identical matK regions, with no interspecific variability among accessions. Similarly, both C. verum and two accessions of C. sinharajaense shared identical matK regions. The intraspecific variation in matK region was only observed in C. capparu-coronde and C. litseifolium while C. litseifolium had unique sequences for each accession (Table 3). C. litseifolium, C. ovalifolium and C. citriodorum had identical trnH-psbA regions with no interspecific variability (Table 3). Interestingly, C. verum and C. sinharajaense had identical trnH-psbA sequences except in one C. sinharajaense accession, collected from Sinharaja rainforest. Interestingly, one C. capparu-coronde accession and one C. dubium accession shared identical trnH-psbA regions.
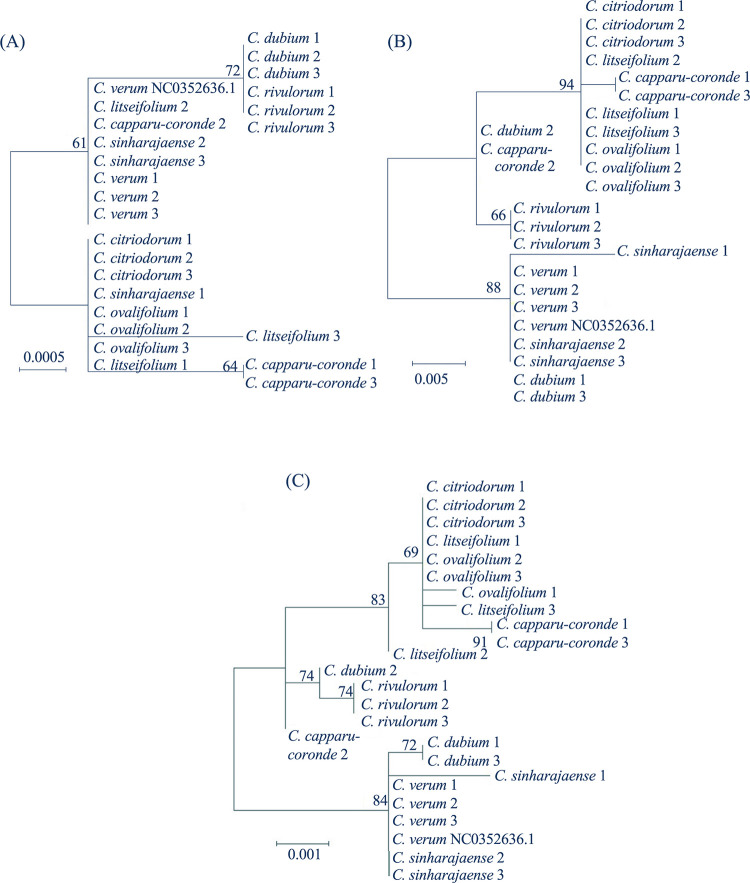
The same relationship was reflected in the unrooted maximum-likelihood trees constructed with complete alignments of matk (Fig 1(A)), trnH-psbA (Fig 1(B)) and all three regions (Fig 1(C)). The phylogeny of Cinnamonum species could not be resolved either with a single sequence barcode of either matK or trnH-psbA or combined barcodes of all three regions. Cinnamomum verum and C. sinharajaense grouped in the combined analysis and the analysis done with trnH-psbA region. However, when the tree was built with matK region, the C.sinharajaense accession (001) did not group with other C. sinharajaense accessions. Cinnamomum dubium accessions grouped only in the tree built with matK region while the accession 002 grouped separately in the phylogenetic trees drawn from trnH-psbA and combined sequences. In the combined analysis, all three accessions of C. citriodorum, C. rivulorum, and C. verum grouped together.
Fig 1. Maximum likelihood trees (unrooted) inferred from DNA barcode data for Cinnamomum species.
(A) matk (B) trnH-psbA (C) combined data sets of rbcL, matK, and trnH-psbA regions. Values on branches represent bootstrap values.
Intraspecific sequence divergence was calculated from the combined sequences of each species. It is calculated as the number of base substitutions per site in averaging all the sequence pairs. This analysis also confirmed no within-species sequence divergence in C. citriodorum, C. rivulorum, and C. verum. The highest intraspecific sequence divergence was observed in C. capparu-coronde and C. dubium (Table 4). The interspecific evolutionary divergence was estimated considering all three accessions from each species as a group (Table 5). The number of base substitutions per site calculated by averaging over all sequence pairs between groups show the highest estimated sequence divergence between C. ovalifolium and the cultivated species C.verum. The lowest sequence divergence was observed between C. sinharajaense and C.verum.
Table 4. Intraspecific sequence divergence.
| Cinnamomum species | Sequence divergence ± Standard Error |
|---|---|
| C. verum | 0 |
| C. sinharajaense | 0.00126±0.0007195 |
| C. rivulorum | 0 |
| C. ovalifolium | 0.00042±0.0003388 |
| C. litseifolium | 0.00084±0.0006215 |
| C. dubium | 0.00253±0.0011095 |
| C. citriodorum. | 0 |
| C. capparu-coronde | 0.00253±0.0009701 |
Table 5. Interspecific sequence divergence between species.
| Cinnamomum species | C. verum | C. sinharajaense | C. rivulorum | C. ovalifolium | C. litseifolium | C. dubium | C. citriodorum. | C. cappru-coronde |
|---|---|---|---|---|---|---|---|---|
| C. verum | 0.0004 | 0.0013 | 0.0018 | 0.0017 | 0.0007 | 0.0018 | 0.0016 | |
| C. sinharajaense | 0.0006 | 0.0014 | 0.0016 | 0.0015 | 0.0008 | 0.0016 | 0.0016 | |
| C. rivulorum | 0.0051 | 0.0057 | 0.0016 | 0.0016 | 0.0010 | 0.0016 | 0.0014 | |
| C. ovalifolium | 0.0066 | 0.0059 | 0.0040 | 0.0003 | 0.0017 | 0.0002 | 0.0009 | |
| C. litseifolium | 0.0063 | 0.0057 | 0.0038 | 0.0006 | 0.0016 | 0.0003 | 0.0009 | |
| C. dubium | 0.0019 | 0.0025 | 0.0032 | 0.0059 | 0.0057 | 0.0016 | 0.0014 | |
| C. citriodorum | 0.0063 | 0.0057 | 0.0038 | 0.0002 | 0.0004 | 0.0057 | 0.0008 | |
| C. capparu-coronde | 0.0063 | 0.0061 | 0.0038 | 0.0019 | 0.0020 | 0.0057 | 0.0017 |
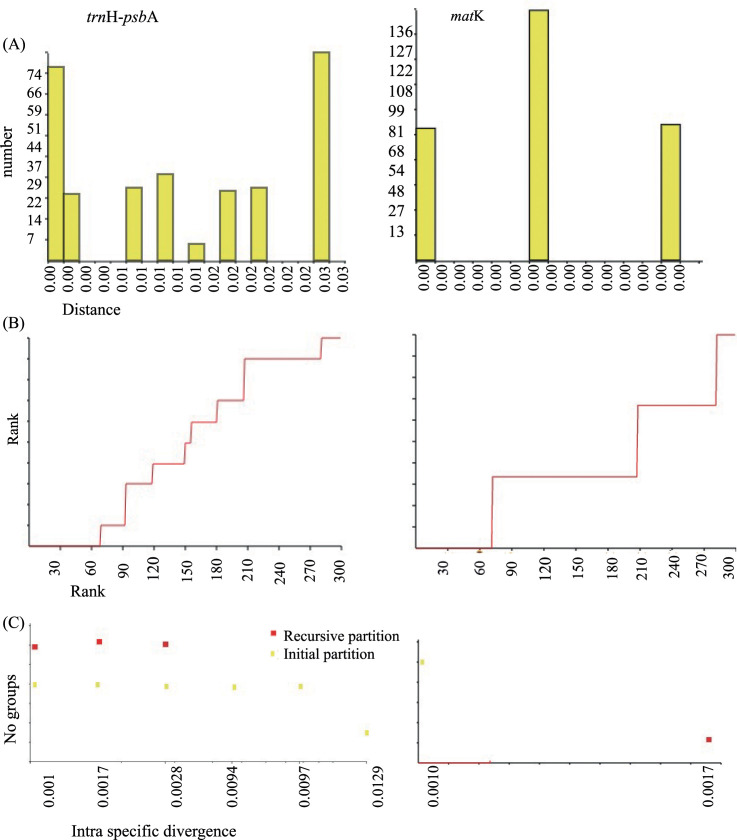
The automatic barcode gap discovery method could not identify clear barcode gap either in matk or in trnH-psbA region. The number of groups ranges from 1 to 6 with trnH-psbA while it ranges from 1 to 5 with matk (Fig 2).
Fig 2. Schematic illustration of Automatic Barcode Gap Discovery (ABGD) with matK and trnH-psbA regions for Cinnamomum species.
(A) Distribution of pairwise differences. Low divergence represent intraspecific divergence, whereas higher divergence represents interspecific divergence (B) Ranked pairwise differences (C) Grouping.
Discussion
All the species included in the study are endemic to Sri Lanka. While the other seven are still in the wild, C. verum has been cultivated since 1500 AC. So far, the species identification and nomenclature have completely been done with morphological traits, highly affected by the environment. Our results also showed the same, for example having significantly different leaves when C. sinharajaense is grown in the Sinharaja rainforest compared to the same grown in the germplasm collection at Matara. Therefore, we wanted to confirm the identity of the collected samples before molecular analysis. We used standard leaf traits as much as possible [22] and further confirmed with the National Herbarium, Sri Lanka. For example, eventhough C. sinharajaense collected from Sinharaja rainforest had significantly larger leaves with longer petiols, qualitative traits, such as leaf apex, base, shape and venation match with typical leaf morphology of the species [22].
DNA barcoding has been used widely in plant biology to resolve phylogenies of related taxa. Nucleotide variation in these standard regions was sufficient to distinguish even closely related taxa. However, in some cases, it was reported that the nucleotide variability in the standard regions was not sufficient to distinguish closely related taxa [33]. It is known that the variability in the rbcL region is relatively lower compared to the matK and trnH-psbA regions [51]. Our data also agree with the claim, having only one variable site in the rbcL region among the local species. It was found only in one of the collected accessions of C. ovalifolium. However it is a synonymous substitution. Since the variable site was present only in one accession, the intraspecific diversity of C.ovalifolium was higher than that of interspecific.
DNA barcoding has been suggested as the method of choice for species differentiation purposes of Cinnamomum. For example, Swetha et al (2014) suggested utilizing the barcoding loci rbcL, matK, and psbA-trnH to detect the presence of adulterants such as C. aromaticum and C. malabathrum (Lam.) J. Presl in traded samples of cinnamon [52].
Nevertheless, our data suggest that the variation in standard barcoding regions rbcL, matK, and trnH-psbA individually or in combination are not sufficient to discriminate Cinnamomum species found in Sri Lanka. While there is only a single base substitution in one C. ovalifolium accession, the other two accessions of the same species and all C. citriodorum accessions consist of identical barcode regions. However, those two species have distinct morphological features such as pinnate venation pattern with no issue in distinguishing at the field level [22]. Both high-performance liquid chromatography (HPLC) and Gas chromatography-mass spectrometry (GC-MS) analysis currently underway show that they also have distinct chemical profiles.
Except for one accession of C. sinharajaense collected from Sinharaja rainforest, the other two accessions of the same species, and all four C. verum accessions included in the analysis had identical barcoding regions considered. Cinnamomum sinharajaense can easily be distinguished from others using leaf traits such as large elliptic leaves and reticulated secondary venation which is visible on both sides of the blade [22]. Prevoius work also suggest such intraspecific morphological diversity depends on the environment [14]. Leaves are larger with more acuminate apex when the plants are grown under shaddy conditions. While C. sinharajaense accession 001 collected from Sinharaja rain forest grown under high shade conditions, accession 002 and 003 were collected from Matara grown under open environment. Germplasm collection in Matara has been established about 10 years ago from seeds collected from Sinharaja forest. Probably both accession 002 and 003 were originated from seeds collected from single mother plant. Our recent work showed that C. zeylanicum offspring resulted from a single cross pollination event are genetically and morphologically divese [53]. Nevertheless, HPLC and GC-MS analysis suggest all C. sinharajaense accessions and C. verum accessions consist of very similar chemical profiles except the differences in concentrations. Such concentration differences are very common in C. zeylanicum [54].
Results suggested high intraspecific diversity in some species. For example, each accession of C. litseifolium has unique matK sequences, and one of them was similar to the matK region of C. ovalifolium. Further, the nucleotide substitution at 119th position resulted change in amino acid from tryptophan to glycine (S1 Fig). However, no intraspecific diversity among three accessions collected from C. citriodorum, C. rivulorum and C. verum. The Cinnamomum flower is naturally adapted for cross-pollination with protogynous dichogamy behavior [2, 55]. The same flower is opened twice, the first day, as a female flower with receptive stigma to be pollinated with mature pollen from another flower/s. The same flower will be opened the next day as a male flower with mature pollen. However, our recent work showed that, overlapping period of 45 min to 1 hr for the two flower types of the same tree [56]. Therefore, self-pollination with pollen from the same plant is also a possibility.
The lowest sequence divergence was recorded between cultivated species, C. verum and C. sinharajaense. Though they are morphologically distinct, they share similar chemical profiles. Cinnamomum sinharajaense is naturally grown only in the Sihnaraja rainforest with limited populations. The highest interspecific divergence was found between C. ovalifolium and C. verum. Interestingly, C. ovalifolium is only found in upcountry rainforests above 1000 m in the central highlands of Sri Lanka. The historical evidence suggests that the cultivated species, C. verum was also naturally grown in upcountry rainforests before domestication.
The barcode gap sorts the sequences into species whenever the divergence among organisms belonging to the same species is smaller than divergence among organisms from different species [50]. The barcode gap of matK or trnH-psbA could not discriminate Cinnamomum species in Sri Lanka.
Overall, the discriminating power of standard barcoding regions selected, rbcL, matK, trnH-psbA are not sufficient even to distinguish morphologically distinct Cinnamomum species found in Sri Lanka and therefore the identity of true cinnamon and its wild relatives couldn’t be assessed. Nevertheless, the analyses suggest genetic closeness among eight Cinnamomum species found in Sri Lanka. Some of the wild species such as C. litseifolium and C. rivulorum are categorized as endangered species in Sri Lanka while all other species are vulnerable [57]. Therefore, correct identification is essential for both conservation and sustainable utilization of valuable genetic resources. The identification of alternative chloroplast barcoding region/s would be important. The complete chloroplast genome sequencing alone or together with fast-evolving genomic regions such as internal transcribed spacer (ITS) regions may provide sufficient molecular data for identifying closely related Cinnamomum species. Complete chloroplast genome analysis could distinguish closely related species such as Pterocarpus [58] and Fritillaria [59]. Therefore, such an approach is proposed for distinguishing Cinnamomum species in Sri Lanka.
Supporting information
Data presented as mean ± standard error of the mean of the three replicates. Mean values represented by different lower case letters within a species in a given column refer to significant differences (P<0.05). LL, leaf length (cm); LW, leaf width (cm); W, leaf weight (g); PL, petiole length (mm); size bar, 10 cm.
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors thank Dr. Ardeshir B. Damania, Department of Plant Science, University of California Davis, USA for helpful comments provided for improving the manuscript. Special thanks to Dr. Ranil Rajapaksha, Department of Crop Sciences, Faculty of Agriculture, University of Peradeniya for his guidance throughout wild cinnamon sample collection. The authors thank the National Cinnamon Research and Training Center, Department of Export Agriculture, Thihagoda, Palolpitiya and Sub Research Station, Department of Export Agriculture, Nillambe, providing cinnamon samples and the Department of Wildlife Conservation, Sri Lanka and the Forest Department for providing permission to collect samples from wild populations in protected areas. The authors would like to thank the staff of the Agricultural Biotechnology Centre, Faculty of Agriculture, University of Peradeniya for the continuous support given.
Data Availability
All relevant data are included in Tables, Figures and Supporting Information.
Funding Statement
PCG is the PI of the grant NSF SP/CIN/2016/01 funded by the Ministry of Primary Industries and Social Empowerment through the National Science Foundation of Sri Lanka (http://www.nsf.ac.lk/) under the special Cinnamon project – Grant No: NSF SP/CIN/2016/01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Willis JC. A dictionary of the flowering plants and ferns. 7th ed New York: Cambridge University press; 1966. [Google Scholar]
- 2.Ravindran PN, Nirmal-Babu K, Shylaja M, editors. Cinnamon and cassia: the genus Cinnamomum. 1st ed CRC press; 2003. [Google Scholar]
- 3.Kostermans AJGH. The New World Species of Cinnamomum Trew (Lauraceae). Reinwardtia. 1961;6: 17–24. [Google Scholar]
- 4.Rohde R, Rudolph B, Ruthe K, Lorea-Hernandez FG, Moraes PL, Li J, et al. Neither Phoebe nor Cinnamomum–the tetrasporangiate species of Aiouea (Lauraceae). Taxon. 2017;66(5): 1085–1111. 10.12705/665.6 [DOI] [Google Scholar]
- 5.Mishra R. Medicinal significance of Cinnamomum spp. International journal of engineering sciences & research technology. 2016;5(7): 749–751. [Google Scholar]
- 6.Chakrabarty T. Second-step lectotypification of the Linnaean name Laurus cassia (Lauraceae) enabling its unambiguous use as Neolitsea cassia. Phytotaxa. 2018. 29;379(3): 274–276. [Google Scholar]
- 7.Jayaprakasha GK, Rao LJM. Chemistry, biogenesis, and biological activities of cinnamomum zeylanicum. Crit Rev Food Sci Nutr. 2011;51(6): 547–562. 10.1080/10408391003699550 [DOI] [PubMed] [Google Scholar]
- 8.Kawatra P, Rajagopalan R. Cinnamon: Mystic powers of a minute ingredient. Pharmacognosy Res. 2015;7(Suppl 1): S1 10.4103/0974-8490.157990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranasinghe P, Pigera S, Premakumara GS, Galappaththy P, Constantine GR, Katulanda P. Medicinal properties of “true” cinnamon (Cinnamomum zeylanicum): A systematic review. BMC Complement Altern Med. 2013;13(1): 275 10.1186/1472-6882-13-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhammad DRA, Dewettinck K. Cinnamon and its derivatives as potential ingredient in functional food—A review. Int J Food Prop. 2017;20(2): 2237–2263. 10.1080/10942912.2017.1369102 [DOI] [Google Scholar]
- 11.Gunawardena D, Govindaraghavan S, Münch G. Anti-Inflammatory Properties of Cinnamon Polyphenols and their Monomeric Precursors. Polyphenols Hum Heal Dis. 2014;1: 409–425. 10.1016/B978-0-12-398456-2.00030-X [DOI] [Google Scholar]
- 12.Singletary K. Cinnamon: Update of Potential Health Benefits. Nutr Today. 2019;54(1): 42–52. 10.1097/NT.0000000000000319 [DOI] [Google Scholar]
- 13.Dassanayake MD, Fosberg FR, Clayton WD eds. A revised handbook to the flora of Ceylon. New Dehli: Amerind Publishing Co. Pvt. Ltd; 1995. vol ix. [Google Scholar]
- 14.Sritharan R. The study of genus Cinnamomum. M.Phil. Thesis, Post Graduate Institute of Agriculture, University of Peradeniya, Sri Lanka. 1984.
- 15.Liyanage AS. Eco-geographic survey of crop wild relatives. Plant Genetic Resources Centre, Gannoruwa, Peredeniya, Sri Lanka. 2010. [Google Scholar]
- 16.Liyanage ASU, Senanayake G. The atlas of selected crop wild relatives in Sri Lanka. Colombo: Department of Agriculture, Sri Lanka, 2010: pp.11–73 [Google Scholar]
- 17.Smerq J, Sharma M. Possible mechanism of Murraya koenigii and Cinnamomum tamala with reference to antioxidants activity. Int J Pharm Sci Drug Res. 2011;3: 260–264. [Google Scholar]
- 18.Ariyarathne HB, Weerasooriya SN, Senarath WT. Comparison of morphological and chemical characteristics of two selected accessions and six wild species of genus Cinnamomum Schaeff. SrLJB. 2018;3(1): 11–23. http://dr.lib.sjp.ac.lk/handle/123456789/8826 [Google Scholar]
- 19.Liyanage T, Madhujith T, Wijesinghe KG. Comparative study on major chemical constituents in volatile oil of true cinnamon (cinnamomum verum presl. syn. c. zeylanicum blum.) and five wild cinnamon species grown in Sri Lanka. Trop Agric Res. 2017;28 (3): 270–280. [Google Scholar]
- 20.Azad R, Senanayake G, Kumara KLW, Pushpakumara DKNG, Geekiyanage S (Team of TURIS 2013 project) Descriptors for Cinnamon (Cinnamomum verum). University of Ruhuna, Sri Lanka: 2016; pp 1–68. [Google Scholar]
- 21.Azad R, Ranawaka RA, Senanayake G, Kumara KW, Pushpakumara DK, Wijesinghe KG, Geekiyanage S. Morphological variation of cinnamon (Cinnamomum verum Presl) germplasm in Matara District of Sri Lanka. IJMFM & AP. 2016;2: 6–14. [Google Scholar]
- 22.Bandusekara BS, Pushpakumara DKNG, Bandaranayake PCG, Wijesinghe KGG, Jayasinghe GG. Field Level Identification of Cinnamomum Species in Sri Lanka Using a Morphological Index. Trop Agric Res. 2020;31(4): 43–53 [Google Scholar]
- 23.Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, et al. A DNA barcode for land plants. Proceedings of the National Academy of Sciences. 2009;106(31): 12794–12977. 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6(5) e19254 10.1371/journal.pone.0019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein PZ, DeSalle R. Review and interpretation of trends in DNA barcoding. Front Ecol Evol. 2019;7: 302 10.3389/fevo.2019.00302 [DOI] [Google Scholar]
- 26.Pang X, Liu C, Shi L, Liu R, Liang D, Li H, et al. Utility of the trnH–psbA intergenic spacer region and its combinations as plant DNA barcodes: a meta-analysis. PloS one. 2012;7(11):e48833 10.1371/journal.pone.0048833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abeysinghe PD, Wijesinghe KGG, Tachida H, Yoshda T. Molecular Characterization of Cinnamon (Cinnamomum Verum Presl) Accessions and Evaluation of Genetic relatedness of Cinnamon species in Sri Lanka based on trnL intron region, intergenic spacers between trnT-rnL, trnL-trnF, trnH-psbA and nuclear ITS. Res J Agric Biol Sci. 2009;5(6): 1079–1088. [Google Scholar]
- 28.Abeysinghe PD, Samarajeewa NG, Li G, Wijesinghe KG. Preliminary investigation for the identification of Sri Lankan Cinnamomum species using randomly amplified polymorphic DNA (RAPD) and sequence related amplified polymorphic (SRAP) markers. JNSF. 2014;42(3): 175–182. 10.4038/jnsfsr.v42i3.7393. [DOI] [Google Scholar]
- 29.Doh EJ, Seung JK, Lee G. Identification and monitoring of Korean medicines derived from Cinnamomum spp. by using ITS and DNA marker. Genes Genomics. 2017;39(1): 101–109. 10.1007/s13258-016-0476-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohwer JG, Trofimov D, Mayland-Quellhorst E, Albach D. Incongruence of morphological determinations and DNA barcode sequences: a case study in Cinnamomum (Lauraceae). Willdenowia. 2019;49(3): 383–400. 10.3372/wi.49.49309 [DOI] [Google Scholar]
- 31.Yang BC, Lee MS, Sun FC, Chao HH, Chang WT, Lin MK, et al. Rapid identification of the indigenous medicinal crop Cinnamomum osmophloeum from various adulterant Cinnamomum species by DNA polymorphism analysis. Pharmacogn Mag. 2020;16(68): 64–68. 10.4103/pm.pm_267_19 [DOI] [Google Scholar]
- 32.Yang HW, Hsu HC, Yang CK, Tsai MJ, Kuo YF. Differentiating between morphologically similar species in genus Cinnamomum (Lauraceae) using deep convolutional neural networks. Comput and Electron in Agric. 2019;162: 739–748. 10.1016/j.compag.2019.05.003 [DOI] [Google Scholar]
- 33.Chen J, Zhao J, Erickson DL, Xia N, Kress WJ. Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol Ecol Resour. 2015;15(2): 337–348. 10.1111/1755-0998.12319 [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Feng Y, Wang XY, Liu B, Lv GH. Failure of DNA barcoding in discriminating Calligonum species. Nord J Bot. 2014;32(4): 511–517. 10.1111/njb.00423 [DOI] [Google Scholar]
- 35.Maia VH, da Mata CS, Franco LO, Cardoso MA, Cardoso SRS, Hemerly AS, et al. DNA barcoding bromeliaceae: Achievements and pitfalls. PLoS One. 2012;7(1): 1–6. 10.1371/journal.pone.0029877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ran JH, Wang PP, Zhao HJ, Wang XQ. A Test of Seven Candidate Barcode Regions from the Plastome in Picea (Pinaceae). J Integr Plant Biol. 2010;52(12): 1109–1126. 10.1111/j.1744-7909.2010.00995.x [DOI] [PubMed] [Google Scholar]
- 37.Roy S, Tyagi A, Shukla V, Kumar A, Singh UM, Chaudhary LB, et al. Universal plant DNA barcode loci may not work in complex groups: A case study with Indian Berberis species. PLoS One. 2010;5(10):e13674 10.1371/journal.pone.0013674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu ZF, Ci XQ, Li L, Li HW, Conran JG, Li J. DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China. PloS one. 2017;17;12(4):e0175788 10.1371/journal.pone.0175788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doyle JJ, Doyle JL. CTAB DNA extraction in plants. Phytochemical Bulletin. 1987;19: 11–15. [Google Scholar]
- 40.Attanayake SR, Kumari SA, Weerakkody WA, Ranil RH, Damania AB, Bandaranayake PC. Molecular diversity and genetic relationships among Sri Lankan pomegranate Punica granatum landraces assessed with inter simple sequence repeat (ISSR) regions. Nord J Bot. 2017. August;35(4): 385–394. [Google Scholar]
- 41.Chandrasekara CHWMRB Bandaranayake PCG, Pushpakumara DKNG. DNA extraction from Cinnamomum zeylanicum cinnamon: a simple and efficient method. In: Proceedings of the 7th YSF Symposium Young Scientists Forum National Science and Technology Commission. 2018. pp. 22–25. [Google Scholar]
- 42.Pathirana CK, Chandrasekara CHWMRB, Attanayake SR, Weerakkody WA, Eeswara JP, Bandaranayake PC. Optimization of DNA extraction and PCR protocols for plants with high Phenolics: Bael, Mango, Pomegranate as examples. CJS. 2018. March 27;47(1):43–48. 10.4038/cjs.v47i1.7485 [DOI] [Google Scholar]
- 43.Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, Zimmer EA, et al. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am J Bot. 2003;90(1): 107–115. 10.3732/ajb.90.1.107 [DOI] [PubMed] [Google Scholar]
- 44.Kress WJ, Erickson DL. A Two-Locus Global DNA Barcode for Land Plants: The Coding rbcL Gene Complements the Non-Coding trnH-psbA Spacer Region. PLoS One. 2007;2(6): e508 10.1371/journal.pone.0000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cueénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am J Bot. 2002;89(1): 132–144. 10.3732/ajb.89.1.132 [DOI] [PubMed] [Google Scholar]
- 46.Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot. 1997;84(8): 1120–1136. 10.2307/2446155 [DOI] [PubMed] [Google Scholar]
- 47.Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proceedings of the National Academy of Sciences. 2009;106(44):18621–18626. 10.1073/pnas.0909820106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3): 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6): 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puillandre N, Lambert A, Brouillet S, ACHAZ G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol. 2012;21(8): 1864–77. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- 51.Yang HQ, Dong YR, Gu ZJ, Liang N, Yang JB. A Preliminary Assessment of matK, rbcL and trnH—psbA as DNA Barcodes for Calamus (Arecaceae) Species in China with a note on ITS. In Annales Botanici Fennici. 2012; pp. 319–330. [Google Scholar]
- 52.Swetha VP, Parvathy VA, Sheeja TE, Sasikumar B. DNA Barcoding for Discriminating the Economically Important Cinnamomum verum from Its Adulterants. Food Biotechnol. 2014;28(3): 183–194. 10.1080/08905436.2014.931239 [DOI] [Google Scholar]
- 53.Liyanage NN, Ranawake AL, Bandaranayake PCG. Cross-pollination effects on morphological, molecular, and biochemical diversity of a selected cinnamon (Cinnamomum zeylanicum Blume) seedling population. J Crop Improv. 2020; 1–17. 10.1080/15427528.2020.1795769 [DOI] [Google Scholar]
- 54.Liyanage NM, Bandusekara BS, Kanchanamala RW, Hathurusinghe HA, Dilhan AM, Pushpakumara DG, et al. Identification of superior Cinnamomum zeylanicum Blume germplasm for future true cinnamon breeding in the world. J Food Comps Anal. 2020; 10.1016/j.jfca.2020.103747 [DOI] [Google Scholar]
- 55.Kumari HRSN, Wijesinghe KGG, Ranawaka RAAK, Study on floral behavior of selected ten cultivars of true cinnamon Cinnamomum verum (Presl.), Syn. C. zeylanicum (Blume), Proceedings of the National Symposium, 2008; pp. 29 [Google Scholar]
- 56.Hathurusinghe HABM, Bandaranayake PCG. Studying Unique Flowering Behaviour of Cinnamomum Zeylanicum: Examples from Two Elite Varieties International Agricultural Research Symposium, edited by Sri Lanka Council for Agricultural Research Policy (SLCARP), Colombo, Sri Lanka: 2018; pp. 22 [Google Scholar]
- 57.MOE. The national red list 2012 of Sri Lanka; conservation status of the fauna and flora. Ministry of Environment, Colombo, Sri Lanka: 2012. [Google Scholar]
- 58.Jiao L, Lu Y, He T, Li J, Yin Y. A strategy for developing high-resolution DNA barcodes for species discrimination of wood specimens using the complete chloroplast genome of three Pterocarpus species. Planta. 2019;250(1): 95–104 10.1007/s00425-019-03150-1 [DOI] [PubMed] [Google Scholar]
- 59.Bi Y, Zhang MF, Xue J, Dong R, Du YP, Zhang XH. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci Rep. 2018;8(1): 1–12. 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data presented as mean ± standard error of the mean of the three replicates. Mean values represented by different lower case letters within a species in a given column refer to significant differences (P<0.05). LL, leaf length (cm); LW, leaf width (cm); W, leaf weight (g); PL, petiole length (mm); size bar, 10 cm.
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are included in Tables, Figures and Supporting Information.