Abstract
Taxon sampling is a central aspect of phylogenetic study design, but it has received limited attention in the context of total-evidence dating, a widely used dating approach that directly integrates molecular and morphological information from extant and fossil taxa. We here assess the impact of commonly employed outgroup sampling schemes and missing morphological data in extant taxa on age estimates in a total-evidence dating analysis under the uniform tree prior. Our study group is Pimpliformes, a highly diverse, rapidly radiating group of parasitoid wasps of the family Ichneumonidae. We analyze a data set comprising 201 extant and 79 fossil taxa, including the oldest fossils of the family from the Early Cretaceous and the first unequivocal representatives of extant subfamilies from the mid-Paleogene. Based on newly compiled molecular data from ten nuclear genes and a morphological matrix that includes 222 characters, we show that age estimates become both older and less precise with the inclusion of more distant and more poorly sampled outgroups. These outgroups not only lack morphological and temporal information but also sit on long terminal branches and considerably increase the evolutionary rate heterogeneity. In addition, we discover an artifact that might be detrimental for total-evidence dating: “bare-branch attraction,” namely high attachment probabilities of certain fossils to terminal branches for which morphological data are missing. Using computer simulations, we confirm the generality of this phenomenon and show that a large phylogenetic distance to any of the extant taxa, rather than just older age, increases the risk of a fossil being misplaced due to bare-branch attraction. After restricting outgroup sampling and adding morphological data for the previously attracting, bare branches, we recover a Jurassic origin for Pimpliformes and Ichneumonidae. This first age estimate for the group not only suggests an older origin than previously thought but also that diversification of the crown group happened well before the Cretaceous-Paleogene boundary. Our case study demonstrates that in order to obtain robust age estimates, total-evidence dating studies need to be based on a thorough and balanced sampling of both extant and fossil taxa, with the aim of minimizing evolutionary rate heterogeneity and missing morphological information. [Bare-branch attraction; ichneumonids; fossils; morphological matrix; phylogeny; RoguePlots.]
Dating phylogenetic trees remains one of the most controversial issues in systematics. Until recently, the so-called “node dating” approach (ND) was the gold standard for dating phylogenetic trees, with fossils providing minimum ages for specific nodes in a phylogeny. ND requires the prior assessment of fossil placement, which is usually far from straightforward, and it can only incorporate the oldest fossil assignable to a particular node in the tree (Donoghue and Benton 2007; Donoghue and Yang 2016). In addition, one must decide on a probability distribution of the node’s age, because minima are insufficient to date molecular trees; although largely arbitrary, these settings determine the outcome of any ND analysis (Warnock et al. 2011; Klopfstein 2020). These issues led to the development of the total-evidence dating (TED) approach, which allows the inclusion of all available fossils as tips while accounting for uncertainty in their age and placement in a tree (Pyron 2011; Ronquist et al. 2012a). The downside of TED is that it requires extensive morphological matrices to be compiled, which inform fossil placements and associated branch lengths. Some concerns were also raised about the potential lack of clock-likeness of morphological data (O’Reilly et al. 2015), and some authors reported what they deemed unrealistically old ages from their TED analyses when compared to the oldest known fossils of the group (Beck and Lee 2014; Arcila et al. 2015).
Several methodological modifications to the TED approach have been suggested, such as replacing the initially introduced uniform tree prior (Ronquist et al. 2012a) with the fossilized birth–death tree prior, which models speciation, extinction and fossilization rates to reconstruct branching events, and thus does not necessarily require morphological data (Heath et al. 2014; Ronquist et al. 2012a; Zhang et al. 2016). It has also become possible to account for fossils possibly being sampled ancestors (Gavryushkina et al. 2014) and for a diversified sampling strategy of extant taxa (Höhna et al. 2011). Finally, some recent studies combined tip and node dating approaches (O’Reilly and Donoghue 2016; Kealy and Beck 2017; O’Hanlon et al. 2018; Travouillon and Phillips 2018). Unfortunately, these different implementations of TED have often reported disagreeing age estimates (Grimm et al. 2015; Herrera and Davalos 2016; Harrington and Reeder 2017; Kealy and Beck 2017; Gustafson et al. 2017), revealing that our understanding of the inner workings of this method is still regrettably patchy (Parins-Fukuchi and Brown 2017).
Incorporating more complex models often requires more and better data for improved estimation of model parameters. With the development of next-generation sequencing technologies, acquiring large amounts of molecular data is no longer a problem (Delsuc et al. 2005; McCormack et al. 2013; Misof et al. 2014), but taxon sampling is still a major limiting factor in phylogenetic study design. Besides the sampling of the focal, ingroup taxa, the choice of outgroup also requires special attention: it should ideally include a sufficient sample of taxa that are closely related to, but clearly different from the ingroup (Wheeler 1990; Nixon and Carpenter 1993; Giribet and Ribera 1998; Graham et al. 2002; Philippe et al. 2011). A poorly chosen outgroup can significantly affect topology estimates in nonclock analyses by introducing or at least exacerbating long-branch attraction (Graham et al. 2002; Holland et al. 2003; Philippe et al. 2011) and/or by increasing compositional heterogeneity of sequences and among-lineage rate variation (Tarrío et al. 2000; Rota-Stabelli and Telford 2008; Borowiec et al. 2019). Numerous simulations and empirical studies have demonstrated positive effects of improved taxon sampling on the estimation of topology (Graybeal 1998; Dunn et al. 2008; Heath et al. 2008; Klopfstein et al. 2017), branch lengths (Fitch and Bruschi 1987; Pick et al. 2010), and parameters of evolutionary models (Zwickl and Hillis 2002; Heath et al. 2008). Nevertheless, thorough taxon sampling is not always easy to achieve, especially in very species-rich groups.
Several studies have investigated the impact of poor taxon sampling on age estimates in molecular dating analysis and demonstrated its severe negative effect. The effect was most pronounced when taxon sampling strategy led to strongly imbalanced phylogenetic trees and/or high among-lineage rate variation, which could not be adequately accommodated by existing rate smoothing algorithms or relaxed molecular-clock models (Milne 2009; Soares and Schrago 2012, 2015; Wertheim et al. 2012; Duchêne et al. 2015, 2014). Duchêne et al. (2015) have shown that the effect of tree imbalance on age estimates is even larger when heterochronous sequences are included, as is the case in molecular tip-dating with ancient DNA or in virus studies; a similar effect can be expected when fossil taxa of different ages are included as tips. Both the tree imbalance and high among-linage rate variation can be introduced through a poorly chosen outgroup, as demonstrated for nonclock analyses (see above), but the outgroup choice notably has received little attention in molecular dating analyses, where a molecular-clock model can infer the root of the tree and thus makes the addition of an outgroup unnecessary. Accordingly, the outgroups then can be excluded from dating analysis to decrease the heterogeneity of evolutionary rates across the tree (Welch and Bromham 2005).
In practice, however, most of the recent TED studies included one to a handful of outgroup taxa, which are thus severely underrepresented compared to the ingroup, both in terms of morphological characters and fossils (Ronquist et al. 2012a; Arcila et al. 2015; Dornburg et al. 2015; Close et al. 2016; Lee 2016; Kittel et al. 2016; Herrera and Davalos 2016; Bannikov et al. 2017; Wang et al. 2018; Paterson et al. 2020). The reasons for including these outgroups, despite their potentially negative effects, is seldom reported in these studies and might vary between authors. They might not trust the clock model enough to rely on it for proper rooting, especially when rate heterogeneity is large, which has led to topological artifacts in the past (Ronquist et al. 2012a). Or they might feel uncomfortable with estimating the root age of their ingroup without providing additional taxa that branch off earlier than that node of main interest. The habit might be another incentive to stick to outgroups, which are standard for rooting nonclock trees. And finally, systematists might simply assume that adding more taxa and thus more data to analysis will overall improve the results. We here aim to systematically test the influence of outgroup sampling on age estimates in TED, using parasitoid wasps of the family Ichneumonidae as a case study.
The Ichneumonidae, or Darwin wasps (Klopfstein et al. 2019b), is the most species-rich family of parasitoid wasps, with more than 25,000 described species (Yu et al. 2016), and at the same time one of the most severely understudied taxa. We focus on Pimpliformes, a monophyletic group comprising nine subfamilies: Acaenitinae, Collyriinae, Cylloceriinae, Diacritinae, Diplazontinae, Orthocentrinae, Pimplinae, Poemeniinae, and Rhyssinae (Wahl and Gauld 1998; Quicke 2014; Klopfstein et al. 2019b). Pimpliformes are especially interesting from a biological perspective since they cover nearly the entire diversity of hosts and parasitoid strategies known from ichneumonids (Broad et al. 2018). They oviposit into (endoparasitoids) or onto (ectoparasitoids) their host, which they either permanently paralyze (idiobionts) or allow to continue developing (koinobionts). Recorded hosts span almost all holometabolous insect orders, as well as spiders (Araneae). Several attempts have been made in the past to reconstruct the evolution of important biological traits in Pimpliformes (Wahl and Gauld 1998; Gauld et al. 2002; Quicke 2014), but their conclusions were highly dependent on the stability and resolution of the pimpliform phylogeny, which is still in part unresolved (Klopfstein et al. 2019b).
Even less-well understood than the sequence of events during the radiation of Pimpliformes is their timing and thus ecological context. The fossil record of ichneumonids is very poorly studied, and most described species come from just a handful of localities (Menier et al. 2004). It starts in the Early Cretaceous with the extinct subfamily Tanychorinae (Kopylov 2010a), but the affiliation of this subfamily with Ichneumonidae is somewhat unclear, as its wing venation is intermediate between Ichneumonidae and their sister family Braconidae (Sharkey and Wahl 1992). The Palaeoichneumoninae (Kopylov 2009), also extinct and of a similar age, are thus usually referred to as the oldest ichneumonids. All remaining ichneumonid fossils from the Cretaceous period have been classified in the extinct subfamilies Labenopimplinae and Novichneumoninae (Kopylov 2010a; Kopylov et al. 2010; Li et al. 2017), except for a single amber fossil that was tentatively placed in the extant subfamily Labeninae (McCormack et al. 2013). The oldest fossil associated with an extant pimpliform subfamily (and an extant genus) is an acaenitine, Phaenolobus arvenus Piton, from the latest Paleocene. However, its placement is questionable due to poor preservation, and more reliable records come from two Early Eocene localities, the Green River Formation and Messel Pit (Spasojevic et al. 2018a,b).
No studies have to date attempted to infer the age of Ichneumonidae as a whole or of Pimpliformes in particular. The only previous studies with some bearing on the question are either concerned with the sister family Braconidae (Whitfield 2002) or with the entire order Hymenoptera (Peters et al. 2017); both studies included a very sparse sample of ichneumonids. They report very different age estimates for the divergence between Ichneumonidae and Braconidae,  138 Ma (Whitfield 2002) and 155–224 (mean 188) Ma (Peters et al. 2017), respectively. In addition to obtaining the first age estimates for Ichneumonidae and Pimpliformes and testing the influence of outgroup sampling on these estimates, we investigate the impact of fossil sampling and phylogenetic placement on the accuracy of age estimates. Finally, we discuss the implications of our findings for taxon sampling in dating studies in general.
138 Ma (Whitfield 2002) and 155–224 (mean 188) Ma (Peters et al. 2017), respectively. In addition to obtaining the first age estimates for Ichneumonidae and Pimpliformes and testing the influence of outgroup sampling on these estimates, we investigate the impact of fossil sampling and phylogenetic placement on the accuracy of age estimates. Finally, we discuss the implications of our findings for taxon sampling in dating studies in general.
Material and Methods
Taxon Sampling
We have included 289 species in our data sets, comprising 210 extant and 79 fossil taxa, with a focus on the pimpliform subfamilies within Ichneumonidae. Among the extant taxa, nine were outside of Ichneumonidae, including seven Braconidae (which together with Ichneumonidae belong to the superfamily Ichneumonoidea) and two more distantly related parasitoid wasps from the superfamilies Chalcidoidea and Evanioidea. The remaining extant taxa consisted of 30 nonpimpliform ichneumonids belonging to 19 subfamilies and an extensive sampling of Pimpliformes, for which we included 142 of the 188 known genera. For most genera, we included a single representative, but additional species were included in some morphologically heterogeneous genera. The complete list of extant taxa is given in Supplementary File S1 available on Dryad at https://dx.doi.org/10.5061/dryad.m0cfxpnzm.
We aimed to get a good representation of fossil taxa from different time periods: from the oldest ichneumonids from the Early Cretaceous to fossils from the latest Oligocene period (Supplementary File S2 available on Dryad). Due to the controversial position of the extinct subfamily Tanychorinae, which is somewhat intermediate in morphology between Ichneumonidae and Braconidae, and poor sampling of the morphological diversity in Braconidae, we excluded the two Tanychorinae fossils from most analyses. However, we also ran two analyses under different outgroup sampling schemes with Tanychorinae included to assess their placement and the impact of their inclusion on age estimates (see below).
Morphological and Molecular Data
We used the morphological matrix from Klopfstein and Spasojevic (2019), but with a strongly expanded taxon sampling. Numerous additional character states were defined to capture the added specimen diversity. The complete morphological matrix consists of 222 morphological characters coded initially for 150 extant and 79 fossil taxa. After observing attraction of fossil taxa to some nonpimpliform ichneumonids for which we had not yet coded any morphological data (“bare-branch attraction,” see Results section), we added another 20 extant taxa to the morphological matrix, increasing the total number of scored extant taxa to 170 (Supplementary File S3 available on Dryad, also available at MorphoBank http://morphobank.org/permalink/?P3821). Morphology was scored for the same species from which we obtained molecular data, with a few exceptions where we scored a closely related, congeneric species for morphology (Supplementary File S1 available on Dryad).
Our molecular data set includes nine nuclear protein-coding genes, which for 57 taxa were extracted from a previously compiled hybrid-capture data set (Klopfstein et al. 2019). For another 89 taxa, we newly obtain sequences of these genes using standard PCR and Sanger sequencing (primers and protocols in Klopfstein et al. 2019). For all taxa, we added the D2/D3 portion of the nuclear rRNA gene 28S and the mitochondrial cytochrome c oxidase (COI; but see below) by Sanger sequencing. We achieved good coverage for these 11 genes for the 146 taxa, with an average of seven genes successfully sequenced per taxon (Supplementary File S4 available on Dryad). The sequences were edited and aligned in Geneious 7.1.3 (https://www.geneious.com, Kearse et al. 2012) using the MAFFT v.7.017 plug-in and the algorithm “E-INS-I” (Katoh and Standley 2013). The “translation alignment” option was used for protein-coding genes. The complete alignment contained 6213 base pairs (bp) (Supplementary File S5 available on Dryad). The newly generated sequences are deposited in GenBank under accession numbers MW048210–MW048331, MW056205–MW056314, and MW123099–MW123898.
Nonclock Analysis
To assess branch length heterogeneity and to examine the power of our combined molecular and morphological data set to resolve ichneumonid relationships, we first ran a nonclock analysis. As the analysis showed convergence issues with parameter estimation for most of the 3rd codon partitions of the nine nuclear protein-coding genes and for the entire COI gene, we excluded those from all further analyses. The final alignment thus contained 10 genes and 4011 bp. We partitioned the data set by gene and codon position and used PartitionFinder 2 (Lanfear et al. 2017) to identify partitions that could be combined (settings: branch lengths  linked, models
linked, models  all, model_selection
all, model_selection  aicc, search
aicc, search  rcluster). In addition, we combined all 1st and all 2nd codon position partitions, respectively, that contained fewer than 30 parsimony informative sites, assuming that substitution model parameter estimates would be very poor for those. The preferred evolutionary model for all partitions was GTR+G+I according to PartitionFinder 2. As MrBayes allows model jumping over the entire GTR subspace, the model parameters for the molecular partitions were set as nst
rcluster). In addition, we combined all 1st and all 2nd codon position partitions, respectively, that contained fewer than 30 parsimony informative sites, assuming that substitution model parameter estimates would be very poor for those. The preferred evolutionary model for all partitions was GTR+G+I according to PartitionFinder 2. As MrBayes allows model jumping over the entire GTR subspace, the model parameters for the molecular partitions were set as nst  mixed and rates
mixed and rates  invgamma, with all substitution model parameters unlinked across partitions. Morphological characters were analyzed under the Mk model (Lewis 2001), accounting for ascertainment bias (“Mkv,” i.e., only variable characters coded), allowing gamma-distributed rate variation across characters, and ordering all the characters where transition only between neighboring states could be assumed (Supplementary File S6 available on Dryad). This model has been identified as the preferred model for the morphological partition in a previous analysis (Klopfstein and Spasojevic 2018). The nonclock analysis included the full set of outgroup taxa (Chalcidoidea, Evanioidea, Braconidae, and nonpimpliform Ichneumonidae), with Gasteruption (Evanioidea) chosen as the functional outgroup.
invgamma, with all substitution model parameters unlinked across partitions. Morphological characters were analyzed under the Mk model (Lewis 2001), accounting for ascertainment bias (“Mkv,” i.e., only variable characters coded), allowing gamma-distributed rate variation across characters, and ordering all the characters where transition only between neighboring states could be assumed (Supplementary File S6 available on Dryad). This model has been identified as the preferred model for the morphological partition in a previous analysis (Klopfstein and Spasojevic 2018). The nonclock analysis included the full set of outgroup taxa (Chalcidoidea, Evanioidea, Braconidae, and nonpimpliform Ichneumonidae), with Gasteruption (Evanioidea) chosen as the functional outgroup.
We ran four independent runs with four Metropolis-coupled chains each for 150 million generations with a sampling frequency of 1000. The heating coefficient was decreased from the default value of 0.1 to 0.05 in order to increase chain swap probabilities. To summarize the result, we used a conservative burn-in of 50%, while the convergence of runs was assessed using typical Markov Chain Monte Carlo (MCMC) diagnostics: the average standard deviation of split frequencies (ASDSF), effective sample size (ESS), and the potential scale reduction factor (PSRF). We also visually inspected the trace plots of the likelihoods and of all parameters for all four runs using Tracer v1.7 (Rambaut et al. 2018). All phylogenetic analyses in this study were carried out using Bayesian inference in MrBayes 3.2.6 (Ronquist et al. 2012b) on the HPC cluster UBELIX of the University of Bern, Switzerland (http://www.id.unibe.ch/hpc). The final data matrices and resulting consensus trees are available at TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S26969).
TED Analysis
In addition to the settings above, in the TED analysis, we used the uniform tree prior and a relaxed clock model with independent gamma rates (IGR). To set priors on the relaxed-clock model parameters, we relied on the calculations from Ronquist et al. (2012a), putting an exponential prior on the IGR variance with a rate of 37.12, and a lognormal prior on the clock rate with a mean on the log scale of  7.08069 and standard deviation of 1; the standard deviation was decreased compared to Ronquist et al. (2012) to put increased weight on lower clock rates and thus older age estimates, assuming that our data set contains enough information from the data to correctly estimate the posterior. The prior on the tree age was set to offsetexp(126, 309), with the offset based on the minimum age of the oldest Evanioidea fossil (Deans et al. 2004), while the mean corresponds to the mean age estimate for Hymenoptera from Ronquist et al. (2012a). In the analyses where nonichneumonid outgroups were excluded (see below), we used the minimum age of the oldest unequivocal ichneumonid fossils (Palaeoichneumoninae, 112.6 Ma) as an offset. We set hard bounds using uniform priors on the age of the fossils according to the range of age estimates for the fossil stratum (Barido-Sottani et al. 2019; Püschel et al. 2020). See Supplementary File S2 available on Dryad for the full list of included fossils and their age intervals with corresponding references.
7.08069 and standard deviation of 1; the standard deviation was decreased compared to Ronquist et al. (2012) to put increased weight on lower clock rates and thus older age estimates, assuming that our data set contains enough information from the data to correctly estimate the posterior. The prior on the tree age was set to offsetexp(126, 309), with the offset based on the minimum age of the oldest Evanioidea fossil (Deans et al. 2004), while the mean corresponds to the mean age estimate for Hymenoptera from Ronquist et al. (2012a). In the analyses where nonichneumonid outgroups were excluded (see below), we used the minimum age of the oldest unequivocal ichneumonid fossils (Palaeoichneumoninae, 112.6 Ma) as an offset. We set hard bounds using uniform priors on the age of the fossils according to the range of age estimates for the fossil stratum (Barido-Sottani et al. 2019; Püschel et al. 2020). See Supplementary File S2 available on Dryad for the full list of included fossils and their age intervals with corresponding references.
To obtain the effective prior implied by our settings, including the clock rate (which is only effective when running with data), we performed an analysis with molecular and morphological data for extant taxa, but without the fossils and thus without temporal information. We ran two independent runs for 100 million generations each and under three different clock rate settings and always obtained a rather flat age distribution for crown-group ichneumonids (Supplementary File S7 available on Dryad). Our preferred setting for the clock rate (lognormal with mean of log values 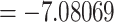 and standard deviation
and standard deviation 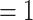 ), for instance, resulted in a median age estimate of 229.3 Ma and a 95% credibility interval (CI) of 51.3–738.6 Ma (Supplementary File S7 available on Dryad). We can thus assume that any more precise age estimates resulting from the analyses with fossils will indeed be informed by the data and not the prior (Parins-Fukuchi and Brown 2017).
), for instance, resulted in a median age estimate of 229.3 Ma and a 95% credibility interval (CI) of 51.3–738.6 Ma (Supplementary File S7 available on Dryad). We can thus assume that any more precise age estimates resulting from the analyses with fossils will indeed be informed by the data and not the prior (Parins-Fukuchi and Brown 2017).
It has been shown that relaxed-clock models can lead to topology artifacts, especially close to the root (Ronquist et al. 2012a). We thus set hard constraints on the monophyly of Braconidae, Ichneumonidae, and Ichneumonoidea (Braconidae 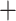 Ichneumonidae), each of which is widely accepted as being monophyletic and have been recovered in previous analyses (Sharkey and Wahl 1992; Dowton and Austin 1994; Peters et al. 2017), as well as in our nonclock analysis. To improve convergence on the clock rate and tree length parameters, we increased the probabilities of the respective MCMC moves (MrBayes command blocks are provided as Supplementary File S8 available on Dryad). MCMC convergence proved much more difficult to attain than in the nonclock analysis and was thus deemed satisfactory when the ASDSF value was below 0.03, ESS values of all scalar parameters were above 100, and PSRF values were below 1.01. The ESS values between 50 and 100 and PSRF values above 1.01 were still accepted for the tree length, tree height, and clock rate parameters in some runs, as it was difficult to get convergence on those even after 150 million generations.
Ichneumonidae), each of which is widely accepted as being monophyletic and have been recovered in previous analyses (Sharkey and Wahl 1992; Dowton and Austin 1994; Peters et al. 2017), as well as in our nonclock analysis. To improve convergence on the clock rate and tree length parameters, we increased the probabilities of the respective MCMC moves (MrBayes command blocks are provided as Supplementary File S8 available on Dryad). MCMC convergence proved much more difficult to attain than in the nonclock analysis and was thus deemed satisfactory when the ASDSF value was below 0.03, ESS values of all scalar parameters were above 100, and PSRF values were below 1.01. The ESS values between 50 and 100 and PSRF values above 1.01 were still accepted for the tree length, tree height, and clock rate parameters in some runs, as it was difficult to get convergence on those even after 150 million generations.
TED Outgroup Settings
We tested five different outgroup sampling strategies (Table 1): i) “full outgroup” (with all outgroup taxa as in the nonclock analysis), ii) “Braconidae” (all braconids and non pimpliform Ichneumonidae, but excluding the two nonichneumonoid taxa), iii) “Braconidae (1)” (a single braconid taxon, Homolobus, the only braconid with both molecular and morphological data, and all nonpimpliform Ichneumonidae), iv) “Xoridinae” (only members of Ichneumonidae as outgroups), and v) “Xorides-only” (a single nonpimpliform Ichneumonidae, Xorides, as outgroup to Pimpliformes). To set the functional outgroup in each of these cases (i.e., nonichneumonoids, braconids, one braconid, Xoridinae, or Xorides), we enforced monophyly of the remaining taxa through a topology constraint. The fossils were in all cases included in the “Ichneumonidae” monophyly constraint, but excluded from any additional topology constraints (see below). Instead, their placement within Ichneumonidae was estimated entirely from the morphological data. In the “Xoridinae” analysis, we applied a partial constraint to enforce the sister relationship of xoridines and the remaining extant ichneumonids, for which there is strong evidence from previous phylogenetic studies (Klopfstein et al. 2019a), and at the same time to allow fossils to attach freely to any branch of the tree.
Table 1.
Summary of outgroup sampling strategies.
| Taxon | Outgroup setting | ||||
|---|---|---|---|---|---|
| “full outgroup” | “Braconidae” | “Braconidae(1)” | “Xoridinae” | “Xorides-only” | |
| Gasteruption (Eva) | x | ||||
| Eupelmophotismus (Cha) | x | ||||
| Aleiodes (Bra) | x | x | |||
| Aphidius (Bra) | x | x | |||
| Cotesia (Bra) | x | x | |||
| Dancusa (Bra) | x | x | |||
| Diaeretus (Bra) | x | x | |||
| Macrocentrus (Bra) | x | x | |||
| Homolobus (Bra) | x | x | x | ||
| Tanychora (Tan) | x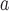
|
x
|
|||
| Kharsutella (Tan) | x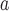
|
x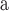
|
|||
| Xorides (Xor) | x | x | x | x | x |
| Odontocolon (Xor) | x | x | x | x | |
| Aplomerus (Xor) | x | x | x | x | |
| Ischnoceros (Xor) | x | x | x | x | |
other nonpimpliform ichneumonids
|
x | x | x | x | |
Notes: Abbreviations in brackets stand for higher level classification of the outgroup taxa: Eva  Evanioidea; Cha
Evanioidea; Cha  Chalcidoidea; Bra
Chalcidoidea; Bra  Braconidae; Tan
Braconidae; Tan  Tanychorinae; Xor
Tanychorinae; Xor  Xoridinae.
Xoridinae.
 Included only in the “full outgroup
Included only in the “full outgroup  Tanychorinae” and “Xoridinae
Tanychorinae” and “Xoridinae  Tanychorinae” analyses.
Tanychorinae” analyses.
 For a list of all included nonpimpliform ichneumonid taxa, see Supplementary File S1 available on Dryad.
For a list of all included nonpimpliform ichneumonid taxa, see Supplementary File S1 available on Dryad.
To assess potential bias when excluding the extinct Tanychorinae, we also performed the “full outgroup” and the “Xoridinae” analyses with two members of this subfamily included. In the “full outgroup” case, we constrained Tanychorinae within Ichneumonoidea, but applied a partial constraint on Braconidae and Ichneumonidae, which allowed Tanychorinae to attach to any crown or stem branch of the two families. In the “Xoridinae” analysis, Tanychorinae were constrained to be the sister group to the remaining taxa, thus effectively rooting the tree with this extinct subfamily.
Fossil Placement
We assumed that an erroneous placement of the oldest included fossils would have the greatest influence, if any, on age estimates. We thus used “RoguePlots” as described in Klopfstein and Spasojevic (2018) to examine the placement of the Cretaceous fossils on 1000 evenly sampled trees from the four runs in relevant analyses (R package available at https://github.com/seraklop/RoguePlots). In the “Xoridinae” outgroup setting, the Cretaceous impression fossils were predominantly placed on some terminal branches leading to nonpimpliform taxa without morphological data (see Results section). We thus ran an additional analysis under the “Xoridinae” outgroup sampling scheme with an improved morphological matrix, which now did not contain any outgroup taxa without morphological data.
Simulations to Study Bare-Branch Attraction
To ascertain that the bare-branch attraction artifact that we observed was not due to some irregularity of our data set, but instead represents a general phenomenon, we conducted simulations. As a simulation tree, we randomly chose one tree from the postburnin sample under the “Xoridinae” outgroup scheme. Because the full tree with 278 taxa would have been too large to obtain a sufficient number of replicates in a reasonable timeframe, we reduced it to 61 extant and 61 fossil taxa that represent all major groups within Ichneumonidae (Supplementary File S9 available on Dryad). To ensure that bare-branch attraction does not only occur in highly unbalanced trees, as with the tree we had produced through our outgroup sampling strategy, but we also removed mostly pimpliform ingroup taxa, which led to a more balanced tree of ichneumonids.
On this simulation tree, which had all the fossils locked in place, we simulated 50 replicate data sets, each containing 300 bp of DNA data simulated under a Jukes–Cantor model and 100 binary characters simulated under equal rates. As an evolutionary rate, we chose 0.25 expected substitutions between the root and any tip for both the molecular and the morphological data partitions. This value has been shown to be near-optimal for phylogenetic inference (Klopfstein et al. 2017), thus potentially producing informative data for TED analyses also. For all fossils, we removed the entire molecular partition, plus the same proportion of morphological data that was missing in the original data set. Fossils thus had on average 27 characters coded (range: 8–61 characters). For the extant taxa, we included all characters, but in each replicate randomly chose 12 of the 61 extant taxa to become “bare” taxa—taxa with only molecular and no morphological data included. Analyses were run in MrBayes with settings identical to the original analyses, except only performing 10 million generations, which was sufficient to obtain ASDSF values below 0.025 while strongly reducing the computational burden.
To assess the impact of bare branches on fossil placement, we calculated for each of the 61 terminal branches leading to extant taxa the cumulative probability that they attracted a fossil, depending on whether they were bare in a given replicate or not. Since the placement of older fossils might be more unstable, for example, due to poorer preservation or weakening of overall morphological signal from tips to root, we tested whether fossil age was the primary determinant for a fossil to be attracted to bare branches by calculating the same statistic separately for the Cretaceous and the Cenozoic fossils. The number of characters coded for a fossil might be another factor, so we also obtained the statistic separately for those fossils that had fewer than 20 characters coded. And finally, we examined the distance of each fossil from its closest extant taxon on the simulation tree, by adding up the relevant branch lengths. We then denoted one-half of the fossils as “close” and one half as “distant,” depending on whether their distance from any extant taxon was below or above the median distance. The R and shell scripts used to conduct these simulations and summarize the results, along with the corresponding simulated data sets, are available in Supplementary File S9 available on Dryad.
Results
Tree Resolution and Topology
The backbone of the majority-rule consensus trees from both the nonclock and TED analyses was unresolved when fossils were included, which was also reflected in the node support values for higher-level relationships. However, when the fossils were excluded from the sampled trees before summarizing them, resolution at the backbone was strongly improved (Fig. 1) and all the basal pimpliform nodes were highly supported (posterior probability  0.95). The exception was the “Xorides-only” TED analysis, where outgroup choice negatively affected the topological resolution of the backbone (Supplementary File S8 available on Dryad). There were only a few topological differences between the nonclock and TED consensus trees, all concerning weakly supported nodes in both analyses (Supplementary Files S6 and S8 available on Dryad). Most of the expected higher-level relationships outside of Pimpliformes were recovered, such as Xoridinae as the sister group to all other ichneumonids (Fig. 1). Within Pimpliformes, Diplazontinae were recovered as the sister group of the remaining subfamilies. The majority of pimpliform subfamilies were recovered as monophyletic (if we disregard a few taxa with alternative placements), the exceptions being Cylloceriinae, Diacritinae, and Pimplinae. Details on alternative placements, taxonomic implications and proposed changes in classification are provided in Supplementary File S10 available on Dryad.
0.95). The exception was the “Xorides-only” TED analysis, where outgroup choice negatively affected the topological resolution of the backbone (Supplementary File S8 available on Dryad). There were only a few topological differences between the nonclock and TED consensus trees, all concerning weakly supported nodes in both analyses (Supplementary Files S6 and S8 available on Dryad). Most of the expected higher-level relationships outside of Pimpliformes were recovered, such as Xoridinae as the sister group to all other ichneumonids (Fig. 1). Within Pimpliformes, Diplazontinae were recovered as the sister group of the remaining subfamilies. The majority of pimpliform subfamilies were recovered as monophyletic (if we disregard a few taxa with alternative placements), the exceptions being Cylloceriinae, Diacritinae, and Pimplinae. Details on alternative placements, taxonomic implications and proposed changes in classification are provided in Supplementary File S10 available on Dryad.
Figure 1.
Majority rule consensus tree of the nonclock analysis of the “full outgroup” setting. The tree contains only extant tips, as fossils were excluded prior to summarizing the tree samples from the Bayesian analysis. Posterior probability values are given only for the nodes of interest.
Impact of Outgroup Sampling on Age Estimates
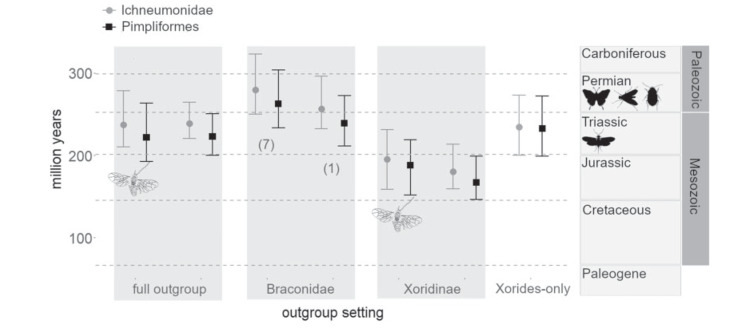
The median age estimates for Pimpliformes varied widely across the different outgroup analyses, ranging from 167 Ma to 263 Ma (Fig. 2, Supplementary File S8 available on Dryad). Age estimates were consistently older and typically less precise when outgroup taxa other than ichneumonids were included (i.e., more distant and more poorly sampled outgroups) and when only a single nonpimpliform ichneumonid was included as the outgroup (Fig. 2, Supplementary Files S8 and S11 available on Dryad). Age estimates were oldest when all seven braconid taxa were used as the outgroup, followed by the “Braconidae (1)” and “Xorides-only” sampling scheme; the latter resulted in the least precise age estimates (Fig. 2, Supplementary File S11 available on Dryad). The inclusion of two representatives of the extinct Tanychorinae in the “full outgroup” and “Xoridinae” analyses also decreased the precision of the age estimates, while the impact on median age estimate was close to zero in the former and the only minor in the latter analysis (Fig. 2).
Figure 2.
Age estimates in million years for Ichneumonidae and Pimpliformes across different analyses. The median and 95% credibility intervals for different outgroup settings are plotted on the y axis. Numbers in brackets indicate the number of included Braconidae species. Dashed horizontal lines indicate boundaries between geological periods. Analyses with Tanychorinae included are denoted by a fossil drawing (modified after Kopylov 2010a). The insect silhouettes denote the estimated time of radiation of the biggest orders of holometabolous insects (according to Tong et al. 2015), which are the main hosts of ichneumoniods: Lepidoptera, Diptera, Coleoptera (Permian), and Hymenoptera (Triassic).
Due to these large differences in age estimates, we assessed the impact of the outgroup setting on the variance of the clock rate to detect any pronounced rate heterogeneities between outgroups and ingroups. The estimated variance for the relaxed clock (IGR) varied considerably across the different outgroup settings (Fig. 3) and was nearly twice as high in the analyses with distant outgroups (“full outgroup” and “Braconidae”) compared to the analysis with only close outgroups (“Xoridinae” and “Xorides-only”). The exception was when a single braconid was included, in which case the terminal branch leading to the single braconid species also obtained a much higher clock rate than the remaining branches, but this was not yet sufficient to increase the average clock variance very much (results not shown). This finding confirms the pronounced rate variation among outgroups as observed already in the nonclock tree (Fig. 1), where especially some Braconidae species showed very long terminal branches compared to the ingroup.
Figure 3.
Variance of the relaxed-clock parameter as estimated under an independent gamma rates model (IGR). Median and 95% credibility intervals are plotted on the y axis across different outgroup settings. Numbers in brackets indicate number of included Braconidae area. Analyses with Tanychorinae included are denoted by a fossil drawing (modified after Kopylov 2010a).
From here onwards, we suppose that the age estimates were biased when more distant outgroups were included, as these outgroups introduced high rate variation, were poorly sampled, and no fossils were included that might have provided timely information for the outgroup branches to accurately estimate the evolutionary rate in this part of the tree. This notion is supported by the smaller clock-rate variance and higher consistency in the age estimates when only close and well-sampled outgroups were included (“Xoridinae” outgroup setting). We also consider age estimates in the “Xorides-only” analysis to be biased, given that it resulted in topological artifacts, poor precision of age estimates, and most importantly, numerous fossils were missing their closest extant relatives under this outgroup setting (see next section). Our preferred analysis is thus the “Xoridinae” outgroup sampling scheme.
Placement of Cretaceous Fossils
Placements of most of the Cretaceous compression fossils in our initial analyses with different outgroup settings were rather similar: they clustered predominantly within crown-groups of nonpimpliform ichneumonid subfamilies (Fig. 4b,c, Supplementary File S12 available on Dryad). The highest placement probabilities were centered around extant Banchinae (Banchus and Apophua) and Tersilochinae (in the case of Labenopimplinae and Tryphopimpla xoridoptera) and/or on the branch leading to Orthopelma (especially in Palaeoichneumoninae), with only a few weakly supported placements in other parts of the tree (less than 10% attachment probability). All Labenopimplinae were placed with the highest probability (34–44%, respectively) on the branch leading to Banchus. The overall support for Labenopimplinae belonging to crown group Banchinae was quite high (46–53%), and it was even higher if we also considered stem Banchinae (63–73%). Tryphopimpla xoridoptera, a fossil with highly uncertain taxonomic affinities, was mostly associated with Apophua (34%), with a total probability of attachment within crown Banchinae of 55%, which increased to 71% when adding stem placements. In contrast, the small undescribed ichneumonid (3311_856b) from Late Cretaceous Yantardakh amber (Rasnitsyn et al. 2016) was attached with very high probability (97%) to the branch leading to the extant Phygadeuontinae genus Gelis. Interestingly, most of the tips to which the Cretaceous fossils attached contained no or only sparse morphological information (Fig. 4b,c), leading us to postulate a “bare-branch attraction” phenomenon—a tendency especially for fossils with uncertain affinities to be attracted to branches leading to extant taxa for which no or very sparse morphological data has been obtained.
Figure 4.
Placement of a) a representative of the Cretaceous fossils (Labenopimpla kasparyan; the image modified after Kopylov (2010b)) in the “Xoridinae” analysis. RoguePlots derived b,c) with and d) without some outgroup taxa missing morphological data (“bare data”). The trees represent the majority-rule consensus tree with fossils excluded. Branches are colored by probability of a fossil attaching to them. Percent values refer to the portion of scored morphological characters for a given extant taxon (c, d) and for the fossil in question (a). The remaining Cretaceous compression fossils had similar attachment patterns as the ones depicted here (Supplementary File S12 available on Dryad).
We thus repeated the analysis with the “Xoridinae” outgroup setting, after completing the scoring of morphological characters for all ichneumonid outgroup taxa, including those on “attracting,” bare branches. The placement of the Cretaceous impression fossils then changed considerably and mostly shifted towards the root of the tree compared to the previous analysis (Fig. 4d). Most of the Cretaceous fossils were now placed on stem branches of both nonpimpliform and pimpliform lineages, with only small placement probabilities for long branches of outgroup taxa (e.g., Orthopelma, Brachycyrtus). All Labenopimplinae now attached with the highest probability to the stem branch of the pimpliform subfamily Diplazontinae (23–39%) and to a nearby long branch leading to the extant tryphonine genera Zagryphus and Thymaris (22–32%). The probabilities of a placement of the Cretaceous impression fossils with crown Banchinae was now close to zero (Fig. 4d, Supplementary File S12 available on Dryad).
Impact of Fossil Placement on Age Estimates
Although the placement of the Cretaceous fossils changed when the improved morphological matrix was employed, the median age estimates remained relatively stable (Fig. 5), but with a consistent improvement in the precision of the age estimates for all but one of the examined nodes. For example, the width of the 95% CI decreased from 53 Myr to 49 Myr for the ancestral node of Pimpliformes and from 63 Myr to 53 Myr for the ancestor of Diplazontinae (Supplementary Files S8 and S11 available on Dryad).
Figure 5.
Age estimates in million years for the nodes of interests in the “Xoridinae” outgroup analyses with and without missing morphological data for some outgroup taxa. The median and 95% credibility intervals are plotted on the y axis. Dashed horizontal lines indicate transitions between major geological periods. The insect silhouettes denote the estimated time of radiation of the biggest orders of holometabolous insects (according to Tong et al. 2015), which are the main hosts of ichneumonids: Lepidoptera, Diptera, Coleoptera (Permian) and Hymenoptera (Triassic).* Pimplinae without the tribe Pimplini.
Bare-Branch Attraction in Simulated Data Sets
Our 50 simulated data sets overall behaved similarly to the original data, in that most nodes of the tree of extant taxa were recovered well, but a few nodes that were subtended by very short branches in the simulation tree were not resolved. Most of the fossils behaved as rogues—taxa with uncertain placement in phylogenetic analyses, as in the original analyses, and including them removed any resolution from the consensus tree. RoguePlots of the fossils showed a heterogeneous picture, with some fossils placed rather firmly, while others shifted around a lot, again reflecting rather well the situation with the empirical data. Analyses of fossil attachment probabilities for the terminal branches leading to extant taxa confirm that bare-branch attraction occurs in simulated data sets as well (Fig. 6). On average, this probability was higher on bare branches than on nonbare ones, although with a lot of variation. The effect was strongest for the one-half of fossils that were more distant from any extant taxon than the others, which were preferentially attaching to bare branches, while the opposite was true for the fossils with rather close ties to at least one extant taxon. Age also seemed to have an impact, but it was far less strong, as was the influence of the number of characters coded for a fossil (Fig. 6).
Figure 6.
Bare-branch attraction in simulated data. Boxplots showing the cumulative probability of a branch leading to an extant taxon to attract a fossil, depending on whether it was bare in the simulation replicate (i.e., it was lacking morphological data) or not. The branches for which morphological data was deleted were chosen randomly for each simulation replicate. The graphs to the right show the same probability, but for subsets of the fossils: the half of the fossils that are more distant versus closer to extant taxa; Cretaceous versus Cenozoic fossils; and finally fossils with fewer than 20 characters coded plotted separately. Distance from any extant taxa thus appears as the most decisive factor here, but beware that we only obtained simulation replicates on a single tree and thus the factors concerning fossil placement are not independent.
Age of Pimpliformes
The age estimates resulting from our preferred analysis, with multiple but only closely related and well-sampled outgroup taxa (“Xoridinae,” Table 2 and Fig. 7), and after filling in the attracting bare branches, suggest that Ichneumonidae originated during the Early Jurassic (95% CI spans most of the Jurassic: 154.0–204.7 Ma) and Pimpliformes during the Middle (95% CI spans most of the Jurassic144.0–193.4 Ma). The start of the radiation of most of the pimpliform subfamilies is estimated as having occurred in the Early Cretaceous.
Table 2.
Age estimates from the preferred analysis with credibility intervals for crown group Ichneumonidae, Pimpliformes, and the pimpliform subfamilies.
| Taxon group | Median | Mean | 95% credibility interval |
|---|---|---|---|
| (millions of years) | |||
| Ichneumonidae | 181.2 | 181.4 | 154.0–204.7 |
| Pimpliformes | 167.8 | 168.7 | 144.0–193.4 |
| Parasitoids of Diptera | 126.2 | 127.8 | 107.3–150.0 |
| Acaenitinae | 127.6 | 129.0 | 105.1–156.7 |
| Diplazontinae | 123.5 | 124.8 | 98.9–152.0 |
| Poemeniinae | 82.7 | 83.5 | 59.1–113.4 |
| Rhyssinae | 96.8 | 97.7 | 69.6–129.9 |
| Delomeristini | 79.7 | 80.6 | 51.1–111.8 |
| Ephialtini | 103.7 | 105.4 | 82.9–128.9 |
| Pimplini | 146.7 | 147.4 | 122.1–175.1 |
| Theroniini | 80.4 | 80.8 | 54.3–106.7 |
Notes: As the subfamily Pimplinae was not recovered as monophyletic, age estimates for the tribes are given (Delomeristini, Ephialtini, Pimplini, and Theroniini). The parasitoids of Diptera here comprise three subfamilies: Cylloceriinae, Diacritinae, and Orthocentrinae (excluding Diplazontinae and the diacritine Ortholaba, which did not form a monophyletic group with them; note that Diacritinae are included in this clade, but there are no host records for this small subfamily).
Figure 7.
Dated majority-rule consensus tree from the total-evidence dating analysis under the “Xoridinae” outgroup setting with improved morphological matrix. The tree contains only extant tips, as the fossils were excluded prior to summarizing the tree samples from the MCMC analysis. Most of the clades are collapsed to depict subfamily-level relationships among Pimpliformes. Arrows indicate nodes for which age estimates are reported in Table 2. The names of the nodes are given if they do not correspond to the names of their tips. Horizontal bars represent 95% credibility intervals and corresponding circles mark median values for age estimates.
Discussion
Impact of Outgroup Sampling on Age Estimates
We show here that with the inclusion of more distantly related and/or poorly sampled outgroups, age estimates for pimpliform parasitoid wasps become older and often less precise. Besides creating a highly unbalanced tree in terms of sampling of both extant and fossil taxa, including distant outgroups in our case introduced large variation in the clock rate, which is most likely the main reason for the observed bias in age estimates (Magall et al. 2013; Magallón 2014; Beck and Lee 2014; Beaulieu et al. 2015; King et al. 2017a). In addition, in the absence of time information in the form of fossils along the branches of distant outgroups together, the age estimates might have been driven by our clock rate prior, resulting in older estimates for crown group Ichneumonidae and Pimpliformes in those analyses (see analysis of effective prior in Materials and Methods). Multiple studies have shown that outgroup choice can greatly affect tree topology estimates, especially in cases with a large heterogeneity of branch lengths and uneven taxon sampling (Puslednik and Serb 2008; Ware et al. 2008; Hayes et al. 2009; Thomas et al. 2013; Kirchberger et al. 2014; Wilberg 2015). However, the influence of outgroup choice on divergence time estimates, especially in the context of total-evidence dating, has scarcely been studied, and only indirectly (Linder et al. 2005; Soares and Schrago 2012; Duchêne et al. 2015; Matschiner 2019).
In the last 8 years since total-evidence dating became established, most of the studies have included outgroups in their taxon sampling (Ronquist et al. 2012a; Arcila et al. 2015; Dornburg et al. 2015; Close et al. 2016; Herrera and Davalos 2016; Kittel et al. 2016; Lee 2016; Bannikov et al. 2017). We here covered most of the outgroup-sampling schemes found in these studies, from including a single closely related taxon (“Xorides-only” outgroup setting), over a few relatively closely related taxa (“Braconidae” and “Braconidae (1)”), to the inclusion of a series of more to less closely related outgroup taxa (“full outgroup”). As in most previous studies, our nonichneumonid outgroups were not only sparsely sampled, but were also missing fossils and morphological data for most of the taxa. All these outgroup settings recovered older and usually less precise age estimates than when we restricted our data set to more closely related outgroup taxa, for which greater and more even effort in sampling both extant and fossil taxa had been applied (“Xoridinae” outgroup setting). Interestingly, when two potentially transitional fossils between Braconidae and Ichneumonidae (the Tanychorinae) were included, the median age estimates and clock variance were only slightly affected, but their precision dropped significantly. This could result from the rather large uncertainty in the age of these fossils themselves, which results from a disagreement over the age of the Khasurty Formation (Kopylov and Rasnitsyn 2017; PaleoBioDB 2020), and the fact that only very few morphological characters were scored for them (9% and 17%, respectively).
Our results might suggest that many of the previous TED analyses would recover younger and likely more accurate age estimates without outgroups or with either more detailed or more restricted, but more even outgroup sampling. Presumably, the effect would be most pronounced for data sets where there are long, unbroken outgroup branches which introduce large rate heterogeneity across the tree. A similar effect has been demonstrated for node dating in the simulation study by Soares and Schrago (2015), where age estimates were significantly biased when there was a combination of large among-lineage rate variation and poor taxon sampling. As in our case, both accuracy and precision were affected in their simulations: the mean age of the node in question was constantly overestimated and precision severely decreased. Some TED studies already conjectured that high evolutionary rate heterogeneity can lead to biased age estimates (Beck and Lee 2014; Lee 2016; King et al. 2017b; Bagley et al. 2018; Luo et al. 2020). Among-lineage rate heterogeneity can already be identified on a nonclock tree by comparing root-to-tip distances of extant taxa and problematic taxa can accordingly be excluded, as we demonstrated here; however, this has rarely been employed in TED studies (but see Grimm et al. 2015).
In addition, more realistic models of morphological evolution might eventually improve the clock-likeness of morphological data sets by improving their match to the underlying evolutionary mechanisms. Some of the aspects for which attempts have already been made to increase realism are asymmetry in transition rates among character states (Lewis 2001; Wright et al. 2016; Pyron 2017), nonstationarity of the evolutionary process (Klopfstein et al. 2015) or even the modeling of undetected character states through hidden Markov models (Tarasov 2019). These models have not been routinely used in TED studies and, at least at present, many of them are either restricted to specific character types or are computationally challenging for large morphological data sets. The latter was also true for our data set when we tried to account for asymmetric transition rates among character states (Lewis 2001; Wright et al. 2016).
Finally, incompletely sampled phylogenies, with respect to fossil and/or extant taxa, could potentially be modeled more adequately with the fossilized birth–death tree prior (FBD) instead of the uniform tree prior used here, but only when model assumptions are satisfied (Zwickl and Hillis 2002). When model assumptions are violated, such as assumptions related to the sampling density of fossil and/or extant taxa, age estimates are quickly compromised (Matschiner 2019; Luo et al. 2020; Püschel et al. 2020). This makes it difficult to apply the FBD prior to the ichneumonid data set at present, as species richness through time even today is still poorly known for this group.
Bare-branch attraction hampering correct fossil placement in TED.—
In TED analyses, the placement of fossils is solely dependent on the available morphological information for both fossil and extant taxa. The quality of fossil placement based on morphological matrices is thus primarily limited by imperfect preservation of fossils, but also by high levels of morphological homoplasy, which has been reported for ichneumonids (Gauld and Mound 1982; Klopfstein and Spasojevic 2019). We here identified another potentially major issue in TED studies, which we called “bare-branch attraction”: the tendency of fossils which are only distantly related to any of the included extant taxa to attach to terminal branches of extant taxa for which no morphological data have been collected. This artifact exposes the dangers of insufficient sampling of extant taxa for morphology, which can distort fossil placement and consequently age estimates in TED analyses.
Many of the previous TED studies included from a few to more than half of the extant taxa without morphological data or at least with high amounts of missing data (Ronquist et al. 2012b; Arcila et al. 2015; Dornburg et al. 2015; Harrington and Reeder 2017). Guillerme and Cooper (2016) addressed this issue in the context of topology reconstruction in TED analyses. In their simulations, topology estimates were more negatively affected by a large percentage of extant taxa with missing morphological data than by any other analyzed parameter. It remains unclear to what extent their results were influenced by bare-branch attraction, as they did not analyze individual fossil placements, but it is likely that the artifact played a role under their scenario as well.
In our study, the bare-branch attraction was most obvious in the compression fossils from the Cretaceous. With the exception of T. xoridoptera, these fossils are all classified in two extinct subfamilies, Palaeoichneumoninae and Labenopimplinae. The phylogenetic position of these subfamilies is unclear, but two options have been suggested: a transitional position between Tanychorinae and extant Ichneumonidae, which would mean they represent stem ichneumonids, or some rather basal position as crown ichneumonids (Kopylov 2009, 2010b). In fact, the name “Labenopimplinae” reflects their similarity to the extant subfamilies Labeninae and Pimplinae (Kopylov 2010b).
We showed that the predominant placement in our initial analysis, of most Labenopimplinae and Paleoichneumoninae with crown group Banchinae and Tersilochinae, was a bare-branch attraction artifact. Among the branches where these fossils attached on the tree, only a single tip (Apophua) contained morphological data, while the remaining taxa were only sampled for molecular characters. Some other, younger fossils with rather labile placement also often attached to these branches, which might suggest that when morphological information for the placement of a fossil is limited, TED analyses tend to place a fossil on “bare branches,” especially if those branches are long. Filling in the missing information among extant outgroup taxa was enough to reverse the initially erroneous placements of the Cretaceous fossils. They then instead ended up on rather basal branches of crown group ichneumonids, in accordance with one of the hypotheses suggested at the time of their original description (Kopylov 2009, 2010b).
Simulations confirm the bare-branch attraction pheno- menon.—
Our simulations have confirmed bare-branch attraction as a general phenomenon, which could be reproduced with simulated molecular and morphological data. The distance in branch lengths between a fossil and its closest extant relative emerged as the strongest factor determining the strength of the artifact, ahead of fossil age and number of morphological characters coded. This makes intuitive sense: fossils that are close to an extant taxon will on average have high information content in their morphological data, even if it is incomplete, while fossils that are distant to any extant taxon are notoriously difficult to place and might thus be more prone to falling victim to the bare-branch attraction.
More extensive simulations are needed to further assess the types of biases arising from the bare-branch attraction. In a recent simulation study concerning the FBD tree prior, Luo et al. (2020) reported difficulties in recovering correct fossil placement when rate variation across branches was high and when morphological data were not included. This might indicate the bare-branch attraction and it remains to be shown how detrimental this artifact is in the contexts of FBD prior. In any case, this phenomenon has the potential to mislead TED analyses and should be accounted for in future studies. Fortunately, the remedy is very simple: morphological data sets of extant taxa should be as complete as possible.
The age of Pimpliformes and the biological context of their diversification.—
Our most credible analysis estimated the median age of the family Ichneumonidae at 181 Ma (95% CI 154.0–204.7 Ma) and of Pimpliformes to 168 Ma (95% CI 144.0–193.4 Ma), with 95% credibility intervals in both cases spanning most of the Jurassic. The estimated age for Ichneumonidae is thus 60–70 Ma older than the oldest certain ichneumonid fossils, which implies a rather long ghost lineage. However, a similar gap exists between the oldest and the second oldest ichneumonid fossils, which is between 26 and 53 Ma, depending on the age of the geological formations in question (Kopylov and Rasnitsyn 2017; PaleoBioDB 2020). This suggests that the ghost range implied by our analysis is not that long after all. Such a gap is even more acceptable if we consider the paucity of Jurassic Hymenoptera fossils in general (Rasnitsyn and Quicke 2002). Furthermore, the last few years have seen unexpected discoveries of fossils that have closed large gaps between much older (molecular) age estimates and the previously known fossil record, for instance in Lepidoptera (Eldijk et al. 2018).
Further insights into the age of ichneumonids come from age estimates for their hosts (nearly exclusively holometabolous insects), which had to originate before their parasitoids could radiate. Initially, the radiations of the most species-rich orders of holometabolous insects, Hymenoptera, Coleoptera, Diptera, and Lepidoptera, were believed to have been associated with the radiation of flowering plants and were dated to the Early Cretaceous (Grimaldi 1999; Misof et al. 2014). However, these estimates were later deemed too young, and a Late Permian origin was suggested based on a reanalysis of a large phylogenomic data set (Misof et al. 2014) with more appropriate calibration points (Tong et al. 2015). This later study implies that the major host groups for ichneumonids were already present during the Jurassic, when both Ichneumonidae and Pimpliformes originated, suggesting that the radiation of these parasitoids might have happened only shortly after the radiation of their host groups.
Conclusions
We have demonstrated that poor outgroup sampling can negatively affect both accuracy and precision of age estimates in total-evidence dating analyses. Even though the exact mechanisms leading to this decrease in performance with the inclusion of increasingly more distant outgroups remain somewhat unclear, the introduction of large among-lineage rate variation and tree imbalance probably play major roles. Thus, to achieve more reliable age estimates, one should consider excluding outgroups altogether from dating analyses, unless they can be sampled adequately. Correct positioning of the root then can be through topology constraints instead, that are based on prior knowledge, or on a nonclock analysis of the data. In case an outgroup needs to be included in the analysis, for whatever reasons, one should sufficiently sample not only molecular but also morphological and fossil data.
We also illustrated the importance of careful consideration of fossil placement in total-evidence dating analyses in order to identify artifacts or biases. The bare-branch attraction artifact that we have discovered here is likely universally problematic for TED. Most concerning is that the artifact most severely affects fossils distant from extant taxa in a phylogeny, such as older and stem-lineage fossils, whose correct placement has proven crucial for reliable time-calibration of phylogenies. Thus, the bare-branch attraction artifact deserves further assessment in the future, through more detailed simulation studies. Nevertheless, it can easily be circumvented by a more complete sampling of morphological data for extant taxa.
Finally, we provided the first age estimate for the extremely diverse group of ichneumonid parasitoid wasps. The Jurassic origin for the family and for Pimpliformes agrees with the timing of the radiation of their major host groups. It remains to be seen how the age estimate for the family will change when more taxa and especially more fossils are included in a TED analysis. As new fossil ichneumonids are being described at a regular pace (Khalaim 2008; Kopylov 2009, 2010b; McKellar et al. 2013; Antropov et al. 2014; Kopylov et al. 2018; Spasojevic et al. 2018a,b), the coming years will certainly provide further evidence for an older age of many groups within this species-rich family. Our insights on taxon sampling will help provide guidance to design increasingly efficient dating studies, not only in Darwin wasps, but across the entire tree of life.
Acknowledgements
For access to fossil specimens, we are grateful to Marsh Finnegan and Alan M. Rulis (Smithsonian National Museum of Natural History, Washington, USA), Talia Karim and David Zelagin (Museum of Natural History, University of Colorado, Boulder, USA), and Ricardo Pérez-de la Fuente (Museum of Comparative Zoology, Harvard University, USA). We also thank Dmitry Kopylov (Borissiak Paleontological Institute, Russian Academy of Sciences, Moscow, Russia) and Sonja Wedmann (Forschungsstation Grube Messel, Senckenberg, Germany) for providing access to additional fossil specimens and for commenting on derived fossil ages. We are grateful to Eric Chapman and Mike Sharkey (Kentucky, USA), Rikio Matsumoto (Osaka, Japan), and the Swedish Malaise Trap project for extensive material for morphological and molecular studies. Remo Ryser (German Centre for Integrative Biodiversity Research, Leipzig, Germany) and Djordje Markovic (Institute of Zoology, University of Belgrade, Belgrade, Serbia) contributed to the lab work. Finally, we thank Simon Ho (editor), Alexander Pyron and two anonymous reviewers for critical and insightful comments which significantly improved the quality of the manuscript.
Funding
This work was supported by the Swiss National Science Foundation [SNSF, PZ00P3_154791 to Seraina Klopfstein]. During the submission and revision process, Tamara Spasojevic was supported by the SNSF [grant number P2BEP3_1882252] and Stanislav Korenko by the Ministry of Education Youth and Sports of Czech Republic [grant number LTAUSA 19084] (program Inter-excellence, Inter-action).
Supplementary Material
Data available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.m0cfxpnzm.
References
-
Antropov A.V., Belokobylskij S.A., Compton S.G., Dlussky G.M., Khalaim A.I., Kolyada V.A., Kozlov M.A., Perfilieva K.S., Rasnitsyn A.P..
2014.
The wasps, bees and ants (Insecta: Vespida
 Hymenoptera) from the Insect Limestone (Late Eocene) of the Isle of Wight, UK. Earth Environ. Sci. Trans. R. Soc. Edinb. 104:335–446. [Google Scholar]
Hymenoptera) from the Insect Limestone (Late Eocene) of the Isle of Wight, UK. Earth Environ. Sci. Trans. R. Soc. Edinb. 104:335–446. [Google Scholar] - Arcila D., Alexander Pyron R., Tyler J.C., Ortí G., Betancur R. R.. 2015. An evaluation of fossil tip-dating versus node-age calibrations in tetraodontiform fishes (Teleostei: Percomorphaceae). Mol. Phylogenet. Evol. 82:131–145. [DOI] [PubMed] [Google Scholar]
- Bagley J.C., Mayden R.L., Harris P.M.. 2018. Phylogeny and divergence times of suckers (Cypriniformes: Catostomidae) inferred from Bayesian total-evidence analyses of molecules, morphology, and fossils. PeerJ. 6:e5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannikov A.F., Tyler J.C., Arcila D., Carnevale G.. 2017. A new family of gymnodont fish (Tetraodontiformes) from the earliest Eocene of the Peri-Tethys (Kabardino-Balkaria, northern Caucasus, Russia). J. Syst. Palaeontol. 15:129–146. [Google Scholar]
- Barido-Sottani J., Aguirre-Fernández G., Hopkins M.J., Stadler T., Warnock R.. 2019. Ignoring stratigraphic age uncertainty leads to erroneous estimates of species divergence times under the fossilized birth–death process. Proc. R. Soc. B Biol. Sci. 286:20190685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J.M., O’Meara B.C., Crane P., Donoghue M.J.. 2015. Heterogeneous rates of molecular evolution and diversification could explain the triassic age estimate for angiosperms. Syst. Biol. 64:869–878. [DOI] [PubMed] [Google Scholar]
- Beck R.M.D., Lee M.S.Y.. 2014. Ancient dates or accelerated rates? Morphological clocks and the antiquity of placental mammals. Proc. R. Soc. B Biol. Sci. 281:20141278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec M.L., Rabeling C., Brady S.G., Fisher B.L., Schultz T.R., Ward P.S.. 2019. Compositional heterogeneity and outgroup choice influence the internal phylogeny of the ants. Mol. Phylogenet. Evol. 134:111–121. [DOI] [PubMed] [Google Scholar]
- Broad G.R., Shaw M.R., Fitton M.G.. 2018. Ichneumonid wasps (Hymenoptera: Ichneumonidae): their classification and biology. Field Studies Council and Royal Entomological Society. United Kingdom, Telford and St Albans. [Google Scholar]
- Close R.A., Johanson Z., Tyler J.C., Harrington R.C., Friedman M.. 2016. Mosaicism in a new Eocene pufferfish highlights rapid morphological innovation near the origin of crown tetraodontiforms. Palaeontology 59:499–514. [Google Scholar]
- Deans A.R., Basibuyuk H.H., Azar D., Nel A.. 2004. Descriptions of two new Early Cretaceous (Hauterivian) ensign wasp genera (Hymenoptera: Evaniidae) from Lebanese amber. Cretac. Res. 25:509–516. [Google Scholar]
- Delsuc F., Brinkmann H., Philippe H.. 2005. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 6:361–375. [DOI] [PubMed] [Google Scholar]
- Donoghue P.C.J., Benton M.J.. 2007. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends Ecol. Evol. 22:424–431. [DOI] [PubMed] [Google Scholar]
- Donoghue P.C.J., Yang Z.. 2016. The evolution of methods for establishing evolutionary timescales. Philos. Trans. R Soc. B. 371:20160020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornburg A., Friedman M., Near T.J.. 2015. Phylogenetic analysis of molecular and morphological data highlights uncertainty in the relationships of fossil and living species of Elopomorpha (Actinopterygii: Teleostei). Mol. Phylogenet. Evol. 89:205–218. [DOI] [PubMed] [Google Scholar]
- Dowton M., Austin A.D.. 1994. Molecular phylogeny of the insect order Hymenoptera: apocritan relationships. Proc. Natl. Acad. Sci. USA 91:9911–9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne D., Duchêne S., Ho S.Y.W.. 2015. Tree imbalance causes a bias in phylogenetic estimation of evolutionary timescales using heterochronous sequences. Mol. Ecol. Resour. 15:785–794. [DOI] [PubMed] [Google Scholar]
- Duchêne S., Lanfear R., Ho S.Y.W.. 2014. The impact of calibration and clock-model choice on molecular estimates of divergence times. Mol. Phylogenet. Evol. 78:277–289. [DOI] [PubMed] [Google Scholar]
- Dunn C.W., Hejnol A., Matus D.Q., Pang K., Browne W.E., Smith S.A., Seaver E., Rouse G.W., Obst M., Edgecombe G.D., Sørensen M.V., Haddock S.H.D., Schmidt-Rhaesa A., Okusu A., Kristensen R.M., Wheeler W.C., Martindale M.Q., Giribet G.. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749. [DOI] [PubMed] [Google Scholar]
- van Eldijk T.J.B., Wappler T., Strother P.K., van der Weijst C.M.H., Rajaei H., Visscher H., van de Schootbrugge B.. 2018. A Triassic-Jurassic window into the evolution of Lepidoptera. Sci. Adv. 4:e1701568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M., Bruschi M.. 1987. The evolution of prokaryotic ferredoxins—with a general method correcting for unobserved substitutions in less branched lineages. Mol. Biol. Evol. 4:381–394. [DOI] [PubMed] [Google Scholar]
- Gauld I.D., Mound L.A.. 1982. Homoplasy and the delineation of holophyletic genera in some insect groups. Syst. Entomol. 7:73–86. [Google Scholar]
- Gauld I.D., Wahl D.B., Broad G.R.. 2002. The suprageneric groups of the Pimplinae (Hymenoptera: Ichneumonidae): a cladistic re-evaluation and evolutionary biological study. Zool. J. Linn. Soc. 136:421–485. [Google Scholar]
- Gavryushkina A., Welch D., Stadler T., Drummond A.J.. 2014. Bayesian inference of sampled ancestor trees for epidemiology and fossil calibration. PLoS Comput. Biol. 10:e1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribet G., Ribera C.. 1998. The position of arthropods in the animal kingdom: a search for a reliable outgroup for internal arthropod phylogeny. Mol. Phylogenet. Evol. 9:481–488. [DOI] [PubMed] [Google Scholar]
- Graham S.W., Olmstead R.G., Barrett S.C.H.. 2002. Rooting phylogenetic trees with distant outgroups: a case study from the commelinoid monocots. Mol. Biol. Evol. 19:1769–1781. [DOI] [PubMed] [Google Scholar]
- Graybeal A. 1998. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst. Biol. 47:9–17. [DOI] [PubMed] [Google Scholar]
- Grimaldi D. 1999. The co-radiations of pollinating insects and angiosperms in the cretaceous. Ann. Mo. Bot. Gard. 86:373–406. [Google Scholar]
- Grimm G.W., Kapli P., Bomfleur B., McLoughlin S., Renner S.S.. 2015. Using more than the oldest fossils: dating Osmundaceae with three Bayesian clock approaches. Syst. Biol. 64:396–405. [DOI] [PubMed] [Google Scholar]
- Guillerme T., Cooper N.. 2016. Effects of missing data on topological inference using a Total Evidence approach. Mol. Phylogenet. Evol. 94, Part A:146–158. [DOI] [PubMed] [Google Scholar]
- Harrington S.M., Reeder T.W.. 2017. Phylogenetic inference and divergence dating of snakes using molecules, morphology and fossils: new insights into convergent evolution of feeding morphology and limb reduction. Biol. J. Linn. Soc. 121:379–394. [Google Scholar]
- Hayes K.A., Cowie R.H., Thiengo S.C.. 2009. A global phylogeny of apple snails: Gondwanan origin, generic relationships, and the influence of outgroup choice (Caenogastropoda: Ampullariidae). Biol. J. Linn. Soc. 98:61–76. [Google Scholar]
- Heath T.A., Hedtke S.M., Hillis D.M.. 2008. Taxon sampling and the accuracy of phylogenetic analyses. J. Syst. Evol. 19:239–257. [Google Scholar]
- Heath T.A., Huelsenbeck J.P., Stadler T.. 2014. The fossilized birth–death process for coherent calibration of divergence-time estimates. Proc. Natl. Acad. Sci. USA 111:E2957–E2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera J.P., Davalos L.M.. 2016. Phylogeny and divergence times of lemurs inferred with recent and ancient fossils in the tree. Syst. Biol. 65:772–791. [DOI] [PubMed] [Google Scholar]
- Höhna S., Stadler T., Ronquist F., Britton T.. 2011. Inferring speciation and extinction rates under different sampling schemes. Mol. Biol. Evol. 28:2577–2589. [DOI] [PubMed] [Google Scholar]
- Holland B.R., Penny D., Hendy M.D., Sullivan J.. 2003. Outgroup misplacement and phylogenetic inaccuracy under a molecular clock—a simulation study. Syst. Biol. 52:229–238. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealy S., Beck R.. 2017. Total evidence phylogeny and evolutionary timescale for Australian faunivorous marsupials (Dasyuromorphia). BMC Evol. Biol. 17:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A.. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaim A.I. 2008. Fossil ichneumon wasps (Hymenoptera: Ichneumonidae) form Biamo (Russia), Oligocene. Alavesia 2:101–112. [Google Scholar]
- King B., Qiao T., Lee M.S.Y., Zhu M., Long J.A.. 2017a. Bayesian morphological clock methods resurrect placoderm monophyly and reveal rapid early evolution in jawed vertebrates. Syst. Biol. 66:499–516. [DOI] [PubMed] [Google Scholar]
- King B., Qiao T., Lee M.S.Y., Zhu M., Long J.A.. 2017b. Bayesian morphological clock methods resurrect placoderm monophyly and reveal rapid early evolution in jawed vertebrates. Syst. Biol. 66:499–516. [DOI] [PubMed] [Google Scholar]
- Kirchberger P.C., Sefc K.M., Sturmbauer C., Koblmüller S.. 2014. Outgroup effects on root position and tree topology in the AFLP phylogeny of a rapidly radiating lineage of cichlid fish. Mol. Phylogenet. Evol. 70:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel R.N., Austin A.D., Klopfstein S.. 2016. Molecular and morphological phylogenetics of chelonine parasitoid wasps (Hymenoptera: Braconidae), with a critical assessment of divergence time estimations. Mol. Phylogenet. Evol. 101:224–241. [DOI] [PubMed] [Google Scholar]
- Klopfstein S. 2020. The age of insects and the revival of minimum age trees. Austral: Entomol; 10.1111/aen.12478. [DOI] [Google Scholar]
- Klopfstein S., Langille B., Spasojevic T., Broad G.R., Cooper S.J.B., Austin A.D., Niehuis O.. 2019a. Hybrid capture data unravel a rapid radiation of pimpliform parasitoid wasps (Hymenoptera: Ichneumonidae: Pimpliformes). Syst. Entomol. 44:361–383. [Google Scholar]
- Klopfstein S., Massingham T., Goldman N.. 2017. More on the best evolutionary rate for phylogenetic analysis. Syst. Biol. 66:769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfstein S., Santos B.F., Shaw M.R., Alvarado M., Bennett A.M.R., Pos D.D., Giannotta M., Florez A.F.H., Karlsson D., Khalaim A.I., Lima A.R., Mikó I., Sääksjärvi I.E., Shimizu S., Spasojevic T., van Noort S., Vilhelmsen L., Broad G.R.. 2019b. Darwin wasps: a new name heralds renewed efforts to unravel the evolutionary history of Ichneumonidae. Entomol. Commun. 1:ec01006. [Google Scholar]
- Klopfstein S., Spasojevic T.. 2019. Illustrating phylogenetic placement of fossils using RoguePlots: an example from ichneumonid parasitoid wasps (Hymenoptera, Ichneumonidae) and an extensive morphological matrix. PLoS One 425090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfstein S., Vilhelmsen L., Ronquist F.. 2015. A nonstationary Markov model detects directional evolution in hymenopteran morphology. Syst. Biol. 64:1089–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylov D.S. 2009. A new subfamily of ichneumonids from the Lower Cretaceous of Transbaikalia and Mongolia (Insecta: Hymenoptera: Ichneumonidae). Paleontol. J. 43:83–93. [Google Scholar]
- Kopylov D.S. 2010a. Ichneumonids of the subfamily Tanychorinae (Insecta: Hymenoptera: Ichneumonidae) from the Lower Cretaceous of Transbaikalia and Mongolia. Paleontol. J. 44:180–187. [Google Scholar]
- Kopylov D.S. 2010b. A new subfamily of ichneumon wasps (Insecta: Hymenoptera: Ichneumonidae) from the Upper Cretaceous of the Russian Far East. Paleontol. J. 44:422–433. [Google Scholar]
- Kopylov D.S., Brothers D.J., Rasnitsyn A.P.. 2010. Two new labenopimpline ichneumonids (Hymenoptera: Ichneumonidae) from the upper cretaceous of Southern Africa. Afr. Invertebr. 51:423–430. [Google Scholar]
- Kopylov D.S., Rasnitsyn A.P.. 2017. New sepulcids (Hymenoptera: Sepulcidae) from the Lower Cretaceous of Asia: II. Ghilarellinae and Trematothoracinae. Paleontol. J. 51:291–303. [Google Scholar]
- Kopylov D.S., Spasojevic T., Klopfstein S.. 2018. New ichneumonids (Hymenoptera, Ichneumonidae) from the Eocene Tadushi Formation, Russian Far East. Zootaxa 4442:319–330. [DOI] [PubMed] [Google Scholar]
- Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34:772–773. [DOI] [PubMed] [Google Scholar]
- Lee M.S.Y. 2016. Multiple morphological clocks and total-evidence tip-dating in mammals. Biol. Lett. 12:20160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.O. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50:913–925. [DOI] [PubMed] [Google Scholar]
- Li L., Kopylov D.S., Shih C., Ren D.. 2017. The first record of Ichneumonidae (Insecta: Hymenoptera) from the Upper Cretaceous of Myanmar. Cretac. Res. 70:152–162. [Google Scholar]
- Linder H.P., Hardy C.R., Rutschmann F.. 2005. Taxon sampling effects in molecular clock dating: an example from the African Restionaceae. Mol. Phylogenet. Evol. 35:569–582. [DOI] [PubMed] [Google Scholar]
- Luo A., Duchêne D.A., Zhang C., Zhu C.-D., Ho S.Y.W.. 2020. A simulation-based evaluation of tip-dating under the fossilized birth–death process. Syst. Biol. 69:325–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S. 2014. A review of the effect of relaxed clock method, long branches, genes, and calibrations in the estimation of angiosperm age. Bot. Sci. 92:1–22. [Google Scholar]
- Magallón S., Hilu K.W., Quandt D.. 2013. Land plant evolutionary timeline: Gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. Am. J. Bot. 100:556–573. [DOI] [PubMed] [Google Scholar]
- Matschiner M. 2019. Selective sampling of species and fossils influences age estimates under the fossilized birth–death model. Front. Genet. 10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack J.E., Hird S.M., Zellmer A.J., Carstens B.C., Brumfield R.T.. 2013. Applications of next-generation sequencing to phylogeography and phylogenetics. Mol. Phylogenet. Evol. 66:526–538. [DOI] [PubMed] [Google Scholar]
- McKellar R.C., Kopylov D.S., Engel M.S.. 2013. Ichneumonidae (Insecta: Hymenoptera) in Canadian Late Cretaceous amber. Foss. Rec. 16:217–227. [Google Scholar]
- Menier J.J., Nel A., Waller A., Ploëg G.D.. 2004.. A new fossil ichneumon wasp from the Lowermost Eocene amber of Paris Basin (France), with a checklist of fossil Ichneumonoidea s.l. (Insecta: Hymenoptera: Ichneumonidae: Metopiinae). Geol. Acta Int. Earth Sci. J. 2:83–94. [Google Scholar]
- Milne R.I. 2009. Effects of taxon sampling on molecular dating for within-genus divergence events, when deep fossils are used for calibration. J. Syst. Evol. 47:383–401. [Google Scholar]
- Misof B., Liu S., Meusemann K., Peters R.S., Donath A., Mayer C., Frandsen P.B., Ware J., Flouri T., Beutel R.G., Niehuis O., Petersen M., Izquierdo-Carrasco F., Wappler T., Rust J., Aberer A.J., Aspöck U., Aspöck H., Bartel D., Blanke A., Berger S., Böhm A., Buckley T.R., Calcott B., Chen J., Friedrich F., Fukui M., Fujita M., Greve C., Grobe P., Gu S., Huang Y., Jermiin L.S., Kawahara A.Y., Krogmann L., Kubiak M., Lanfear R., Letsch H., Li Y., Li Z., Li J., Lu H., Machida R., Mashimo Y., Kapli P., McKenna D.D., Meng G., Nakagaki Y., Navarrete-Heredia J.L., Ott M., Ou Y., Pass G., Podsiadlowski L., Pohl H., Reumont B.M. von, Schütte K., Sekiya K., Shimizu S., Slipinski A., Stamatakis A., Song W., Su X., Szucsich N.U., Tan M., Tan X., Tang M., Tang J., Timelthaler G., Tomizuka S., Trautwein M., Tong X., Uchifune T., Walzl M.G., Wiegmann B.M., Wilbrandt J., Wipfler B., Wong T.K.F., Wu Q., Wu G., Xie Y., Yang S., Yang Q., Yeates D.K., Yoshizawa K., Zhang Q., Zhang R., Zhang W., Zhang Y., Zhao J., Zhou C., Zhou L., Ziesmann T., Zou S., Li Y., Xu X., Zhang Y., Yang H., Wang J., Wang J., Kjer K.M., Zhou X.. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Nixon K.C., Carpenter J.M.. 1993. On outgroups. Cladistics 9:413–426. [DOI] [PubMed] [Google Scholar]
- O’Hanlon S.J., Rieux A., Farrer R.A., Rosa G.M., Waldman B., Bataille A., Kosch T.A., Murray K.A., Brankovics B., Fumagalli M., Martin M.D., Wales N., Alvarado-Rybak M., Bates K.A., Berger L., Böll S., Brookes L., Clare F., Courtois E.A., Cunningham A.A., Doherty-Bone T.M., Ghosh P., Gower D.J., Hintz W.E., Höglund J., Jenkinson T.S., Lin C.-F., Laurila A., Loyau A., Martel A., Meurling S., Miaud C., Minting P., Pasmans F., Schmeller D.S., Schmidt B.R., Shelton J.M.G., Skerratt L.F., Smith F., Soto-Azat C., Spagnoletti M., Tessa G., Toledo L.F., Valenzuela-Sánchez A., Verster R., Vörös J., Webb R.J., Wierzbicki C., Wombwell E., Zamudio K.R., Aanensen D.M., James T.Y., Gilbert M.T.P., Weldon C., Bosch J., Balloux F., Garner T.W.J., Fisher M.C.. 2018. Recent Asian origin of chytrid fungi causing global amphibian declines. Science 360:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly J.E., Donoghue P.C.J.. 2016. Tips and nodes are complementary not competing approaches to the calibration of molecular clocks. Biol. Lett. 12:20150975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly J.E., dos Reis M., Donoghue P.C.J.. 2015. Dating tips for divergence-time estimation. Trends Genet. 31:637–650. [DOI] [PubMed] [Google Scholar]
- PaleoBioDB. 2020. The paleobiology database. Available from: https://paleobiodb.org. [Google Scholar]
- Parins-Fukuchi C., Brown J.W.. 2017. What drives results in Bayesian morphological clock analyses? bioRxiv: 219048. [Google Scholar]
- Paterson R.S., Rybczynski N., Kohno N., Maddin H.C.. 2020. A total evidence phylogenetic analysis of pinniped phylogeny and the possibility of parallel evolution within a monophyletic framework. Front. Ecol. Evol. 7:1–16. [Google Scholar]
- Peters R.S., Krogmann L., Mayer C., Donath A., Gunkel S., Meusemann K., Kozlov A., Podsiadlowski L., Petersen M., Lanfear R., Diez P.A., Heraty J., Kjer K.M., Klopfstein S., Meier R., Polidori C., Schmitt T., Liu S., Zhou X., Wappler T., Rust J., Misof B., Niehuis O.. 2017. Evolutionary history of the hymenoptera. Curr. Biol. 27:1013–1018. [DOI] [PubMed] [Google Scholar]
- Philippe H., Brinkmann H., Lavrov D.V., Littlewood D.T.J., Manuel M., Wörheide G., Baurain D.. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9:e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick K.S., Philippe H., Schreiber F., Erpenbeck D., Jackson D.J., Wrede P., Wiens M., Alié A., Morgenstern B., Manuel M., Wörheide G.. 2010. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol. Biol. Evol. 27:1983–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Püschel H.P., O’Reilly J.E., Pisani D., Donoghue P.C.J.. 2020. The impact of fossil stratigraphic ranges on tip-calibration, and the accuracy and precision of divergence time estimates. Palaeontology 63: 67–83. [Google Scholar]
- Puslednik L., Serb J.M.. 2008. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol. Phylogenet. Evol. 48:1178–1188. [DOI] [PubMed] [Google Scholar]
- Pyron R.A. 2011. Divergence time estimation using fossils as terminal taxa and the origins of lissamphibia. Syst. Biol. 60:466–481. [DOI] [PubMed] [Google Scholar]
- Pyron R.A. 2017. Novel approaches for phylogenetic inference from morphological data and total-evidence dating in squamate reptiles (lizards, snakes, and amphisbaenians). Syst. Biol. 66:38–56. [DOI] [PubMed] [Google Scholar]
- Quicke D.L.J. 2014. The braconid and ichneumonid parasitoid wasps: biology, systematics, evolution and ecology. Chichester, United Kingdom, John Wiley & Sons, Ltd. [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A., Susko E.. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67:901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnitsyn A.P., Bashkuev A.S., Kopylov D.S., Lukashevich E.D., Ponomarenko A.G., Popov Yu.A., Rasnitsyn D.A., Ryzhkova O.V., Sidorchuk E.A., Sukatsheva I.D., Vorontsov D.D.. 2016. Sequence and scale of changes in the terrestrial biota during the Cretaceous (based on materials from fossil resins). Cretac. Res. 61:234–255. [Google Scholar]
- Rasnitsyn A.P., Quicke D.L.. 2002. History of insects. Springer Science & Business Media; Dordrecht, Netherlands, Springer. [Google Scholar]
- Ronquist F., Klopfstein S., Vilhelmsen L., Schulmeister S., Murray D.L., Rasnitsyn A.P.. 2012a. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61:973–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Lartillot N., Phillips M.J.. 2016. Closing the gap between rocks and clocks using total-evidence dating. Philos. Trans. R. Soc. B 371:20150136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P.. 2012b. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O., Telford M.J.. 2008. A multi criterion approach for the selection of optimal outgroups in phylogeny: recovering some support for Mandibulata over Myriochelata using mitogenomics. Mol. Phylogenet. Evol. 48:103–111. [DOI] [PubMed] [Google Scholar]
- Saladin B., Leslie A.B., Wueest R.O., Litsios G., Conti E., Salamin N., Zimmermann N.E.. 2017. Fossils matter: improved estimates of divergence times in Pinus reveal older diversification. BMC Evol. Biol. 17:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M.J., Wahl D.B.. 1992. Cladistics of the Ichneumonoidea (Hymenoptera). J. Hymenopt. Res. 1:15–24. [Google Scholar]
- Soares A.E.R., Schrago C.G.. 2012. The influence of taxon sampling and tree shape on molecular dating: an empirical example from mammalian mitochondrial genomes. Bioinforma. Biol. Insights 6:BBI.S9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares A.E.R., Schrago C.G.. 2015. The influence of taxon sampling on Bayesian divergence time inference under scenarios of rate heterogeneity among lineages. J. Theor. Biol. 364:31–39. [DOI] [PubMed] [Google Scholar]
- Spasojevic T., Broad G.R., Bennett A.M.R., Klopfstein S.. 2018a. Ichneumonid parasitoid wasps from the Early Eocene Green River Formation: five new species and a revision of the known fauna (Hymenoptera, Ichneumonidae). PalZ 92:35–63. [Google Scholar]
- Spasojevic T., Wedmann S., Klopfstein S.. 2018b. Seven remarkable new fossil species of parasitoid wasps (Hymenoptera, Ichneumonidae) from the Eocene Messel Pit. PLoS One 13:e0197477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrío R., Rodríguez-Trelles F., Ayala F.J.. 2000. Tree rooting with outgroups when they differ in their nucleotide composition from the ingroup: the Drosophila saltans and Willistoni groups, a case study. Mol. Phylogenet. Evol. 16:344–349. [DOI] [PubMed] [Google Scholar]
- Thomas J.A., Trueman J.W.H., Rambaut A., Welch J.J.. 2013. Relaxed phylogenetics and the palaeoptera problem: resolving deep ancestral splits in the insect phylogeny. Syst. Biol. 62:285–297. [DOI] [PubMed] [Google Scholar]
- Tong K.J., Duchêne S., Ho S.Y.W., Lo N.. 2015. Comment on “Phylogenomics resolves the timing and pattern of insect evolution.” Science 349:487–487. [DOI] [PubMed] [Google Scholar]
- Travouillon K.J., Phillips M.J.. 2018. Total evidence analysis of the phylogenetic relationships of bandicoots and bilbies (Marsupialia: Peramelemorphia): reassessment of two species and description of a new species. Zootaxa 4378:224. [DOI] [PubMed] [Google Scholar]
- Wahl D., Gauld I.. 1998. The cladistics and higher classification of the Pimpliformes (Hymenoptera: Ichneumonidae). Syst. Entomol. 23:265–298. [Google Scholar]
- Wang X., Grohé C., Su D.F., White S.C., Ji X., Kelley J., Jablonski N.G., Deng T., You Y., Yang X.. 2018. A new otter of giant size, Siamogale melilutra sp. nov. (Lutrinae: Mustelidae: Carnivora), from the latest Miocene Shuitangba site in north-eastern Yunnan, south-western China, and a total-evidence phylogeny of lutrines. J. Syst. Palaeontol. 16:39–65. [Google Scholar]
- Ware J.L., Litman J., Klass K.-D., Spearman L.A.. 2008. Relationships among the major lineages of Dictyoptera: the effect of outgroup selection on dictyopteran tree topology. Syst. Entomol. 33:429–450. [Google Scholar]
- Warnock R.C.M., Yang Z., Donoghue P.C.J.. 2011. Exploring uncertainty in the calibration of the molecular clock. Biol. Lett. 8:156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.J., Bromham L.. 2005. Molecular dating when rates vary. Trends Ecol. Evol. 20:320–327. [DOI] [PubMed] [Google Scholar]
- Wertheim J.O., Fourment M., Pond K., L S.. 2012. Inconsistencies in estimating the age of HIV-1 subtypes due to heterotachy. Mol. Biol. Evol. 29:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler W.C. 1990. Nucleic acid sequence phylogeny and random outgroups. Cladistics 6:363–367. [DOI] [PubMed] [Google Scholar]
- Whitfield J.B. 2002. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc. Natl. Acad. Sci. USA 99:7508–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilberg E.W. 2015. What’s in an outgroup? The impact of outgroup choice on the phylogenetic position of thalattosuchia (Crocodylomorpha) and the origin of Crocodyliformes. Syst. Biol. 64:621–637. [DOI] [PubMed] [Google Scholar]
- Wright A.M., Lloyd G.T., Hillis D.M.. 2016. Modeling character change heterogeneity in phylogenetic analyses of morphology through the use of priors. Syst. Biol. 65:602–611. [DOI] [PubMed] [Google Scholar]
- Yu D.S.K., van Achterberg C., Horstmann K.. 2016. Taxapad 2016, Ichneumonoidea 2015. Database on flash-drive. Nepean, Ontario, Canada. [Google Scholar]
- Zhang C., Stadler T., Klopfstein S., Heath T.A., Ronquist F.. 2016. Total-evidence dating under the fossilized birth–death process. Syst. Biol. 65:228–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D.J., Hillis D.M.. 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst. Biol. 51:588–598. [DOI] [PubMed] [Google Scholar]