ABSTRACT
The notochord is a defining feature of the chordates. The transcription factor Brachyury (Bra) is a key regulator of notochord fate but here we show that it is not a unitary master regulator in the model chordate Ciona. Ectopic Bra expression only partially reprograms other cell types to a notochord-like transcriptional profile and a subset of notochord-enriched genes is unaffected by CRISPR Bra disruption. We identify Foxa.a and Mnx as potential co-regulators, and find that combinatorial cocktails are more effective at reprogramming other cell types than Bra alone. We reassess the network relationships between Bra, Foxa.a and other components of the notochord gene regulatory network, and find that Foxa.a expression in the notochord is regulated by vegetal FGF signaling. It is a direct activator of Bra expression and has a binding motif that is significantly enriched in the regulatory regions of notochord-enriched genes. These and other results indicate that Bra and Foxa.a act together in a regulatory network dominated by positive feed-forward interactions, with neither being a classically defined master regulator.
KEY WORDS: Brachyury, Ciona, FoxA, GRN, Feedforward, Notochord
Summary: In the simple chordate Ciona, the notochord-specific transcription factor Brachyury acts together with Foxa.a as part of a densely interlocked FGF-dependent feed-forward network.
INTRODUCTION
The notochord is the earliest chordate organ to form and plays essential structural and signaling roles in establishing the chordate body plan. The ascidian chordate Ciona has a particularly small and simple embryo in which invariant cell lineages give rise to a notochord of only 40 cells under the control of a compact ∼130 Mb genome (Passamaneck and Di Gregorio, 2005; Satoh et al., 2003). As in all chordates, the Ciona notochord expresses the T-box transcription factor Bra (Corbo et al., 1997a) and loss of Bra function gives rise to major defects in notochord morphogenesis (Chiba et al., 2009; Satou et al., 2001; Yamada et al., 2003). Unlike in vertebrates, ascidian Bra is expressed only in the notochord and does not have broader roles in mesoderm induction (Chiba et al., 2009; Corbo et al., 1997a; Yasuo and Satoh, 1993, 1998). In one of the earliest efforts to systematically define a tissue-specific transcriptional profile, Bra misexpression outside the notochord was used together with subtractive hybridization to identify 40 notochord-enriched genes induced directly or indirectly downstream of Bra (Hotta et al., 1999; Takahashi et al., 1999). As Bra misexpression leads to the ectopic expression of multiple notochord-specific markers, it has been proposed that Bra expression is sufficient to transform other cell types to notochord (Hotta et al., 1999; Takahashi et al., 1999; Yasuo and Satoh, 1998). Given that Ciona Bra is specifically expressed in the developing notochord, that loss of function leads to notochord defects and that misexpression leads to the ectopic expression of a broad range of notochord target genes, it is potentially a canonical master regulator gene that is not only necessary but also sufficient for notochord fate. Bra has also been proposed to act as a master regulator in human chordoma: a rare but serious notochord-derived tumor (Nelson et al., 2012).
There is strong evidence that Brachyury is a key regulator of Ciona notochord fate, but the evidence that it can fully transform other cell types to notochord is weaker. Bra misexpression in the endodermal, A-line neural and notochord lineages under the control of the Foxa.a enhancer does not typically give rise to a larger notochord with additional intercalated cells, but instead gives rise to a mass of cells with little morphological differentiation (Takahashi et al., 1999). Yasuo and Satoh (1998) proposed that ectopic Bra expression in Halocynthia gives rise to extra notochord cells, but noted uncertainty as to whether the transformation to notochord was complete. Several notochord-enriched genes have been identified that are not induced by ectopic Bra expression (José-Edwards et al., 2011; Kugler et al., 2008), and several notochord enhancers have been identified that drive early notochord expression via essential Fox-binding sites but with no recognizable Bra sites (José-Edwards et al., 2015).
We have previously used RNAseq on flow-sorted Ciona notochord cells to identify a larger and more comprehensive set of ∼1300 transcripts enriched in the notochord during key stages of intercalation and elongation (Reeves et al., 2017). We also misexpressed Bra under the control of the Foxa.a enhancer similar to Takahashi et al. (1999), but using RNAseq as a more sensitive and quantitative readout. We identified over 900 upregulated genes, but found there was only modest overlap between the genes upregulated by ectopic Bra and the genes enriched in the notochord at comparable stages. This prompted the experiments described here, in which we use both gain-of-function and loss-of-function strategies coupled with modern transcriptional profiling to revise the gene regulatory network models for Ciona notochord fate specification. We find that Bra acts together with Foxa.a as part of a densely interlocked positive feedforward network.
RESULTS
Bra misexpression only induces a subset of notochord-enriched genes
Different tissues vary in their transcriptional and epigenetic states, so we misexpressed Bra in a broader range of cell types to determine whether Bra could be more effective at inducing a notochord-like transcriptional profile in other contexts. In addition to the previously used Foxa.a enhancer, which drives expression in endoderm, A-line neural, notochord and mesenchyme cells (Di Gregorio et al., 2001; Reeves et al., 2017; Takahashi et al., 1999), we also used a pan-neural Etr1 (Celf3.a) enhancer (Sierro et al., 2006) and a muscle- specific Tbx6-r.b enhancer (Kugler et al., 2010) (Fig. 1A). Embryos were harvested for whole-embryo RNAseq during notochord intercalation at early tailbud stage (Hotta stage 19) in triple biological replicates, and gene expression was compared with control embryos electroporated with Bra>GFP. Reads were aligned using TopHat (Trapnell et al., 2012) and differential expression between conditions was tested by DESeq2 (Love et al., 2014) using a P-value threshold adjusted for multiple comparisons of 0.05. We quantified transgene expression using the Venus tag on each Bra misexpression construct and found that all three were expressed within a twofold range (Foxa.a, 136 FPKM; Etr1, 77 FPKM; Tbx6-r.b, 83 FPKM). Although we did not precisely define the earliest stage at which transgene expression could first be detected, endogenous Foxa.a and Tbx6-r.b are both strongly expressed by the 16-cell stage and Etr1 by the 110-cell stage, well before our early tailbud stage endpoint.
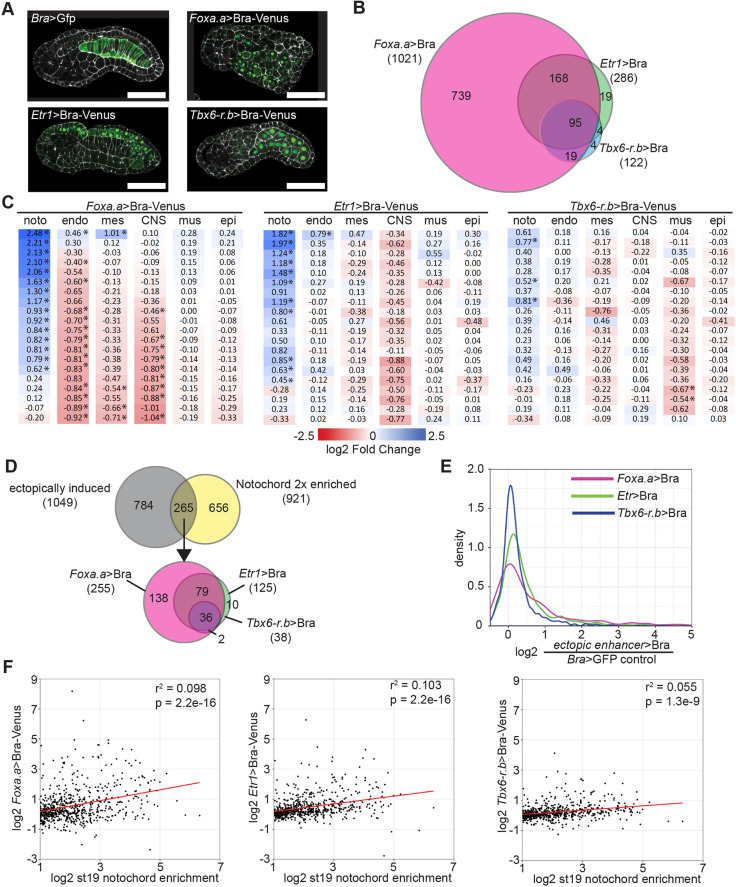
Fig. 1.
Ectopic expression of Bra upregulates a subset of notochord-enriched genes. (A) Venus-tagged Bra was misexpressed in Ciona embryos using the Foxa.a, Etr1 or Tbx6-r.b enhancers. Bra>GFP plasmid was used for control embryos. Phalloidin is in white; Venus is in green. Scale bars: 50 μm. (B) Differential expression was calculated using DESeq2. The overlap of all genes with significantly increased expression is shown. (C) Effects of Bra misexpression on the expression of tissue-specific markers. Marker genes highly enriched in notochord (noto), endoderm (endo), mesenchyme (mes), central nervous system (CNS), muscle (mus) and epidermis (epi) were identified using the scRNAseq data of Cao et al. (2019). Log fold changes of Bra misexpression compared with control are shown. *Padj≤0.05. (D) Overlap of genes with at least twofold enriched notochord expression (Reeves et al., 2017) and genes upregulated by Bra misexpression. (E) Kernel density plot of the increase in expression by ectopic Bra for twofold notochord-enriched genes. (F) Scatter plots of notochord enrichment at stage 19 in wild-type embryos (Reeves et al., 2017) versus the fold change in response to Bra misexpression driven by the different constructs. Only genes with a statistically significant stage 19 notochord enrichment of at least twofold are shown. The red lines show the best fit linear regressions.
We found that Bra misexpression using the neural and muscle drivers led to considerably fewer genes being identified as upregulated (Foxa.a>Bra, 1021; Etr1>Bra, 286; Tbx6-r.b>Bra, 122). These ectopic expression constructs induced largely overlapping sets of genes (Fig. 1B, Table S1), with the genes upregulated by the muscle and neural drivers representing a small subset of the genes induced by Foxa.a>Bra.
If Bra is a master regulator of notochord fate, then misexpression outside the notochord should lead to a downregulation of tissue-specific markers in the ectopically expressing tissues and an upregulation of notochord-enriched genes. To test this, we used recent Ciona single cell RNAseq data (Cao et al., 2019) to identify the 20 genes that are most differentially expressed in each target tissue at stage 19 (Table S2). Although many target tissue markers were downregulated to some extent, the fold changes were very variable. Many showed minimal change, and of those that were downregulated many were not statistically significant. A few were paradoxically upregulated (Fig. 1C). Notochord marker genes tended to be upregulated but, again, many were not strongly affected. This suggests that, unlike with a classically defined master regulator gene, ectopic misexpression of Bra can induce only a partial reprogramming of other tissues.
When we looked at the full set of notochord-enriched genes previously identified by FACS-seq (Reeves et al., 2017), relatively few were upregulated by ectopic Bra expression. Only 293 of the 1372 genes (21%) that are significantly notochord enriched at either stage 16 and/or stage 19 were statistically significantly upregulated by any of the ectopic Bra misexpression constructs (Table S3). The 1372 notochord-enriched genes span a broad range of enrichment levels, including many with expression that is only slightly higher in the notochord compared with other cell types. If we restrict this comparison to genes that are at least twofold enriched in notochord, then 265 of the 921 two fold-enriched genes (29%) were ectopically induced (Fig. 1D,E), but the majority showed no statistically significant change. There is a relationship between strength of notochord enrichment and ectopic upregulation by Bra, but there were still many highly enriched notochord genes that were not upregulated by ectopic Bra expression (Fig. 1F and Table S3). This correlation between notochord enrichment and the degree of ectopic upregulation by Bra was statistically significant for all three constructs, but the correlation coefficients were very low (0.06-0.1).
Bra expression outside the notochord induces many genes that are not notochord enriched by FACS-seq. We previously tested a sample of these from our earlier Foxa.a>Bra dataset by in situ hybridization and confirmed that these genes do not have obvious notochord expression that was undetected by FACS-seq, and instead tend to be either diffusely or undetectably expressed (Reeves et al., 2017). Bra misexpression in diverse tissues can upregulate many notochord genes, but it is not sufficient to upregulate expression of all notochord-enriched genes, including some of the most highly enriched. Given that expression levels of the three transgenes are similar, it is likely that the differences in reprogramming ability observed reflect transcriptional or epigenetic differences between the different cell types in which Bra was misexpressed. We cannot exclude the possibility that there is some cell type or temporal context in which Bra misexpression would lead to a more extensive transformation towards a notochord-like transcriptional profile, but Etr1>Bra and Tbx6.r.b>Bra show less reprogramming than Foxa.a>Bra, not more.
Some notochord-enriched genes are independent of Brachyury
Even if Bra expression is not sufficient to completely transform other cell types to notochord, it could still be a unitary regulator alone at the top of the transcriptional cascade for notochord differentiation. To address the necessity of Bra function in the expression of notochord-enriched genes, we used CRISPR-Bra RNP egg injections to disrupt Bra and examined the effect on gene expression at early neurula stage (Hotta stage 14). A germline Bra mutant line exists, but the CRISPR method allowed us to quantify early changes in gene expression before homozygous mutant embryos can reliably be identified. We used a targeting RNA specific to the third exon of Bra that is capable of replicating the tail elongation defect of homozygous mutants (Chiba et al., 2009) (Fig. 2A). TIDE analysis (Brinkman et al., 2014) confirmed a somatic mutation efficiency ranging from 67% to 77% in the three biological replicates harvested for RNAseq (Fig. 2B). It was impractical to flow-sort notochord cells from injected embryos, so we used RNA from whole embryos. Assuming that Bra inhibition will only affect notochord expression and not expression elsewhere in the embryo, the maximum possible fold decrease for a given gene is a function of its notochord enrichment levels, which we have previously estimated by FACS-seq (Reeves et al., 2017). The red line in Fig. 2C indicates the theoretical maximum decrease across the range of stage 16 notochord enrichment levels, as described in the Materials and Methods.
Fig. 2.
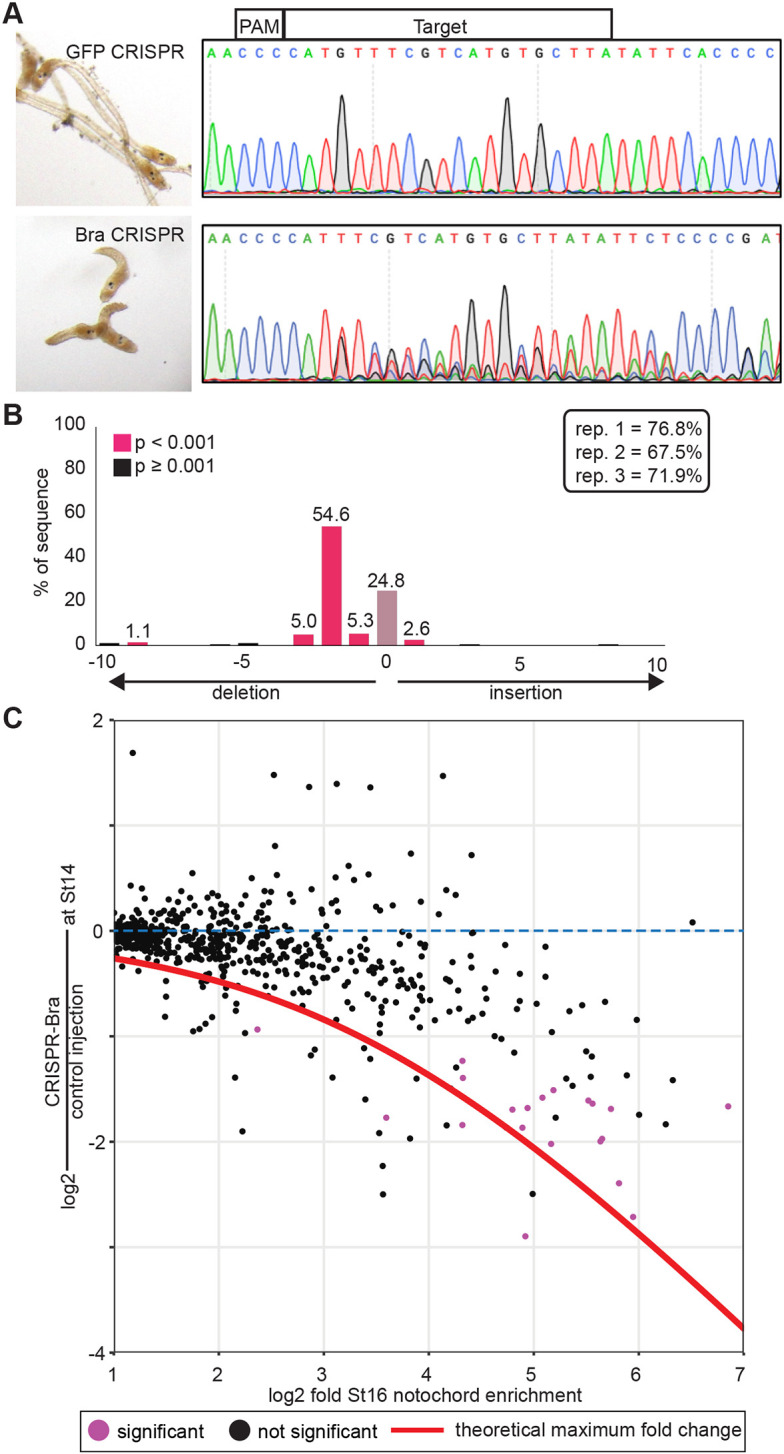
Somatic CRISPR disruption of Bra. (A) Eggs were injected with RNP-guide RNA complexes targeting either GFP (control, top) or Bra (bottom). Representative embryos at late tailbud are shown on the left with sequencing results used for TIDE analysis of the Bra locus on the right. (B) TIDE results from one replicate, showing efficient mutagenesis giving rise largely to a 2 bp deletion in injected embryos. TIDE-estimated indel rates for the three RNAseq replicates are shown in the inset. (C) Notochord enrichment at stage 16 (Reeves et al., 2017) compared with change in expression at stage 14 in CRISPR-Bra-injected embryos versus controls. Only genes with a statistically significant stage 16 notochord enrichment of at least twofold are shown. Red line indicates the theoretical maximum decrease observable in whole-embryo RNAseq for a given notochord enrichment value. Genes with a statistically significant decrease in expression in CRISPR-Bra embryos (Padj≤0.05) are purple.
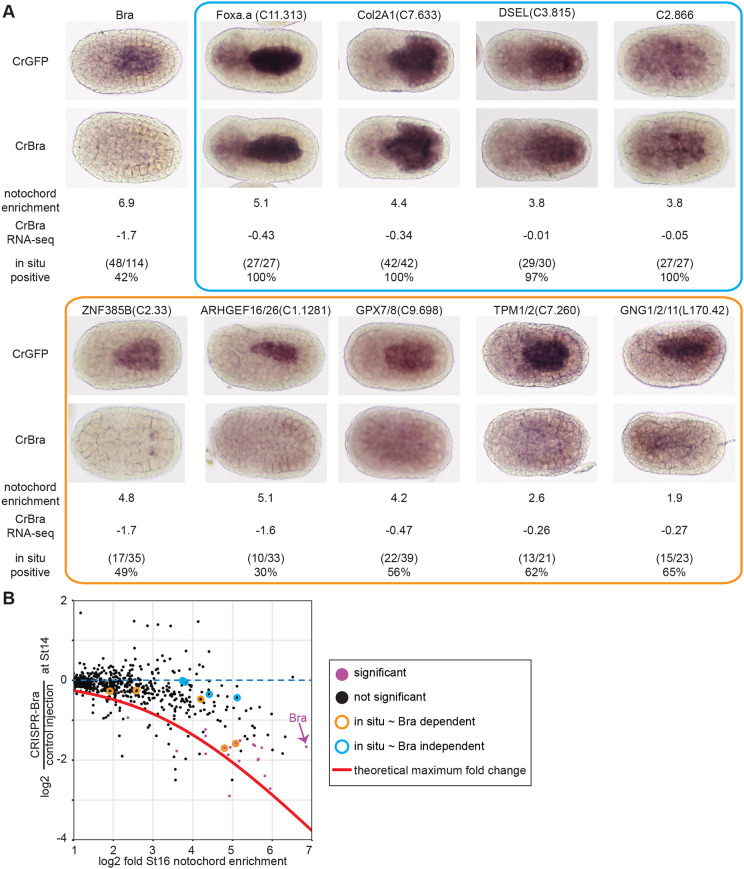
As expected, expression of notochord-enriched genes tended to be decreased, with genes that are more notochord enriched showing larger responses (Fig. 2C). There was, however, a broad range of fold changes, with some highly notochord-enriched genes showing little or no response to Bra disruption. To validate the RNAseq results, we selected several notochord genes spanning a range of notochord enrichment values and fold changes in response to Bra CRISPR and examined their expression using in situ hybridization in Bra crispant and control embryos (Fig. 3). As expected, notochord expression was lost for strongly notochord-enriched genes showing large decreases in the Bra CRISPR RNAseq (Fig. 3, orange box or circles). We included a Bra probe as a control in these experiments and found that the loss of Bra expression was similar in penetrance to the loss of these highly Bra-dependent transcripts. A similar loss of notochord expression was also seen for several moderately notochord-enriched genes with smaller responses to Bra CRISPR (Fig. 3, orange box or circles). Normal notochord expression was confirmed, however, for several highly notochord-enriched genes showing minimal changes in response to Bra CRISPR, including the notochord-enriched transcription factor Foxa.a (Fig. 3, blue box or circles). This indicates that some aspects of notochord-enriched gene expression are independent of Bra.
Fig. 3.
Expression patterns of notochord-enriched genes in Bra crispant embryos. (A) Injected embryos were fixed at stage 16 and stained by in situ hybridization using probes for the indicated genes. As mosaic expression was not observed in control embryos, Bra crispant embryos were scored as positive if they had uniform expression in the notochord and as negative if there was either a total loss of notochord expression or a mosaic loss in a subset of notochord cells. Representative embryos are shown along with log2 notochord enrichment at stage 16 (Reeves et al., 2017) and log2 fold change in Bra crispant embryos. The blue and orange boxes indicate nominally Bra-independent genes and Bra-dependent genes, respectively. (B) Scatter plot of notochord enrichment and Bra CRISPR fold change comparable with Fig. 2C, but now highlighting the specific genes tested by in situ hybridization. Genes with a statistically significant decrease in expression in CRISPR-Bra embryos (Padj≤0.05) by RNAseq are purple.
Foxa.a and Mnx are candidate factors to act in parallel with Bra
Given that Bra is neither necessary in CRISPR loss-of-function assays nor sufficient in ectopic expression assays for the expression of all notochord-enriched genes, there are likely to be additional transcription factors working in parallel to Bra at the top of the notochord GRN. Our inferences based on genome-wide transcriptional profiling are also supported by previous observations. Kugler et al. (2008) identified six genes expressed in notochord after gastrulation that were neither induced by Foxa.a>Bra misexpression nor repressed by expression of a Bra-Engrailed repressor fusion protein. The LMX-like transcription factor has also been shown to retain some notochord expression in Bra homozygous mutant embryos (José-Edwards et al., 2011).
In our misexpression studies, the extent of notochord gene upregulation varied greatly between different target tissues, with the Foxa.a>Bra construct having the greatest effect (Fig. 1D). One possibility is that these tissues are uniquely sensitive to Bra misexpression because Foxa.a is a co-regulator with Bra of notochord fate. Foxa.a is required for notochord fate, but it is difficult to assess whether it acts at least in part in parallel to Bra because it is also an upstream regulator of Bra expression (Imai et al., 2006; Kumano et al., 2006). Foxa.a is an essential and direct upstream regulator of ZicL (Imai et al., 2002b), and ZicL is an essential and direct upstream regulator of Bra (Imai et al., 2002b; Yagi et al., 2004). Important FoxA sites have been found in several notochord enhancers (José-Edwards et al., 2015; Passamaneck et al., 2009), but specific roles of Foxa.a after the induction of ZicL at the 32-cell stage are not well understood. Given its strong, Bra-independent notochord expression in neurula stage embryos, it could easily be acting in parallel to Bra as well as upstream. Overexpression studies in both Xenopus and Halocynthia suggest that Bra and FoxA act synergistically in promoting notochord fate (O'Reilly et al., 1995; Shimauchi et al., 2001).
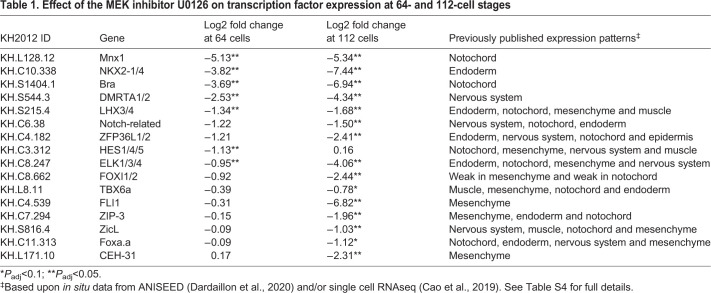
Notochord fate and Bra expression are dependent on vegetal FGF signaling (Imai et al., 2002a, 2006; Yasuo and Hudson, 2007), so we hypothesized that other parallel regulators of notochord fate might share this same dependence. To identify such genes, we inhibited FGF signaling with the MEK inhibitor U0126 at the 32-cell stage and then performed RNAseq at the 64 and 112-cell stages to catch the earliest changes in gene expression between drug-treated and control embryos. A total of 152 genes had significantly decreased expression at either stage (14 genes at the 64 cell stage and 150 at the 112 cell stage), including 16 transcription factors (Table 1, Table S4). We compared these results with published in situ hybridization and single cell RNAseq data and found that 4 of the 16 are specifically expressed in other FGF-dependent tissues, like endoderm, nerve cord or mesenchyme, and thus are not likely involved in notochord specification. Nine transcription factors were expressed broadly in the endoderm, nervous system, mesenchyme and notochord of the early embryo. Of the remaining genes, Foxa.a was slightly decreased (P-value<0.1) at the 112-cell stage in U0126-treated embryos, and both Bra and Mnx were strongly and significantly decreased at both stages (P-value<0.05). During gastrulation, Mnx is expressed in medial row I of the neural plate and primary muscle (Hudson et al., 2007), and becomes specific to the visceral ganglion after neurulation (Imai et al., 2004, 2009). However, it also has specific but transient expression in the notochord as it becomes fate-restricted at the 64- to 112-cell stages (Imai et al., 2004, 2006). Mnx is a homeodomain transcription factor previously hypothesized to play a role in repressing neural genes in the early notochord (Kodama et al., 2016).
Table 1.
Effect of the MEK inhibitor U0126 on transcription factor expression at 64- and 112-cell stages
Combined cocktails are more effective at reprogramming neural plate to notochord
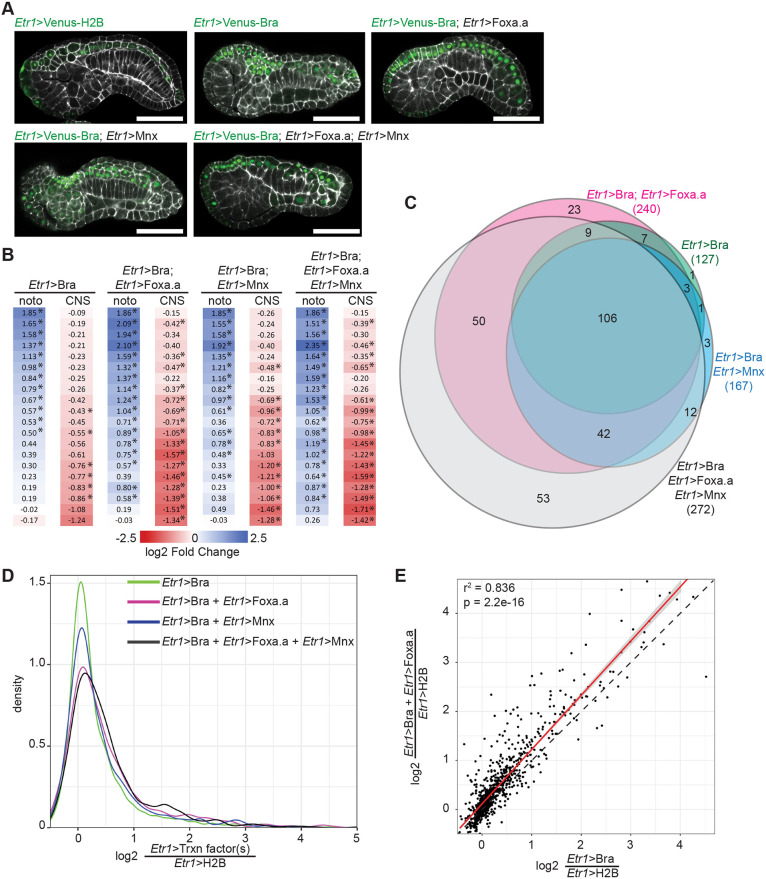
Foxa.a and Mnx are both strong candidates for transcription factors that could function in parallel with Bra at the top of the notochord GRN. We used the neural enhancer Etr1 to express Mnx and/or Foxa.a in combination with Bra (Fig. 4A) to determine whether combinatorial expression of these transcription factors was more efficacious than Bra alone in upregulating notochord-enriched genes. We chose this tissue-specific enhancer because Etr1>Bra misexpression had an intermediate effect on gene expression profiles compared with Foxa.a>Bra and Tbx6-r.b>Bra. Etr1-expressing neural tissues were less refractory to reprogramming than muscle (Fig. 1), and only partially overlap with the endogenous expression of Foxa.a. Whole-embryo expression at stage 19 was measured by RNAseq and compared with expression in embryos electroporated with Etr1>H2B-Venus (Table S5). As predicted, joint misexpression of Foxa.a and/or Mnx with Bra had a greater effect on the induction of notochord markers and repression of neural markers than Bra alone (Fig. 4B, Table S2), although some markers still remained largely unchanged.
Fig. 4.
Combinatorial misexpression of Bra, Foxa.a and Mnx. (A) Representative confocal images of embryos expressing different combinations of Bra-Venus, Foxa.a-myc and/or Mnx-HA fusion proteins under the control of the pan-neural Etr1 enhancer. Etr1>H2B-Venus was expressed in control embryos. Embryos were collected for RNAseq and confocal imaging at stage 19. Phalloidin is shown in white; Venus is in green. Scale bars: 50 μm. (B) Effect of transcription factor misexpression on tissue-specific markers derived from Cao et al. (2019) as in Fig. 1. noto, notochord; CNS, central nervous system. *Padj≤0.05. (C) Overlap of notochord twofold enriched genes whose expression is significantly upregulated by misexpression of the indicated transcription factors. (D) Kernel density plot of the fold-change of expression for twofold enriched notochord genes in response to different ectopic transcription factor combinations. (E) Comparison of differential expression due to Bra misexpression alone versus the Bra/Foxa.a combination. Best fit linear regression shown in red flanked by the 95% confidence interval. The dotted line indicates a perfect 1:1 relationship.
We next looked at the 921 genes that are enriched at least twofold in notochord according to our earlier FACS-RNAseq experiment (Reeves et al., 2017). Joint misexpression of Mnx and Bra had a significant effect on fewer notochord-enriched genes than the Foxa.a/Bra combination (Fig. 4C,D, Table S3), suggesting a more limited or gene-specific role for Mnx in the regulation of notochord gene expression. There were only 15 notochord genes, for example, uniquely upregulated by the Mnx/Bra cocktail versus 73 for the Foxa.a/Bra cocktail and 42 that were upregulated by both combinations (Fig. 4C, Table S3). However, as Mnx is normally expressed in notochord for only a short window around the 64/112 cell stages (Imai et al., 2004, 2006), it is possible that a short pulse of misexpression might have different effects than the protracted misexpression performed here. Additional work will be necessary to fully test the potential roles of Mnx in notochord fate induction.
In contrast, joint misexpression of Foxa.a and Bra had a larger effect on the upregulation of notochord-enriched genes. Bra/Foxa.a-misexpressing embryos had almost twice as many notochord-enriched genes upregulated compared with Bra alone (240 versus 127; 26% of twofold notochord-enriched genes versus 14%) (Fig. 4C,D). Their co-expression also increased the expression of most genes already upregulated by Etr1>Bra alone (Fig. 4E). There was considerable overlap between the 240 notochord genes induced by the Etr1>Bra/Etr>Foxa.a cocktail and the 255 genes induced by Foxa.a>Bra, with 193 being significantly upregulated in both treatments and most of the remainder having increased expression in both but only passing the test of statistical significance in one. This supports the idea that Bra/Foxa.a co-expression leads to a more notochord-like transcriptional profile than Bra alone.
We saw little synergy when simultaneously misexpressing all three genes. The triple Bra/Foxa.a/Mnx cocktail produced similar effects to Bra/Foxa.a, but with the addition of 12 genes that had been specifically upregulated in Bra/Mnx embryos and 53 genes unique to the triple cocktail. Simultaneous mixexpression of Bra and Foxa.a in the neural plate did not upregulate all notochord-enriched genes, but was considerably more potent than Bra alone. These results support the hypothesis that Foxa.a is not just an indirect upstream activator of Bra expression, but also acts in parallel to Bra to regulate notochord target genes. The role of Mnx is not clear, but it is less important than Foxa.a in misexpression assays. FoxA is known to be a key regulator of notochord fate in both vertebrates (Ang and Rossant, 1994; Weinstein et al., 1994) and tunicates (Imai et al., 2006; José-Edwards et al., 2015; Kumano et al., 2006), but the details of the regulatory networks connecting FoxA, Bra and downstream targets are not well understood in any model. We pursued several parallel strategies to clarify these relationships.
Notochord Foxa.a expression depends on vegetal FGF signaling
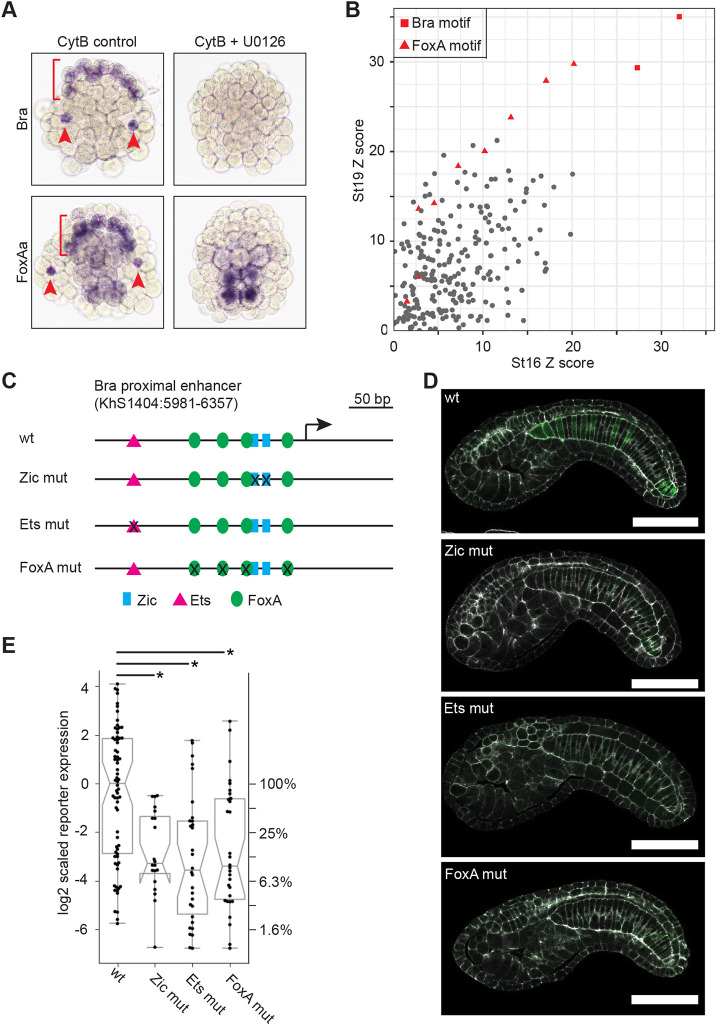
Foxa.a is a major determinant of anterior vegetal fates, with a complex expression pattern in A-line neural, notochord and endodermal cells. Foxa.a expression is initially induced throughout the vegetal region by β-catenin signaling at the 16-cell stage (Imai et al., 2006). By early gastrulation, it is expressed most strongly in notochord, a pattern that continues through neurulation (Corbo et al., 1997b; Di Gregorio et al., 2001) and early tailbud stages (Imai et al., 2004). In contrast, early CNS expression of Foxa.a is lost by late gastrula stage, and then turned back on in only the ventral-most neuroepithelium during neurulation (Corbo et al., 1997b). Expression in the endodermal strand is weak but stable throughout embryonic development (Corbo et al., 1997b; Di Gregorio et al., 2001). Inhibition of FGF signaling by a MEK inhibitor at the 32-cell stage results in a modest decrease in Foxa.a expression by whole embryo RNAseq at the 112 cell stage (see above/Table 1). Although FGF dependence was not previously seen in Ciona by whole-embryo RT-PCR (Imai et al., 2006), notochord expression of FoxA in Halocynthia was reduced at the 64-cell, but not 32-cell, stage by injection of a FGF9/16/20 morpholino (Kumano et al., 2006). This raises the question of whether its later notochord expression in Ciona is comparable with Bra in being dependent on the vegetal FGF signals at the 32-cell stage that are required for notochord fate induction. We treated embryos with U0126 at the 32 cell stage and then cytochalasin B at the 112 cell stage, to arrest cell division and allow for clear identification of cell types in older embryos. Major cell fates, including notochord, are known to be established correctly in cleavage-arrested embryos (Crowther and Whittaker, 1986; Whittaker and Meedel, 1989) and we have previously shown that cytochalasin B treatment does not have a significant effect on expression of notochord-enriched genes (Reeves et al., 2017). At stage 16 (mid-notochord intercalation in untreated embryos), Foxa.a expression has been lost specifically in the notochord but not in the presumptive endoderm (Fig. 5A). This indicates that notochord-enriched Foxa.a expression in Ciona has essential direct or indirect inputs from MAPK signaling specifically in the notochord lineage and distinct from its MAPK-independent regulation at early cleavage stages. We have not mapped out precise start or end points to this dependency, but it identifies an important new edge in the Ciona notochord GRN. We speculate that it may be similar to the regulation of Bra expression by FGF, where distinct temporally separable roles in initiation and maintenance have been found for FGF9/16/20 and FGF8/17/18 (Yasuo and Hudson, 2007).
Fig. 5.
Cis-regulatory interactions involving Foxa.a and Bra. (A) In situ hybridization for Bra and Foxa.a with and without inhibition of early FGF signaling. Embryos were treated at the 32-cell stage with the MEK inhibitor U0126 or DMSO vehicle control, then cleavage arrested at the 112-cell stage with cytochalasin B before fixing at a timepoint equivalent to stage 16. Red arrowheads indicate secondary notochord and the red bracket indicates the arc of primary notochord. (B) Transcription factor-binding site enrichment analysis for putative regulatory regions of genes enriched at least twofold in notochord at stage 16 or stage 19 compared with the putative regulatory regions of genes depleted in the notochord. Z-scores for all enriched TFBSs are shown, with Bra and FoxA motifs highlighted in red. (C,D) Quantitative reporter assay analysis of specific predicted transcription factor binding site mutations in the proximal Bra enhancer region. (C) A map of the Bra upstream region with predicted binding sites for Ets (magenta triangle), Zic (blue box) and FoxA (green oval) labeled. (D) Representative confocal images for the wild-type and TFBS mutation reporter constructs. White, phalloidin; green, reporter expression. Scale bars: 50 µm. The 2D images shown are computationally flattened slices extracted along the midline of each notochord in the Z dimension from 3D confocal stacks. (E) Reporter expression for the different TFBS mutation constructs. Each point represents a different embryo imaged by 3D confocal microscopy with notochord expression of the reporter quantified as described in the Materials and Methods. This confocal-based assay has high dynamic range and there is considerable embryo-to-embryo variation due to transgene mosaicism. The reporter activity scores are normalized to give the control construct a log-transformed median of zero. A separate y-axis is used on the right to show linear-scaled percent decreases with respect to the median of the control plasmid. The bar and whisker plots show the medians (horizontal lines), quartiles (upper and lower edges of the boxes) and minima/maxima (vertical bars). The notches on the boxes indicate the confidence interval of the median, which is outside the lower quartile for the Zic mut construct. Asterisks indicate statistical significance comparing TFBS mutation constructs to the wild-type control (Wilcoxon rank sum test, P<0.0005).
FoxA and Bra sites are both enriched near Ciona notochord genes
An alternate strategy for inferring cell-type specific transcriptional regulators is to identify transcription factor binding motifs enriched in the putative regulatory regions of genes with tissue-specific expression patterns. If Foxa.a is an important co-regulator of notochord gene expression acting not just upstream of Bra but also in parallel, then FoxA-binding motifs should be over-represented in the enhancers of notochord-enriched genes. José-Edwards et al. (2015) previously found that several CRMs capable of driving notochord expression of a reporter at mid-tailbud frequently contained functionally important Bra- and Fox-binding sites, but did not address whether these or other binding motifs are statistically enriched across notochord genes overall. Transcription factor-binding site (TFBS) enrichment analysis also provides an alternate means of predicting regulators that may have been overlooked based on complex or imperfectly annotated expression patterns.
We focused again on genes enriched at least twofold in notochord during neurulation (stage 16, 693 genes) as well as early tailbud (stage 19, 658 genes). Ciona enhancers tend to be near to the transcriptional start site (Irvine, 2013), so we used a window 1500 bp upstream and downstream from the transcription start site of each gene model from which we subtracted predicted exons (see Materials and Methods for full details). Putative regulatory regions of notochord-enriched genes were tested for overrepresented TFBSs using the 2018 JASPAR vertebrate TFBS profile set (Khan et al., 2018) and the oPOSSUM 3.0 Single-Site Analysis algorithm (Kwon et al., 2012), with the enhancers from the 1000 most notochord depleted genes at each stage as the control set. As expected, T-box-binding sites predicted to be bound by Bra had the highest Z-scores, representing a statistical test for enrichment in the number of TFBS occurrences in the notochord versus control set (Fig. 5B). FoxA-binding sites were the next most significantly over-represented motif in the notochord-enriched set at both stages, consistent with a key role for Foxa.a in regulating notochord gene expression. We also saw modest enrichment for several other motifs whose associated transcription factors have putative roles in notochord differentiation, including TBX2 (José-Edwards et al., 2013), Fos/Jun related bZIPs (José-Edwards et al., 2011) and some homeodomain motifs (José-Edwards et al., 2011; Katikala et al., 2013), but Bra and FoxA sites were by far the most enriched. Mnx is a homeodomain TF with a very short and common consensus motif, so enrichment would not necessarily be expected. Owing to false positives and false negatives at the level of individual TFBS predictions, TFBS enrichment analysis does not provide a reliable list of which specific genes are direct targets of Bra and/or FoxA, but it does provide strong support for both these TFs having important regulatory roles in the notochord transcriptome.
Foxa.a is a direct activator of Ciona Brachyury expression
Foxa.a acts indirectly upstream of Bra via induction of ZicL, and also acts in parallel to Bra in the induction of downstream notochord genes, but it is also possible that it could also be acting directly on Bra. This is supported by work in Halocynthia, suggesting that FoxA and Zic act at least partly in parallel as competence factors for induction of Bra expression by FGF (Kumano et al., 2006). Potential FoxA-binding sites have been identified in a region of the Halocynthia Bra enhancer, but their mutation did not decrease Bra>reporter expression in a LacZ reporter assay (Matsumoto et al., 2007). Potential roles for Foxa.a sites in the cis-regulatory control of Ciona Bra have not previously been tested. To determine whether Foxa.a is a direct activator of Ciona Bra expression, we quantified the effects of mutations in Ets-, Zic- and FoxA-binding sites in the 377 bp Bra proximal enhancer (KhS1404:5981-6357) using a sensitive, high-dynamic range assay based on confocal microscopy. Ets and Zic were used as positive controls as they have previously been shown to be essential and direct positive regulators of Bra expression (Farley et al., 2016; Matsumoto et al., 2007; Yagi et al., 2004). Wild-type and mutant enhancers were cloned upstream of a basal FOG promoter and Venus reporter (Harder et al., 2019; Stolfi et al., 2015), and electroporated into embryos in multiple biological replicates (Fig. 5C). At least 10 embryos were imaged for each construct in each biological replicate, and reporter expression was quantified using a method similar to that of Harder et al. (2019). As seen previously, Zic and Ets sites were crucial for strong expression, with mutations reducing median expression down to 8-10% of that seen with a wild-type reporter (Fig. 5D,E). Mutation of four FoxA sites within the enhancer similarly reduced median expression to 9.5% of wild-type levels, supporting the hypothesis that Bra expression is directly Foxa.a dependent.
Feed-forward loops in the Ciona notochord GRN
Although we did not identify any entirely new nodes (trans-acting factors) in the Ciona notochord GRN, we identified multiple new edges (regulatory interactions) (Fig. 6). These demonstrate that notochord fate specification is dominated by interlocked coherent feed-forward interactions. FGF is not only a direct activator of Bra, but also an activator of Foxa.a. Foxa.a is not only an indirect activator of Bra via ZicL but also a direct activator. Notochord target genes are induced not only by Bra but also by other factors, including Foxa.a. All of the major regulators of notochord fate specification act positively at multiple hierarchical levels.
Fig. 6.
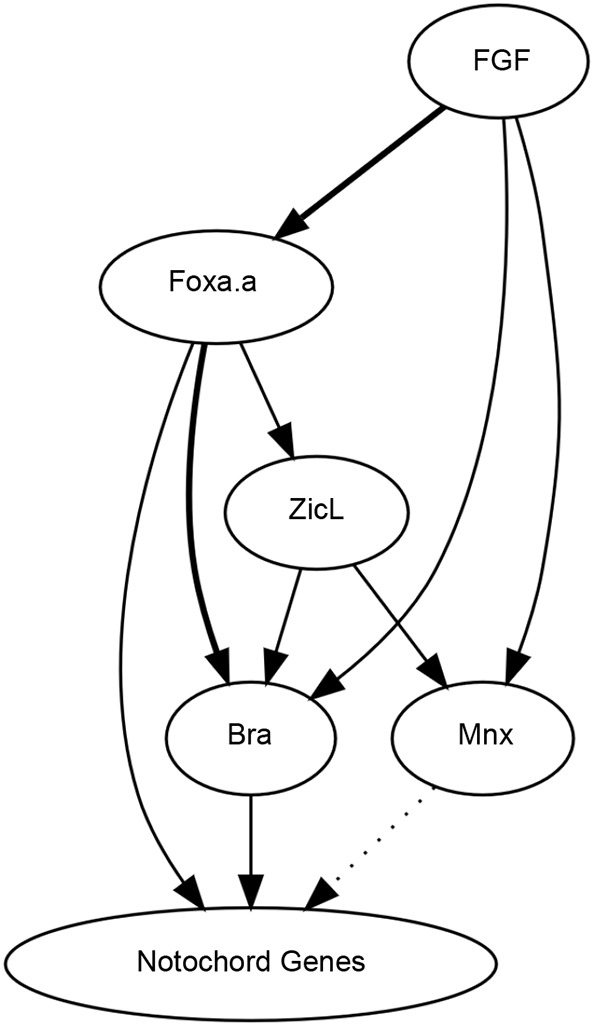
A gene regulatory network for Ciona notochord fate specification. Bold lines indicate new cis-regulatory relationships in the Ciona notochord GRN identified or validated here. The dotted arrow from Mnx is supported by overexpression results but remains to be confirmed by loss of function. Other regulatory interactions in the Ciona notochord GRN have been described previously (Imai et al., 2002a,b, 2006; Yagi et al., 2004; Yasuo and Hudson, 2007).
DISCUSSION
Coherent feed-forward loops where A induces B and then A and B jointly induce C are common in biological networks. Comparable loops have been identified in diverse models of cell fate specification, including sea urchin endomesoderm (Peter and Davidson, 2017), C. elegans gut development (Maduro, 2015) and secondary cell wall synthesis in vascular plants (Zhong and Ye, 2014). Coherent feed-forward loops have multiple potential functions. One potential role is to introduce a deliberate delay into a developmental program (Mangan et al., 2003), although that seems unlikely in this case given the extremely rapid pace of Ciona fate specification. Another potential role for feed-forward loops is to act as persistence detectors and noise filters to prevent stochastic fluctuations in transcription and/or signaling from inadvertently triggering unintentional and irrevocable cell fate decisions (Peter and Davidson, 2017). This role seems more likely here, especially given the extreme stereotypy of ascidian cell fate specification. A similar feed-forward loop involving FGF signaling has recently been identified in the Ciona cardiopharyngeal mesoderm (Razy-Krajka et al., 2018). FGF signaling plays a key role in numerous Ciona cell fate decisions, and more work will be needed to test the ubiquity of feed-forward network motifs across different FGF-dependent cell types and more explicitly test the persistence detector/noise filter hypothesis.
The master regulator concept is used inconsistently in the literature, but it typically refers to a transcription factor that sits alone atop the regulatory cascade for the differentiation of a particular cell type that is not only necessary for the development of that cell type but also sufficient to transform other cell types to that fate when ectopically expressed (Chan and Kyba, 2013). Many putative master regulator TFs have been identified, including eyeless in Drosophila retinal cells (Gehring, 1996), MyoD for myoblasts (Tapscott et al., 1988; Weintraub et al., 1989) and a variety of TFs each controlling a specific CD4+ T cell lineage (Oestreich and Weinmann, 2012). More recently, however, there has been a growing recognition that these developmental cell fate decisions typically involve more complex regulatory networks with multiple essential regulators connected via feedback and/or feed-forward loops (Davis and Rebay, 2017; Desplan, 1997; Kassar-Duchossoy et al., 2004; Wang et al., 2015). Ciona Brachyury satisfies the master regulator criteria in terms of being specifically expressed in the notochord and required, as tested in loss-of-function experiments, for notochord fate, but it has not been clear whether it is truly capable of transforming other cell types into notochord. Bra misexpression induces expression of numerous notochord-specific markers (Hotta et al., 1999; Imai et al., 2006; Takahashi et al., 1999) suggesting that it is indeed sufficient for notochord fate, but subsequent studies have suggested that this might not be definitive (José-Edwards et al., 2011; Kugler et al., 2008; Reeves et al., 2017). Here, we have misexpressed Bra in multiple tissues and found that in all cases it led to only a partial reprogramming towards a notochord-like transcriptional profile. We also disrupted Bra by CRISPR and found that some notochord genes are unaffected. Although it is indisputably a key regulator of Ciona notochord fate, these results show that Bra alone is neither necessary nor sufficient to control expression of all notochord-enriched genes. Based upon ectopic expression studies, CRISPR gene disruption and cis-regulatory analysis, we propose a modified notochord GRN in which Bra and Foxa.a act through a feed-forward loop in which neither acts as a canonical unitary master regulator (Fig. 6). This is consistent with the observation in mouse embryos that, although FoxA is an obligate upstream inducer of Bra expression (Ang and Rossant, 1994), it also has a temporally separable role later in notochord morphogenesis (Yamanaka et al., 2007). Bra and FoxA are co-expressed in axial tissues even in basal metazoans (Fritzenwanker et al., 2004), so it is possible that this feed-forward network may be evolutionarily ancient. FoxA is intriguing in this context in that it has a well-established pioneer ability to bind to the closed nucleosome-bound chromatin that Bra lacks (Fernandez Garcia et al., 2019; Iwafuchi-Doi and Zaret, 2014). The role of epigenetics in Ciona notochord fate specification is not currently understood, but we speculate that Fox family pioneer activity may be important for the increased reprogramming ability of the Bra/Foxa.a cocktail compared with Bra alone.
Decades of research in developmental genetics support the idea that individual cell fate decisions are dominated by a small number of key transcription factors. Models in which single cell type-specific transcription factors are uniquely necessary and sufficient to establish distinct cell states are appealing, as they are straightforward to understand and carry important implications for the evolvability of body plans via conceptually simple cis-regulatory mutations in single tissue-specific regulators. Despite the importance of Bra for Ciona notochord fate, our results here show that it does not act alone at the top of the transcriptional cascade for notochord differentiation but instead as part of a feed-forward network. We speculate that feed-forward interactions between small but non-singular sets of regulatory TFs may be characteristic features of cell fate decisions, allowing a balance between the evolvability of body plans and the robustness of development to stochastic transcriptional noise.
MATERIALS AND METHODS
Ciona husbandry and embryology
Ciona robusta (formerly known as Ciona intestinalis type A) (Pennati et al., 2015) were collected in San Diego and shipped to KSU by Marine Research and Educational Products (M-REP, San Diego, CA). Adult Ciona, which are hermaphrodites, were maintained in a recirculating aquarium. Standard fertilization, dechorionation and electroporation protocols were used (Veeman et al., 2011). Staging is based upon the series of Hotta et al. (2007).
Electroporations and RNA prep
Fertilized dechorionated eggs were electroporated with either 40 µg of enhancer>Venus-Bra plasmid or 40 µg of control plasmid (Bra>GFP) in 800 µl volumes. The Etr1 and Tbx6-r.b enhancers were tested identically to our previously described test of the Foxa.a enhancer (Reeves et al., 2017). For each of the three replicates, 400 experimental and 400 control embryos were collected at stage 19.5 into RNA lysis buffer and stored at −80°C until RNA purification. RNA was purified with the Zymogen Quick-RNA miniprep kit (R1054), genomic DNA was removed using the in-column DNAse treatment step of the kit and purified RNA was eluted in 50 μl water, at a concentration of 52-60 ng/µl. Libraries were constructed with the TruSeq stranded mRNA library kit (Illumina) using standard protocols. Libraries were quality checked with an Agilent Tapestation and quantified by qPCR. Single end (1×100) sequencing was performed on an Illumina HiSeq 2500 at the KU Genome Sequencing Core (GSC) with a read depth of 33.5-38.5 million reads per sample for Foxa.a>Venus-Bra and its matched control or 13.5-15.4 million reads per sample for Etr1>Venus-Bra, Tbx6-r.b >Venus-Bra and their matched control.
For transcription factor combinations in the Etr1> plasmids, 33 μg of each plasmid was used per electroporation. The control plasmid was Etr1>H2B-Venus. Embryo collection and RNA preparation were as described above. Libraries were constructed with the NEBNext UltraII stranded mRNA library kit (NEB) using standard protocols and quality checks as above. Rapid Read single end (1×75) sequencing was performed on an Illumina NextSeq 550 at the KU GSC with a read depth of 26.6-39.6 million reads per sample.
CRISPR-RNP injection and RNAseq
The guide RNA sequence used for Bra CRISPR was selected using scores calculated by CRISPOR (http://crispor.tefor.net) (Haeussler et al., 2016) and IDT (Integrated DNA Technologies). For GFP CRISPR, the sequence was obtained from Addgene (51762, https://www.addgene.org/) (Shalem et al., 2014). The sequences were TAAGCACATGACGAAACATG for Bra CRISPR and GGCCACAAGTTCAGCGTGTC for GFP CRISPR (PAM sequence not shown). crRNAs, tracrRNAs and Cas9 protein were purchased from IDT (Alt-R CRISPR-Cas9 crRNA, Alt-R CRISPR-Cas9 tracrRNA and Alt-R S.p. Cas9 Nuclease 3NLS or V3). The RNP mix was prepared immediately before injection by combining cr/tracrRNA, Cas9 protein and centrifuged HEPES/KCl buffer at final concentrations of 15 µM, 30 µM or 15 µM, and 10 mM/75 mM, respectively, and then activated at 37°C for 5 min. RNP mix was injected into unfertilized eggs according to standard procedure (Yasuo and McDougall, 2018). Approximately 50 to 85 normal developing crispant embryos or control embryos were collected at early neurula (stage 14) and fixed by adding 350 µl of buffer RLT Plus, included in AllPrep DNA/RNA Mini Kit (80204, Qiagen). The solution was then split in half before extracting genomic DNA and total RNA using the manufacturer's protocol.
To evaluate the CRISPR mutagenesis efficiency, the Bra genomic locus was amplified (forward primer, TACGGCGCACTTTCAACAAA; reverse primer, TTTGGTAGGGCGGGGTAATT) and Sanger sequenced from two directions (forward sequence primer, GAGTGTGATTTGGAGGCAGA; reverse sequence primer, ATGTGGATCCTGGGTTCGTA). The results were compared with ones from CRISPR GFP embryos using TIDE (https://tide.deskgen.com) (Brinkman et al., 2014) with default settings. The lower value for each sample (sequenced from the forward or reverse primer) was used as the samples TIDE score. Three replicates with high TIDE scores (∼70%) were used for constructing RNA libraries. RNA libraries were constructed, tested and sequenced as described for the Bra/Mnx/Foxa.a ectopic expression experiment. Read depth was 19.2-23.4 million reads per sample.
Calculation of theoretical maximum decrease by Bra CRISPR
We previously showed using RNAseq on FACS-sorted notochord and not-notochord cells that there is a smooth distribution of notochord enrichment values and not a bimodal distribution of ‘notochord-specific’ versus ‘notochord-enriched’ genes. In our Bra CRISPR experiments, we assume that knocking out Bra will affect gene expression in notochord cells but not in other cell types. The fold change observed by whole-embryo RNAseq for a given gene is thus a function of not only the degree to which that gene is dependent on Bra but also the degree to which it is enriched in the notochord. As the total expression of a notochord-enriched gene in whole-embryo RNAseq experiments is equal to total notochord expression+total non-notochord expression, then total expression=aN+(1-a)E, where N is equal to expression in notochord cells, E is the expression in non-notochord cells and a is the fraction of the embryo comprised of notochord cells. The fold perturbation effect (Fp) is equal to total expression in Bra crispant embryos/total expression in control embryos. The maximum fold change of perturbation expected in a Bra CRISPR experiment using whole-embryo RNAseq will be achieved if the notochord expression is eliminated, leaving only the background expression outside the notochord:
We have previously estimated notochord fold enrichment (Fe) by FACS-Seq, with Fe=N/E or N=Fe*E. Therefore, maxFp=(1−a)E/[a*Fe*E+(1−a)E], which can be simplified to (1−a)/(A*Fe+1−a). All 10 notochord founder cells are specified by the 112-cell stage, so we estimate that 10/112 (∼9%) of the embryo by mass at later stages consists of notochord cells:
Drug treatments
For the RNAseq experiment, U0126 (Sigma-Aldrich 662005) was dissolved in DMSO at 4 mM and used at a final concentration of 4 µM. DMSO alone (at 1:1000 dilution) was used as a control. Embryos were treated at the 32-cell stage, then collected at the 64-cell stage and 112-cell stage. RNA purification, library preparation and sequencing was as described for Bra/Mnx/Foxa.a ectopic expression experiments. Read depth was 18.7-23 million reads per sample.
For in situ hybridization, embryos were treated with 10 µM cytochalasin B (Sigma-Aldrich C2732) at the 112-cell stage after U0126/DMSO treatment at the 32-cell stage, then fixed at stage 16 as described by Reeves et al. (2014).
RNAseq analysis and bioinformatics
Tophat (Trapnell et al., 2012, 2013) was used to align reads to the Ciona KH2012 gene models (KH.KHGene.2012.gff3, retrieved from ANISEED, http://www.aniseed.cnrs.fr/) (Brozovic et al., 2016; Dardaillon et al., 2020). The standard DEseq2 pipeline (Love et al., 2014) was used to calculate read counts and test for differential expression. Statistical significance was reported as adjusted P-values corrected for multiple comparisons.
TFBS enrichment analysis
We used bedtools (Quinlan and Hall, 2010) and custom scripts to extract putative regulatory regions near the predicted transcriptional start sites of both notochord-enriched and control gene sets. We performed separate analyses using our FACS RNAseq data from both stage 16 and stage 19. For each timepoint, the notochord-enriched gene set included all genes enriched at least twofold in notochord (693 genes at stage 16 and 658 genes at stage 19). As control sequences, we used the 1000 most notochord-depleted genes at each stage. Putative regulatory regions were defined as DNA sequences 1500 bp upstream and downstream of transcription start sites. For genes predicted to be expressed in operons, we used the operon TSS. We then subtracted coding exon sequences and removed any features smaller than 100 bp. We removed any sequences from analysis that were present in both sets at a given stage. TFBS enrichment was calculated using the 2018 JASPAR vertebrate TFBS profile set (JASPAR; http://jaspar.genereg.net) (Khan et al., 2018) and the oPOSSUM 3.0 Single-Site Analysis algorithm (Kwon et al., 2012) with default settings. The Z-scores reported indicate enrichment in the rate of occurrence of a given TFBS in the test set versus the control set.
Analysis of mutated Bra enhancer constructs
The Bra reporter TFBS mutation analysis made use of a 377 bp fragment matched to an ATAC-seq peak over the upstream region and first exon of the Bra locus (KhS1404:5981-6357), cloned into a vector containing a minimal promoter from Ciona Friend of Gata (bpFOG) and a Venus YFP reporter [pX2+bpFOG>UNC76:Venus (Stolfi et al., 2015)]. Predicted transcription factor-binding sites were identified using JASPAR 2018 PWMs and the FIMO scanner (http://meme-suite.org/doc/fimo.html) (Grant et al., 2011) using a P-value threshold of 0.001. Mutant sequences designed to disrupt specific binding sites were rescanned by FIMO to ensure that they did not affect nearby sites or inadvertently introduce new sites. Control and mutant sequences were all synthesized as gBlocks (IDT) and verified by Sanger sequencing after Gibson cloning into the reporter vector. All constructs were electroporated in two (Zic mutant) or three (Ets and Fox mutants) different biological replicates in different combinations, at 50 µg each. All experiments included the wild-type control plasmid as one of the electroporated constructs. For each experiment, embryos were fixed at stage 21, immunostained for reporter expression and cleared in Murray's Clear. The reporter immunostaining used a polyclonal rabbit anti-GFP antibody (Fisher A11122) and an Alexa555 anti-rabbit secondary antibody (Fisher A21429), both at 1:1000. Alexa488-conjugated phalloidin (Fisher A12379) was used at 1:100.
At least 10 embryos were imaged per construct per replicate using a 40×1.3 NA objective on a Zeiss 880 confocal microscope using uniform imaging settings. These were randomly selected without inspecting reporter staining from embryos showing normal embryonic morphology by phalloidin staining. 3D image stacks were used to reconstruct a computationally straightened slice along the length of the notochord as described by Harder et al. (2019) and a thick line trace along the notochord was summed to quantify a reporter expression value for each embryo. Expression of electroporated transgenes in Ciona is mosaic, so expression at the level of individual embryos is quite variable. This represents both embryo to embryo variation in the number of expressing cells and how brightly they are expressing, as well as variation in transfection efficiency between different electroporations. We controlled for this by imaging multiple randomly chosen embryos for each replicate and electroporating multiple replicates for each construct as described above. The data were normalized at the level of the entire dataset to give the control plasmid a median log-transformed value of 0.
In situ hybridization and probes
Probe synthesis, embryo collection, in situ hybridization and imaging were performed as described previously (Reeves et al., 2014). Probes for Bra (Reeves et al., 2014), C7.633 and C1.1281 (Reeves et al., 2017) have been described previously. Probe sequences for C11.313, C3.815, C2.866, C2.33, C9.698, C7.260 and L170.42 were amplified from cDNA, then cloned into pBSII SK(-). Primers used are provided in Table S6.
Plasmid construction details
Foxa.a>Bra-Venus has been described previously (Reeves et al., 2017; Takahashi et al., 1999). For Etr1>Bra-Venus, the Foxa.a enhancer was removed by XhoI/XbaI digest, and replaced with PCR amplified enhancers for Etr1 (forward primer, TCGACCACGGAGTTAATTG; reverse primer, TCTGGATAAAGCAATACATACGAG) and Tbx6-r.b (forward primer, AATGTAGCGTCGCTTCAC; reverse primer, GGAATCATATTCGCCATAGTC) by Gibson cloning (NEB kit). For Etr1>Mnx-HA, full-length Mnx-coding sequence (PCR amplified from genomic DNA) was Gibson cloned upstream of the HA tag in SwaI-Rfa-HA, replacing the Gateway cloning cassette, and then the Etr enhancer was cloned into XhoI/XbaI as above. For Etr1>Foxa.a-Myc we used three-fragment Gibson cloning to combine the Etr1 enhancer (PCR amplified from Etr1<Bra-Venus), the Foxa.a-coding sequence (PCR amplified from genomic DNA with myc tag added in primer) and the plasmid backbone of SwaI-Venus-Rfa (ref) outside the Venus-Rfa region.
Supplementary Material
Acknowledgements
We thank the University of Kansas Center for Molecular Analysis of Disease Phenotypes Genome Sequencing Core, the Kansas State University Integrated Genomics Facility, the Kansas State University College of Veterinary Medicine Confocal Core and the Kansas State University Beocat supercomputing cluster.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: W.M.R., K.S., M.T.V.; Methodology: W.M.R., K.S., M.T.V.; Software: W.M.R., M.T.V.; Formal analysis: W.M.R., K.S., K.M.W., M.T.V.; Investigation: W.M.R., K.S.; Writing - original draft: W.M.R., K.S., M.T.V.; Writing - review & editing: W.M.R., K.S., K.M.W., M.T.V.; Supervision: W.M.R., M.T.V.; Funding acquisition: M.T.V.
Funding
This work was supported by the National Institutes of Health (1R01HD085909 to M.T.V.). Deposited in PMC for release after 12 months.
Data availability
The new RNAseq data generated for this publication have been deposited in GEO under accession number GSE160548. Our original Foxa.a>Bra RNAseq data from Reeves et al. (2017) are available in GEO under accession number GSE102142.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.195230.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.195230.reviewer-comments.pdf
References
- Ang, S.-L. and Rossant, J. (1994). HNF-3β is essential for node and notochord formation in mouse development. Cell 78, 561-574. 10.1016/0092-8674(94)90522-3 [DOI] [PubMed] [Google Scholar]
- Brinkman, E. K., Chen, T., Amendola, M. and van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168-e168 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozovic, M., Martin, C., Dantec, C., Dauga, D., Mendez, M., Simion, P., Percher, M., Laporte, B., Scornavacca, C., Di Gregorio, A.et al. (2016). ANISEED 2015: a digital framework for the comparative developmental biology of ascidians. Nucleic Acids Res. 44, D808-D818. 10.1093/nar/gkv966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, C., Lemaire, L. A., Wang, W., Yoon, P. H., Choi, Y. A., Parsons, L. R., Matese, J. C., Wang, W., Levine, M. and Chen, K. (2019). Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349-354. 10.1038/s41586-019-1385-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. S.-K. and Kyba, M. (2013). What is a master regulator? J. Stem Cell Res. Ther. 3, 680-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, S., Jiang, D., Satoh, N. and Smith, W. C. (2009). Brachyury null mutant-induced defects in juvenile ascidian endodermal organs. Development 136, 35-39. 10.1242/dev.030981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo, J. C., Levine, M. and Zeller, R. W. (1997a). Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589-602. [DOI] [PubMed] [Google Scholar]
- Corbo, J. C., Erives, A., Di Gregorio, A., Chang, A. and Levine, M. (1997b). Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development 124, 2335-2344. [DOI] [PubMed] [Google Scholar]
- Crowther, R. J. and Whittaker, J. R. (1986). Differentiation without cleavage: Multiple cytospecific ultrastructural expressions in individual one-celled ascidian embryos. Dev. Biol. 117, 114-126. 10.1016/0012-1606(86)90354-4 [DOI] [PubMed] [Google Scholar]
- Dardaillon, J., Dauga, D., Simion, P., Faure, E., Onuma, T. A., DeBiasse, M. B., Louis, A., Nitta, K. R., Naville, M., Besnardeau, L.et al. (2020). ANISEED 2019: 4D exploration of genetic data for an extended range of tunicates. Nucleic Acids Res. 48, D668-D675. 10.1093/nar/gkz955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, T. L. and Rebay, I. (2017). Master regulators in development: views from the Drosophila retinal determination and mammalian pluripotency gene networks. Dev. Biol. 421, 93-107. 10.1016/j.ydbio.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan, C. (1997). Eye development: governed by a dictator or a junta? Cell 91, 861-864. 10.1016/S0092-8674(00)80475-4 [DOI] [PubMed] [Google Scholar]
- Di Gregorio, A., Corbo, J. C. and Levine, M. (2001). The regulation of forkhead/HNf-3 beta expression in the Ciona embryo. Dev. Biol. 229, 31-43. 10.1006/dbio.2000.9964 [DOI] [PubMed] [Google Scholar]
- Farley, E. K., Olson, K. M., Zhang, W., Rokhsar, D. S. and Levine, M. S. (2016). Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc. Natl. Acad. Sci. USA 113, 6508-6513. 10.1073/pnas.1605085113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Garcia, M., Moore, C. D., Schulz, K. N., Alberto, O., Donague, G., Harrison, M. M., Zhu, H. and Zaret, K. S. (2019). Structural features of transcription factors associating with nucleosome binding. Mol. Cell 75, 921-932.e6. 10.1016/j.molcel.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzenwanker, J. H., Saina, M. and Technau, U. (2004). Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev. Biol. 275, 389-402. 10.1016/j.ydbio.2004.08.014 [DOI] [PubMed] [Google Scholar]
- Gehring, W. J. (1996). The master control gene for morphogenesis and evolution of the eye. Genes Cells 1, 11-15. 10.1046/j.1365-2443.1996.11011.x [DOI] [PubMed] [Google Scholar]
- Grant, C. E., Bailey, T. L. and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017-1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler, M., Schönig, K., Eckert, H., Eschstruth, A., Mianné, J., Renaud, J.-B., Schneider-Maunoury, S., Shkumatava, A., Teboul, L., Kent, J.et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder, M., Reeves, W., Byers, C., Santiago, M. and Veeman, M. (2019). Multiple inputs into a posterior-specific regulatory network in the Ciona notochord. Dev. Biol. 448, 136-146. 10.1016/j.ydbio.2018.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta, K., Takahashi, H., Erives, A., Levine, M. and Satoh, N. (1999). Temporal expression patterns of 39 Brachyury-downstream genes associated with notochord formation in the Ciona intestinalis embryo. Dev. Growth Differ. 41, 657-664. 10.1046/j.1440-169x.1999.00467.x [DOI] [PubMed] [Google Scholar]
- Hotta, K., Mitsuhara, K., Takahashi, H., Inaba, K., Oka, K., Gojobori, T. and Ikeo, K. (2007). SPECIAL ISSUE RESEARCH ARTICLE a web-based interactive developmental table for the ascidian ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Biol. 236, 1790-1805. 10.1002/dvdy.21188 [DOI] [PubMed] [Google Scholar]
- Hudson, C., Lotito, S. and Yasuo, H. (2007). Sequential and combinatorial inputs from Nodal, Delta2/Notch and FGF/MEK/ERK signalling pathways establish a grid-like organisation of distinct cell identities in the ascidian neural plate. Development 134, 3527-3537. 10.1242/dev.002352 [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Satoh, N. and Satou, Y. (2002a). Early embryonic expression of FGF4/6/9 gene and its role in the induction of mesenchyme and notochord in Ciona savignyi embryos. Development 129, 1729-1738. [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Satou, Y. and Satoh, N. (2002b). Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development 129, 2723-2732. [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Hino, K., Yagi, K., Satoh, N. and Satou, Y. (2004). Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047-4058. 10.1242/dev.01270 [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Levine, M., Satoh, N. and Satou, Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183-1187. 10.1126/science.1123404 [DOI] [PubMed] [Google Scholar]
- Imai, K. S., Stolfi, A., Levine, M. and Satou, Y. (2009). Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136, 285-293. 10.1242/dev.026419 [DOI] [PubMed] [Google Scholar]
- Irvine, S. Q. (2013). Study of Cis-regulatory elements in the ascidian ciona intestinalis. Curr. Genomics 14, 56-67. 10.2174/138920213804999192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi, M. and Zaret, K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679-2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Edwards, D. S., Kerner, P., Kugler, J. E., Deng, W., Jiang, D. and Di Gregorio, A. (2011). The identification of transcription factors expressed in the notochord of Ciona intestinalis adds new potential players to the brachyury gene regulatory network. Dev. Dyn. 240, 1793-1805. 10.1002/dvdy.22656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Edwards, D. S., Oda-Ishii, I., Nibu, Y. and Di Gregorio, A. (2013). Tbx2/3 is an essential mediator within the Brachyury gene network during Ciona notochord development. Development 140, 2422-2433. 10.1242/dev.094227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- José-Edwards, D. S., Oda-Ishii, I., Kugler, J. E., Passamaneck, Y. J., Katikala, L., Nibu, Y. and Di Gregorio, A. (2015). Brachyury, Foxa2 and the cis-regulatory origins of the notochord. PLoS Genet. 11, 1-16. 10.1371/journal.pgen.1005730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy, L., Gayraud-Morel, B., Gomès, D., Rocancourt, D., Buckingham, M., Shinin, V. and Tajbakhsh, S. (2004). Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431, 466-471. 10.1038/nature02876 [DOI] [PubMed] [Google Scholar]
- Katikala, L., Aihara, H., Passamaneck, Y. J., Gazdoiu, S., José-Edwards, D. S., Kugler, J. E., Oda-Ishii, I., Imai, J. H., Nibu, Y. and Di Gregorio, A. (2013). Functional brachyury binding sites establish a temporal read-out of gene expression in the ciona notochord. PLoS Biol.. 11, e1001697 10.1371/journal.pbio.1001697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A., Fornes, O., Stigliani, A., Gheorghe, M., Castro-Mondragon, J. A., van der Lee, R., Bessy, A., Chèneby, J., Kulkarni, S. R., Tan, G.et al. (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260-D266. 10.1093/nar/gkx1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, H., Miyata, Y., Kuwajima, M., Izuchi, R., Kobayashi, A., Gyoja, F., Onuma, T. A., Kumano, G. and Nishida, H. (2016). Redundant mechanisms are involved in suppression of default cell fates during embryonic mesenchyme and notochord induction in ascidians. Dev. Biol. 416, 162-172. 10.1016/j.ydbio.2016.05.033 [DOI] [PubMed] [Google Scholar]
- Kugler, J. E., Passamaneck, Y. J., Feldman, T. J., Beh, J., Regnier, T. W. and Di Gregorio, A. (2008). Evolutionary conservation of vertebrate notochord genes in the ascidian Ciona intestinalis. Genesis 46, 697-710. 10.1002/dvg.20403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler, J. E., Gazdoiu, S., Oda-Ishii, I., Passamaneck, Y. J., Erives, A. J. and Di Gregorio, A. (2010). Temporal regulation of the muscle gene cascade by Macho1 and Tbx6 transcription factors in Ciona intestinalis. J. Cell Sci. 123, 2453-2463. 10.1242/jcs.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano, G., Yamaguchi, S. and Nishida, H. (2006). Overlapping expression of FoxA and Zic confers responsiveness to FGF signaling to specify notochord in ascidian embryos. Dev. Biol. 300, 770-784. 10.1016/j.ydbio.2006.07.033 [DOI] [PubMed] [Google Scholar]
- Kwon, A. T., Arenillas, D. J., Worsley Hunt, R. and Wasserman, W. W. (2012). oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2, 987-1002. 10.1534/g3.112.003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro, M. F. (2015). Developmental robustness in the Caenorhabditis elegans embryo. Mol. Reprod. Dev. 82, 918-931. 10.1002/mrd.22582 [DOI] [PubMed] [Google Scholar]
- Mangan, S., Zaslaver, A. and Alon, U. (2003). The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 334, 197-204. 10.1016/j.jmb.2003.09.049 [DOI] [PubMed] [Google Scholar]
- Matsumoto, J., Kumano, G. and Nishida, H. (2007). Direct activation by Ets and Zic is required for initial expression of the Brachyury gene in the ascidian notochord. Dev. Biol. 306, 870-882. 10.1016/j.ydbio.2007.03.034 [DOI] [PubMed] [Google Scholar]
- Nelson, A. C., Pillay, N., Henderson, S., Presneau, N., Tirabosco, R., Halai, D., Berisha, F., Flicek, P., Stemple, D. L., Stern, C. D.et al. (2012). An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J. Pathol. 228, 274-285. 10.1002/path.4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly, M. A., Smith, J. C. and Cunliffe, V. (1995). Patterning of the mesoderm in Xenopus: dose-dependent and synergistic effects of Brachyury and Pintallavis. Development 121, 1351-1359. [DOI] [PubMed] [Google Scholar]
- Oestreich, K. J. and Weinmann, A. S. (2012). Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat. Rev. Immunol. 12, 799-804. 10.1038/nri3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck, Y. J. and Di Gregorio, A. (2005). Ciona intestinalis: chordate development made simple. Dev. Dyn. 233, 1-19. 10.1002/dvdy.20300 [DOI] [PubMed] [Google Scholar]
- Passamaneck, Y. J., Katikala, L., Perrone, L., Dunn, M. P., Oda-Ishii, I. and Di Gregorio, A. (2009). Direct activation of a notochord cis-regulatory module by Brachyury and FoxA in the ascidian Ciona intestinalis. Development 136, 3679-3689. 10.1242/dev.038141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennati, R., Ficetola, G. F., Brunetti, R., Caicci, F., Gasparini, F., Griggio, F., Sato, A., Stach, T., Kaul-Strehlow, S., Gissi, C.et al. (2015). Morphological differences between larvae of the ciona intestinalis species complex: hints for a valid taxonomic definition of distinct species. PLoS ONE 10, e0122879 10.1371/journal.pone.0122879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, I. S. and Davidson, E. H. (2017). Assessing regulatory information in developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA 114, 5862-5869. 10.1073/pnas.1610616114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A. R. and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razy-Krajka, F., Gravez, B., Kaplan, N., Racioppi, C., Wang, W. and Christiaen, L. (2018). An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. Elife 7, 1-27. 10.7554/eLife.29656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, W., Thayer, R. and Veeman, M. (2014). Anterior-posterior regionalized gene expression in the Ciona notochord. Dev. Dyn. 243, 612-620. 10.1002/dvdy.24101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, W. M., Wu, Y., Harder, M. J. and Veeman, M. T. (2017). Functional and evolutionary insights from the Ciona notochord transcriptome. Development 144, 3375-3387. 10.1242/dev.156174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, N., Satou, Y., Davidson, B. and Levine, M. (2003). Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 19, 376-381. 10.1016/S0168-9525(03)00144-6 [DOI] [PubMed] [Google Scholar]
- Satou, Y., Imai, K. S. and Satoh, N. (2001). Action of morpholinos in Ciona embryos. Genesis 30, 103-106. 10.1002/gene.1040 [DOI] [PubMed] [Google Scholar]
- Shalem, O., Sanjana, N. E., Hartenian, E., Shi, X., Scott, D. A., Mikkelsen, T. S., Heckl, D., Ebert, B. L., Root, D. E., Doench, J. G.et al. (2014). Genome-scale CRISPR-Cas9 Knockout screening in human cells. Science (80-.) 343, 84-87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimauchi, Y., Chiba, S. and Satoh, N. (2001). Synergistic action of HNF-3 and Brachyury in the notochord differentiation of ascidian embryos. Int. J. Dev. Biol. 45, 643-652. [PubMed] [Google Scholar]
- Sierro, N., Kusakabe, T., Park, K.-J., Yamashita, R., Kinoshita, K. and Nakai, K. (2006). DBTGR: a database of tunicate promoters and their regulatory elements. Nucleic Acids Res. 34, D552-D555. 10.1093/nar/gkj064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi, A., Ryan, K., Meinertzhagen, I. A. and Christiaen, L. (2015). Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527, 371-374. 10.1038/nature15758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Hotta, K., Erives, A., Di Gregorio, A., Zeller, R. W., Levine, M. and Satoh, N. (1999). Brachyury downstream notochord differentiation in the ascidian embryo. Genes Dev. 13, 1519-1523. 10.1101/gad.13.12.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott, S. J., Davis, R. L., Thayer, M. J., Cheng, P. F., Weintraub, H. and Lassar, A. B. (1988). MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405-411. 10.1126/science.3175662 [DOI] [PubMed] [Google Scholar]
- Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., Pimentel, H., Salzberg, S. L., Rinn, J. L. and Pachter, L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562-578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Hendrickson, D. G., Sauvageau, M., Goff, L., Rinn, J. L. and Pachter, L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46-53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman, M. T., Chiba, S. and Smith, W. C. (2011). Ciona genetics. In Vertebrate Embryogenesis (F. J. Pelegri, ed), pp. 401-422. Springer [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Collins, M. and Kuchroo, V. K. (2015). Effector T cell differentiation: are master regulators of effector T cells still the masters? Curr. Opin. Immunol. 37, 6-10. 10.1016/j.coi.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Weinstein, D. C., Ruiz i Altaba, A., Chen, W. S., Hoodless, P., Prezioso, V. R., Jessell, T. M. and Darnell, J. E. (1994). The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78, 575-588. 10.1016/0092-8674(94)90523-1 [DOI] [PubMed] [Google Scholar]
- Weintraub, H., Tapscott, S. J., Davis, R. L., Thayer, M. J., Adam, M. A., Lassar, A. B. and Miller, A. D. (1989). Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA 86, 5434-5438. 10.1073/pnas.86.14.5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker, J. R. and Meedel, T. H. (1989). Two histospecific enzyme expressions in the same cleavage-arrested one-celled ascidian embryos. J. Exp. Zool. 250, 168-175. 10.1002/jez.1402500208 [DOI] [PubMed] [Google Scholar]
- Yagi, K., Satou, Y. and Satoh, N. (2004). A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development 131, 1279-1288. 10.1242/dev.01011 [DOI] [PubMed] [Google Scholar]
- Yamada, L., Shoguchi, E., Wada, S., Kobayashi, K., Mochizuki, Y., Satou, Y. and Satoh, N. (2003). Morpholino-based gene knockdown screen of novel genes with developmental function in Ciona intestinalis. Development 130, 6485-6495. 10.1242/dev.00847 [DOI] [PubMed] [Google Scholar]
- Yamanaka, Y., Tamplin, O. J., Beckers, A., Gossler, A. and Rossant, J. (2007). Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev. Cell 13, 884-896. 10.1016/j.devcel.2007.10.016 [DOI] [PubMed] [Google Scholar]
- Yasuo, H. and Hudson, C. (2007). FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev. Biol. 302, 92-103. 10.1016/j.ydbio.2006.08.075 [DOI] [PubMed] [Google Scholar]
- Yasuo, H. and McDougall, A. (2018). Practical guide for ascidian microinjection: Phallusia mammillata. In Transgenic Ascidians (ed. Sasakura Y.), pp. 15-24. Singapore: Springer. [DOI] [PubMed] [Google Scholar]
- Yasuo, H. and Satoh, N. (1993). Function of vertebrate T gene. Nature 364, 582-583. 10.1038/364582b0 [DOI] [PubMed] [Google Scholar]
- Yasuo, H. and Satoh, N. (1998). Conservation of the developmental role of Brachyury in notochord formation in a Urochordate, the ascidian Halocynthia roretzi. Dev. Biol. 200, 158-170. 10.1006/dbio.1998.8958 [DOI] [PubMed] [Google Scholar]
- Zhong, R. and Ye, Z.-H. (2014). Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci. 229, 193-207. 10.1016/j.plantsci.2014.09.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.