Plants with altered cell walls have expanded cells that compress adjacent tissues to stimulate stress hormone production.
Abstract
Despite the vital roles of jasmonoyl-isoleucine (JA-Ile) in governing plant growth and environmental acclimation, it remains unclear what intracellular processes lead to its induction. Here, we provide compelling genetic evidence that mechanical and osmotic regulation of turgor pressure represents a key elicitor of JA-Ile biosynthesis. After identifying cell wall mutant alleles in KORRIGAN1 (KOR1) with elevated JA-Ile in seedling roots, we found that ectopic JA-Ile resulted from cell nonautonomous signals deriving from enlarged cortex cells compressing inner tissues and stimulating JA-Ile production. Restoring cortex cell size by cell type–specific KOR1 complementation, by isolating a genetic kor1 suppressor, and by lowering turgor pressure with hyperosmotic treatments abolished JA-Ile signaling. Conversely, hypoosmotic treatment activated JA-Ile signaling in wild-type plants. Furthermore, constitutive JA-Ile levels guided mutant roots toward greater water availability. Collectively, these findings enhance our understanding on JA-Ile biosynthesis initiation and reveal a previously undescribed role of JA-Ile in orchestrating environmental resilience.
INTRODUCTION
In higher plants, the phytohormone (+)-7-iso-jasmonoyl-l-isoleucine (JA-Ile) is a decisive coordinator of growth and stress responses (1). Induced JA-Ile signaling secures a successful execution of reproductive development; regulates the acclimation to unfavorable conditions such as drought, salt, cold, elevated ozone, and ultraviolet light; and is essential to protect plants against insect herbivory, necrotrophic pathogens, and RNA viruses (1–3). It is estimated that insect herbivory alone affects >20% of global net plant productivity, and plants unable to synthesize JA-Ile, such as the allene oxide synthase (aos) mutant of the model plant Arabidopsis thaliana, are unprotected against chewing insects (4, 5). Rising hormone levels elicit nuclear JA-Ile perception and signaling, leading to the increased expression of JA-Ile marker genes, such as JASMONATE-ZIM DOMAIN 10 (JAZ10), and up-regulation of JA-Ile–induced defense transcripts such as VEGETATIVE STORAGE PROTEIN 2 (VSP2) and PLANT DEFENSIN 1.2 (PDF1.2) (1, 6, 7). As mounting defense responses are accompanied by reduced vegetative growth, JA-Ile levels are tightly regulated and normally induced only when required (1, 8). Despite the broad and critical functions of the jasmonate (JA) pathway in plant growth and environmental acclimation, the intracellular events triggering JA-Ile production remain poorly understood (1, 9, 10).
Several molecular elicitors of JA-Ile biosynthesis have been proposed, including herbivore-, microbe-, and damage-associated molecular patterns generated during environmental stresses. These elicitors would be recognized by putative pattern recognition receptors at the cell surface that transduce the signal intracellularly and initiate de novo JA-Ile synthesis, reviewed in (9, 11). However, conclusive genetic evidence for ligand-based induction of JA-Ile biosynthesis is still missing. Mechanical wounding is another potent activator of JA-Ile biosynthesis and is widely used to stimulate endogenous hormone production (6, 10). In parallel to necrotroph and herbivorous insect attacks, mechanical wounding causes the disruption of cell walls that surround each plant cell and serve as the immediate contact surface with the extracellular environment. Plant cell walls are elaborate structural networks consisting predominantly of complex polysaccharides (cellulose, hemicelluloses, and pectins), a lower amount of proteins, and other soluble and phenolic materials (12). Notably, chemical or genetic inhibition of cellulose biosynthesis by isoxaben application or by mutations in CELLULOSE SYNTHASE (CesA) genes leads to increased JA-Ile levels, e.g., (11, 13, 14). Consistently, mutations in KORRIGAN1 (KOR1), a CesA-interacting membrane protein with endo-1,4-β-d-glucanase activity involved in cellulose biosynthesis, exhibit a slight basal increase in the JA-Ile precursor JA (15–19). Nevertheless, it remains unclear how cellulose deficiency stimulates JA-Ile production and in which plant tissues the hormone accumulates.
Plant cells have a high intracellular turgor pressure deriving from a gradient in osmotic potential across the plasma membrane, which is counterbalanced by their cell walls, with cellulose microfibrils serving as a load-bearing component (12). If osmotic conditions change, then plant cells may experience mechanical stress and even deform their shape depending on the extent of the stimulus, their geometry, and the properties of their cell walls. Hence, they regulate water and ionic fluxes across the plasma membrane and remodel their cell wall until a new osmotic equilibrium is reached and turgor pressure is reestablished (20, 21). Cell walls with compromised cellulose microfibrils may be inefficient at counteracting the high intracellular turgor pressure, resulting in enlarged cells. Cell swelling is, in fact, a typical feature observed in cellulose-deficient mutants or isoxaben-treated plants (22, 23). Mechanical stress arising from turgor pressure changes might be therefore involved in activating JA-mediated stress responses (24). In fact, cotreatments of liquid-grown seedlings with isoxaben and sorbitol serving as osmotica nullify the increased JA levels induced by isoxaben alone (23, 25), linking osmoregulation to the JA pathway.
Here, we investigated how cues derived from perturbed cellulose biosynthesis integrate with JA-Ile production. First, we identified kor1 alleles with increased JA-Ile levels in cell type–specific contexts of the primary root. While elevated JA-Ile signaling in kor1-4 was confined to inner root tissues, restoring KOR1 function in adjacent cortex cells complemented the JA phenotype. Transversal sections revealed a pronounced enlargement of cortex cells, likely exerting mechanical pressure on physically constrained inner tissues. Reducing cortex cell size by genetic and chemical means fully abolished expanded kor1 cortex cells and constitutive JA-Ile signaling, indicating that JA biosynthesis is triggered by turgor-driven mechanical compression. Consistently, increasing turgor pressure with hypoosmotic treatment activated JA-Ile signaling in inner root tissues of wild-type (WT) plants. While constitutive JA-Ile signaling did not affect kor1 root growth rate nor the expression of JA-dependent defense genes, it was critical to guide root growth toward greater water availability.
RESULTS
Cellulose-deficient kor1 mutants have increased root JA-Ile levels
In a forward genetic screen aimed at identifying negative regulators of JA signaling (6), we isolated two alleles of KOR1, kor1-4 and kor1-5, exhibiting ectopic expression of the JA-responsive reporter JAZ10p:GUSPlus (JAZ10p:GUS, JGP). Basal JGP expression in the WT and in JA-deficient aos plants is very weak, whereas kor1 alleles showed JGP activation in the primary root (Fig. 1A and fig. S1A). Allelism tests with a transfer DNA (T-DNA) insertion kor1-6 mutant in the JGP background and complementation by transformation with untagged (KOR1p:KOR1) and N-terminal CITRINE (CIT)–tagged KOR1 (KOR1p:CIT-KOR1) constructs, both restoring the kor1 short root length, confirmed the causative mutations of the observed JGP phenotype (Fig. 1, A and B). Ethyl methanesulfonate (EMS) kor1 alleles harbored single amino acid exchanges within the extracellular GLYCOSYL HYDROLASE 9 (GH9) domain of KOR1 (L573F in kor1-4 and P172L in kor1-5), while kor1-6 resulted in lower KOR1 transcript levels (fig. S1, B and C). Among the three kor1 alleles, kor1-5 showed the most severe JA and root elongation phenotypes and kor1-6 the mildest (Fig. 1, A and C, and fig. S1, A and D), implying that all mutants still retain partial KOR1 function. This is in line with previous reports proposing that full KOR1 knockouts are lethal (16). Given the milder growth defects and robust JA phenotype of kor1-4, we used this allele for further analyses.
Fig. 1. kor1 mutant roots display increased JA-Ile levels and signaling.
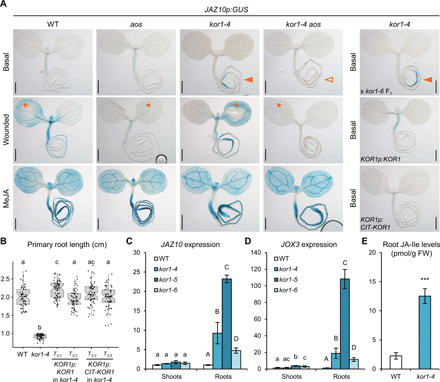
(A) Representative JAZ10p:GUS (JGP) reporter activity in 5-do seedlings of WT, aos, kor1-4, kor1-4 aos at basal conditions, 2 hours after cotyledon wounding (orange asterisks), 2 hours after 25 μM MeJA treatment, and in kor1-4 x kor1-6 F1 (allelism test), kor1-4 complemented with KOR1p:KOR1 and KOR1p:CIT-KOR1 lines. Note the increased JAZ10p:GUS reporter activity in kor1-4 and allelism test (orange arrowheads) and its absence from kor1-4 aos (empty arrowhead). Scale bars, 0.5 mm. (B) Primary root length box plot summary in 7-do WT, kor1-4, and two independent T3 lines for each complementing KOR1p:KOR1 and KOR1p:CIT-KOR1 construct. Both constructs restored the kor1-4 short root phenotype to WT length. Medians are represented inside the boxes by solid lines, and circles depict individual measurements (n = 59 to 61). (C and D) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) of basal (C) JAZ10 and (D) JOX3 expression in shoots and roots of WT and three kor1 mutant alleles. JAZ10 and JOX3 transcript levels were normalized to those of UBC21. Bars represent the means of three biological replicates (±SD), each containing a pool of ~60 organs from 5-do seedlings. (E) Absolute JA-Ile content in WT and kor1-4 roots. Bars represent the means of three biological replicates (±SD), each containing a pool of ~600 roots from 5-do seedlings. Letters and asterisks denote statistically significant differences among samples as determined by analysis of variance (ANOVA) followed by Tukey’s honest significant difference (HSD) test (P < 0.05) in (B) to (D) and by Student’s t test (P = 0.0005) in (E).
Despite the broad KOR1 expression domain and stunted shoot growth phenotypes of characterized mutants (26), constitutive JGP activation was not detected in aerial tissues of kor1 alleles (Fig. 1A and fig. S1A). Shoot wounding and exogenous MeJA treatment promptly induced JGP reporter expression across kor1 tissues, validating the root specificity of basal JGP activity (Fig. 1A and fig. S1A). Quantitative analysis of JAZ10 transcripts further supported the specific activation of JA signaling in roots but not shoots of all three kor1 alleles (Fig. 1C). This phenotype was dependent on JA-Ile production, as increased reporter activity and JAZ10 transcript levels were abolished in JA-deficient kor1 aos double mutants (Fig. 1A and fig. S1, A, E, and F). In addition to JAZ10, kor1-4 exhibited increased root expression of JA marker transcripts JASMONATE OXYGENASE 3 (JOX3) and JAZ3 in a JA-dependent manner, as well as increased levels of the bioactive JA-Ile conjugate (Fig. 1, D and E, and fig. S1G). Collectively, our data indicate that KOR1 is a negative regulator of root JA-Ile biosynthesis. Hence, kor1 mutants represent valuable genetic tools to study how cell wall–derived signals are integrated with intracellular hormone production and uncover JA-Ile functions in moderating root responses to cellulose deficiency.
Cortex-specific KOR1 expression complements ectopic JA signaling in kor1 endodermis and pericycle
To map the precise tissues and cell types displaying ectopic JA signaling in kor1-4 roots, we used a transcriptional reporter expressing three VENUS (3xVEN) fluorescent proteins fused N-terminally to a NUCLEAR LOCALIZATION SIGNAL (NLS) under the control of JAZ10p (JAZ10p:NLS-3xVEN) (27). Similar to JGP, JAZ10p:NLS-3xVEN expression was not detectable at basal conditions but increased notably after MeJA treatment in both WT and aos roots, while mechanical wounding triggered JAZ10p:NLS-3xVEN induction in the WT but not in aos (Fig. 2A and fig. S2). In contrast to the WT, kor1-4 exhibited constitutive JAZ10p:NLS-3xVEN expression predominantly in the early differentiation zone of the primary root, mostly confined to endodermal and pericycle cells (Fig. 2, A to C). The extent of activated JA signaling was quantified by evaluating the presence of JAZ10p:NLS-3xVEN along longitudinal cell files and displaying the resulting frequency in a heatmap for each cell layer (Fig. 2D). Weak reporter activation coincided with the onset of cell elongation and proceeded longitudinally into the early differentiation zone for approximately 30 cells before ceasing. While only <10% of kor1-4 roots showed sporadic JAZ10p:NLS-3xVEN expression in a few epidermal or cortex cells, the majority of individuals displayed consistent JA signaling in a stretch of 10 to 15 endodermal and pericycle cells in the root early differentiation zone (Fig. 2D).
Fig. 2. Cortex-specific expression of CIT-KOR1 complements increased JA signaling in endodermis and pericycle of kor1-4.
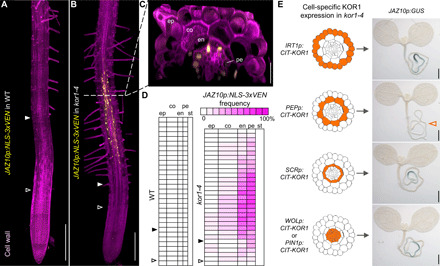
(A to C) JAZ10p:NLS-3xVEN expression in 5-do (A) WT and (B and C) kor1-4 roots cleared with ClearSee, counterstained with the cellulose dye Direct Red 23, and visualized as three-dimensional Z-stacks. (C) Orthogonal view from the epidermis to the vascular cylinder of kor1-4. The onset of elongation is indicated by empty arrowheads (first elongated cortex cell), and that of differentiation by filled arrowheads (appearance of root hairs). ep, epidermis; co, cortex; en, endodermis; pe, pericycle. (D) Heatmap of JAZ10p:NLS-3xVEN frequency in individual cells from WT and kor1-4 primary roots (n = 21). Presence or absence of the reporter was evaluated from the onset of elongation in individual cells along consecutive longitudinal files for each tissue layer. Reporter expression was not observed in the WT nor in kor1-4 vascular tissues of the stele (st). (E) Cell-specific complementation of JAZ10p:GUS (JGP) activity in kor1-4 by expressing CIT-KOR1 in either epidermis (IRT1p:CIT-KOR1), cortex (PEPp:CIT-KOR1), endodermis (SCRp:CIT-KOR1), or stele (WOLp:CIT-KOR1 or PIN1p:CIT-KOR1). Note that cortex-expressed CIT-KOR1 complements JAZ10p:GUS activity in kor1-4, as indicated by lack of the JAZ10p:GUS reporter in representative images from T3 lines (empty orange arrowhead). A minimum of 30 T3 individuals were analyzed for each line. Scale bars, 200 μm (A and B), 30 μm (C), and 0.5 mm (E).
Because JA-Ile precursors can relocate across tissues (28, 29), increased JA signaling in different cell types could result from cell-autonomous or non–cell-autonomous signals. To discriminate between these two possibilities and identify the source tissue responsible for increased JA-Ile biosynthesis, we drove the expression of a functional CIT-KOR1 fusion protein under the control of cell type–specific promoters and evaluated its capacity to complement ectopic JGP expression in kor1-4. As expected, when driven by its endogenous promoter (KOR1p), CIT-KOR1 expression was detectable across the entire root, and respective cell type–specific promoters resulted in CIT-KOR1 expression at the intended locations: IRON-REGULATED TRANSPORTER 1 (IRT1p) in the epidermis, PLASTID ENDOPEPTIDASE (PEPp) in the cortex, SCARECROW (SCRp) in the endodermis, and WOODEN LEG 1 (WOL1p) or PIN-FORMED 1 (PIN1p) in the stele, which includes the pericycle (fig. S3, A to E) (26, 30). Intriguingly, ectopic JGP expression in kor1-4 was complemented only when expressing CIT-KOR1 in the cortex (PEPp:CIT-KOR1), but not in the epidermis (IRT1p:CIT-KOR1), nor in endodermal (SCRp:CIT-KOR1) or pericycle (WOLp:CIT-KOR1 or PIN1p:CIT-KOR1) cells that exhibited high JA signaling (Fig. 2E). Furthermore, the short root kor1-4 phenotype was ameliorated by expressing CIT-KOR1 in the cortex, while it remained unchanged when expressing the construct in the epidermis, and it was even slightly exacerbated by cell type–specific expression in the endodermis or pericycle (fig. S3F). These results suggest that maintaining KOR1 functionality in the cortex is critical for impeding the activation of JA-Ile production in adjacent endodermal and pericycle cells.
Mutations in ESMD1 suppress elevated JA signaling levels in kor1
To identify genetic regulators of JA-Ile production in kor1, we performed an EMS suppressor screen in kor1-4 JGP for lack of JA reporter activation. Mapping by whole-genome-sequencing of bulk segregants isolated an allele in ESMERALDA1 (ESMD1), which we named esmd1-3. The esmd1-3 allele abolished the constitutive JGP expression and high JAZ10 levels in kor1-4 roots while retaining the capacity to induce JAZ10 transcripts after wounding (Fig. 3, A and B, and fig. S4, A to C). An ESMD1-mTURQUOISE2 (mT) fusion protein expressed under the ESMD1p native promoter restored the ectopic JA signaling in kor1-4 esmd1-3 (Fig. 3A and fig. S4A). Consistently, introgressing another esmd1-1 EMS allele (31) into kor1-4 partially suppressed the elevated JAZ10 levels in kor1-4 (Fig. 3B), confirming the suppressor’s identity and causative amino acid R373C substitution in esmd1-3 (fig. S4D). As we could not retrieve homozygous mutants from a segregating T-DNA population in ESMD1 (esmd1-4) (fig. S4D), and esmd1-1 was a weaker JA suppressor than esmd1-3, it is likely that full loss of ESMD1 function leads to lethality and that both EMS alleles used here are partial loss of function. ESMD1 is a member of the plant-specific glycosyltransferase GT106 family, with putative O-fucosyltransferase activity involved in regulating pectin homeostasis (31, 32). Specifically, esmd1 was identified as a suppressor of the stunted growth phenotypes of mutants deficient in the major pectin constituent homogalacturonan (HG) (31). Because esmd1 restored the cell-to-cell adhesion defects without complementing the low HG content of pectin mutants quasimodo1 (qua1, a putative galacturonosyltransferase of the glycosyltransferase GT8 family) and quasimodo2 (qua2, a methyltransferase) (31, 33–35), we wondered whether, in addition to suppressing JA signaling, esmd1-3 was able to revert other kor1-4 phenotypes.
Fig. 3. ESMD1 is a positive regulator of ectopic JA-Ile production in kor1.
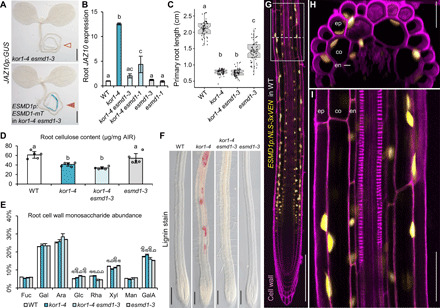
(A) JAZ10p:GUS expression is absent in kor1-4 esmd1-3 (empty arrowhead) and present in its ESMD1p:ESMD1-mTurquoise (mT) complementation line (orange arrowhead). (B) qRT-PCR of basal JAZ10 expression in roots of indicated genotypes. JAZ10 transcript levels were normalized to UBC21. Bars represent the means of three biological replicates (±SD), each containing ~60 roots from 5-do seedlings. (C) Primary root length box plots from 7-do seedlings of indicated genotypes. Medians are represented inside the boxes by solid lines, and circles depict individual measurements (n = 58 to 66). (D) Crystalline cellulose content from alcohol-insoluble residue (AIR) extracted from roots of indicated genotypes. Bars represent means of five biological replicates depicted as circles (±SD), each consisting of pools from ~300 roots from 12-do seedlings. (E) Cell wall monosaccharide composition analysis from root-extracted AIR of indicated genotypes. Bars represent the means of three biological replicates (±SD). Only statistically significant differences among genotypes indicated with letters and assessed for each individual sugar are shown above bars. Fuc, fucose; Gal, galactose; Ara, arabinose; Glc, glucose; Rha, rhamnose; Xyl, xylose; Man, mannose; GalA, galacturonic acid. (F) Lignin deposition visualized by phloroglucinol stain (fuchsia) in primary roots of indicated genotypes. (G to I) ESMD1p:NLS-3xVEN expression in 5-do WT primary roots counterstained with PI. (H) Orthogonal view from epidermis to vascular cylinder of a Z-stack section in the early differentiation zone through (G) (dotted line). (I) Increased magnification from (G) (boxed). ep, epidermis; co, cortex; en, endodermis. Letters in (B) to (E) denote statistically significant differences among samples as determined by ANOVA followed by Tukey’s HSD test [P < 0.05 and P < 0.001 in (E)]. Scale bars, 0.5 mm (A), 200 μm (F and G), 25 μm (H), and 50 μm (I).
Contrary to ameliorating qua1 and qua2 growth phenotypes (31), esmd1-3 did not restore the short root length nor the cellulose deficiency in shoots and roots of kor1-4 seedlings (Fig. 3, C and D, and fig. S4E). On the basis of previous reports (31), we expected similar neutral and acidic sugar profiles from cell walls of genotypes with esmd1-3. Instead, monosaccharide analysis of cell walls from shoots and roots revealed some significant changes in matrix polysaccharide composition between WT and certain mutants (Fig. 3E and fig. S4F). Most notably, the esmd1-3 mutation reduced rhamnose abundance (by 32 to 38% in roots and 15% in shoots) with respect to the WT (Fig. 3E and fig. S4F). Because cellulose deficiency often leads to ectopic lignification (36), we next evaluated lignin deposition in our mutant set. Phloroglucinol staining revealed patches of lignified cells across the primary root of kor1-4 that were absent in WT and aos plants (Fig. 3F and fig. S4G). The ectopic lignification in kor1-4 was not a consequence of elevated JA levels as kor1-4 aos still exhibited lignified cells, and was entirely abolished in kor1-4 esmd1-3. Overall, the cell wall analysis indicated an elaborate compensatory network in kor1 esmd1. The kor1 mutation was epistatic to esmd1 in terms of short root length and cellulose deficiency, while esmd1 was epistatic to kor1 with respect to lack of increased JA signaling, reduced rhamnose abundance, and lack of ectopic lignification. The results therefore suggest that increased JA signaling in kor1 may be due to indirect consequences of cellulose deficiency on cell wall architecture and mechanical properties, rather than to altered levels of a specific cell wall component.
To gain further insights into how ESMD1 may influence JA responses in kor1, we concentrated on its localization in the primary root. Although an ESMD1–green fluorescent protein fusion protein was expressed in the Golgi when transiently overexpressed in leaf epidermal cells of Nicotiana benthamiana (31), we were unable to visualize the functional ESMD1p:ESMD1-mT construct nor ESMDp:ESMD1-CIT in WT Arabidopsis roots, suggesting that ESMD1 levels are very low and/or tightly regulated. Instead, ESMD1p:NLS-3xVEN expression was mapped to older parts of the root as expected (31) and to the epidermis, cortex, and endodermis of the early differentiation zone (Fig. 3, G to I).
Enlarged kor1 cortex cells affect JA production in inner tissues
Having found that ESMD1 expression includes our zone of interest, we further noticed that kor1-4 esmd1-3 roots are considerably thinner than kor1-4 (Fig. 4, A and B). Transversal sections across the root early differentiation zone coinciding with the sites of JA production in kor1-4 revealed that root thickness in the mutant was twice that of the WT and resulted from enlarged areas of all cell types examined (Fig. 4, C and D, and fig. S5, A to F). The relatively small expansion of kor1-4 epidermal cell area (1.1-fold) was counterbalanced by a significant increase in their cell number (fig. S5, A and C). Although aberrant cell divisions were also occasionally observed in kor1-4 cortex and endodermal cells, the overall cell number in these cell types did not differ significantly from the WT (fig. S5A). The largest augmentation in kor1-4 cell area was found for cortex cells, being 2.6-fold larger than WT, while endodermal and pericycle cells displayed an expansion of 2- and 2.4-fold, respectively (Fig. 4, C and D, and fig. S5, C to F). These kor1-4 phenotypes were almost completely restored to WT levels in the kor1-4 esmd1-3 suppressor, which showed a diminished root thickness; a full complementation of epidermal cell numbers; and reduced cell areas of cortex, endodermis, and pericycle cells (Fig. 4, A to D, and fig. S5, A to F). Notably, the large expansion of kor1-4 cortex cells was drastically reduced in kor1-4 esmd1-3 (Fig. 4, C and D), leading us to hypothesize that enlarged cortex cells may impinge upon inner tissues.
Fig. 4. Cortex cell enlargement in kor1 correlates with constitutive JA production.
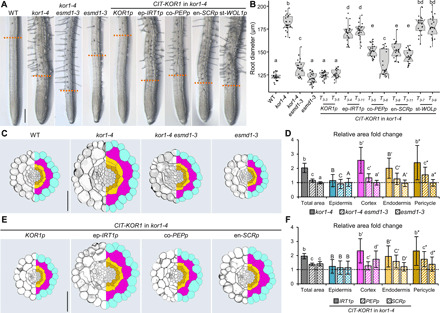
(A) Representative primary root images of WT, kor1-4, kor1-4 esmd1-3, esmd1-3, and kor1-4 complemented with CIT-KOR1 under the control of its native KOR1p promoter, or epidermis (ep-IRT1p)–, cortex (co-PEPp)–, endodermis (en-SCRp)–, or stele (st-WOL1p)–specific promoters. Orange dashed lines define the initiation of the differentiation zone, as indicated by the appearance of root hairs in 7-do seedlings. (B) Box plot summary of primary root diameter at the onset of differentiation in 7-do seedlings of indicated genotypes. Medians are represented inside the boxes by solid lines, and circles depict individual measurements (n = 22 to 23). (C to F) Anatomy and cell size comparisons from transverse sections across the early differentiation zone of the primary root in (C and D) WT, kor1-4, kor1-4 esmd1-3, and esmd1-3, and in (E and F) kor1-4 complemented with cell type–specific promoters. (C and E) Representative split images from cross sections (left) and respective cell segmentations (right). Segmented cell types are color-coded as: epidermis, turquoise; cortex, magenta; endodermis, yellow; pericycle, mustard; stele, gray. (D and F) Fold change in total and cell-specific areas from segmented transversal root sections in indicated genotypes. Bars represent mean fold changes of individual cellular areas from 10 to 11 roots (±SD), normalized to values of the (D) WT or (F) KOR1p:CIT-KOR1 indicated by dashed lines (individual measurements are available in fig. S5). Letters in (B), (D), and (F) denote statistically significant differences among samples as determined by ANOVA followed by Tukey’s HSD test (P < 0.05). Scale bars, 200 μm (A) and 50 μm (C and E).
To verify whether kor1-4 enlarged cortex cells indeed affected JA-Ile production in inner epidermal and pericycle tissues, we analyzed our transgenic lines expressing CIT-KOR1 under cell type–specific promoters in transversal root sections coinciding with ectopic JA production (Fig. 2). As expected, expressing CIT-KOR1 under its native promoter fully restored kor1-4 root thickness to WT dimensions, while epidermis-, cortex-, and endodermis-specific CIT-KOR1 expression reduced kor1-4 root thickness to various degrees. Stele-specific CIT-KOR1 expression (st-WOLp) did not have any significant effect and was thus excluded from further analysis (Fig. 4, A and B). Compared to the KOR1p:CIT-KOR1 complemented kor1-4 line, epidermal CIT-KOR1 expression (ep-IRT1p) still exhibited typical kor1-4 features of twofold enlarged root area; increased epidermal cell number; and enlarged areas of cortex, endodermal, and pericycle cells (Fig. 4, E and F, and fig. S5, G to L). Conversely, expressing CIT-KOR1 in either cortex (co-PEPp) or endodermal (en-SCRp) cell layers rendered kor1-4 phenotypes more similar to the KOR1p:CIT-KOR1 complemented kor1-4 line by showing a 1.4-fold increase in total root area and only a milder enlargement of cortex, endodermis, and pericycle cells (Fig. 4, E and F, and fig. S5, G to L). While the most prominent effect of expressing CIT-KOR1 in cortex or endodermis was to reduce the area of cells in which the fusion protein was localized, the strongest correlation between JA signaling and cell type area was again found for the cortex cell layer. Expressing CIT-KOR1 in the cortex (co-PEPp) abolished kor1-4 JGP expression and restored cortex cell area to almost WT levels (1.2-fold) without fully recovering cellular enlargement of endodermal and pericycle cells, which persisted being 1.6- and 1.7-fold larger than the KOR1p:CIT-KOR1 complemented line (Figs. 2E and 4, E and F). In turn, endodermal CIT-KOR1 expression (en-SCRp) did not abolish elevated JA levels in kor1-4 nor led to a drastic reduction in cortex expansion that remained 1.7-fold larger, albeit almost completely restoring endodermal and pericycle cell areas (Figs. 2E and 4, E and F). As KOR1 activity in cortex cells is important to regulate their size and JGP expression in adjacent inner tissues, and ESMD1 is expressed in both cortex and endodermal cells, it is conceivable that the cortex-endodermis interface is critical for governing JA-Ile production in kor1.
Osmotically driven turgor pressure changes regulate JA-Ile signaling in kor1 and WT
JA responses can be triggered by exogenous treatment with specific pectin- and cellulose-derived fragments serving as ligands binding putative plasma membrane receptors and activating intracellular signaling, reviewed in (11). More notoriously, JA-Ile biosynthesis is swiftly activated by mechanical stress (24). The morphological changes in cellular areas across root transversal sections (Fig. 4) led us to hypothesize that increased JA-Ile levels in kor1 may result from enlarged cortex cells “squeezing” spatially constrained inner tissues, rather than from cell wall–derived elicitors. To test this hypothesis, we decreased intracellular turgor pressure by growing kor1-4 seedlings under hyperosmotic conditions to withdraw water from their cells. All tested substances acting as osmotica [mannitol, sorbitol, polyethylene glycol (PEG), hard agar] effectively abolished JA signaling in kor1-4 roots (Fig. 5, A and B, and fig. S6A). Mannitol-grown seedlings still responded to wounding, reinforcing the tissue specificity of the JGP reporter (fig. S6B). Furthermore, while WT root length decreased under hyperosmotic conditions, kor1-4 root elongation exhibited a tangible amelioration (Fig. 5C). This was also reflected in the reduced area of kor1-4 transversal root sections grown on mannitol reverting to WT size (Fig. 5, D and E). Consistently, mannitol treatment restored kor1-4 epidermal cell number as well as enlarged epidermal, cortex, endodermis, and pericycle cell size (Fig. 5, D and E, and fig. S6, C to H). Notably, the hyperosmotic treatment fully recovered the enlarged kor1-4 cortex cell area to WT levels, while endodermal and pericycle cells were still 1.2-fold larger with respect to the WT (Fig. 5, D and E). Thus, the activation of JA production in kor1 is consistent with inner tissues being mechanically stressed by enlarged cortex cells.
Fig. 5. Osmotic support abolishes constitutive JA production in kor1, while hypoosmotic treatment triggers JA-Ile signaling in WT.
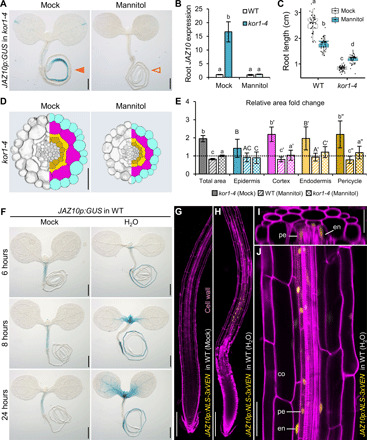
(A) Constitutive JAZ10p:GUS activity in kor1-4 seedlings (orange arrowhead) is abolished following growth in 3% mannitol-supplemented media (empty arrowhead). (B) qRT-PCR of basal JAZ10 root expression of indicated genotypes grown without (mock) or with mannitol. JAZ10 levels were normalized to UBC21. Bars represent the means of three biological replicates (±SD), each containing pools of ~60 5-do roots. (C) Root length box plots of 7-do WT and kor1-4 grown without or with mannitol. Medians are solid lines inside the boxes, and circles depict n = 59 to 61. Mannitol reduced WT root length by −0.78 cm, whereas it increased kor1-4’s by +0.35 cm (linear model, P = 2 × 10−6). (D) Representative split images from kor1-4 root cross sections (left) and respective cell segmentations (right) grown without or with mannitol. (E) Total and cell-specific area fold change from segmented transversal root sections of WT and kor1-4 grown without or with mannitol. Bars represent mean fold changes of individual cellular areas from 10 roots (±SD), normalized to mock-treated WT indicated by a dashed line (individual measurements are in fig. S6). (F) JAZ10p:GUS expression in WT seedlings submerged in isotonic solution (liquid MS, mock) or deionized water (H2O) for the indicated time. (G to J) JAZ10p:NLS-3xVEN expression in WT roots submerged in (G) mock solution or (H) H2O for 24 hours and counterstained with PI. (I) Orthogonal and (J) longitudinal view from the root early differentiation zone in (H). co, cortex; en, endodermis; pe, pericycle. Letters in (B), (C), and (E) denote statistically significant differences among samples as determined by ANOVA followed by Tukey’s HSD test (P < 0.05). Scale bars, 0.5 mm (A and F), 50 μm (D and J), 200 μm (G and H), and 30 μm (I).
We then reasoned that changes in turgor pressure leading to cellular enlargement, such as those driven by hypoosmotic treatments that stimulate cellular water uptake, may cause mechanical stress within tissues and result in JA-Ile biosynthesis even in WT plants. Transferring WT JGP reporter seedlings to deionized water indeed resulted in reporter activation, which increased over time and encompassed the root and portions of the shoot apical meristem (Fig. 5F). Because the JGP reporter is suited to reveal JA-Ile signaling in tissues but not to map cellular expression sites (6), we verified which cells activated JA-Ile signaling with the JAZ10p:NLS-3xVEN reporter. While a 24-hour treatment in isotonic mock solution did not affect JAZ10p:NLS-3xVEN expression, hypoosmotic treatment severely damaged the root apical meristem (RAM) as indicated by propidium iodide (PI) penetration and activated the reporter in internal root tissues (Fig. 5, G and H). Notably, although JAZ10p:NLS-3xVEN expression can be induced in every cell type of the primary root (fig. S2), the hypoosmotic treatment triggered JA-Ile signaling predominantly in endodermal and pericycle cells starting at the root early differentiation zone (Fig. 5, H to J). Overall, our data indicate that osmotically driven turgor pressure changes have a remarkable impact on JA-Ile production.
Heightened JA levels guide kor1 roots toward greater water availability
The activation of JA signaling often leads to an induction in defense responses accompanied by a reduction in organ growth (37). kor1-4 root meristem length was significantly shorter than the WT, while its cell number remained unchanged (fig. S7, A to C). It is thus possible that increased JA production in kor1 contributes to slow down the mutant’s root growth rate to better adjust to cellulose deficiency and concomitantly protect the mutant from potential threats found in the soil. However, blocking JA production in kor1-4 did not alter root elongation nor the expression of JA-dependent defense marker genes, as the mutant had the same root growth rate as its JA-deficient kor1-4 aos counterpart and similar low basal levels of VSP2 and PDF1.2 transcripts (Fig. 6A and fig. S7, D and E).
Fig. 6. Increased JA production facilitates kor1 response to root hydrotropism.
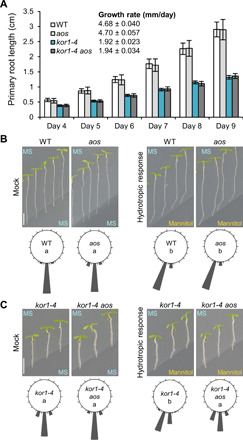
(A) Primary root elongation of indicated genotypes between 4 and 9 days after germination. Bars represent the means of 40 to 50 plants. Data were used to determine the root growth rate in millimeter per day by linear regression. (B and C) Root hydrotropic response of (B) WT and aos and (C) kor1-4 and kor1-4 aos seedlings. Representative images and circular histograms summarizing root curvatures of indicated genotypes 24 hours after transfer to split-agar Murashige and Skoog (MS) plates under mock (MS/MS) or hydrotropism-inducing (MS/400 mM mannitol) conditions. Bars indicate the percentage of seedlings exhibiting a root bending angle assigned to one of the 1820° sectors on the circular diagram [individual measurements are available in fig. S7 (D and E)], n = 42. Letters indicate statistically significant differences as determined by two-way ANOVA followed by Tukey’s HSD test (P < 0.05). Scale bars, 5 mm.
As the stunted kor1-4 root growth was alleviated by continuous growth in mannitol (Fig. 5C), we tested whether the mutant might preferentially grow toward hyperosmotic conditions in a split-agar assay used for measuring root hydrotropic responses among genotypes elongating at similar growth rates (38). Following transfer to split-agar plates with mannitol harboring asymmetric water availability, both WT and aos seedling roots readjusted their growth away from the hyperosmotic media in search for greater water availability (Fig. 6B and fig. S7, F and G). Contrary to our expectations, kor1-4 roots also bent away from mannitol with a positive hydrotropic root curvature despite their slower growth rate. To our surprise, the JA-deficient kor1-4 aos double mutant failed to effectively readjust its root growth direction toward a greater water availability and instead grew into the mannitol media (Fig. 6C and fig. S7H). This effect was not a consequence of differential root thickness or the double mutant’s inability to undergo general root bending responses such as those triggered by root gravitropism, but rather to a specific insensitivity to root hydrotropism (fig. S7, I to L). Consistently, while esmd1-3 roots showed a normal root hydrotropic response, the kor1-4 esmd1-3 double mutant with abolished constitutive root JA-Ile production was unable to redirect its root growth away from the mannitol media (fig. S7, M to P). Our data thus unveiled that the constitutive activation of JA-Ile production and signaling in kor1-4 serves to facilitate root water foraging.
DISCUSSION
Plant cell walls are defining plant structures that serve a wide range of biological functions including shaping cell morphology, providing mechanical support for growth, and connecting cells within tissues. Changes in cell wall structure and assembly may have pronounced repercussions on intracellular and whole-plant responses (21). Here, we investigated how cues derived from perturbed cell walls integrate with the production of the stress hormone JA-Ile. The identification of cellulose-deficient kor1 alleles displaying elevated JA-Ile levels specifically in seedling roots prompted us to examine cell wall–triggered induction of hormone biosynthesis in cell type–specific contexts. By restoring KOR1 function in specific layers of the primary root, we showed that ectopic JA-Ile signaling in endodermal and pericycle cells was caused by cell-nonautonomous signals deriving from adjacent and markedly enlarged cortex cells. Swollen kor1 cortex cells could exert an increased mechanical pressure upon both external epidermal cells and inner tissues. As JA-Ile production is readily triggered by mechanical stress, why was the hormone increase observed only in inner tissues?
A possibility is that epidermal cells could dissipate the cortex-exerted mechanical pressure by expanding toward the available outer space and by stimulating cell division. Mechanical cues are known to influence cell cycle progression in animals, and although this has not yet been demonstrated in plants, they guide the orientation of plant cell division planes (20, 39). Contrary to outer tissues, endodermal and pericycle cells are physically constrained and may become mechanically compressed by enlarged cortex cells. Reducing the swollen kor1 cortex cell size in three independent manners fully abolished increased JA-Ile signaling in inner tissues. First, restoring KOR1 activity in the cortex complemented both cortex cell size and JA-Ile marker gene expression. Second, we identified a genetic suppressor of kor1 that affected cell wall properties in which cortex cell size and JA-Ile levels were also restored. Third, by reducing kor1 turgor pressure with hyperosmotic treatments, enlarged cortex cells and JA signaling were fully recovered. Our findings might explain why wounding single cells by laser ablation in living roots does not induce JA-Ile signaling (27). The disintegration of a target cell provokes adjacent cells to bulge toward the vacuity left by the ablated cell, in a process compatible from turgor pressure changes in adjacent cells due to the vanished support from the damaged cell (40, 41). However, similarly to epidermal cells in kor1, wound-adjacent cells are not subjected to compression and might be able to compensate for the increased mechanical stress by expanding toward the void space without activating JA production. In addition to mechanical compression, a concomitant cause of JA-Ile production may reside in the mechanical tension generated between adjacent cells during cell elongation, possibly causing stretching imbalances in growing cells. In fact, JA-Ile signaling in kor1-4 was observed immediately after the elongation zone and included sporadic activation in epidermal and cortical cells undergoing anisotropic extension. Collectively, the data demonstrate that the JA pathway is triggered by turgor pressure–driven mechanical changes in response to cellulose deficiency.
Increases in turgor pressure by hypoosmotic treatment caused the induction of JA-Ile signaling also in inner tissues of WT roots, implying that turgor pressure–driven mechanical stress may be a general elicitor of JA-Ile production. Mechanical wounding and chewing insect herbivores squash tissues and possibly compress spatially constrained cells adjacent to wounds. Because cells within tissues are connected through their cell walls, mechanical stress signals arising from turgor pressure changes may propagate over distances depending on the extent of damage (42). Consistently, it has been proposed that mechanical tissue damage causes sudden pressure changes in vascular xylem vessels, leading to the compression of cells adjacent to vessels where JA biosynthesis is then initiated (24, 43). Hence, turgor pressure changes generating mechanical compression may be a crucial elicitor of JA-Ile biosynthesis in circumstances extending beyond cell wall perturbations, with putative osmo- and mechanosensors located across different subcellular compartments (44). However, measuring mechanical stress patterns in vivo, particularly from cells embedded in inner tissues that are not physically accessible, remains challenging (45). Emerging tools are offering encouraging prospects to assess mechanobiological processes in vivo in future studies (46).
In our search to identify genetic regulators of ectopic JA-Ile production in kor1 with potential roles in stress signals arising from turgor pressure changes, we did not retrieve known components involved in osmoregulation such as mechanically activated ion channels OSCA1 or MSL10, nor cell wall integrity sensors such as THESEUS1 whose loss of function was insensitive to isoxaben-induced JA production (25, 47–49). This might be due to the screen not being saturated or to genetic redundancy. Instead, the kor1 suppressor screen recovered an allele in ESMD1, a putative O-fucosyltransferase involved in pectin homeostasis (31). Although ESMD1’s substrate and biochemical function are unknown, it was proposed that it may O-fucosylate cell wall integrity sensor proteins and thereby contribute to cell signaling (31). Recently, two enzymes belonging to the same GT106 family were shown to be rhamnosyltransferases involved in the biosynthesis of the pectin component rhamnogalacturonan I (RG-I) (32, 50). Given that our monosaccharide composition analysis in genotypes with esmd1-3 showed a substantial rhamnose reduction, which in Arabidopsis derives mostly from RG-I (51), and in agreement with other GT106 family members (32, 50), ESMD1 may be directly involved in pectin biosynthesis. Although it remains unclear how esmd1 restores cell shape and JA-Ile levels in kor1, RG-I can have significant and specific contacts with cellulose microfibrils (52). As turgor pressure is counterbalanced by cell walls being under tension (53), it is plausible that the diminished cell wall tensile strength in kor1 is compensated by lack of ESMD1. The identification of ESMD1 also indicates that cell size and mechanical compression on inner tissues can be alleviated by remodeling cell wall properties and thus reequilibrate turgor pressure.
An unexpected observation was that constitutive JA-Ile signaling enabled kor1 roots to modify their growth toward greater water availability. This finding highlights the importance of characterizing cell wall mutants as they may provide opportunities to uncover more subtle functions of the JA pathway. While the root hydrotropic response of the JA-deficient aos mutant did not differ from the WT, the suppression of constitutive JA-Ile production in kor1-4 aos and kor1-4 esmd1-3 prevented roots to grow away from the media with reduced water availability. These findings imply that while JA deficiency per se does not affect root hydrotropism, ectopic JA-Ile signaling in precise tissues and cell types is beneficial for root water foraging. The heightened activation of JA signaling in kor1 did not result in typical JA-mediated growth inhibition responses (54, 55), suggesting that manipulating JA biosynthesis in specific cell types might confer acclimation advantages without hampering overall growth. As abscisic acid (ABA) perception in root cortical cells is required for WT root hydrotropic responses (38), ABA biosynthesis or signaling may be compromised in kor1-4 cortex and counteracted by constitutive JA-Ile production to redirect root growth. Hyperosmotic mannitol treatments are also used to study plant responses to water deficiency, and applying exogenous JA to various plants renders them more tolerant to drought, reviewed in (56). Accordingly, Arabidopsis mutants impaired in JA production or signaling are more drought-susceptible (57). Although it is unknown how constitutive JA signaling guides kor1 roots away from mannitol, future studies deciphering the molecular mechanisms governing this effect could have beneficial impacts on breeding programs aimed at increasing plant drought tolerance.
MATERIALS AND METHODS
Plant material and growth conditions
A. thaliana accession Columbia (Col) was used for all experiments. JAZ10p:GUSPlus (JGP) transgenic lines in Col (WT) and in the JA-deficient aos mutant background were described previously (6). JAZ10p:NLS3xVEN was as in (27) but cloned and crossed into aos de novo. kor1-6 (SALK_075812) and esmd1-4 (GABI_216D03) were obtained from the Nottingham Arabidopsis Stock Centre (NASC), and kor1-6 was crossed into JGP and aos JGP. esmd1-1 was described in (31). For in vitro assays, seeds were sterilized and stratified 2 days at 4°C in the dark. Seedlings were grown on 0.5× solid Murashige and Skoog (MS; Duchefa) medium supplemented with MES hydrate (0.5 g/liter; Sigma-Aldrich) and 0.7 or 0.85% plant agar (AppliChem) for horizontal or vertical growth, respectively. Horizontally grown seedlings were germinated on a nylon mesh placed on top of the MS media as described (6). Controlled growth conditions were set at 21°C under light (100 μE m−2 s−1), with a 14-hour light/10-hour dark photoperiod. For soil propagation, transformation, and crossing, plants were grown with the same temperature (T) and light intensity, but under continuous light.
Histochemical detection of GUS activity and lignin deposition
GUS stainings were performed as described (58) on a minimum of 20 individuals across experiments, and seedlings were photographed with a Leica M165 FC stereomicroscope fitted with a Leica MC170 HD camera. Lignin deposition was visualized by submerging seedlings in acidified phloroglucinol solution (1% phloroglucinol in 18% HCl) for 5 min, washing in 1× phosphate-buffered saline buffer, mounting in 10% glycerol, and imaging using differential interference contrast optics on a Leica DM6B microscope fitted with a Leica DMC6200 camera.
Plant treatments
Single cotyledon wounding of seedlings and MeJA (Sigma-Aldrich) treatments to assess JGP reporter activity were performed as described (6). To evaluate the JA response of the JAZ10p:NLS-VEN reporter in individual seedlings grown vertically, primary roots were mounted in mock or 10 μM MeJA 0.5× MS with PI (30 μg/ml; Sigma-Aldrich) solution, imaged immediately (t = 0) and after 2 hours (n = 10). For osmotic support experiments, plant growth media was supplemented with either 3% mannitol (165 mM; J&K Scientific), 3% sorbitol (165 mM; Carl Roth), 3% PEG 6000 (Serva), or 3% plant agar (AppliChem). For hypoosmotic treatments, seedlings were grown vertically for 5 or 7 days on MS media and then transferred to liquid isotonic solution (MS) or deionized water for the indicated times before GUS staining (n = 20) or confocal laser scanning microscopy (LSM) imaging (n = 10).
Gene expression analyses
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR) of JAZ10 (At5g13220), JOX3 (At3g55970), VSP2 (At5g24770), and UBC21 (At5g25760) were performed as described (29, 58). Similarly, the expression of JAZ3 (At3g17860) was assayed with GACTCGGAGCCACAAAAGC and TACGCTCGTGACCCTTTCTTTG and of PDF1.2 (At5g44420) with TTTGCTGCTTTCGACGCAC and GCATGATCCATGTTTGGCTCC primers. RT-PCR of KOR1 was performed with the primer pair AGATGCTGAAGCCAGAGCAG and TGTCATGGAGAGGTAATTCTGG.
JA-Ile quantification
5-do roots were excised beneath the collet region and flash-frozen to yield approximately 50-mg FW for each biological replicate. Extraction and quantitative measurements were performed as described (29). The limit of JA-Ile quantification (LOQ = 3× limit of detection) was determined from an Arabidopsis matrix as 0.49 pmol/g FW.
Cloning and generation of transgenic lines
All transcriptional and translational reporter constructs were generated by double or triple Multisite Gateway Technology (Thermo Fisher Scientific). ENTRY plasmids containing cell type–specific promoters (pEN-L4-IRT1p-L3, pEN-L4-PEPp-L3, pEN-L4-SCRp-L3, pEN-L4-WOLp-L3, and pEN-L4-PIN1p-L3) were described in (30) and obtained from NASC. pEN-L4-JAZ10p-R1, pEN-L1-NLS-3xVEN-L2, and pEN-R2-CIT-L3 were as in (6, 58). CIT and mTurquoise (mT) fluorophores were subcloned into pDONR221 or pDONR-P2R-P3 to obtain pEN-L1-CIT-L2 and pEN-R2-mT-L3. Promoters were amplified from WT genomic DNA with oligonucleotides containing adequate restriction sites for KOR1p (GATGATGCTCTCTGATAAAGC and AAGTCTTTTGGGAGCTGCAA; 2.132 kb) and ESMD1p (ATCGACAGATCTCAATCTC and GGACGAGGACATCCTTGGTA; 2.168 kb) and cloned into pUC57 to create pEN-L4-promoter-R1 clones, as described (58). Coding DNA sequences of KOR1 (ATGTACGGAAGAGATCCATG and TCAAGGTTTCCATGGTGCTG) and ESMD1 (ATGCTAGCGAAGAATCGG and GGTGGCAGGAGGTGGTCTC) were amplified from WT complementary DNA with oligonucleotides specified in parenthesis containing appropriate att sites and recombined with pDONR221 or pDONR-P2R-P3 to obtain pEN-L1-KOR1-L2, pEN-L1-ESMD1-L2, and pEN-R2-KOR1-L3. For transcriptional reporters, pEN-L4-JAZ10p-R1 and pEN-L4-ESMD1-R1 were recombined with pEN-L1-NLS-3xVEN-L2 into pEDO097, as described (58). pEN-L4-KOR1p-R1 was also recombined with pEN-L1-KOR1-L2 into pEDO097 for complementation analysis. For translational reporters, pEN-L4-KOR1p-R1 or cell type–specific promoters were recombined with pEN-L1-CIT-L2 and pEN-R2-KOR1-L3 into a modified pH7m34gw vector named pFR7m34gw, which harbors seed red fluorescent protein (RFP) expression (OLE1p:RFP) instead of Hg resistance for in planta selection, to generate promoter:CIT-KOR1 constructs. Similarly, to obtain ESMD1p:ESMD1-mT and ESMD1p:ESMD1-CIT, we recombined pEN-L4-ESMD1p-R1, pEN-L1-ESMD1-L2, and pEN-R2-mT-L3 or pEN-R2-CIT-L3 into pFR7m34gw (in both cases, fluorescence signals were undetectable, although the constructs were functionally complementing the mutant phenotype). All constructs were verified by Sanger sequencing, and transgenic plants were generated by floral dip with Agrobacterium tumefaciens strain GV3101. Transformed seeds expressing RFP in T1, T2, and T3 generations were selected by fluorescence microscopy, and segregation analysis was performed in >12 independent T2 lines. A minimum of two independent T3 transgenic lines were used for each construct to perform experiments and verify reproducibility.
Confocal microscopy
Confocal LSM imaging was performed on Zeiss LSM 700 or LSM 880 instruments. For live imaging, 5-do vertically grown seedling roots were mounted in 0.5× MS with PI (30 μg/ml). As kor1 roots are thick and recalcitrant to PI penetration, kor1 genotypes were fixed in 4% paraformaldehyde, cleared with ClearSee, and stained with Direct Red 23 (Sigma-Aldrich) as described (59). Excitation/detection ranges were set as follows: VENUS (VEN) and CIT: 514/515 to 545 nm; mT: 458/460 to 510 nm; Direct Red 23: 514/580 to 615 nm; PI: 561/600 to 700 nm. All images shown within one experiment were taken with identical settings and by analyzing at least 10 individuals per line. Image processing was performed in Fiji. Z-stacks were displayed as texture-based volume renderings using the 3D Viewer plugin of Fiji.
Suppressor screen and mapping by next-generation sequencing
Approximately 5000 seeds (0.1 g) of kor1-4 JGP were mutagenized with EMS (Sigma-Aldrich) as described (6). Resulting M1 plants were either harvested individually (n = 1243) or in pools of 12 (n = 230). Twenty M2 seedlings were screened from individually harvested plants, and 480 M2s were screened from each pool to enlarge both the screening breadth and depth. A total of 135,260 M2 seedlings, from 4003 M1 plants, were assayed for lack of JGP activity in 5-do kor1-4 seedlings by live GUS staining as described (6). To increase the screen stringency and avoid the recovery of false positives, M2 seedlings were shifted from 21° to 26°C 24 hours prior GUS staining as kor1 mutants are known exacerbate their phenotypes at higher T (15). Putative M2 suppressors were transferred to soil and crossed to JA-deficient (aos, opr3, and jar1) and JA-insensitive (coi1) mutants to avoid the recovery of expected genes, and backcrossed to kor1-4 JGP for segregation analysis, phenotype confirmation, and mapping population development. esmd1-3 was identified as a JGP suppressor of kor1-4 by pooling 120 individuals lacking JGP reporter activity from a BC1F2 population and sequencing the bulk segregants by whole-genome sequencing. Genomic DNA extraction was as in (6). Library preparation (Illumina Shotgun TruSeq DNA PCR-free) and Illumina paired-end (PE) sequencing on a HiSeq X platform with 150-base pairs per read was performed by Macrogen. The output of 15 Gb resulted in an average of 118 sequencing depth for each base across the genome. EMS-generated single-nucleotide polymorphisms (SNPs) were identified as described (6), with updated software tools. Sequence reads were mapped to the TAIR10 A. thaliana genome with bowtie2 aligner (v. 2.3.1, parameter end-to-end). Alignment files were converted to BAM with SAMtools (v 1.8), and SNP calling was performed with GATK tool (v.4.1.0.0). Common SNPs with the kor1-4 JGP parental line were filtered out with the intersectBed tool from BEDTools utilities (v.2.22.1). The SNPEff tool (v.2.0.4 RC1) was used to predict the effect of the SNPs in coding regions. SNP frequencies (the number of reads supporting a given SNP over the total number of reads covering the SNP location) were extracted using the Unix command awk and plotted with R. Candidate SNPs were identified in genomic regions with high SNP frequencies (0.5 to 1) linked to the causal SNP, which had the expected frequency of 1. Validation of the candidate SNP was done by allelism test and by complementation by transformation.
Cell wall composition analysis
Alcohol-insoluble residue (AIR) was extracted from shoots and roots of 12-do seedlings as previously described (60). Each biological replicate consisted of ~100 shoots (~150-mg FW) or ~300 roots (~80-mg FW). A slurry solution of AIR [water (1 mg/ml)] was prepared for each sample and homogenized using a ball mill followed by sonication. Matrix polysaccharide composition of 300 μg of AIR after 2 M trifluoroacetic acid hydrolysis was analyzed via high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), similar to (60), but on a 940 Professional IC Vario ONE/ChS/PP/LPG instrument (Metrohm) equipped with Metrosep Carb2 250/4.0 analytical and guard columns. Each run consisted of neutral sugar separation (22 min; 2 mM sodium hydroxide and 3.4 mM sodium acetate isocratic gradient), followed by uronic acid separation (23 min; 100 mM sodium hydroxide and 170 mM sodium acetate), and reequilibration (14 min; starting eluents) steps.
Cellulose was quantified on the basis of the two-step sulfuric acid hydrolysis method described by (61), with some modifications. Aliquots of AIR (200 μg each) were first pretreated with concentrated sulfuric acid (to swell cellulose) or were directly used for Seaman hydrolysis (to measure noncrystalline glucose) using ribose as an internal standard. Hydrolyzed glucose was quantified using the HPAEC-PAD system described above but with a shorter run: 2 mM sodium hydroxide and 3.4 mM sodium acetate isocratic gradient (22 min), followed by a 3-min rinse with 80 mM sodium hydroxide and 136 mM sodium acetate and a 4-min reequilibration with starting eluent.
Sectioning, segmentation, and cell analysis
Roots from vertically grown 5-do seedlings were vacuum infiltrated and fixed in glutaraldehyde:formaldehyde:50 mM sodium phosphate buffer (pH 7.2) (2:5:43, v/v/v) for 1 hour, dehydrated through an EtOH series, and embedded in Technovit 7100 resin (Heraeus Kulzer) as described (58). Samples were sectioned on a Microm HM355S microtome with a carbide knife (Histoserve) into 5-μm sections and mounted in 10% glycerol, and cell walls were visualized under dark field of a Zeiss AxioImager microscope fitted with an AxioCam MRm camera. TIF (tag image file format) images were segmented with PlantSeg (62) using preset parameters of the prediction model “lightsheet_unet_bce_dice_ds1x,” which empirically segmented our images most accurately. For display purposes only, dark-field images were inverted, and segmented images were recolored in Photoshop to visualize different cell types more easily.
Root growth and tropism assays
Primary root length was evaluated in 7-do seedlings as described (6), and root growth rate was determined by measuring primary root length in 4-do seedlings for 6 consecutive days. Root diameter was assessed in 7-do seedlings by imaging vertically grown seedlings on a Leica M165 FC stereomicroscope fitted with a Leica MC170 HD camera, measuring the root thickness in the early differentiation zone (marked by the appearance of root hairs). For RAM measurements, 5-do seedlings were mounted in chloral hydrate:glycerol:water (8:2:1) solution and observed with a Leica DM6B microscope fitted with a Leica DMC6200 camera. The number of cells in the meristematic zone was counted in the cortex cell file between the quiescent center and the first elongating cell, as described (6). Root gravitropism assays were performed on vertically grown 5-do seedlings by rotating the plates by 90° and evaluating root bending angles 24 hours after the rotation on scanned images with Fiji. For root hydrotropism assays, 5-do seedlings were transferred to split-agar plates containing either mock (MS/MS) or 400 mM mannitol (MS/mannitol) by aligning root tips 3 mm from the split-media boundary (63). Root bending angles were evaluated 24 hours after transfer to split-agar plates by analyzing scanned images with Fiji as described (63).
Statistical analysis
Box plots, multiple comparisons [analysis of variance (ANOVA) followed by Tukey’s honest significant difference test].
Acknowledgments
We thank E. E. Farmer for enabling the initiation of this work by gifting the kor1-4 and kor1-5 EMS alleles originally identified in his lab and for critical comments on the manuscript. We also thank G. Mouille for sharing esmd1-1 seeds, I. F. Acosta for gifting the pFR7m34gw plasmid, C. Wagner for assistance with AIR sample preparation, B. Yang for HPAEC-PAD maintenance, C. Wasternack and members of the Gasperini lab for critical discussions. Funding: This work was supported by the Leibniz Institute of Plant Biochemistry from the Leibniz Association, the German Research Foundation (DFG grant GA2419/2-1) to D.G. and (DFG grant 414353267) to C.V., and a Marie Skłodowska-Curie postdoctoral fellowship to M.K.M. Author contributions: S.M. and D.G. designed research. S.M, M.Z., M.K.M., and D.G. performed research. S.M., M.K.M., R.D., and D.G. analyzed data. H.S. and B.H. quantified JA-Ile levels. C.V. performed cell wall composition analyses. S.M. and D.G. wrote the manuscript with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/7/eabf0356/DC1
REFERENCES AND NOTES
- 1.Howe G. A., Major I. T., Koo A. J., Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69, 387–415 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Browse J., Wallis J. G., Arabidopsis flowers unlocked the mechanism of jasmonate signaling. Plants 8, 285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z., Huang Y., Yang J., Yao S., Zhao K., Wang D., Qin Q., Bian Z., Li Y., Lan Y., Zhou T., Wang H., Liu C., Wang W., Qi Y., Xu Z., Li Y., Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28, 89–103.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Agrawal A. A., Current trends in the evolutionary ecology of plant defence. Funct. Ecol. 25, 420–432 (2011). [Google Scholar]
- 5.Goodspeed D., Chehab E. W., Min-Venditti A., Braam J., Covington M. F., Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U.S.A. 109, 4674–4677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acosta I. F., Gasperini D., Chetelat A., Stolz S., Santuari L., Farmer E. E., Role of NINJA in root jasmonate signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 15473–15478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penninckx I. A., Thomma B. P., Buchala A., Métraux J. P., Broekaert W. F., Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Q., Major I. T., Howe G. A., Resolution of growth-defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 44, 72–81 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Campos M. L., Kang J.-H., Howe G. A., Jasmonate-triggered plant immunity. J. Chem. Ecol. 40, 657–675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasternack C., Feussner I., The oxylipin pathways: Biochemistry and function. Annu. Rev. Plant Biol. 69, 363–386 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Mielke S., Gasperini D., Interplay between plant cell walls and jasmonate production. Plant Cell Physiol. 60, 2629–2637 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Cosgrove D. J., Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 67, 463–476 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Denness L., McKenna J. F., Segonzac C., Wormit A., Madhou P., Bennett M., Mansfield J., Zipfel C., Hamann T., Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis C., Karafyllidis I., Wasternack C., Turner J. G., The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane D. R., Wiedemeier A., Peng L., Höfte H., Vernhettes S., Desprez T., Hocart C. H., Birch R. J., Baskin T. I., Burn J. E., Arioli T., Betzner A. S., Williamson R. E., Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126, 278–288 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei L., Zhang T., Strasser R., Lee C. M., Gonneau M., Mach L., Vernhettes S., Kim S. H., Cosgrove D. J., Li S., Gu Y., The jiaoyao1 mutant is an allele of korrigan1 that abolishes endoglucanase activity and affects the organization of both cellulose microfibrils and microtubules in arabidopsis. Plant Cell 26, 2601–2616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Cruz J., Finiti I., Fernández-Crespo E., Crespo-Salvador O., García-Agustín P., González-Bosch C., Absence of endo-1,4-β-glucanase KOR1 alters the jasmonate-dependent defence response to Pseudomonas syringae in Arabidopsis. J. Plant Physiol. 171, 1524–1532 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Peng L., Kawagoe Y., Hogan P., Delmer D., Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 295, 147–150 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Vain T., Crowell E. F., Timpano H., Biot E., Desprez T., Mansoori N., Trindade L. M., Pagant S., Robert S., Höfte H., Gonneau M., Vernhettes S., The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 165, 1521–1532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamant O., Haswell E. S., Life behind the wall: Sensing mechanical cues in plants. BMC Biol. 15, 59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polko J. K., Kieber J. J., The regulation of cellulose biosynthesis in plants. Plant Cell 31, 282–296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng L., Hocart C. H., Redmond J. W., Williamson R. E., Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Chaudhary A., Chen X., Gao J., Lesniewska B., Hammerl R., Dawid C., Schneitz K., The Arabidopsis receptor kinase STRUBBELIG regulates the response to cellulose deficiency. PLOS Genet. 16, e1008433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer E. E., Gasperini D., Acosta I. F., The squeeze cell hypothesis for the activation of jasmonate synthesis in response to wounding. New Phytol. 204, 282–288 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Engelsdorf T., Gigli-Bisceglia N., Veerabagu M., McKenna J. F., Vaahtera L., Augstein F., Van der Does D., Zipfel C., Hamann T., The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci. Signal. 11, eaao3070 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Rips S., Bentley N., Jeong I. S., Welch J. L., von Schaewen A., Koiwa H., Multiple N-glycans cooperate in the subcellular targeting and functioning of Arabidopsis KORRIGAN1. Plant Cell 26, 3792–3808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marhavý P., Kurenda A., Siddique S., Tendon V. D., Zhou F., Holbein J., Hasan M. S., Grundler F. M. W., Farmer E. E., Geldner N., Single-cell damage elicits regional, nematode-restricting ethylene responses in roots. EMBO J. 38, e100972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M., Wang F., Li S., Yu G., Wang L., Li Q., Zhu X., Li Z., Yuan L., Liu P., Importers drive leaf-to-leaf transmission of jasmonic acid in wound induced systemic immunity. Mol. Plant 13, 1485–1498 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Schulze A., Zimmer M., Mielke S., Stellmach H., Melnyk C. W., Hause B., Gasperini D., Wound-induced shoot-to-root relocation of JA-Ile precursors coordinates Arabidopsis growth. Mol. Plant 12, 1383–1394 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Marquès-Bueno M. M., Morao A. K., Cayrel A., Platre M. P., Barberon M., Caillieux E., Colot V., Jaillais Y., Roudier F., Vert G., A versatile multisite gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J. 85, 320–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verger S., Chabout S., Gineau E., Mouille G., Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development 143, 2536–2540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takenaka Y., Kato K., Ogawa-Ohnishi M., Tsuruhama K., Kajiura H., Yagyu K., Takeda A., Takeda Y., Kunieda T., Hara-Nishimura I., Kuroha T., Nishitani K., Matsubayashi Y., Ishimizu T., Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat. Plants 4, 669–676 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Bouton S., Leboeuf E., Mouille G., Leydecker M.-T., Talbotec J., Granier F., Lahaye M., Höfte H., Truong H.-N., QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14, 2577–2590 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J., Kirui A., Huang S., Wang L., Barnes W. J., Kiemle S. N., Zheng Y., Rui Y., Ruan M., Qi S., Kim S. H., Wang T., Cosgrove D. J., Anderson C. T., Xiao C., Mutations in the pectin methyltransferase QUASIMODO2 influence cellulose biosynthesis and wall integrity in Arabidopsis thaliana. Plant Cell 32, 3576–3597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouille G., Ralet M.-C., Cavelier C., Eland C., Effroy D., Hématy K., Cartney L. M., Truong H. N., Gaudon V., Thibault J.-F., Marchant A., Höfte H., Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 50, 605–614 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Hématy K., Sado P.-E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., Pelletier S., Renou J.-P., Höfte H., A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17, 922–931 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Yang D.-L., Yao J., Mei C.-S., Tong X.-H., Zeng L.-J., Li Q., Xiao L.-T., Sun T.-p., Li J., Deng X.-W., Lee C. M., Thomashow M. F., Yang Y., He Z., He S. Y., Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dietrich D., Pang L., Kobayashi A., Fozard J. A., Boudolf V., Bhosale R., Antoni R., Nguyen T., Hiratsuka S., Fujii N., Miyazawa Y., Bae T.-W., Wells D. M., Owen M. R., Band L. R., Dyson R. J., Jensen O. E., King J. R., Tracy S. R., Sturrock C. J., Mooney S. J., Roberts J. A., Bhalerao R. P., Dinneny J. R., Rodriguez P. L., Nagatani A., Hosokawa Y., Baskin T. I., Pridmore T. P., De Veylder L., Takahashi H., Bennett M. J., Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat. Plants 3, 17057 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Fernández-Sánchez M. E., Barbier S., Whitehead J., Béalle G., Michel A., Latorre-Ossa H., Rey C., Fouassier L., Claperon A., Brullé L., Girard E., Servant N., Rio-Frio T., Marie H., Lesieur S., Housset C., Gennisson J.-L., Tanter M., Ménager C., Fre S., Robine S., Farge E., Mechanical induction of the tumorigenic β-catenin pathway by tumour growth pressure. Nature 523, 92–95 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Hoermayer L., Montesinos J. C., Marhava P., Benkova E., Yoshida S., Friml J., Wounding-induced changes in cellular pressure and localized auxin signalling spatially coordinate restorative divisions in roots. Proc. Natl. Acad. Sci. U.S.A. 117, 15322–15331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W., Lozano-Torres J. L., Blilou I., Zhang X., Zhai Q., Smant G., Li C., Scheres B., A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177, 942–956.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Boudaoud A., An introduction to the mechanics of morphogenesis for plant biologists. Trends Plant Sci. 15, 353–360 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Kurenda A., Nguyen C. T., Chételat A., Stolz S., Farmer E. E., Insect-damaged Arabidopsis moves like wounded Mimosa pudica. Proc. Natl. Acad. Sci. U.S.A. 116, 26066–26071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu D., Haswell E. S., Plant mechanosensitive ion channels: An ocean of possibilities. Curr. Opin. Plant Biol. 40, 43–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bidhendi A. J., Geitmann A., Methods to quantify primary plant cell wall mechanics. J. Exp. Bot. 70, 3615–3648 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Michels L., Gorelova V., Harnvanichvech Y., Borst J. W., Albada B., Weijers D., Sprakel J., Complete microviscosity maps of living plant cells and tissues with a toolbox of targeting mechanoprobes. Proc. Natl. Acad. Sci. U.S.A. 117, 18110–18118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu D., Haswell E. S., The mechanosensitive ion channel MSL10 potentiates responses to cell swelling in Arabidopsis seedlings. Curr. Biol. 30, 2716–2728.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Murthy S. E., Dubin A. E., Whitwam T., Jojoa-Cruz S., Cahalan S. M., Mousavi S. A. R., Ward A. B., Patapoutian A., OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7, e41844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan F., Yang H., Xue Y., Kong D., Ye R., Li C., Zhang J., Theprungsirikul L., Shrift T., Krichilsky B., Johnson D. M., Swift G. B., He Y., Siedow J. N., Pei Z.-M., OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Wachananawat B., Kuroha T., Takenaka Y., Kajiura H., Naramoto S., Yokoyama R., Ishizaki K., Nishitani K., Ishimizu T., Diversity of Pectin Rhamnogalacturonan I Rhamnosyltransferases in Glycosyltransferase Family 106. Front. Plant Sci. 11, 997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettolino F. A., Walsh C., Fincher G. B., Bacic A., Determining the polysaccharide composition of plant cell walls. Nat. Protoc. 7, 1590–1607 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Wang T., Park Y. B., Cosgrove D. J., Hong M., Cellulose-pectin spatial contacts are inherent to never-dried arabidopsis primary cell walls: Evidence from solid-state nuclear magnetic resonance. Plant Physiol. 168, 871–884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson S., Burian A., Couturier E., Landrein B., Louveaux M., Neumann E. D., Peaucelle A., Weber A., Nakayama N., Mechanical control of morphogenesis at the shoot apex. J. Exp. Bot. 64, 4729–4744 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Campos M. L., Yoshida Y., Major I. T., de Oliveira Ferreira D., Weraduwage S. M., Froehlich J. E., Johnson B. F., Kramer D. M., Jander G., Sharkey T. D., Howe G. A., Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7, 12570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Major I. T., Guo Q., Zhai J., Kapali G., Kramer D. M., Howe G. A., A phytochrome B-independent pathway restricts growth at high levels of jasmonate defense. Plant Physiol. 183, 733–749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruan J., Zhou Y., Zhou M., Yan J., Khurshid M., Weng W., Cheng J., Zhang K., Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20, 2479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J.-M., To T. K., Matsui A., Tanoi K., Kobayashi N. I., Matsuda F., Habu Y., Ogawa D., Sakamoto T., Matsunaga S., Bashir K., Rasheed S., Ando M., Takeda H., Kawaura K., Kusano M., Fukushima A., Endo T. A., Kuromori T., Ishida J., Morosawa T., Tanaka M., Torii C., Takebayashi Y., Sakakibara H., Ogihara Y., Saito K., Shinozaki K., Devoto A., Seki M., Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 3, 17097 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Gasperini D., Chételat A., Acosta I. F., Goossens J., Pauwels L., Goossens A., Dreos R., Alfonso E., Farmer E. E., Multilayered organization of jasmonate signalling in the regulation of root growth. PLOS Genet. 11, e1005300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ursache R., Andersen T. G., Marhavý P., Geldner N., A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 93, 399–412 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Voiniciuc C., Schmidt M. H.-W., Berger A., Yang B., Ebert B., Scheller H. V., North H. M., Usadel B., Günl M., MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in arabidopsis seed mucilage. Plant Physiol. 169, 403–420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeats T. H., Sorek H., Wemmer D. E., Somerville C. R., Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol. 171, 110–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolny A., Cerrone L., Vijayan A., Tofanelli R., Barro A. V., Louveaux M., Wenzl C., Strauss S., Wilson-Sánchez D., Lymbouridou R., Steigleder S. S., Pape C., Bailoni A., Duran-Nebreda S., Bassel G. W., Lohmann J. U., Tsiantis M., Hamprecht F. A., Schneitz K., Maizel A., Kreshuk A., Accurate and versatile 3D segmentation of plant tissues at cellular resolution. eLife 9, e57613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Antoni R., Dietrich D., Bennett M. J., Rodriguez P. L., Hydrotropism: Analysis of the root response to a moisture gradient. Methods Mol. Biol. 1398, 3–9 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/7/eabf0356/DC1


