Blood vessels induce epidermal proliferating clusters in the vicinity of activating stem cells to populate expanding skin.
Abstract
Stem cell (SC) proliferation and differentiation organize tissue homeostasis. However, how SCs regulate coordinate tissue scaling in dynamic organs remain unknown. Here, we delineate SC regulations in dynamic skin. We found that interfollicular epidermal SCs (IFESCs) shape basal epidermal proliferating clusters (EPCs) in expanding abdominal epidermis of pregnant mice and proliferating plantar epidermis. EPCs consist of IFESC-derived Tbx3+–basal cells (Tbx3+-BCs) and their neighboring cells where Adam8–extracellular signal–regulated kinase signaling is activated. Clonal lineage tracing revealed that Tbx3+-BC clones emerge in the abdominal epidermis during pregnancy, followed by differentiation after parturition. In the plantar epidermis, Tbx3+-BCs are sustained as long-lived SCs to maintain EPCs invariably. We showed that Tbx3+-BCs are vasculature-dependent IFESCs and identified mechanical stretch as an external cue for the vasculature-driven EPC formation. Our results uncover vasculature-mediated IFESC regulations, which explain how the epidermis adjusts its size in orchestration with dermal constituents in dynamic skin.
INTRODUCTION
Skin, an essential barrier, is a dynamic organ that expands and shrinks flexibly in response to physiological changes in body shape. Skin consists of stratified epidermis and underlying dermis where blood and lymphatic vessels, peripheral nerves, and skin appendages form complex networks (1, 2). Skin epidermis is compartmentalized into two main areas: hair follicles (HFs) and their surrounding interfollicular epidermal (IFE) (3). To replenish IFE for homeostasis, IFE stem cells (IFESCs) residing in the basal layer balance self-renewal and differentiation (4). Extensive studies using clonal lineage-tracing techniques and mathematical modeling have proposed models that explain the IFESC composition and dynamics in homeostasis and wound repair. They include the single type of stem cell/progenitor model, in which IFESCs/progenitors undergo self-renewal and differentiation stochastically (4–6), and the stem cell–to–committed progenitor hierarchical model (7, 8) and multiple IFESC lineages model (9–11) in which each cell lineage can be distinguished by individual markers or their differential proliferation capacities. In addition, recent studies of single-cell transcriptomes in both murine and human epidermis have revealed multiple IFE basal cell (BC) clusters on the basis of their transcriptional heterogeneity (12–15). Beyond homeostasis and wound repair, we have shown that the epidermal basal layer of rapidly expanding abdominal skin of pregnant mice harbors highly proliferating IFESC progeny that expresses T-box3 (Tbx3) necessary for their proliferation and skin expansion (16).
Here, we investigated functions and cell fate of Tbx3+-BCs in epidermal remodeling. We showed that Tbx3+-BCs shape the distinct epidermal proliferating clusters (EPCs) to populate expanding epidermis, including abdominal skin of pregnant mice, planter skin, and stretched skin. In addition, we found that Tbx3+-BCs are vasculature-induced IFESCs, which undergo differentiation after vascular regression.
RESULTS
Tbx3+-BCs shape EPCs in abdominal epidermis of pregnant mice
To assess the function of Tbx3+-BCs in epidermal expansion during pregnancy, we used a histone 2B (H2B)–green fluorescent protein (GFP) pulse-chase assay using K5-tTA (tet-off);pTREH2B-GFP mice in which doxycycline administration turns off the H2B-GFP gene transcription, leading to dilution of the GFP fluorescence intensity at each cell division (17). Skin sections from doxycycline-treated pregnant mice showed that non–label-retaining cells (non-LRCs) were enriched and forming clusters in the abdominal skin (fig. S1A). Non-LRCs were either Tbx3+ cells or their neighboring Tbx3− cells (fig. S1B). This raised the possibility that Tbx3+-BCs not only proliferate themselves but also promote proliferation of neighboring cells. To examine this possibility, we explored the Tbx3-dependent transcriptomes in abdominal IFEBCs of pregnant mice by RNA sequencing (RNA-seq) of fluorescence-activated cell sorting (FACS)–isolated IFEBCs from the abdominal skin of wild-type (WT) nonpregnant (NP) mice, pregnant mice at days post-coitum (dpc16), and K14creERT2;Tbx3flox/flox (Tbx3 cKO) pregnant mice at dpc16 (Fig. 1A and fig. S1C). We detected 356 differentially expressed genes with 170 up-regulated and 186 down-regulated in dpc16 mice versus NP mice, whereas there were 765 differentially expressed genes with 460 up-regulated and 305 down-regulated in Tbx3 cKO pregnant mice versus WT pregnant mice at dpc16 (>1.5-fold changes) (fig. S1D). Among pregnancy signature genes (356 genes), 113 genes (31%) were Tbx3 signature genes (Fig. 1B, P < 2.2 × 10−16, and table S1), demonstrating a crucial role of Tbx3 in regulating the pregnancy-associated transcriptome. Gene ontology (GO) analysis of the 113 dual signature genes revealed that they were associated with tissue morphology, organ development, and hair and skin development and function, reflecting their potential roles in epidermal remodeling during pregnancy (Fig. 1C). Next, we performed network analysis of the 113 dual signature genes and found that mitogen-activated protein kinase interactomes were linked to Tbx3 interactomes (fig. S1E). Consistently, IFEBCs with the activated form of extracellular signal–regulated kinase (ERK) (p-ERK+–IFEBCs) were increased significantly at dpc16, which was attenuated in Tbx3 cKO pregnant mice (Fig. 1, D to F, and fig. S1F), indicating that Tbx3 induces ERK activation to promote IFEBC proliferation. To further explore the Tbx3-ERK axis, we focused on one of the dual signature genes, A disintegrin and metalloproteinase domain-containing protein 8 (Adam8), a member of the ADAM cell-surface metalloproteinase family that is known to be up-regulated in various tumors and activates ERK signaling (18–21). Consistent with RNA expression patterns (fig. S1G), ADAM8+-IFEBCs were increased at dpc16, which was attenuated in Tbx3 cKO pregnant mice (Fig. 1, G and H). In addition, ADAM8 knockout (KO) mice manifested attenuated ERK activation (Fig. 1, D and E) and reduced IFEBC division (Fig. 1, I and J) at dpc16 without affecting the Tbx3+-BC population (fig. S1, H and I). Heparin-binding epidermal growth factor–like growth factor (HB-EGF), a member of the EGF family whose membrane-anchored form is cleaved by ADAM-mediated ectodomain shedding to release the soluble form (22–24), was included in the pregnancy–up-regulated signature genes (Fig. 1B and fig. S1G). Furthermore, conditional deletion of Hbegf in epidermis decreased IFEBC proliferation at dpc16 (Fig. 1, K and L), implying that ADAM8–HB-EGF–ERK signaling occurs in EPCs. Notably, ADAM8 was expressed in not only Tbx3+-BCs but also their neighboring cells (Fig. 1M), indicating that Tbx3+-BCs promote ADAM8 expression in their surroundings. These results demonstrate that Tbx3+-BCs shape EPCs in their vicinity by activating ADAM8-EGF-ERK signaling in neighboring cells.
Fig. 1. Tbx3+-BCs shape EPCs in expanding abdominal epidermis of pregnant mice.
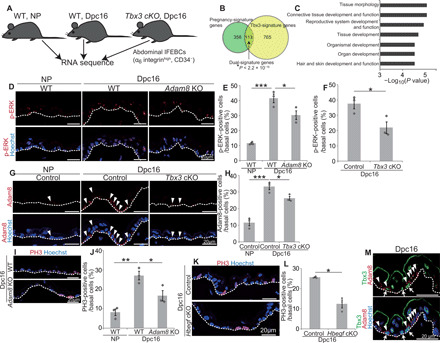
(A) Experimental design for RNA-seq. (B) Venn diagram of pregnancy and Tbx3 signature genes. *P < 2.2 × 10−16 analyzed by Fisher’s exact test. (C) GO analysis of dual signature genes (113 genes). (D and E) Staining and quantification of p-ERK+–BCs in abdominal epidermis (n > 150 cells, three mice). (F) Quantification of p-ERK+–BCs (n > 150 cells, three mice). (G and H) Staining and quantification of Adam8+-BCs (n > 140 cells, three mice). (I and J) Staining and quantification of PH3+-BCs (n > 190 cells, three mice). (K and L) Staining and quantification of PH3+-BCs (n > 100 cells, three mice). (M) Tbx3/Adam8 staining in abdominal epidermis. Arrows indicate Tbx3+-BCs. Arrowheads indicate ADAM8+-BCs. (E, F, H, J, and L) Error bars show SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 by Dunnett’s multiple comparison test (E, H, and J) and the two-tailed Student’s t test (F and L). (D, G, I, K, and M) White dashed lines indicate basement membranes (BMs).
Tbx3+-BCs are differentiated and eliminated from epidermis after parturition
Tbx3+-BCs and proliferating IFEBCs were increased at dpc16, followed by decreases after parturition at day postpartum (dpp) 40 (fig. S2, A to D), indicating that EPCs were induced transiently during pregnancy. To explore this dynamicity, we determined the cell fate of Tbx3+-BCs after parturition. To this end, we label-traced Tbx3+-BC clones by generating Tbx3creERT2 mouse lines (fig. S3, A and B) that were further crossed with R26H2B-EGFP mice (fig. S3, C to F). The obtained mice (Tbx3creERT2;R26H2B-EGFP) were treated with tamoxifen at dpc14 to dpc16, and the abdominal skin was harvested at dpc18, dpp14, dpp30, and dpp180 (Fig. 2A). We confirmed that Tbx3cre-EGFP labeled Tbx3+-BCs in the abdominal skin of pregnant mice (fig. S3G). For the control, enhanced GFP (EGFP)–labeled cells were traced in K14creERT2;R26H2B-EGFP mice (K14cre-labeled clones) (Fig. 2A). Then, the number of surviving clone fractions and their basal clone size were measured. Consistent with previous studies (7, 25), K14cre-labeled clones survived in the basal layer at dpp180 with a growing divergence in clone size (Fig. 2, B, D, and F). In contrast, Tbx3cre-labeled clones, which survived in the basal layer up to dpp30, were no longer detected in the epidermis at dpp180, (Fig. 2, C, E, and G), indicating that Tbx3+-BCs emerge during pregnancy and their clones are eliminated from the epidermis after parturition. To resolve the cell fate of Tbx3+-BCs by transcription levels at the single-cell resolution, we performed single-cell RNA-seq (scRNA-seq) of epidermal cells isolated from the abdominal skin of Tbx3creERT2;R26H2B-EGFP mice treated with tamoxifen during pregnancy. Then, the abdominal skin epidermis was harvested at dpc16, dpp6, and dpp40 (Fig. 2H). For the control, abdominal skin epidermis was harvested from NP mice treated with tamoxifen for 3 days. After dissociation of the epidermis into single cells, we performed scRNA-seq using the chromium system. We profiled 36,912 cells (NP, 6485 cells; dpc16, 12,846 cells; dpp6, 9029 cells; dpp40, 8,552 cells) with a range of 20,000 to 40,000 mean reads per cell in each sample, whereby approximately 1400 to 1600 median genes per cell were detected in each sample. Unsupervised evaluation of clustering-based cell identities of total samples from whole epidermis classified the cells into 14 clusters (fig. S4A) based on the differentially expressed gene signatures (table S2) and marker genes projected onto t-distributed Stochastic Neighbor Embedding (t-SNE) plots (fig. S4B). Next, we selected all EGFP-expressing cells from IFE clusters and reconstructed pseudo-time differentiation trajectories. The resultant trajectory displayed a single linear pattern of the cell lineage along which cells in each cluster were plotted in order according to their differentiation state (Fig. 2I). In addition, the proportion of the IFE1 undifferentiated cluster was increased at dpc16 and decreased at dpp6 and dpp40 with a concomitant increase in the granular cell cluster IFE5 (Fig. 2J). These results indicate that the Tbx3+-BC clones undergo differentiation after parturition. Consistently, apoptotic cells were barely detected in abdominal epidermal basal layers in NP, pregnant, or postpartum mice (NP, 0 of 4821 cells; dpc16, 0 of 6972 cells; dpp6, 0 of 4292 cells; dpp40, 0 of 5119 cells), supporting the conclusion that Tbx3+-BCs undergo differentiation rather than apoptosis after parturition.
Fig. 2. Tbx3+-BCs undergo differentiation to be eliminated from the epidermis after parturition.
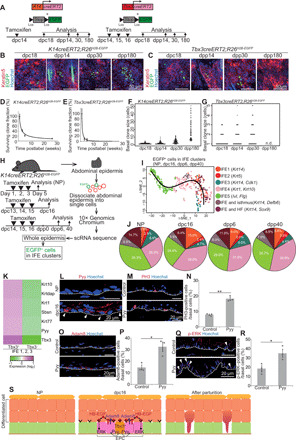
(A) Tracing K14cre- and Tbx3cre-labeled clones. (B and C) Whole-mount images of K14cre- and Tbx3cre-labeled clones in abdominal epidermis in tamoxifen-treated K14creERT2;R26H2B-EGFP and Tbx3creERT2;R26H2B-EGFP mice. (D and E) Quantification of clone survivability. (F and G) Distribution of the basal clone size. n.d., not detected. (H) Experimental design for scRNA-seq. (I) Pseudo-time reconstruction of differentiation of EGFP+ cells in IFE clusters. (J) Pie charts of the IFE clusters. (K) Heatmap of the Tbx3+ cell–enriched genes in IFE1 to IFE3 clusters. (L) Pyy staining in abdominal epidermis. Arrowheads indicate Pyy+-BCs. (M and N) Staining and quantification of PH3+-BCs in abdominal epidermis of control and Pyy-injected NP mice (n > 140 cells, three mice). (O and P) Staining and quantification of Adam8+-BCs (n > 170 cells, three mice). (Q and R) Staining and quantification of p-ERK+–BCs (n > 130 cells, three mice). (S) Schematic of EPCs. (D to G, N, P, and R) Error bars indicate SEM. (N, P, and R) *P < 0.05 and **P < 0.01, by two-tailed Student’s t test. (L, M, O, and Q) White dashed lines indicate BMs.
To further explore the function of Tbx3+-BCs, we separated the cells from IFEBC clusters (IFE1, IFE2, and IFE3) into Tbx3+-BC and Tbx3−-BC populations and compared their gene expression profiles. We found that epidermal differentiation genes [Keratin10 (Krt10), Keratinocyte differentiation associated protein (Krtdap), Krt1, Suprabasin (Sbsn), and Krt77] were up-regulated in the Tbx3+-BC population (>1.5-fold changes), which suggested that Tbx3+-BCs harbored the property of primed stem cells that are prone to differentiation (Fig. 2K and fig. S4C). Peptide YY (PYY), a member of the neuropeptide family of hormones (26), which was included in pregnancy and Tbx3 dual signature genes (Fig. 1B, fig. S1G, and table S1), was up-regulated in Tbx3+-BCs (Fig. 2K and fig. S4C). Consistently, PYY+-IFEBCs were increased at dpc16 (Fig. 2L). Furthermore, intradermal injection of PYY1–36 peptides into the abdominal skin induced phospho-histone h3 (PH3)+-BCs, Adam8+-BCs, and p-ERK+–BCs in NP mice (Fig. 2, M to R). These results demonstrate that Tbx3+-BCs secrete PYY that functions as a paracrine signal to neighboring cells to shape EPCs in their vicinity during pregnancy (Fig. 2S).
Tbx3+-BCs and EPCs are maintained in plantar epidermis invariably
There is regional variability in the epidermis throughout the body (14). Because Tbx3+-BCs and EPCs emerge in highly proliferating abdominal epidermis of pregnant mice but are barely present in slowly proliferating dorsal epidermis (16), we determined whether they constitutively exist in the anatomical area where a high rate of IFEBC proliferation occurs for homeostasis. We focused on the plantar skin that exhibits a thickened epidermis attributed to prominent cell proliferation (27) (Fig. 3, A and B) and a high rate of basal-to-suprabasal transition (Fig. 3, C and D). Notably, EPC components (Tbx3+-BCs, Adam8+-BCs, and p-ERK+-BCs) and PYY+-BCs were enriched significantly in the plantar skin (Fig. 3, E to K). In addition, Tbx3 cKO mice (Fig. 3, L and M) and Hbegf cKO mice (Fig. 3, N and O) manifested attenuated cell proliferation, indicating that Tbx3+-BCs and EPCs are constitutively maintained in the plantar skin. Next, we investigated the stemness properties of Tbx3+-BCs in the plantar skin. Unlike abdominal skin, Tbx3cre-labeled clones persisted in the epidermis for up to a year with growing divergence in the basal clone size (Fig. 3, P to S), which generated clones over multiple IFE layers (Fig. 3T), indicating that Tbx3cre-labeled clones maintain stemness in the plantar skin for homeostasis. Together, Tbx3+-BCs are latent IFESCs that generate EPCs in a distinct anatomical area with a high rate of epidermal proliferation.
Fig. 3. Tbx3+-BCs and EPCs are maintained in proliferating plantar epidermis for homeostasis.
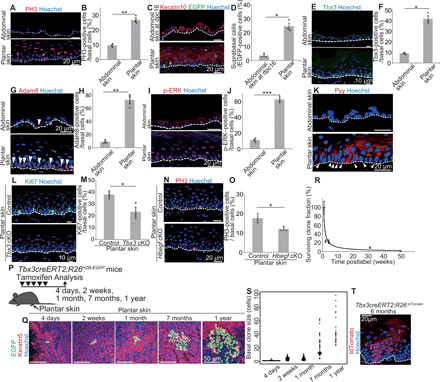
(A and B) Staining and quantification of PH3+-BCs in abdominal and plantar epidermis (n > 300 cells, three mice). (C) Tracing K14cre-labeled BCs. (D) Quantification of basal-to-suprabasal transition of K14cre-labeled BCs (n > 110 cells, three mice). (E and F) Staining and quantification of Tbx3+-BCs (n > 330 cells, three mice). (G and H) Staining and quantification of Adam8+-BCs (n > 100 cells, three mice). (I and J) Staining and quantification of p-ERK+–BCs (n > 120 cells, three mice). (K) Pyy staining. Arrowheads indicate Pyy+-BCs. (L and M) Staining and quantification of plantar epidermal Ki67+-BCs in control and Tbx3 cKO mice (n > 230 cells, three mice). (N and O) Staining and quantification of plantar epidermal PH3+-BCs in control and Hbegf cKO mice (n > 110 cells, three mice). (P) Tracing Tbx3cre-labeled clones. (Q) Whole-mount images of Tbx3cre-labeled clones in plantar epidermis. (R) Quantification of Tbx3cre-labeled clone survivability. (S) Distribution of the basal clone size. (T) tdTomato staining of Tbx3creERT2;R26tdTomato mice. (B, D, F, H, J, M, O, R, and S) Error bars show SEM. (B, D, F, H, J, M, and O) *P < 0.05, **P < 0.01, and ***P < 0.001, by two-tailed Student’s t test. (A, C, E, G, I, K, L, N, and T) White dashed lines indicate BMs.
Tbx3+-BCs are induced by vasculatures
Next, we explored the cue that induces or maintains Tbx3+-BCs. Our previous study had shown that dermal signals are involved in the induction of Tbx3+-BCs during pregnancy (16). Therefore, we explored dermal dynamics by scRNA-seq of whole dermis isolated from the abdominal skin of NP mice, pregnant mice at dpc16, and postpartum mice at dpp42 using the chromium system. A total of 6020 cells (NP, 1289 cells; dpc16, 3650 cells; dpp42, 1081 cells) were profiled with a range of 4500 to 5400 mean reads per cell in each sample, whereby approximately 1200 to 1500 median genes per cell were detected in each sample. Unsupervised evaluation for clustering-based cell identities of total samples from whole dermis classified the cells into 13 clusters (fig. S5A) based on the differentially expressed gene signatures (table S3) and marker genes projected onto t-SNE plots (fig. S5B). Among them, the vascular cell cluster (CL-7; Pecam1high and Flt1high) was further subjected to a second round of unsupervised clustering, which resulted in classification of the cells into four subclusters (endothelial cell clusters 1 to 3 and a vascular smooth muscle cell cluster) (Fig. 4A) on the basis of differentially expressed gene signatures (table S4) and marker genes projected onto t-SNE plots (fig. S5C). We found that the vascular cell cluster exhibited an altered cluster proportion at dpc16, where the ratio of endothelial cluster 3 was increased at dpc16 and decreased at dpp42 (Fig. 4, A and B). GO analysis showed that genes related to vasculature development, morphogenesis, and angiogenesis were enriched in endothelial cluster 3 signature genes (Fig. 4C), suggesting that vascular remodeling occurs in the abdominal skin during pregnancy. Whole-mount immunofluorescence and three-dimensional reconstruction of dermal blood vessels, which were positive for panendothelial marker CD31 and negative for lymphatic vessel endothelial hyaluronan receptor-1, showed that the blood vessel density (total vessel volume and length per field) was larger in the abdominal skin of pregnant mice at dpc16 than that of NP mice and postpartum mice at dpp180 (Fig. 4, D to F, and fig. S6A). In addition, blood vessel branching points and endothelial tip cells, the leading cells of vascular sprouts during angiogenesis and positive for endothelial cell–specific molecule-1 (Esm1) (28), were increased at dpc16 (Fig. 4, G to I). Furthermore, the expression of VEGF-A mRNA was increased in the dermis at dpc16 (Fig. 4, J and K). Thus, angiogenesis occurs in the abdominal skin during pregnancy. The blood vessel density in the plantar skin of male mice was as large as that in the abdominal skin of pregnant mice (Fig. 4, D to F), indicating that the blood vessel density was correlated to the appearance of Tbx3+-BCs and EPCs. To assess the functional relevance of vasculature in Tbx3+-BC emergence, we injected a blocking antibody against vascular endothelial growth factor receptor 2 (VEGFR2) (29) intradermally into the abdominal skin of pregnant mice. We found that both Tbx3+-BCs and IFEBC cell proliferation were decreased significantly by the antibody injection (Fig. 4, L to O). Note that Kdr (VEGFR2) was expressed specifically in vasculature and lymphatic cell clusters and showed no or low level of expression in epidermal cells during pregnancy (fig. S5F), indicating that VEGFR2 signaling acts mainly in vasculatures. In addition, Tbx3+-BCs emerged during wound healing in WT mice (16), which were significantly decreased in angiogenesis-defective VE-cadherin-creERT2;VEGFR2flox/flox (VEGFR2 cKO) mice (Fig. 4, P and Q, and fig. S6C), indicating that vasculatures are necessary for Tbx3+-BC induction. Furthermore, ectopic induction of angiogenesis in skin tissue using K14 promoter–driven VEGF-A transgenic mice (30) (fig. S6D) increased Tbx3+-BCs compared with WT mice (Fig. 4, R and S), indicating that angiogenesis evokes Tbx3+-BCs. Thus, blood vessels play a pivotal role in the emergence of Tbx3+-BCs, which leads to the generation of EPCs to populate expanding epidermis.
Fig. 4. Vasculatures induce Tbx3+-BCs.
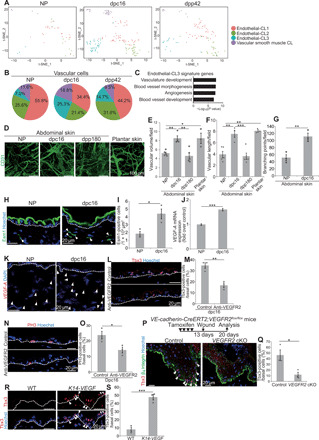
(A) t-SNE plots for vascular cluster cells (NP, n = 43; dpc16, n = 154; dpp42, n = 95). (B) Pie charts of the vascular subclusters. (C) GO analysis of endothelial-CL3 signature genes. (D) Whole-mount images of CD31 (Lyve1−) in abdominal and plantar dermis. (E and F) Quantification of vascular volume and length (n > 3 areas, three mice). (G) Quantification of vascular branching points (n = 3 areas, three mice). (H and I) Staining and quantification of dermal Esm1+ cells (n > 10 areas, three mice). (J) VEGF-A quantitative reverse transcription polymerase chain reaction (qRT-PCR) (n = 4). (K) VEGF-A fluorescence in situ hybridization (FISH). DAPI, 4cence in situ hybridization ( (L and M) Staining and quantification of Tbx3+-BCs in control and anti-VEGFR2–injected abdominal skin at dpc16 (n > 200 cells, three mice). (N and O) Staining and quantification of PH3+-BCs (n > 200 cells, three mice). (P) Tbx3/β4 integrin staining in wounded back skin of control and VEGFR2 cKO mice. Arrowheads indicate Tbx3+-BCs. (Q) Quantification of Tbx3+-BCs (n > 100 cells, three mice). (R and S) Staining and quantification of Tbx3+-BCs (arrowheads) in WT and K14-VEGF back skin (n > 200 cells, three mice). (E, F, G, I, J, M, O, Q, and S) Error bars show SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by Tukey’s multiple comparison tests (E and F) and two-tailed Student’s t test (G, I, J, M, O, Q, and S). (H, K, L, N, and R) White dashed lines indicate BMs.
Mechanical stretch induces vasculature-driven EPC formation
Next, we examined whether mechanical stretch is relevant to the vasculature-EPC axis. To this end, we stretched the abdominal skin of male mice by sandwiching the abdominal skin with clips that were fastened on the back (Fig. 5A). Exposing the skin to 50% uniaxial static stretch for four consecutive days increased the blood vessel density (Fig. 5, B to D, and fig. S6B), blood vessel branching points (Fig. 5E), Esm1+ endothelial tip cell populations (Fig. 5, F and G), and VEGF-A mRNA expression (Fig. 5, H and I) in the dermis. In addition, EPC components [Tbx3+-BCs (Fig. 5, J and K), Adam8+-BCs (Fig. 5, L and M), and p-ERK+–BCs (Fig. 5, N and O)] and IFEBC proliferation (Fig. 5, P and Q) were increased under stretched conditions. Moreover, PYY+-BCs were increased under the stretched condition (fig. S6E). Stretch-induced EPCs (Fig. 5, R to W) and dermal angiogenesis (Fig. 5, X and Y) were attenuated in VEGFR2 cKO mice, indicating a pivotal role of blood vessels in EPC induction. Together, mechanical stretch recapitulates pregnancy-associated skin remodeling by inducing dermal angiogenesis that mediates the generation of EPCs to drive epidermal expansion.
Fig. 5. Mechanical stretch induces vasculature-mediated EPC formation.
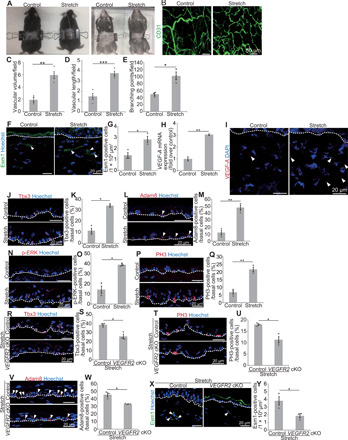
(A) Methods for stretching abdominal skin. (B) Whole-mount images of CD31 (Lyve1−) in abdominal dermis. (C and D) Quantification of vascular volume and length (n > 3 areas, three mice). (E) Quantification of vascular branching points (n = 3 areas, three mice). (F and G) Staining and quantification of dermal Esm1+ cells (n > 10 areas, three mice). (H) VEGF-A qRT-PCR (n = 3). (I) VEGF-A FISH. (J and K) Staining and quantification of Tbx3+-BCs (n > 150 cells, three mice). (L and M) Staining and quantification of Adam8+-BCs (n > 100 cells, three mice). (N and O) Staining and quantification of p-ERK+–BCs (n > 120 cells, three mice). (P and Q) Staining and quantification of PH3+-BCs (n > 100 cells, three mice). (R and S) Staining and quantification of Tbx3+-BCs in control and VEGFR2 cKO abdominal skin under stretching (n > 150 cells, three mice). (T and U) Staining and quantification of PH3+-BCs (n > 150 cells, three mice). (V and W) Staining and quantification of Adam8+-BCs (n > 200 cells, three mice). (X and Y) Staining and quantification of dermal Esm1+ cells (n > 8 areas, three mice). (C, D, E, G, H, K, M, O, Q, S, U, W, and Y) Error bars show SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, by two-tailed Student’s t test. (F, I, J, L, N, P, R, T, V, and X) White dashed lines indicate BMs. Photo credit: Ryo Ichijo, Institute for Frontier Life and Medical Sciences, Kyoto University.
Last, we investigated how blood vessels induced EPCs. Dermal angiogenesis occurred in Tbx3 cKO mice as the control during pregnancy (fig. S6, F and G), which excluded the possibility that Tbx3+-BCs induced angiogenesis. Our previous study had shown that α–smooth muscle actin–positive (αSMA+)/vimentin+ cells in the dermis, which are either myofibroblasts or vascular smooth muscle cells, were increased during pregnancy and Secreted frizzled-related protein 1 (Sfrp1) and Insulin like growth factor binding protein 2 (Igfbp2) that induce Tbx3+-BCs (16). We found that αSMA+/vimentin+ cells were also enriched in the dermis of both stretched and plantar skin (Fig. 6, A to D). In addition, inhibition of angiogenesis decreased dermal αSMA+/vimentin+ cells in both abdominal skin of pregnant mice (Fig. 6, E and F) and stretched skin (Fig. 6, G and H). Next, using the scRNA-seq data of vascular clusters, we examined genes up-regulated and down-regulated in the endothelial cell clusters and the vascular smooth muscle cell cluster during pregnancy (fig. S5D). We found that expression of Fstl1, a secreted protein that induces myofibroblasts (31), was increased at dpc16 in both endothelial cell clusters and the vascular smooth muscle cell cluster (fig. S5E). In addition, Igfbp2 was increased at dpc16 in the vascular smooth muscle cell cluster (fig. S5E). Furthermore, stretched skin had increased expression of Igfbp2 in the dermis (Fig. 6, I and J), which was attenuated in VEGFR2 cKO mice (Fig. 6, K and L). These results demonstrated that blood vessels induced Tbx3+-BCs and EPCs by secreting Igfbp2 from vascular smooth muscle cells or by activating myofibroblasts.
Fig. 6. Vasculatures induce dermal-to-epidermal signals.
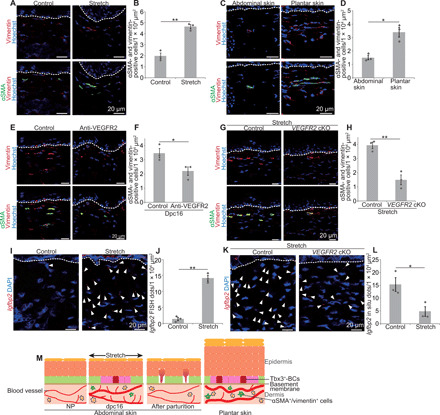
(A and B) Staining and quantification of dermal αSMA+/vimentin+ cells in the abdominal skin under control or stretched conditions. (C and D) Staining and quantification of dermal αSMA+/vimentin+ cells in the abdominal and plantar skin (n > 10 areas, three mice). (E and F) Staining and quantification of dermal αSMA+/vimentin+ cells in control and anti-VEGFR2–injected abdominal skin at dpc16 (n > 10 areas, three mice). (G and H) Staining and quantification of dermal αSMA+/vimentin+ cells in the abdominal skin of control and VEGFR2 cKO mice under stretched conditions (n > 10 areas, three mice). (I) Igfbp2 FISH in control or stretched abdominal skin. (J) Quantification of Igfbp2 FISH dots (n > 4 areas, three mice). (K) Igfbp2 FISH in the abdominal skin of control and VEGFR2 cKO mice under stretched conditions. (L) Quantification of Igfbp2 FISH dots (n > 4 areas, three mice). (M) Summary illustrating vasculature-induced EPC formation in the dynamic skin. (B, D, F, H, J, and L) Error bars indicate SEM. *P < 0.05 and **P < 0.01, by two-tailed Student’s t test. (A, C, E, G, I, and K) White dashed lines indicate BMs.
DISCUSSION
Our results uncovered the vasculature-mediated IFESC adaptation mechanisms for epidermal expansion in the dynamic skin. Upon an increase in the blood vessel density, IFESCs transit their cell state from a homeostatic stem cell state to a Tbx3+-primed stem cell state to generate EPCs in their vicinity, but no longer maintain their stemness when vasculature regression occurs (Fig. 6M).
We found that Tbx3+-BCs induced ADAM8 expression in neighboring cells. ADAM8 expression is highly relevant to tumorigenesis and cancer progression in which ADAM8 cleaves various tumor-associated molecules including extracellular matrix proteins, cytokine receptors, and adhesion molecules (19–21). ADAM8 may shape the microenvironment for epidermal cell proliferation of EPCs through ectodomain shedding of these substrates in addition to HB-EGF. The expression of ADAM8 in IFEBCs was induced by PYY. PYY is a well-known anorectic hormone that is mainly secreted from neuroendocrine L cells in the lower intestinal tract into circulating plasma in response to food intake (32). PYY has been reported to be expressed in the epidermal skin of humans (33), although its function in the skin has not been explored so far. There are two active forms of PYY, the full-length protein PYY1–36 and truncated form PYY3–36. The latter is abundant in circulating plasma and functions as an anorectic hormone. In this study, we injected PYY1–36 into the skin to induce EPCs, which suggested that PYY1–36 functioned as a paracrine signaling molecule in tissues that induced epidermal remodeling in the dynamic skin.
Vasculatures are induced by mechanical stretch and play a pivotal role in the generation of Tbx3+-BCs. This would explain the transient appearance of Tbx3+-BCs during pregnancy. The increase in mechanical tension on the abdominal skin during pregnancy induces angiogenesis to drive Tbx3+-BCs emergence, followed by vasculature regression and Tbx3+-BC differentiation with reduced tension after parturition. In a skin squamous cell carcinoma mouse model, interplay between tumors and vasculature creates a perivascular niche for cancer stem cells and regulates their stemness and pathogenesis (34, 35). In expanding abdominal skin of pregnant mice and plantar skin, it is unlikely that the vasculature itself is a niche for Tbx3+-BCs, because no vasculatures are located next to Tbx3+-BCs. The scRNA-seq analysis showed an increase in Igfbp2 and Fstl1 in vascular cells during pregnancy, suggesting that vasculatures secrete the signal to epidermis in a paracrine manner or affect dermal cell dynamics by inducing myofibroblasts, which mediate the Tbx3+-BC induction. Skin vasculatures, including lymphatic vessels, have been identified as an HFSC niche to maintain HFSC quiescence (36, 37). As HFSCs enter quiescence during pregnancy (38), vasculatures may regulate epidermal dynamics in a dual manner: (i) directly acting on HFSCs to delay the hair cycle and (i) indirectly on IFESCs to enhance IFEBC proliferation. This dual opposite regulation of distinct stem cells by dermal vasculatures in different tissue compartments would ensure orchestrating skin remodeling throughout a tissue. Molecular interplay between vasculatures and dermal cells will need to be determined in the future, as is the nature of mechanosensory machineries that induce vascularization.
MATERIALS AND METHODS
Mice
Mice were maintained on a C57BL/6 genetic background. Eight- to 12-week-old male mice were used for plantar skin experiments and skin stretch assays. All other experiments were performed using 8- to 12-week-old female mice. The Tbx3creERT2 strain was generated as described below. RosatdTomato (39), K5-tTA (40), pTRE-H2B-GFP (17), K14creERT2 (25), K14–VEGF-A (30), and VEGFR2flox/flox (41) strains were obtained from the Jackson laboratories. The R26H2B-EGFP strain (42) (CDB0203K; www2.clst.riken.jp/arg/reporter_mice.html) was obtained from RIKEN Center for Developmental Biology. The Hbegf flox/flox (43) (RBRC01308) strain was provided by RIKEN BioResource Research Center through the National Bio-Resource Project of Ministry of Education, Culture, Sports, Science and Technology, Japan. VE-cadherin-CreER strain was generated previously (44). Adam8 KO (18) and Tbx3flox/flox (45) strains were provided by J. W. Bartsch (Philipps University Marburg) and A. Moon (University of Utah), respectively. All experiments were performed in accordance with the guidelines of the Kyoto University Regulation on Animal Experimentation. The animal experiments were approved by the Committee for Animal Experiments of the Institute for Frontier Life and Medical Sciences, Kyoto University. The sample size was chosen by experimental consideration and not a statistical method. The experiments were not randomized. Investigators were not blinded to allocation during experiments and outcome assessment.
Generation of Tbx3creERT2 mice
To construct the donor plasmid, we amplified the chromosome sequence flanking the stop codon of the Tbx3 locus as homology arms. The 5′- and 3′-homologous arms were amplified using the following primer pairs: 5′-homologous arm, atcgataagcttgattcaagtgcctcagtatcctg and ccctccggtgagccactgcaagacctgtctggcttgg; 3′-homologous arm, aaacaagaaaaacaaaatcgccc and ctgcaggaattcgatacaattcaataaattaaagtt. We introduced silent mutations to remove the CRISPR-targeting site within the 5′-homologous arm. The 2A-CreERT2 sequence was amplified using pCAG-CreERT2 (Addgene plasmid no. 14797) as a template with the following primers: tggctcaccggagggcagaggaagtcttctaacatgcggtgacgtggaggagaatcccggccctatgtccaatttactgacc and ttgtttttcttgttttcaagctgtggcagggaaac. We ligated these three fragments into the Eco RV site of the pBluescriptII SK(−) vector using an In-Fusion HD Cloning kit (Clontech). The plasmid expressing hCas9 and single guide RNA (sgRNA) was prepared by ligating an oligonucleotide (46) into the Bbs I site of px330 (Addgene plasmid no. 42230). All plasmids were purified using a PureLink HiPure Plasmid Midiprep kit (Invitrogen). Donor plasmids were digested by Spe I and Sal I and purified using a Gel Extraction kit (QIAGEN) to remove the vector backbone. The donor cassette and px330-Tbx3 sgRNA plasmid were injected into pronuclear stage eggs that were then transferred into the oviducts of pseudo-pregnant Institute of Cancer Research (ICR) female mice.
Southern blotting
Genomic DNA was digested by Stu I and separated on an agarose gel, followed by denaturation and neutralization of the gel. DNA was transferred to a hybond membrane (GE Healthcare) and hybridized with the probe amplified from the donor plasmid using the PCR DIG Labeling Mix (Roche) with the following primers: tggaagatactaagatactgtgtgc and tcagtgtccttgtcacccaa. The hybridized DNA bands were visualized using anti-digoxigenin–Alkaline phosphatase (AP) Fab fragments and CDP-Star chemiluminescence substrate (Roche), followed by image acquisition with an ImageQuant LAS 4000 mini (GE Healthcare).
H2B-EGFP pulse chasing and lineage tracing
For the H2B-EGFP pulse-chase assay, K5-tTA (tet-off);pTREH2B-GFP pregnant mice were fed with doxycycline-containing chow (1 g of doxy/1 kg) from dpc2 to dpc16. For lineage tracing, Tbx3creERT2;R26H2B-EGFP and K14creERT2;R26H2B-EGFP mice were treated with 3 and 0.2 mg of tamoxifen (Sigma-Aldrich)/25-g body weight, respectively. Tamoxifen was injected intraperitoneally into mice, and then skin pieces were prepared at the indicated time points. To measure the surviving clone fraction, the number of labeled clones in the basal layer was counted in an area of 0.34 mm2. The data were normalized to the initial clone density.
Whole-mount immunofluorescence
Pieces of abdominal and plantar skin were incubated in 20 mM EDTA at 37°C for 3 hours to peel the epidermis from the dermis. The epidermal sheets were fixed in 4% paraformaldehyde overnight. For immunofluorescence staining, epidermal pieces were blocked by incubation in 0.5% dry skim milk and 0.5% Triton X-100 on a shaker for 1 hour. The samples were incubated with primary antibodies at 4°C overnight. After washing the samples in phosphate-buffered saline (PBS), secondary antibodies and Hoechst were added. Images were acquired using an SP8 confocal microscope (Leica).
Immunofluorescence staining
Skin tissues were cryoprotected in 20% sucrose/PBS and frozen in optimal cutting temperature compound. The samples were sectioned and subjected to immunostaining. Tissue samples were fixed with 4% paraformaldehyde, followed by permeabilization with 0.5% Triton X-100 in tris-buffered saline for 10 min at room temperature. The sections were blocked with 5% bovine serum albumin at room temperature for 1 hour, incubated with primary antibodies at 4°C overnight, washed, and then incubated for 1 hour with secondary antibodies (Alexa Fluor 488– or cy3-conjugated goat anti-rabbit and anti-rat anti-chicken; Jackson ImmunoResearch). The following primary antibodies were used: anti-Tbx3 (rabbit, 1:200; Abcam, ab99302), anti-GFP (chicken, 1:1000; Abcam, ab13970), anti–keratin 5 (rabbit, 1:1000; BioLegend, 905504), anti–keratin 10 (mouse, 1:500; Santa Cruz Biotechnology, sc-23877), anti-vimentin (rabbit, 1:500; MBL, JM-3634-100), anti-αSMA (mouse, 1:500; Dako, M0851), anti-Adam8 (rabbit, 1:100; Biorbyt, orb389340), anti–β4 integrin [rat, 1:500; Becton, Dickinson and Company (BD), 553745], anti-CD31 (rat, 1:500; BD, 553370), anti-Lyve1 (rabbit, 1:500; Abcam, ab14917), anti-Ki67 (rabbit, 1:500; Novus, NB600-1209), anti-Esm1 (goat, 1:200; R&D Systems, AF1999), anti-PYY (rabbit, 1:1000; Abcam, ab22663), anti–p-ERK (rabbit, 1:500; Cell Signaling Technology, 9101), and anti–phospho-histone H3 (rabbit, 1:1000; Cell Signaling Technology, 9701). All images were acquired under the SP8 confocal microscope.
Administration of synthetic PYY peptides
NP mice were injected intradermally in the abdominal skin with a synthetic PYY1–36 peptide (0.8 μg/g of body weight) (Bachem, 4031137) or PBS for four consecutive days. The mice were analyzed the day after the injection.
Quantification of the blood vessel volume and length
Whole-mount immunofluorescence images of CD31 and Lyve1 in abdominal and plantar skin pieces were obtained. CD31+ Lyve1+ signals were removed from the images to exclude lymphatic vasculatures. CD31+ Lyve1− signals, autofluorescence of HFs, and the compensated volume and length of blood vessels were quantified by Volocity (PerkinElmer). The volume of blood vessels was calculated by the following formula: (CD31+ Lyve1− signals) − (autofluorescence of HFs) + (compensated volume of blood vessels) in a volume of 1100 μm by 1100 μm by 100 μm.
Quantification of blood vessel branching points
The numbers of blood vessel branching points were counted in the whole-mount images of skin blood vessels (CD31+ Lyve1−) in a volume of 1162.5 μm by 1162.5 μm by 100 μm obtained under the SP8 confocal microscope.
Administration of an anti-VEGFR2 antibody and wounding
Pregnant mice were injected intradermally in the abdominal skin with an anti-VEGFR2 antibody (25 μg/g of body weight) (Bio X Cell) (29) or PBS from dpc11 to dpc15. Wounding was induced by punching a round hole (5 mm in diameter) in the surface of back skin with a Biopsy Punch (Kai Medical). Isoflurane was used to anesthetize mice. Control (VEGFR2flox/flox) and VEGFR2 cKO mice were treated with tamoxifen for four consecutive days. Wounds were introduced on the back skin at 13 days after tamoxifen treatment and analyzed at 20 days.
Skin stretching experiment
The abdominal skin was sandwiched by clips that were fastened on the back, so that the abdominal skin was exposed to 50% uniaxial static stretch for 4 days. In control mice, the back skin was sandwiched by clips without stretching. VEGFR2 cKO mice were treated with tamoxifen every day for four consecutive days. A stretch assay was applied to the skin at 7 days after tamoxifen treatment.
Fluorescence in situ hybridization
Fresh embedded skin sections (15 μm) were processed for RNA in situ detection using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics), according to the manufacturer’s instructions. The RNAscope probes used were VEGF-A (436961) and Igfbp2 (405951).
RNA extraction from dermis and quantitative reverse transcription polymerase chain reaction
Subcutaneous fat was removed from skin with a scalpel, and the whole skin was incubated for 30 min in 0.25% trypsin (Nacalai) to remove the epidermis. Total RNA was purified from dermis pieces with the RNeasy Micro Kit (QIAGEN), according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed with random primers, and the obtained complementary DNA was subjected to quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis using the KAPA SYBR FAST Universal qPCR Kit. The primer sequences were as follows: VEGF-A, 5′-TGGGCTCTTCTCGCTCCGTAGTAG-3′ (forward) and 5′-GCCGCCTCACCCGTCCAT-3′ (reverse); β-actin, 5′-CCAGCCTTCCTTCTTGGGTAT-3′ (forward) and 5′-TGTTGGCATAGAGGTCTTTACGG-3′ (reverse).
RNA-seq of IFEBCs
Subcutaneous fat was removed from the skin with a scalpel, and the whole skin was trypsinized at 37°C for 1 hour to remove the dermis. The cell suspensions were filtered through strainers (70 μm; BD Falcon), collected by centrifugation (300g for 5 min), and resuspended in Dulbecco’s modified Eagle’s medium (Ca−). Cell suspensions were incubated with Pacific Blue anti-human/mouse CD49f (eBioscience) and eFluor660 anti-mouse CD34 antibodies (BioLegend) for 30 min on ice, washed twice with PBS, and then subjected to flow cytometric analysis. Cell isolation was performed on a FACSAria III sorter. RNA was extracted from CD49f+ and CD34− cells using the RNeasy Micro Kit. Quality RNA samples were selected (RNA integrity number, >7). RNA-seq libraries were generated using a TruSeq Stranded mRNA library prep kit (Illumina). The libraries were sequenced on a NextSeq 500 (Illumina), according to the manufacturer’s instructions. The sequenced reads were mapped to the mm10 mouse reference genome using TopHat2 (version 2.2.1) (47) with the GENCODE M15 annotation gene feature format (GTF) file after trimming adaptor sequences and low-quality bases by cutadapt-1.14 (48). Mapped reads with high mapping quality (MAPQ, ≥20) were used for further analyses. Fragments per kilobase of transcript per million mapped read values, fold changes, and q values were calculated using Cuffdiff (49) within the Cufflinks version 2.2.1 package and GENCODE M15 annotation file.
scRNA-seq of abdominal epidermis and dermis
Subcutaneous fat was removed from the skin with a scalpel, and the whole skin was incubated for 30 min in 0.25% trypsin to separate into the epidermis and dermis. The dermal tissue was cut into small pieces and treated with 0.2% collagenase type I (Gibco) at 37°C for 2 hours (50). Epidermal and dermal cells were dissociated into single cells by pipetting and passing through 40-μm (Falcon) and 20-μm [pluriSelect (PLS)] strainers. Single-cell suspensions were resuspended in 1% bovine serum albumin/PBS at 1000 cells/μl and barcoded with a 10× Chromium Controller (10× Genomics). RNA from the barcoded cells of each sample was subsequently reverse transcribed, and sequencing libraries were constructed with reagents from a Chromium Single Cell v2 reagent kit (10× Genomics), according to the manufacturer’s instructions. Sequencing was performed with the NextSeq 500, according to the manufacturer’s instructions. The 10× Genomics Cell Ranger pipeline (version 2.2.0) was used to perform sample demultiplexing, alignment to the mm10 reference genome and GFP sequence, barcode/Unique Molecular Identifier (UMI) processing, and gene counting for each cell. The Seurat package (v.2.3.4) (51, 52) and R-3.5.1 were used for quality checking, filtering, normalization, clustering analyses, and visualization. The slingshot package (v.1.0.0) (53) was used for pseudo-time analysis.
GO analysis
GO analyses for scRNA-seq and RNA-seq were performed using the DAVID web tool (54) and Ingenuity Pathway Analysis (IPA)/Metascape (55), respectively. Statistically significant enrichments corresponded to P values of less than 0.05.
Statistics and reproducibility
All experiments with or without quantification were independently performed at least three times with different mice. The statistical analysis used for each quantification is indicated in figure legends.
Acknowledgments
We thank R. Morita, T. Nakamura, T. Kondo, and M. Eiraku for technical advice and help with the scRNA-seq analysis. We thank J. W. Bartsh and A. Sehara for Adam8 KO mice and A. Moon for Tbx3flox/flox mice. We also thank M. Arico from the Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript. Funding: This work was supported by JSPS KAKENHI 16H06280 (to R.Ic.), 17H06809 (to R.Ic.), 19K17796 (to R.Ic.), 17H05640 (to F.T.), 16H06279 (to F.T.), and 19H03681 (to F.T.); AMED under grant number JP20gm5810029 (to F.T.); the KAO Foundation for Arts and Sciences (to R.Ic.); the Takeda Science Foundation (to F.T.); the Cell Science Research Foundation (to F.T.); and the Joint Usage/Research Center Program of Institute for Frontier Life and Medical Sciences Kyoto University (to A.S.). Author contributions: R.Ic. and F.T. conceived the project and designed the experiments. R.Ic. performed most experiments. M.K. and T.Y performed sequencing experiments and bioinformatics analyses. R.Is. and K.A. generated Tbx3creERT2 transgenic mice. Y.K. provided VE-cadherin-CreER mice. A.S. conducted experiments using K5-tTA (tet-off);pTREH2B-GFP mice. H.F. and K.T. contributed to dermal scRNA-seq. H.Ku. contributed to RNA-seq analysis. Y.I. contributed to plantar skin experiments. S.K. and H.M. performed in vitro fertilization experiments. H.Ki. and F.M. performed vasculature-related experiments. R.Ic. and F.T. wrote the manuscript, with input from T.Y., R.Is., and H.M. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The VE-cadherin-CreER mouse can be provided by Y.K. pending scientific review and a completed material transfer agreement. Requests for the VE-cadherin-CreER mice should be submitted to Y.K. All software used in this study is freely or commercially available. All data needed to evaluate the conclusions in the study are present in the article and/or Supplementary Materials. All RNA-seq data were deposited in the Gene Expression Omnibus under the accession numbers GSE136747 and GSE151212. The reviewers can access our private data using the following link. The data will become available to the public after publication of our manuscript: www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE136747 and www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151212. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/7/eabd2575/DC1
REFERENCES AND NOTES
- 1.Zimmerman A., Bai L., Ginty D. D., The gentle touch receptors of mammalian skin. Science 346, 950–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rognoni E., Watt F. M., Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. 28, 709–722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanpain C., Fuchs E., Epidermal homeostasis: A balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 10, 207–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rompolas P., Mesa K. R., Kawaguchi K., Park S., Gonzalez D., Brown S., Boucher J., Klein A. M., Greco V., Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton E., Doupé D. P., Klein A. M., Winton D. J., Simons B. D., Jones P. H., A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Lim X., Tan S. H., Koh W. L. C., Chau R. M. W., Yan K. S., Kuo C. J., van Amerongen R., Klein A. M., Nusse R., Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science 342, 1226–1230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mascré G., Dekoninck S., Drogat B., Youssef K. K., Broheé S., Sotiropoulou P. A., Simons B. D., Blanpain C., Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257–262 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Sánchez-Danés A., Hannezo E., Larsimont J.-C., Liagre M., Youssef K. K., Simons B. D., Blanpain C., Defining the clonal dynamics leading to mouse skin tumour initiation. Nature 536, 298–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez C., Chua W., Miremadi A., Quist S., Headon D. J., Watt F. M., The interfollicular epidermis of adult mouse tail comprises two distinct cell lineages that are differentially regulated by Wnt, Edaradd, and Lrig1. Stem Cell Rep. 1, 19–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy E., Neufeld Z., Cerone L., Wong H. Y., Hodgson S., Livet J., Khosrotehrani K., Bimodal behaviour of interfollicular epidermal progenitors regulated by hair follicle position and cycling. EMBO J. 35, 2658–2670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sada A., Jacob F., Leung E., Wang S., White B. S., Shalloway D., Tumbar T., Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat. Cell Biol. 18, 619–631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan D. W. M., Jensen K. B., Trotter M. W. B., Connelly J. T., Broad S., Watt F. M., Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development 140, 1433–1444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joost S., Zeisel A., Jacob T., Sun X., Manno G. L., Lönnerberg P., Linnarsson S., Kasper M., Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 3, 221–237.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng J. B., Sedgewick A. J., Finnegan A. I., Harirchian P., Lee J., Kwon S., Fassett M. S., Golovato J., Gray M., Ghadially R., Liao W., White B. E. P., Mauro T. M., Mully T., Kim E. A., Sbitany H., Neuhaus I. M., Grekin R. C., Yu S. S., Gray J. W., Purdom E., Paus R., Vaske C. J., Benz S. C., Song J. S., Cho R. J., Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep. 25, 871–883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghahramani A., Donati G., Luscombe N. M., Watt F. M., Epidermal Wnt signalling regulates transcriptome heterogeneity and proliferative fate in neighbouring cells. Genome Biol. 19, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichijo R., Kobayashi H., Yoneda S., Iizuka Y., Kubo H., Matsumura S., Kitano S., Miyachi H., Honda T., Toyoshima F., Tbx3-dependent amplifying stem cell progeny drives interfollicular epidermal expansion during pregnancy and regeneration. Nat. Commun. 8, 508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E., Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly K., Hutchinson G., Nebenius-Oosthuizen D., Smith A. J. H., Bartsch J. W., Horiuchi K., Rittger A., Manova K., Docherty A. J. P., Blobel C. P., Metalloprotease-disintegrin ADAM8: Expression analysis and targeted deletion in mice. Dev. Dyn. 232, 221–231 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Romagnoli M., Mineva N. D., Polmear M., Conrad C., Srinivasan S., Loussouarn D., Barillé-Nion S., Georgakoudi I., Dagg Á., McDermott E. W., Duffy M. J., McGowan P. M., Schlomann U., Parsons M., Bartsch J. W., Sonenshein G. E., ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis. EMBO Mol. Med. 6, 278–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlomann U., Koller G., Conrad C., Ferdous T., Golfi P., Garcia A. M., Höfling S., Parsons M., Costa P., Soper R., Bossard M., Hagemann T., Roshani R., Sewald N., Ketchem R. R., Moss M. L., Rasmussen F. H., Miller M. A., Lauffenburger D. A., Tuveson D. A., Nimsky C., Bartsch J. W., ADAM8 as a drug target in pancreatic cancer. Nat. Commun. 6, 6175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrad C., Benzel J., Dorzweiler K., Cook L., Schlomann U., Zarbock A., Slater E. P., Nimsky C., Bartsch J. W., ADAM8 in invasive cancers: Links to tumor progression, metastasis, and chemoresistance. Clin. Sci. 133, 83–99 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Goishi K., Higashiyama S., Klagsbrun M., Nakano N., Umata T., Ishikawa M., Mekada E., Taniguchi N., Phorbol ester induces the rapid processing of cell surface heparin-binding EGF-like growth factor: Conversion from juxtacrine to paracrine growth factor activity. Mol. Biol. Cell 6, 967–980 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi Y., Hirata M., Hasuwa H., Iwamoto R., Umata T., Miyado K., Tamai Y., Kurisaki T., Sehara-Fujisawa A., Ohno S., Mekada E., A metalloprotease–disintegrin, MDC9/meltrin-γ/ADAM9 and PKCδ are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. EMBO J. 17, 7260–7272 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreymueller D., Pruessmeyer J., Groth E., Ludwig A., The role of ADAM-mediated shedding in vascular biology. Eur. J. Cell Biol. 91, 472–485 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Vasioukhin V., Degenstein L., Wise B., Fuchs E., The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U.S.A. 96, 8551–8556 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holzer P., Reichmann F., Farzi A., Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut–brain axis. Neuropeptides 46, 261–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Y., Boncompagni A. C., Kim S. S., Gochnauer H. R., Zhang Y., Loots G. G., Wu D., Li Y., Xu M., Millar S. E., Regional control of hairless versus hair-bearing skin by Dkk2. Cell Rep. 25, 2981–2991.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha S. F., Schiller M., Jing D., Li H., Butz S., Vestweber D., Biljes D., Drexler H. C. A., Nieminen-Kelhä M., Vajkoczy P., Adams S., Benedito R., Adams R. H., Esm1 modulates endothelial tip cell behavior and vascular permeability by enhancing VEGF bioavailability. Circ. Res. 115, 581–590 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Witte L., Hicklin D. J., Zhu Z., Pytowski B., Kotanides H., Rockwell P., Böhlen P., Monoclonal antibodies targeting the VEGF receptor-2 (Flk1/KDR) as an anti-angiogenic therapeutic strategy. Cancer Metastasis Rev. 17, 155–161 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Detmar M., Brown L. F., Schön M. P., Elicker B. M., Velasco P., Richard L., Fukumura D., Monsky W., Claffey K. P., Jain R. K., Increased microvascular density and enhanced leukocyte rolling and adhesion in the skin of VEGF transgenic mice. J. Invest. Dermatol. 111, 1–6 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Maruyama S., Nakamura K., Papanicolaou K. N., Sano S., Shimizu I., Asaumi Y., van den Hoff M. J., Ouchi N., Recchia F. A., Walsh K., Follistatin-like 1 promotes cardiac fibroblast activation and protects the heart from rupture. EMBO Mol. Med. 8, 949–966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batterham R. L., Cowley M. A., Small C. J., Herzog H., Cohen M. A., Dakin C. L., Wren A. M., Brynes A. E., Low M. J., Ghatei M. A., Cone R. D., Bloom S. R., Gut hormone PYY(3-36) physiologically inhibits food intake. Nature 418, 650–654 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Dumont Y., Bastianetto S., Duranton A., Breton L., Quirion R., Immunohistochemical distribution of neuropeptide Y, peptide YY, pancreatic polypeptide-like immunoreactivity and their receptors in the epidermal skin of healthy women. Peptides 70, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Beck B., Driessens G., Goossens S., Youssef K. K., Kuchnio A., Caauwe A., Sotiropoulou P. A., Loges S., Lapouge G., Candi A., Mascre G., Drogat B., Dekoninck S., Haigh J. J., Carmeliet P., Blanpain C., A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature 478, 399–403 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Oshimori N., Oristian D., Fuchs E., TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K. N., Jain P., He C. H., Eun F. C., Kang S., Tumbar T., Skin vasculature and hair follicle cross-talking associated with stem cell activation and tissue homeostasis. eLife 8, e45977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gur-Cohen S., Yang H., Baksh S. C., Miao Y., Levorse J., Kataru R. P., Liu X., de la Cruz-Racelis J., Mehrara B. J., Fuchs E., Stem cell-driven lymphatic remodeling coordinates tissue regeneration. Science 366, 1218–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein J., Fletcher S., Roth E., Wu C., Chun A., Horsley V., Calcineurin/Nfatc1 signaling links skin stem cell quiescence to hormonal signaling during pregnancy and lactation. Genes Dev. 28, 983–994 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diamond I., Owolabi T., Marco M., Lam C., Glick A., Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J. Invest. Dermatol. 115, 788–794 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Hooper A. T., Butler J. M., Nolan D. J., Kranz A., Iida K., Kobayashi M., Kopp H., Shido K., Petit I., Yanger K., James D., Witte L., Zhu Z., Wu Y., Pytowski B., Rosenwaks Z., Mittal V., Sato T. N., Rafii S., Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell 4, 263–274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe T., Kiyonari H., Shioi G., Inoue K., Nakao K., Aizawa S., Fujimori T., Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis 49, 579–590 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Iwamoto R., Yamazaki S., Asakura M., Takashima S., Hasuwa H., Miyado K., Adachi S., Kitakaze M., Hashimoto K., Raab G., Nanba D., Higashiyama S., Hori M., Klagsbrun M., Mekada E., Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. U.S.A. 100, 3221–3226 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okabe K., Kobayashi S., Yamada T., Kurihara T., Tai-Nagara I., Miyamoto T., Mukouyama Y., Sato T. N., Suda T., Ema M., Kubota Y., Neurons limit angiogenesis by titrating VEGF in retina. Cell 159, 584–596 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Frank D. U., Carter K. L., Thomas K. R., Burr R. M., Bakker M. L., Coetzee W. A., Tristani-Firouzi M., Bamshad M. J., Christoffels V. M., Moon A. M., Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc. Natl. Acad. Sci. U.S.A. 109, E154–E163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Li J., Xiang J., Wen B., Mu H., Zhang W., Han J., Highly efficient generation of biallelic reporter gene knock-in mice via CRISPR-mediated genome editing of ESCs. Protein Cell 7, 152–156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kechin A., Boyarskikh U., Kel A., Filipenko M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 49.Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., Pachter L., Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.K. Tsutsui, H. Machida, R. Morita, A. Nakagawa, K. Sekiguchi, J. H. Miner, H. Fujiwara, Mapping the molecular and structural specialization of the skin basement membrane for inter-tissue interactions. bioRxiv 2020.04.27.061952 [Preprint]. 28 April 2020. 10.1101/2020.04.27.061952. [DOI] [PMC free article] [PubMed]
- 51.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A. H., Tanaseichuk O., Benner C., Chanda S. K., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/7/eabd2575/DC1


