Abstract
Trypsin inhibitors from tamarind seed have been studied in vitro and in preclinical studies for the treatment of obesity, its complications and associated comorbidities. It is still necessary to fully understand the structure and behaviour of these molecules. We purifed this inhibitor, sequenced de novo by MALDI-TOF/TOF, performed its homology modelling, and assessed the interaction with the trypsin enzyme through molecular dynamics (MD) simulation under physiological conditions. We identified additional 75 amino acid residues, reaching approximately 72% of total coverage. The four best conformations of the best homology modelling were submitted to the MD. The conformation n°287 was selected considering the RMSD analysis and interaction energy (–301.0128 kcal.mol−1). Residues Ile (54), Pro (57), Arg (59), Arg (63), and Glu (78) of pTTI presented the highest interactions with trypsin, and arginine residues were mainly involved in its binding mechanism. The results favour bioprospecting of this protein for pharmaceutical health applications.
Keywords: Tamarind, antitryptic, homology modelling, computational methods, protein-protein interaction
Introduction
Proteolytic enzymes are critical elements for several biological events, such as digestion, healing, viral replication and the blood clotting cascade, and these processes must be regulated with great precision1,2. Within proteolytic enzymes, more than a third are serine proteases (EC 3.4.21), which are subdivided into families according to catalytic mechanisms and specificity, especially the S1 peptidase family, with trypsin (EC 3.4.21.11) as the most representative member being mainly related to the digestive process3–5.
The proteolytic enzymes are regulated by protease inhibitors (PIs) and these natural regulators are present in multiple forms in the animals, plants and microorganisms1,2. The purification and characterisation of PIs biomolecules may lead to a molecule with structure and functions and used as a potential biomolecule for herbal medicine nutraceutical or even a medicine6–8.
The trypsin inhibitors from tamarind seed have already been studied from the perspective of biotechnology and health applications in vitro and in preclinical studies9–14. In our previous studies, eutrophic Wistar rats fed with partially purified trypsin inhibitor from Tamarind seeds (TTI) increased serum levels of the cholecystokinin (CCK) hormone, reduced food consumption and weight gain without altering true digestibility13. On the other hand, Costa et al.15, evaluating the effect of TTI on plasma CCK and leptin in Wistar rats with diet-induced obesity, found no increases in CCK with TTI treatment, despite decreased leptin concentrations. The treatment with TTI in Wistar rats with diet-induced obesity and metabolic syndrome (MS) reduced food consumption and the plasma concentration of tumour necrosis factor-alpha (TNF-α) regardless of weight loss, characterising TTI as a molecule with anti-inflammatory characteristics11.
To increase its potential effect, TTI was nanoencapsulated in chitosan and whey-protein (ECW), and results revealed an improvement in the function and stability of TTI12. In another study, ECW induced a significant reduction in fasting blood glucose in Wistar rats fed with a high glycemic index and glycemic load diet (HGLI)16. In addition, nanoencapsulation protected TTI and promoted the controlled release in vitro simulations of digestion under physiological conditions16. Also, TTI nanoparticles revealed no cytotoxic effect in intestinal cell lines (Caco-2 and CDD18-Co), nor significant changes in hematological parameters, liver and kidney functions monitored by blood markers in Wistar rats under a HGLI diet17, representing a promising formulation for the clinical application.
Another study purified the trypsin inhibitor from Tamarind seeds (pTTI), verifying it was a competitive inhibitor with 19.578 kDa, resistant to high temperatures and extreme pH10. Medeiros et al.10 also evaluated the effect of pTTI on plasma CCK and leptin in Wistar rats with diet-induced obesity, and results similar to the partially purified TTI were found15, without altering CCK concentrations, but reducing leptin, suggesting an improvement in the leptin resistance profile, a characteristic present in obesity and SM. Further, Carvalho et al.18 indicated pTTI, was able to reduce molecular and plasm levels of TNF-α (tumour necrosis factor-α), VLDL-C (very-low-density lipoprotein-cholesterol), and TG (triglycerides) in Wistar rats with diet-induced dyslipidemia. Thus, we have a protein with beneficial biological effects in experimental studies applied in the context of obesity and associated complications.
However, information about a better understanding of the structure and physical-chemical properties of pTTI are important for pTTI function’s enlightenment. Once the variety of bioactive compounds, such as PIs, in nature is limited, synthetic peptides presenting similar biological activity may be an alternative to overcome this barrier19,20.
Comprehension of the antitrypsin activity mechanism and its structure, through molecular dynamics (MD) under physiological conditions, may contribute to future research, such as in the synthesis of a bioactive peptide from pTTI. It is of great scientific interest to understand the interactions between biomolecules that are involved in biological reactions, especially those that are related to diseases at a global level, such as obesity, metabolic syndrome and its associated comorbidities; and to achieve this study objective, MD appear as an important computational tool21.
According to several discoveries in preclinical studies, biological activities attributed to TTI and considering this molecule is limited in nature, it is important to expand the protein identification to provide technical and scientific advances, such as the generation of bioactive peptides derived from the pTTI that attract the interest of the pharmaceutical industry. In this present work, the protein sequence of pTTI was extended, homology modelling was performed, and its interaction with the enzyme trypsin was validated using MD simulation.
Materials and methods
Plant material
The tamarind (Tamarindus indica L.) fruit, was obtained from the seed bank in Natal-RN, Brazil, and registered in the National System for the Management of Genetic Heritage and Associated Traditional Knowledge under the number AF6CE9C. The pulp was removed, the seeds were peeled and ground, and used for the procedures.
Purification and sequencing of the tamarind seed trypsin inhibitor
pTTI purification and preparation for MSMS analysis
The purification process was performed according to Medeiros et al.10. The disulphide bridges from cys were reduced, and the free thiol groups alkylated to obtain the protein’s secondary structure. The reduction and alkylation steps were carried out according to Medeiros et al.10 and the reduced and alkylated pTTI fractions were subjected to chromatographic analysis. The procedure was performed on reverse-phase HPLC on an analytical column (Pharmacia Biotech µRCP C2/C18 ST 4.6/100 mm, 120 Å, cod. No.175057–01). Solvent A (analytical water grade + 0.1% of trifluoroacetic acid (TFA)) and B (acetonitrile (ACN) + 0.1% TFA) were used. The pTTI purification was performed using the following gradient steps: three minutes with 5% of solvent B, to desalinate and remove the surfactant from the sample, a linear gradient of 5–95% of solvent B in 22 min (4.09% B.min−1), and a final step, of 5 min at 95% of solvent B, with a flow of 1 mL.min−1, monitored by UV detection at 216 nm for peptide detection and 280 nm for aromatic ring detection, in a run of 30 min.
Proteolytic digestion of pTTI and MALDI-TOF/TOF MS/MS mass spectrometry
The cleavage of the reduced and alkylated pTTI was performed with two sequencing grade proteolytic enzymes, according to the manufacturer’s instructions: trypsin (Sequencing Grade Modified Trypsin, Promega trypsin, v511A), with the use of surfactant (RapiGest SF, Water, Part 186001861) and endoproteinase GluC (Staphylococcus aureus Protease V8–Typical GluC Digest, P8100).
The products generated by the enzymatic cleavages with trypsin and GluC were centrifuged at 24,500 × g and 6 °C for 30 min, and the supernatants were eluted in a linear gradient of 5–95% solvent B in a C18 analytical column (Pharmacia Biotech µRCP C2/C18 ST 4.6/100 mm, 120 Å, cod. No.175057–01) with a flow of 1 mL.min−1. Each fraction was collected manually and analysed by mass spectrometry using MALDI-TOF/TOFMS/MS in positive mode.
Then, they were analysed in an ultrafleXtreme mass spectrometer (Bruker Daltonics, Bremen–Germany) after mixing 1:3 (v/v) with saturated solution of α-cyano-4-hydroxycinnamic acid (α-CHCA) (5 mg CHCA, 250 μL of ACN, 50 μL TFA 3% and 200 μL of deionised H2O) and application in MALDI plate. The MS and spectra were obtained in positive reflected mode in the range of m/z between 700 and 4500 and MS/MS spectra were acquired in the same mass range in LIFTTM method after external calibration using calibration standards (Protein Calibration Standard I and II) (Bruker Daltonics, Bremen–Germany). The new sequencing of the pTTI triptych fragments was performed by signalling and manual interpretation of the spectra, using the FlexAnalysis 3.4 software (Bruker Daltonics, Bremen–Germany).
Computational methods
Multiple alignments
The pTTI protein sequence obtained by MS/MS was compared with other sequences present in the protein database of NCBI (National Centre for Biotechnology Information) (https://www.ncbi.nlm.nih.gov/) and PDB (Protein Data Bank proteins) files sing the BLASTp algorithm (Basic Local Alignment Search Tool–protein)22 (https://blast.ncbi.nlm.nih.gov/Blast.cgi), with multiple alignments of protein sequences on the Clustal Omega server23 (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Homology modelling and validation of predicted models
The three-dimensional structure of the pTTI was determined based on multiple alignments with the four proteins, of vegetable origin, with the highest percentage identity (% ID) deposited on the NCBI/PDB24 database server: 4AN7_B, 4AN6_A14, 4J2K_A25 and 1TIE_A26 (https://www.rcsb.org/). Amino acids from the sequence with higher % ID protein sequence were used (4AN7_B) to fill the gaps14. Homology modelling was performed in the Modeller27 9.21 program (https://salilab.org/modeller/) to create 150 models, and models with the highest negative modules Discrete Optimised Protein Energy (DOPE) values were selected28 and validated by MolProbity server29 (http://molprobity.biochem.duke.edu/), according to the following parameters: residues with bad bonds, residues with bad angles, MolProbity score and Ramachandran plot. The model showing better stability and less steric impediment was chosen as the three-dimensional structure of the pTTI.
Conformations
The best homology model of the pTTI, was used to generate 500 different conformations through CONCOORD (from CONstraints to COORDinates)30 version 2.1.2 (https://www3.mpibpc.mpg.de/groups/de_groot/concoord/), according to geometric restrictions, which are based on the strength of the interaction and with a set of upper and lower geometric limits, for all pairs of atoms in interaction30. From all conformations generated by the program, four were selected based on the sum of violations, numerical value related to the correction of the distances of the positions of atoms that are involved in interatomic distances that violate the upper or lower limit of distance. The correction process is interrupted when the sum of violations is zero30.
Molecular dynamics (MD) simulation
From the four best pTTI conformations generated by CONCOORD, MD simulations were performed between each conformation and trypsin enzyme (PDB ID 2PTN)31. MD simulations were performed using GROningen MAchine for Chemical Simulations (GROMACS)32 version 2018.4 package implemented with the CHARMM36 force field33. The pTTI conformers and trypsin were placed 10 Å away before the simulations started. The transferable intramolecular potential with 3 points (TIP3P)34 water molecules were used to solvate the simulated systems. The systems neutralisation was achieved through the addition of counter ions.
The systems’ geometry was minimised by the steepest descent algorithm35 for 50,000 steps with a tolerance of 100 kJ mol−1 nm−1 followed by conjugate gradient algorithm36 for 10,000 steps with a tolerance of 100 kJ mol−1 nm−1.
The Leap-Frog algorithm37 was applied to integrate the motion equation with time step of 2.0 fs. The long-range interactions were modelled using particle-mesh Ewald sum (PME)38 with a cut-off of 1.2 nm. The van der Waals interactions were also calculated using the same threshold. Bonds involving hydrogen atoms were restrained using LINCS algorithm39. The Nosé–Hoover thermostat40,41 was used to fix the system temperature (310 K) in all production simulations, while the system pressure was controlled using a Parrinello–Rahman barostat42 in the NPT simulations. Four shorts 200 ps equilibrium dynamics with NVT and NPT ensembles were performed. Finally, 200 ns production MD simulation using NVT ensemble was carried out for each system to determine its interaction with trypsin.
After MD simulations, the most stable interaction complex model between the pTTI conformers and trypsin was analysed through the Interaction Potential Energy (IPE) (in kcal.mol−1)43, which can be defined as the total interaction energy between two groups (the sum of van der Waals and electrostatic contributions), calculated according to the Equation (1):
| (1) |
where IPEi,j is the interaction energy between a group of atoms i and a group of atoms j, and Ni and Nj are the total number of atoms on groups i and j, Velec and VvdW are the terms corresponding to electrostatic and van der Waals contribution, respectively. This parameter is often used to evaluate interaction energies in protein–ligand and protein–protein systems44.
Results
Purification and sequencing of the tamarind seed trypsin inhibitor
Purification and determination of molecular weight of the trypsin inhibitor of purified tamarind seed
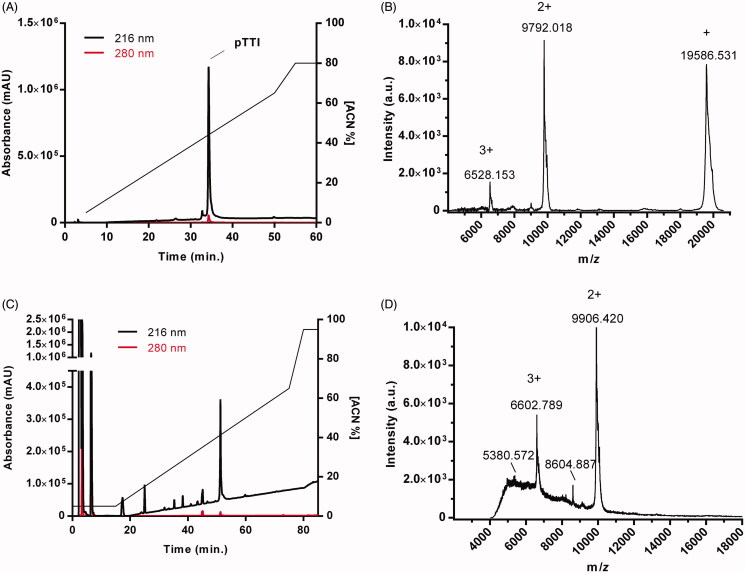
The chromatogram demonstrates the protein profile of the partially purified inhibitor (Figure 1(A)). The major peak was collected and named pTTI, eluted in 35 min in 45% of solvent B (ACN/TFA 0.1%). The pTTI presented different ions with m/z values [M + H]+ = 19,586 Da; [M + 2H]+ = 9792 Da e [M + 3H]+ = 6528 Da (Figure 1(B)).
Figure 1.
Identification of native pTTI and postreduction and alkylation by reverse-phase high-performance liquid chromatography and mass spectrometry. (A) Chromatographic profile of the TTI in the C18 Vydac reverse-phase analytical column. (B) Mass spectrum with the pTTI MALDI-TOF ionisation source. (C) Chromatographic profile of the reduced and alkylated pTTI in a C2/C18 Pharmacia Biotech µRCP analytical column. (D) Mass spectrum with reduced and alkylated pTTI MALDI-TOF ionisation source. TTI: Partially purified trypsin inhibitor from tamarind seeds. pTTI: Purified trypsin inhibitor from tamarind seeds.
The pTTI, when reduced and alkylated, showed a protein peak with greater absorbance with a retention time (RT) of 51 min and solvent concentration B of approximately 41% (Figure 1(C)). Consequently, pTTI was analysed by MALDI-TOF, verifying the reduction and alkylation reaction. The pTTI presented two predominant ions with m/z values [M + 3H]+ = 6602,789 Da and m/z [M + 2H]+ = 9,906,420 Da (Figure 1(D)), which indicates pTTI. It can be inferred that the pTTI is reduced and alkylated, since the difference in mass observed shows the presence of the acetamide group in the reduced thiol groups.
Proteolytic digestion of pTTI
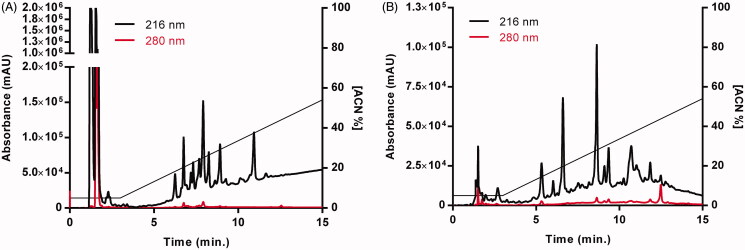
The chromatograms of the digestion profiles of both enzymes are shown in Figure 2. Within the initial 3 min of chromatography after enzymatic digestion, the digestion salts are eluted (Figure 2(A,B)), and in the case of trypsin digestion, the surfactant is also eluted (Figure 2(A)), followed by fractions resulting from digestion.
Figure 2.
Chromatograms of the reduced and alkylated pTTI by reverse-phase high-performance liquid chromatography treated with digestive enzymes with degree sequencing for mass spectrometry. (A) Chromatographic profile of pTTI digested with trypsin in a C2/C18 Pharmacia Biotech µRCP analytical column. (B) Chromatographic profile of pTTI digested with GluC in a C2/C18 Pharmacia Biotech µRCP analytical column. pTTI: Purified trypsin inhibitor from tamarind seeds.
Sequencing and analysis by mass spectrometry by MALDI-TOF/TOF MS/MS
The peptide sequences from reduced and alkylated pTTI determined were show in Table 1, with peptides number 1, 2, 4 and 6 originating from digestion with trypsin and peptides number 3 and 5 coming from digestion with GluC. The ions acquired by MALDI-TOF/TOF, of each proteolytic digestion, with sequential signals are highlighted in Supplementary Fig. S1. The mass spectra of the peptides with the y and b-ions series assigned are shown in Supplementary Fig. S2.
Table 1.
Sequences of peptide fragments of the reduced and alkylated pTTI determined by MALDI TOF/TOF in LIFT method.
| Peptide | Sequence | [M + H]+ Theoretica |
[M + H]+ (MALDI) |
|---|---|---|---|
| 1 | DTVHDTDGQVP(I/L)NNAQGYY(I/L) (I/L)PAQQGK | 2942.42 Da | 2942.46 Da |
| 2 | (I/L)FDEQSSEKGYTPVK | 1727.84 Da | 1727.84 Da |
| 3 | QGYT(PV)K(I/L)SDDFSSAAPFK (I/L)KQFEE | 2832.37 Da | 2832.30 Da |
| 4 | QGFEEDYK(I/L)VYC*SK | 1765.77 Da | 1765.96 Da |
| 5 | DYK(I/L)VYC*SKSE | 1391.65 Da | 1391.62 Da |
| 6 | (I/L)V(I/L)KEGDPFKVKFKKVDEES | 2335.29 Da | 2335.25 Da |
C*: Carbamidomethylcysteine; pTTI: Purified trypsin inhibitor from tamarind seeds.
The sequencing identified additional 75 amino acid residues, expanding the coverage percentage of the sequencing from 28% (with the sequencing of 53 previously published residues) to 72% in total, comprising 128 amino acid residues elucidated from the pTTI. Peptide 4 showed a glycine residue, which is not found in peptide number 3 that contains part of the same fragment, this may occur due to the presence of pTTI isoform fragment. It is not possible to distinguish leucine and isoleucine residues and glutamine and lysine, by mass spectrometry because their molecular masses are identical, with 113 Da and 128 Da, respectively.
Computational methods
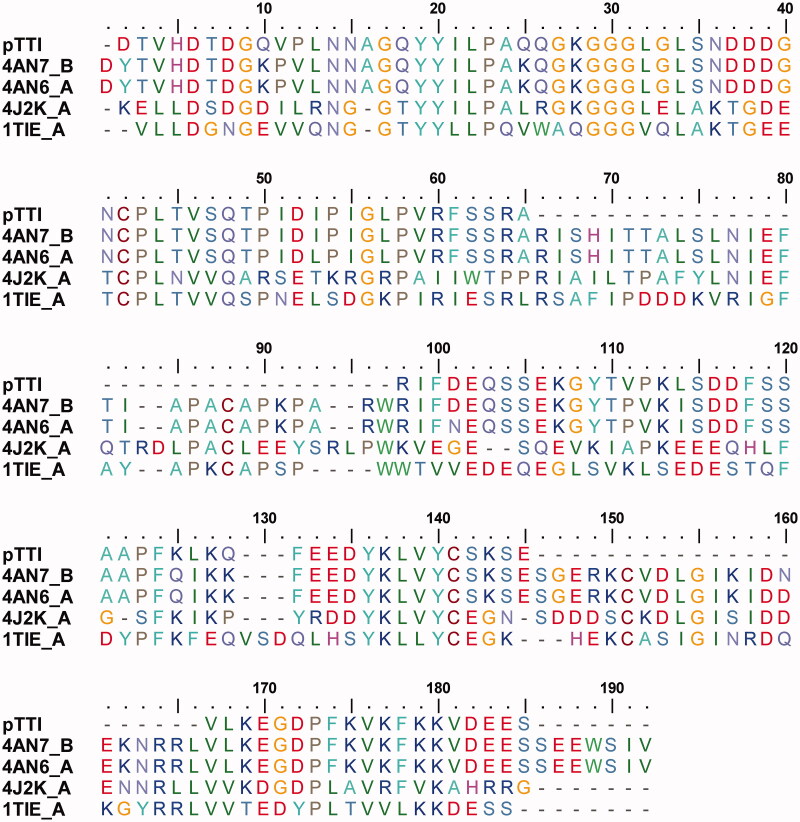
Table 2 shows the proteins with the highest identity with the pTTI. According to Figure 3, it is possible to observe the alignment of the primary structure of the pTTI with the sequences obtained in the BLASTp database, which were used to perform the homology modelling. Among the acquired sequences, it was possible to fill in the gaps remaining in the sequencing of the pTTI with the sequence of the highest percentage of identity, 4AN7_B, which comprises: 28 residues between numbers 66 and 93, 21 residues from number 138 to 158 and 7 residues in C’-terminal portion 178 to 184; totalling the addition of 56 amino acid residues.
Table 2.
Values obtained from the protein sequences selected in the BLASTp search.
| Accession | Max Score | Total Score | Query Cover | E value | %ID | Chain |
|---|---|---|---|---|---|---|
| 4AN7_B | 191.0 | 191.0 | 99% | 7e–63 | 66.48% | β |
| 4AN6_A | 188.0 | 188.0 | 99% | 7e–62 | 65.34% | α |
| 4J2K_A | 64.3 | 64.3 | 93% | 2e–13 | 30.77% | α |
| 1TIE_A | 59.7 | 59.7 | 78% | 1e–11 | 33.08% | α |
BLASTp: Basic Local Alignment Search Tool–protein; %ID: percentage identity.
Figure 3.
Multiple alignment of the primary partial sequence of pTTI with other sequences deposited in the tool Protein BLAST/NCB tooI. pTTI: Purified trypsin inhibitor from tamarind seeds.
The proteins mentioned above were used as a basic model for homology construction, and the five models with the highest DOPE score are shown in Table 3.
Table 3.
The five best three-dimensional models generated by Modeller and their respective DOPE values.
| Model Number | DOPE |
|---|---|
| 37 | –13092.01855 |
| 56 | –13057.27832 |
| 63 | –13081.18359 |
| 94 | –13165.39648 |
| 148 | –13107.49219 |
DOPE: Discrete Optimised Protein Energy.
The results of the validation in MolProbity to validate the predicted models are shown in Table 4.
Table 4.
MolProbity selection criteria for the five best models generated by Modeller.
| Model Number | Ramachandran favoured | Residues with bad angles | Residues with bad bonds | MolProbity score |
|---|---|---|---|---|
| 37 | 86.81% | 0.00% | 1.56% | 3.77 |
| 56 | 87.36% | 0.00% | 1.11% | 3.44 |
| 63 | 86.26% | 0.00% | 1.81% | 3.63 |
| 94 | 89.01% | 0.00% | 1.86% | 3.70 |
| 148 | 87.91% | 0.00% | 1.36% | 3.67 |
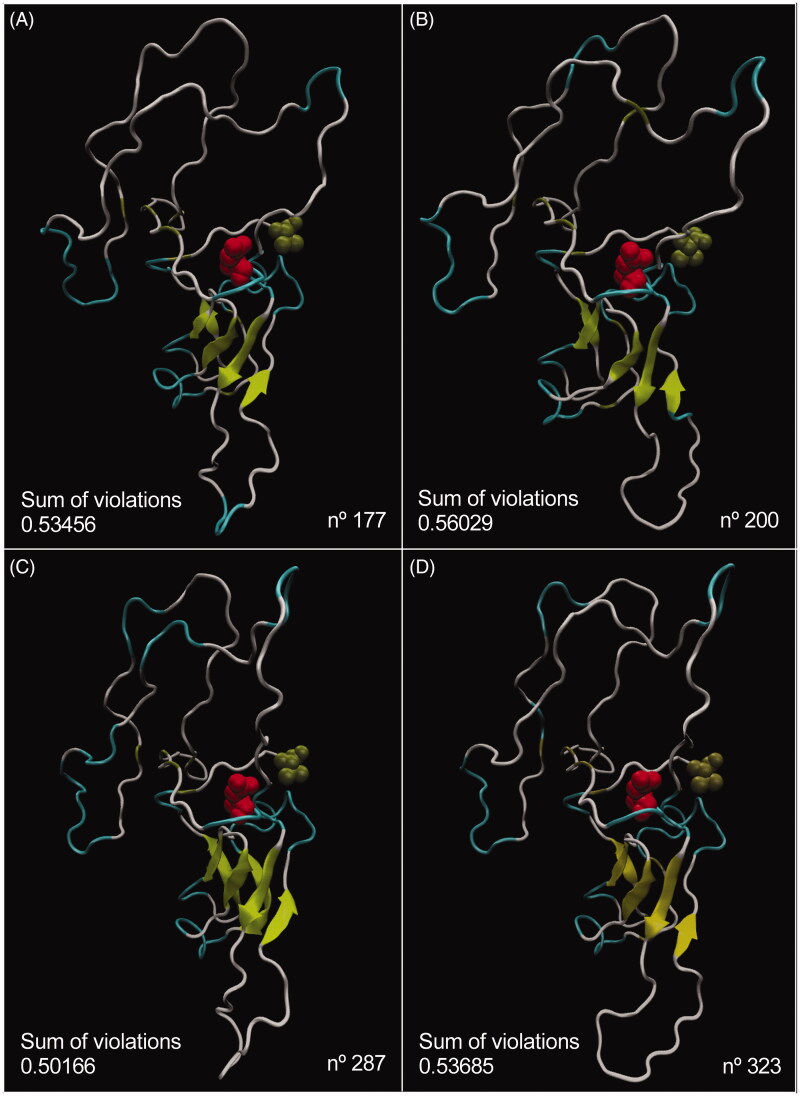
Due to a set of factors, such as residues with bad bonds and MolProbity score, the best result was achieved for model number 56, hence this model was used to generate 500 possible conformations for pTTI structure. among these conformations, four were selected according to the sum of the violations and the geometric restrictions (Figure 4).
Figure 4.
Visualisation of the four best conformations generated by CONCOORD of model n. 56 of the pTTI and their respective values of the sum of violations for conformations number 177 (A), 200 (B), 287 (C), and 323 (D). The N′-terminal portion which is a residue of Asp is highlighted in red and the C’-terminal portion, which is a residue of Val in tan, in all images. (The colours: yellow, β-sheet; tan, β-bridge; white, coil; cyan, turn; red, N’-terminal portion; tan, C’-terminal portion). pTTI: Purified trypsin inhibitor from tamarind seeds.
The best conformation of the pTTI obtained from model number 56 was subjected to MD simulations to analyse pTTI interactions with trypsin enzyme. The MD revealed a Root Mean Square Deviation (RMSD) plot as a function of time and in an aqueous system (Supplementary Fig. S2). Regarding the total energy of interaction, taking as a minimum and maximum point the interval between sudden changes, the results demonstrated the interaction conformation number 287 has the lowest interaction energy value (Table 5), suggesting, the conformation n. 287 was the best model and, therefore, may represent the putative pTTI structure.
Table 5.
Interaction potential energy of the molecular dynamics between the four best conformations of the pTTI with the enzyme trypsin.
| Conformer number | IPE (kcal.mol–1) |
|---|---|
| 177 | –172.8038 |
| 200 | –235.1159 |
| 287 | –301.0128 |
| 323 | –96.5838 |
IPE: interaction potential energy.
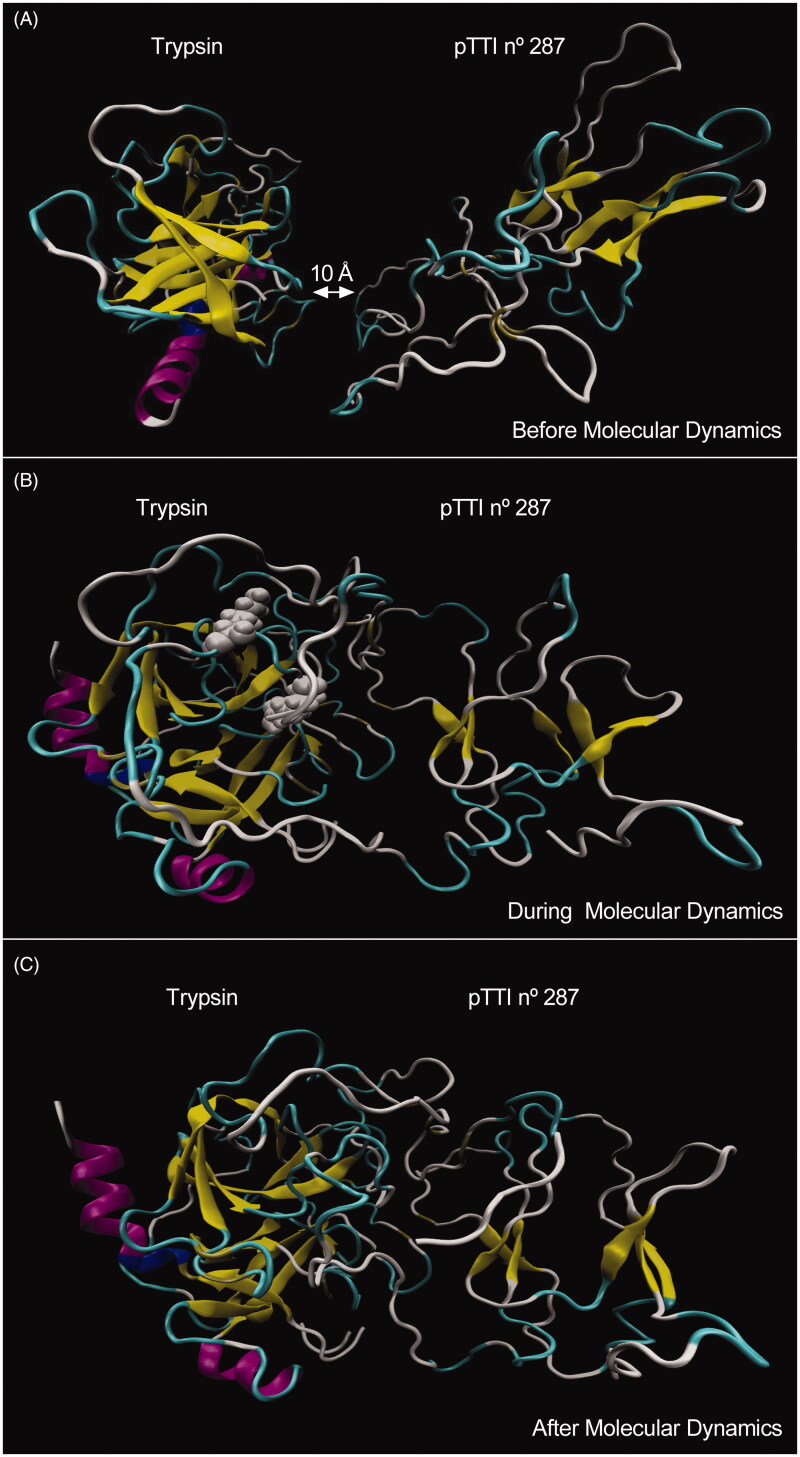
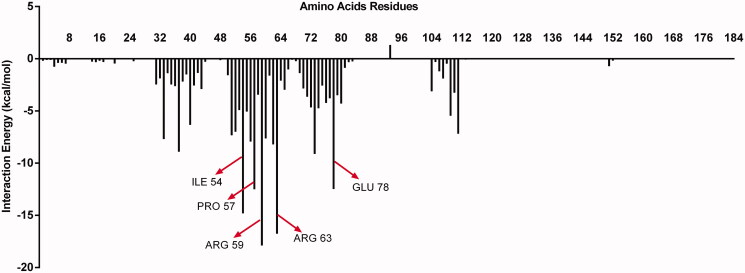
Figure 5 shows the 287 pTTI conformer and tripsin before, during and after the MD simulation. The amino acid residues of pTTI that most interacted with trypsin were Arg (59), Arg (63), Ile (54), Pro (57) and Glu (78) (Figure 6), which showed lower IPE. Among the residues, Arg (59) and (63) are those that have the greatest interaction with trypsin, and both are highlighted in grey (above Arg (63) and below Arg (59)) when MD occurs (Figure 5(B)).
Figure 5.
Visualisation of the stages of molecular dynamics simulation between model number 56 in conformation number 287 of pTTI with the enzyme trypsin. (A) Immediately before the MD, with the molecules positioned 10 A apart. (B) During MD, arginine residues are discarded, above Arg 63 and below Arg 59 (in grey, both). (C) Immediately after MD. (The colours: yellow, β-sheet; tan, β-bridge; white, coil; cyan, turn; blue, 310-helix; purple, α-helix). pTTI: Purified trypsin inhibitor from tamarind seeds.
Figure 6.
Representative graph of the interaction energy of each amino acid residue of the pTTI and enzyme trypsin. The highlighted residues have the lowest interaction energy, which represents those that have the greatest interaction with trypsin, among them Arg (59), Arg (63), Ile (54), Pro (57) and Glu (78). pTTI: Purified trypsin inhibitor from tamarind seeds.
Gathering all information obtained through CONCOORD and MD, the model number 56 in its conformation number 287 may represent the pTTI structure and its is interaction with trypsin, in a system under physiological conditions.
Discussion
Countless biological processes involving ligand-protein interactions and substances with potential bioactivity are investigated concerning healthcare applications45. Considering previously studies carried out with TTI and its bioactive properties in preclinical studies10,11,13,15,16,18, it is crucial to understand acutely this biomolecule.
In the current work, we pursue more information regarding a trypsin inhibitor’s identity extracted from a vegetal source of the tamarind fruit seeds’10,11. The protein sequence obtained through mass spectrometry considerably increased the coverage percentage compared to the previously known protein sequence. The former coverage sequence was nearby 52 amino acid residues10 and our efforts expanded the sequence to a total of 128 amino acid residues, representing 72% of the total pTTI molecular mass (19,586 Da).
Analysis regarding the protein identification pTTI exhibited high identity to tamarind Kunitz inhibitor–TKI14 (66% of identity–represents 127 hits of the 128 sequenced pTTI residues). The pTTI protein sequence’s missing gaps were manually completed considering the TKI protein sequence identity deposited in BLASTp database. Nevertheless, even with high similarity between both sequences, a difference of 989 Da in molecular masses between pTTI and TKI remains. This difference may be because they are isoforms.
The pTTI model was built up from other proteins recognised as trypsin inhibitors and also extracted from seeds, including two proteins from Tamarindus indica14, one from Enterolobium contortisiliquum25, and lastly one from Erythrina caffra26. All proteins described were extracted from plants belonging to the Fabaceae family, which impacts the relation of protein identity between them. In silico modelling demonstrated to be an extremely useful tool for hypotheses application, especially in molecular biology/biochemistry providing clearance regarding putative active sites and protein-ligand interactions and specificity of purified or designed biomolecules for drug discovery46. The three-dimensional structure model of the pTTI provided constructive information concerning pTTI interaction with trypsin, reinforcing the need for future studies with other molecules that may interact with pTTI, with applications in the healthcare system.
The MD performed in this study was between two large proteins under physiological conditions, unusual computational parameters, nonetheless with great relevance and application in the healthcare area. The best model fitting the MD parameters revealed arginine residue in two positions, which is positively charged; the isoleucine and proline residues, which have nonpolar and aliphatic characteristics; and glutamate, which has a negative charge47. Considering the two residues that presented the most significant interaction, Arg (59) and Arg (63), it is possible to evaluate the biomolecule dynamics and infer that the active site of trypsin for interaction with pTTI has a negative character. Thus, electrostatic interactions are observed between pTTI and trypsin, and these residues should receive attention as the target of further studies about pTTI.
A recently published study proposed a model system that measures the binding mechanism between trypsin and its inhibitor bovine pancreatic trypsin inhibitor (BPTI)–an inhibitor type Kunitz, known as aprotinin48 – using MD approach. The electrostatic bonding is mainly performed by Van der Waals force, with the trypsin binding cavity being negative and BPTI positively charged. BPTI Arg (17) is a crucial residue that requires conformational rearrangement to bind into trypsin’s active site perfectly and is responsible for the protein complex maintainance49. Therefore, the residues Arg (59) and Arg (63) of pTTI may also be critical for the pTTI inhibition mechanism.
Furthermore, the importance of the aqueous medium during the interaction process’ between proteins is also discussed. In the trypsin and BPTI interaction, the solvation layer is an agent that enhances the electrostatic force between the molecules, bringing the molecules closer together and displacing the water molecules throughout protein-protein interactions49. Consequently, an MD under physiological conditions is a crucial factor for the effective study of understanding the interaction, considering water in the environment through in vivo/in vitro reactions. These insights are essential to study the mechanism of interaction between proteins, which is suggested to be similar between several protein-protein complexes49 and may be extrapolated to the interaction between pTTI and trypsin.
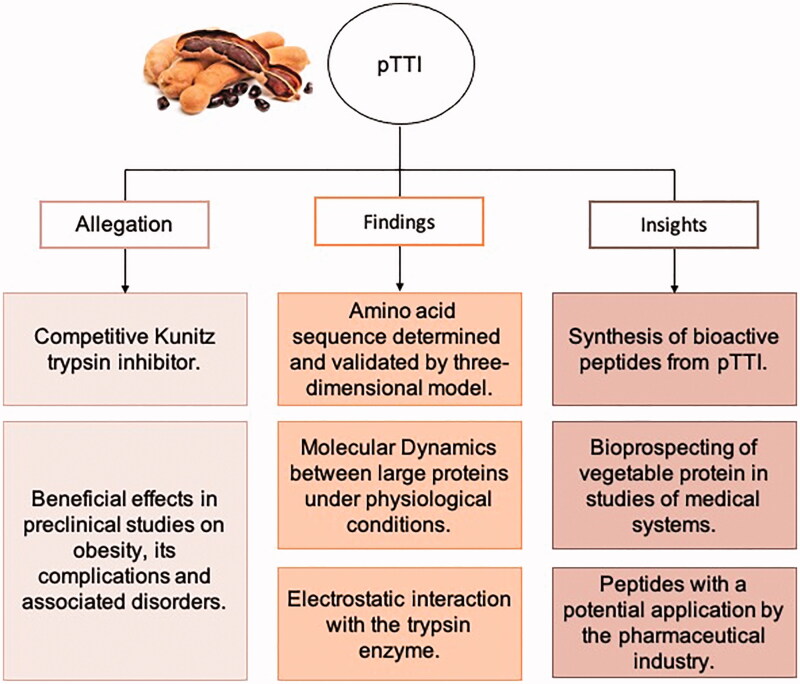
Previous studies have shown the activities and health application perspectives of Tamarind enzyme inhibitors50, and with pTTI, findings and insights are gathered in Figure 7. The exploration of computational technologies is a handling tool for unravelling the pTTI structure and innovation towards the bioprospecting of active molecules from plant origin, useful for the biotechnology industry. Designed bioactive peptides, MD studies with other proteins related to energy metabolism (membrane receptor), and MD with other compounds support new application perspectives, whether in silico, in vitro, or in vivo studies. Therefore, the study of these molecules directly influences future proposals for application in healthcare-related to obesity, its complications, and associated comorbidities, thus linked to a possibility of interest to the pharmaceutical industry.
Figure 7.
Summary of claims in pre-clinical studies already documented, findings and insights related to the biochemical characterisation of pTTI presented in this paper. pTTI: Purified trypsin inhibitor from tamarind seeds.
Supplementary Material
Acknowledgements
We thank the support of Dr. Carlos Bloch Jr. and Dr. Maura Vianna Prates, researchers at the Laboratory of Spectrometry Mass of Embrapa Brazilian Agricultural Research Corporation (Embrapa)–Genetic Resources and Biotechnology (Cenargen). We also thank Dr. José de Lima Cardozo Filho for data interpretation and conducting the mass spectrometry experiments. The authors thank Embrapa-Cenargen and High-Performance Computing Center (NPAD) at Federal University of Rio Grande do Norte (UFRN), the National High-Performance Processing Center of the Federal University of Ceará (UFC) for providing computational resources.
Funding Statement
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [CAPES–Finance Code 001] and the Conselho Nacional de Desenvolvimento Cientítico e Tecnológico [CNPq–Award Number: 426116/2018–6].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Farady CJ, Craik CS.. Mechanisms of macromolecular protease inhibitors. Clin Lymphoma 2010;11:19–222341–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laskowski, M, Jr, Kato I.. Protein inhibitors. Annu Rev Biochem 1980;49223:593–626. [DOI] [PubMed] [Google Scholar]
- 3.Page MJ, Di Cera E.. Serine peptidases: classification, structure and function. Cell Mol Life Sci 2008;65:1220–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Cera E. Serine proteases. IUBMB Life 2009;61:510–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos EA, Oliveira AS, Arajo Rablo LM, Ferreira A, Arajo Morais AH. Affinity chromatography as a key tool to purify protein protease inhibitors from plants. In: Affinity chromatography. InTech; 2012. p. 35. http://www.intechopen.com/books/affinity-chromatography/affinity-chromatography-as-a-key-tool-to-purify-protease-inhibitors-from-plants.
- 6.Ryan CA. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol 1990;28:425–49. [Google Scholar]
- 7.Fan S-G, Wu G-J.. Characteristics of plant proteinase inhibitors and their applications in combating phytophagous insects. Bot Bull Acad Sinica 2005;46:273–92. [Google Scholar]
- 8.Oliveira AS, Pereira RA, Lima LM, et al. Activity toward bruchid pest of a Kunitz-type inhibitor from seeds of the algaroba tree (Prosopis juliflora D.C.). Pestic Biochem Phys 2002;72:122–32. [Google Scholar]
- 9.Fook J, Macedo LLP, Moura GEDD, et al. A serine proteinase inhibitor isolated from Tamarindus indica seeds and its effects on the release of human neutrophil elastase. Life Sci 2005;76:2881–91. [DOI] [PubMed] [Google Scholar]
- 10.Medeiros AF, Costa I de S, Carvalho FMC, et al. Biochemical characterisation of a Kunitz-type inhibitor from Tamarindus indica L. seeds and its efficacy in reducing plasma leptin in an experimental model of obesity. J Enzyme Inhib Med Chem 2018;33:334–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho FMCC, Lima VCOO, Costa IS, et al. A trypsin inhibitor from tamarind reduces food intake and improves inflammatory status in rats with metabolic syndrome regardless of weight loss. Nutrients 2016;8:544–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Queiroz JLC, Costa ROA, Matias LLR, et al. Chitosan-whey protein nanoparticles improve encapsulation efficiency and stability of a trypsin inhibitor isolated from Tamarindus indica L. Food Hydrocolloids 2018;84:247–56. [Google Scholar]
- 13.Ribeiro J, Serquiz A, Silva P, et al. Trypsin inhibitor from Tamarindus indica L. seeds reduces weight gain and food consumption and increases plasmatic cholecystokinin levels. Clinics 2015;70:136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patil DN, Chaudhary A, Sharma AK, et al. Structural basis for dual inhibitory role of tamarind Kunitz inhibitor (TKI) against factor Xa and trypsin. FEBS J 2012;279:4547–64. [DOI] [PubMed] [Google Scholar]
- 15.Costa IS, Medeiros AF, Carvalho FMC, et al. Satietogenic protein from tamarind seeds decreases food intake, leptin plasma and CCK-1r gene expression in Obese Wistar Rats. Obes Facts 2018;11:440–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matias LLR, Costa ROA, Passos TS, et al. Tamarind trypsin inhibitor in chitosan–whey protein nanoparticles reduces fasting blood glucose levels without compromising Insulinemia: a preclinical study. Nutrients 2019;11:2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa RO de A, Matias LLR, Passos TS, et al. Safety and potential functional of nanoparticles loaded with a trypsin inhibitor isolated from tamarind seeds. Future Foods 2020;1–2:100001. [Google Scholar]
- 18.Carvalho FMC, Lima VCO, Costa IS, et al. Anti-TNF-α agent tamarind Kunitz trypsin inhibitor improves lipid profile of wistar rats presenting dyslipidemia and diet-induced obesity regardless of PPAR-γ induction. Nutrients 2019;11:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daliri E, Oh D, Lee B.. Bioactive peptides. Foods 2017;6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez Espitia PJ, de Fátima Ferreira Soares N, dos Reis Coimbra JS, et al. Bioactive peptides: synthesis, properties, and applications in the packaging and preservation of food. Compr Rev Food Sci Food Safety 2012;11:187–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Singh N, Li W, Molecular dynamics simulation of biomolecular interactions. In: Systems Medicine. Amsterdam: Elsevier; 2021:182–189. [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 23.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Sys Biol 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman HM, Battistuz T, Bhat TN, et al. The protein data bank. Acta Crystallogr D Biol Crystallogr 2002;58:899–907. [DOI] [PubMed] [Google Scholar]
- 25.Zhou D, Lobo YA, Batista IFC, et al. Crystal structures of a plant trypsin inhibitor from Enterolobium contortisiliquum (EcTI) and of its complex with bovine trypsin. PLoS One 2013;8:e62252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onesti S, Brick P, Blow DM.. Crystal structure of a Kunitz-type trypsin inhibitor from Erythrina caffra seeds. J Mol Biol 1991;217:153–76. [DOI] [PubMed] [Google Scholar]
- 27.Webb B, Sali A.. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 2016;54:139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Šali A, Blundell TL.. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993;234:779–815. [DOI] [PubMed] [Google Scholar]
- 29.Williams CJ, Headd JJ, Moriarty NW, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci 2018;27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groot BL, Van Aalten DMF, Scheek RM, et al. Prediction of protein conformational freedom from distance constraints. Pro Struc Funct Genet 1997;29:240–51. [DOI] [PubMed] [Google Scholar]
- 31.Jung SH, Kim C-K, Lee G, et al. Structural analysis of recombinant human preproinsulins by structure prediction, molecular dynamics, and protein-protein docking. Genom Inform 2017;15:142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berendsen HJC, van der Spoel D, van Drunen R.. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 1995;91:43–56. [Google Scholar]
- 33.Huang J, Mackerell AD.. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 2013;34:2135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys 1983;79:926–35. [Google Scholar]
- 35.Arfken GB, Weber HJ, Harris FE.. Mathematical methods for physicists. Amsterdam: Elsevier Inc.; 2013. [Google Scholar]
- 36.Hestenes MR, Stiefel E.. Methods of conjugate gradients for solving linear systems. J Res Natl Bureau Standards 1952;49:409. [Google Scholar]
- 37.Van Gunsteren WF, Berendsen HJC.. A leap-frog algorithm for stochastic dynamics. Mol Simulat 1988;1:173–85. [Google Scholar]
- 38.Darden T, York D, Pedersen L.. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J Chem Phys 1993;98:10089–92. [Google Scholar]
- 39.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM.. LINCS: a linear constraint solver for molecular simulations. J Comput Chem 1997;18:1463–72. [Google Scholar]
- 40.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A Gen Phys 1985;31:1695–7. [DOI] [PubMed] [Google Scholar]
- 41.Nosé S, Klein ML.. Constant pressure molecular dynamics for molecular systems. Mol Phys 1983;50:1055–76. [Google Scholar]
- 42.Parrinello M, Rahman A.. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 1981;52:7182–90. [Google Scholar]
- 43.Amorim-Carmo B, Daniele-Silva A, Parente AMS, et al. Potent and broad-spectrum antimicrobial activity of analogs from the scorpion peptide stigmurin. Int J Mol Sci 2019;20:623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ribeiro LF, Tullman J, Nicholes N, et al. A xylose-stimulated xylanase-xylose binding protein chimera created by random nonhomologous recombination. Biotechnol Biofuels 2016;9:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salmaso V, Moro S.. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front Pharmacol 2018;9:923–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang Z. Advances in homology protein structure modeling. Curr Pro Pept Sci 2006;7:217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson DL, Cox MM, Lehninger principles of biochemistry. 6th ed. New York: W.H. Freeman and Company; 2013. [Google Scholar]
- 48.Ascenzi P, Bocedi A, Bolognesi M, et al. The bovine basic pancreatic trypsin inhibitor (Kunitz Inhibitor): a milestone protein. Curr Pro Pept Sci 2003;4:231–51. [DOI] [PubMed] [Google Scholar]
- 49.Kahler U, Kamenik AS, Waibl F, et al. Protein-protein binding as a two-step mechanism: preselection of encounter poses during the binding of BPTI and Trypsin. Biophys J 2020;119:652–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carvalho F. d, Maciel BLL, Morais AH, de Tamarind A.. Enzymatic inhibitors: activities and health application perspectives. Food Rev Int 2020;20:1–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.