Abstract
Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) is an accurate and convenient method for mRNA quantification. Selection of optimal reference gene(s) is an important step in RT-qPCR experiments. However, the stability of housekeeping genes in spinach (Spinacia oleracea) under various abiotic stresses is unclear. Evaluating the stability of candidate genes and determining the optimal gene(s) for normalization of gene expression in spinach are necessary to investigate the gene expression patterns during development and stress response. In this study, ten housekeeping genes, 18S ribosomal RNA (18S rRNA), actin, ADP ribosylation factor (ARF), cytochrome c oxidase subunit 5C (COX), cyclophilin (CYP), elongation factor 1-alpha (EF1α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone H3 (H3), 50S ribosomal protein L2 (RPL2), and tubulin alpha chain (TUBα) from spinach, were selected as candidates in roots, stems, leaves, flowers, and seedlings in response to high temperature, CdCl2, NaCl, NaHCO3, and Na2CO3 stresses. The expression of these genes was quantified by RT-qPCR and evaluated by NormFinder, BestKeeper, and geNorm. 18S rRNA, actin, ARF, COX, CYP, EF1α, GAPDH, H3, and RPL2 were detected as optimal reference genes for gene expression analysis of different organs and stress responses. The results were further confirmed by the expression pattern normalized with different reference genes of two heat-responsive genes. Here, we optimized the detection method of the gene expression pattern in spinach. Our results provide the optimal candidate reference genes which were crucial for RT-qPCR analysis.
1. Introduction
Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) is an accurate and convenient method of quantifying mRNA levels of gene expression [1]. Selection of appropriate reference genes is crucial for validating accurate gene expression [2, 3]. Improper reference genes used in data processing may lead to inaccurate and even wrong results [4]. The commonly used reference genes in RT-qPCR analysis are the housekeeping genes because they are usually expressed steadily at mRNA levels in any organs under various conditions [5, 6]. Although 18S ribosomal RNA (18S rRNA), actin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are usually taken as reference genes [5], their mRNAs are not always stable in any cases [7–10]. Therefore, selection and optimization of reference genes are important steps in RT-qPCR experiments.
Some statistical algorithms, such as BestKeeper, NormFinder, and geNorm, are widely used for analyses of reference genes for RT-qPCR. BestKeeper analyzes the stability by calculating the standard deviation (SD) of the quantification cycle (Cq) values of candidate reference genes [11]. NormFinder compares the variation within and between sample groups of candidate genes and calculates the stability value of each gene based on the 2−ΔCq of genes [12]. geNorm evaluates the stability of candidate genes through calculating the stability value based on the geometric mean of 2−ΔCq of genes and mean pairwise variation in sample groups, as well as providing the optimal numbers of reference genes under each condition [13].
Spinach (Spinacia oleracea) belongs to the Amaranth family and is rich in carotenoid, vitamins, and minerals. It is a favorite vegetable all over the world. In our previous study, the draft of the spinach genome has been sequenced, with 25,495 encoding genes predicted [14]. Moreover, the patterns of gene expression and protein abundance have been reported in spinach in response to diverse stresses (e.g., heat, salinity, heavy metal, and virus) using molecular genetic, transcriptomic, and proteomic approaches [15–23].
To date, there are only a few reports on gene characterization and function analyses in spinach in response to stresses. A recent study reported that the S. oleracea heat shock 70 (SoHSC70) was induced by heat stress, and overexpression of SoHSC70 enhanced the heat tolerance in spinach calli [24]. Besides, spinach cytochrome P450 85A1 (SoCYP85A1) was upregulated in response to Phytophthora nicotianae infection, and 35S-promoted SoCYP85A1 overexpression conferred resistance to P. nicotianae pathogen inoculation in tobacco [25]. In addition, the spinach nonsymbiotic hemoglobin family gene (SoHb) was induced by several stress treatments (i.e., polyethylene glycol, NaCl, H2O2, salicylic acid, and nitric oxide) but suppressed by a nitric oxide scavenger, nitrate reductase inhibitor, and nitric oxide synthase inhibitor. Overexpression of SoHb in Arabidopsis resulted in the decreases in nitric oxide level and sensitivity to nitrate stress [26]. In these studies, some housekeeping genes in spinach were used as reference genes in RT-qPCR experiments for normalization analysis. 18S rRNA was used as a reference gene to detect the expression patterns of chilling-/drought-responsive SoCAP85 (85 kD cold acclimation protein) [27], drought-/salt-/oxidative stress-responsive SoHb [26], 13 heat-responsive genes (including SoHSFB2b and SobZIP9) [22], 15 nitrate transport and assimilation-related genes [28], and anthocyanin biosynthesis-related genes in various spinach germplasms [29]. In addition, actin, GAPDH, and ubiquitin 5 (UBQ5) were used to normalize the expression levels of several genes in response to various stresses, such as drought [30], biotic stress [25, 31], gibberellic acid (GA) treatment, and gender-specific condition [32]. However, only one study evaluated the stability of these reference genes [27]. Five commonly used housekeeping genes (i.e., GAPDH, actin, 16S rRNA, tubulin alpha chain (TUBα), and 18S rRNA) were evaluated between partially/fully hydrated versus dry seeds of spinach under chilling, desiccation, and optimum conditions. Among them, 18S rRNA appeared to be most stable, but still fluctuated under several treatments [27].
Some other housekeeping genes, such as ADP ribosylation factor (ARF), cytochrome c oxidase (COX), cyclophilin (CYP), elongation factor 1-alpha (EF1α), histone H3 (H3), and 50S ribosomal protein L2 (RPL2), were widely used as reference genes for RT-qPCR analysis in many plants [33–40]. ARFs encode small GTP-binding proteins (GTPases) for regulating various biological processes (e.g., cell division, cell expansion, and cellulose biosynthesis) [41–43], and COX is the terminal oxidase for mitochondrial oxidative metabolism and ATP synthesis [44]. CYP is critical in facilitating protein folding, which is involved in diverse cellular processes, such as apoptosis [45], immune response [46], and spliceosome assembly [47]. In addition, the histones and RPL are the components of the chromosome and ribosome, respectively [48, 49], and EF1α is involved in the protein synthesis [50]. However, there is still little evaluation on these reference genes in spinach.
In this study, ten candidate genes (i.e., 18S rRNA, actin, ARF, COX, CYP, EF1α, GAPDH, H3, RPL2, and TUBα) were selected for reference genes in spinach. Three Excel programs (i.e., BestKeeper, geNorm, and NormFinder) were used to evaluate the stability of these candidate genes in different organs, as well as stresses of heat, heavy metal, NaCl, Na2CO3, and NaHCO3. Optimal reference genes for each condition were verified. In addition, the two stable reference genes, ARF and actin, and the commonly used reference gene TUBα were selected to normalize the mRNA levels of two representative heat-responsive genes (SobZIP9 and SoHSFB2b) on references.
2. Materials and Methods
2.1. Plant Materials and Growth Condition
A heat-resistant sibling inbred line of spinach, Sp75, was used in this study. Spinach plants were placed in a growth chamber with a temperature regime of 22/18°C, 10/14 h day/night cycle, and a relative humidity of 60%. The top third and fourth leaves, stems, roots, male flowers, and female flowers of plants with uniform growth were sampled at 50 days after planting. Seedlings were sampled at 10 days after planting. These samples were flash-frozen in liquid nitrogen and stored at -80°C for further experiment. Three biological replicates were taken for each organ, and at least three plants were used for each replicate.
2.2. Stress Treatment
Fifty-day-old spinach seedlings with uniform growth were used for stress treatment. For heat treatments, the plants were moved into a chamber (37°C) at 0, 1, 2, 4, 6, 8, 12, 24, and 48 h before they were sampled [15]. For the treatments with heavy metal, salt, and alkali, the seedlings were watered with 200 μmol/L CdCl2 [51], 200 mmol/L NaCl [52], 200 mmol/L NaHCO3 [53], and 100 mmol/L Na2CO3 [54, 55], and the treatment times are 0, 1, 3, 6, 12, 24, and 48 h [56]. The top third and fourth leaves as well as roots were sampled for RNA extraction. Three biological replicates were taken for each time point of all stress treatments, and at least three plants were used for each replicate.
2.3. RNA Isolation and First-Strand cDNA Synthesis
Plant samples were ground in liquid nitrogen with a mortar and a pestle. Total RNA was isolated from 100 mg sample powder with TRIzol™ LS Reagent (Invitrogen, USA). The RNA samples with 260/280 ratios ranging from 1.8 to 2.1 were used for the following experiment. Total RNA was also examined by electrophoresis with 1% agarose gel to ensure the integrity. One microgram of total RNA was used for first-strand cDNA synthesis in a 20 μL total volume with a mixture of oligo dT primer and Random 6-mer in PrimeScript™ RT reagent (Takara, Japan) according to the manufacturer's instructions.
2.4. Candidate Reference Gene Selection
Ten candidate genes (18S rRNA, actin, ARF, COX, CYP, EF1α, GAPDH, H3, RPL2, and TUBα) were selected for this study according to their homologous gene stability in other plant species, such as sweet potato (Ipomoea batatas) [38], radish (Raphanus sativus) [57], potato (Solanum tuberosum) [39], soybean (Glycine max) [40], kenaf (Hibiscus cannabinus) [34], corn poppy (Papaver rhoeas) [37], bladder campion (Silene vulgaris) [36], Achyranthes bidentata [35], and Baphicacanthus cusia [33]. The sequences of these candidate homologous genes in spinach were obtained from SpinachBase (http://www.spinachbase.org) [14, 58, 59].
2.5. Primer Design and Evaluation
The primer pairs of each reference gene were designed according to their sequences by using the online program Primer3Plus (Table 1) (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi) [60]. The mixed cDNA from all samples was used as a template in primer evaluation. The PCR amplification products of each primer pair were checked by 2% agarose gel electrophoresis and sequencing, and then, the specificity of each pair of primers was evaluated by melting curve analysis followed by the amplification in RT-qPCR. Standard curves of each primer pair were established using a 5-fold dilution series ([1/1], [1/5], [1/25], [1/125], [1/625], and [1/3125]) of template cDNA [61]. The amplification efficiencies (E) of these primer pairs were calculated by the slope of standard curves (E = 10 − 1/slope), and the correlation coefficients (R2) were acquired from the standard curves as well [62].
Table 1.
Primers used in comparison of candidate reference genes in Spinach.
| Gene name1 | Gene ID2 | Sequence of primer (forward/reverse) | Size of PCR product (bp) | Amplification efficiency (%) | Correlation coefficient (R2) |
|---|---|---|---|---|---|
| 18S rRNA | Spo14194 | GATTCCGACGAACAACTGCG | 141 | 98.96 | 0.998 |
| AAGTAACATCCGCCGATCCC | |||||
| Actin | Spo23599 | TGTTCACGACATCAGCCGAA | 138 | 99.36 | 0.999 |
| CGTCGGGTAGCTCGTAGTTC | |||||
| ARF | Spo09845 | CCGATAAGCTTGGCCTCCAT | 125 | 92.53 | 0.997 |
| AGCCTTGCTAGCGATGTTGT | |||||
| COX | Spo27102 | AGGTTGCTCATGCTGTCTTGA | 168 | 94.85 | 0.991 |
| CAACGACACTGATCTGGCCT | |||||
| CYP | Spo15438 | TCCTTTCCATGGCCAATGCT | 132 | 93.11 | 0.999 |
| CCCTAACAACGTCCATGCCT | |||||
| EF1α | Spo03008 | ACCTCTCAGGCTGATTGTGC | 173 | 102.80 | 0.995 |
| GAGTACTTGGGAGTGGTGGC | |||||
| GAPDH | Spo24687 | GGCTGCCATCAAGGAGGAAT | 129 | 93.56 | 0.999 |
| GCAATTCCAGCCTTGGCATC | |||||
| H3 | Spo20638 | AAGAAGCCTCACCGTTACCG | 178 | 94.48 | 0.998 |
| CCTCCTGAAGGGCCAAAACA | |||||
| RPL2 | Spo08157 | TTCTCGTCCGTCTCCCTTCT | 198 | 101.18 | 0.997 |
| TACCCTCACCACCACCATGA | |||||
| TUBα | Spo15071 | TAATGCCGCTGTTGCTACCA | 137 | 94.90 | 0.998 |
| CTCTCTGCACCTTGGCAAGA |
1Full names of these 10 genes are 18S ribosomal RNA (18S rRNA), actin, ADP ribosylation factor (ARF), cytochrome c oxidase (COX), cyclophilin (CYP), elongation factor 1-alpha (EF1α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone H3 (H3), 50S ribosomal protein L2 (RPL2), and tubulin alpha chain (TUBα). 2These sequences and IDs were obtained from SpinachBase (http://www.spinachbase.org).
2.6. Real-Time Quantitative PCR
RT-qPCR analysis was performed in 0.2 mL tubes with the Applied Biosystems 7500 Real-Time PCR System (ABI, USA). Each reaction contained 1 μL cDNA (5-time diluted), 10 μL AceQ Universal SYBR qPCR Master Mix (Vazyme, China), 0.5 μL of primer (500 μmol/L), and 8 μL deionized water. The PCR was carried out as the following program: predegeneration at 95°C for 3 min; 40 cycles of degeneration at 95°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s; and melting curve analysis at 65°C-95°C. RT-qPCR of each cDNA sample was carried out three times as technical replicates.
2.7. Stability Evaluation of Candidate Reference Genes
The Cq values of 10 candidate genes were put in an Excel sheet, and a boxplot of these Cq values was generated. The stability of these 10 genes was evaluated by three statistical Excel macro programs, including BestKeeper [11], NormFinder [12], and geNorm [13]. The Cq values of each gene in roots, stems, leaves, flowers, and seedlings as well as leaves and roots in response to various stresses (i.e., high temperature, NaCl, Na2CO3, NaHCO3, and CdCl2) were used in this evaluation. In BestKeeper, the standard deviation (SD) of the Cq values of each candidate gene was calculated, and the gene with the lowest SD was taken as the most stable gene [11, 63]. In NormFinder, the expression stability (M1) was calculated by Cq values obtained by RT-qPCR of candidate genes and ranked in each sample set. The lowest M1 indicates that the gene is most stable [12]. In geNorm, the stability of candidate genes was evaluated by relative expression levels (Q) transformed from the Cq values for each sample according to the formula of Q = 2−ΔCq(ΔCq = Cq value of each sample − the minimum Cq value in each set). This formula works under the precondition that the efficiency of primers should range from 90% to 105% [64, 65]. An average expression stability value (M2) of each candidate was calculated to demonstrate their stability. The gene with the lowest M2 value was regarded as the most stable expression. Besides, the geNorm software also determines the optimal number of reference genes required for RT-qPCR data normalization under each condition by pairwise variation (Vn/Vn+1) between the normalization factors NFn and NFn+1. If Vn/Vn+1 < 0.15, the first n is the optimal number of genes required for this condition [13].
2.8. Normalization of SobZIP9 and SoHSFB2b
The expression pattern of two heat-responsive genes SobZIP9 and SoHSFB2b [22] in spinach was detected in response to heat stress (37°C). The mRNA levels of SobZIP9 and SoHSFB2b were then normalized by the most stable genes suggested by NormFinder and BestKeeper, as well as TUBα, a commonly used reference gene. Primer pairs for SobZIP9 (qSoHSFB2b-F: TCTTTCCACACTCGCTCTGT, qSoHSFB2b-R: CGGATTACAAGAAGGCAGGC) and SoHSFB2b (qSobZIP9-F: TGCTGGAAACCCTAGGACTG, qSobZIP9-R: CTTCTGGTGCTTCTAGGCCT) [22] were used in this experiment. A linear ANOVA was used for evaluation of the variation of each gene in response to heat stress.
3. Results
3.1. Analysis of Primer Specificity and PCR Amplification
Ten candidates were chosen as reference genes for RT-qPCR analysis. They are 18S rRNA, actin, ARF, COX, CYP, EF1α, GAPDH, H3, RPL2, and TUBα. To evaluate the specificity of designed primers for the ten genes, the analyses of PCR, gel electrophoresis, and melting curves were performed. The gel electrophoresis showed a single band with expected size of each pair of primers (Figure 1), and the melting curves of each primer pair exhibited a single peak (Supplementary Figure S1), indicating the specificity of these primer pairs of candidate genes. The target amplicons were sequenced, and the results were consistent with their gene sequences in SpinachBase [14, 30, 58]. The standard curves indicated that the RT-qPCR amplification efficiency of candidate genes ranged from 92.53% (ARF) to 102.80% (EF1α), and the correlation coefficients varied from 0.991 (COX) to 0.999 (actin, CYP, and GAPDH) (Table 1 and Supplementary Figure S2). Thus, these primers are specific for their respective genes and can be used in RT-qPCR analysis.
Figure 1.
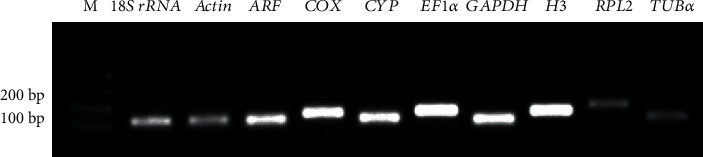
Specificity of primers for ten candidate reference genes in PCR and their amplicon sizes. The names of ten candidate reference genes including 18S ribosomal RNA (18S rRNA), actin, ADP ribosylation factor (ARF), cytochrome c oxidase (COX), cyclophilin (CYP), elongation factor 1-alpha (EF1α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), histone H3 (H3), 50S ribosomal protein L2 (RPL2), and tubulin alpha chain (TUBα) are noted on each lane. M represents 100 DNA ladder.
3.2. Expression Profiles of Ten Candidate Genes in Spinach
Analysis of the expression levels of ten candidates was performed in young seedlings, roots, stems, leaves, male flowers, and female flowers, as well as leaves and roots in response to stresses of heat, heavy metal, NaCl, Na2CO3, and NaHCO3. Cq values of ten candidates in various samples obtained by RT-qPCR were shown in a boxplot (Figure 2). The Cq values varied from 10.19 (18S rRNA in organs/seedlings) to 35.79 (TUBα in heat treatment) indicating that these candidate genes present different expression levels. The Cq value range reveals variability among the candidate genes. 18S rRNA shows the minimal range of Cq values in organs/seedlings (2.08, Figure 1(a)) and under NaCl treatment (0.86, Figure 1(d)), while actin (3.07, Figure 1(c)), ARF (1.64, Figure 1(c)), GAPDH (3.09, Figure 1(e)), and CYP (1.38, Figure 1(f)) show the minimal range of Cq values under heat, heavy metal (CdCl2), NaHCO3, and Na2CO3 treatments, respectively. These minimal ranges of Cq values indicated that these genes are more stable than others in each condition. However, further analyses are needed, because the comparison of the range of Cq values along is deficient to reveal the stability of the candidate genes. Thus, statistical macro programs were then used in this study.
Figure 2.
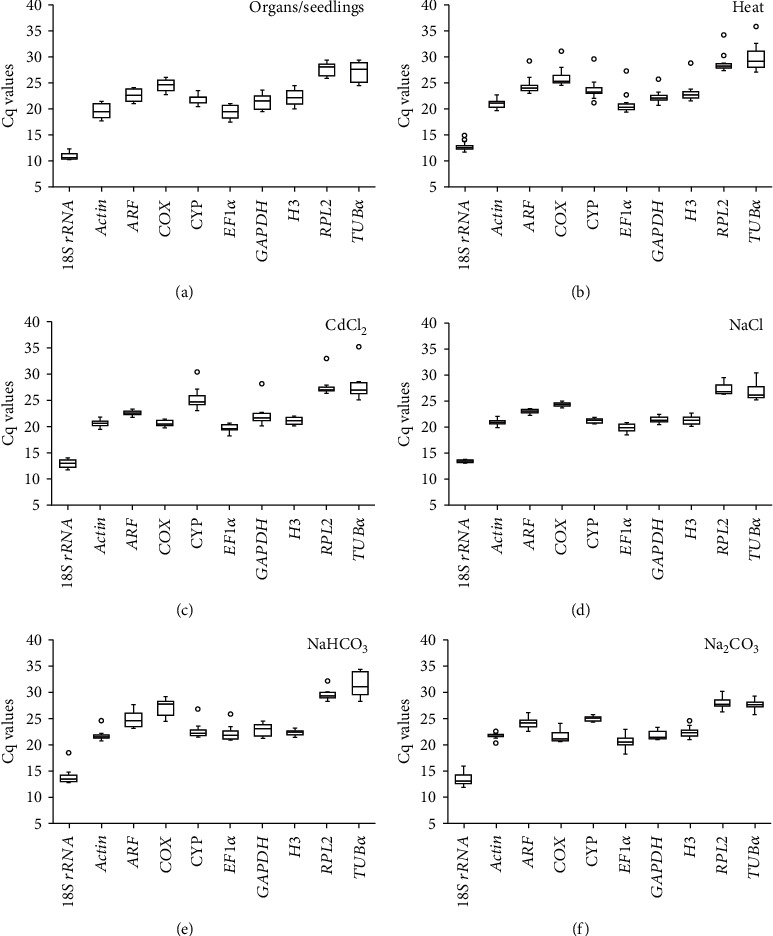
Cq values of ten candidate reference genes in spinach detected by RT-qPCR. The medians of Cq values for these genes are represented by the line in the boxes, and the upper and lower quartiles of Cq values are represented by the upper and lower boundaries of the boxes. The whiskers represent the ranges for the bottom 25% and the top 25% of these Cq values. Small circles represent outliers of these Cq values: (a) Cq values in organs including leaves, stems, roots, flowers, and seedlings, (b) Cq values in the leaves and roots under heat stress, (c) Cq values in the leaves and roots under heavy metal stress, (d) Cq values in the leaves and roots under NaCl, (e) Cq values in the leaves and roots under NaHCO3, and (f) Cq values in the leaves and roots under Na2CO3. Ten candidate reference genes refer to Figure 1. Three biological replicates were taken for different organs/seedlings, as well as leaves and roots at each time point of all stress treatments.
3.3. Expression Stability of Ten Candidate Genes
Three statistical Excel macro programs (i.e., BestKeeper, NormFinder, and geNorm) were used to evaluate the stability of ten candidate genes, in order to find optimal reference genes in spinach for RT-qPCR normalization (Table 2, Figures 3 and 4). The stability of candidate genes was evaluated in 6 data sets, organs/seedlings, leaves, and roots under heat, heavy metal, NaCl, NaHCO3, and Na2CO3, respectively.
Table 2.
The evaluation of candidate reference genes with NormFinder and BestKeeper1.
| Method | Ranking order (better-good-average) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Organs/seedlings2 | |||||||||||
| NormFinder | Gene3 | ARF | EF1α | COX | GAPDH | RPL2 | Actin | H3 | 18S rRNA | Tubα | CYP |
| M1 | 0.131 | 0.203 | 0.365 | 0.373 | 0.382 | 0.389 | 0.427 | 0.582 | 0.678 | 0.703 | |
| BestKeeper | Gene3 | 18S rRNA | CYP | COX | Actin | RPL2 | ARF | EF1α | H3 | GAPDH | Tubα |
| SD | 0.58 | 0.72 | 0.94 | 1.07 | 1.07 | 1.08 | 1.10 | 1.13 | 1.28 | 1.58 | |
| Heat | |||||||||||
| NormFinder | Gene3 | ARF | RPL2 | EF1α | CYP | H3 | GAPDH | COX | 18S rRNA | Actin | Tubα |
| M1 | 0.212 | 0.228 | 0.238 | 0.387 | 0.424 | 0.454 | 0.500 | 0.757 | 0.850 | 0.928 | |
| BestKeeper | Gene3 | Actin | 18S rRNA | GAPDH | RPL2 | ARF | H3 | EF1α | CYP | COX | Tubα |
| SD | 0.57 | 0.60 | 0.77 | 0.92 | 0.93 | 0.95 | 1.10 | 1.11 | 1.16 | 1.67 | |
| CdCl2 | |||||||||||
| NormFinder | Gene3 | CYP | RPL2 | ARF | EF1α | H3 | GAPDH | COX | Actin | 18S rRNA | Tubα |
| M1 | 0.625 | 0.65 | 0.75 | 0.751 | 0.765 | 0.791 | 0.836 | 0.91 | 0.987 | 1.621 | |
| BestKeeper | Gene3 | ARF | EF1α | CYP | Actin | 18S rRNA | RPL2 | H3 | GAPDH | COX | Tubα |
| SD | 0.37 | 0.49 | 0.53 | 0.54 | 0.62 | 0.98 | 1.05 | 1.22 | 1.33 | 2.09 | |
| NaCl | |||||||||||
| NormFinder | Gene3 | EF1α | ARF | CYP | GAPDH | H3 | COX | Actin | 18S rRNA | RPL2 | Tubα |
| M1 | 0.201 | 0.213 | 0.251 | 0.275 | 0.304 | 0.324 | 0.426 | 0.492 | 0.496 | 0.852 | |
| BestKeeper | Gene3 | 18S rRNA | ARF | COX | EF1α | Actin | GAPDH | CYP | H3 | RPL2 | Tubα |
| SD | 0.24 | 0.32 | 0.34 | 0.38 | 0.39 | 0.45 | 0.61 | 0.66 | 0.86 | 1.24 | |
| NaHCO3 | |||||||||||
| NormFinder | Gene3 | RPL2 | Actin | CYP | GAPDH | H3 | COX | ARF | EF1α | Tubα | 18S rRNA |
| M1 | 0.253 | 0.454 | 0.48 | 0.485 | 0.489 | 0.57 | 0.832 | 1.184 | 1.273 | 1.407 | |
| BestKeeper | Gene3 | Actin | RPL2 | H3 | GAPDH | COX | CYP | 18S rRNA | ARF | EF1α | Tubα |
| SD | 0.64 | 0.74 | 0.86 | 0.89 | 1.02 | 1.06 | 1.11 | 1.22 | 1.45 | 2.13 | |
| Na2CO3 | |||||||||||
| NormFinder | Gene3 | ARF | H3 | RPL2 | CYP | EF1α | Actin | Tubα | 18S rRNA | COX | GAPDH |
| M1 | 0.284 | 0.297 | 0.298 | 0.366 | 0.413 | 0.432 | 0.455 | 0.469 | 0.473 | 0.523 | |
| BestKeeper | Gene3 | COX | Actin | ARF | GAPDH | H3 | Tubα | CYP | RPL2 | EF1α | 18S rRNA |
| SD | 0.36 | 0.38 | 0.68 | 0.7 | 0.76 | 0.85 | 0.89 | 0.89 | 0.92 | 1.03 | |
1Lower stability value M1 obtained by NormFinder or SD obtained by BestKeeper indicates higher stability of housekeeping genes under each condition and is marked with bold text. 2These samples include leaves, stems, roots, flowers, and seedlings, as well as the leaves and roots under heat (37°C for 0, 1, 2, 4, 6, 8, 12, 24, and 48 h), heavy metal (200 μM CdCl2 for 0, 1, 3, 6, 12, 24, and 48 h), salt (200 mM NaCl for 0, 1, 3, 6, 12, 24, and 48 h), and alkali (200 mM NaHCO3 or 100 mM Na2CO3, treated for 0, 1, 3, 6, 12, 24, and 48 h) treatments. Three biological replicates were taken for different organs/seedlings, as well as leaves and roots at each time point of all stress treatments. 3Names of candidate reference genes refer to Table 1.
Figure 3.
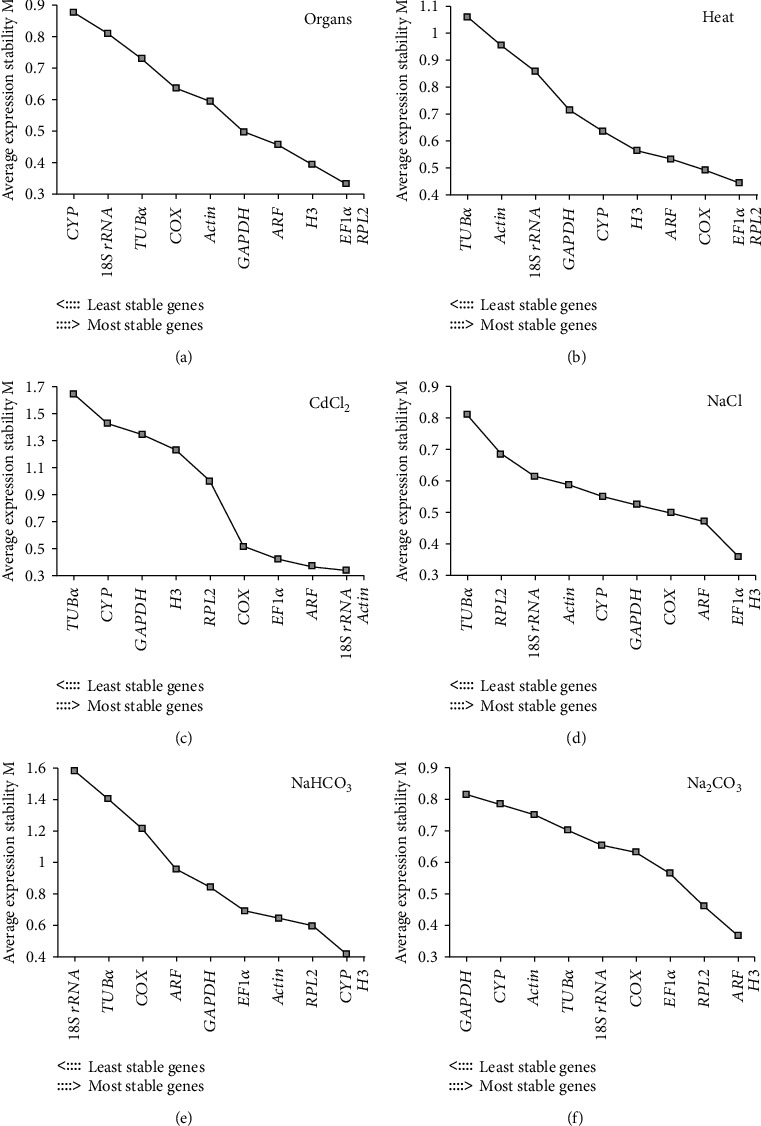
Average expression stability values (M2) in different data sets obtained from the software geNorm. The stability values (M2) and stability rank were obtained from geNorm. A lower M2 value suggests higher stability: (a) stability rank obtained in organs including leaves, stems, roots, flowers, and seedlings, (b) stability rank obtained in the leaves and roots under heat stress, (c) stability rank obtained in the leaves and roots under heavy metal stress, (d) stability rank obtained in the leaves and roots under NaCl, (e) stability rank obtained in the leaves and roots under NaHCO3, and (f) stability rank obtained in the leaves and roots under Na2CO3. Ten candidate reference genes refer to Figure 1. Three biological replicates were taken for different organs/seedlings, as well as leaves and roots at each time point of all stress treatments.
Figure 4.
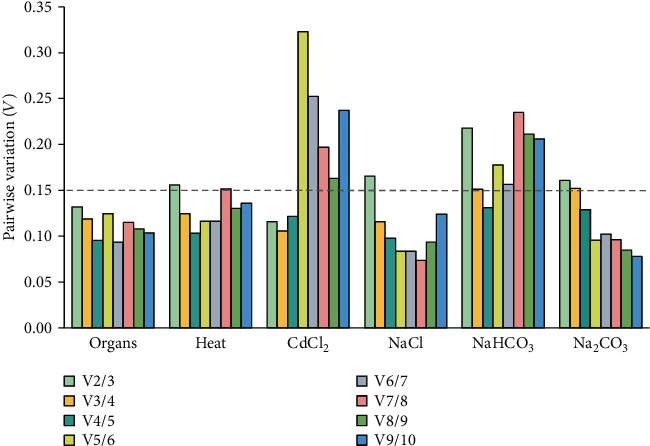
Pairwise variation (V) analysis of the candidate reference genes. The geNorm software was used to analyze the pairwise variation (Vn/Vn+1) between the normalization factors (NF) NFn and NFn+1 in order to determine the optimal number of candidate reference genes required for RT-qPCR data normalization. If Vn/Vn+1 < 0.15 (gray dotted line), the minimum value of n is the optimal number of genes required.
The stability of candidate genes was analyzed with BestKeeper (Table 2), which can calculate the standard deviation (SD) on the basis of the Cq values of all candidate reference genes [11]. The reference genes exhibiting the lowest standard deviation (SD) were taken as the most stable genes [63]. In the samples of different organs, 18S rRNA (SD = 0.58) and CYP (SD = 0.72) were regarded as the optimal reference genes. Under the heat (37°C) treatment, the expression levels of actin (SD = 0.57) and18S rRNA (SD = 0.60) were more stable than those of other genes. Under heavy metal (200 μM CdCl2) treatment, ARF (SD = 0.37) and COX (SD = 0.49) displayed stable expression. Under NaCl (200 mM) treatment, 18S rRNA (SD = 0.24) and ARF (SD = 0.32) showed shared and stable expression. Actin (SD = 0.64) and RPL2 (SD = 0.74), as well as CYP (SD = 0.24) and actin (SD = 0.32), were the stable genes in 200 mM NaHCO3 and 100 mM NaCO3 treatments, respectively, while 18S rRNA (SD = 1.03) was the least stable gene under 100 mM NaCO3 treatment (Table 2). Importantly, the TUBα was identified as the least stable gene in organs (SD = 1.58), as well as under treatments of heat (SD = 1.67), NaCl (SD = 1.24), and NaHCO3 (SD = 2.13).
The expression stability of ten genes was also analyzed using the NormFinder software (Table 2), which can provide a stability value (M1) for each gene by comparing the variation within and between sample groups. For the organs/seedlings, there were six sample groups, including young seedlings, roots, stems, leaves, male flowers, and female flowers. For the heat-treated samples of leaves and roots, there were nine sample groups, which were samples under heat (37°C) stresses for 0, 1, 2, 4, 6, 8, 12, 24, and 48 h, respectively. In addition, for the samples under heavy metal, NaCl, Na2CO3, and NaHCO3 treatment, there were six sample groups for each treatment (i.e., 0, 1, 3, 6, 12, 24, and 48 h) and totally 24 sample groups. The variation of each candidate gene in each case was compared, respectively. The gene stability was evaluated by M1 values.
The gene with lower M1 indicated that it was more stable [12]. For the samples from different organs, ARF (M1 = 0.131) and EF1α (M1 = 0.228) were regarded as the optimal reference genes, while CYP (M1 = 0.703) was the least stable gene. Under the heat (37°C) treatment, the expression of ARF (M1 = 0.212) and RPL2 (M1 = 0.228) was more stable than that of others, while TUBα (M1 = 0.928) was the least stable gene. Under heavy metal (200 μM CdCl2) treatment, EF1α (M1 = 0.625) and RPL2 (M1 = 0.650) were suggested to normalize the expression level of other genes, and TUBα (M1 = 1.621) seemed to be inappropriate as a reference gene. Under NaCl (200 mM) treatment, COX (M1 = 0.201) and ARF (M1 = 0.213) showed the common and highest stable expression, and TUBα (M1 = 0.852) was considered the least stable gene. In addition, RPL2 (M1 = 0.253) and actin (M1 = 0.454), as well as ARF (M1 = 0.284) and H3 (M1 = 0.297) were stable genes, but 18S rRNA (M1 = 1.407) and GAPDH (M1 = 0.523) were the least stable genes under NaHCO3 and Na2CO3 treatments, respectively (Table 2).
In addition, geNorm was used for gene stability analysis. geNorm evaluates the stability of candidate genes based on the geometric mean of these genes and mean pairwise variation in sample groups [13]. As mentioned above, there were six, nine, and 24 sample groups for organs/seedlings, heat stress, and other stresses, respectively. The stability value (M2) (Figure 3) and pairwise variation (Vn/Vn+1) (Figure 4) in the results given by geNorm revealed the gene stability and optimal gene numbers in certain case. The genes with the lowest M2 values were regarded as the most stable ones in each case (Figure 3). Besides, when pairwise variation (Vn/Vn+1) was less than 0.15, the minimum value of n was the optimal number of genes required for such condition (Figure 4). And the suitable gene pair included the genes from rank 1 to rank n in each condition [13]. According to this validation method, we recommended several reference genes for RT-qPCR analyses in organs and stress response. In different organs, EF1α/RPL2 (V2/V3 = 0.132) was a suitable gene pair for mRNA level normalization (Figures 3(a) and 4). EF1α, RPL2, and COX (V3/V4 = 0.125) were identified as an appropriate gene set under heat treatment (Figures 3(b) and 4). Besides, the gene pair of 18S rRNA/actin (V2/V3 = 0.116) was recommended for heavy metal stress (200 μM CdCl2) (Figures 3(c) and 4), while EF1α/H3/ARF (V3/V4 = 0.106) were suggested as reference genes under NaCl (200 mM) (Figures 3(d) and 4) treatment. In addition, CYP/H3/RPL2/actin (V4/V5 = 0.131) and ARF/H3/RPL2/EF1α (V4/V5 = 0.129) were selected as reference gene sets under 200 mM NaHCO3 (Figures 3(e) and 4) and 100 mM Na2CO3 (Figures 3(f) and 4), respectively.
3.4. Validation Test of Candidate Genes
To prove the feasibility of reference genes for RT-qPCR in spinach, mRNA levels of two heat-responsive genes, spinach basic region-leucine zipper 9 (SobZIP9) and heat stress transcription factor 2b (SoHSFB2b) [22], were detected in spinach under heat treatment and normalized by heat-stable actin and ARF, as well as commonly used TUBα. Under heat stress, the expression level of SobZIP9 was reduced in spinach after 2-12 h heat treatment when normalized by actin and also reduced after 2 h heat treatment when normalized by ARF. However, when normalized by TUBα, it exhibited an increase under heat treatment (Figure 5(a)). Besides, SoHSFB2b was increased about 10-fold after 1 h heat stress normalized by actin and ARF, but it exhibited 24-time increase when being normalized by TUBα (Figure 5(b)). This implies that the SoHSFB2b expression level was overestimated when normalized by TUBα. In addition, when normalized by actin and ARF, the levels of heat-reduced SobZIP9 and heat-induced SoHSFB2b were consistent with those in spinach under 35°C for 30 min and 5 h based on results from a previous transcriptomic study [22], which showed that actin and ARF are the appropriate reference genes in spinach for heat response analysis.
Figure 5.
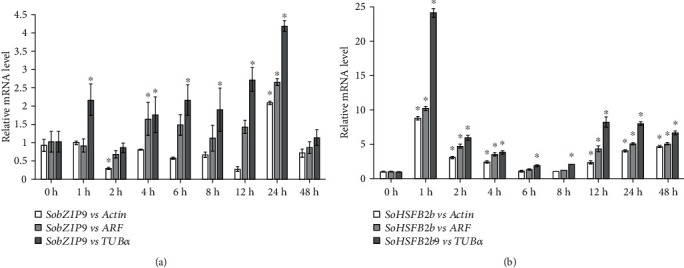
Expression pattern of SobZIP9 and SoHSFB2b normalized by different genes in response to heat stress. Expression patterns of two heat-induced genes, basic region-leucine zipper 9 (bZIP9, a) and heat stress transcription factor 2 b (HSFB2b, b), were normalized with actin, ADP ribosylation factor (ARF), and tubulin alpha chain (TUBα). Three biological replicates were taken for leaves at each time point of heat treatments. Data represent average ± SD, and ∗ indicates significant difference (p < 0.05).
4. Discussion
Diverse PCR approaches have been applied in the evaluation of gene expression levels, such as semiquantitative PCR, RT-qPCR, and digital PCR (dPCR). About ten years ago, digital PCR (dPCR) was developed as a novel technology for mRNA quantitation at a single molecular level [66, 67]. The dPCR can detect absolute copy numbers of certain gene expression without reference genes and standard curve [68], and it is more sensitive than qPCR for detecting the smaller copy number variation of genes [69]. However, dPCR is labour-intensive and expensive and has relatively low throughput when compared with qPCR [67], which leads to being not popularly used for the evaluation of gene expression in plant molecular labs. RT-qPCR is a convenient and accurate method to detect the mRNA levels of certain genes. The accuracy requires one or more stable reference genes for calculating the gene expression levels using the 2−ΔΔCq method [64]. Transcriptomic results suggest that there is always more than one gene stable in each set of samples, but none of them remains stable under all the conditions [5]. To date, 18S rRNA [22, 28, 29], actin [25], GAPDH [27, 31], and ubiquitin [32] were used as reference genes in previous studies in spinach, although their stability was not validated yet.
One study in spinach seeds evaluated the stabilities of GAPDH, actin, 16s rRNA, TUBα, and 18S rRNA, and 18S rRNA was taken as a relative stable reference gene [27]. In this report, we have determined 18S rRNA as the optimal reference gene in roots, stems, leaves, flowers, and seedlings, as well as in leaves and roots in response to salinity stress.
TUB (TUBα or TUBβ) was commonly used as a reference gene in some plant species, such as Raphanus sativus, Baphicacanthus cusia, and Hibiscus cannabinus [33, 34, 57]. However, in this report, TUBα was regarded as the most unstable reference gene in spinach in many cases. This is similar to the situation in Achyranthes bidentata and Amaranthaceae. In A. bidentata under hormone treatments (e.g., indole-3-butytric acid and methyl jasmonate), NaCl, and drought, TUB was also determined to be an unstable reference gene when compared with other candidate references (e.g., 18S rRNA, actin, APT1, EF1α, GAPDH, TUBβ, UBC, and UBQ) [35]. Besides, EF1α was regarded as the optimal reference gene in spinach under NaCl treatment. Similarly, EF1α was also the optimum gene in A. bidentate under NaCl and drought stresses [35]. It should be noted that EF1α was considered relatively unstable in several other species, such as Silene vulgaris and Papaver rhoeas [37, 57]. This implies that homologous genes in different plant species may not always exhibit similar expression levels under different conditions.
The recommended reference genes from NormFinder and BestKeeper look different (Table 2), because of their different mathematical models [11, 12]. BestKeeper uses Cq to calculate SD of candidate genes [11], but NormFinder uses 2−ΔCq to calculate the expression stability [12]. In our results, the expression patterns of heat-responsive genes (SobZIP9 or SoHSFB2b) are consistent when using the SoARF and SoActin as reference genes, which are recommended from NormFinder and BestKeeper, respectively (Figure 5). Importantly, the heat-responsive patterns of SobZIP9 or SoHSFB2b normalized by SoARF and SoActin are also similar with previous reports [22], but these heat-responsive patterns are absolute opposite or exaggerated when using TUBα as a reference gene that is not recommended by BestKeeper and NormFinder. This indicates that the optimal genes obtained in response to heat stress by NormFinder and BestKeeper are reliable for normalization of gene expression.
Due to different expression levels of genes, the Cq values of some reference genes are high (Cq > 30, Figure 2), or the ΔCq between the reference gene and target genes is relatively large. In this condition, even if the amplification efficiency of both primers of the reference gene and target genes is close to 100%, the error of gene expression level is probably enlarged, due to using the 2−ΔΔCq method [64, 70]. Thus, in this case, it is better to normalize the mRNA levels using the values of primer amplification efficiency rather than 100%.
In this study, optimal reference genes of each sample set were determined in spinach. 18S rRNA and ARF were validated as internal reference genes in different organs. Actin and ARF, instead of 18S rRNA, were the most suitable genes in leaves and roots under heat treatment. For reference genes under various abiotic stresses, EF1α and ARF were suitable for CdCl2, 18S rRNA and COX for NaCl, RPL2 and actin for NaHCO3, and ARF and CYP for Na2CO3. These results demonstrate that different reference genes should be used under different conditions [9]. Taken together, actin should be a reference gene for evaluating gene expression across all organs under various stress conditions, because actin has relatively better rank and M/SD values among all cases (Table 2).
5. Conclusion
It is important that optimal genes should be used for certain conditions when RT-qPCR is conducted to determine the normalized gene expression pattern using the 2−ΔΔCq method. Commonly used housekeeping genes in plant species may not be suitable under all the conditions or in certain species. In this study, the optimal genes were determined for gene expression normalization in spinach organs and in response to stresses of heat, CdCl2, NaCl, NaHCO3, and Na2CO3. The data provide a list of useful reference genes for future studies of gene expression patterns in spinach.
Acknowledgments
The project was supported by grants from the Shanghai Science and Technology Committee Foundation, China (No. 17391900600), the Fund of Shanghai Engineering Research Center of Plant Germplasm Resources (No. 17DZ2252700), the Heilongjiang Province Natural Science Foundation (No. ZD2019C003), and the Fundamental Research Funds for the Central Universities (Nos. 2572019CT03, 2572017ET01, and 2572017EA05).
Contributor Information
Ying Li, Email: ly7966@163.com.
Shaojun Dai, Email: daishaojun@hotmail.com.
Data Availability
The data used to support the findings of this study are included in this published article and its supplementary information files.
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
SD, YL, and HX designed the experiments. HX, BL, YC, YZ, XH, YL, and QW performed the experiments. HX, SG, and YM analyzed the data. HX, SD, and SC wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Materials
Figure S1: specificity of primers used in RT-qPCR experiment. Figure S2: standard curves of ten candidate reference genes.
References
- 1.Higuchi R., Dollinger G., Walsh P. S., Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology. 1992;10(4):413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 2.Bustin S. A., Benes V., Garson J. A., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 3.Taylor S., Wakem M., Dijkman G., Alsarraj M., Nguyen M. A practical approach to RT-qPCR – publishing data that conform to the MIQE guidelines. Methods. 2010;50(4):S1–S5. doi: 10.1016/j.ymeth.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Lanoix D., Lacasse A., St-Pierre J., et al. Quantitative PCR pitfalls: the case of the human placenta. Molecular Biotechnology. 2012;52(3, article 9539):234–243. doi: 10.1007/s12033-012-9539-2. [DOI] [PubMed] [Google Scholar]
- 5.Kozera B., Rapacz M. Reference genes in real-time PCR. Journal of Applied Genetics. 2013;54(4):391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenberg E., Levanon E. Y. Human housekeeping genes, revisited. Trends in Genetics. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Cruz F., Kalaoun S., Nobile P., et al. Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Molecular Breeding. 2009;23(4):607–616. doi: 10.1007/s11032-009-9259-x. [DOI] [Google Scholar]
- 8.Mallona I., Lischewski S., Weiss J., Hause B., Egea-Cortines M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biology. 2010;10(1) doi: 10.1186/1471-2229-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong Z., Gao Z., Wang F., Zhou J., Zhang Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Molecular Biology. 2009;10(1) doi: 10.1186/1471-2199-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Li H., Li X., et al. Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiologiae Plantarum. 2015;37(2) doi: 10.1007/s11738-015-1784-0. [DOI] [Google Scholar]
- 11.Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26(6):509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 12.Andersen C. L., Jensen J. L., Ørntoft T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 13.Vandesompele J., de Preter K., Pattyn F., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C., Jiao C., Sun H., et al. Draft genome of spinach and transcriptome diversity of 120 _Spinacia_ accessions. Nature Communications. 2017;8(1, article 15275) doi: 10.1038/ncomms15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Q., Chen W., Bian J., et al. Proteomics and phosphoproteomics of heat stress-responsive mechanisms in spinach. Frontiers in Plant Science. 2018;9 doi: 10.3389/fpls.2018.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi A., Mou B. Genetic diversity and association analysis of leafminer (Liriomyza langei) resistance in spinach (Spinacia oleracea) Genome. 2016;59(8):581–588. doi: 10.1139/gen-2016-0075. [DOI] [PubMed] [Google Scholar]
- 17.Bagheri R., Bashir H., Ahmad J., Iqbal M., Qureshi M. I. Spinach (Spinacia oleracea L.) modulates its proteome differentially in response to salinity, cadmium and their combination stress. Plant Physiology and Biochemistry. 2015;97:235–245. doi: 10.1016/j.plaphy.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Maruthachalam K., Klosterman S. J., Anchieta A., Mou B., Subbarao K. V. Colonization of spinach by Verticillium dahliae and effects of pathogen localization on the efficacy of seed treatments. Phytopathology. 2013;103(3):268–280. doi: 10.1094/PHYTO-05-12-0104-R. [DOI] [PubMed] [Google Scholar]
- 19.Duressa D., Rauscher G., Koike S. T., et al. A real-time PCR assay for detection and quantification of Verticillium dahliae in spinach seed. Phytopathology. 2012;102(4):443–451. doi: 10.1094/PHYTO-10-11-0280. [DOI] [PubMed] [Google Scholar]
- 20.Fagioni M., Zolla L. Does the different proteomic profile found in apical and basal leaves of spinach reveal a strategy of this plant toward cadmium pollution response? Journal of Proteome Research. 2009;8(5):2519–2529. doi: 10.1021/pr8011182. [DOI] [PubMed] [Google Scholar]
- 21.Fagioni M., D’Amici G. M., Timperio A. M., Zolla L. Proteomic analysis of multiprotein complexes in the thylakoid membrane upon cadmium treatment. Journal of Proteome Research. 2009;8(1):310–326. doi: 10.1021/pr800507x. [DOI] [PubMed] [Google Scholar]
- 22.Yan J., Yu L., Xuan J., Lu Y., Lu S., Zhu W. De novo transcriptome sequencing and gene expression profiling of spinach (Spinacia oleracea L.) leaves under heat stress. Scientific Reports. 2016;6(1, article 19473) doi: 10.1038/srep19473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S., Yu J., Li Y., et al. Heat-responsive proteomics of a heat-sensitive spinach variety. International Journal of Molecular Sciences. 2019;20(16) doi: 10.3390/ijms20163872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi C., Lin X., Li S., et al. SoHSC70 positively regulates thermotolerance by alleviating cell membrane damage, reducing ROS accumulation, and improving activities of antioxidant enzymes. Plant Science. 2019;283:385–395. doi: 10.1016/j.plantsci.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Duan F., Song W. Overexpression of SoCYP85A1 increases the accumulation of castasterone and confers enhanced black shank tolerance in tobacco through modulation of the antioxidant enzymes’ activities. Frontiers in Plant Science. 2019;10(349) doi: 10.3389/fpls.2019.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai X., Long J., He X., et al. Overexpression of spinach non-symbiotic hemoglobin in Arabidopsis resulted in decreased NO content and lowered nitrate and other abiotic stresses tolerance. Scientific Reports. 2016;6, article 26400 doi: 10.1038/srep26400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K., Fessehaie A., Arora R. Dehydrin metabolism is altered during seed osmopriming and subsequent germination under chilling and desiccation in Spinacia oleracea L. cv. Bloomsdale: Possible role in stress tolerance. Plant Science. 2012;183:27–36. doi: 10.1016/j.plantsci.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang X., Cai X., Xu C., Wang S., Dai S., Wang Q. Nitrate accumulation and expression patterns of genes involved in nitrate transport and assimilation in spinach. Molecules. 2018;23(9, article 2231) doi: 10.3390/molecules23092231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai X., Lin L., Wang X., Xu C., Wang Q. Higher anthocyanin accumulation associated with higher transcription levels of anthocyanin biosynthesis genes in spinach. Genome. 2018;61(7):487–496. doi: 10.1139/gen-2017-0261. [DOI] [PubMed] [Google Scholar]
- 30.Chen J., Shang Y. T., Wang W. H., et al. Hydrogen sulfide-mediated polyamines and sugar changes are involved in hydrogen sulfide-induced drought tolerance in Spinacia oleracea seedlings. Frontiers in Plant Science. 2016;7, article 1173 doi: 10.3389/fpls.2016.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.She H., Qian W., Zhang H., et al. Fine mapping and candidate gene screening of the downy mildew resistance gene RPF1 in spinach. Theoretical and Applied Genetics. 2018;131(12):2529–2541. doi: 10.1007/s00122-018-3169-4. [DOI] [PubMed] [Google Scholar]
- 32.West N. W., Golenberg E. M. Gender-specific expression of GIBBERELLIC ACID INSENSITIVE is critical for unisexual organ initiation in dioecious Spinacia oleracea. New Phytologist. 2018;217(3):1322–1334. doi: 10.1111/nph.14919. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y., Tan H., Yu J., et al. Stable internal reference genes for normalizing real-time quantitative PCR in Baphicacanthus cusia under hormonal stimuli and UV irradiation, and in different plant organs. Frontiers in Plant Science. 2017;8 doi: 10.3389/fpls.2017.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou B., Chen P., Khan A., et al. Candidate reference genes selection and application for RT-qPCR analysis in kenaf with cytoplasmic male sterility background. Frontiers in Plant Science. 2017;8, article 1520 doi: 10.3389/fpls.2017.01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Han X., Wang C., et al. Validation of suitable reference genes for RT-qPCR data in Achyranthes bidentata Blume under different experimental conditions. Frontiers in Plant Science. 2017;8 doi: 10.3389/fpls.2017.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koloušková P., Stone J. D., Štorchová H. Evaluation of reference genes for reverse transcription quantitative real-time PCR (RT-qPCR) studies in Silene vulgaris considering the method of cDNA preparation. PLoS One. 2017;12(8, article e183470) doi: 10.1371/journal.pone.0183470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarabel L., Milani A., Panozzo S., Rasori A. Suitable reference genes for accurate gene expression analysis in Papaver rhoeas under 2,4-D herbicide stress. Pesticide Biochemistry and Physiology. 2017;143:66–72. doi: 10.1016/j.pestbp.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Park S. C., Kim Y. H., Ji C. Y., et al. Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLoS One. 2012;7(12, article e51502) doi: 10.1371/journal.pone.0051502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang X., Zhang N., Si H., Calderon-Urrea A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods. 2017;13(1) doi: 10.1186/s13007-017-0238-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan Q., Chen S., Shan Z., et al. Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS One. 2017;12(12, article e189405) doi: 10.1371/journal.pone.0189405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 42.Boman A. L., Zhang C. J., Zhu X., Kahn R. A. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Molecular Biology of the Cell. 2000;11(4):1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gebbie L. K. Genes encoding ADP-ribosylation factors in Arabidopsis thaliana L. Heyn.; genome analysis and antisense suppression. Journal of Experimental Botany. 2005;56(414):1079–1091. doi: 10.1093/jxb/eri099. [DOI] [PubMed] [Google Scholar]
- 44.Srinivasan S., Avadhani N. G. Cytochrome c oxidase dysfunction in oxidative stress. Free Radical Biology and Medicine. 2012;53(6):1252–1263. doi: 10.1016/j.freeradbiomed.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin D. T., Lechleiter J. D. Mitochondrial targeted cyclophilin D protects cells from cell death by peptidyl prolyl isomerization. Journal of Biological Chemistry. 2002;277(34):31134–31141. doi: 10.1074/jbc.M112035200. [DOI] [PubMed] [Google Scholar]
- 46.Wiederrecht G., Hung S., Chan H. K., et al. Characterization of high molecular weight FK-506 binding activities reveals a novel FK-506-binding protein as well as a protein complex. Journal of Biological Chemistry. 1992;267(30):21753–21760. doi: 10.1016/s0021-9258(19)36676-1. [DOI] [PubMed] [Google Scholar]
- 47.Horowitz D. S., Lee E. J., Mabon S. A., Misteli T. A cyclophilin functions in pre-mRNA splicing. EMBO Journal. 2002;21(3):470–480. doi: 10.1093/emboj/21.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henneman B., van Emmerik C., van Ingen H., Dame R. T. Structure and function of archaeal histones. PLoS Genetics. 2018;14(9, article e1007582) doi: 10.1371/journal.pgen.1007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X., Liao W., Liao J., Liao P., Lu H. Ribosomal proteins: functions beyond the ribosome. Journal of Molecular Cell Biology. 2015;7(2):92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carvalho M. D., Carvalho J. F., Merrick W. C. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Archives of Biochemistry and Biophysics. 1984;234(2):603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Liu L., Qi J., Dang P., Xia T. Cadmium activates ZmMPK3-1 and ZmMPK6-1 via induction of reactive oxygen species in maize roots. Biochemical and Biophysical Research Communications. 2019;516(3):747–752. doi: 10.1016/j.bbrc.2019.06.116. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Q., Zhang H., Wang T., Chen S., Dai S. Proteomics-based investigation of salt-responsive mechanisms in plant roots. Journal of Proteomics. 2013;82:230–253. doi: 10.1016/j.jprot.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 53.Yin Z., Zhang H., Zhao Q., et al. Physiological and comparative proteomic analyses of saline-alkali NaHCO3-responses in leaves of halophyte Puccinellia tenuiflora. Plant and Soil. 2019;437(1-2):137–158. doi: 10.1007/s11104-019-03955-9. [DOI] [Google Scholar]
- 54.Yu J., Chen S., Wang T., Sun G., Dai S. Comparative proteomic analysis of Puccinellia tenuiflora leaves under Na2CO3 stress. International Journal of Molecular Sciences. 2013;14(1):1740–1762. doi: 10.3390/ijms14011740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang W., Liu J., Zhang Y., et al. A high-quality genome sequence of alkaligrass provides insights into halophyte stress tolerance. Science China-Life Sciences. 2020;63(9):1–14. doi: 10.1007/s11427-020-1662-x. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H., Han B., Wang T., et al. Mechanisms of plant salt response: insights from proteomics. Journal of Proteome Research. 2012;11(1):49–67. doi: 10.1021/pr200861w. [DOI] [PubMed] [Google Scholar]
- 57.Duan M., Wang J., Zhang X., et al. Identification of optimal reference genes for expression analysis in radish (Raphanus sativus L.) and its relatives based on expression stability. Frontiers in Plant Science. 2017;8, article 1605 doi: 10.3389/fpls.2017.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu C., Jiao C., Zheng Y., et al. De novo and comparative transcriptome analysis of cultivated and wild spinach. Scientific reports. 2016;5, article 17706(1) doi: 10.1038/srep17706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins K., Zhao K., Jiao C., et al. SpinachBase: a central portal for spinach genomics. Database. 2019;2019, article baz072 doi: 10.1093/database/baz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Untergasser A., Cutcutache I., Koressaar T., et al. Primer3 – new capabilities and interfaces. Nucleic Acids Research. 2012;40(15, article e115) doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Zhang Q., Jiang Y., Li Y., Zhang H., Li R. Reference genes identification for normalization of qPCR under multiple stresses in Hordeum brevisubulatum. Plant Methods. 2018;14(1) doi: 10.1186/s13007-018-0379-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Derveaux S., Vandesompele J., Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50(4):227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 63.De Spiegelaere W., Dern-Wieloch J., Weigel R., et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One. 2015;10(3, article e122515) doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters. 2003;339(1):62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 66.Vogelstein B., Kinzler K. W. Digital PCR. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perkel J. M. Life science technologies: the digital PCR revolution. Science. 2014;344(6180):212–214. doi: 10.1126/science.344.6180.212. [DOI] [Google Scholar]
- 68.Hoshino T., Inagaki F. Molecular quantification of environmental DNA using microfluidics and digital PCR. Systematic and Applied Microbiology. 2012;35(6):390–395. doi: 10.1016/j.syapm.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Whale A. S., Huggett J. F., Cowen S., et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Research. 2012;40(11):p. e82. doi: 10.1093/nar/gks203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9):45e–445. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: specificity of primers used in RT-qPCR experiment. Figure S2: standard curves of ten candidate reference genes.
Data Availability Statement
The data used to support the findings of this study are included in this published article and its supplementary information files.


