Abstract
COVID-19 and its causative pathogen SARS-CoV-2 have rushed the world into a staggering pandemic in a few months, and a global fight against both has been intensifying. Here, we describe an analysis procedure where genome composition and its variables are related, through the genetic code to molecular mechanisms, based on understanding of RNA replication and its feedback loop from mutation to viral proteome sequence fraternity including effective sites on the replicase-transcriptase complex. Our analysis starts with primary sequence information, identity-based phylogeny based on 22,051 SARS-CoV-2 sequences, and evaluation of sequence variation patterns as mutation spectra and its 12 permutations among organized clades. All are tailored to two key mechanisms: strand-biased and function-associated mutations. Our findings are listed as follows: 1) The most dominant mutation is C-to-U permutation, whose abundant second-codon-position counts alter amino acid composition toward higher molecular weight and lower hydrophobicity, albeit assumed most slightly deleterious. 2) The second abundance group includes three negative-strand mutations (U-to-C, A-to-G, and G-to-A) and a positive-strand mutation (G-to-U) due to DNA repair mechanisms after cellular abasic events. 3) A clade-associated biased mutation trend is found attributable to elevated level of negative-sense strand synthesis. 4) Within-clade permutation variation is very informative for associating non-synonymous mutations and viral proteome changes. These findings demand a platform where emerging mutations are mapped onto mostly subtle but fast-adjusting viral proteomes and transcriptomes, to provide biological and clinical information after logical convergence for effective pharmaceutical and diagnostic applications. Such actions are in desperate need, especially in the middle of the War against COVID-19.
Keywords: SARS-CoV-2, Nucleotide composition, Mutation spectrum, Viral replication
Introduction
COVID-19, a novel pneumonia epidemic causing an outbreak first identified and reported in Dec. 2019 from China [1] and subsequently spread to other countries swiftly, has been posing enormous professional, economic, and political challenges to global health services. As of 12 June 2020, there have been 7,410,510 confirmed cases and 418,294 deaths reported [2]. COVID-19 is of great contagiousness and has lower mortality to our current understanding [3], [4], [5]. The novel betacoronavirus identified through de novo sequencing from patients with COVID-19 is designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by International Committee on Taxonomy of Viruses (ICTV) [1], [6], [7].
The recent threats from SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) are different from those from earlier human coronaviruses (CoVs), including alphacoronaviruses, such as hsa-CoV-229E, hsa-CoV-NL63, hsa-CoV-OC43, and hsa-CoV-HKU1 [8], [9], [10] in at least two aspects. First, the recent groups of betacoronaviruses appear to come more frequently in the past two decades as compared to the early comers where new members may be discovered as technologies become more efficient and accurate [11]. The current SARS-CoV-2 is also different from both SARS-CoV and MERS-CoV as its genome composition is most closely for “living with mammals and humans”, where a much lower G + C content has been evolved and is closer to two other human-adapted CoVs (hsa-CoV-229E and hsa-CoV-OC43) than its members of the recent group, although it shares higher sequence identities with the two new CoVs, 80.12% for SARS-CoV and 60.06% for MERS-CoV, respectively [11]. Second, it has been infecting far larger populations, as compared to the two recent outbreaks, with variable yet more complex symptoms [12]. The causative factors of such an unprecedented disease potency remain to be elucidated for the days and months to come [1], [3], [4], [5], [6], [7].
Genomes of CoVs mutate in a unique way where signatures of DNA pairing and repairing mechanisms are absent, and instead, they possess an error-prone synthesis of single-stranded full or partial genomic sequences, catalyzed by a multi-component membrane-associated enzymatic structure known as replicase-transcriptase complexes (RTCs) and double-membrane vesicles (DMVs), although the viruses do have certain enzymatic activity resembling cellular repair mechanisms, such as proofreading [13] and other possible cellular mechanisms may also be involved, such as RNA editing as recently proposed [14], [15]. Here we define a series of displays to understand compositional dynamics or variability that ultimately interconnects to proteomic variability including RTCs and DMVs (of course also other omics) through the organization of the genetic code [16], [17], [18], [19]. We subsequently compare SARS-CoV-2 with other human CoVs for between-lineage variation analysis to point out that it is not a direct descendant of all previous human-infecting CoVs. We finally make efforts to decipher the SARS-CoV-2 clades in terms of its variations, suggesting that what we have seen now is not the natural picture of the pandemics and the missing-links are not among human population but possibly the wildlife close to human habitats in Southeast Asian territories, islands or shorelines, not just limited to bats and pangolins. We also show how to examine clade-associated permutation variations and relate genetic variations to protein. Nailing down a single animal of human origin of the virus will not be the goals of this genomics-based study but to provide information for smarter drug design, effective vaccine development, and accurate diagnosis.
Results and discussion
Compositional dynamics and its parameters are essential for evaluating the evolutionary status of SARS-CoV-2
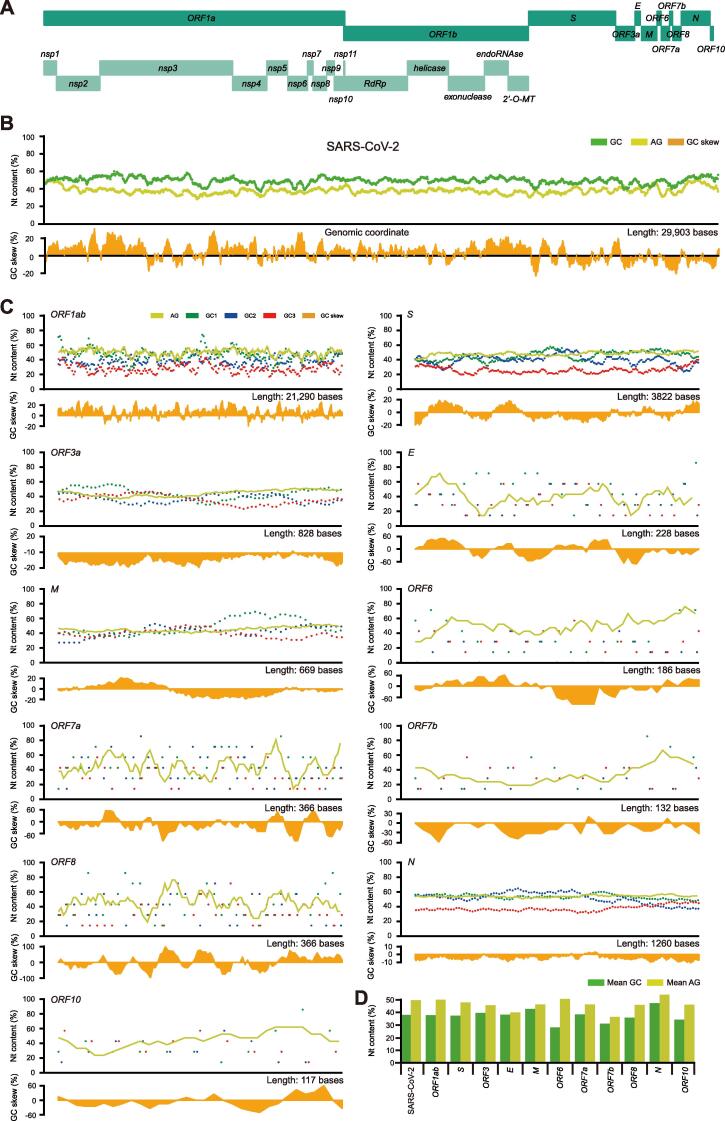
As a positive-sense single-stranded RNA virus, SARS-CoV-2 has a genome length of 29,903 nucleotides (nt) (GenBank: NC_045512.2). It encodes two large polypeptides, ORF1a and ORF1b, along with their processed products, 15 non-structure proteins (nsps). In order to propagate and complete the life cycle, its positive-sense genome is first replicated to synthesize full-length negative-sense antigenomes and 10 shorter subgenomes (sgRNAs), executed by RTCs and DMVs. Those sgRNAs encode four structural proteins (S, spike; E, envelope; M, membrane; and N, nucleocapsid) and six accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10) arranged among structural proteins (Figure 1A), depending on the current annotation (GenBank: NC_045512.2).
Figure 1.
A display of genome compositional dynamics of SARS-CoV-2
A. The complete genome sequence of SARS-CoV-2 (GenBank: NC_045512.2), including both structural and non-structural components. B. We use a 300-nt sliding window with a 21-nt step to show dynamic changes of genomic G + C, purine, and G + C contents at three codon positions (denoted as GC1, GC2, and GC3) as well as GC skews (G-C/G + C) over the entire genome. C. A similar procedure as described above is applied to individual ORFs and proteins. Note that GC skews are not uniform over the genome length and the ORFs based plots are not in the same scale as what for the genome-wide GC skews. D. Mean G + C and purine contents of SARS-CoV-2 genome, individual ORFs and proteins.
Traditionally, three basic plots were used to display composition dynamics based on primary genomic parameters over genome length: G + C content at three codon positions (GC1, GC2, and GC3), purine content (A + G content), and GC skew [the content of (G-C)/(G + C)]. Here, we use a 300-nt sliding window with a step size of 21 nt, as the majority of viral sequences are protein-coding, to illustrate the dynamics of the composition parameters, G + C and purine contents (Figure 1B). The G + C and purine contents of SARS-CoV-2 vary in a narrow but significant window of 22.00% (28.33%–50.33%) and 23.33% (36.67%–60.00%), respectively. The GC skew of the SARS-CoV-2 genome indicates the G + C ratio in structural proteins is relatively higher than that in ORF1ab, and this imbalance is a signature of distinct mutational biases caused by viral replication machinery, known as RTCs. And a frequent shift toward negative values is often seen in individual ORFs and defined proteins (Figure 1C and D), suggesting either mutation or selection events which are species- or isolate-specific. Differences are still obvious in the SARS-CoV-2 closely-related bat and pangolin CoVs (raf-betaCoV-RaTG13 and mja-betaCoV-P4L; Figure S1A and B) and the last two human-infecting CoV outbreaks (SARS-CoV and MERS-CoV; Figure S1C and D). The G + C content of different codon positions is also very informative (Figure 1C), where GC3 is a characteristic of mutation pressure as it is obvious that nearly all GC3 values of the viral structure proteins are biased toward lower G + C contents. GC3-associated mutations often reflect directional mutation patterns as observed strongly in certain lineages of plants and warm-blooded vertebrates as negative gradients from the transcription starts, and such trends are attributable to a special DNA repair mechanism, transcription-coupled DNA repair [20], [21], [22]. The notion here is to remind ourselves that transcription-centric mutations may be accounted for some of the mutation events in RNA viruses in their replication-transcription processes. Occasional twists from the trend often indicate selective pressures, such as in the case of S, M, and N proteins, and weaker GC3 or stronger GC1 or GC2 selections. Codon-associated G + C content trends are less informative for small ORFs, such as the case of ORF10. Most of the sequence signatures are indicative rather than proven functional relevance of proteins but very useful for providing clues of sequence signature and anomaly.
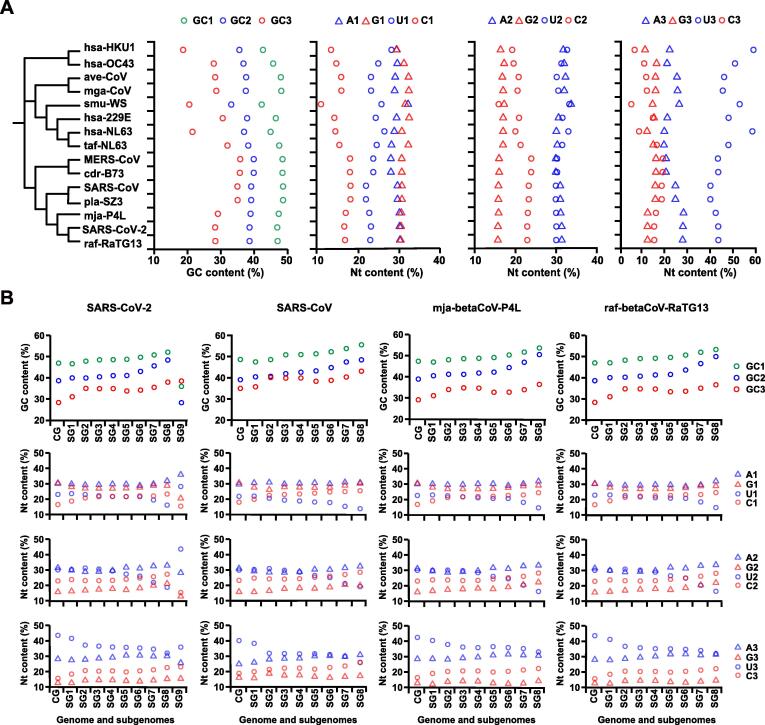
RNA genomics is rather different from DNA genomics in several ways [11]. For studying RNA viral genomes, in addition to previously-defined parameters, we introduce the concept of single nucleotide (A, U, G, and C) contents at three codon positions (such as U1, U2, and U3 for uridines) (Figure 2). As shown in a phylogenetic tree constructed based on 15 representative CoVs (Figure 2A), the nucleotide contents of SARS-CoV-2 are most similar to those of raf-betaCoV-RaTG13 and mja-betaCoV-P4L, which are considered to be distantly related but most closely related so-far-found host of SARS-CoV-2. While other known zoonotic and corresponding human counterpart CoVs are rather close to each other in their compositions. We have made a few interesting observations here. First, the single nucleotide content is more informative than G + C content, especially for genome analysis on RNA viruses. The former points out only how G + C content drifts toward richness or poorness but the latter narrows it down to single nucleotide effect. In our case, U stands out at codon position 3 (CP3), which alters the overall nucleotide contents, and it drives the G + C content so low that even its partner A content has gone to the same extremity. If the organization principles are considered here, half of the codons are not sensitive to CP3 changes, and most of them are smaller amino acids (Figure S2; [16], [17], [18], [19]). Second, at the codon position 1 (CP1), G and C contents are both pulled apart toward extremity but not A or U, while the two pyrimidines and two purines appear stretched to separate directions; these trends suggest strong selective pressure at the first codon position over the entire genome. It is indeed that CP1 codons shoulder the most mutation pressures since they fall into all 4 negative-sense strand permutations (known as R1-derived permutations, C-to-U, G-to-A, U-to-C, and A-to-G). Third, the codon position (CP2) contents are most row-flipping changes referenced to the genetic code organization [18]. These alterations are very useful for alternating chemical characteristics between related amino acids, and in terms of flexibility, CP2 codons are less stringent than CP3 but more flexible than CP1. Finally, it is conclusive that the more similar the CoVs in composition dynamic parameters, the closer they are genetically and phylogenetically in principle. However, primary parameters, such as G + C and purine contents are necessary but may not be sufficient. For instance, there has been a CoV genome isolate from a wild vole captured in northeastern China, whose G + C and purine contents overlap with SARS-CoV-2 completely (RtMruf-CoV-2/JL2014; G + C = 0.380, A + G = 0.496, which completely overlaps with those of SARS-CoV-2 in both parameters; [23]) but its genome sequence is different (sharing 61.87% identity with SARS-CoV-2). Therefore, we have yet to find any immediate animal hosts of SARS-CoV-2 albeit best similarity of composition dynamics seen among them.
Figure 2.
Nucleotide contents of genomes and subgenomes of SARS-CoV-2 and related CoVs
A. A schematic phylogenetic tree is used to cluster genome sequences and compositional variables (15 CoVs genome sequences, from top to bottom, are: hsa-betaCoV-HKU1, hsa-betaCoV-OC43, ave-gamaCoV, mga-gamaCoV, smu-alphaCoV-WS, hsa-alphaCoV-229E, hsa-alphaCoV-NL63, taf-alphaCoV-NL63, MERS-CoV, cdr-betaCoV-B73, SARS-CoV, pla-betaCoV-SZ3, mja-betaCoV-P4L, SARS-CoV-2, and raf-betaCoV-RaTG13). These compositional variables include GC1, GC2, and GC3 and single nucleotide contents at three codon positions (A1, A2, A3; U1, U2, U3; G1, G2, G3; and C1, C2, C3). Nucleotides are labeled in different shapes: purines, triangles; pyrimidines, open circles. A and U or G and C are colored blue or red, respectively. It becomes obvious that the two closely-related CoV genomes to SARS-CoV-2, the reported bat (raf-betaCoV-RaTG13) and the pangolin (mja-betaCoV-P4L), have very similar codon G + C contents as well as base contents. The CP1 (codon position 1) base content appears most characteristic of balanced purine content of SARS-CoV-2 and its close relatives. The CP2 (codon position 2) base content of SARS-CoV-2 and all other CoVs has higher and relatively balanced A + U content. The older human CoVs have either lowest or higher G + C content and unbalanced purine content. G + C content represents a single measure but single nucleotide content demonstrates trends of all four nucleotides. B. The G + C and single nucleotide contents at different codon positions of complete genomes (labeled as “CG”) and subgenomes (labeled as “SG”) of SARS-CoV-2, SARS-CoV, mja-betaCoV-P4L, and raf-betaCoV-RaTG13 are displayed to illustrate the driving force for G + C content decrease towards 3’ end of the genome, which is rather a result of, in terms of mechanism, the increased U content and C-to-U permutation. The negative gradient of U is also obvious from the 5′ end to the 3′ end.
We further compare the compositional dynamics for both the complete genome and subgenomes of SARS-CoV-2 and its closely related viral sequences (Figure 2B). For compositional dynamics of RNA genomes, uridine is the star nucleotide, and A + U content becomes the most important. Just for the sake of convenience, we would like to keep the concept of G + C content since it has been known to be a useful variable for DNA compositional dynamics [24] and provide an approximation for less selected nucleotide position. It is interesting to see uniformity among all codon position contents of all 4 CoV genomes, and increased G + C content from complete genome to subgenomes due to stronger selection over structural proteins. SARS-CoV-2 has an exceptionally short subgenome 9 (sg9) which only contains ORF10, but we have no evidence that it is either functional or non-functional. These results collectively remind us that SARS-CoV-2 and its most-closely-related CoVs, unlike in the case of many other known CoVs, have a unique genome composition and similar dynamics to the early-adapted human CoVs [11] and CoV-borne bats and other mammals of the same lineage (such as the vole [23]) may already coexist with ability to jump on to humans and domestic animals but only limited by environmental and geographic constraints.
Mutation spectrum is composed of permutations that are distinct according to their strand specificity
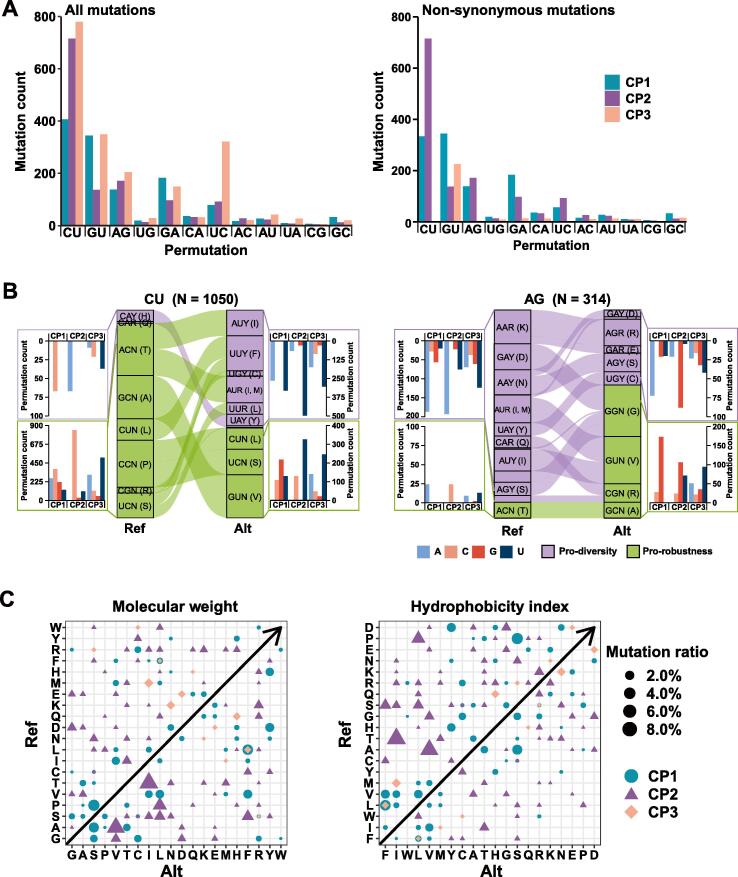
We use 12 permutations (Figure 3) to represent directional mutations and classify them according to strand-specific replication mechanisms since they are readily related to codons [11] (Figure S2). The permutations are categorized into R1 (C-to-U, G-to-A, U-to-C, and A-to-G), R2 (C-to-A, U-to-G, A-to-C, and G-to-U), and R12 (C-to-G, U-to-A, A-to-U, and G-to-C) according to their occurrence tailored to RTC-directed strand synthesis: R1 occurring mainly in replication of the first negative-sense strand as transition mutations, R2 occurring mainly in replication of positive-sense strand (when abasic sites obscured negative-sense strand replication for viral particle packaging), and R12 maybe occurring in both strand replication (R1 plus R2). From a total of 5054 point mutations (identified from 22,051 public sequences as of 12 June 2020), the most abundant permutations are four R1 permutations and one R2 permutation, G-to-U. And there are 1416, 1497, and 2141 mutations falling on codon position 1, 2, and 3, respectively (Figure 3A). What we have shown here is how sensitive is nucleotide content of CP3 to selective pressure, and most CP3 permutations disappear except the G-to-U permutation at CP3, where all changes are transversions and half of all codons (all pro-diversity changes) are sensitive to them (Figure S2). Similar results are observed in our analysis on SARS-CoV and MERS-CoV (Figure S3A and B). There are slightly different patterns, the higher U-to-C permutation, among SARS-CoV and MERS-CoV and their within-lineage bats and mammalian hosts. The predominate C-to-U represents the driving force of variation, and it manifests why both G + C and purine contents of SARS-CoV-2 appear relatively lower against MERS-CoV and SARS-CoV and even more when compared to the human CoVs, such as 229E and OC43 [11].
Figure 3.
Mutation spectra of SARS-CoV-2 in a context of codon positions
A. The SARS-CoV-2 mutation spectrum is composed of 12 permutations and they are divided by codon positions among all mutations. For all mutations, C-to-U (CU), U-to-C (UC), A-to-G (AG), G-to-A (GA), and G-to-U (GU), are always dominant due to two principles; one is that the first four permutations occur when positive-sense genome is synthesized, and the other is that a G-by-A replacement is always preferred by RTCs so that G-to-U permutation is the most dominant when the antigenome serves as a template. For non-synonymous mutations, C-to-U permutations at CP3 diminish among non-synonymous mutations and this phenomenon indicates that most protein composition relevant variations are CP1 and CP2 variations. The remaining non-synonymous mutations in G-to-U (GU) permutation may be a result of biased strand synthesis. B. Displays of permutation-to-codon changes among non-synonymous mutations. The codon table is divided into two halves: the pro-diversity half (purple) whose CP3 is sensitive to transitional change and the pro-robust half (green) whose CP3 position is insensitive to any change. Two examples, C-to-U (1051 in counts) and A-to-G (314 in counts) permutations are shown here. When a codon has a C-to-U change, the codon position varies, results of such changes relative to codon positions are summarized on both sides of the codon flow chart. Note that CP1 and CP2 changes appear more than those of CP3. C. All permutations are plotted against the reference genome sequence to show how changes are related to amino acids. In the molecular weight index, most CP1 and CP2 changes are showing an obvious increasing trend. In the hydrophobicity index, most CP1 and CP2 changes toward less hydrophobicity.
Since most CP1 and CP2 related permutations are sensitive to selection, we further examine how individual permutations correlate to codon rearrangements in the two halves: pro-diversity and pro-robustness (Figure 3B) [16], [17], [18]. Only two examples, C-to-U and A-to-G, are shown here and the rest are summarized in Figure S3C. Several observations are worthy of in-depth discussion. First, it is known that three amino acids and their codons are unique in balancing one of two purine-sensitive halves; they are leucine (Leu), arginine (Arg), and serine (Ser) [16], [17], [18], [19]. The most abundant amino acid in protein coding sequences (known as codon usage) is Leu and it buffers C-to-U|U-to-C mutations at CP1. Arg and Ser are also abundant as they both are 6-fold degenerate codons; Arg appears buffering A-to-G|G-to-A at CP1 and Ser carries two: U-to-A|A-to-U at CP1 and G-to-C|C-to-G at CP2. Second, amino acid exchanges are permissive in physiochemical properties [20], [21], [22]. For instance, Ser has a very similar size to alanine (Ala) so that G + C content increase is buffered by the two amino acids as G-to-U|U-to-G permutations. Third, other examples are codon alterations among hydrophobic amino acids, as they are mostly C-to-U changes at CP2 among the pro-robustness half. The overall effects are displayed together in Figure 3C. It is rather clear that changes toward lower G + C content and near the balanced purine content are both beneficial for CoVs, especially SARS-CoV-2, as these changes are pro-diversity, in favor of larger and more hydrophilic amino acids.
Clade-associated biased mutation trend in SARS-CoV-2 reveals physiochemical features of replication machinery
Difficulties for analyzing CoV genomes are multifold. Since we have yet to identify the direct natural and intermediate mammalian hosts, if there are any, this massive dataset has to be analyzed by stratifying the data into structured and non-structured clades. The next is even more troublesome. Assuming that we have 5 or more genome sequences per CoV isolate and variations identified among them are still a miniscule fraction of the total virions produced in a patient body (means and medians of variations per CoV isolate among C01 to C09, see Table S1), since the viral load per patient sample, such as sputum [25], [26] is equivalent to a 5-person or more sampling of the entire human population on earth, 1 out of 109. Even so, we have still been able to find shared variations among patient samples occasionally and even more lucky to have some clade structures, by and large due to the relatedness of the patients in the transmission network.
Here, we have constructed a somewhat stable phylogenetic tree-and-branch structure (Figure 4A, Figure S4), and it is composed of 8 monophyletic clades and 1 non-monophyletic clade based on both orders of sample collection date and highly-shared mutations. Among the clades, C02 shares two landmark mutations, 8782C > U in ORF1ab and 28144C > U in ORF8, and earlier date (2019/12/30). C04 shares three more mutations (17747C > U, 17858A > G, and 18060C > U in ORF1ab) than what C02 has, and a late collection date (2020/02/20). Clades C03, C05, and C07 are also distinguishable by some major mutations, so are C06, C08, and C09; the latter clades are clustered together based on four shared and other clade-associated mutations. The leftover isolates that lack all landmark mutations are grouped into C01, which have the earliest collection date on 2019/12/24. According to the literature and our discussion, we have further grouped the clades into three clusters, S (C02 and C04), G (C06, C08, and C09), and L (all the rest), since phylogeny shows clear divergence among them. We have several notions about this imperfect hierarchical structure. First, our within- and between-clade analysis of variations with high major allele frequency (MAF) reveals that some clade-associated signature mutations are also shared among other clades. For instance, 14805C > U in ORF1ab and 23403A > G in S have recurred in other clades of different clusters, which are excellent landmarks for subclade definition. Another notion is that within-clade mutations with higher MAF (such as MAF > 0.2 in one clade) are mostly non-synonymous mutations, indicating selection at work (Figure 4B). Our neighbor-joining tree based on distances from 9 clades suggests that SARS-CoV-2 appears originated from a closely-related zoonotic population of a single lineage (Figure 4B). In addition, our classification rationales are largely in agreement with published reports [27]; for example, Cluster S is in accordance with previously defined S type [28] Cluster G is in line with the G clade defined by Global Initiative on Sharing All Influenza Data (GISIAD) [29] and Cluster L is similar to the V and L clades combined, of GISAID.
Figure 4.
Sequence-variation-based phylogenies of SARS-CoV-2
A. SARS-CoV-2 genomes are divided into clades and clade clusters based on high-frequency mutations among the genome sequences. The variations are labeled with positions and nucleotide variations that are all referenced to the SARS-CoV-2 genome (GenBank: NC_045512.2). The thin vertical bars at the bottom displays positions and relative frequencies of variations. The dates when each clade started are also indicated. B. Signature site information and frequency table of star mutations in each clade, with a neighbor-joining tree based on the frequency data in the table. Star mutations are mutations with MAF greater than 0.02 in all 22,051 SARS-CoV-2 sequences. The frequency table represents mutation frequency within each clade. Frequencies greater than 0.5 and those in the range of 0.1 to 0.5 are marked in red and yellow, respectively. The “Mutation type” column indicates synonymous mutation (labeled as “SM”) or non-synonymous mutation (labeled as “NSM”).
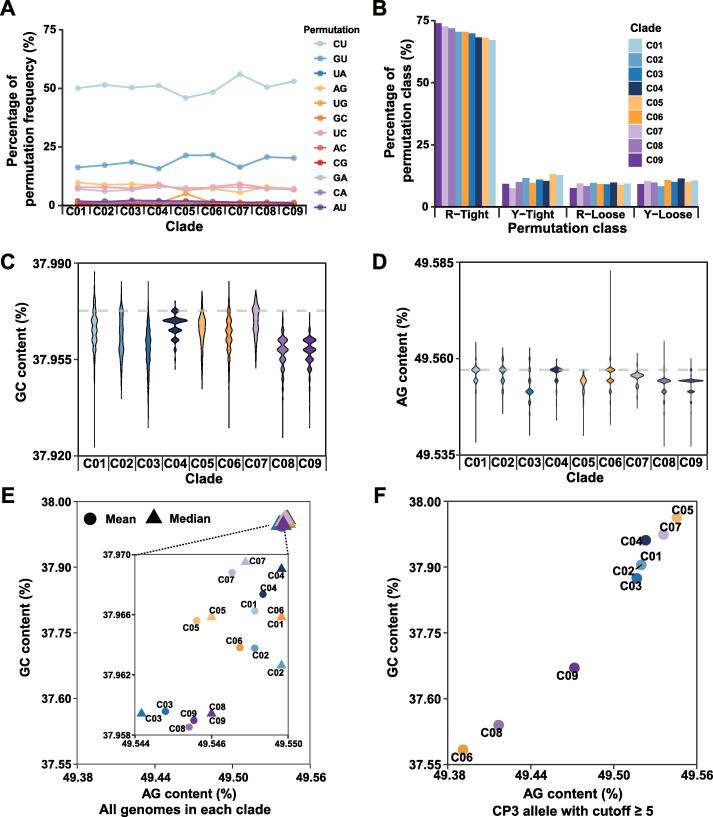
To look for clade-associated compositional and functional features, we have first built a consensus sequence for each clade and subsequently calculated frequencies for each within-clade permutation (Figure 5A; Table S2). A key assumption behind this is that certain functional mutations may have clade-specific effects on mutation spectrum, to close a loop where sequence mutations through genetic coding principles alter the viral proteome function. Our observations are of importance in establishing logics about compositional dynamics between nucleic acids and proteins. First, permutations among clades are indeed variable according to their proportions calculated from genome variants, and aside from 5 high-proportion permutations, 4 R1 and 1 R2 permutations, two other R2 and one R12 permutations appear also joining in, which are U-to-G and A-to-C, as well as A-to-U, respectively. Second, the variable permutations, where some may represent effect of mutation pressure and others may exaggerate selection pressure, are unique to clades and clade clusters. For instance, clade cluster S has the lowest G-to-U fraction as compared to those of L and G; in addition, among the S clades, C04 has the lowest value of G-to-U. Similarly, C03, C05, C06, C08, and C09 have relatively higher G-to-U permutations (Figure 5A). Third, based on the disparity of permutations or simply mutation spectra, we have taken a rather radical step to assume RTC statuses in favor of either tight or loose statuses for binding to purines and pyrimidines (Figure 5B, Figure S5). Since purines are larger than pyrimidines in size, the purine- or R-tight must be different from pyrimidine- or Y-tight. The results are strikingly predictable in that the R-tight status suggests a tighter binding pocket where a descending trend for tight permutations (C-to-U, G-to-U, and U-to-A) reverses into the opposite trend for Y-tight permutations. It indicates that the RTC structure and conformation variables may be definable in principle. At this point, we do not have discrete definitions for these so-called tight statuses but the less trendy R-loose and Y-loose statuses also support a similar idea [11].
Figure 5.
Mutation spectrum and composition dynamics among 9 SARS-CoV-2 clades
A. Plots showing permutation variation of each clade. Aside from the 5 dominant permutations (C-to-U (R1 permutation), U-to-C (R1 permutation), A-to-G (R1 permutation), G-to-A (R1 permutation), and G-to-U (R2 permutation)), A-to-C (R2 permutation), U-to-G (R2 permutation), and A-to-U (R12 permutation) changes appear also significant; such an increase in proportion of R2 and R12 permutations often indicates copy number (synthesis) bias between the two strands. B. When permutations are grouped based on structure-conformation model (Figure S5) into tight and loose groups (a four-parameter model), their trends of changes become obvious. The R-tight discourages A-by-G replacement but encourages C-by-U replacement when the genome is replicated. The loose statuses, regardless R-loose or Y-loose, place no pressure on permutation variability. C. Violin plots showing the G + C content of genomes among clades. D. Violin plots showing the purine content of genomes among the clades. C08 and C09 have been drifting both contents toward lower ends. E. The mean (solid circles) and median (solid triangles) of G + C and purine contents among clades. The same two more expressive clades (C08 and C09), as seen in (C) and (D), are indeed obvious (inset). F. The compositional dynamics of CP3 nucleotides that are less selected and with a stringent cutoff value (≥ 5).
We further examined the compositional subtleties among the clades and clusters with a focus on the variability of G + C (Figure 5C) and purine contents (Figure 5D) as both contents appear drifting toward optima in SARS-CoV-2 and its relatives. Different clades exhibit distinct compositional features and such dynamics are very indicative for the existence of feedback loops connecting RNA variables to protein variables. Two directions have to be advised for understanding these features. The first direction is driven by strong mutations, perhaps coupled to tight-loose switches in the catalytic pocket of RNA-dependent RNA polymerases (RdRps) in RTCs. It is clear that except C01, the G or C06-C08-C09 cluster has the lowest G + C content (e.g., 0.37929 of a C08 CoV sampled in Australia) and the lowest purine content (0.49527, based on a C08 CoV collected in Bangladesh and a C09 CoV collected in England). Both lower G + C and purine contents are indicative of mutation pressure and signal of this fast-evolving cluster of CoVs due to the older human-adapted CoVs (hsa-alphaCoV-229E, hsa-betaCoV-OC43, hsa-betaCoV-HKU1, and hsa-alphaCoV-NL63) have lower G + C and purine contents [11]. Since cluster G has the largest collection of CoVs, it is also not surprising to see a more complex median diversification within clades (Figure 5E). The second direction is the drive from mutation or both mutation and selection in balance or imbalance, as well as in modes of fine-tuning or quick-escaping. For instance, G + C and purine contents at CP3 are informative for mutation drives, displaying both lower G + C and purine contents of cluster G (Figure 5F).
Based on our clade and clade-cluster analysis, it is tempting for us to speculate that there are plenty of rooms for further investigations into mutation spectra among large clades and even smaller subclades or closely related individual CoV genomes. First, all high-frequency mutations should be identified and classified, and these variations are candidates for highly selected mutations. Second, all minor but not rare within-clade mutations, such as those of mutations with MAF in a range of 0.01% to 10% should also be identified; they provide basis of within-clade sequence analysis. Third, all non-structured CoV genomes must be also classified based on shared variations, as they are not only valuable for within-clade but also for clade-cluster analyses.
Within-clade variations and their implications for future SARS-CoV surveillance
Within-clade compositional dynamics can also be very informative, especially for covering and predicting future functional changes, such as identifying mutated and diversified forms of CoVs for drug and vaccine designs. It is also of essence for nucleic acid-based diagnostics, such as clade-specific identifications. Within-population variations can be identified based on clade consensus sequence after alignment and extracted from datasets that have hundreds and thousands of genome sequences. The analysis of within-population variations relies on structured phylogeny and proportion change of permutations, which can be classified into either copy number-related or RTC-specificity related, or sometimes both.
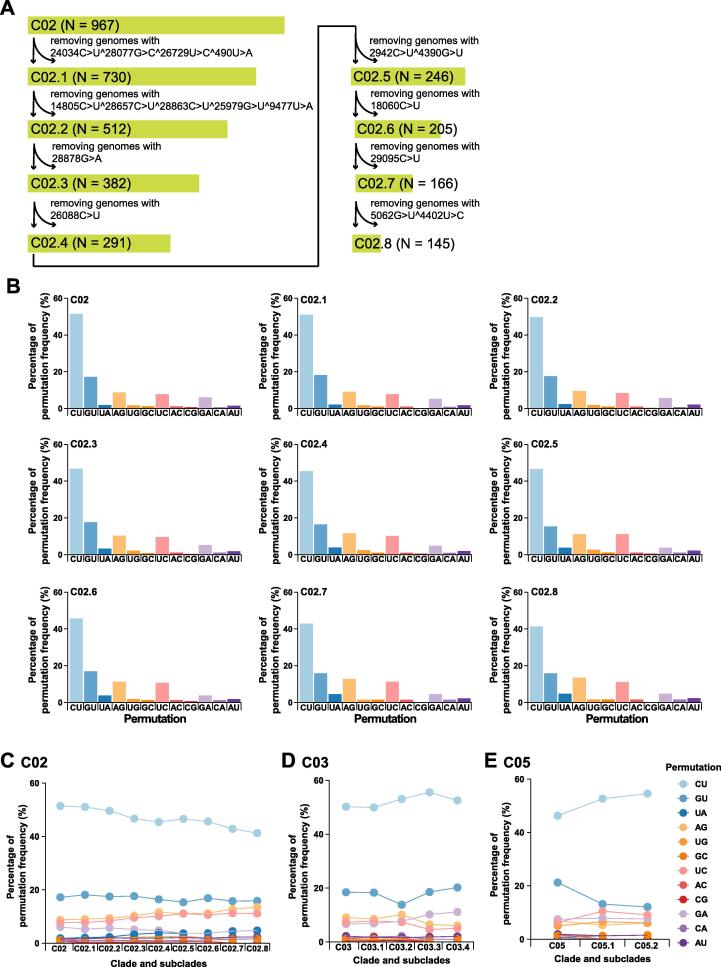
To distinguish the underlining mechanisms, we first identify key mutations based on MAF of mutations with a consideration of relatively even distribution among subclades, and then name the subclades in a sequential order based on the absence of a subset (Figure 6A). We further plot out permutations to track changes among subclades. For instance, clade C02 can be divided into 8 subclades and its variable permutation fractions are clearly recognizable. An immediate discovery is the trends of descending C-to-U, ascending A-to-G, and wavy G-to-U that initially goes up with A-to-G but rides down with C-to-U afterward (Figure 6B and C). Taking the two smaller clades, such as C03 and C05, as examples (Figure 6D and E), we first find that their trends of permutation variables show opposite directions, where the increasing C-to-U accompanies with the decreasing G-to-U. A closer examination reveals that the increasing C-to-U in C03 is also accompanied by descending U-to-C. The only permutation showing an increasing trend in C03 is G-to-A. Other less variable within-clade permutation changes were shown in Figure S6. The take-home message from these trends is that RNA synthesis of subclades in C02 is biased toward producing more negative-sense strands, which also means the mutation spectrum exhibits increasing mutations generated during the negative-sense strand synthesis (less production of positive-sense strands results in descending C-to-U). Such analysis can be carried out continuously when more CoV genome data become available.
Figure 6.
Within-clade permutation variations are excellent indicators of functional mutations
A. The identification pipeline of subclades in clade C02. The number of SARS-CoV-2 genomes in each clade and subclade is indicated in the parentheses. B. Permutation variation of each subclade in clade C02. The clear trends are two-fold. First, decreased C-to-U permutation is coupled with increased A-to-G and decreased G-to-U permutations. Second, A-to-U permutation is also increased as expected based on the model shown in Figure S5. These trends of permutation changes suggest irrelevant to the ratio of strand-biased synthesis (positive sense vs negative sense) but possible structural and/ or conformational variations in the RTCs. C.–E. show within-clade permutation changes of C02, C03, and C05. In each display, the first column of the x-axis shows the proportion of permutations calculated for each clade. Two opposite trends of permutation variations are seen between C03 and C05, which has a rather wavy pattern.
Several precautions are worth noting in such analysis. The most noticeable weakness is the fact that we assume function-related mutations are discovered in our dataset. As we have proposed an analogy before, chances are slim, dozens out of millions or even billions. Furthermore, even if we see drastic changes in permutations and mutation spectra, the mutations we identified still need validation empirically and based on different data types or sources albeit rare and precious. Finally, most frequently encountered situations are those that multiple mutations exhibit confounding (or genetically linking) effects for a phenotypically identified functional or structural feature, and undoubtedly, more and deep-sequencing data are still invaluable and irreplaceable.
Conclusion
This COVID-19 pandemic provides once-in-a-lifetime opportunity for the fields of biomedicine and other life sciences to work together on it as many facets as possible albeit exchanging with lives and other massive losses. If lessons told, we had learned things in serious ways in the last two CoV epidemics and we did prepare ourselves with vaccines and medication since we would not have suffered this much this time. If one assumes that the last two outbreaks of SARS-CoV and MERS-CoV came surely by chance, this time SARS-CoV-2 is here for real, and a worst-case scenario is that it may stay with us forever or until effective vaccination is developed. Nevertheless, it certainly will stay with us for quite a while for many reasons [11]. First, at least it and other within-lineage CoVs may come again because we have not been able to trace its origin and its transmission route from the very beginning. Next, this particular virus, SARS-CoV-2, has evolved to a composition status where some of its natural yet genetically distant hosts or possible intermediate mammalian hosts also have acquired similar status [30], [31]. Furthermore, we do not yet have enough data to really map out the phylogenetic position that allows us to pinpoint its natural hosts, geographic origins, and animal-to-human transmission routes.
The number one need for us is data, genomic and clinical data, which should be as complete as possible and with characteristics including high-quality and high-coverage at single-molecule resolution. We currently have been acquiring genomic data and the specialized databases have collections over ten thousand non-redundant sequence variations, but still not enough to address a few possible functional changes of some key protein components [32], [33], [34], [35] let alone understanding mutation-centric cellular mechanisms. Based on median and mean estimates, we have on average a mutation accumulation rate of half a dozen per patient. Although there have been data reported from single-molecule sequencing platform, they are low in coverage [36].
Our final notion is to emphasize the importance of analysis strategies and supporting platforms. Since questions always overwhelm what we can possibly address [37] prioritizing task is of essence together with choices of strategies. The first platform to be established concerns mutation-to-function interpretation, where we have presented one in this report. Another to be considered is mathematic modeling, such as cellular and disease transmissions [38], [39], [40], [41], [42], [43] and viral mutation-selection paradigm, for testing and evaluating different parameters and prioritizing what kind of data to be acquired with high priorities. In addition, cellular and molecular data, including different omics studies [44] all need to be incorporated into a COVID-19 knowledgebase, where information from multi-disciplinary studies are managed, organized, and mined.
Materials and methods
SARS-CoV-2 and other related CoV sequences
We used the public SARS-CoV-2 data collected worldwide among several major databases, including National Genomics Data Center (NGDC) [45] China National GeneBank DataBase (CNGBdb) [46] GISAID [29] NCBI GenBank [47] and National Microbiology Data Center (NMDC) [48] on 12 June 2020. To ensure authenticity and reliability, our datasets must meet the following criteria: 1) The genome sequence is labeled as complete that covers all coding regions of the reference genome (GenBank: NC_045512.2). 2) It has no more than 15 uncertain bases substituted as “N”s. 3) It has no more than 50 degenerate bases that labeled as discrete nucleotides (e.g., R represents A or G and Y represents C or T). These high-quality genomes were aligned to the reference using MUSCLE (version 3.8.31) with default parameter settings [49]. Further analyses of SARS-CoV-2 and related CoV genomes were referenced to genome annotation of the same reference genome (GenBank: NC_045512.2) and other genomes accessed from NCBI RefSeq or GenBank.
Other closely related CoV genome sequences, including hsa-betaCoV-HKU1, hsa-betaCoV-OC43, ave-gamaCoV, mga-gamaCoV, smu-alphaCoV-WS, hsa-alphaCoV-229E, hsa-alphaCoV-NL63, taf-alphaCoV-NL63, MERS-CoV (from human and camel hosts), cdr-betaCoV-B73, SARS-CoV (from human and civet hosts), pla-betaCoV-SZ3, mja-betaCoV-P4L, and raf-betaCoV-RaTG13, were retrieved from NCBI GenBank. A full list of our sequences including virus genus, strain name, accession number and sources was provided in Table S3.
Calculation of genomic composition parameters
Several genomic composition dynamics and its parameters (G + C content, A + G content and GC skew) were displayed using different sliding windows. The first 300 nt are grouped as an initial window, and subsequent windows are uniformly shifted in a 21-nt step. Within these displays, the G + C contents referenced to the three codon positions of each open reading frame (ORF) are measured by adjusting the sliding window according to the ORF lengths within viral genomes. As for ORFs longer than 2000 nt, a relatively large window size (300 nt) is adopted, and the step size is calculated via a custom formula where denotes the length of ORF and varies from zero to two bases to make sure the window size is divisible by 3; for ORFs with a medium size (longer than 500 nt and shorter than 2000 nt), the window size is defined as , while the step size is simply defined as 3 nt; as for those small ORFs (shorter than 500 nt) such as structural proteins, a constant 21-nt window size and 3-nt step size is used for calculating genomic composition frequency.
The criteria for choosing the representative CoV genome sequences for constructing a representative phylogenetic tree (Figure 2) are multi-fold. First, all 7 human- infecting CoVs were included for the analysis, namely, SARS-CoV, SARS-CoV-2, MERS-CoV, hsa-alphaCoV-229E, hsa-betaCoV-OC43, hsa-betaCoV-HKU1, and hsa-alphaCoV-NL63 (a prefix hsa- stands for Homo sapiens to label the unfamiliar human-infecting CoVs). Second, all human-infecting CoVs were categorized into 4 lineages for simplicity: SARS-CoV-2, SARS-CoV, MERS-CoV, and the older human CoV lineages. Therefore, their related CoVs in the literature were also selected for the analysis, including a single closely-related CoV for each lineage (based on sequence identity): SARS-CoV-related (pla-betaCoV-SZ3), MERS-CoV-related (cdr-betaCoV-B73), SARS-CoV-2-related (raf-betaCoV-RaTG13), and NL63-related (taf-alphaCoV-NL63; both species and CoV genera were labeled for clarity). Third, more informative CoV genome sequences were also added to enrich lineage-associated information, which are a pangolin CoV genome (mja-betaCoV-P4L) reported to be closed to SARS-CoV-2 and 3 non-betacoronaviruses that infect animals (e.g., ave-gamaCoV from gammacoronavirus genus and smu-alphaCoV-WS from alphacoronavirus genus). Fourth, only complete protein-coding sequences from the CoVs were used to construct the phylogenetic tree and to calculate genome parameters. The sequences were aligned with MUSCLE and the UPGMA tree was constructed by MEGA-X [50]. The G + C and single nucleotide contents of each virus genome at three codon positions were also calculated. Subgenomes of SARS-CoV were obtained from Marra et al. [51] and we annotated the subgenomes of SARS-CoV-2, mja-betaCoV-P4L, and raf-betaCoV-RaTG13 based on the annotation of NCBI (GenBank: NC_045512.2, MT040333.1, and MN996532.1, respectively). In addition, G + C and single nucleotide contents of the complete genome and its subgenomes of these four viruses at three codon positions were displayed to serve as sequence composition references.
Variation detection and categorization
All sequence variations were identified and categorized based on comparisons between the query and the reference genomes, and files were generated by using an in-hoc Perl script based on alignment results. The tailored annotation (gene, location, and consequence on the protein sequence) of each variant was determined with VEP (version 99.0) [52]. Since a large number of gaps and low-quality sequences at the 3′ and 5′ ends, variations (substitutions, insertions, and deletions or indels) occurring 50 nt each at 5′- and 3′-ends of the genome were not considered. Since the higher quartile of variations per genome among SARS-CoV-2 populations is 9 (based on the 22,051 sequences we analyzed in this study), we filtered out the problematic genomes that exceed 50 variations as compared to the reference genome. CoV genome sequences have at least one mutation were used in this study. A full list of variations among coding regions identified in this study was provided in Table S4.
All continuously updated mutation files of the SARS-CoV-2 populations in variant call format (version 4.2) were deposited at the “Genome Variations” page of the 2019nCoVR database [53] contributed by NGDC (https://bigd.big.ac.cn/ncov/variation/).
Mutation spectrum analysis
A mutation spectrum for within-population variations is composed of two lines of information; one concerns mutations that are referenced to a population consensus built based on the entire collection, and the other contains frequencies of all mutations and their directional changes, i.e., permutations. To reduce pitfalls of sequencing errors, we only selected mutations that occur more than twice in the whole collection of SARS-CoV-2 populations (clades or clade clusters that are often defined based on phylogenetic analysis). In theory, there are 16 possible permutations but 4 of them (C-to-C, A-to-A, U-to-U, and G-to-G) are unrecognizable so that 12 permutations (C-to-U, A-to-G, U-to-C, G-to-A, G-to-U, U-to-G, A-to-C, C-to-A, U-to-A, G-to-C, C-to-G, and A-to-U) are there as an informative set. When the number of CoV genomes collected is limited, such as SARS-CoVs and MERS-CoVs, entire data sets are pooled together without clades. In our analyses on SARS-CoVs and MERS-CoVs (Figure S3A and B), we aligned sequences from these two lineages to their reference genomes (GenBank: NC_004718.3 and NC_019843.3 for SARS-CoV and MERS-CoV, respectively) to call variations. When aligned on overlapping sequences, due to large deletions or additional ORFs, we always chose the largest ORFs to represent the segment. For example, in the SARS-CoV lineage, if a mutation falls into the overlapping region of ORF9a (encoding the N protein) and ORF9b, we only used the ORF9a annotations to avoid redundancy.
Phylogeny construction
Given the scale of SARS-CoV-2 sequence collections, we focused on genomes with unique information contributing to phylogenetic analysis. First, mutations (including single-nucleotide substitution and indel) at frequencies equal or greater than 10 were selected. FastTree (version 2.1.11) [54] was used to construct maximum likelihood phylogeny in Figure S4 based on 5121 genomes that met our criteria, and iTol [55] an interactive web server, was employed for setting an unrooted format and annotating samples.
For Figure 4B, the neighbor-joining method was used for constructing phylogeny from the Euclidean distance of the mutation frequency matrix of clades, and the tree was generated and visualized by R packages, phangorn [56] and ggtree [57].
Estimation of G + C and purine (or A + G) contents of SARS-CoV-2 genome sequences
G + C and A + G contents of SARS-CoV-2 genomes in general vary in a narrow range, and therefore, subtleties among the content changes have to be scrutinized with low-quality sequences excluded. A more sensitive approach was used in this study where two points were assumed; all genomes are full-length and variant alleles in coding sequences are the varied composition. The absolute frequencies of A + G and G + C contents were defined as:
| (1) |
and
| (2) |
where 8954, 5492, 5863, and 29,903 are the frequencies of A, G, C, and total length of the SARS-CoV-2 reference, respectively. For any sequence compared with the reference, and measure the deleted and inserted nucleotides of this sequence, respectively, and that is why means the variation of sequence length. For all the variant sites in this sequence, in Equation (1) measures the number of A and G variations in compared sequence, where and denote the number of nucleotides mutated to A or G while and represent the number of nucleotides mutated from A or G. Similarly, in Equation (2) represents the varied number of G and C in compared sequences.
Clade subgrouping
To detect trend followers and disrupters in mutation spectra, a pipeline was developed to select such genomes and mutations within clades iteratively. The first step includes locating high-frequency mutations (major alleles, MA) in a clade and extracting all genomes without this MA mutation to form a subset of the clade. The second step is, within the new subclade, to iterate the process until such mutations are thoroughly identified and no more mutations exceed a manually set threshold of MAF. Since the number of unique variations among clades has been varying significantly over time, the thresholds were 0.05 in C01, C04, C06, C08, and C09, and 0.1 in C02, C03, C05, and C07. The proportion of permutations in each subclade and the located gene and mutation type (synonymous or non-synonymous) of subclade-defining mutations were provided in Table S5.
CRediT author statement
Xufei Teng: Data curation, Methodology, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Qianpeng Li: Data curation, Methodology, Formal analysis, Visualization, Writing - review & editing. Zhao Li: Data curation, Formal analysis, Visualization. Yuansheng Zhang: Methodology, Formal analysis, Visualization. Guangyi Niu: Formal analysis. Jingfa Xiao: Methodology, Resources. Jun Yu: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Supervision. Zhang Zhang: Methodology, Resources, Writing - original draft, Writing - review & editing, Supervision. Shuhui Song: Conceptualization, Methodology, Resources, Writing - original draft, Writing - review & editing, Supervision. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported by grants from The Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA19090116 to SS, Grant No. XDA19050302 to ZZ), National Key R&D Program of China (Grant Nos. 2020YFC0848900 and 2017YFC0907502), 13th Five-year Informatization Plan of Chinese Academy of Sciences (Grant No. XXH13505-05), K. C. Wong Education Foundation to ZZ, and International Partnership Program of the Chinese Academy of Sciences (Grant No. 153F11KYSB20160008). The Youth Innovation Promotion Association of Chinese Academy of Science (Grant No. 2017141 to SS), National Natural Science Foundation of China (Grant No. 31671350 to JY) and the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (Grant No. QYZDY-SSW-SMC017 to JY). We thank our colleagues and students for their hard working on the 2019nCoVR (https://bigd.big.ac.cn/ncov). We acknowledge Dr. Lina Ma, Dr. Lili Hao, and Dr. Meng Zhang (Cixi Institutes for Life Science Innovation, Cixi, Zhejiang 315999, China) for direct involvement in discussion and professional advice.
Handled by Kai Ye
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2020.10.003.
Contributor Information
Jun Yu, Email: junyu@big.ac.cn.
Zhang Zhang, Email: zhangzhang@big.ac.cn.
Shuhui Song, Email: songshh@big.ac.cn.
Supplementary material
The following are the Supplementary data to this article:
A display of genome compositional dynamics of raf-betaCoV-RaTG13, mja-betaCoV-P4L, SARS-CoV, and MERS-CoV. We use a 300-bp sliding window with a 21-bp step to show dynamic changes of genomic G+C, purine, GC1 to GC3, GC skew (G-C/G+C) contents, and mean G+C and purine contents over the entire genomes, individual ORFs, and proteins of raf-betaCoV-RaTG13 (A), mja-betaCoV-P4L (B), SARS-CoV (C), and MERS-CoV (D). The complete genome sequences and data sources are listed in Table S1.
A genetic table linked permutations to codons. A genetic code table is shown how nucleotide permutations are related to codons. The table is divided into two halves (purple and green backgrounds) and CP1 and CP2 relative to their permutation changes are indicated with half parentheses with color-coding: U-to-C|C-to-U and A-to-G|G-to-A, red; U-to-G|G-to-U and A-to-C|C-to-A, blue; and A-to-U|U-to-A and G-to-C|C-to-G, green. Note that CP1 and CP2 are sensitive to column and row codon swaps, respectively. CP3 is in a unique position where only half of the codons are sensitive to its changes, and the other half is so organized that some codons are more permissive than others.
Mutation spectra of MERS-CoV, SARS-CoV, and SARS-CoV-2 in a context of codon positions. A. MERS-CoV and SARS-CoV mutation spectra are composed of 12 permutations and they are divided by codon positions. C-to-U (CU) permutations are always as dominant as what SARS-CoV-2 shows. B. C-to-U permutations at CP3 diminish among non-synonymous mutations and this phenomenon indicates that most protein composition relevant variations are CP1 and CP2 variations. C. Displays of permutation-to-codon changes among non-synonymous mutations. The permutations shown here contain U-to-C, G-to-A, G-to-U, U-to-G, C-to-A, A-to-C, U-to-A, G-to-C, A-to-U, and C-to-G.
High-frequency within-clade mutations. The current collection shows 9 clades (C01 to C09) in three clusters (L, S, and G). An unrooted phylogenic tree of the clades and clusters (color-coded) is displayed with the tree scale at 0.01.
A table illustrates how 12 permutations are related to G+C and purine contents and the subtle RTC specificity of CoVs. Mutations occur when the positive-sense or the negative-sense RNA genomes are replicated. R1 permutations are those mutations created when the negative-sense genome is synthesized, and all of them are the transition type. R2 permutations are those mutations when negative-sense genome is used as template for replication, where abasic nucleotides have occurred before synthesis, and all of them are transversion type. R12 permutations are those assumed to occur in a more complex process as compared to both R1 and R2 permutations since they are even rarer. C-to-U (or CU) through a G-by-A replacement in the synthesis of the negative-sense genome is strongly preferred when RTC is in a status that encourages a large-to-small substrate exchange in a higher ratio than the opposite, small-to-large substrate exchange. We term this preferred exchange as tight status. This status is also divided into R-tight (R, purine; to indicate that the mechanism is a G-by-A replacement; G is 15-dalton larger that A) and Y-tight (Y, pyrimidine; to indicate that the mechanism is a U-by-C replacement; U is 1-dalton larger than C). When an exchange of substrate happens from small-to-large, it is referred as a loose status that is also divided into two, R-loose and Y-loose accordingly. Note that some permutations are not sensitive to G+C and purine contents but others are sensitive. The arrow-headed lines indicate the changing trends of G+C and A+G content. This figure is adopted from Figure 3B in [11].
Within-clade permutation variations are excellent indicators of functional mutations. (A)–(F) show within-clade permutation changes of C01, C04, C06, C07, C08, and C09, respectively.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Coronavirus disease (COVID-2019) situation report - 144. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (Jun 12 2020, date last accessed).
- 3.He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 4.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 6.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 11.Yu J. From mutation signature to molecular mechanism in the RNA world: a case of SARS-CoV-2. Genomics Proteomics Bioinformatics. 2020;18:625–637. doi: 10.1016/j.gpb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmonds P. Rampant C→U Hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5:e00408–20. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Giorgio S, Martignano F, Torcia MG, Mattiuz G, Conticello SG. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv 2020;6:eabb5813. [DOI] [PMC free article] [PubMed]
- 16.Xiao J., Yu J. A scenario on the stepwise evolution of the genetic code. Genomics Proteomics Bioinformatics. 2007;5:143–151. doi: 10.1016/S1672-0229(08)60001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J. A content-centric organization of the genetic code. Genomics Proteomics Bioinformatics. 2007;5:1–6. doi: 10.1016/S1672-0229(07)60008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Yu J. On the organizational dynamics of the genetic code. Genomics Proteomics Bioinformatics. 2011;9:21–29. doi: 10.1016/S1672-0229(11)60004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Yu J. The pendulum model for genome compositional dynamics: from the four nucleotides to the twenty amino acids. Genomics Proteomics Bioinformatics. 2012;10:175–180. doi: 10.1016/j.gpb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui P., Lin Q., Ding F., Hu S., Yu J. The transcript-centric mutations in human genomes. Genomics Proteomics Bioinformatics. 2012;10:11–22. doi: 10.1016/S1672-0229(11)60029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui P., Ding F., Lin Q., Zhang L., Li A., Zhang Z. Distinct contributions of replication and transcription to mutation rate variation of human genomes. Genomics Proteomics Bioinformatics. 2012;10:4–10. doi: 10.1016/S1672-0229(11)60028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong G.K.S., Wang J., Tao L., Tan J., Zhang J., Passey D.A. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z., Lu L., Du J., Yang L., Ren X., Liu B. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 2018;6:178. doi: 10.1186/s40168-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 25.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang X., Wu C., Li X., Song Y., Yao X., Wu X. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. 2020;30 doi: 10.1016/j.cub.2020.09.030. 2196-203 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 2020;74 doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniloski Z., Jordan T.X., Ilmain J.K., Guo X., Bhabha G., tenOever B.R. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021;10:e65365. doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korber B., Fischer W., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. Nat Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M., Fu A., Hu B., Tong Y., Liu R., Gu J. Nanopore targeted sequencing for the accurate and comprehensive detection of SARS-CoV-2 and other respiratory viruses. Small. 2020;16 doi: 10.1002/smll.202002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teymoori-Rad M., Samadizadeh S., Tabarraei A., Moradi A., Shahbaz M.B., Tahamtan A. Ten challenging questions about SARS-CoV-2 and COVID-19. Expert Rev Respir Med. 2020;14:881–888. doi: 10.1080/17476348.2020.1782197. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q., Zhao S., Shi C.M., Song S.H., Zhu S., Su Y. Population genetics of SARS-CoV-2: disentangling sampling bias and clustering infections. Genomics Proteomics Bioinformatics. 2020;18:640–647. doi: 10.1016/j.gpb.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cotten M., Watson S.J., Kellam P., Al-Rabeeah A.A., Makhdoom H.Q., Assiri A. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gire S.K., Goba A., Andersen K.G., Sealfon R.S.G., Park D.J., Kanneh L. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemey P., Suchard M., Rambaut A. Reconstructing the initial global spread of a human influenza pandemic: a bayesian spatial-temporal model for the global spread of H1N1pdm. PLoS Curr. 2009;1:RRN1031. doi: 10.1371/currents.RRN1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith G.J.D., Vijaykrishna D., Bahl J., Lycett S.J., Worobey M., Pybus O.G. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 43.Yu W.B., Tang G.D., Zhang L., Corlett R.T. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2/HCoV-19) using whole genomic data. Zool Res. 2020;41:247–257. doi: 10.24272/j.issn.2095-8137.2020.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanders W., Fritch E.J., Madden E.A., Graham R.L., Vincent H.A., Heise M.T. Comparative analysis of coronavirus genomic RNA structure reveals conservation in SARS-like coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.06.15.153197. [DOI] [Google Scholar]
- 45.National Genomics Data Center Members and Partners Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020;48:D24–33. doi: 10.1093/nar/gkz913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B., Liu F., Zhang E.C., Wo C.L., Chen J., Qian P.Y. The China national GeneBank horizontal line owned by all, completed by all and shared by all. Hereditas (Beijing) 2019;41:761–772. doi: 10.16288/j.yczz.19-148. (in Chinese with an English abstract) [DOI] [PubMed] [Google Scholar]
- 47.Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I. GenBank. Nucleic Acids Res. 2020;48:D84–D86. doi: 10.1093/nar/gkz956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu L., Sun Q., Desmeth P., Sugawara H., Xu Z., McCluskey K. World data centre for microorganisms: an information infrastructure to explore and utilize preserved microbial strains worldwide. Nucleic Acids Res. 2017;45:D611–D618. doi: 10.1093/nar/gkw903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 52.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song S., Ma L., Zou D., Tian D., Li C., Zhu J. The global landscape of SARS-CoV-2 genomes, variants, and haplotypes in 2019nCoVR. Genomics Proteomics Bioinformatics. 2020;18:749–759. doi: 10.1016/j.gpb.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu G. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics. 2020;69 doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A display of genome compositional dynamics of raf-betaCoV-RaTG13, mja-betaCoV-P4L, SARS-CoV, and MERS-CoV. We use a 300-bp sliding window with a 21-bp step to show dynamic changes of genomic G+C, purine, GC1 to GC3, GC skew (G-C/G+C) contents, and mean G+C and purine contents over the entire genomes, individual ORFs, and proteins of raf-betaCoV-RaTG13 (A), mja-betaCoV-P4L (B), SARS-CoV (C), and MERS-CoV (D). The complete genome sequences and data sources are listed in Table S1.
A genetic table linked permutations to codons. A genetic code table is shown how nucleotide permutations are related to codons. The table is divided into two halves (purple and green backgrounds) and CP1 and CP2 relative to their permutation changes are indicated with half parentheses with color-coding: U-to-C|C-to-U and A-to-G|G-to-A, red; U-to-G|G-to-U and A-to-C|C-to-A, blue; and A-to-U|U-to-A and G-to-C|C-to-G, green. Note that CP1 and CP2 are sensitive to column and row codon swaps, respectively. CP3 is in a unique position where only half of the codons are sensitive to its changes, and the other half is so organized that some codons are more permissive than others.
Mutation spectra of MERS-CoV, SARS-CoV, and SARS-CoV-2 in a context of codon positions. A. MERS-CoV and SARS-CoV mutation spectra are composed of 12 permutations and they are divided by codon positions. C-to-U (CU) permutations are always as dominant as what SARS-CoV-2 shows. B. C-to-U permutations at CP3 diminish among non-synonymous mutations and this phenomenon indicates that most protein composition relevant variations are CP1 and CP2 variations. C. Displays of permutation-to-codon changes among non-synonymous mutations. The permutations shown here contain U-to-C, G-to-A, G-to-U, U-to-G, C-to-A, A-to-C, U-to-A, G-to-C, A-to-U, and C-to-G.
High-frequency within-clade mutations. The current collection shows 9 clades (C01 to C09) in three clusters (L, S, and G). An unrooted phylogenic tree of the clades and clusters (color-coded) is displayed with the tree scale at 0.01.
A table illustrates how 12 permutations are related to G+C and purine contents and the subtle RTC specificity of CoVs. Mutations occur when the positive-sense or the negative-sense RNA genomes are replicated. R1 permutations are those mutations created when the negative-sense genome is synthesized, and all of them are the transition type. R2 permutations are those mutations when negative-sense genome is used as template for replication, where abasic nucleotides have occurred before synthesis, and all of them are transversion type. R12 permutations are those assumed to occur in a more complex process as compared to both R1 and R2 permutations since they are even rarer. C-to-U (or CU) through a G-by-A replacement in the synthesis of the negative-sense genome is strongly preferred when RTC is in a status that encourages a large-to-small substrate exchange in a higher ratio than the opposite, small-to-large substrate exchange. We term this preferred exchange as tight status. This status is also divided into R-tight (R, purine; to indicate that the mechanism is a G-by-A replacement; G is 15-dalton larger that A) and Y-tight (Y, pyrimidine; to indicate that the mechanism is a U-by-C replacement; U is 1-dalton larger than C). When an exchange of substrate happens from small-to-large, it is referred as a loose status that is also divided into two, R-loose and Y-loose accordingly. Note that some permutations are not sensitive to G+C and purine contents but others are sensitive. The arrow-headed lines indicate the changing trends of G+C and A+G content. This figure is adopted from Figure 3B in [11].
Within-clade permutation variations are excellent indicators of functional mutations. (A)–(F) show within-clade permutation changes of C01, C04, C06, C07, C08, and C09, respectively.