Abstract
Actinobacteria are among the most prolific sources of medically and agriculturally important compounds, derived from their biosynthetic gene clusters (BGCs) for specialized (secondary) pathways of metabolism. Genomics witnesses that the majority of actinobacterial BGCs are silent, most likely due to their low or zero transcription. Much effort is put into the search for approaches towards activation of silent BGCs, as this is believed to revitalize the discovery of novel natural products. We hypothesized that the global transcriptional factor AdpA, due to its highly degenerate operator sequence, could be used to upregulate the expression of silent BGCs. Using Streptomyces cyanogenus S136 as a test case, we showed that plasmids expressing either full-length adpA or its DNA-binding domain led to significant changes in the metabolome. These were evident as changes in the accumulation of colored compounds, bioactivity, as well as the emergence of a new pattern of secondary metabolites as revealed by HPLC-ESI-mass spectrometry. We further focused on the most abundant secondary metabolite and identified it as the polyene antibiotic lucensomycin. Finally, we uncovered the entire gene cluster for lucensomycin biosynthesis (lcm), that remained elusive for five decades until now, and outlined an evidence-based scenario for its adpA-mediated activation.
Subject terms: Microbiology, Antimicrobials, Microbial genetics
Introduction
Over decades, prokaryotes were one of the most prolific sources of antibiotics, rivaled only by lower fungi1. Among antibiotic-producing prokaryotes, the classes Actinobacteria and Myxobacteria currently dominate2,3, although exploration of the other taxa could change this view significantly in the near future4–6. The actinobacterial genus Streptomyces probably still is the most successful provider of drugs for healthcare and agriculture1. Beginning with seminal works of Selman Waksman7,8, the majority of known antibiotic compounds from Streptomyces were isolated through classical screening approaches. However, due to high rediscovery rates, this approach has gradually lost its efficiency9. More recently, the sequencing of Streptomyces spp. genomes revealed a large hidden potential for the production of antibiotics and other secondary metabolites. Exemplary studies of the fully sequenced genomes of S. coelicolor and S. avermitilis revealed many more biosynthetic gene clusters (BGCs) than compounds previously known to be produced by these species10,11. Ever since, each new sequenced Streptomyces genome gave rise to the idea that most of the actinobacterial BGCs are silent, i.e. that they are not associated with the production of certain compounds under typical cultivation conditions, most likely due to low or zero transcription12,13.
Today’s relative simplicity of in silico identification of BGCs within genomic sequences14,15 prompted the search for approaches to activate silent BGCs. Certain mutations in genes for ribosomal proteins and RNA polymerases are capable of eliciting the expression of the silent secondary metabolome16,17. Extracellular triggers, like xenobiotics, rare earth elements or co-cultivation with other bacteria were also shown to activate the production of novel antibiotics by streptomycetes18–22. Manipulations of either regulatory elements within the putative BGCs23,24, or BGC-situated regulatory genes23,25,26 often lead to the activation of silent BGCs. As antibiotic production in streptomycetes is tightly intertwined with morphogenesis, global regulators that coordinate both aforementioned processes can be useful tools for activation of silent BGCs for known or cryptic biosynthetic pathways27–32. Such an approach could be especially useful for activation of the BGCs lacking cluster-situated regulatory genes.
The AraC family transcriptional factor (TF) AdpA is a global regulator of secondary metabolism and morphogenesis, and was successfully used to activate secondary metabolism29,30. AdpA appears as an appealing tool in this regard because it binds to a highly degenerate operator sequence33 and thus possesses an enormous regulon34. The overexpression of such a promiscuous TF could enforce binding to numerous (probably unrecognizable by AdpA under physiological conditions) sites on the chromosome, impacting the expression of multiple genes and operons. The biosynthesis of many antibiotics in streptomycetes is AdpA-regulated and AdpA TFs are almost identical across different Streptomyces species and therefore likely display the same properties35. AdpA-binding sites are common in silent BGCs and overexpression of adpA genes could be used for high-throughput activation of such BGCs in strains of unknown genomic background.
In this work we used Streptomyces cyanogenus S136 as a proof-of-concept to probe the above conjecture. This strain is known to produce deeply colored angucyclines, polyketides from the landomycin family, in a medium-dependent fashion; furthermore, sequencing revealed the presence of > 30 BGCs in its genome. We show that the secondary metabolome of S. cyanogenus actively responds to the overexpression of heterologous adpA genes with the production of novel secondary metabolites. Here we focused on the genetic basis of the production of the most abundant compound, lucensomycin, known for its antifungal activity. From our results we deduced a scheme for AdpA-mediated regulation of lucensomycin biosynthesis. Finally, we explored the functional significance of different parts of the AdpA protein for its ability to induce lucensomycin production. Our results expand the current knowledge of the secondary metabolome of S. cyanogenus and offer new insights into the ways of activation of silent BGCs in Streptomyces.
Results
Genome of Streptomyces cyanogenus encodes sizable secondary metabolome that responds to adpA
In the genome of S. cyanogenus S136, in addition to the lan cluster coding for landomycin biosynthesis, at least 33 putative BGCs were found under the default antiSMASH search conditions. From these, 16 BGCs shared significant similarities to known BGCs (ESM Fig. S1). These were the BGCs for informatipeptin, pimaricin, naphtomycin, chlorothricin and different RiPPs, furthermore for terpenoids hopene, isorenieratene and albaflavenone, the siderophore desferrioxamine B, ectoine, melanins and the spore pigment of polyketide nature. It is worth mentioning that most of the antibiotic BGCs putatively encode the biosyntheses of polyketide compounds (e.g. pimaricin, naphtomycin, chlorothricin).
In S. cyanogenus S136, the gene SCY4743 codes for a nonfunctional AdpA, and expression of heterologous adpA genes activated landomycin biosynthesis under conditions where S136 normally does not produce this antibiotic36. This observation prompted us to look deeper into the secondary metabolome of S. cyanogenus strains carrying heterologous versions of adpA. Initially, we chose a landomycin-nonproducing mutant ΔlanI737 for these experiments. The mutant strain was chosen in order to eliminate the competition for precursors (acyl-CoA, carbohydrates) of the landomycin pathway with potentially activated BGCs directing the production of biogenetically related compounds. Plasmids pGM4181 and pGM4181d carrying full adpA of S. albus J1074 and its DNA-binding domain (DBD), respectively, under the control of moeE5p promoter, were introduced into S. cyanogenus ΔlanI7. This promoter allows strong constitutive gene expression in S. cyanogenus36.
S. cyanogenus ΔlanI7 pGM4181+, but not pGM4181d+, started producing a dark brown compound when grown on TSA plates or in TSB media (Fig. 1a). However, we were not able to extract this compound from culture broths or solid agar media with common solvents. Likewise, mass spectrometry of the culture broth failed to identify distinct mass peaks. We suspected that the observed pigmentation was due to accumulation of tyrosine-based polymers, melanins, because in several Streptomyces species production of the latter is known to be upregulated by AdpA38. The structure elucidation of these polymers remains elusive due to their heterogeneity and poor solubility39,40. We therefore tentatively suggest that melanin-type compounds are accumulated in S. cyanogenus in presence of adpA. Indeed, its genome carries two melC homologs responsible for melanin biosynthesis, and their expression is activated upon pGM4181 introduction (ESM Fig. S2).
Figure 1.
The adpA gene activates silent secondary metabolome of S. cyanogenus. (a) Phenotypes of S. cyanogenus ΔlanI7 (1), S. cyanogenus ΔlanI7 pGM4181d+ (2; DBD of adpA) and S. cyanogenus ΔlanI7 pGM4181+(3; full-length adpA) after 168 h of growth on TSB agar; B. cereus (b) and D. hansenii (c) growth inhibition assays using agar plugs
taken from 120 h old ISP5 lawns of S. cyanogenus ΔlanI7 (1), S. cyanogenus ΔlanI7 pGM4181d+ (2) and S. cyanogenus ΔlanI7 pGM4181+(3). (d) LC–MS analysis of the secondary metabolites produced in liquid YMPG medium by S. cyanogenus ΔlanI7 pGUSmoeE5script+ (control) and S. cyanogenus ΔlanI7 pGM4181d+; four new mass peaks appeared in the chromatogram for the latter strain.
S. cyanogenus ΔlanI7 displayed weak activity against Gram-positive bacteria when grown on ISP5 agar. Such an activity was significantly increased in S. cyanogenus ΔlanI7 pGM4181+ and pGM4181d+ (Fig. 1b). Neither initial nor recombinant strains showed activity against E. coli (data not shown). The adpA expression in S. cyanogenus ΔlanI7 elicited strong antifungal activity, apparently absent in the control strain (Fig. 1c). Encouraged by this finding, we tested a number of agar media and found that many of them (but not all, see ESM Fig. S3) supported the production of antifungal compound(s) by S. cyanogenus ΔlanI7 pGM4181+ and pGM4181d+. We noted that for certain media the expression of DBD of AdpA is sufficient to induce the occurrence of antifungal activity (ESM Fig. S3), but the full-length gene was found to be more efficient. YMPG medium supported the highest level of antifungal activity, and was used for further analysis of adpA effects on the S. cyanogenus metabolome.
We grew ΔlanI7 and its pGM4181+ derivative in liquid YMPG medium and analyzed the biomass and supernatant extracts with LC–MS; the same was done in parallel for wild type (S136) and its pGM4181+ derivative. The peaks for landomycin A and its precursors dominated the chromatograms of the extracts from S136 and S136 pGM4181+; no qualitative differences were found between the HPLC chromatograms of these strains (ESM Fig. S4). In contrast, at least four new mass peaks occurred in the HPLC chromatograms of ΔlanI7 carrying pGM4181 (Fig. 1d). The m/z values corresponding to the less abundant mass peaks had no matches in the databases of natural compounds. The molecular mass of the most prominent “activated” ion peak (708.35 Da, [M + H]+) matched to that of the polyene antibiotic lucensomycin (Lcm)41,42, and thus we focused our attention on this peak. Lcm is known as antifungal compound, and its molecular mass peak is strongly prevailing in the HPLC–MS analytics of the metabolome of the ΔlanI7 pGM4181+ strain. We suppose therefore that Lcm is responsible for the antifungal activity exhibited by this strain. Nevertheless, we cannot exclude the contribution of the other, as yet unknown, compounds accumulated by ΔlanI7 pGM4181+ to antifungal activities of its extracts and agar plugs, although this contribution (if present) is likely insignificant.
Optimized conditions for expression of antifungal activity in S. cyanogenus ΔlanI7
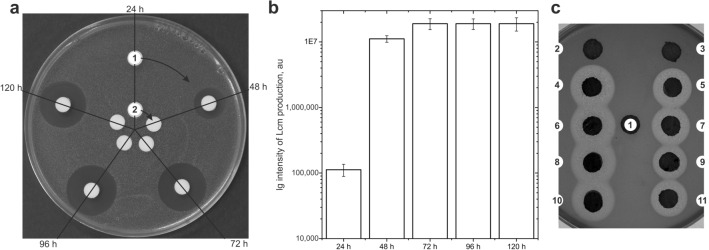
We set out to identify the optimal conditions for the production of the antifungal compound by S. cyanogenus ΔlanI7 pGM4181+ prior to its detailed structural verification. In liquid YMPG the production of an antifungal compound became detectable after 48 h of growth and reached its peak at 96 h (Fig. 2a). The bioassay data correlated with the abundance of the 708.35 Da peak in HPLC chromatograms of the same extracts (Fig. 2b). No antifungal activity was observed in the extracts of ΔlanI7 biomass harvested at 24 h, 48 h, 72 h, 96 h and 120 h (Fig. 2a).
Figure 2.
Antifungal activity of S. cyanogenus ΔlanI7 pGM4181+ as a function of cultivation time (a, b) and origin of adpA gene (c). For D. hansenii growth inhibition assay (a) the extracts were prepared from YMPG-grown ΔlanI7 pGM4181+ (1) and ΔlanI7 (2) at timepoints shown in the figure. The initial strain ΔlanI7 (2) showed no activity at all investigated time points. LC–MS quantification of the Lcm mass peak in the same extracts (b) was in agreement with the results of the bioassay. Data represent mean values of three independent experiments ± 2SD. (c) D. hansenii growth inhibition assay showing that adpA genes (and corresponding DNA-binding domains, DBD), other than XNR_4181, were able to activate Lcm production in S. cyanogenus ΔlanI7: (4,5)—ΔlanI7 pGMSCO+ and pGMSCOd+, overexpressing S. coelicolor adpA and its DBD, respectively; (6,7)—ΔlanI7 pGMSCLA+ and pGMSCLAd+, S. clavuligerus adpA and its DBD, respectively; (8,9)—ΔlanI7 pGM4181+ and pGM4181d+, S. albus adpA and its DBD, respectively; (10,11)—ΔlanI7 pOOB95d+ and pGMSGHd+, S. ghanaensis adpA and its DBD, respectively. Initial strain (1) as well as strains carrying native adpA from S. cyanogenus (ΔlanI7 pGMSCY+) and its DNA-binding domain (ΔlanI7 pGMSCYd+) do not inhibit growth of D. hansenii. Agar plugs were cut from the lawns grown on YMPG agar for 120 h.
Subsequently, we introduced into S. cyanogenus ΔlanI7 a set of full-length adpA genes and corresponding DBDs—adpAdbd—from different Streptomyces species. This was carried out to determine whether and to what extent different adpA genes induce the antifungal activity. All heterologous adpA genes were shown to be strong inducers, and adpAdbd’s were less active in this regard (Fig. 2c). The overexpression of neither native adpA (SCY4743) nor its adpAdbd led to the activation of antifungal activity, once again pointing to the non-functionality of SCY4743 in S. cyanogenus.
The AdpA-induced compound corresponds to the polyene antibiotic lucensomycin
Using the optimized production conditions, we were able to purify 40 mg of target antifungal compound from ΔlanI7 pGM4181+ grown in 2 L of YMPG (shaken flasks). In disk diffusion assay the compound showed activity only against fungal species (ESM Table S2, ESM Fig. S5). High-resolution mass spectrometric (MS) measurements (calculated for Lcm: 706.3444 Da ([M − H]−), 708.3590 Da ([M + H]+); observed: 706.3447 ([M − H]−) and 708.3586 ([M + H]+); mass tolerance ≤ 5 mmu) and the fragmentation pattern of the compound (ESM Fig. S6) are in full agreement with the structure of Lcm42,43.
Description of the lucensomycin (lcm) BGC in S. cyanogenus
A survey of BGCs within the S. cyanogenus genome (see above) readily led us to recognize that one BGC shared significant similarity to the pimaricin (also known as natamycin) BGC (pim cluster)44–46. As Lcm and pimaricin are structural homologs41, we suggested that activation of this particular BGC results in Lcm accumulation by S. cyanogenus ΔlanI7 pGM4181+ and pGM4181d+. We therefore refer to this BGC as the lcm (lucensomycin) BGC (accession number MW175861). Besides the lcm BGC, a few other genes for type I polyketide synthases were found within the genome of S. cyanogenus (ESM Fig. S1). There was an overall synteny between the lcm BGC and the BGC already proposed to control Lcm production in S. achromogenes NRRL 312547. However, the Lcm BGC from NRRL 3125 appeared to be incomplete (see below) and its nucleotide sequence is absent in publicly available databases; this precludes their detailed comparison. Nineteen ORFs were annotated within the lcm BGC (Table 1; Fig. 3a), among which 18 are similar to genes found in the other polyene BGCs (Table 1). We took advantage of detailed knowledge of the biosynthesis of pimaricin and structural similarities between the latter and Lcm to propose a tentative biochemical pathway leading to Lcm.
Table 1.
Description of the ORFs discovered in lcm gene cluster.
| lcm | The closest homologue from antibiotic BGCs | AA sequence identity(%)/E-value | Putative product | Putative function in Lcm biosynthesis |
|---|---|---|---|---|
| lcm1 | N/A | – | – | |
| lcm2 | AviE3 (avilamycin BGC) | 61.92/5e−111 | GDP-D-mannose 4,6-dehydratase (COG1089) | Transformation of GDP-D-mannose into GDP-4-keto-6-deoxy-D-mannose |
| lcm3 | FscTE (candicidin BGC) | 60,08/5e−70 | Type II thioesterase (COG3208) | Removal of aberrant intermediates |
| lcmB | PKS modules 2–5 | Lcm aglycone assembly line | ||
| lcmA | PKS initiation and 1 modules | Lcm aglycone assembly line | ||
| lcm4 | AmphDI (amphotericin B BGC) | 58,89/4e−156 | GDP-mycosamine glycosyltransferase (COG1819) | Glycosylation of Lcm aglycone with mycosamine |
| lcm5 | FscMII (candicidin BGC) | 74,43/3e−154 | GDP-3-keto-6-deoxy-D-mannose C-3 aminotransferase (COG0399) | Transformation of GDP-3-keto-6-deoxy-D-mannose into GDP-mycosamine |
| lcm6 | PimG (pimaricin BGC) | 64,78/5e−153 | Cytochrome P450 monooxygenase (COG2124) | Formation of carboxyl group at C-12 |
| lcm7 | RimH (rimocidin BGC) | 66,13/8e−13 | Ferredoxin (COG1141) | Electron transfer in P450 system |
| lcmE | PKS module 12 | Lcm aglycone assembly line | ||
| lcmD | PKS module 11 | Lcm aglycone assembly line | ||
| lcmC | PKS modules 5–10 | Lcm aglycone assembly line | ||
| lcm8 | PimA (pimaricin BGC) | 57,71/3e−147 | ABC transporter (COG1132) | Export of Lcm |
| lcm9 | AmphG (amphotericin B BGC) | 56,20/1e−171 | ABC transporter (COG1132) | Export of Lcm |
| lcm10 | NysL (nystatin BGC) | 42,46/3e−81 | Cytochrome P450 monooxygenase (COG2124) | Epoxidation of Lcm aglycone at C4-C5 |
| lcmRI | NysRIV (nystatin BGC) | 39,38/1e−29 | HTH-LuxR domain containing TF | Regulation of Lcm biosynthesis |
| lcmRII | PimM(pimaricin BGC) | 40,1/2e−32 | HTH-LuxR domain containing TF | Regulation of Lcm biosynthesis |
| lcm11 | PimE | 79,34/0 | Cholesterol oxidase (COG2303) | “Fungal sensor” for ergosterol detection |
| lcmRIII | PimR | 77,9/0 | Transcriptional regulator | Regulation of Lcm biosynthesis |
Figure 3.
A scheme of Lcm biosynthetic gene cluster (lcm BGC) (a) and proposed sequence for the Lcm PKS assembly line (b) with the tailoring reactions (c). For more details, please see the main text and Table 1.
Assembly of Lcm aglycone. There are five PKS genes within the lcm BGC, four of which (lcmB, lcmC, lcmD and lcmE) are colinear to pimS1, pimS2, pimS3 and pimS4. It is therefore reasonable to propose analogous functions for pim and lcm homologs in the polyene aglycone assembly (Fig. 3b). However, the lcmA gene encodes for a PKS in which the initiation module is apparently fused to the first PKS module. This kind of organization is unprecedented in the biosynthesis of polyenes, although widely known for other type I PKS-derived products, such as erythromycin48. Most likely, this reflects the unique structure of Lcm featuring a n-propyl side chain. From analysis of the LcmA domain order, we propose that KS0 would activate methylmalonyl-CoA as the starter substrate, and the second module (featuring full reductive loop) would incorporate malonyl-CoA, thus leading to a fully reduced C5 unit. Indeed, by a number of in silico tools KS0 and KS1 are reliably predicted to bind methylmalonyl- and malonyl-CoA, respectively, lending support for our conjecture. The identified type II thioesterase Lcm3, a homolog of FscTE from the biosynthesis of polyene candicidin49, would be assigned responsible for the proofreading in Lcm aglycone formation.
Post-PKS modifications of Lcm aglycone. This is achieved in three sequential tailoring reactions. First, a carboxy group is installed at position C-12 (Fig. 3), most probably via action of monooxygenase Lcm6 (Table 1). In the next tailoring reaction aminosugar mycosamine is attached. Genes responsible for the biosynthesis of mycosamine and its attachment to the aglycone are present within all polyene BGCs, and the lcm gene cluster is no exception to this finding. Gene lcm2 encodes a GDP-D-mannose 4,6-dehydratase, most probably responsible for the transformation of GDP-D-mannose into GDP-4-keto-6-deoxy-D-mannose. The latter undergoes spontaneous isomerization forming GDP-3-keto-6-deoxy-D-mannose (GDPDM). GDPDM is converted by aminotransferase Lcm5 into GDP-mycosamine. From the latter, the mycosamine residue is transferred onto polyene scaffold by glycosyltransferase Lcm4. Epoxidation is the last tailoring reaction, and Lcm10 is the most likely candidate for catalysis of this reaction50.
Pathway-specific regulation and export. All antifungal polyene BGCs described to date carry genes for transcriptional regulators45. The pim cluster, for instance, contains two such genes, PimR and PimM51,52, the nystatin BGC four: NysRI, NysRII, NysRIII and NysRIV53. PimR is a large multidomain protein known so far to have a single target within the pim cluster: pimM. PimM consists of a N-terminal effector PAS-like domain and a C-terminal LuxR-like DNA binding domain. PimM upregulates structural pim genes. In nystatin biosynthesis NysRI, NysRII and NysRIII are paralogs co-orthologous to PimR that play a redundant role in the activation of nysRIV, a pimM ortholog. The aforementioned pathway-specific regulators are crucial for the biosynthesis of respective compounds. Of note is that regulatory genes apparently are not within the sequenced part of the Lcm BGC of S. achromogenes NRRL 312547. In our case the flank of the lcm gene cluster possesses three genes for the pathway-specific transcriptional regulators: lcmRI, lcmRII and lcmRIII (Table 1). LcmRI is a NysRIV homolog, LcmRII and LcmRIII are homologs of PimM and PimR. There are also genes for ABC-transporters within the lcm cluster, lcm8 and lcm9, encoding homologs of known ABC transporters involved in the export of pimaricin and amphotericin54. Finally, lcm11 encodes a PimE homologue, which is functionally assigned to a cholesterol oxidase. The role of PimE remains obscure. It is crucial for pimaricin production and is believed to be a sensor which detects ergosterol in fungal membrane and somehow triggers the polyene export55–57.
Relationships between AdpA, lucensomycin and landomycin biosynthetic pathways
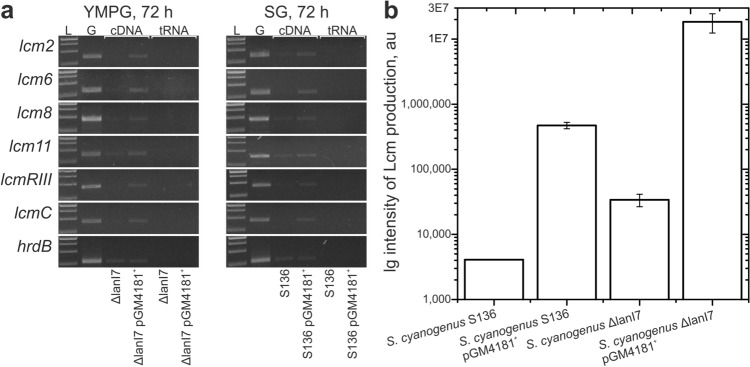
To understand the mechanism of AdpA-mediated activation of Lcm biosynthesis, we analyzed the transcription of selected lcm genes in S. cyanogenus strains S136, S136 pGM4181+, ΔlanI7 and ΔlanI7 pGM4181+ by means of semi-quantitative RT-PCR. Chosen genes code for GDP-D-mannose 4,6-dehydratase (lcm2), monooxygenase (lcm6), ABC transporter (lcm8), fungal sensor (lcm11), key pathway-specific regulator (lcmRIII) and PKS (lcmC). Their products are involved in different steps of Lcm biosynthesis.
First, we checked lcm gene expression in S. cyanogenus ΔlanI7 and S. cyanogenus ΔlanI7 pGM4181+ grown in YMPG for 72 h. The lcm transcripts were absent in S. cyanogenus ΔlanI7 and present in ΔlanI7 pGM4181+ strain (Fig. 4a). Next we analyzed the Lcm biosynthesis and lcm gene expression in landomycin-producing strains S. cyanogenus S136 and S136 pGM4181+ cultivated for 72 h in SG medium (optimal for landomycin A production) and YMPG (supports Lcm synthesis). In bioassays, strain S136 showed no signs of Lcm accumulation when grown on either SG or YMPG agar; weak antifungal activity was detected in case of YMPG-grown S136 pGM4181+ (ESM Fig. S7a). Although Lcm production by S136 pGM4181+ in SG was below detection levels (as evident from data in ESM Fig. S7b) the lcm genes were transcribed in the latter (Fig. 4a). We quantified the accumulation of Lcm by mass spectrometry in the extracts of different S. cyanogenus strains grown in liquid YMPG. The Lcm molecular mass signal was barely detectable in S136 and ΔlanI7. In S136 pGM4181+ and ΔlanI7 pGM4181+ Lcm increased roughly 100-fold (to (5.2 ± 0.5) × 105 au) and 450-fold (to (1.9 ± 0.6 × 107 au), respectively, as compared to their parental strains (Fig. 4b). Hence, in the wild type (S136) AdpA transcriptionally upregulates both Lcm (see above) and landomycin36 BGCs, yet only traces of Lcm can be detected. When the landomycin biosynthesis pathway is shut off (strain ΔlanI7), AdpA leads to abundant production of Lcm. Substantial quantitative differences in the levels of activation of Lcm production by S136 and ΔlanI7 most likely underlie qualitative differences in the bioassay results.
Figure 4.
(a) Transcription of lcm genes involved in different stages of Lcm production (as well as house-keeping gene hrdB) in S. cyanogenus ΔlanI7 and ΔlanI7 pGM4181+ grown in YMPG medium for 72 h; S. cyanogenus S136 and S136 pGM4181+ grown in landomycin production SG medium. Genomic DNA isolated from S136 strain served as a positive control for the PCR-reactions (G). Total RNA samples from ΔlanI7, ΔlanI7 pGM4181+, S136 and S136 pGM4181+ were negative controls of the PCR reactions; (L) was 1 kbp DNA Ladder (Thermo Fisher Scientific, USA). (b) Quantification of Lcm production by S. cyanogenus S136, S136 pGM4181+, ΔlanI7 and ΔlanI7 pGM4181+ grown in YMPG for 120 h. Areas of Lcm mass peaks (708.35 Da (M + H)+) were integrated and represented as arbitrary units (au); please note logarithmic scale of the y-axis. Data represent mean values of three independent experiments ± 2SD.
Mechanism of AdpA-mediated regulation of lucensomycin production
The above findings show that AdpA is required for lcm gene expression and Lcm production in S. cyanogenus. To identify possible targets of AdpA, we analyzed the lcm cluster in silico for putative AdpA operator sites. Two adjacent high-scoring binding sites (TGGCGGAAAC, TGGCCGATAG; p < 10−4) were found within the lcmRIII promoter region. This, along with data on transcriptional activity of lcm genes (see above), allows to suggest that AdpA is likely to activate transcription of lcm genes by directly turning on lcmRIII. As in the case of pimaricin biosynthesis, LcmRIII should activate transcription of either lcmRI or lcmRII, or both. Since LcmRI and LcmRII are PimM/NysRIV-like regulators, at present it is difficult to deduce which one is responsible for the activation of lcm genes.
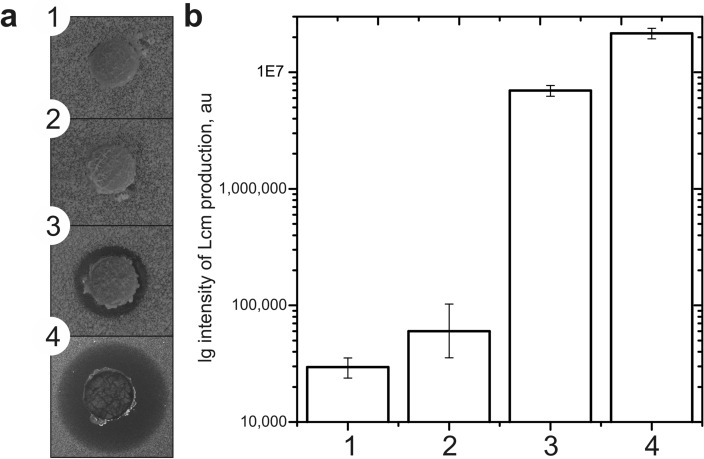
To clarify the role of regulatory lcm genes, we overexpressed them individually under the control of ermEp in S. cyanogenus ΔlanI7 and determined Lcm production levels in the resulting strains. Lcm was observed in S. cyanogenus ΔlanI7 lcmRIII+ and lcmRII+, but not in S. cyanogenus ΔlanI7 lcmRI+ (Fig. 5a, b). We therefore assume that AdpA has no targets in the lcm cluster other than lcmRII and lcmRIII. This assumption remains speculative and requires further investigations on AdpA binding to the lcmRIII promoter region. Gene lcmRI likely encodes a nonfunctional lcmRII paralog.
Figure 5.
D. hansenii growth inhibition assay (a) revealed the activation of Lcm production by S. cyanogenus ΔlanI7 upon introduction of extra copy of lcm cluster-situated transcriptional regulators. Agar plugs were cut from the lawns of S. cyanogenus ΔlanI7 (1), ΔlanI7 lcmRI+ (2), ΔlanI7 lcmRII+ (3) and ΔlanI7 lcmRIII+ (4) grown on YMPG agar for 120 h. Lcm production by the same strains in liquid YMPG was quantified via LC–MS (b) in the three recombinant strains as compared to the initial one. Data represent mean values of three independent experiments, error bars are mean values ± 2SD.
If low Lcm titers observed in the adpA-carrying S136 strain are caused by the concurrent production of landomycins, then a pathway-specific activation of the Lcm BGC should provide a competitive advantage for Lcm production, irrespective of the cultivation medium. Indeed, upon introduction of a lcmRIII expression construct into S136 we were able to detect significant antifungal activity of the latter when grown in media that support either landomycin (SG) or Lcm (YMPG) production, as well as on tryptic soy agar unfavorable for either biosynthetic route (Fig. S8a). At the same time, introduction of lcmRIII did not impact landomycin production by S136 (Fig. S8b).
Variable termini of AdpA and a rare TTA codon are involved in high-level activation of Lcm biosynthesis
Results presented here and previously35 show that the isolated DNA binding domain of AdpA is capable of turning on gene expression. Yet, the contribution of different structural parts of AdpA to the efficiency of its function as an “activation tool” was not quantified. We have decided to build and test the allele of adpA lacking variable termini. Amino acid sequences of XNR_4181 (AdpA ortholog from S. albus J1074; 406 amino acids) and its 45 orthologs from the other Streptomyces were aligned, allowing for the identification of the most conserved region spanning from 15 to 350th aa residue of XNR_4181 (Fig. S9). We have cloned the sequence of XNR_4181 coding the 15-350-aa stretch (XNR_4181i; Fig. S10) into pmoeE5script generating pGM4181i. Next, we cloned synthetic version of XNR_4181 with TTA → CTC substitution58 into pmoeE5script generating pGM4181tta-. The recombinant S. cyanogenus ∆lanI7 strains carrying engineered adpA alleles produced Lcm on solid media as judged by the bioassays. Quantitative analysis of liquid cultures showed that removal of variable termini (pGM4181i) led to over tenfold decrease in Lcm production in comparison to ∆lanI7 pGM4181+. The Lcm production level of ∆lanI7 pGM4181tta-+ increased fivefold as compared to ∆lanI7 pGM4181i+, yet it still was two times lower than in ∆lanI7 pGM4181+ (Fig. S11).
Discussion
Over the last two decades of investigations it has become clear that the vast majority of secondary metabolite BGCs within actinobacterial genomes is silent59,60. As summarized in the introductory part, the obstructed transcription appears to be the key reason for the BGCs to be silent, and researchers proposed numerous approaches to overcome this obstacle. There is a recent report on the activation of silent BGCs in actinomycetes via knockouts of genes for active secondary metabolic pathways, implying that precocious production of one metabolite diminishes the chances for the other to be produced61. Our study, summarized in Fig. 6, describes a more complex case of successful activation of a silent pathway requiring its transcriptional activation, elimination of the competing active pathway, and appropriate cultivation medium. A few aspects of our findings merit further comments.
Figure 6.
Proposal for the AdpA-mediated regulation of Lcm and the landomycin A biosynthetic pathways in S. cyanogenus. Both biosynthetic pathways compete for PKS precursor supply. AdpA, presumably through direct binding to the lcmRIII promoter region, activates the expression of this lcm cluster-situated transcriptional regulator. Both LcmRII and LcmRIII are positive regulators of Lcm production. The role of LcmRI in the regulation of the Lcm biosynthesis is unclear and awaits further investigations.
Our previous results36 showed that overexpression of heterologous adpA genes in S. cyanogenus S136 enhances the transcription of lan genes and improves landomycin A production. The above mentioned study and the results reported here support the idea that native AdpA of S. cyanogenus is non-functional. The landomycin A biosynthetic route benefits from functional AdpA, but is not dependent on it. In contrast, the Lcm biosynthetic pathway remains silent in the absence of functional adpA. RT-PCR analysis shows that in the wild type S. cyanogenus S136 (as well as in S. cyanogenus ∆lanI7) the transcription of lcm genes is enhanced upon introduction of functional adpA. Yet in case of S. cyanogenus S136 (and unlike in ∆lanI7) this did not cause Lcm production high enough to suppress fungal growth in the bioassays. This phenomenon might be explained by the competition between landomycin and Lcm biosynthetic pathways for the common pool of acyl-CoA units. If the landomycin biosynthetic route is silenced (as it is the case for mutant ∆lanI7), the precursor pool becomes available for the Lcm pathway. Such scenario, however, does not explain the absence of Lcm production in S. cyanogenus S136 pGM4181+, where lcm genes are expressed at the same level as in ∆lanI7 pGM4181+ and Lcm is actively produced. The reasons for and mechanisms of such “preferences” for the landomycin biosynthetic route in S. cyanogenus at the expense of the other secondary metabolic pathways remain obscure and require further investigations. These findings demonstrate that transcription of genes for secondary metabolism might be inconsequential to the production of secondary metabolites themselves. Altogether our results caution against overt reliance on transcriptome and translatome data as a proxy to phenotypic expression of actinobacterial BGCs13,62,63.
AdpA-activated expression of lcm BGC is mediated with the activation of lcmRIII, coding for a lcm cluster-situated transcriptional regulator. Transcription of this gene appears to be strictly dependent on adpA and putative AdpA binding sites were found within its promoter region. We note that AdpA was also shown to be involved in regulation of two other polyene antibiotics, pimaricin and candicidin64,65. Nevertheless, the location of AdpA binding sites relative to coding sequences cannot serve as a predictor of its involvement into or a sign of effects on secondary metabolic pathways66,67. Thus, more experiments are required to unambiguously prove that lcmRIII is a major and direct target of AdpA in context of Lcm biosynthesis. These experiments are underway in our laboratories.
Besides the importance of a DNA-binding domain for AdpA function, little is known about other structural features of AdpA that influence its regulatory capabilities. Here we show that removal of N- and C-terminal variable regions of AdpA affected its ability to upregulate Lcm production. One possibility is that AdpA with shortened termini is not able to form dimers. This inability would not block regulatory properties of AdpA (as the expression of separate DNA-binding domain of AdpA showed), but may lower the affinity of its binding to operator sequences. Also, the shortened AdpA might not be able to efficiently recruit RNA polymerase to form the open complex at the promoter regions. The adpA allele featuring synonymous TTA → CTC substitution was also less efficient in the activation of Lcm biosynthesis. The TTA codon is thought to be a part of a regulatory mechanism that limits the level of AdpA protein at the translational level68. For a number of TTA-containing regulatory genes of Streptomyces the substitution of this codon for a more abundant synonym caused earlier and more abundant production of respective regulatory protein69. Nevertheless, the role of the TTA codon in adpA remains elusive. AdpA is not produced in mutants devoid of tRNA for the UUA codon70–72, yet strains expressing TTA+ and TTG+ adpA alleles did not differ in temporal patterns of AdpA accumulation34. The rather detrimental effect of TTA → CTC substitution on Lcm production suggests that TTA could be more than a switch to control the onset/level of AdpA production. For example, rare codons within interdomain linkers of bacterial mRNAs could slow down protein synthesis of the ribosome and thus optimize co-translational protein folding73.
Activation of Lcm BGC in S. cyanogenus led to the first description of the entire biosynthetic machinery behind the assembly of this natural product. It is similar to other antifungal polyene (e.g. pimaricin, nystatin, rimocidin etc.) BGCs in many respects, making it straightforward to ascribe the function to most lcm genes. The organization of initiation module LcmA protein is a distinguishing feature of Lcm PKS. We tentatively suggest that this module is responsible for the generation of the side propyl chain of the lucensomycin aglycone. Further investigations are in progress to verify the proposed mechanism and to understand as to whether LcmA can be used to modify the other polyene antibiotics.
The identity of the other novel mass peaks observed in adpA-expressing ∆lanI7 strain remains unknown. This emphasizes one another formidable challenge in the area of natural product research: signals of novel chemical compounds detected in biological samples are often not pursued if there are no ways for their robust production in reasonable quantities for downstream analysis. Our report stands in a long line of the other “success stories” where a single product was activated in a particular strain (in response to a certain procedure), leaving out of focus the factors that limit the expression of the rest BGCs in that strain. Nevertheless, our work provides a rationale for a new research venue to solve this issue. Particularly, much like the elimination of expression of the landomycin BGC helped us to produce and characterize Lcm, the engineered silencing of Lcm BGC may lead to increased production of the other as-yet-unknown metabolites. We believe that a more systematic investigation of the effects of active and activated BGCs on the silent metabolome of single Streptomyces strain is worth the effort. This will yield new insights into complex interactions between the pathways at genetic and metabolic levels, eventually leading to a fuller exploitation of chemical diversity hidden in bacterial genomes.
Methods
Bacterial strains, plasmids and cultivation conditions
Bacterial strains and plasmids, used in this work, are described in ESM Table S1. For intergeneric conjugations, S. cyanogenus strains were grown on ISP3 or SFM agar74 at 30 °C. Characterization of antimicrobial activities of recombinant S. cyanogenus ΔlanI7 derivatives was done on different solid media described in ESM. The same amounts of S. cyanogenus mycelia ((0.8 ÷ 1) × 107 c.f.u.) were plated to produce lawns for the bioassays. For submerged production of Lcm, S. cyanogenus strains were grown in YMPG medium (please see ESM). Where required, the strains were maintained in presence of apramycin sulphate (50 μg/mL).
Bioassays
Agar plug antibiotic diffusion assay was used to determine antimicrobial activities of S. cyanogenus strains grown on agar media listed in ESM. In all cases plates contained 30 mL of solid medium. To assay antifungal activity, agar plugs (Ø 5 mm) were cut from S. cyanogenus lawns and placed on the surface of TSB agar plates with freshly seeded 106 cells of D. hansenii. The antibacterial activity was similarly tested; here plugs were stacked onto modified minimal medium (g/L: KH2PO4—3, K2HPO4—7, sodium citrate × 4H2O—0.5, MgSO4 × 7H2O—0.1, (NH4)2SO4—1, glucose—2, bacto peptone—0.3, agar—16;75) with B. cereus ATCC 19637 spores (107 per plate); or LB agar with E. coli DH5α cells (107 per plate). Disc diffusion assay was also employed. Here 5 mm Whatman paper discs were soaked into methanol extracts containing lucensomycin and placed on the surface of TSB agar plates with 106 cells of D. hansenii. Unless otherwise stated, in all cases the extracts were obtained from the same amount of biomass (400 mg, wet weight). Bioassay plates were incubated at 37 °C (30 °C in the case of D. hansenii), halos of growth inhibition around the plugs or discs were analyzed after 20 h of incubation.
Submerged production, purification and LC–MS of Lcm
250-mL flasks containing 50 mL of TSB were inoculated with agar slices cut from 168 h old S. cyanogenus lawns grown on ISP3 agar. The flasks were incubated for 72 h on the rotary shaker at 30 °C, 200 rpm. 1.5 mL of the resulting culture was transferred into 250-mL flasks containing 50 mL of YMPG medium. Main cultures were grown for up to 120 h at 30 °C, 200 rpm. For analytical purposes, 1 g of wet biomass was washed three times with deionized water and extracted with 2 mL of methanol. To determine optimal fermentation time for Lcm production, biomass was collected from the main culture at five time points. Extracts were concentrated to the volume of 100 μL and 40 μL of were subjected to paper disc diffusion assay (see above).
Scaled-up purification of Lcm was done as follows. Four 2-L flasks with steel springs containing 500 mL of YMPG were inoculated with 8 mL of preculture (S. cyanogenus ∆lanI7 pGM418+; prepared as described above) and grown for 72 h on the rotary shaker at 30 °C, 200 rpm. Biomass and spent medium were separated with centrifugation. Cells from 2 L of the cultures were washed with water and resuspended in 160 mL of methanol:acetone (1:1). The resulting mixture was sonicated for 15 min, spun down; supernatant was filtered through the 3 mm paper. The spent medium was mixed with ethyl acetate (1:1) by inversion a few times, then the mixture was left to allow for the phases to separate. The organic extracts from cells and supernatant were combined and evaporated in vacuo. Dry residue was dissolved in methanol and separated on preparative HPLC column (conditions were the same as for analytical LC–MS, see below). Fractions having an absorption maximum at λ = 310 nm (characteristic for polyene compounds) eluted at 7.4- ÷ 8.4 min. The presence of the 708.35 Da (Lcm) mass peak in this fraction was verified with LC–MS. The collected fraction was evaporated to dryness, giving approximately 100 mg of pale pink solid. Upon addition of 4 mL methanol a white precipitate was formed, which was shown via LC–MS to consist of pure 708.35 Da compound. As a result, we were able to purify 40 mg of Lcm from 2-L flask fermentation. Whenever possible, all plastic- and glassware was wrapped in aluminum foil to eliminate light-induced degradation of Lcm.
Methanol-dissolved Lcm samples were analyzed with HPLC–MS on a HiRes Extractive Oribtrap MassSpectrometer (Thermo Fisher Scientific). The solvent system was water + 0.1% HFo (solvent A); acetonitrile + 0.1% HFo (solvent B); 95% A to 0% A in 8 min, then 1.5 at 0% A, then back to 95% A in 1.5 min. Data analysis was carried out using Xcalibur 5.1 software (Thermo Fisher Scientific).
Plasmid construction
Fragment of the XNR_4181 coding for DNA-binding domain (700–1245 bp) was amplified using xnr4181araC_up and xnr4181_EcoRIrp primers and cloned into SpeI/EcoRI sites of pmoeE5script76 yielding pGM4181d. Gene fragments coding for DNA-binding domains of adpAsco, adpAscla and adpAgh were cloned similarly using primers described in Table 2, generating pGMSCOd, pGMSCLAd, pGMSGHd. To construct pGM4181i, fragment of XNR_4181 coding sequences from 46 to 1019 bp was amplified using iAdpA_F/R primer pair from pGM4181 and cloned into SpeI/EcoRI sites of pmoeE5script. pGM4181tta- was constructed in a similar fashion, using xnr4181_XbaIup/xnr4181_EcoRIrp primer pair to amplify XNR_4181tta- coding sequence from pUCXNR. Pathway-specific regulatory genes lcmRI, lcmRII and lcmRIII were amplified from the chromosome DNA of S. cyanogenus S136 using primers listed in Table 2 and cloned into XbaI/EcoRI sites of pTES77, yielding pTES22, pTES23 and pTES25 respectively.
Table 2.
Primers used in this work.
| Primer | Nucleotide sequence (5′–3′), recognition sites for restriction endonucleases are underlined in primer sequence | Purpose |
|---|---|---|
| iAdpA_F | AAATCTAGACGAGCAACGGAGGTACGGACATGAAACTCTCCGGGCGGCGCC | XNR_4181i cloning |
| iAdpA_R | AAAGAATTCTCACGGACGGCGGGCCCGGTAGG | -//- |
| xnr4181_XbaIup | AAATCTAGAGGGGGGCTTAGTCACATG | XNR_4181tta- cloning |
| xnr4181_EcoRIrp | AAAGAATTCGGAGCTGTCCTCTCTCAGAC | -//- |
| Xnr4181araC_up | AAATCTAGACGAGCAACGGAGGTACGGACATGCCGGAGGAGATCGGCGCCGAC | adpAscydbd/ adpAsghdbd/ adpAscodbd cloning |
| SCLAd_F | AAATCTAGACGAGCAACGGAGGTACGGACATGCCAGAGGAAATCGGGTCGGAC | adpAscladbd cloning |
| SCLAVADPA_R | AAAGAATTCCATGCGACTACCTTATGG | adpAscladbd cloning |
| SCOADPA_R | AAAGAATTCGCCGTCTGCTCACCTCACGG | adpAscodbd cloning |
| SGHADPA_R | AAAGATATCGCCTCCGGCCCCGTCCGGTGT | adpAsghdbd cloning |
| gbc22_F | TTTTCTAGAGGAGGGTCGGAATGGCGGCGCTGG | lcmRI cloning |
| gbc22_R | TTTGAATTCGGGGTTTCAGCCGGACTCG | -//- |
| gbc23_F | TTTTCTAGAGGAGGCACACCGTGATTGACGAAAC | lcmRII cloning |
| gbc23_R | TTTGAATTCTCCGCTCCCTGTCGTCCTC | -//- |
| gbc25_F | TTTTCTAGAGGAGGACCGTCATGCCCGTACCCC | lcmRIII cloning |
| gbc25_R | TTTGAATTCGACCGCGCGAGCGGTGGCTC | -//- |
| amR_F | CGGGGTCTGACGCTCAGTGGA | aac(3)IV amplification |
| amR_R | AGCGTCTGCTCCGCCATTCG | -//- |
| rt_hrdB_fp | GCTGGCCAAGGAGCTCGAC | 278 bp fragment of hrdB (RT-PCR) |
| rt_hrdB_rp | CGTCGAGGGTCTTCGGCTG | -//- |
| L2_F | ACGCCGGTCCCGATCACGTA | 355 bp fragment of lcm2 (RT-PCR) |
| L2_R | TCGGGAAGGTCGCGGAGACT | -//- |
| L6_F | CGAGTCGCGCACCAAAACGC | 312 bp fragment of lcm6 (RT-PCR) |
| L6_R | TAGCGCAGCAGCTCGTCCAC | -//- |
| L8_F | GGTCGGCCGCGATCTGAGGT | 446 bp fragment of lcm8 (RT-PCR) |
| L8_R | CCCTGCGGGTCAGCACGAAC | -//- |
| L11_F | CTGCGCTGCCGGATCGTCTT | 380 bp fragment of lcm11 (RT-PCR) |
| L11_R | TTCGAGGAGGTGCTGCCCCA | -//- |
| LRIII_F | CGTCACGGATCGCGGGCAC | 430 bp fragment of lcmRIII (RT-PCR) |
| LRIII_R | CGTGCTTTCCAGACGTGCGG | -//- |
| lPKS_F | TGTTGCCCGAGAGCCTCACG | 420 bp fragment of lcmC (RT-PCR) |
| lPKS_R | CTACGGCCGCAACCGCCCC | -//- |
Recognition sites of the restriction endonucleases used for the cloning are underlined.
Generation of recombinant Streptomyces strains
All constructs were transferred into S. cyanogenus ΔlanI7 by intergeneric conjugation with E. coli ET12567 pUZ8002+, as described elsewhere74. S. cyanogenus and transconjugants carrying pmoeE5script- and pTES-based plasmids were selected on plates overlaid with 50 μg/mL of apramycin sulphate and 30 μg/mL of nalidixic acid. Apramycin-resistant transconjugants were verified with PCR and primers complementary to marker aac(3)IV gene (Table 2).
Total RNA isolation and semi-quantitative RT-PCR analysis
Samples of S. cyanogenus mycelium were collected from 5 mL of culture broth by centrifugation (6000 rpm, 5 min, 4 °C). RNA isolation and cDNA synthesis was performed as described previously36. PCR was performed using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific) and primer pairs specific to each individual gene (Table 2). As a positive control for gene expression, the primer pair specific to S. cyanogenus house-keeping gene hrdB was used. PCR products were analyzed by electrophoresis in 2% TAE agarose gel.
DNA sequencing and in silico analysis
Initially, S. cyanogenus genome was sequenced using Illumina approach as described in36. However, the initial sequence of lcm cluster contained large gaps in the one of the lcm PKS genes, known to be challenging target to sequence due to their highly repetitive nature. Therefore, in this work S. cyanogenus S136 genome was re-sequenced using a combination of HiSeq Illumina and GridION ONT technologies, essentially as described in78. Final S136 genome (8773 kbp) was manually curated and annotated using Prokka annotation pipeline79. Full sequence of S136 genome will be published separately.
Annotation of putative BGCs on the chromosome of S. cyanogenus was achieved with antiSMASH14. Comparison of the lcm cluster with already described polyene antibiotic BGCs was made with the assistance of DoBISCUIT database80. PKS modules were predicted using PKS/NRPS Analysis website81. Search for conserved motifs within the lcm cluster was carried out with the help of the MEME suite82.
Supplementary Information
Acknowledgements
This work was supported by grants Bg-80F, M/26-2018 (to B.O.) and BG-09F (to V.F.), all from Ministry of Education and Science of Ukraine. B.O. was supported by DAAD fellowship. R.D.S. and B.O. thank the Bundesministerium für Bildung und Forschung (BMBF) for funding the project 021DK17013 AWAKEDRUGS. B.O. thanks staff of the Collection of microorganisms-producers of antibiotics at Lviv University for the provision of fungal strains.
Author contributions
O.Y., J.K., V.F., R.D.S. and B.O. conceived the experiments, O.Y., I.O., I.V., E.M. and M.D. conducted the experiments, O.Y., B.O. and R.D.S. wrote the manuscript, all authors reviewed and approved the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Roderich D. Süssmuth, Email: suessmuth@chem.tu-berlin.de
Bohdan Ostash, Email: bohdan.ostash@lnu.edu.ua.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82934-6.
References
- 1.Bérdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012;65:385–395. doi: 10.1038/ja.2012.54. [DOI] [PubMed] [Google Scholar]
- 2.Genilloud O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017;34:1203–1232. doi: 10.1039/c7np00026j. [DOI] [PubMed] [Google Scholar]
- 3.Diez J, et al. Myxobacteria: Natural pharmaceutical factories. Microb. Cell. Fact. 2011;11:52. doi: 10.1186/1475-2859-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letzel A-C, Pidot SJ, Hertweck C. A genomic approach to the cryptic secondary metabolome of the anaerobic world. Nat. Prod. Rep. 2013;30:392–428. doi: 10.1039/c2np20103h. [DOI] [PubMed] [Google Scholar]
- 5.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 6.Challinor VL, Bode HB. Bioactive natural products from novel microbial sources. Ann. NY Acad. Sci. 2015;1354:82–97. doi: 10.1111/nyas.12954. [DOI] [PubMed] [Google Scholar]
- 7.Waksman SA, Woodruff HB. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 1940;40:581–600. doi: 10.1128/JB.40.4.581-600.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waksman SA, Woodruff HB. Actinomyces antibioticus, a new soil organism antagonistic to pathogenic and non-pathogenic bacteria 1. J. Bacteriol. 1941;42:231–249. doi: 10.1128/JB.42.2.231-249.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 10.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 11.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spohn M, et al. Overproduction of ristomycin A by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents. Chemother. 2014;58:6185–6196. doi: 10.1128/AAC.03512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed Y, Rebets Y, Tokovenko B, Brötz E, Luzhetskyy A. Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci. Rep. 2017;7:9784. doi: 10.1038/s41598-017-10316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber T, et al. antiSMASH 30-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic. Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alanjary M, et al. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids. Res. 2017;45:W42–W48. doi: 10.1093/nar/gkx360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosaka T, et al. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009;27:462–464. doi: 10.1038/nbt.1538. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka Y, et al. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J. Bacteriol. 2013;195:2959–2970. doi: 10.1128/JB.00147-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riedlinger J, et al. Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium Streptomyces strain AcH 505. Appl. Environ. Microbiol. 2006;72:3550–3557. doi: 10.1128/AEM.72.5.3550-3557.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka Y, Hosaka T, Ochi K. Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2) J. Antibiot. Tokyo. 2010;63:477–481. doi: 10.1038/ja.2010.53. [DOI] [PubMed] [Google Scholar]
- 20.Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011;77:400–406. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onaka H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J. Antibiot. Tokyo. 2017;70:865–870. doi: 10.1038/ja.2017.51. [DOI] [PubMed] [Google Scholar]
- 22.Okada BK, Seyedsayamdost MR. Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2016;41:19–33. doi: 10.1093/femsre/fuw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olano C, et al. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014;7:242–256. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang MM, et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017;13:607–609. doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano E, et al. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 2005;56:465–479. doi: 10.1111/j.1365-2958.2005.04543.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen R, et al. Genome mining and activation of a silent PKS/NRPS gene cluster direct the production of totopotensamides. Org. Lett. 2017;19:5697–5700. doi: 10.1021/acs.orglett.7b02878. [DOI] [PubMed] [Google Scholar]
- 27.Gao C, Hindra A, Mulder D, Yin C, Elliot MA. Crp is a global regulator of antibiotic production in Streptomyces. mBio. 2012;3:e00407-12. doi: 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H, et al. Activation of a plasmid-situated type III PKS gene cluster by deletion of a wbl gene in deepsea-derived Streptomyces somaliensis SCSIO ZH66. Microb. Cell. Factor. 2016;15:116. doi: 10.1186/s12934-016-0515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, et al. Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017;292:19708–19720. doi: 10.1074/jbc.M117.809145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becerril A, et al. Uncovering production of specialized metabolites by Streptomyces argillaceus: activation of cryptic biosynthesis gene clusters using nutritional and genetic approaches. PLoS ONE. 2018;13:e0198145. doi: 10.1371/journal.pone.0198145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigali S, et al. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gessner A, et al. Changing biosynthetic profiles by expressing bldA in Streptomyces strains. Chembiochem. 2015;16:2244–2252. doi: 10.1002/cbic.201500297. [DOI] [PubMed] [Google Scholar]
- 33.Yamazaki H, Tomono A, Ohnishi Y, Horinouchi S. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 2004;53:555–572. doi: 10.1111/j.1365-2958.2004.04153.x. [DOI] [PubMed] [Google Scholar]
- 34.Wolanski M, et al. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 2011;193:6358–6365. doi: 10.1128/JB.05734-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabyk M, Yushchuk O, Rokytskyy I, Anisimova M, Ostash B. Genomic insights into evolution of AdpA family master regulators of morphological differentiation and secondary metabolism in Streptomyces. J. Mol. Evol. 2018;86:204–215. doi: 10.1007/s00239-018-9834-z. [DOI] [PubMed] [Google Scholar]
- 36.Yushchuk O, et al. Heterologous AdpA transcription factors enhance landomycin production in Streptomyces cyanogenus S136 under a broad range of growth conditions. Appl. Microbiol. Biotechnol. 2018;102:8419–8428. doi: 10.1007/s00253-018-9249-1. [DOI] [PubMed] [Google Scholar]
- 37.Rebets Y, et al. Function of lanI in regulation of landomycin A biosynthesis in Streptomyces cyanogenus S136 and cross-complementation studies with Streptomyces antibiotic regulatory proteins encoding genes. Arch. Microbiol. 2008;189:111–120. doi: 10.1007/s00203-007-0299-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhu D, He X, Zhou X, Deng Z. Expression of the melC operon in several Streptomyces strains is positively regulated by AdpA, an AraC family transcriptional regulator involved in morphological development in Streptomyces coelicolor. J. Bacteriol. 2005;187:3180–3187. doi: 10.1128/JB.187.9.3180-3187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pralea IE, et al. From extraction to advanced analytical methods: the challenges of melanin analysis. Int. J. Mol. Sci. 2019;20:3943. doi: 10.3390/ijms20163943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nosanchuk JD, Stark RE, Casadevall A. Fungal melanin: what do we know about structure? Front. Microbiol. 2015;6:1463. doi: 10.3389/fmicb.2015.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaudiano G, Bravo P, Quilico A, Golding BT, Rickards RW. The structure of lucensomycin. Part II. Tetrahedron. Lett. 1966;30:3567–3571. doi: 10.1016/S0040-4039(01)82829-X. [DOI] [PubMed] [Google Scholar]
- 42.Kim JD, Kang JE, Kim BS. Postharvest disease control efficacy of the polyene macrolide lucensomycin produced by Streptomyces plumbeus strain CA5 against gray mold on grapes. Postharvest. Biol. Technol. 2020;162:111115. doi: 10.1016/j.postharvbio.2019.111115. [DOI] [Google Scholar]
- 43.Lin N, et al. Structure elucidation of polyene macrolide antibiotic lucensomycin. Chin. J. Magn. Reson. 2008;25:514–522. [Google Scholar]
- 44.Aparicio JF, Colina AJ, Ceballos E, Martín JF. The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. A new polyketide synthase organization encoded by two subclusters separated by functionalization genes. J. Biol. Chem. 1999;274:10133–10139. doi: 10.1074/jbc.274.15.10133. [DOI] [PubMed] [Google Scholar]
- 45.Aparicio JF, Caffrey P, Gil JA, Zotchev SB. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 2003;61:179–188. doi: 10.1007/s00253-002-1183-5. [DOI] [PubMed] [Google Scholar]
- 46.Martín JF, Aparicio JF. Enzymology of the polyenes pimaricin and candicidin biosynthesis. Methods Enzymol. 2009;459:215–242. doi: 10.1016/S0076-6879(09)04610-2. [DOI] [PubMed] [Google Scholar]
- 47.Dejong CA, et al. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol. 2016;12:1007–1014. doi: 10.1038/nchembio.2188. [DOI] [PubMed] [Google Scholar]
- 48.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Y, et al. Selective removal of aberrant extender units by a type II thioesterase for efficient FR-008/candicidin biosynthesis in Streptomyces sp. strain FR-008. Appl. Environ. Microbiol. 2008;74:7235–42. doi: 10.1128/AEM.01012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendes MV, Antón N, Martín JF, Aparicio JF. Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin. Biochem. J. 2005;386:57–62. doi: 10.1042/BJ20040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antón N, Mendes MV, Martín JF, Aparicio JF. Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J. Bacteriol. 2004;186:2567–2575. doi: 10.1128/JB.186.9.2567-2575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antón N, et al. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology. 2007;53:3174–3183. doi: 10.1099/mic.0.2007/009126-0. [DOI] [PubMed] [Google Scholar]
- 53.Sekurova ON, et al. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 2004;186:1345–1354. doi: 10.1128/jb.186.5.1345-1354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendez C, Salas JA. The role of ABC transporters in antibiotic producing organisms: Drug secretion and resistance mechanisms. Res. Microbiol. 2001;152:341–350. doi: 10.1016/S0923-2508(01)01205-0. [DOI] [PubMed] [Google Scholar]
- 55.de Kruijff B, Demel RA. Polyene antibiotic-sterol interactions in membranes of Acholeplasma laidlawii cells and lecithin liposomes. 3 .Molecular structure of the polyene antibiotic-cholesterol complexes. Biochim. Biophys. Acta. 1974;339:57–70. doi: 10.1016/0005-2736(74)90332-0. [DOI] [PubMed] [Google Scholar]
- 56.Hammond SM. Biological activity of polyene antibiotics. Prog. Med. Chem. 1977;14:105–179. doi: 10.1016/s0079-6468(08)70148-6. [DOI] [PubMed] [Google Scholar]
- 57.Mendes MV, et al. Cholesterol oxidases act as signaling proteins for the biosynthesis of the polyene macrolide pimaricin. Chem. Biol. 2007;14:279–290. doi: 10.1016/j.chembiol.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Koshla O, et al. Gene miaA for post-transcriptional modification of tRNAXXA is important for morphological and metabolic differentiation in Streptomyces. Mol. Microbiol. 2019;112:249–265. doi: 10.1111/mmi.14266. [DOI] [PubMed] [Google Scholar]
- 59.Ward AC, Allenby NE. Genome mining for the search and discovery of bioactive compounds: The Streptomyces paradigm. FEMS Microbiol. Lett. 2018;365:fny240. doi: 10.1093/femsle/fny240. [DOI] [PubMed] [Google Scholar]
- 60.Hoskisson PA, Seipke RF. Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. mBio. 2020;11:e02642-20. doi: 10.1128/mBio.02642-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Culp EJ, et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 2019;37:1149–1154. doi: 10.1038/s41587-019-0241-9. [DOI] [PubMed] [Google Scholar]
- 62.Jeong Y, et al. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nat. Commun. 2016;7:11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bucca G, et al. Translational control plays an important role in the adaptive heat-shock response of Streptomyces coelicolor. Nucleic. Acids Res. 2018;46:5692–5703. doi: 10.1093/nar/gky335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du YL, et al. The pleitropic regulator AdpAch is required for natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis. Microbiology. 2011;157:1300–1311. doi: 10.1099/mic.0.046607-0. [DOI] [PubMed] [Google Scholar]
- 65.Liu X, et al. Dynamic changes of metabolomics and expression of candicidin biosynthesis gene cluster caused by the presence of a pleiotropic regulator AdpA in Streptomyces ZYJ-6. Bioprocess Biosyst. Eng. 2019;42:1353–1365. doi: 10.1007/s00449-019-02135-4. [DOI] [PubMed] [Google Scholar]
- 66.Pan Y, Liu G, Yang H, Tian Y, Tan H. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 2009;72:710–723. doi: 10.1111/j.1365-2958.2009.06681.x. [DOI] [PubMed] [Google Scholar]
- 67.Higo A, Hara H, Horinouchi S, Ohnishi Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012;19:259–273. doi: 10.1093/dnares/dss010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takano E, et al. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 2003;50:475–486. doi: 10.1046/j.1365-2958.2003.03728.x. [DOI] [PubMed] [Google Scholar]
- 69.Rebets YV, Ostash BO, Fukuhara M, Nakamura T, Fedorenko VO. Expression of the regulatory protein LndI for landomycin E production in Streptomyces globisporus 1912 is controlled by the availability of tRNA for the rare UUA codon. FEMS Microbiol. Lett. 2006;256:30–37. doi: 10.1111/j.1574-6968.2005.00087.x. [DOI] [PubMed] [Google Scholar]
- 70.Higo A, Horinouchi S, Ohnishi Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 2011;81:1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- 71.Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 2003;185:7291–7296. doi: 10.1128/jb.185.24.7291-7296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.López-García MT, Santamarta I, Liras P. Morphological differentiation and clavulanic acid formation are affected in a Streptomyces clavuligerus adpA-deleted mutant. Microbiology. 2010;156:2354–2365. doi: 10.1099/mic.0.035956-0. [DOI] [PubMed] [Google Scholar]
- 73.Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. John Innes Foundation: Norwich; 2000. [Google Scholar]
- 75.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 1961;81:741–746. doi: 10.1128/JB.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Makitrynskyy R, et al. Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol. 2013;3:130121. doi: 10.1098/rsob.130121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrmann S, et al. Site-specific recombination strategies for engineering actinomycete genomes. Appl. Environ. Microbiol. 2012;78:1804–1812. doi: 10.1128/AEM.06054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bekiesch P, et al. Viennamycins: lipopeptides produced by a Streptomyces sp. J. Nat. Prod. 2020;83:2381–2389. doi: 10.1021/acs.jnatprod.0c00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 80.Ichikawa N, et al. DoBISCUIT: A database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013;41:D408–D414. doi: 10.1093/nar/gks1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bachmann BO, Ravel J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 82.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.