Abstract
We present GSD_1.0, a high-quality domestic dog reference genome with chromosome length scaffolds and contiguity increased 55-fold over CanFam3.1. Annotation with generated and existing long and short read RNA-seq, miRNA-seq and ATAC-seq, revealed that 32.1% of lifted over CanFam3.1 gaps harboured previously hidden functional elements, including promoters, genes and miRNAs in GSD_1.0. A catalogue of canine “dark” regions was made to facilitate mapping rescue. Alignment in these regions is difficult, but we demonstrate that they harbour trait-associated variation. Key genomic regions were completed, including the Dog Leucocyte Antigen (DLA), T Cell Receptor (TCR) and 366 COSMIC cancer genes. 10x linked-read sequencing of 27 dogs (19 breeds) uncovered 22.1 million SNPs, indels and larger structural variants. Subsequent intersection with protein coding genes showed that 1.4% of these could directly influence gene products, and so provide a source of normal or aberrant phenotypic modifications.
Subject terms: Genomics, Genome assembly algorithms
Here, the authors report an improved de novo reference genome for the domestic dog based on a female German Shepherd named Mischka. The new genome increases contiguity 55-fold over the previous dog assembly and uncovers functional elements that were not previously identifiable.
Introduction
Domestic dogs have lived alongside humans for at least 10,000 years1,2, and during this time, they have adapted to a shared environment and diet, while being selectively bred for traits such as morphology3 and behaviour4. Humans and dogs also share orthologous genes, genomic architecture and disease sets, placing the dog as an important comparative species for human genetics and genomics. Taking advantage of pet dog medical records, within breed homogeneity and disease risk enrichment, it has been possible to provide insights into both rare and common spontaneous disease. The Online Mendelian Inheritance in Animals website (OMIA, June 2020, omia.org) currently catalogues 774 canine traits with linked genetic associations, 234 of which are likely causative in the canine models for human disease. The types of canine variants implicated in disease range from single-nucleotide polymorphisms (SNPs) (e.g. a missense variation in SOD1 leading to degenerative myelopathy5) through complex genomic rearrangements (e.g. a deletion in the repetitive interferon alpha gene cluster associated with hypothyroidism6), and were identified with canine SNP chips, e.g., CanineHD BeadChip (Illumina), genotyping complemented with imputation7 or genome and transcriptome sequencing of individuals, families8 or large populations3. Clearly, genome contiguity as well as gene and regulatory element annotation from a range of diverse breeds and tissues are all required to translate association to causation.
The current canine reference genome, CanFam3.1, is based on a 2005 7.4× Sanger sequencing framework9, improved in 2014 with multiple methods to better resolve euchromatic regions and annotate transcripts from gross tissues10. However, it still contains 23,876 gaps, with 19.6% of these within gene bodies, and a further 9.8% located a mere 5 kb upstream of predicted gene start sites. These gaps result from the accumulation of regions that are difficult to sequence, and are in part due to the loss of PRDM9 which leads to genomic sections with very high GC content11. The consequence of this is the loss of promoters, CpG islands and other regulatory elements from the reference; sequences which may hold the key to deciphering complex traits12,13.
To drive canine comparative genomics forward, we generated a high-quality canine reference assembly using a combination of Pacific Biosciences (PacBio) long read sequencing, 10x Genomics Chromium Linked Reads (henceforth called 10x) and HiC proximity ligation. The new reference, UU_CFam_GSD_1.0/canFam4 (henceforth called GSD_1.0), was subsequently annotated with both novel and published whole-genome sequencing (WGS), assay for transposase-accessible chromatin (ATAC) and RNA sequencing to enhance gene models and variant annotation. A liftover of gap regions from CanFam3.1 showed 23,251/23,836 elements contain uniquely anchored sequences in GSD_1.0, and annotation of the new reference resulted in 159 thousand transcripts across 29,583 genes. This novel data open the door to the identification of functional variants underlying complex traits, especially in difficult to sequence, and often biologically important regions.
Results and discussion
De novo assembly
Mischka, a 12-year-old female German Shepherd, was selected as the source for our high-quality reference genome assembly. Mischka was free of known genetic disorders, and when compared with additional German Shepherd sourced from within Sweden, was found to be genetically representative of the breed (Supplementary Fig. 1). We sequenced the genome using ~100× coverage PacBio long reads and assembled these in contigs with the standard FALCON method14. Further scaffolding using 94× of 10x and 48× of HiC linked reads resulted in 39 single-scaffold chromosomes (total 2.35 Gb) and 2159 unplaced scaffolds (total 128.5 Mb; Fig. 1a). The latter contigs predominantly contain segmental duplications (58.1%) and centromeric repeats (30.1%; Supplementary Fig. 2).
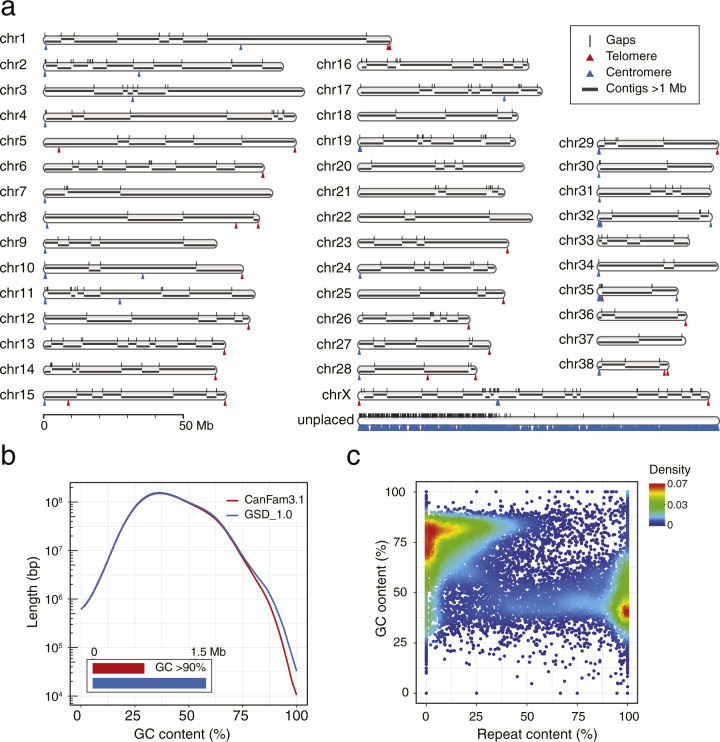
Fig. 1. Features of the novel canine assembly.
a GSD_1.0 ideogram showing chromosomes, contigs, gaps, centromere and telomere repeats. All unplaced sequences were concatenated into a single scaffold (segmental duplications, 58.1%; centromeric repeats, 30.1%). b Comparison of GC content (50 bp window) between GSD_1.0 and CanFam3.1. c Sequence characteristics of filled CanFam3.1 gaps in GSD_1.0. These are predominately high in GC or repeat content.
Reference benchmarking
Compared to CanFam3.1, the contiguity of GSD_1.0 has been improved 55-fold, reaching a contig N50 of 14.8 Mb (Supplementary Fig. 3), with only 367 gaps in the chromosome (chr) scaffolds (Table 1 and Fig. 1a). The identified sequence with extreme GC content (>90% in 50 bp windows) increased from 0.8 to 1.7 Mb (Fig. 1b), leading to a 14% increase in the average length of CpG islands (1056 vs 926 bp, P = 8.4 × 10−4, t-test). Meanwhile, we examined the CanFam3.1 gaps that could be considered closed (23,251/23,836 gap elements from CanFam3.1 have sequence in GSD_1.0; see “Methods”), and found that these regions have either high GC or high repeat content (Fig. 1c).
Table 1.
Assembly statistics of GSD_1.0 compared to CanFam3.1.
| GSD_1.0 | CanFam3.1 | |
|---|---|---|
| Number of contigs | 2783 | 27,104 |
| N50 (L50) contig | 14,840,767 bp (57) | 267,478 bp (2436) |
| Number of scaffolds | 2198 | 3268 |
| N50 (L50) scaffolds | 64,299,765 bp (15) | 63,241,923 bp (15) |
| Number of Gaps | 585 | 23,876 |
| Gap density (gaps/Mb) | 0.24 | 9.9 |
| Total bases | 2,482,000,080 bp | 2,410,976,875 bp |
| Total ungapped bases | 2,481,941,580 bp | 2,392,715,236 bp |
Repeat structure
Approximately 42.7% of the genome is repetitive sequence, with the three major categories being LINEs (504 Mb), SINEs (253 Mb) and LTRs (120 Mb) (Supplementary Fig. 4 and Supplementary Table 1). Long read technology allowed for the further resolution of centromeric repeats, and based on their positions, the orientation of chr 27 and 32 were reversed compared to CanFam3.1. These two chromosomal re-orientations were further supported by published recombination rate patterns and fluorescence in situ hybridization experiments15. In addition, the q-arms of 21 autosomes now begin with centromeric repeats, and 17 autosomes end in telomeric repeats (Fig. 1a). As expected, the sub-metacentric chr X has telomeric repeats at each end, and a clear centromeric signal at 49.4–49.9 Mb. Throughout the genome we found 10 internal centromeric and 7 internal telomeric repeats. These may indicate ancient centromere and telomere positions prior to chromosomal rearrangements and most were also present in the previous reference genome assembly.
Functional annotation
To resolve transcript complexity and account for the CanFam3.1 gap closures in GSD_1.0, we generated more than 70 M nanopore and PacBio full-length cDNA reads from 40 tissues (including 15 brain regions; Supplementary Table 2), and combined this with 24 billion public RNA-seq paired reads (Supplementary Data 1). The annotation consisted of 159 thousand transcripts in 29,583 genes; of which 20,654 had an open reading frame (ORF) of at least 100 amino acids and 19,691 genes had a significant BLAST hit against proteins in Swissprot or ENSEMBL. Further, 7725 were defined as long noncoding genes. Compared to proteins extracted from CanFam3.1, our new GSD1.0 annotation has a higher number of genes with BLAST hits and the number of genes with a full-length match has increased by 11% (Supplementary Fig. 5). Gene predictions and non-dog refSeq alignments were used to identify potentially missed genes that did not overlap with our annotation, yielding an additional 874 protein-coding genes with BLAST evidence. Using a combination of new miRNA-seq reads and public data we identified a conservative set of 719 miRNAs, similar to the set found for CanFam3.116. Among the novel miRNAs, a copy of the highly expressed Mirlet-7i was identified in a filled CanFam3.1 gap region (Supplementary Fig. 6). This miRNA has been implicated in several human diseases, including multiple sclerosis17, gastric cancer18 and breast cancer19, but has yet to be extensively studied in dogs.
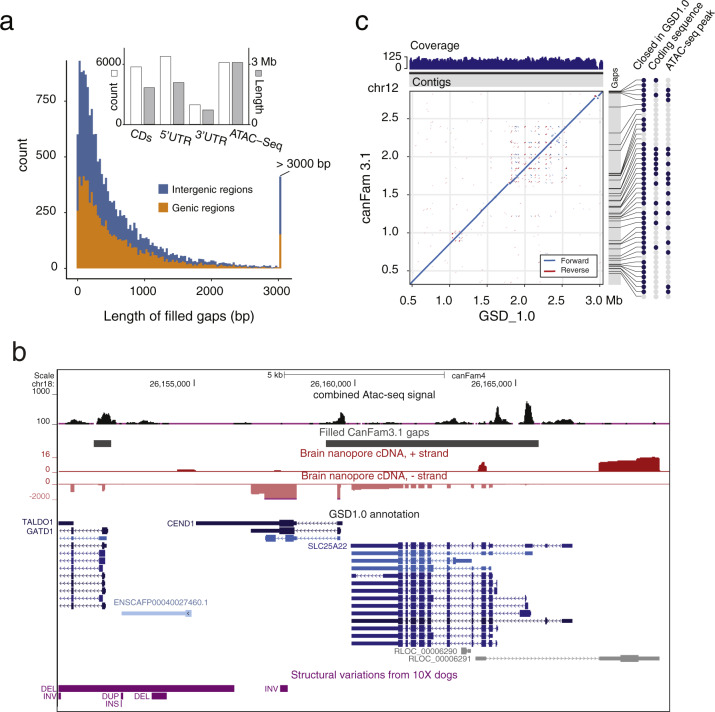
We identified 7468 closed CanFam3.1 gaps containing either an exon or promoter sequence as defined by ATAC-seq peaks, accounting for 5743 unique coding exons which were missing in CanFam3.1 (Fig. 2a). Notably, eight genes with expression across multiple tissues were completely absent or represented by pseudogenes in CanFam3.1 but were now available for interrogation (PSMA4, CDHR5, SCT, PAOX, UTF1, EFNA2, GPX4 and SLC25A22). These genes have diverse functions ranging from embryonic stem cell co-activator (UTF1) to osmoregulation (SCT). Both CDHR5 and SLC25A22 (Fig. 2b) have been investigated as biomarkers for either renal20 or colorectal21 cancers.
Fig. 2. Filled CanFam3.1 gaps and functionality.
a Size distribution and overlap with exons and promoters for the filled CanFam3.1 gaps. b Representative GSD_1.0 annotation from the UCSC track hub highlighting available data and an example of a gene hidden in CanFam3.1. c Sequence comparison of DLA on chr 12 between CanFam3.1 and GSD_1.0. The blue indicates a forward alignment and the red indicates a reverse alignment.
Implications for research
We assessed the chromosomal order and contiguity of regions essential to the study of cancer and immunological disease. Using the human COSMIC22 gene list as a baseline, we affirmed that 282 tier1 and 78 tier2 genes are now completely captured, including HOXD13 and KLF4 (Supplementary Table 3). Both have been implicated in human breast cancer; HOXD13 methylation status functions as a prognostic indicator23 and deubiquitination of KLF4 promotes metastasis24 (Supplementary Fig. 7). Two main dog leucocyte antigen (DLA) regions on chr 12 (Fig. 2c) and 35 (Supplementary Fig. 8a) are contiguous in GSD_1.025 (covering 2.58 and 0.61 Mb, respectively) and contain new coding and potential regulatory sequences absent in CanFam3.1 gaps. Meanwhile, small DLA regions on two other chromsomes26 (chr7, 1 kb, C1PG-26 and chr 18, 3Kb, DLA-79) remain contiguous in GSD_1.0. Contiguous sequence was also reported for both the T cell receptor alpha (TRA) and T cell receptor beta (TRB) loci on chr 8 and 16, respectively (Supplementary Fig. 8b, c).
Comparison to canine assemblies
Four additional canine genome assemblies have recently been deposited in NCBI (Supplementary Table 4). For each assembly, we compared BUSCO27 scores and mappability using in-house Iso-Seq cDNA alignments generated above from a beagle dog (Supplementary Table 2). With GSD_1.0 it was possible to map >5% more bases from 25,609 of Iso-Seq reads compared to CanFam3.1 (4.8% of total reads; Supplementary Fig. 9). This was a higher fraction than for the other assemblies (Supplementary Table 5 and Supplementary Fig. 10). GSD_1.0 had the second highest BUSCO score for complete genes (95.5%), but each canine assembly is of value to the community and may serve different experimental goals.
Genome variation
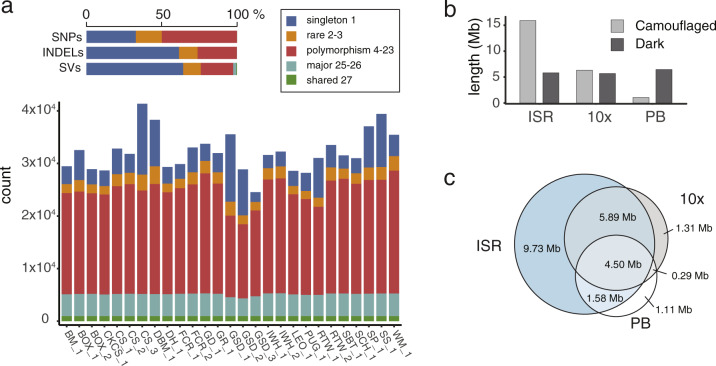
Polymorphisms detected in 27 dogs (19 breeds) were extracted from 10x sequencing data to facilitate the investigation of genome features and across-breed variant segregation (Supplementary Table 6). We identified 14,953,199 SNPs, 6,958,645 indels and 217,951 structural variants (SV, average 2.4 kb; Fig. 3a). Of these, 42.1% were private, 57.9% polymorphic across multiple individuals and 1.4% overlapped with protein-coding regions (295,112 SNPs and 16,654 SVs). Intersection with existing SV catalogues based on either SNP or aCGH arrays28–30 showed between 12.6 and 39.0% agreement, but these numbers are likely a reflection of within project breed and detection technology. 10x sequencing allowed for the detection of many novel SVs with small to medium size (≥30 kb) with accurate breakpoints.
Fig. 3. Genome variation and dark/camouflaged regions.
a SNPs, indels and structural variations shared among Mischka and the 27 10x sequenced dogs. b The total length of dark and camouflaged regions detected from Illumina short reads (ISRs), 10x and PacBio sequencing. c Intersection of merged dark and camouflaged regions from different datasets.
Genome “dark” regions unmasked
The majority of publicly available dog WGSs were generated with short read technologies. To facilitate the reanalysis of these resources with GSD_1.0 we aimed to identify the genome’s “dark” regions31; those sections either not adequately covered due to sequencing method (dark by depth, dark) or to which unique alignment is not possible (camouflaged regions, camouflaged). We defined GSD_1.0 dark and camouflaged regions for Illumina short reads (ISRs), 10x, and PacBio (PB) sequencing (see “Methods”). Dark regions comprised 5.8, 5.7 and 6.4 Mb, respectively, while camouflaged regions comprised 15.9, 6.4 and 1.0 Mb (Fig. 3b). Intersection showed that while 10x could rescue 11.3 Mb dark and camouflaged regions not seen with ISR (9.73 + 1.56 Mb), more than half of this again (5.9 Mb) could be further recovered by PacBio (Fig. 3c). We noted six tier1 & 2 COSMIC genes that contained either dark or camouflaged regions (EPHA3, RALGDS, LRP1B, CSMD3, ZMYM2, PTEN; 0.8–6.6% of coding region hidden), potentially masking drivers of disease. Due to the nature of dark and camouflaged regions, default practices will not allow for the mapping of ISR reads to, and subsequent variant extraction from, these positions. Instead, we extracted variants overlapping annotated dark and camouflaged regions from our “healthy” 10x dataset, and in doing so, identified 51,994 SNPs and indels, including 19,340 intronic and 2074 exonic variants. Many of these variants were embedded in genes that may be important for morphology or associated with disease. For example, 14 variants were found within seven intronic TYRP1 ISR dark/camouflaged regions (Supplementary Fig. 11a): a gene linked to brown colour in dogs32 and melanoma in humans33,34. Likewise, 76 variants were found in ADCY2 ISR dark/camouflaged regions (Supplementary Fig. 11b). Polymorphisms in this gene have previously been associated with psychiatric and neurological disorders (bipolar disorder35 and Alzheimer’s disease36), and response to associated drug therapies of schizophrenia37 in humans.
Chromosome mis-assembly resolved
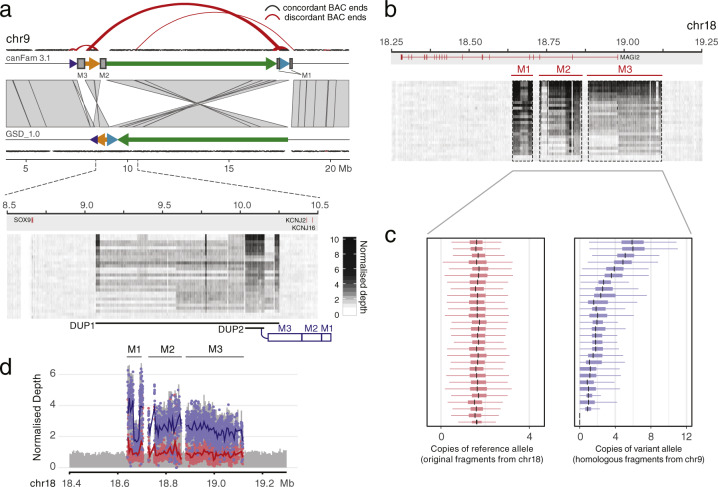
A direct comparison of CanFam3.1 and GSD_1.0 revealed a complex ~10 Mb inverted region on chr 9 that harboured SOX9 and was previously implicated in canine XX disorder of sex development (DSD)38–40. Three polymorphic regions homologous to parts of MAGI2 on chr 18 (M1, M2, M3) have been inserted upstream of SOX9 (Fig. 4a, b). In DSD, having multiple copies of a copy number variation (CNV) overlapping M239 was shown to be associated with altered SOX9 function during gonadal development. Using HiC and BAC end sequencing data, we confirmed that the inverted GSD_1.0 orientation was correct and refined the placement of regions M1, M2 and M3 (Fig. 4a). These chr 9 insertions are missing from GSD_1.0, but allelic depth analysis revealed that most 10x dogs (26/27) carry between 2 and 6 chr 9 copies (Fig. 4c, d), similar to the estimates reported for non-DSD dogs40. Recently it was shown that the DSD phenotype presents in a breed-specific manner, and is influenced by the combination of an SNP and CNVs in this region38,40. However, as this inversion contains numerous genes and regulatory elements, this rearrangement, including multiple CNV expansions, has the potential to impact additional canine traits.
Fig. 4. Correction of an inverted region in chromosome 9.
a Four fragments from the region were rearranged in GSD_1.0. The order was further confirmed using CanFam3.1 BAC clone (CH82) end sequences. Forty-nine discordant end pairs (red curves; >500 kb or not in a forward–reverse direction) were found at the edge of rearranged fragments in CanFam3.1, whereas these were properly mapped in GSD_1.0. From this region, three homologous chr 18 fragments spanning MAGI2 (M1, M2 and M3) were present on chr 9 of CanFam3.1, but missing in the GSD_1.0. We proposed that those homologous fragments should be located together with a duplication (DUP2, chr 9: 10.03–10.16 Mb) within a large duplicated region (DUP1, chr 9: 9.07–10.25 Mb). b Reads from both original and homologous M1, M2 and M3 fragments were mapped to chr 18 of GSD_1.0. c Mischka and all 10x dogs have only two original chr 18 copies M1, M2 and M3, but carry between 0 and 6 copies of the chr 9 homologous fragments. d The example plot of normalised depth illustrates how the copy number of the reference alleles and variant alleles were measured to distinguish the original (red) and homologous (blue) of M1, M2 and M3.
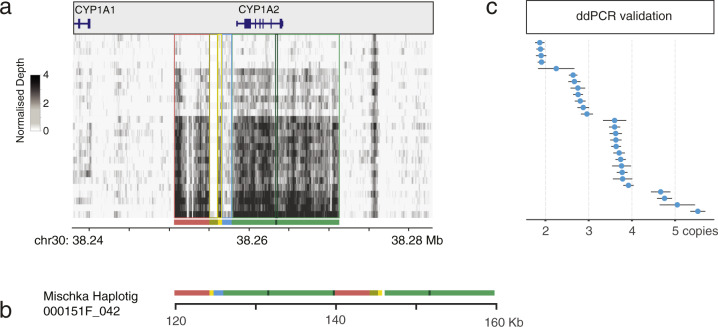
CYP1A2 locus variation
To further investigate the impact of SVs on coding genes, we examined the 16.2 kb copy number locus which encompassed CYP1A2 (Fig. 5a, b). Dogs are used as comparative models for human xenobiotic metabolism, and while a CYP1A2 premature stop codon (rs852922442 C>T) has been reported41,42, the CNV locus expansion has not. The homozygous T genotype can be found in multiple breeds43 and results in an array of pharmacokinetic effects, including reduced hepatic drug metabolism44. The T allele was observed in 4/27 10x dogs, but in heterozygous form and not segregating with CNV count (2–5 copies; Fig. 5c). Differential gene expression analyses for this and neighbouring genes outside the locus were performed using either liver or spleen tissue from additional individuals (Supplementary Data 2 and Supplementary Table 2). After accounting for CYP1A2 SNP rs852922442-T, no significant relative gene expression difference was observed, leaving the phenotypic consequence of this expansion unresolved (CNV 3 vs >3; Supplementary Table 7). It may be that the effect in this region is subtle, and so not detectable with qPCR; however, CYP1A2 is an inducible gene and so the true outcome may only be observed after a drug challenge45.
Fig. 5. Copy number expansion encompassing CYP1A2.
a A duplication identified on chr 30 consists of six segments and contains the CYP1A2 gene. b The individual pieces from the reference are plotted as they appear in the alternative haplotig sequence (000151F_042) for Mischka (CNV = 3). Sequence was extracted from the FALCON assembly. Full-length CYP1A2 sits within copies of the green fragment. c The duplication was validated in the 10x sequenced individuals using ddPCR.
Conclusion
Through the combination of sequencing technologies, PacBio (~100X) long read, 10x and HiC proximity ligation, we have generated a contiguous, chromosome length scaffolded GSD_1.0 canine reference genome. GSD_1.0 has a 55-fold increased contiguity compared with its predecessor CanFam3.1. This brings the canine reference genome quality in line with other key mammalian species, e.g. human46, mouse47, and gorilla48. For both human and mouse projects, the de novo sequence assembly of multiple individuals from different population backgrounds has revealed novel sequence not found in the single (hybrid in the case of human) species reference, and facilitated the search for population-specific variants which likely contribute to traits of interest, including within the highly polymorphic immune gene clusters46,47. While this type of de novo collection is on-going within the canine community, GSD_1.0 is the first genome of reference quality that is further annotated with novel long read RNA sequencing data, allowing for the resolution of transcript complexity through regions with high GC context, or “dark” regions31.
The resolution and placement of repeats in GSD_1.0, including non-LTR retrotransposons, will facilitate the study of gene and genome evolution and the process of neofunctionalization across mammalian lineages to an extent not possible previously. Over more recent timespans, these mobile elements can allow for genome slippage, and to the accumulation of within and across population SVs. In human clinical genomics, SVs spanning coding and/or noncoding sequence have been responsible for a range of maladies including cardiac anomalies (OMIM 192430) and intellectual delay and autism (OMIM 608 636). Accordingly, this source of variation is of keen interest in canine genetics, and should facilitate similar lines of investigation. The technology used to read across repeats was also successful in reading into regions of constitutive heterochromatin, allowing for the correction of chromosomal direction (chr 27 and 32) and revealing novel centromeric and telomeric sequences.
Perhaps the largest gain offered by the contiguity of GSD_1.0 is to the accelerating field of low pass genotyping and imputation for trait mapping7. The completion of key regions to the investigation of immunological disease and cancer, e.g. DLA and TCR, when combined with large reference populations, will facilitate the more accurate genotyping of these regions and hopefully fast track the process from association to causation. We believe that the catalogues generated here (extended gene models, dark/camouflaged regions, within and across-breed variation), based on the GSD_1.0 framework, will propel the comparison of canine and human genetic disease forward by leaps and bounds.
Methods
Reference individual
Mischka, a 12-year-old female German Shepherd, was born and raised in Sweden with known ancestral background and no medical history of genetic disease. Mischka was genotyped with the CanineHD BeadChip (Illumina) and compared to a population of 260 German Shepherds from a previous study49. Mischka was assessed to be representative of the population via expected inbreeding value (F = 0.037) and multiple dimensional scaling genetic distance measures (PLINK v1.9) and selected for the genome assembly. High molecular weight (HMW) DNA was extracted from blood with MagAttract HMW DNA Kit (Qiagen).
Genome sequencing
The assembly used multiple sequencing technologies. Long read libraries were prepared with SMRTbell Template Prep Kit 1.0 and 70 SMRT cells were sequenced on the PacBio Sequel system with v2.1 chemistry (Pacific Biosciences; 276.86 Gb data). Linked reads were sequenced from HMW DNA with Chromium libraries (10x Genomics) on an Illumina HiSeq X (2 × 150 bp; 269.75 Gb of data). Dovetail Genomics prepared three HiC libraries which were sequenced on an Illumina HiSeq X (2 × 150 bp paired-end reads; 121.47 Gb data, Supplementary Table 8).
Assembly construction
De novo assembly used PacBio subreads (>8 kb) with the standard FALCON50 v0.5.0 method. After Arrow50 (v2.3.3) polishing, the assembly yielded 3656 contigs with an N50 and mean length of 4.66 Mb and 677 kb, respectively. ARCS51 v1.05 and LINKS52 v1.8.6, with the recommended link ratio (-a) 0.9, were used to scaffold contigs with 10x reads. In all, 1170 FALCON contigs were joined in this step, increasing the scaffold N50 to 18.5 Mb.
Conflict resolution
Scaffolding correctness was evaluated by aligning scaffold sequences onto the high-density canine linkage map15. In all, 21,278 of 22,362 markers (95%) were unambiguously mapped to the assembly by BLAT53 v36. Synteny of genetic and physical location of markers was further compared with Chromonomer54 v1.0, which showed 207 scaffolds were anchored correctly, but that four had conflicting markers. These four scaffolds were split after careful sequence review confirmed that each discrepancy arose from incorrect inter-chromosomal joining.
Gap filling and assembly polishing
PBjelly from PBSuite55 v15.8.24 was used with PacBio subreads to close 648 gaps. An initial QC scan showed no putative wrong joins, and so long-distance interaction information from HiC (HiRise, Dovetail Genomics) was used to successfully extend scaffolds to chromosome level (scaffold N50: 64.3 Mb). These results were evaluated with the JUICER56 pipeline; HiC reads were mapped back to the HiRise assembly and HiC map with intra- and inter-chromosomal interactions visualised. We identified and manually adjusted contigs placed in either the wrong order or orientation (chr 6, 14, 17, 26 and X), and joined separated contigs from the same chromosome (chr 8 and 18). A second round of PBjelly gap filling closed another 110 gaps. The assembly was polished with Arrow (PacBio subreads) and Pilon57 v1.22(10x Genomics reads, BWA58 v0.7.15 mem mapping). A FreeBayes-based method was applied to further correct indel errors59. SNPs and indels were called from short reads aligned to the polished assembly (FreeBayes60 v1.1.0). The reference base was replaced with the variant allele at 149,264 positions where 10x sequencing depth was at least 30× and the variant allele ratio was >90% using FastaAlternateReferenceMaker from GATK61 v4.1.1.0. A final round of Pilon short read polishing was completed prior to the removal of 68 unplaced contigs with suspected bacterial contamination (Kraken262 v2.0.8).
The correctness of a large rearranged region on chr 9 of GSD1.0 was confirmed through comparison to end sequences from original CanFam BAC clones (CH82 library; NCBI TraceDB). BAC sequences were mapped as paired reads (BWA58 mem default setting), to GSD_1.0 and CanFam3.1. End pairs that mapped to both assemblies were compared and defined as concordant when they aligned in forward and reverse direction with a distance <500 kb.
GC content and repetitive elements
GC content (%) was assessed in 50 bp windows (NUC from BEDTools63 v2.29.2). CpG islands were detected with the “cpg_lh” script from UCSC utilities (http://hgdownload.soe.ucsc.edu/admin/exe/linux.x86_64.v369/), a modified method from Gardiner-Garden64. The unique mappability of GSD_1.0 was tested with different k-mers (50/150/250 bp in GEM-Tools65 v1.71). Repetitive elements were annotated by Repeat Masker v4.0.8 in a sensitive mode (http://www.repeatmasker.org) with a combined library (dc20171107-rb20181026). Telomere repeats, “TTAGGG”, were highlighted on both strands with fuzznuc (EMBOSS66 v6.6.0). Putative telomere sequences were defined as at least 12 consecutive repeats with less than 11 variant bases between each, and multiple sequences were merged if within 100 bp. Centromeric regions were defined based on satellite repeat67 (CarSat1/Carsat2/SAT1_CF) content in 5 kb windows. Putative centromere sequences were annotated if the repeat content was >80%.
RNA preparation and long read cDNA sequencing
Multiple RNA samples from Beagles were used for RNA sequencing (Supplementary Table 2). First, total RNA from hypothalamus (RIN > 8; Zyagen) was purchased for sequencing via PacBio Iso-Seq express protocol. Two libraries were run on two separate SMRT cells using the Sequel system, and yielded ~500,000 reads each with mean read lengths of 2452 and 451 bp. Total RNA from a further 24 tissues (including 15 brain regions; Supplementary Table 2) was extracted using a standard TRIzol protocol (Invitrogen) and used for nanopore cDNA and Illumina miRNA-sequencing. The PCR strand-switch protocol and the SQK-LSK109 kit were used for MinION sequencing (Nanopore). All tissue samples were amplified with PBC096 barcoding for 8–10 cycles with both LongAmp (female samples, 62 °C annealing; NEB) and PrimeSTAR GXL (both sexes, 64°C annealing; Takara Bio), with a 10 minutes extension time. The retina sample was sequenced using both the nanopore direct cDNA sequencing kit SQK-DCS109 and as stranded 2 × 150 bp reads on a NovaSeq 6000 S4 lane (Illumina). Reads were base called with the high accuracy model in guppy (v3.6 for direct cDNA and v3.3 for amplified samples). Qcat and pychopper (https://github.com/nanoporetech/) were used to demultiplexed reads and to identify and orient fully sequenced reads. Mapping accuracy was increased by only using reads with a quality value above 15. For PacBio, full-length circular consensus sequencing (CCS) reads with at least three passes were selected. The long read cDNA runs were mapped with Minimap268 (v2.17) with the options -x splice -G 500000 and --junc-bed with splice junctions identified from the Illumina alignments. These settings improved mapping both to genes with long introns and to short exons. MicroRNA libraries were made with the NEXTFLEX small RNA library kit v3 (PerkinElmer) and 25 million reads were generated with a NextSeq500 instrument (75 bp high-output kit v2.5 in paired-end mode; Illumina).
Gene annotation
Public Illumina stranded RNA-seq runs with paired reads of at least 100 bp were downloaded from NCBI using the SRA-Explorer (https://sra-explorer.info/). Samples were selected to cover a diverse set of dog tissues and breeds (Supplementary Data 1). Reads from the same study and tissue were combined and adaptors were trimmed with BBmap. HISAT266 and RSeQC69 were used on a small subset of reads for each sample to infer library type. Stringtie267 superreads module was used to assemble and merge transcripts from Illumina reads, with setting -f 0.05 as the threshold for isoform expression. Stringtie2 assemblies were made both for individual samples and with combined samples from the same tissue type. PacBio iso-seq alignments were combined with alignments of nanopore full-length cDNA reads for assembly with Stringtie2 with options “-L -c 3 -s 10 -f 0.05” to suppress low-coverage transcript models from internal priming and partially spliced mRNAs. Stringtie2 was further used to merge transcripts from the individual assemblies of long and short reads. Assembled transcripts were processed with TAMA tools68 for ORF detection and BLAST parsing to identify coding regions based on hits against a database of curated proteins from Uniprot_Swissprot and proteins from the latest ENSEMBL dog annotation (v100, Great Dane assembly). The id of the protein was determined from the longest BLAST hit from the top five hits with an E-value below 10−10. We found the Stringtie assembly sometimes missed low-coverage genes that were close to, but not overlapping, highly expressed genes. To make the assembly as complete as possible we therefore combined the output from multiple runs, used TAMA to assemble long reads not overlapping with Stringtie2 transcripts and included novel transcripts if they were multi-exonic and had a blast hit covering at least 50% of the target. Additional filtering was applied to remove transcripts that, (1) were long single exon transcripts (>10 kb and <10% intronic sequence) or (2) originated from genomic polyA/T regions. Gffread70 was used to re-group transcripts into genes, retaining only one transcript per unique CDS region. Finally, transcripts which had either >2 exons downstream of the stop codon, or a bad BLAST classification (<50% hit) were removed if they belonged to a group with high scoring transcripts (Supplementary Fig. 12). Long noncoding genes were defined as having at least two exons, a length of >200 bases, no ORF longer than 100 amino acids and no overlap with protein-coding exons on the same strand.
miRNA identification
Public microRNA-seq samples (Supplementary Data 1) were combined with the above brain microRNA-seq reads (Total reads, 1.3 billion). Reads were included if they were between 20 and 30 bases after adaptor trimming. Bowtie alignments of unique sequences were used for MiRDeep271 analysis and compared to known dog and human miRNAs (miRBase) in order to identify the position of both known and novel miRNAs.
ATAC-seq analysis
Reads from BARKbase72 (Supplementary Data 1) were aligned with BWA mem and peaks called with Genrich (https://github.com/jsh58/Genrich). BedGraph files were produced with BEDTools.
CanFam3.1 gap comparison
CanFam3.1 gaps were defined as any continuous ambiguous”N” bases, and for each, 1 kb flanking sequences were extracted and mapped as pairs to GSD_1.0 (BWA mem). CanFam3.1 gaps were considered closed when (1) flanking sequence pairs could be mapped properly in the same scaffold with mapping quality >20; (2) the distance between pairs was less than 100 kb; and (3) no GSD_1.0 gap was present in the sequence between pairs. This approach identified the sequence for 18,649 of 19,553 (95.4%) gaps from assembled chromosomes, and 1563 of 4323 (36.2%) gaps from unplaced scaffolds of CanFam3.1 in GSD_1.0. The flanking sequences of 3072 gaps overlapped each other in GSD_1.0, suggesting artificial gaps in CanFam3.1 that can be considered closed in GSD_1.0. For the other closed gaps, we extracted the filled sequences from GSD_1.0 and calculated GC and repeat content. BEDTools was used to intersect exons, miRNA and ATAC-seq peaks mapped above with filled CanFam3.1 gaps. Specifically, we looked for novel genes from the filled CanFam3.1 gaps. A novel gene was defined if it (1) had at least 80% of the gene body identified from the filled CanFam3.1 gaps; (2) was not a pseudogene; (3) had not been annotated in the unplaced scaffolds of CanFam3.1; and (4) did not have the duplicated/homologous fragment in another region of the genome. With these thresholds, we found eight novel genes from the filled CanFam3.1 gaps, and all located in regions with good synteny of human hg38 assembly.
Region comparison
We compared dog DLA, TRA and TRB regions between GSD_1.0 and CanFam3.1 by NUCMER73. By lifting the human major histocompatibility complex regions from the genome reference consortium, two main DLA regions were found in GSD_1.0: chr 12: 0.45–3.05 Mb (TRIM39 → SYNGAP1), chr35: 27.0–27.9 Mb (GPX6 → TRIM26 gene). Two additional DLA regions, chr7:59.69 Mb (1 kb, C1PG-26) and chr 18: 41.56 Mb (3 kb, DLA-79) were identified by the previous study26.
Assembly benchmark with Busco and Iso-Seq data
BUSCO27 v3.0.2b was run with the mammalia_odb9 dataset. Mappability was assessed with Iso-Seq data using only PacBio CCS reads supported by >10 subreads (483,702 reads). CCS reads were mapped with minimap2 v2.17, and the percentage of mapped bases per read calculated according to the “difference string” in cs tag. With these methods, GSD_1.0, CanFam3.1 and four newly released canine assemblies, Luka (Basenji), Nala74 (German Shepherd), Zoey75 (Great Dane) and Scarlet76,77 (Golden Retriever, Supplementary Table 4).
10x and standard ISR mapping
HMW DNA was extracted from the blood of 27 additional dogs (19 breeds), and Chromium library preparation and sequencing completed as per “Genome sequencing”. Sequencing depth ranged between 30 and 93× (Supplementary Table 6). Unplaced GSD_1.0 scaffolds were concatenated into a single scaffold with 500 “N” base spacers and 10x reads were mapped to each with the Long Ranger v2.2.2 WGS pipeline (10x Genomics). 10x breed-matched ISR data were downloaded for 25 individuals (Supplementary Table 9) and mapped to GSD_1.0 (BWA mem, default settings). SNPs and short indels were detected in 10x and ISR dataset using appropriate modules from GATK4. Variants were called from alignment by HaplotypeCaller, and further merged by the CombineGVCFs and GentoypesGVCFs. The SNPs and indels were filtered by SelectVariants with “QD < 2.0| | FS > 60.0| | MQ < 40.0| | MQRankSum < −12.5| | ReadPosRankSum < −8.0” and “QD < 2.0| | FS > 200.0| | ReadPosRankSum < −20.0”, respectively.
Dark and camouflaged region detection
Both depth and mapping quality were calculated for each sample in each 10x or ISR dataset. For sequencing coverage, bamCoverage (Deeptools78 v3.3.2) with a 25 bp window was used, with unmapped reads and secondary alignments excluded from the analysis. For the same windows, the proportion of reads with mapping quality >10 was also assessed. Regions dark by depth (dark) were defined as windows with coverage ≤5×, with threshold adjusted for sequencing depth. A lower cutoff was applied in low-coverage samples to select a maximum of 60 Mb (Supplementary Data 3). The individual dark regions were merged, and the dark fraction for each window was assessed for both ISR and 10x datasets: windows with Fdark > 0.9 (90% individuals, in at least 23 ISR dogs or 25 10x dogs) retained as the candidate dark regions. Camouflaged regions (camouflaged) were defined if the coverage was ≥10× and the proportion of high mapping quality reads was less than 10%. We searched for and merged the genomic windows that reached the threshold from each dog. As the camouflaged regions detected in one individual could have been assigned as dark in others, we excluded those dark dogs before we calculated the fraction of camouflaged bases for each window. Any window with Fcamouflaged > 0.9 was selected as a candidate.
Structural variation (SV) detection
We scanned the genomes of 27 10x dogs using four SV callers. The first, Long Ranger, was used to call the SVs in two size ranges. Medium SVs spanning from 50 to 30 kb were detected by examining the haplotype-specific coverage drops and discordant reads pairs. Larger-scale SVs, >30 kb, were identified as regions where paired coverage of genomic loci shared many more barcodes than expected by chance. Candidate SVs were further refined and categorised (DEL deletion, CNV copy number variant, INV inversion) by comparing the layout of reads and barcodes around the breakpoints. Three additional callers were adapted to discover other types of median size SVs (50 bp–30 kb). GridSS79 and Manta80 are assembly-based callers which have been reported to have a good performance in different studies81,82. Both detected SVs using evidence from split and paired reads, and also assembled the sequences of breakpoints to accurately estimate these positions. The type of SVs called by GridSS was determined by the orientation of reads from the breakpoints using a R script (https://github.com/PapenfussLab/StructuralVariantAnnotation). From the three callers above, only high-quality SV calls marked as “PASS” in vcfs were kept for analysis. Lastly, CNVnator83 predicted CNVs by a read-depth (RD) approach. A 150 bp bin size was used for screening, and retained SVs were required to have a p value <0.05 for a RD t-test statistic (“e-val1”) and the probability of RD frequency <0.05 in a gaussian distribution of (“e-val2”). The result was converted into VCF form using the “cnvnator2VCF.pl” script from the CNVnator package. For each 10x sample, the filtered median SVs from all four callers were merged by the SURVIVOR84, and combined with the large size SVs called from Long Ranger. Chr X SVs that were only supported by CNVnator were pruned as the algorithm lacks the right model sex chromosome. SVs were further merged across individuals into a nonredundant SVs set.
SV validation and genotyping
Four DELs and four CNVs which overlapped protein-coding genes that were polymorphic within the 10x dataset (>3/27 individuals) were selected (Supplementary Data 2). SV breakpoints were confirmed with Sanger sequencing where possible. PCR was performed with either PrimeSTAR GXL DNA Polymerase (Takara) or AmpliTaq Gold DNA Polymerase (Applied Biosystems) according to the manufacturer’s recommendations. PCR fragments were cloned using either Zero Blunt or TOPO TA Cloning Kit (Invitrogen) depending on PCR overhang. Plasmid DNA was extracted using QIAprep Spin Miniprep Kit (Qiagen), PCR products and plasmids sequenced using the Mix2Seq service (Eurofins Genomics) and analysed using CodonCode Aligner v6.0.2 (CodonCode). For CYP1A2 CNV genotyping, ddPCR absolute quantification (BioRad) was performed and quantified as before85. CYP1A2 C1117T was genotyped according to a published method86. New Primers and probes were designed using Primer3 v0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and collated in Supplementary Data 2.
Gene expression
Total RNA was extracted from liver and spleen tissues using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to the manufacturer’s specification and including on-column DNaseI treatment (Supplementary Data 4). In total, 1000 ng of total RNA was reverse transcribed using the Advantage RT-for-PCR Kit (Takara) and qPCR performed in quadruplet using SYBR Green PCR Master Mix (Thermo Fisher Scientific) and 900 nM primers in a QuantStudio 6 Real-Time system (Thermo Fisher Scientific) with standard cycling and dissociation curve analysis. Two housekeeper primer sets (RPS19 and RPS5) were assessed for stability (Normfinder87 R package) and used in combination to calculate relative gene expression88. These calculations included primer specific efficiencies and used the average Ct from all control samples for initial delta Ct normalisation. wilcox.test in R was used to assess the significance of between genotypic class gene expression changes.
Statistics and reproducibility
Statistical analysis was performed by R v3.6.0 with algorithms and packages as described.
Ethics approval and consent to participate
Approval was obtained from dog owners before collecting the biological samples at veterinary clinics. Ethical approvals for sampling were granted by Uppsala Animal Ethical Committee and Swedish Board of Agriculture (C139/9, C2/12, C12/15). Importation of canine tissues was approved by Jordbruksverket (6.7.18-14513/17).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Mischka’s owners who kindly allowed us to collect blood and tissues for scientific purposes, Susanne Gustafsson from the SLU Canine Biobank for the management of these and other canine samples used throughout the project and Anna Darlene van der Heiden for generating retina data. We would like to acknowledge Mats Pettersson, Olga Vinnere Pettersson and Ignas Bunikis for helpful suggestions. We thank Erez Lieberman (Golden Retriever assembly), Wes Warren and Gary Johnson (Basenji assembly) for allowing access to the additional assemblies used in this study. Nex-generation sequencing was made possible with assistance from the Uppsala Genome Center (PacBio) and the SNP&SEQ Technology Platform (10x Chromium). Both fall under the umbrella of National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory, Sweden and themselves are supported by RFI/VR and the Swedish Research Council and the Knut and Alice Wallenberg Foundation respectively. The computations and data handling were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX, Projects SNIC 2019/8-266, 2018/3-677, 2017/7-384, 2017/7-385 and 2020/5-190) partially funded by the Swedish Research Council through grant agreement no. 2018-05973. .KL.-T. is a Distinguished Professor at the Swedish Research Council. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA225755, The Knut and Alice Wallenberg Foundation and Agria och Svenska Kennelklubben Forskningsfond (https://www.skk.se/sv/Agria-SKK-Forskningsfond/, grant numbers: P2012-0015, N2013-0020, P2014-0018, P2015-0012).
Author contributions
K.L.-T., J.R.S.M. and M.L.A. conceived the study and designed the experiments. G.R.P. and M.L.A. collected the samples with the help of J.H., Å.O., S.S., H.R., I.L., S.M., J. Häggström and Å.H. M.L.A., Å.K. and Å.O. performed the DNA/RNA extractions. Å.K., E.S. and J.R.S.M. performed the validation of structural variation, genotyping and expression analyses. C.W., O.W., M.-L.A., J.N. and K.L.-T. contributed to the data analysis of the genome assembly. O.W. performed the gene annotation with the help of T.F.B. and S.M. J.R.S.M. and K.L.-T. oversaw and interpreted the results together with C.W., O.W., M.L.A. and E.S., C.W., OW, J.R.S.M. and K.L.-T. wrote the manuscript with input from all authors.
Funding
Open Access funding provided by Uppsala University.
Data availability
The PacBio long reads, HiC, and Illumina 10x data of Mischka are available in SRA under BioProject PRJNA587469. The Illumina 10x data of 27 dogs are available in SRA under BioProject PRJNA588624. miRNA & RNA sequencing data are available in SRA under BioProject PRJNA657719. The canFam_GSD_1.0 assembly is deposited in DDBJ/ENA/GenBank under JAAHUQ000000000, and also available in UCSC browser (http://genome-euro.ucsc.edu/cgi-bin/hgTracks?db=canFam4).
Code availability
Scripts used in the study are available at the GitHub repository (https://github.com/Chao912/Mischka/).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jennifer R. S. Meadows, Kerstin Lindblad-Toh.
Contributor Information
Chao Wang, Email: chao.wang@imbim.uu.se.
Kerstin Lindblad-Toh, Email: kersli@broadinstitute.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01698-x.
References
- 1.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 2.Freedman AH, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plassais J, et al. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 2019;10:1489. doi: 10.1038/s41467-019-09373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedrich J, et al. Genetic dissection of complex behaviour traits in German Shepherd dogs. Heredity. 2019;123:746–758. doi: 10.1038/s41437-019-0275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awano T, et al. Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA. 2009;106:2794–2799. doi: 10.1073/pnas.0812297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi M, et al. Whole-genome genotyping and resequencing reveal the association of a deletion in the complex interferon alpha gene cluster with hypothyroidism in dogs. BMC Genomics. 2020;21:307. doi: 10.1186/s12864-020-6700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedenberg SG, Meurs KM. Genotype imputation in the domestic dog. Mamm. Genome. 2016;27:485–494. doi: 10.1007/s00335-016-9636-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver JAC, Ricketts SL, Kuehn MH, Mellersh CS. Primary closed angle glaucoma in the Basset Hound: genetic investigations using genome-wide association and RNA sequencing strategies. Mol. Vis. 2019;25:93–105. [PMC free article] [PubMed] [Google Scholar]
- 9.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 10.Hoeppner, M. P. et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE9, e91172 (2014). [DOI] [PMC free article] [PubMed]
- 11.Axelsson E, et al. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin C-S, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong AK, et al. A comprehensive linkage map of the dog genome. Genetics. 2010;184:595–605. doi: 10.1534/genetics.109.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penso-Dolfin L, et al. An improved microRNA annotation of the canine genome. PLoS ONE. 2016;11:e0153453. doi: 10.1371/journal.pone.0153453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura K, et al. Circulating exosomes suppress the induction of regulatory T cells via let-7i in multiple sclerosis. Nat. Commun. 2018;9:17. doi: 10.1038/s41467-017-02406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y, et al. Down-regulation of the let-7i facilitates gastric cancer invasion and metastasis by targeting COL1A1. Protein Cell. 2019;10:143–148. doi: 10.1007/s13238-018-0550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Anda-Jáuregui G, Espinal-Enríquez J, Drago-García D, Hernández-Lemus E. Nonredundant, highly connected microRNAs control functionality in breast cancer networks. Int. J. Genomics. 2018;2018:9585383. doi: 10.1155/2018/9585383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bläsius FM, et al. Loss of cadherin related family member 5 (CDHR5) expression in clear cell renal cell carcinoma is a prognostic marker of disease progression. Oncotarget. 2017;8:75076–75086. doi: 10.18632/oncotarget.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong CC, et al. SLC25A22 promotes proliferation and survival of colorectal cancer cells with KRAS mutations and xenograft tumor progression in mice via intracellular synthesis of aspartate. Gastroenterology. 2016;151:945–960.e6. doi: 10.1053/j.gastro.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Sondka Z, et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018;18:696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong Z, et al. HOXD13 methylation status is a prognostic indicator in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:10716–10724. [PMC free article] [PubMed] [Google Scholar]
- 24.Zou H, Chen H, Zhou Z, Wan Y, Liu Z. ATXN3 promotes breast cancer metastasis by deubiquitinating KLF4. Cancer Lett. 2019;467:19–28. doi: 10.1016/j.canlet.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Yuhki N, Beck T, Stephens R, Neelam B, O’Brien SJ. Comparative genomic structure of human, dog, and cat MHC: HLA, DLA, and FLA. J. Hered. 2007;98:390–399. doi: 10.1093/jhered/esm056. [DOI] [PubMed] [Google Scholar]
- 26.Wagner JL. Molecular organization of the canine major histocompatibility complex. J. Hered. 2003;94:23–26. doi: 10.1093/jhered/esg002. [DOI] [PubMed] [Google Scholar]
- 27.Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 28.Berglund J, et al. Novel origins of copy number variation in the dog genome. Genome Biol. 2012;13:R73. doi: 10.1186/gb-2012-13-8-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molin A-M, Berglund J, Webster MT, Lindblad-Toh K. Genome-wide copy number variant discovery in dogs using the CanineHD genotyping array. BMC Genomics. 2014;15:210. doi: 10.1186/1471-2164-15-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholas TJ, et al. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res. 2009;19:491–499. doi: 10.1101/gr.084715.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebbert MTW, et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol. 2019;20:97. doi: 10.1186/s13059-019-1707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmutz SM, Berryere TG, Goldfinch AD. TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm. Genome. 2002;13:380–387. doi: 10.1007/s00335-001-2147-2. [DOI] [PubMed] [Google Scholar]
- 33.Gilot D, et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat. Cell Biol. 2017;19:1348–1357. doi: 10.1038/ncb3623. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein AM, et al. Rare germline variants in known melanoma susceptibility genes in familial melanoma. Hum. Mol. Genet. 2017;26:4886–4895. doi: 10.1093/hmg/ddx368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mühleisen TW, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat. Commun. 2014;5:3339. doi: 10.1038/ncomms4339. [DOI] [PubMed] [Google Scholar]
- 36.Silver M, et al. Identification of gene pathways implicated in Alzheimer’s disease using longitudinal imaging phenotypes with sparse regression. NeuroImage. 2012;63:1681–1694. doi: 10.1016/j.neuroimage.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jajodia A, et al. Evaluation of genetic association of neurodevelopment and neuroimmunological genes with antipsychotic treatment response in schizophrenia in Indian populations. Mol. Genet. Genom. Med. 2016;4:18–27. doi: 10.1002/mgg3.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyers-Wallen VN, et al. XX disorder of sex development is associated with an insertion on chromosome 9 and downregulation of RSPO1 in dogs (Canis lupus familiaris) PLoS ONE. 2017;12:e0186331. doi: 10.1371/journal.pone.0186331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowacka-Woszuk J, et al. Deep sequencing of a candidate region harboring the SOX9 gene for the canine XX disorder of sex development. Anim. Genet. 2017;48:330–337. doi: 10.1111/age.12538. [DOI] [PubMed] [Google Scholar]
- 40.Nowacka-Woszuk J, et al. Association between polymorphisms in the SOX9 region and canine disorder of sex development (78,XX; SRY-negative) revisited in a multibreed case-control study. PLoS ONE. 2019;14:e0218565. doi: 10.1371/journal.pone.0218565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenmizu D, Endo Y, Noguchi K, Kamimura H. Identification of the novel canine CYP1A2 1117 C>T SNP causing protein deletion. Xenobiotica Fate Foreign Compd. Biol. Syst. 2004;34:835–846. doi: 10.1080/00498250412331285436. [DOI] [PubMed] [Google Scholar]
- 42.Mise M, et al. Polymorphic expression of CYP1A2 leading to interindividual variability in metabolism of a novel benzodiazepine receptor partial inverse agonist in dogs. Drug Metab. Dispos. Biol. Fate Chem. 2004;32:240–245. doi: 10.1124/dmd.32.2.240. [DOI] [PubMed] [Google Scholar]
- 43.Court MH. Canine cytochrome P-450 pharmacogenetics. Vet. Clin. North Am. Small Anim. Pract. 2013;43:1027–1038. doi: 10.1016/j.cvsm.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mise M, Hashizume T, Komuro S. Characterization of substrate specificity of dog CYP1A2 using CYP1A2-deficient and wild-type dog liver microsomes. Drug Metab. Dispos. Biol. Fate Chem. 2008;36:1903–1908. doi: 10.1124/dmd.108.022301. [DOI] [PubMed] [Google Scholar]
- 45.Graham RA, et al. In vivo and in vitro induction of cytochrome P450 enzymes in beagle dogs. Drug Metab. Dispos. Biol. Fate Chem. 2002;30:1206–1213. doi: 10.1124/dmd.30.11.1206. [DOI] [PubMed] [Google Scholar]
- 46.Ameur, A. et al. De novo assembly of two Swedish genomes reveals missing segments from the human GRCh38 reference and improves variant calling of population-scale sequencing data. Genes9, 486 (2018). [DOI] [PMC free article] [PubMed]
- 47.Lilue J, et al. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat. Genet. 2018;50:1574–1583. doi: 10.1038/s41588-018-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon D, et al. Long-read sequence assembly of the gorilla genome. Science. 2016;352:aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tengvall K, et al. Genome-wide analysis in German Shepherd dogs reveals association of a locus on CFA 27 with atopic dermatitis. PLoS Genet. 2013;9:e1003475. doi: 10.1371/journal.pgen.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin C-S, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 51.Yeo S, Coombe L, Warren RL, Chu J, Birol I. ARCS: scaffolding genome drafts with linked reads. Bioinformatics. 2018;34:725–731. doi: 10.1093/bioinformatics/btx675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warren RL, et al. LINKS: scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience. 2015;4:35. doi: 10.1186/s13742-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ. BLAT-the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catchen, J., Amores, A. & Bassham, S. Chromonomer: a tool set for repairing and enhancing assembled genomes through integration of genetic maps and conserved synteny. G3-Genes Genom. Genet. 10, 4115–4128 (2020). [DOI] [PMC free article] [PubMed]
- 55.English AC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand NC, et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kronenberg, Z. N. et al. High-resolution comparative analysis of great ape genomes. Science360, eaar6343 (2018). [DOI] [PMC free article] [PubMed]
- 60.Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907 (2012).
- 61.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 65.Derrien T, et al. Fast computation and applications of genome mappability. PLoS ONE. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 67.Hayden KE, Willard HF. Composition and organization of active centromere sequences in complex genomes. BMC Genomics. 2012;13:324. doi: 10.1186/1471-2164-13-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 70.Pertea, G. & Pertea, M. GFF Utilities: GffRead and GffCompare. F1000Research9, ISCB Comm J-304 (2020). [DOI] [PMC free article] [PubMed]
- 71.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Megquier, K. et al. BarkBase: epigenomic annotation of canine genomes. Genes10, 433 (2019). [DOI] [PMC free article] [PubMed]
- 73.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Field, M. A. et al. Canfam_GSD: de novo chromosome-length genome assembly of the German Shepherd Dog (Canis lupus familiaris) using a combination of long reads, optical mapping, and Hi-C. GigaScience9, giaa027 (2020). [DOI] [PMC free article] [PubMed]
- 75.Halo, J. V. et al. Long-read assembly of a Great Dane genome highlights the contribution of GC-rich sequence and mobile elements to canine genomes. Preprint at bioRxiv10.1101/2020.07.31.231761 (2020). [DOI] [PMC free article] [PubMed]
- 76.Dudchenko, O. et al. The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. Preprint at bioRxiv10.1101/254797 (2018).
- 77.Dudchenko O, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cameron DL, et al. GRIDSS: sensitive and specific genomic rearrangement detection using positional de Bruijn graph assembly. Genome Res. 2017;27:2050–2060. doi: 10.1101/gr.222109.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 81.Cameron DL, Di Stefano L, Papenfuss AT. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat. Commun. 2019;10:3240. doi: 10.1038/s41467-019-11146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kosugi S, et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117. doi: 10.1186/s13059-019-1720-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeffares DC, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017;8:14061. doi: 10.1038/ncomms14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olsson M, et al. Absolute quantification reveals the stable transmission of a high copy number variant linked to autoinflammatory disease. BMC Genomics. 2016;17:299. doi: 10.1186/s12864-016-2619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mise M, Hashizume T, Matsumoto S, Terauchi Y, Fujii T. Identification of non-functional allelic variant of CYP1A2 in dogs. Pharmacogenetics. 2004;14:769–773. doi: 10.1097/00008571-200411000-00008. [DOI] [PubMed] [Google Scholar]
- 87.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 88.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The PacBio long reads, HiC, and Illumina 10x data of Mischka are available in SRA under BioProject PRJNA587469. The Illumina 10x data of 27 dogs are available in SRA under BioProject PRJNA588624. miRNA & RNA sequencing data are available in SRA under BioProject PRJNA657719. The canFam_GSD_1.0 assembly is deposited in DDBJ/ENA/GenBank under JAAHUQ000000000, and also available in UCSC browser (http://genome-euro.ucsc.edu/cgi-bin/hgTracks?db=canFam4).
Scripts used in the study are available at the GitHub repository (https://github.com/Chao912/Mischka/).