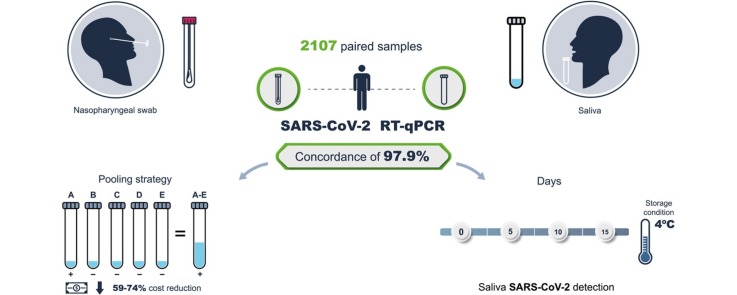
Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Diagnostic test, Saliva testing, Pooling strategy
Abstract
Objectives
The aim of this study was to investigate the feasibility of saliva sampling as a non-invasive and safer tool to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and to compare its reproducibility and sensitivity with nasopharyngeal swab samples (NPS). The use of sample pools was also investigated.
Methods
A total of 2107 paired samples were collected from asymptomatic healthcare and office workers in Mexico City. Sixty of these samples were also analyzed in two other independent laboratories for concordance analysis. Sample processing and analysis of virus genetic material were performed according to standard protocols described elsewhere. A pooling analysis was performed by analyzing the saliva pool and the individual pool components.
Results
The concordance between NPS and saliva results was 95.2% (kappa 0.727, p = 0.0001) and 97.9% without considering inconclusive results (kappa 0.852, p = 0.0001). Saliva had a lower number of inconclusive results than NPS (0.9% vs 1.9%). Furthermore, saliva showed a significantly higher concentration of both total RNA and viral copies than NPS. Comparison of our results with those of the other two laboratories showed 100% and 97% concordance. Saliva samples are stable without the use of any preservative, and a positive SARS-CoV-2 sample can be detected 5, 10, and 15 days after collection when the sample is stored at 4 °C.
Conclusions
The study results indicate that saliva is as effective as NPS for the identification of SARS-CoV-2-infected asymptomatic patients. Sample pooling facilitates the analysis of a larger number of samples, with the benefit of cost reduction.
Introduction
The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) worldwide has generated considerable demand for medical supplies for use in fighting the pandemic. Among other problems, this has resulted in a shortage of nasopharyngeal swabs (NPS) and tests for the detection of SARS-CoV-2. Scarce consumables and invasive sample collection, which can expose medical personnel to biohazards, are obstacles to effective mass screening of the population to identify infected individuals. Mass screening is essential to identify and isolate infected individuals during reopening. Additionally, fast massive effective screening is essential in the event of a coronavirus disease 2019 (COVID-19) resurgence and for the safe return to productive activities, an approach that has been implemented by several governments around the globe. Although this situation has been addressed using different innovative approaches, such as three-dimensional printing of NPS (Callahan et al., 2020), additional solutions for sample collection that are easier and less invasive, with minimal risk to health professionals, together with strategies aiming to maximize the number of samples analyzed, must be explored.
The gold standard test for the diagnosis of SARS-CoV-2 infection involves sample collection via NPS, followed by viral RNA extraction and detection by real-time polymerase chain reaction (RT-qPCR). Recent reports have indicated that saliva is a viable option for testing with several potential advantages over NPS, including that it is a less invasive procedure, making it more viable for repeated testing. Furthermore, saliva can be self-collected by the patient with minimal guidance and intervention from healthcare personnel (Azzi et al., 2020). SARS-CoV-2 can be detected in more than 95% of saliva samples, and the virus can be cultured from saliva samples (To et al., 2020b). Detection of the virus in saliva has also been used to monitor viral load dynamics over time, indicating that the highest viral load in saliva presents during the first week after symptom onset and then declines over time (To et al., 2020a). Recently, the Food and Drug Administration (FDA) in the United States approved the first diagnostic test with the option for saliva sampling for SARS-CoV-2 detection (U.S. Food and Drug Administration, 2020a). Another study found that the home-based collection method of saliva, supervised by a clinician, performed similarly to or even better than NPS for the detection of infection (Noah et al., 2020). These findings were confirmed by recent studies, which found that saliva is more sensitive for SARS-CoV-2 detection than NPS in patients with COVID-19 (Wyllie et al., 2020). In another report, 229 paired samples from 95 patients also showed a high concordance and no significant temporal variation in viral load between the two sample types (Cheuk et al., 2020).
The combined advantages offered by saliva sampling and sample pooling result in an inexpensive diagnostic procedure suitable for assaying large numbers of samples, as has been required during the current pandemic (Abdalhamid et al., 2020, Yelin et al., 2020). Sample pooling has proven its efficacy in different applications, including retrospective testing (Hogan et al., 2020) and, more importantly, in large-scale screening of asymptomatic populations (Ben-Ami et al., 2020, Lohse et al., 2020, U.S. Food and Drug Administration, 2020b). There is work showing that pooling saliva samples for the detection of SARS-CoV-2 provides a mechanism to support testing for a greater number of individuals with substantial cost savings, especially at lower prevalence levels (Pasomsub et al., 2020a, Pasomsub et al., 2020b, Watkins et al., 2020). Mirimus Clinical Labs in their SalivaClear test already use the pooling strategy to monitor and detect infections in groups of symptomatic and asymptomatic individuals (SalivaClear by Mirimus Clinical, 2020).
This study was performed to compare the reproducibility, accuracy, and feasibility of saliva sampling using NPS followed by RT-qPCR for the detection of SARS-CoV-2 in paired samples from asymptomatic clinical and laboratory personnel working in two Mexico National Institutes of Health laboratories and from asymptomatic office workers (N = 2107 individuals). This study presents evidence that saliva sample pooling is a reliable and inexpensive method that allows for the screening of a large number of samples.
Materials and methods
Participants
A cross-sectional study design was used to collect samples from personnel engaged in clinical and laboratory activities at Mexico’s National Cancer Institute and National Institute of Genomic Medicine. Consecutive asymptomatic subjects were sampled after signing an informed consent form. The study was approved by the ethics and research committees of both institutes (CEI/1479/20 and CEI 2020/21). Paired saliva and NPS samples were collected from 2107 asymptomatic healthcare and office workers to compare the two sample sources for SARS-CoV-2 detection. Additionally, saliva samples were collected from 3983 asymptomatic office workers, 2126 asymptomatic healthcare personnel, and 846 symptomatic office workers to detect SARS-CoV-2.
Sample collection
NPS were collected by a trained clinician with a flexible nylon swab that was inserted through the patient’s nostrils to reach the posterior nasopharynx. It was left in place for several seconds and slowly removed while rotating. The swab was then placed in 3 mL of sterile viral transport medium. Swabs from both nostrils were deposited in a single viral transport tube. Saliva samples were self-collected by the individuals without any stimulation and without rinsing the mouth before sample collection. Five milliliters of saliva was collected in a 50-ml sterile conical centrifuge tube without preservatives. Sample collection was done within the same facilities where the viral diagnosis laboratory is located. They were also collected from nearby hospitals. As a result, the swabs and the saliva samples were processed for viral RNA extraction within 5 h after collection.
SARS-CoV-2 RNA extraction and detection
Total nucleic acid was extracted from 300 μL of viral transport medium from the NPS or 300 μL of whole saliva using the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific) and eluted into 75 μL of elution buffer. For SARS-CoV-2 RNA detection, 5 μL of RNA template was tested using the US CDC real-time RT-qPCR primer/probe sets for 2019-nCoV_N1 and 2019-nCoV_N2 and human RNase P (RP) as an extraction control. Samples were classified as positive for SARS-CoV-2 when both the N1 and N2 primer/probe sets were detected with a Ct value of less than 40 (Centers for Disease Control and Prevention, 2020). If only one of these genes was detected, the sample was labeled inconclusive. All tests were run on Thermo Fisher ABI QuantStudio 5 or QuantStudio 7 real-time thermal cyclers.
Validation of saliva performance in independent laboratories and different extraction and detection methods
For validation purposes, 60 samples that were analyzed in our laboratory were also processed in two independent authorized laboratories (30 samples in each laboratory: Instituto de Investigaciones Biomédicas and Facultad de Química, Universidad Nacional Autónoma de México) using two additional RNA extraction methods and detection systems. The additional extraction methods consisted of spin-column-based RNA extraction (Total RNA Purification Kit, Jena Biosciences) and the use of a quick extraction solution from Lucigen. The two additional methods for SARS-CoV-2 detection were conducted using the GoTaq Probe 1-Step RT-qPCR System from Promega on a 7500 ABI system and the Star Q One-step RT-qPCR from Genes2Life.
Viral copy number analysis
Copies of the SARS-CoV-2 virus were quantified using a standard curve with serial dilutions (10-fold) using the 2019-nCoV_N and Hs_RPP30 positive controls synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA), with the same detection protocol as the clinical samples. The Ct values obtained from each dilution were used to interpolate the Ct value of each gene from the samples and to calculate viral copy numbers.
Stability assay
The stability of viral RNA in saliva for the detection of SARS-CoV-2 over time after sampling was assessed. A second RNA and an RT-qPCR extraction were performed from 150 SARS-CoV-2-positive saliva samples (stored at 4 °C) at 5, 10, and 15 days after the first positive result.
Sample pooling
SARS-CoV-2 detection in the pooling strategy was performed using the DAAN-Gene Kit following the manufacturer’s instructions. Briefly, the kit detects the ORF1ab and N genes of the virus. Five microliters of total RNA were used in the RT-qPCR reaction, and Ct values less than 40 were considered positive.
The evaluation of sample pools was conducted for both viral transport medium with NPS and saliva samples collected in parallel. To test the sensitivity of the pooling strategy, several pools were prepared from saliva and NPS. In the first pooling approach, one positive saliva sample (Ct values of 19.6 and 28.0 for the N viral gene) and its paired positive NPS (Ct values of 18.0 and 36.3 for the N viral gene) were mixed with five and nine known negative samples, respectively. Five hundred microliters of saliva was used from each sample to obtain the pool. This volume was necessary to obtain a homogeneous mixture in the saliva pool, given the differences in viscosity between different samples.
Based on the results obtained from the 10 sample pools and with the premise that asymptomatic individuals might have lower Ct values, which might result in false-negatives in the 10-sample pool, we generated five-sample pools from NPS and saliva from asymptomatic non health-related workers. For NPS, 51 pools made out of 255 individuals were evaluated. For saliva, 26 pools made out of 130 individuals were evaluated.
Statistical analysis
The accuracy of SARS-CoV-2 saliva detection, including sensitivity, sensitivity, predictive values, and likelihood ratios, was determined using RT-qPCR in NPS as the ‘gold standard’. Other statistical analyses were performed using GraphPad Prism 7.0 and IBM SPSS Statistics version 24 software. One-tailed parametric (Student t-test) and non-parametric (Mann–Whitney U-test) statistical tests were used to determine the significance of the data, considering a statistically significant value of p ≤ 0.05. The kappa coefficient was used to estimate the concordance between saliva and NPS results (McHugh, 2012).
Results and discussion
The design of this study was intended to compare the reproducibility, accuracy, and feasibility of saliva sampling followed by RT-qPCR to identify SARS-CoV-2 and to evaluate the use of saliva in sample pooling strategies. It was a priori accepted that the use of NPS followed by RT-qPCR is the gold standard for identification of the virus, despite current studies showing marked variation in the accuracy of this test.
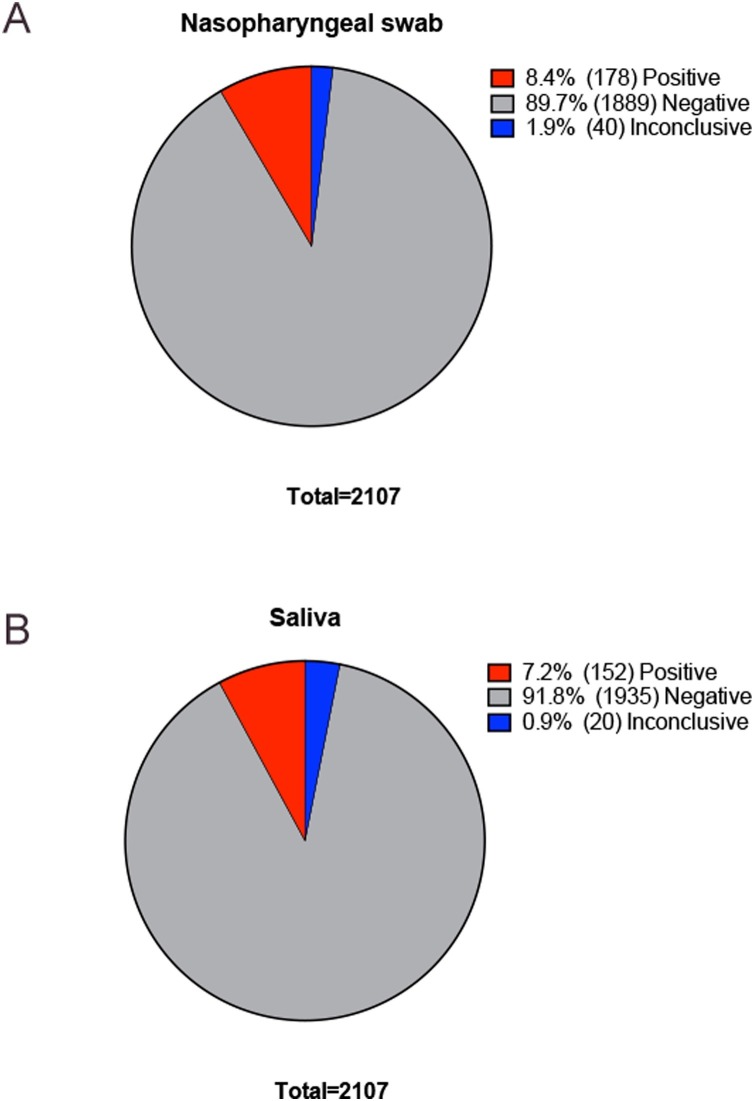
A total of 2107 paired NPS and saliva samples were included in the analysis. The distribution of the results is described in Figure 1 . Concordance between saliva and NPS results was statistically significant (Cohen’s kappa 0.727, standard error 0.025; p = 0.0001; Table 1 A). Concordance improved when inconclusive samples were removed from the analysis (Cohen’s kappa 0.852, standard error = 0.022; p = 0.0001). Overall, 2006 out of 2050 tests (98%) showed the same results in both saliva and NPS (Table 1B). Saliva had a lower number of inconclusive results than NPS (0.9% vs 1.9%) (Table 1 and Fig. 1).
Figure 1.
Frequencies and percentages of positive, negative, and inconclusive samples in 2107 paired nasopharyngeal swab and saliva samples: (A) nasopharyngeal swabs; (B) saliva samples.
Table 1.
Detection of SARS-CoV-2 by RT-qPCR between nasopharyngeal swab and saliva samples: (A) positive, negative, and inconclusive samples; (B) only positive and negative samples.
| A | Nasopharyngeal swab |
||||
|---|---|---|---|---|---|
| Positive | Negative | Inconclusive | Total | ||
| Saliva | Positive | 139 (6.6%) | 10 (0.5%) | 3 (0.1%) | 152 (7.2%) |
| Negative | 34 (1.6%) | 1867 (88.6%) | 34 (1.6%) | 1935 (91.8%) | |
| Inconclusive | 5 (0.2%) | 12 (0.6%) | 3 (0.1%) | 20 (0.9%) | |
| Total | 178 (8.4%) | 1889 (89.7%) | 40 (1.9%) | 2107 (100%) | |
| B | Nasopharyngeal swab |
|||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Saliva | Positive | 139 (6.8%) | 10 (0.5%) | 149 (7.3%) |
| Negative | 34 (1.7%) | 1867 (91.1%) | 1901 (92.7%) | |
| Total | 173 (8.4%) | 1877 (91.6%) | 2050 (100%) | |
Prevalence positive test = 8.44% (95% CI 7.27–9.73%), sensitivity = 80.35% (95% CI 73.63–85.99%), specificity = 99.47% (95% CI 99.02–99.74%), positive predictive value = 93.29% (95% CI 88.18–96.28%), negative predictive value = 98.21% (95% CI 97.60–98.67%), positive likelihood ratio = 150.81 (95% CI 80.92–281.06), negative likelihood ratio = 0.20 (95% CI 0.15–0.27).
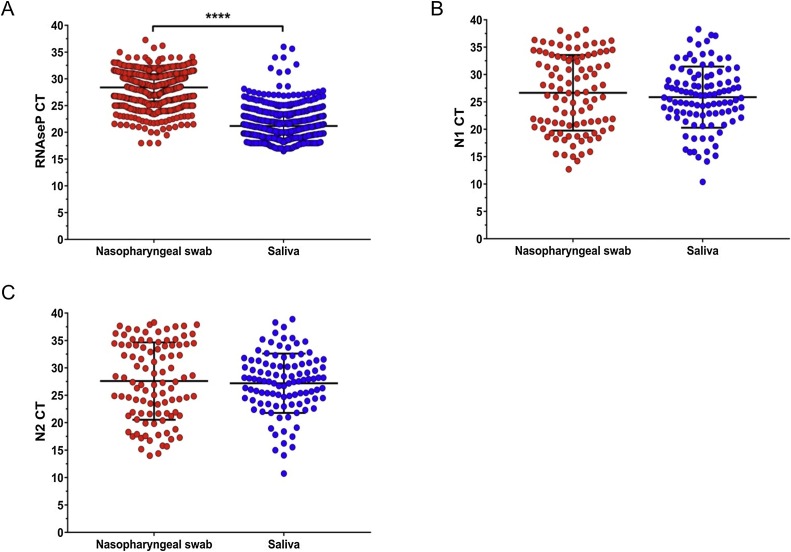
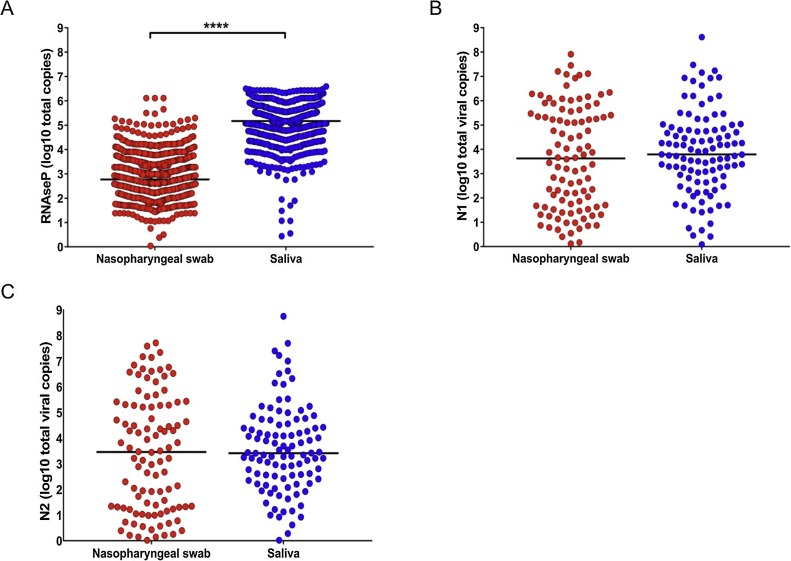
The concordance between the RT-qPCR results from viral RNA obtained from saliva and NPS was statistically significant, indicating that saliva is at least as sensitive as NPS for SARS-CoV-2 detection. Comparison of both the Ct values and the transcript copies of RNAse P showed a significantly higher total RNA concentration in saliva than in NPS (Figures 2A and 3 A ). However, when the two viral genes in the positive samples were analyzed, saliva and NPS did not show significant differences in viral load (Figure 2B, C and Figure 3B, C). Spearman correlation analysis of viral copies confirmed that saliva and NPS are both reliable sources for SARS-CoV-2 detection (N1: r = 0.4217, p = 0.0001; N2: r = 0.4261, p = 0.0001).
Figure 2.
Cycle threshold values (Ct) in nasopharyngeal swab versus saliva. (A) RNAse P gene in all samples; (B) N1 in SARS-CoV-2-positive samples; (C) N2 in SARS-CoV-2-positive samples. RNAse P had a significantly higher concentration of total RNA in saliva compared to nasopharyngeal swab; p < 0.00001 (t-test).
Figure 3.
Total viral copies in nasopharyngeal swab versus saliva. (A) RNAse P gene in all samples; (B) N1 in SARS-CoV-2-positive samples; (C) N2 in SARS-CoV-2-positive samples. RNAse P had significantly higher copies in saliva than nasopharyngeal swab; p < 0.00001 (Mann–Whitney U-test).
Saliva and paired NPS, which were previously analyzed in our laboratory (60 paired samples), were sent to two independent laboratories for extraction and SARS-CoV-2 detection and processed using different extraction and detection kits. Each laboratory processed 30 paired samples. A 100% concordance was observed in the results between our laboratory and the Instituto de Investigaciones Biomédicas (27 negative and three positive both in saliva and NPS), while 96.7% of the samples sent to Facultad de Química had the same result as in our laboratory (28 negative, one positive, and one discordant). This independent validation is an initial and exploratory assessment.
The accuracy of the saliva test is useful for clinical purposes. The positive likelihood ratio strongly supports its use as a reliable clinical test. A statistically significant correlation and concordance of the RT-qPCR detection of the virus in the saliva samples compared to NPS was identified, and a high concordance between the two types of samples was observed (Table 2 A). Given the high number of paired samples analyzed, the results clearly indicate that saliva is as good as NPS for viral detection in the diagnosis of COVID-19, as it has been shown in other studies in hospitalized patients (Table 2B) The data also demonstrated that saliva is stable even without the use of any preservative during sample collection and that a positive SARS-CoV-2 sample can be detected 5, 10, and 15 days after collection when the sample is stored at 4 °C: variation in Ct values in the viral N gene was 0.88 ± 1.92 at 5 days, −0.93 ± 3.01 at 10 days, and −0.76 ± 2.12 at 15 days. Other studies have also demonstrated the stability of saliva for the detection of SARS-CoV-2, with storage for 10–25 days at room temperature (Uwamino et al., 2021) without buffers or stabilizers (Ott et al., 2020).
Table 2.
Detection of SARS-CoV-2 in samples of saliva and nasopharyngeal swabs: (A) paired samples; (B) saliva samples only; (C) saliva only in the present study.
| A | Paired samples |
||||
|---|---|---|---|---|---|
| Country | Study population | Paired samples | Viral genes | Concordance % | Reference |
| Australia | Ambulatory patients | 522 | ORF1a, ORF8 | 84.6 | (Williams et al., 2020) |
| Canada | Hospitalized patients | 91 | RdRp, E, N | 61.0 | (Jamal et al., 2020) |
| China | Ambulatory patients | 229 | E | 76.0 | (Cheuk et al., 2020) |
| China | Hospitalized patients | 58 | RdRp/Hel, E, N2 | 84.5 | (Chen et al., 2020) |
| China | Patients from 12 independent cohorts | 944 | S, E, ORF1ab, N, RdRp, 5′UTR | 92.1 | (Zhu et al., 2020) |
| China | Hospitalized patients | 95 | E, RdRp | 78.9 | (Leung Chi‐man et al., 2021) |
| Japan | Ambulatory patients | 76 | N | 97.4 | (Iwasaki et al., 2020) |
| Mexico | Ambulatory patients | 253 | E | 78.6 | (Moreno-Contreras et al., 2020) |
| Thailand | Hospitalized patients | 200 | ORF1ab, N | 97.5 | (Pasomsub et al., 2020a) |
| USA | Hospitalized patients and asymptomatic healthcare workers | 29 | N1, N2 | 79.0 | (Wyllie et al., 2020) |
| USA | Ambulatory patients | 91 | N1, N2 | 94.0 | (Miller et al., 2020) |
| Mexico | Asymptomatic healthcare and office workers | 2107 | N1, N2 | 97.9 | Our study |
| B | Saliva samples only |
||||
|---|---|---|---|---|---|
| Country | Study population | Saliva samples | Viral genes | Positivity % | Reference |
| China | Hospitalized patients | 12 | S | 91.7 | (To et al., 2020a) |
| China | Hospitalized patients | 18 | E | 84.0 | (Hung et al., 2020) |
| Italy | Hospitalized patients | 25 | 5′UTR | 100.0 | (Azzi et al., 2020) |
| Japan | Hospitalized patients | 103 | N1, N2, ORF1, E | 93.4 | (Ikeda et al., 2020) |
| C | Saliva only in the present study |
|||||
|---|---|---|---|---|---|---|
| Setting | Total samples | Number of tests | Positive samples | Positivity (%) | Reduction in testing costs (%)a | Reduction in total sample collection direct costs (USD)b, c |
| Asymptomatic office workers | 3983 | 1032 | 26 | 0.65 | 74 | $10 754.50 |
| Asymptomatic healthcare personnel | 2126 | 870 | 98 | 4.6 | 59 | $5740.20 |
| Symptomatic office workers | 846 | 846 | 67 | 7.9 | 0 | $2284.20 |
RdRp, RNA-dependent RNA polymerase; RdRp/Hel, RNA -dependent RNA polymerase/helicase; ORF1, open reading frame 1 (a,b); ORF8, open reading frame 8; E, envelope; N, nucleocapsid.
Cost reduction was calculated considering the number of tests necessary to identify the positive individuals in the positive pools.
Sample collection direct cost: 3 USD vs 0.3 USD, nasopharyngeal and saliva, respectively.
The sample cost includes both direct and indirect costs.
Sample pooling
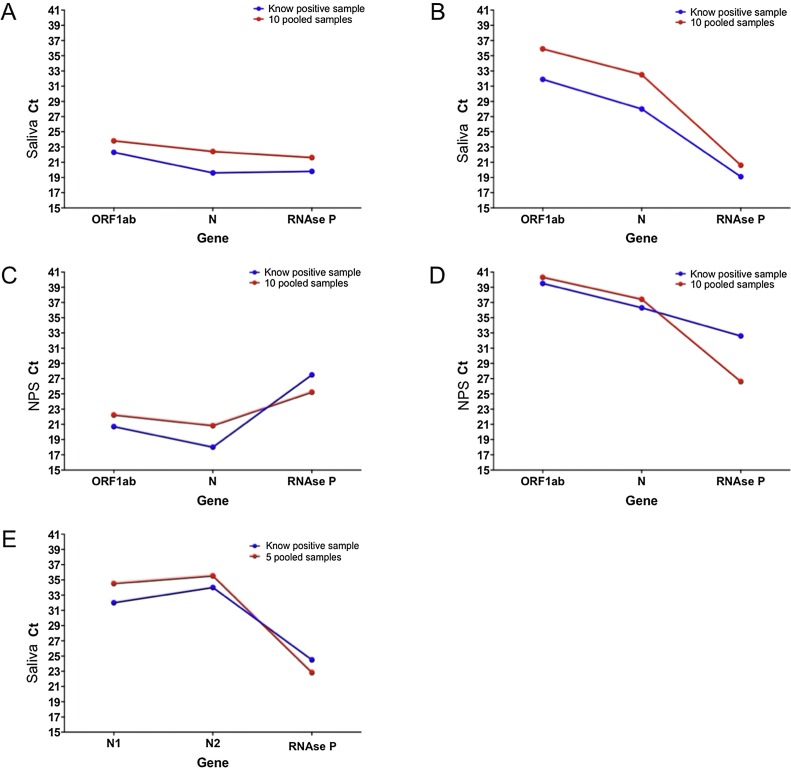
Positive samples were selected according to their RT-qPCR results, representing low and high Ct values, to evaluate the effect of a 1:10 pooling with negative samples in the detection capacity of the test. In the first set of saliva samples, the initial Ct values for the positive sample were 22.3 for ORF1ab, 19.6 for N, and 19.8 for RNAse P. As expected, after pooling with the additional nine negative samples, the Ct values increased to 23.8 for ORF1ab, 22.4 for N, and 21.6 for RNAse P, showing that pooling did not affect the detection capacity of the test. A similar situation was observed in the NPS sample pool. In the second saliva pool, the positive sample had higher Ct values (31.9 ORF1ab, 28 for N, and 19.1 for RNAse P). After pooling, an increase in four Ct values in both viral genes was observed. Even though this result is still within the acceptable range for detecting the positive sample in the pool (Figure 4 ), samples with a higher Ct value might become false-negatives if analyzed by pooling; for this reason, the subsequent experiments were focused on the analysis of five-sample pools.
Figure 4.
Analysis of pooled (1:5 and 1:10) saliva and swab paired samples. Saliva and nasopharyngeal swab pools were generated by mixing one positive sample with four/nine known negative samples. Positive samples with early and late Ct of the N gene were selected to evaluate the impact of dilution on its detection. (A) and (B) show saliva pooled 1:10; (C) and (D) show nasopharyngeal swab pooled 1:10; (E) shows saliva pooled 1:5.
A total of 130 individuals were tested in 26 NPS pools with five samples each, identifying 20 positive cases (15.4%). All positive cases identified in the pools were confirmed through the analysis of the individual samples used to generate the pool. In the case of saliva, 255 individuals were grouped into 26 pools with five samples each. In this case, two positive cases were identified (7.7%), which were also confirmed through analysis of the individual samples.
Additionally, asymptomatic office and healthcare personnel were tested, as well as office workers presenting mild symptoms who were suspected of being SARS-CoV-2 carriers. Only saliva was used and five samples were pooled in the first two groups. Table 2C shows the positivity among the three groups, which was increased in healthcare personnel and symptomatic office workers. Substantial reductions in direct costs for sampling compared with NPS and in the costs by testing pools instead of individual samples were observed.
These results showed that it is feasible to apply pooling strategies using saliva. However, some considerations should be taken into account, including the use of 500 μL of saliva to generate the pool to obtain a homogeneous mixture. Dilution of one positive sample with nine negative samples showed that, even though positive results can still be obtained in the pool, samples with a low viral load might become difficult to detect. Therefore, we suggest pooling no more than five samples, even though other reports indicate that pooling strategies of 16 and 24 samples are useful in high prevalence populations (≥10%) (Verwilt et al., 2020).
Concluding remarks
The study data indicate that saliva is a reliable source for the detection of SARS-CoV-2 infection. However, several aspects must be addressed to successfully use saliva testing: (1) Sample collection: even though saliva self-collection might be easier than NPS sampling, proper biosafety and risk evaluation protocols must be followed by medical personnel to minimize contagions due to the production of potential aerosols during saliva collection. (2) Sample handling: the application of proven and standardized methods for the inactivation and handling of a saliva sample should be considered, and saliva samples must always be regarded as potentially infected. The packaging and cold-chain protocols used for NPS samples must be followed. (3) RNA extraction and RT-qPCR: it has been well documented that several components of saliva can inhibit PCR, highlighting the importance of using viral RNA extraction systems that have been tested and approved by regulatory agencies that generate pure and high-quality RNA for RT-qPCR analysis. We did not use any preservative for saliva samples and suggest that samples should be stored at 4 °C after collection and processed within 4 days post collection.
Given the situations mentioned above, the use of saliva represents a viable option for SARS-CoV-2 detection. Thus, saliva and the pooling strategy presented here are effective options for the analysis of samples in well-controlled cohorts, which provide a cost-effective screening tool in asymptomatic populations. The cost reduction was calculated considering the number of tests necessary to identify the positive individuals in the positive pool. This is particularly suitable, for example, in office workers, faculty, or other groups where testing is necessary on a periodic basis to identify and isolate infected individuals. The implementation of testing for SARS-CoV-2 infection by RT-qPCR using saliva as a source for viral RNA constitutes an easy, non-invasive, inexpensive, and less risky option compared to NPS, without compromising the accuracy of the test. The combination of saliva sampling and pooling represents a viable and useful method for population-based studies that will be necessary for a safe return to economic activities.
Funding
This work was funded by the Secretaría de Educación, Ciencia, Tecnología e Innovación de la Ciudad de México (SECTEI).
Conflict of interest
The authors do not have an association that might pose a conflict of interest.
Acknowledgments
The authors acknowledge M.C. Isabel Gracia Mora for providing BSL2 facilities at UNAM; Facultad de Química-UNAM, Patronato de la Facultad de Química-UNAM for providing all materials used in the RT-qPCR analyses; and CONACyT 314298 for project funding.
References
- Abdalhamid B., Bilder C.R., McCutchen E.L., Hinrichs S.H., Koepsell S.A., Iwen P.C. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol. 2020;153 doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81 doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect. 2020;26 doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan C.J., Lee R., Zulauf K.E., Tamburello L., Smith K.P., Previtera J. Open development and clinical validation of multiple 3D-printed sample-collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. MedRxiv. 2020 doi: 10.1101/2020.04.14.20065094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. (2019-nCoV), CDC 2019-Novel Coronavirus Panel, Real-Time RT-PCR Diagnostic.https://www.fda.gov/media/134922/download [Google Scholar]
- Chen J.H.K., Yip C.C.Y., Poon R.W.S., Chan K.H., Cheng V.C.C., Hung I.F.N. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect. 2020;9 doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheuk S., Wong Y., Tse H., Siu H.K., Kwong T.S., Chu M.Y. Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis. 2020;71:2939–2946. doi: 10.1093/cid/ciaa797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C.A., Garamani N., Sahoo M.K., Huang C.H., Zehnder J., Pinsky B.A. Retrospective screening for SARS-CoV-2 RNA in California, USA, late 2019. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2610.202296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung D.L.-L., Li X., Chiu K.H.-Y., Yip C.C.-Y., To K.K.-W., Chan J.F.-W. Early-morning vs spot posterior oropharyngeal saliva for diagnosis of SARS-CoV-2 infection: implication of timing of specimen collection for community-wide screening. Open Forum Infect Dis. 2020;7 doi: 10.1093/ofid/ofaa210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T. Clinical evaluation of self-collected saliva by RT-qPCR, direct RT-qPCR, RT-LAMP, and a rapid antigen test to diagnose COVID-19. MedRxiv. 2020;58 doi: 10.1101/2020.06.06.20124123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81 doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Liu A.X., Paterson A. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) MedRxiv. 2020 doi: 10.1101/2020.05.01.20081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Chi-man E., Chow Chi-ying V., Lee Kin-ping M., Lai Wai-man R. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS-CoV-2. J Med Virol. 2021;93 doi: 10.1002/jmv.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse S., Pfuhl T., Berkó-Göttel B., Rissland J., Geißler T., Gärtner B. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020;20 doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh M.L. Interrater reliability: The kappa statistic. Biochem Medica. 2012;22 doi: 10.11613/bm.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Jansen M., Bisignano A., Mahoney S., Wechsberg C., Albanese N. Validation of a Self-administrable, Saliva-based RT-qPCR Test Detecting SARS-CoV-2. MedRxiv. 2020 doi: 10.1101/2020.06.05.20122721. [DOI] [Google Scholar]
- Moreno-Contreras J., Espinoza M.A., Sandoval-Jaime C., Cantú-Cuevas M.A., Barón-Olivares H., Ortiz-Orozco O.D. Saliva sampling and its direct lysis, an excellent option to increase the number of SARS-CoV-2 diagnostic tests in settings with supply shortages. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01659-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah K., Fred T., Vlad S., Agatha B., Laura D., Siri K. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. MedRxiv. 2020 doi: 10.1101/2020.04.11.20062372. [DOI] [Google Scholar]
- Ott I.M., Strine M.S., Watkins A.E., Boot M., Kalinich C.C., Harden C.A. Simply saliva: Stability of SARS-CoV-2 detection negates the need for expensive collection devices. MedRxiv. 2020 doi: 10.1101/2020.08.03.20165233. [DOI] [Google Scholar]
- Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasomsub Ekawat, Watcharananan S.P., Watthanachockchai T., Rakmanee K., Tassaneetrithep B., Kiertiburanakul S. Saliva sample pooling for the detection of SARS-CoV-2. J Med Virol. 2020 doi: 10.1002/jmv.26460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SalivaClear by Mirimus Clinical . 2020. The SalivaClear Solution.https://www.salivaclear.com/ [Google Scholar]
- To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Tsang O.T.Y., Yip C.C.Y., Chan K.H., Wu T.C., Chan J.M.C. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration . 2020. Coronavirus (COVID-19) update: FDA authorizes first diagnostic test using at-home collection of saliva specimens.https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-diagnostic-test-using-home-collection-saliva [Google Scholar]
- U.S. Food and Drug Administration . 2020. Coronavirus (COVID-19) update: FDA authorizes first diagnostic test for screening of people without known or suspected COVID-19 infection authorization is also second to allow testing of pooled samples. [Google Scholar]
- Uwamino Y., Nagata M., Aoki W., Fujimori Y., Nakagawa T., Yokota H. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2. J Clin Pathol. 2021;74 doi: 10.1136/jclinpath-2020-206972. [DOI] [PubMed] [Google Scholar]
- Verwilt J., Mestdagh P., Vandesompele J. Evaluation of efficiency and sensitivity of 1D and 2D sample pooling strategies for diagnostic screening purposes. MedRxiv. 2020 doi: 10.1101/2020.07.17.20152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins A.E., Fenichel E.P., Weinberger D.M., Vogels C.B.F., Brackney D.E., Casanovas-Massana A. Pooling saliva to increase SARS-CoV-2 testing capacity. MedRxiv. 2020 doi: 10.1101/2020.09.02.20183830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of sars-cov-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. MedRxiv. 2020 doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- Yelin I., Aharony N., Tamar E.S., Argoetti A., Messer E., Berenbaum D. Evaluation of COVID-19 RT-qPCR Test in Multi sample Pools. Clin Infect Dis. 2020;71 doi: 10.1093/cid/ciaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Guo J., Xu Y., Chen X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J Infect. 2020;81 doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]