The long noncoding RNA DLGAP1‐AS2 facilitates the malignant progression of cholangiocarcinoma via miR‐505 and GALNT10 and may have potential as a novel target for cholangiocarcinoma therapy.
Keywords: cholangiocarcinoma, DLGAP1‐AS2, GALNT10, migration, miR‐505, viability
Abstract
Cholangiocarcinoma (CCA) is a highly invasive malignant tumor with high mortality. Most cases of CCA are already advanced when they are detected, resulting in poor prognosis. As such, there is an ongoing need for the identification of effective biomarkers for CCA. The long noncoding RNA DLGAP1‐AS2 has been reported to have prognostic value in glioma and Wilms' tumor. Here, we investigated the function of DLGAP1‐AS2 in CCA. The differential expression of DLGAP1‐AS2 in CCA tissues and normal tissues was first examined using data from the The Cancer Genome Atlas database and then in CCA cell lines by quantitative RT‐PCR (qRT‐PCR). The target gene was predicted by bioinformatics analysis, and the binding sites were confirmed using luciferase assay. DLGAP1‐AS2 is up‐regulated in CCA, and high DLGAP1‐AS2 expression promotes cell viability and is associated with poor prognosis. Notably, DLGAP1‐AS2 acts as a sponge to suppress miR‐505 expression, and miR‐505 reduces the expression of N‐acetylgalactosaminyltransferase 10 (GALNT10) in CCA cells. Biofunctional experiments revealed that a miR‐505 inhibitor almost completely removed the inhibitory effect of si‐DLGAP1‐AS2 on CCA cell malignant progression, whereas the malignant phenotype of cells cotransfected with si‐DLGAP1‐AS2 and si‐GALNT10 was significantly reduced as compared with the control. In summary, the DLGAP1‐AS2/miR‐505/GALNT10 axis may contribute to regulating the malignant progression of CCA and may have potential as a novel target for CCA therapy.
Abbreviations
- CCA
cholangiocarcinoma
- CCK‐8
Cell Counting Kit‐8
- GALNT
N‐acetylgalactosaminyltransferase 10
- lncRNA
long noncoding RNA
- NC
negative control
- qRT‐PCR
quantitative RT‐PCR
- SD
standard deviation
- TCGA
The Cancer Genome Atlas
Cholangiocarcinoma (CCA) is a highly invasive malignant tumor with high mortality, which originated from the epithelial cells of bile ducts [1]. In recent decades, despite the difference in incidence rate and etiology of CCA in the world, the incidence rate of CCA has been presented as an upward trend worldwide [2]. Until now, radical resection was the most valid treatment for CCA [3]. However, because of the insidious onset and rapid progress of CCA, most patients with CCA are in advanced stage when they are diagnosed clinically, thus losing the opportunity of radical operation, and have a poor prognosis [4, 5]. Hence it is important to master the molecular mechanism of CCA development for the study of effective biomarkers and therapeutic strategies.
Recent studies have found that long noncoding RNA (lncRNA) is a class of macromolecular noncoding RNA that mainly regulated gene expression in the form of RNA from three levels: transcription level, posttranscriptional regulation and epigenetics [6, 7, 8]. Importantly, lncRNA has been confirmed to act as a regulatory factor in tumor formation, invasion, metastasis and drug resistance mechanisms by affecting the expression of oncogenes [9, 10], including CCA. For example, lncRNA‐EPIC1 is up‐regulated in CCA and accelerates the malignant biological behaviors of CCA by targeting Myc [11]. lncRNA‐RMRP contributes to accelerating the tumorigenesis and development of CCA by sponging miR‐217 [12]. Hence the development of lncRNA will provide the feasible targets for the targeted therapy of CCA. lncRNA DLGAP1‐AS2 is located on human chromosome 18p11, and its role in the physiological process is rarely reported. Recently, two studies have confirmed its prognostic value in glioma [13] and Wilms' tumor [14]. Wherein, Miao et al. [13] indicated that DLGAP1‐AS2 was highly expressed in gliomas and promoted the malignant phenotype of glioma cells by targeting YAP1. Nevertheless, the expression and function of DLGAP1‐AS2 in CCA have not been studied.
Herein, we researched the level of DLGAP1‐AS2 in CCA and assessed the impact of DLGAP1‐AS2 on the malignant biological behaviors of CCA cells in vitro, as well as its potential molecular mechanism, which laid a theoretical foundation for the application of DLGAP1‐AS2 as a therapeutic target of CCA.
Materials and methods
Data acquisition
We downloaded the RNA‐seq data from The Cancer Genome Atlas (TCGA) database. These data were applied to analyze the differential expression of DLGAP1‐AS2, miR‐505 and N‐acetylgalactosaminyltransferase 10 (GALNT10), which contains 36 patients with CCA and nine healthy control subjects. Moreover, the prognostic value of DLGAP1‐AS2 was also assessed using data from TCGA database. Furthermore, patient information was extracted from TCGA database.
Cell culture and transfection
CCA cell lines HUCCT1 and RBE were obtained from the Shanghai Cell Bank (Shanghai, China). Normal bile duct cell HEBEpics was purchased from the ScienCell Research Laboratories (Carlsbad, CA, USA). Dulbecco's modified Eagle's medium supplemented with 10% FBS and 100 U·mL−1 penicillin–streptomycin was used to culture the cells at 37 °C with 5% CO.
DLGAP1‐AS2 negative control (si‐NC, 5′‐TTCTCCGAACGTGTCACGT‐3′), si‐DLGAP1‐AS2#1 (5′‐CUGGCUGCGAAGUAAUAAAUU‐3′), si‐DLGAP1‐AS2#2 (5′‐GGCAGGAAAUUGUGUUUGU UU‐3′), pcDNA3.1‐DLGAP1‐AS2, miR‐505 inhibitor, miR‐505 mimic, negative control (miR‐NC) and si‐GALNT10 (5′‐ATGAGTACGCAGAGTACAT‐3′) were synthesized by Dharmacon. HUCCT1 and RBE cells were transiently transfected with these sequences by Lipofectamine 2000 as directed by the supplier. After 48 h, the transfection efficiency was measured using qRT‐PCR. The transfected cells were used for subsequent experiments.
Total RNA extraction and qRT‐PCR
RNeasy Mini Kit (Qiagen, Dusseldorf, Germany) was used to isolate the total RNA from cells. One microgram of total RNA was used as a template, and cDNA was formed using the reverse transcription reaction system following the standard of the RT‐PCR kit. Then, qRT‐PCR was conducted to assess the mRNA levels of DLGAP1‐AS2 and GALNT10 with the SYBR Premix Ex Tap II kit (Takara, Dalian, China) following the supplier's instructions, and the data were normalized to GAPDH. The primer sequences of DLGAP1‐AS2 and GALNT10 were presented as follows: DLGAP1‐AS2: forward (F), 5′‐TTCCTGTCTTTCAGGATGAATGCC‐3′ and reverse (R), 5′‐TGGTAGCCTGTGGCGAGTTGAA‐3′; GALNT10: F, 5′‐GAGCCTTTGACTGGGAGATGTAC‐3′ and R, 5′‐GAGTTCCCAGAACCACTTCCGA‐3′; and GAPDH: F, 5′‐TAGATGACACCCGTCCCTGA‐3′ and R, 5′‐ACCTCCACCTGTCCTTAGTG‐3′.
Western blotting
The cells were lysed with radioimmunoprecipitation assay buffer (containing protease inhibitor) to extract protein. Then, the proteins were separated with 10% SDS/PAGE gels and then transferred to polyvinylidene difluoride membranes. The membranes were sealed in 5% skimmed milk powder for 1 h, which then incubated with primary antibodies at 4 °C overnight. The primary antibodies were as follows: GALNT10 and GAPDH (1 : 1000; Abcam, Cambridge, MA, USA). After that, HRP‐conjugated secondary antibody was used to incubate the membranes for 2 h. Finally, Electro‐Chemiluminescence substrate was used to measure the blots.
Bioinformatics prediction
LncBase (http://carolina.imis.athena‐innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex) was applied to predict miRNAs regulated by DLGAP1‐AS2, and the target molecule of miR‐505 was predicted by starBase (http://starbase.sysu.edu.cn/).
Luciferase assay
To confirm the results of bioinformatics prediction, we chose 293T cells with high transfection efficiency for luciferase activity detection. The DLGAP1‐AS2 sequence was infixed with the pGL3 Basic vector. The luciferase reporter plasmid and miR‐505 inhibitor/NC were cotransfected into cells and cultured for 48 h. Then, the luciferase activity was detected using the Promega dual‐luciferase reporter gene detection kit following the supplier's standards. Also, according to the earlier steps, the luciferase reporter assay was conducted to assess the regulatory effect between GALNT10 and miR‐505 as well.
Proliferation assays
Cell Counting Kit‐8 (CCK‐8) and colony formation assays were carried out to assess the proliferative capacity of cells as described previously [15, 16]. In CCK‐8 assay, the cells were seeded into 96‐well plates (4 × 103 cells per well) and cultured for 0, 24, 48 and 72 h. Then, CCK‐8 kit was used to detect the OD value at the specified time point as directed by supplier. Herein, 2 h before the detection, 10 μL CCK‐8 solution was appended to each well for culture.
For colony formation assay, in brief, about 400 cells were seeded into each medium and cultured at 37 °C with 5% CO2 for 7–14 days until the visible colonies appeared. Then, the cells were fixed with 4% paraformaldehyde and stained with crystal violet. Finally, we counted the number of colonies.
Invasion and migration assays
Transwell assay was performed to appraise the ability of cell invasion and migration as mentioned earlier, with minor modifications [17]. The upper surface of the Transwell chambers was precoated with (invasion assay) or without (migration assay) Matrigel. Cells were inoculated to the upper chamber with the serum‐free medium, and the lower chamber was perfused with 500 μL complete medium. After 48 h, the remaining cells were removed from the upper surface, while the invaded or migrated cells on the lower surface were fixed with 4% paraformaldehyde and stained with crystal violet. Five visual fields were randomly selected to photograph and count under the microscope.
Statistical analysis
spss 21.0 (IBM, New York, USA) statistical software was applied for the data analysis. All assays were performed three times, and the data were represented as mean ± standard deviation (SD) using the t‐test (two groups) and one‐way ANOVA (multiple groups) for comparison. The correlations between DLGAP1‐AS2 and miR‐505, as well as GALNT10 and DLGAP1‐AS2/miR‐505, were analyzed using Pearson’s correlation analysis. The survival curves were plotted using Kaplan–Meier survival analysis. A P‐value <0.05 represented that the difference was significant.
Results
Anomalous overexpression of lncRNA DLGAP1‐AS2 in CCA tissues and cell lines
To detect whether DLGAP1‐AS2 is abnormally expressed in CCA, we measured the level of DLGAP1‐AS2 in CCA tissues and normal tissues based on the TCGA database. Patient information was displayed in Table S1. As presented in Fig. 1A, the DLGAP1‐AS2 level was markedly enhanced in CCA tissues compared with that in normal tissues. Then, qRT‐PCR was conducted to confirm the DLGAP1‐AS2 expression in CCA cell lines. Consistently, DLGAP1‐AS2 presented the high level in CCA cell lines HUCCT1 and RBE compared with that in HEBEpics cells (Fig. 1B). Moreover, Kaplan–Meier survival analysis indicated that high DLGAP1‐AS2 expression was related to poor outcomes of patients with CCA (Fig. 1C).
Fig. 1.
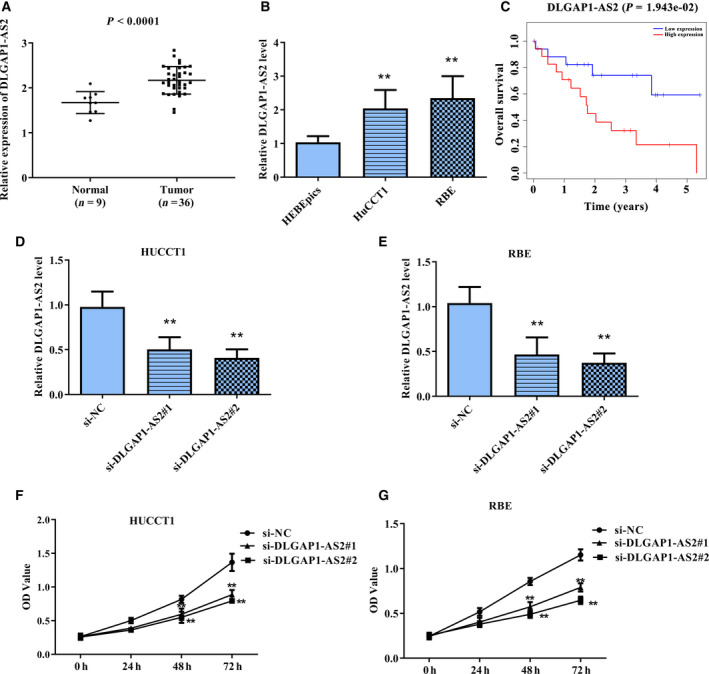
lncRNA DLGAP1‐AS2 was up‐regulated in CCA and contributes to promoting cell viability of CCA cells. (A) Bioinformatics analysis of DLGAP1‐AS2 expression in CCA tissues and normal tissues. P < 0.0001. (B) The levels of DLGAP1‐AS2 in CCA cell lines HUCCT1 and RBE, as well as normal HEBEpics cells. (C) Kaplan–Meier survival analysis of the relevance of DLGAP1‐AS2 expression and survival rate of patients with CCA. P = 0.0019. (D,E) The transfection efficiency of si‐DLGAP1‐AS2#1 and si‐DLGAP1‐AS2#2 in HUCCT1 and RBE cells was measured using qRT‐PCR. (F,G) The impacts of si‐DLGAP1‐AS2#1 and si‐DLGAP1‐AS2#2 on cell viability of HUCCT1 and RBE cells were detected using CCK‐8 assay. **P < 0.01. Error bar represents the mean ± SD derived from three independent experiments. Comparisons between groups were analyzed using t‐tests (two‐sided). OD, absorbance.
Considering the abnormal expression of DLGAP1‐AS2 in CCA and its prognostic value, we speculate that DLGAP1‐AS2 is involved in the malignant progression of CCA. First, we transfected si‐DLGAP1‐AS2#1 and si‐DLGAP1‐AS2#2 into HUCCT1 and RBE cells, respectively, to knock down DLGAP1‐AS2, and then qRT‐PCR was performed to detect the transfection efficiency. As presented in Fig. 1D,E, si‐DLGAP1‐AS2#1 and si‐DLGAP1‐AS2#2 both markedly decreased the DLGAP1‐AS2 level, and si‐DLGAP1‐AS2#2 had the higher efficiency. Then, CCK‐8 assay was performed to assess the cell viability. It can be seen from Fig. 1F,G that knockdown of DLGAP1‐AS2 led to a marked decline in the cell viability of both HUCCT1 and RBE cells. These findings revealed that DLGAP1‐AS2 may contribute to promoting the malignant progression of CCA.
lncRNA DLGAP1‐AS2 targeted miR‐505
The potential targets of DLGAP1‐AS2 were predicted using miRDB and obtained 199 miRNAs. Then, the predicted miRNAs were intersected with the down‐regulated miRNAs analyzed in TCGA and obtained three common miRNAs (Fig. 2A), namely, hsa‐miR‐3163, hsa‐miR‐4705 and hsa‐miR‐505. Apparently, miR‐505 was lowly expressed in CCA tissues (Fig. 2B). Furthermore, Pearson’s correlation analysis indicated that the miR‐505 expression was negatively correlated with DLGAP1‐AS2 (Fig. 2C). In view of the earlier results, we selected miR‐505 for the follow‐up experiments. After transfection of the miR‐505 inhibitor, the level of miR‐505 decreased significantly, which indicated that the miR‐505 inhibitor actually inhibits miR‐505 (Fig. 2D). Based on the bioinformatics prediction, the binding site of DLGAP1‐AS2 and miR‐505 was presented in Fig. 2E. Subsequently, we conducted luciferase reporter assay to validate this prediction. As displayed in Fig. 2F, the luciferase activity of the WT‐DLGAP1‐AS2 reporter was markedly enhanced in the miR‐505 inhibitor group, while that of the MUT‐DLGAP1‐AS2 reporter with mutated binding site was almost unchanged in the miR‐505 inhibitor group. Moreover, we detected the effect of the miR‐505 inhibitor on DLGAP1‐AS2 level using qRT‐PCR. Interestingly, inhibition of miR‐505 led to a marked increase on DLGAP1‐AS2 level (Fig. 2G). These data indicated that DLGAP1‐AS2 directly bound to miR‐505.
Fig. 2.
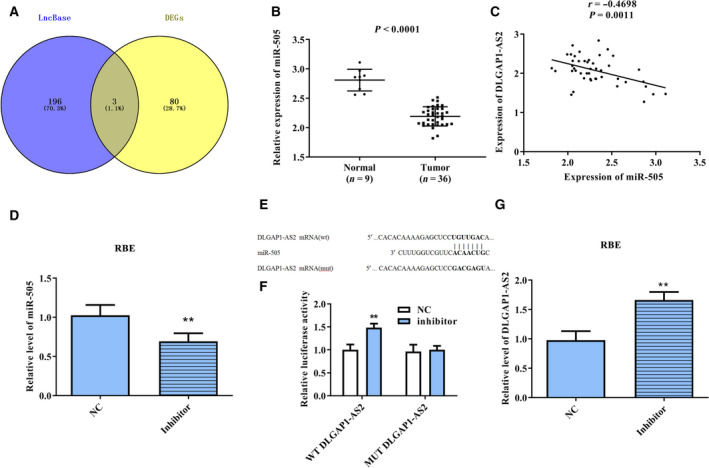
DLGAP1‐AS2 targeted miR‐505. (A) Wayne diagram was applied to ascertain the possible targets of DLGAP1‐AS2. (B) Bioinformatics analysis of miR‐505 expression in CCA tissues and normal tissues. P < 0.0001. (C) The correlation between miR‐505 and DLGAP1‐AS2. R = −0.4698; P = 0.0011. (D) The transfection efficiency of the miR‐505 inhibitor in RBE cells was measured using qRT‐PCR. (E) Comparison of miR‐505 with DLGAP1‐AS2 sequences. (F) Dual‐luciferase reporter assay was performed to confirm that miR‐505 directly binds to the 3′‐UTR of DLGAP1‐AS2. (G) The effect of the miR‐505 inhibitor on DLGAP1‐AS2 level was measured using qRT‐PCR. **P < 0.01. Error bar represents the mean ± SD derived from three independent experiments. Comparisons between groups were analyzed using t‐tests (two‐sided).
GALNT10, a downstream molecule of miR‐505, was regulated via DLGAP1‐AS2
Next, starBase was used to predict the downstream molecules of miR‐505 and obtained 1488 target genes. Then, we analyzed the coexpression genes of DLGAP1‐AS2. Under the condition of P < 0.05 and correlation coefficient ≥ 0.3, 2646 genes with positive correlation were obtained. The predicted targets of miR‐505 and the coexpression genes of DLGAP1‐AS2 were intersected with the up‐regulated mRNA analyzed in TCGA database and obtained four common genes (Fig. 3A), namely, ENTPD6, MECOM, GALNT10 and HEY1. The up‐regulated GALNT10 (Fig. 3B) with the largest difference and the strongest correlation (Fig. 3C) was selected for the study. The binding site of GALNT10 and miR‐505 was presented in Fig. 3D. Then, luciferase reporter assay revealed that the relative luciferase activity was markedly increased in the WT+miR‐505 inhibitor group compared with NC or mutant (MUT) groups (Fig. 3E). Our results verified that GALNT10 is a downstream molecule of miR‐505.
Fig. 3.
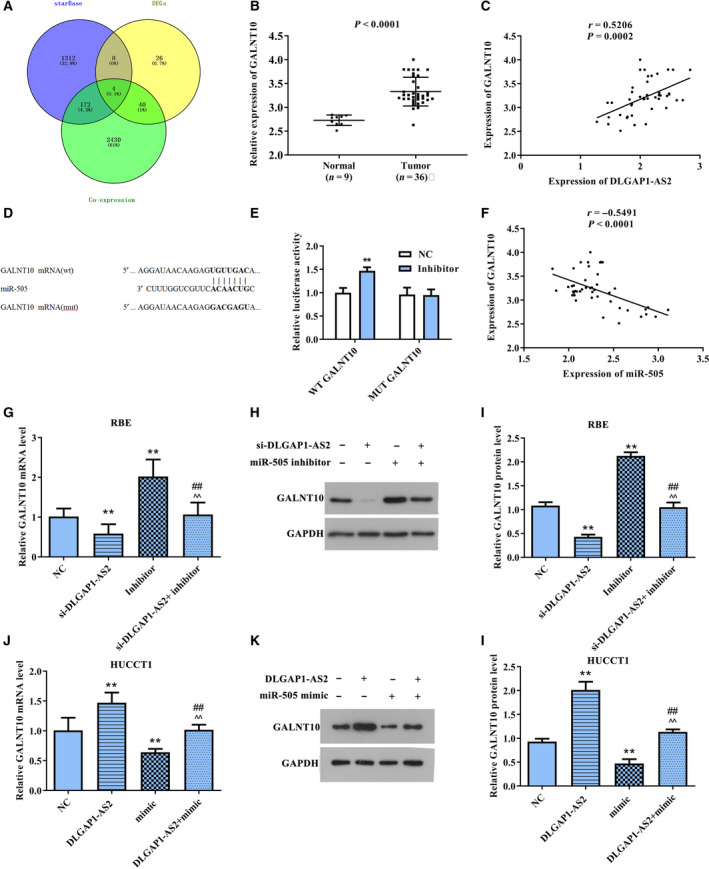
miR‐505 targeted GALNT10. (A) Wayne diagram was used to ascertain the possible targets of miR‐505. (B) Bioinformatics analysis of GALNT10 expression in CCA tissues and normal tissues. P < 0.0001. (C) The correlation between GALNT10 and DLGAP1‐AS2. R = 0.5206, P = 0.0002. (D) Comparison of miR‐505 with GALNT10 sequences. (E) Dual‐luciferase reporter assay was performed to confirm that miR‐505 directly binds to the 3′‐UTR of GALNT10. **P < 0.01. (F) The correlation between GALNT10 and DLGAP1‐AS2. R = 0.5206, P = 0.0002. (G–I) The impacts of si‐DLGAP1‐AS2, miR‐505 inhibitor and si‐DLGAP1‐AS2+miR‐505 inhibitor on GALNT10 mRNA and protein levels were detected using qRT‐PCR and western blotting. **P < 0.01 versus NC group, ## P < 0.01 versus si‐DLGAP1‐AS2 group, ^^P < 0.01 versus miR‐505 inhibitor group. (J–L) The impacts of DLGAP1‐AS2, miR‐505 mimic and DLGAP1‐AS2+miR‐505 mimic on GALNT10 mRNA and protein levels were detected using qRT‐PCR and western blotting. **P < 0.01 versus NC group, ## P < 0.01 versus DLGAP1‐AS2 group, ^^P < 0.01 versus miR‐505 mimic group. Error bar represents the mean ± SD derived from three independent experiments. Comparisons between groups were analyzed using t‐tests (two‐sided).
Because Pearson’s correlation analysis also revealed that GALNT10 expression was negatively interrelated to miR‐505 expression and positively correlated with DLGAP1‐AS2 (Fig. 3F), we further measured levels of GALNT10 in the si‐DLGAP1‐AS2 group, miR‐505 inhibitor group and si‐DLGAP1‐AS2+miR‐505 inhibitor group, as well as in the DLGAP1‐AS2 group, miR‐505 mimic group and DLGAP1‐AS2+miR‐505 mimic. As presented in Fig. 3G–I, si‐DLGAP1‐AS2 restrained GALNT10 expression, whereas miR‐505 inhibitor up‐regulated its expression. Importantly, the inhibitory effect caused by down‐regulation of DLGAP1‐AS2 was attenuated after cells cotransfected with si‐DLGAP1‐AS2 and miR‐505 inhibitors. Contrarily, the GALNT10 expression was markedly increased in DLGAP1‐AS2 overexpressed cells but was obviously decreased in miR‐505 up‐regulated cells. Also, overexpression of miR‐505 can reverse the DLGAP1‐AS2‐induced promotion of GALNT10 levels (Fig. 3J–L). These outcomes revealed that GALNT10 levels were mainly mediated by the modulation of miR‐505 through DLGAP1‐AS2.
DLGAP1‐AS2 promotes the malignant biological behaviors of CCA cells by increasing the GALNT10 level via restraining miR‐505
In view of the differential expression of DLGAP1‐AS2, miR‐505 and GALNT10, as well as their regulatory relationship, we conducted the functional experiments in vitro to verify the biological significance of the DLGAP1‐AS2/miR‐505/GALNT10 axis. CCK‐8 (Fig. 4A,B), colony formation (Fig. 4C,D) and Transwell assays (Fig. 4E,F) revealed that suppression of miR‐505 can markedly accelerate cell proliferation, migration and invasion compared with the NC group, whereas the phenotype of cells cotransfected with si‐DLGAP1‐AS2 and miR‐505 inhibitor was almost no different compared with the NC group. Also, after cells cotransfected with si‐DLGAP1‐AS2 and si‐GALNT10, the malignant biological behaviors of both HUCCT1 and RBE cells were significantly reduced compared with that of the NC group. These results demonstrated that DLGAP1‐AS2 facilitates the malignant progression of CCA cells via regulating the miR‐505/GALNT10 cascade.
Fig. 4.
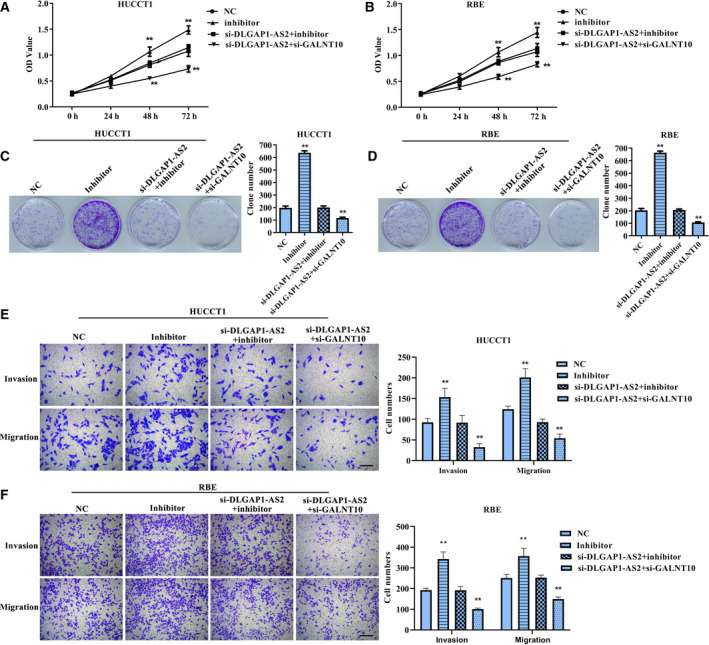
The impacts of miR‐505 inhibitor, si‐DLGAP1‐AS2+miR‐505 inhibitor and si‐DLGAP1‐AS2+si‐GALNT10 on malignant phenotype of HUCCT1 and RBE cells. (A,B) CCK‐8 assay for viability of HUCCT1 and RBE cells. **P < 0.01 versus NC group. (C,D) Colony formation assay for proliferation of HUCCT1 and RBE cells. **P < 0.01 versus NC group. (E,F) Transwell assay for migration and invasion of HUCCT1 and RBE cells. **P < 0.01 versus NC group. Scale bar: 200 µm. Error bar represents the mean ± SD derived from three independent experiments. Comparisons between groups were analyzed using t‐tests (two‐sided). OD, absorbance.
Discussion
To our knowledge, most early‐stage CCAs are asymptomatic [1]. The lack of effective biomarkers leads to the loss of the best treatment opportunity for most patients. Herein, we expounded the possibility of the DLGAP1‐AS2/miR‐505/GALNT10 axis as a diagnostic marker and therapeutic target for CCA. As a result, high expression of DLGAP1‐AS2 in both CCA tissues and cell lines was associated with poor prognosis. Also, knockdown of DLGAP1‐AS2 decreased the viability of CCA cells. Importantly, miR‐505 was negatively regulated by DLGAP1‐AS2, and it inhibited GALNT10 expression. Then, functional experiments in vitro revealed that inhibition of miR‐505 could reverse the facilitating effects of si‐DLGAP1‐AS2 on cell phenotype, while the malignant biological behaviors of cells cotransfected with si‐DLGAP1‐AS2 and si‐GALNT10 were significantly inhibited compared with that of the NC group.
Extensive research reported that the abnormal expression of lncRNA in malignant tumors is closely related to the tumorigenesis, development and prognosis of cancer [18]. Some lncRNAs are highly expressed in tumors, which presented the characteristics of oncogenes [19, 20, 21]. As a little‐reported lncRNA, DLGAP1‐AS2 was up‐regulated in CCA tissues and cell lines, and down‐regulation of DLGAP1‐AS2 significantly decreased the cell viability of CCA cell lines HUCCT1 and RBE. These findings suggest that DLGAP1‐AS2 has tumor‐promoting properties.
To further explore the functional mechanism of DLGAP1‐AS2, we searched for its target and found that DLGAP1‐AS2 directly targets miR‐505. In recent years, multiple studies indicated that numerous microRNAs are aberrantly expressed in CCA and functioned as oncogenes or tumor suppressors in the proliferation, apoptosis, migration and invasion of CCA. For example, miR‐21 is highly expressed in CCA, which enhances the proliferation but restrains the apoptosis of CCA cells [22]. Contrarily, miR‐186 is down‐regulated in CCA, and overexpression of miR‐186 can suppress the proliferation of CCA cells [23]. Recently, miR‐505 has been widely studied in cancer. It is generally considered that miR‐505 has tumor‐suppressor properties, which has been confirmed to be down‐regulated in colorectal cancer [24], gastric cancer [25], endometrial cancer [26], osteosarcoma [27] and hepatocellular carcinoma [28]. Nevertheless, the role of miR‐505 in CCA has not been elucidated. In this work, we have reached a consistent conclusion. miR‐505 was down‐regulated in CCA tissues based on public data, and inhibition of miR‐505 led to a marked increase in cell proliferation, migration and invasion. Importantly, inhibition of miR‐505 can also reverse the si‐DLGAP1‐AS2‐induced inhibitory effect on cell viability. These data revealed that the DLGAP1‐AS2/miR‐505 axis contributes to regulating the malignant progression of CCA.
Interestingly, we found a negative correlation between miR‐505 and GALNT10 expression in CCA tissues. GALNT10, a member of the GALNT family, has been studied in a variety of cancers [29]. GALNT10 can be applied as an independent prognostic factor for advanced ovarian serous carcinoma, which can predict poor prognosis and promote tumor progression [30]. Moreover, GALNT10 is up‐regulated via HBV‐mediated inhibition of miR‐122 and functioned as an oncogene in the progression of hepatocellular carcinoma [31]. Our data indicated that GALNT10 was overexpressed in CCA tissues and related to the poor outcomes of patients with CCA. Notably, a biological function experiment revealed that knockdown of GALNT10 and DLGAP1‐AS2 together inhibited the proliferation, migration and invasion of tumor cells.
In conclusion, we elucidated the new function of lncRNA DLGAP1‐AS2 in facilitating the malignant progression of CCA cells by modulating the miR‐505/GALNT10 axis. These findings revealed the significance of the lncRNA DLGAP1‐AS2/miR‐505/GALNT10 axis in the malignant progression of CCA and supply a novel target for the CCA diagnosis and therapy.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
ZL, LP and XD designed the study and collected data. ZL and XY analyzed the data. ZL and LP wrote the manuscript. ZL and XD reviewed and edited the manuscript. All authors read and approved the final manuscript.
Supporting information
Table S1. Patient information.
Data accessibility
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Razumilava N and Gores GJ (2014) Cholangiocarcinoma. Lancet 383, 2168–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chiang NJ, Shan YS, Hung W‐C and Chen L‐T (2015) Epigenetic regulation in the carcinogenesis of cholangiocarcinoma. Int J Biochem Cell Biol 67, 110–114. [DOI] [PubMed] [Google Scholar]
- 3. Rizvi S, Khan SA, Hallemeier CL, Kelley RK and Gores GJ (2018) Cholangiocarcinoma ‐ evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 15, 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khan SA, Toledano MB and Taylor‐Robinson SD (2008) Epidemiology, risk factors, and pathogenesis of cholangiocarcinoma. HPB 10, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doherty B, Nambudiri VE and Palmer WC (2017) Update on the diagnosis and treatment of cholangiocarcinoma. Curr Gastroenterol Rep 19, 2. [DOI] [PubMed] [Google Scholar]
- 6. Dykes IM and Emanueli C (2017) Transcriptional and post‐transcriptional gene regulation by long non‐coding RNA. Genom Proteomics Bioinformatics 15, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quinn JJ and Chang HY (2016) Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet 17, 47–62. [DOI] [PubMed] [Google Scholar]
- 8. Ransohoff JD, Wei Y and Khavari PA (2018) The functions and unique features of long intergenic non‐coding RNA. Nat Rev Mol Cell Biol 19, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F and Ochiya T (2018) Emerging roles of long non‐coding RNA in cancer. Cancer Sci 109, 2093–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi Y, Wang D, Wang J, Yu W and Yang J (2019) Long non‐coding RNA in the pathogenesis of cancers. Cells 8, 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Cai Q, Li W, Feng F and Yang L (2018) Long non‐coding RNA EPIC1 promotes cholangiocarcinoma cell growth. Biochem Biophys Res Commun 504, 654–659. [DOI] [PubMed] [Google Scholar]
- 12. Tang L, Wang Y, Wang H, Xu B, Ji H, Xu G, Ge X, Li Q and Miao L (2019) Long noncoding‐RNA component of mitochondrial RNA processing endoribonuclease is involved in the progression of cholangiocarcinoma by regulating microRNA‐217. Cancer Sci 110, 2166–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miao W, Li N, Gu B, Yi G, Su Z and Cheng H (2020) LncRNA DLGAP1‐AS2 modulates glioma development by up‐regulating YAP1 expression. J Biochem 167, 411–418. [DOI] [PubMed] [Google Scholar]
- 14. Ren P and Hu M (2019) A three long non‐coding RNA signature to improve survival prediction in patients with Wilms' tumor. Oncol Lett 18, 6164–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao L, Zhang X, Guo H, Liu M and Wang L (2020) LOXL1‐AS1 contributes to non‐small cell lung cancer progression by regulating miR‐3128/RHOXF2 Axis. Onco Targets Ther 13, 6063–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeng M, Zhu L, Li L and Kang C (2017) miR‐378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett 22, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhen S, Hua L, Liu YH, Sun XM, Jiang MM, Chen W, Zhao L and Li X (2017) Inhibition of long non‐coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget 8, 9634–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhan A, Soleimani M and Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77, 3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G and Song H (2018) LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol 96, 38–43. [DOI] [PubMed] [Google Scholar]
- 20. Jiang N, Meng X, Mi H, Chi Y, Li S, Jin Z, Tian H, He J, Shen W, Tian H et al (2018) Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin Chim Acta 486, 26–33. [DOI] [PubMed] [Google Scholar]
- 21. Zhang YX, Yuan J, Gao ZM and Zhang ZG (2018) LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci 22, 443–449. [DOI] [PubMed] [Google Scholar]
- 22. Wang LJ, He CC, Sui X, Cai MJ, Zhou CY, Ma JL, Wu L, Wang H, Han SX and Zhu Q (2015) MiR‐21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget 6, 5932–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang M, Shi B and Zhang K (2019) miR‐186 suppresses the progression of cholangiocarcinoma cells through inhibition of Twist1. Oncol Res 27, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Y, Bian L and Zhang Y (2018) MiR‐505 mediates methotrexate resistance in colorectal cancer by targeting RASSF8. J Pharm Pharmacol 70, 937–951. [DOI] [PubMed] [Google Scholar]
- 25. Tian L, Wang ZY, Hao J and Zhang XY (2018) miR‐505 acts as a tumor suppressor in gastric cancer progression through targeting HMGB1. J Cell Biochem 120, 8044–8052. [DOI] [PubMed] [Google Scholar]
- 26. Chen S, Sun KX, Liu BL, Zong ZH and Zhao Y (2016) MicroRNA‐505 functions as a tumor suppressor in endometrial cancer by targeting TGF‐α. Mol Cancer 15, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu YJ, Li W, Chang F, Liu JN, Lin JX and Chen DX (2018) MicroRNA‐505 is downregulated in human osteosarcoma and regulates cell proliferation, migration and invasion. Oncol Rep 39, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Ren L, Yao Y, Wang Y and Wang S (2019) MiR‐505 suppressed the growth of hepatocellular carcinoma cells via targeting IGF‐1R. Biosci Rep 39, BSR20182442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu L, Xiong Y, Xi W, Wang J, Qu Y, Lin Z, Chen X, Yao J, Xu J and Guo J (2017) Prognostic role of N‐Acetylgalactosaminyltransferase 10 in metastatic renal cell carcinoma. Oncotarget 8, 14995–15003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang G, Lu J, Yang M, Wang Y, Liu H and Xu C (2020) Elevated GALNT10 expression identifies immunosuppressive microenvironment and dismal prognosis of patients with high grade serous ovarian cancer. Cancer Immunol Immunother 69, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, Yang L, Fu Q, Xu J‐J and Gu J‐X (2015) Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA‐122 (miR‐122) axis in hepatitis B virus‐associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem 290, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


