Abstract
Background
Comorbid chronic conditions are common among people with type 2 diabetes. We developed an artificial intelligence algorithm, based on reinforcement learning (RL), for personalized diabetes and multimorbidity management, with strong potential to improve health outcomes relative to current clinical practice.
Methods
We modeled glycemia, blood pressure, and cardiovascular disease (CVD) risk as health outcomes, using a retrospective cohort of 16,665 patients with type 2 diabetes from New York University Langone Health ambulatory care electronic health records in 2009–2017. We trained an RL prescription algorithm that recommends a treatment regimen optimizing patients’ cumulative health outcomes using their individual characteristics and medical history at each encounter. The RL recommendations were evaluated on an independent subset of patients.
Results
The single-outcome optimization RL algorithms, RL–glycemia, RL–blood pressure, and RL–CVD, recommended consistent prescriptions as that observed by clinicians in 86.1%, 82.9%, and 98.4% of the encounters, respectively. For patient encounters in which the RL recommendations differed from the clinician prescriptions, significantly fewer encounters showed uncontrolled glycemia (A1c > 8% in 35% of encounters), uncontrolled hypertension (blood pressure > 140 mmHg in 16% of encounters), and high CVD risk (risk > 20% in 25% of encounters) under RL algorithms compared with those observed under clinicians (43%, 27%, and 31% of encounters, respectively; all p < 0.001).
Conclusions
A personalized RL prescriptive framework for type 2 diabetes yielded high concordance with clinicians’ prescriptions, and substantial improvements in glycemia, blood pressure, and CVD risk outcomes.
Electronic supplementary material
The online version of this article (10.1007/s40265-020-01435-4) contains supplementary material, which is available to authorized users.
Key Points
| Artificial intelligence (AI) prescription algorithms have been successfully applied to single disease problems, but previous applications have not considered comorbid conditions, pharmacological treatments, treatment histories, and other individual characteristics that are important for personalized diabetes management. |
| We trained and evaluated a series of AI algorithms to optimize patients’ glycemia, blood pressure, and CVD risk outcomes, either individually or jointly, using a retrospective cohort of type 2 diabetes patients from an ambulatory care electronic health records database (2009–2017). |
| When optimizing glycemia, blood pressure, and CVD risk individually, the algorithms consistently recommended prescriptions with clinicians’ decisions in 86.1%, 82.9%, and 98.4% of patient encounters. In cases where the AI recommendation differed from the clinicians’ prescriptions, health outcomes were significantly improved. |
| The RL algorithm can be integrated into electronic health record platforms to assist physicians with dynamic real-time suggestions on personalized treatment paths. |
Introduction
Comorbid chronic conditions are common among people with type 2 diabetes (T2DM) [1]. Hypertension (HTN) and atherosclerotic cardiovascular disease (CVD) are the two most common multimorbidities for T2DM patients [2]; Therefore, the need to address comorbid chronic conditions, in addition to patients’ diabetes-specific treatment goals [3], poses a substantial challenge for effective T2DM management. Although improvements in glycemic monitoring and control have been documented in several large systems of care, and more widespread use of treatments such as angiotensin-converting enzyme (ACE) inhibitors and aspirin have decreased patients’ risk of cardiovascular death, the current commonly used standard of care and guidelines are usually built around single diseases [4]. Despite the increasing numbers of patients with multimorbidity, such patients are usually excluded from randomized controlled trials [5–7]. A systematic review of managing patients with multimorbidity identified only 10 randomized trials worldwide and highlighted the paucity of research into interventions to improve outcomes for patients with multimorbidity [8]. On the other hand, there is a large volume of evidence suggesting that the response to T2DM treatment, HTN treatment, and CVD prevention differs between population subgroups [9, 10]. Therefore, the need for an individualized approach is especially pressing given the variety of comorbid conditions, pharmacological treatments, individual treatment histories, and other individual characteristics that may inform treatment selection.
We provide an artificial intelligence (AI) prescription algorithm, based on reinforcement learning (RL), which is able to dynamically suggest personalized optimal treatments for patients with T2DM to manage their multimorbidity based on evidence from patients’ electronic health records (EHRs). RL has been successfully applied in the past to single disease problems, such as blood glucose control [11], HIV therapy [12], cancer treatment [13], anemia treatment in hemodialysis patients [14], treatment strategies for sepsis in intensive care [15], and a personalized regimen of sedation dosage and ventilator support for patients in intensive care units (ICUs) [12]. Prescriptive algorithms using regression trees and nearest neighbors (kNN) have previously shown great potential in personalized diabetes management [16, 17].
Our approach leverages the power of RL and abundant data in the EHR system to dynamically recommend treatment prescriptions, which are personalized based on patient characteristics, including age, sex, race, body mass index (BMI), blood pressure (BP), laboratory tests, duration of T2DM, and treatment history. In our setting, we first applied RL to optimize glycemic control, BP control, and CVD prevention separately, and then studied the potential of RL for multimorbidity management by optimizing all three outcomes jointly. We evaluated the effectiveness of the personalized treatment recommendations made by RL against the observed clinicians’ treatment by estimating patient outcomes based on the outcomes of similar patients in the EHR database.
Research Design and Methods
Study Design and Participants
We used ambulatory care EHR samples for T2DM patients from New York University Langone Health (NYULH–EHR) to derive and validate the RL algorithm. Eligible patients had had at least one encounter with an NYULH ambulatory primary care physician between 2009 and 2017 and had been selected by a T2DM rule-based phenotyping algorithm, defined as the following criteria: (1) had at least two encounters with an International Classification of Diseases, Tenth Revision (ICD-10) code for T2DM; (2) had two or more abnormal hemoglobin A1c (A1c; ≥ 6.5%) and at least one encounter with an ICD-10 code for T2DM; or (3) had a prescription for a T2DM medication, excluding metformin and acarbose. We excluded patients seen for consultation only and patients in emergency department, inpatient, or specialist settings, as these lacked consistent documentation of T2DM across encounters. We randomly selected 60% of the eligible patients as the training cohort to develop the RL algorithm, and reserved the remaining 40% of patients as the test cohort to evaluate the performance of the RL algorithm. This study was approved by the NYULH Institutional Review Board, and the data were de-identified to ensure anonymity.
For each patient, we had access to demographic data, including age, sex, race, ethnicity, and smoking status, as well as the following biomarkers: systolic BP (SBP), diastolic BP (DBP), BMI, HbA1c, total cholesterol (TC), low-density lipoprotein (LDL), high-density lipoprotein (HDL), creatinine, triglycerides, and estimated glomerular filtration rate (eGFR). In NYULH–EHR, 1% of samples had missing vitals, including BPs and BMI, 8% had missing HbA1c, 5–32% had missing renal function biomarkers, and 13% had missing lipid biomarkers. Following on from the work of Lundberg et al. [18], we imputed the missing patients’ biomarkers based on the observed values measured in previous encounters.
Medication prescriptions were first grouped by therapeutic class codes of antihyperglycemic, antihypertensive, and lipid-lowering, then analyzed by pharmacologic subclass. The antihyperglycemic therapeutic class contains nine pharmacologic subclasses, including the peroxisome proliferator-activated receptor (PPAR) agonist thiazolidinedione (PPARg), insulin-release stimulant type (INSR), incretin mimetic (glucagon-like peptide 1 receptor agonist; GLP1), dipeptidyl peptidase-4 (DPP4) inhibitor and biguanide (DPP4-BIG), DPP-4 inhibitors (DPP4), biguanide type (BIG), insulin-release stimulant and biguanide (INSR-BIG), sodium-glucose cotransporter-2 inhibitors (SGLT2), and insulins (INSO). The antihypertensive therapeutic class contains 10 pharmacologic subclasses, including angiotensin receptor antagonists (ARAs), potassium-sparing diuretics in combination (PSD), α/β-adrenergic blocking agents (ABAB), ACE inhibitor with thiazide or thiazide-like diuretic (ACE-TD), ARAs with thiazide diuretic (ARA-TD), ACE inhibitors (ACE), thiazide and related diuretics (TD), β-adrenergic blocking agents (BAB), calcium channel blocking agents (CCB), and ARAs with CCBs (ARA-CCB). The antihyperlipidemic therapeutic class contains five pharmacologic subclasses, including bile salt sequestrants (BSS), HMG-CoA reductase inhibitors (HMG), HMG-CoA reductase inhibitors and cholesterol absorption inhibitors (HMG-CA), proprotein convertase subtilisin/kexin type 9 inhibitors (PCSK9), and lipotropics (LIP).
Overview of the Reinforcement Learning Algorithm
RL algorithms model the course of patients' EHR histories, which includes prescriptions, biomarkers, and health outcomes changing over time using a Markov decision process with key elements, including state, action, and reward [15, 19]. In this setting, ‘state’ refers to the observed patient demographics, laboratory test results at the current encounter, and their histories of laboratories tests and prescriptions. ‘Action’ refers to the prescribed treatment regimen at the current encounter, which are pharmacologic subclasses or their combinations. The result of an action is a numerical reward representing the improvement of health outcomes compared with the previous encounter. The cumulative reward is defined as the sum of the rewards along the course of EHR encounter records. RL has been well-established as an efficient AI learning algorithm to maximize cumulative reward by selecting an optimal action at each encounter through a learning algorithm called Deep Q Networks [20, 21] with a multilayer (deep) neural network. An important advantage of RL is that the action in every encounter is personalized to the patient's individual characteristics as they are observed, in a way that optimizes the cumulative reward. In this paper, we focus on glycemia control (lowering A1c towards 6.5%), BP control (lowering SBP towards 120 mmHg), and CVD prevention (minimizing CVD risk). We first optimized each outcome individually using three separate RL algorithms, referred to as RL–glycemia, RL–BP, and RL–CVD. We then trained a multimorbidity management RL algorithm (RL–multimorbidity) to optimize glycemia, BP and CVD risk simultaneously. The details of state, action, and reward are described as follows
State: A list of observed patient characteristics, including age, sex, race, smoking status; vitals and laboratory test values at current encounter and in the past 6 months, including BMI, weight, SBP, DBP, triglycerides, TC, HDL, LDL, A1c, and creatinine; prescription history in the past 6 months; and encounter histories, including days since the previous encounter and days since the first encounter.
Action: The action space consists of the pharmacologic subclasses and their combinations, referred to as the treatment regimen. The action space of RL–glycemia contains nine pharmacologic subclasses in the antihyperglycemic therapeutic class, or their combinations; the action space of RL–BP contains 10 pharmacologic subclasses in the antihypertensive therapeutic class, or their combinations; the action space of RL–CVD contains five pharmacologic subclasses in the antihyperlipidemic therapeutic class, or their combinations; and the action space of RL–multimorbidity contains pharmacologic subclasses in all three therapeutic classes, or their combinations.
Reward: The reward of a prescription is a numeric measure of treatment efficacies between two consecutive encounters. For RL–glycemia, if A1c <5.6% in both encounters, their rewards are zero, otherwise the reward is defined by the reduction in A1c. For RL–BP, if patients have no HTN symptoms (< 120 mmHg) in both encounters, the reward is zero, otherwise it is equal to the decrease in SBP. For RL–CVD, the reward is the reduction in global CVD Framingham Risk Score (FRS) [22], which is a function of age, TC, HDL, SBP, treatment for HTN, smoking, and T2DM status (all yes). Sex-specific risk equations were applied to males and females separately. For RL–multimorbidity, the reward is defined as the average of standardized rewards values of RL–BP, RL–glycemia, and RL–CVD (model and training details are shown in the electronic supplementary materials).
Model Evaluation
We evaluated the RL-recommended therapy by comparing its effect with the observed clinicians’ prescriptions on the test cohort of NYULH-EHR samples. In each encounter, the RL algorithm recommends a treatment regimen for the patient. If the recommendation is the same as the observed clinicians’ prescriptions in the data, we noted that RL is ‘consistent’ with the clinicians’ prescriptions. When RL is discrepant with the clinicians’ prescriptions, the efficacy of the RL-recommended treatment is not directly observed. For this reason, we imputed the outcome of the RL-recommended treatment using kNN regression, an approach commonly used for causal inference in observational studies [23]. In short, the imputation works by averaging the outcomes of the most similar patient encounters, in terms of patient characteristics, in which the RL-recommended therapy had been administered by clinicians. The similarity between patient encounters was estimated by Euclidean distance, as in the study by Bertsimas et al. [16]. To assess the performance of the imputation, we first compared imputed outcomes with observed outcomes under clinicians’ treatments, and found 87–95% correlation between them, indicating that the imputation algorithm can effectively estimate unobserved health outcomes (Table 1). We varied the number k of nearest neighbors and found the performance of the imputation (for any of the three health outcomes) was insensitive when k was between 8 and 10. We estimated the efficacy of the recommendations made by RL, first in the whole set of test samples, and then for individual sex, racial, and age subgroups.
Table 1.
Counterfactual outcome versus true clinical outcome comparison based on kNN regression
| Biomarkers | Counterfactual outcome | True outcome | Pearson correlation |
|---|---|---|---|
| BP systolic | 128.55 (0.017) | 128.68 (0.022) | 0.89 |
| BP diastolic | 74.27 (0.010) | 74.29 (0.014) | 0.89 |
| Triglycerides | 152.85 (0.118) | 153.74 (0.14) | 0.87 |
| Total cholesterol | 174.05 (0.056) | 174.31 (0.061) | 0.93 |
| HDL cholesterol | 51.37 (0.022) | 50.37 (0.024) | 0.95 |
| LDL Cholesterol | 92.73 (0.047) | 92.85 (0.052) | 0.92 |
| A1c | 7.02 (0.002) | 7.05 (0.002) | 0.92 |
kNN k nearest neighbor, BP blood pressure, HDL high-density lipoprotein, LDL low-density lipoprotein
Feature Importance
To better understand which features have the most impact on treatment recommendations, we used SHAP (SHapley Additive exPlanations) [24, 25] to estimate and rank the contributions of clinician features explaining RL and clinician prescriptions.
Results
Overall, 16,665 patients in NYULH ambulatory care EHR samples had a query-based T2DM diagnosis in 2009 to 2017, with 1,278,785 encounters (median 12 encounters per patient). The number of T2DM patients was robust to variations in the T2DM phenotyping algorithm resulting from changes in the required number of encounters with T2DM ICD-10 codes and the medications. The demographic and clinic characteristics of the analysis cohort are shown in Table 2. Overall, patients were 65.6 years of age and comprised 8278 females (54.6%). On average, T2DM patients showed A1c of 7.1% and SBP of 128.9 mmHg. Antihyperglycemic, antihypertensive, and antihyperlipidemic medications were prescribed in 665,768 (52.1%), 849,328 (66.4%), and 428,427 (33.5%) encounters, respectively. The median follow-up time was 2.6 years since T2DM diagnosis (interquartile range [IQR] 1.9–3.9 years). We first trained the RL algorithms using 530,786 (60%) T2DM patient encounters, and then assessed their performance using the remaining 394,447 (40%) T2DM patient encounters.
Table 2.
Demographics and clinical characteristics of NYULH-EHR patients with type 2 diabetes
| Demographics and clinical characteristics | No. of patients [N = 16,665] |
|---|---|
| Age, years [mean (SD)] | 65.62 (13.66) |
| Male [n (%)] | 6876 (45.37) |
| Race [n (%)] | |
| African American | 5146 (33.96) |
| Native American | 55 (0.36) |
| Asian | 692 (4.57) |
| Caucasian (White) | 7888 (52.05) |
| Smoker, ever and current [n (%)] | 1043 (6.88) |
| Systolic blood pressure, mmHg [mean (SD)] | 128.93 (14.60) |
| Diastolic blood pressure, mmHg [mean (SD)] | 74.19 (8.88) |
| Body mass index, kg/m2 [mean (SD)] | 31.56 (6.86) |
| Triglycerides, mg/dL [mean (SD)] | 155.06 (91.97) |
| Creatinine, mg/dL [mean (SD)] | 1.02 (0.44) |
| Total cholesterol, mg/dL [mean (SD)] | 173.37 (39.82) |
| Low-density lipoproteins, mg/dL [mean (SD)] | 91.99 (33.53) |
| High-density lipoproteins, mg/dL [mean (SD)] | 51.00 (15.25) |
| A1c, % [mean (SD)] | 7.11 (1.46) |
| Medications | No. of patient encounters (n = 1,278,785) |
|---|---|
| Antihyperglycemic class [n (%)] | 665,768 |
| Biguanide type (BIG) | 250,438 (37.62) |
| Insulin-release stimulant type (INSR) | 110,139 (16.54) |
| Insulins (INSO) | 106,356 (15.97) |
| DPP-4 inhibitors (DPP4) | 64,090 (9.63) |
| DPP-4 inhibitor and biguanide (DPP4-BIG) | 53,337 (8.01) |
| Incretin mimetic (GLP-1 receptor agonist; GLP1) | 35,696 (5.36) |
| Sodium-glucose cotransporter-2 inhibitors (SGLT2) | 23,021 (3.46) |
| PPAR agonist thiazolidinedione (PPARg) | 12,573 (1.89) |
| Insulin-release stimulant and biguanide (INSR-BIG) | 10,118 (1.52) |
| Antihypertensive class [n (%)] | 849,328 |
| β-adrenergic blocking agents (BAB) | 200,114 (23.56) |
| Calcium channel blocking agents (CCB) | 151,701 (17.86) |
| ACE inhibitors (ACE) | 149,561 (17.61) |
| Angiotensin receptor antagonists (ARA) | 138,705 (16.33) |
| Angiotensin receptor antagonists with thiazide diuretic (ARA-TD) | 67,964 (8.00) |
| α/β-adrenergic blocking agents (ABAB) | 57,426 (6.76) |
| Thiazide and related diuretics (TD) | 57,196 (6.73) |
| ACE inhibitor with thiazide or thiazide-like diuretic (ACE-TD) | 14,486 (1.71) |
| Potassium-sparing diuretics in combination (PSD) | 6246 (0.74) |
| Angiotensin receptor antagonists with calcium channel blocking agents (ARA-CCB) | 5929 (0.70) |
| Lipid-lowering class [n (%)] | 428,427 |
| HMG-CoA reductase inhibitors (HMG) | 379,924 (88.68) |
| Lipotropics (LIP) | 40,173 (9.38) |
| Bile salt sequestrants (BSS) | 5262 (1.23) |
| HMG-CoA reductase inhibitors and cholesterol absorption inhibitors (HMG-CA) | 2286 (0.53) |
| Proprotein convertase subtilisin/kexin type 9 inhibitors (PCSK9) | 782 (0.18) |
Categorical variables are expressed as frequency (%) unless otherwise indicated, and continuous variables are expressed as the mean (SD) of biomarkers
NYULH-EHR New York University Langone Health electronic health record, SD standard deviation, DPP-4 dipeptidyl peptidase-4, GLP-1 glucagon-like peptide 1, PPAR peroxisome proliferator-activated receptor, ACE angiotensin-converting enzyme
The performance of the RL algorithms on the test dataset is summarized in Table 3. The RL–glycemia algorithm was consistent with clinicians' prescriptions in 86.1% of encounters. In the remaining 15,578 (13.9%) encounters, the mean A1c under clinician prescription was 8.09% (95% confidence interval [CI] 8.06–8.12), while the mean A1c under RL–glycemia was 7.80% (95% CI 7.78–7.82), showing a 0.30% (95% CI 0.28–0.32) reduction (p < 0.001). Significantly fewer encounters showed uncontrolled A1c (A1c >8%) under RL–glycemia than under clinicians (35% vs. 43%, p < 0.001). The RL–BP algorithm was consistent with clinicians’ prescriptions in 82.9% of encounters. In the remaining 20,251 encounters (17.1%) with discrepant recommendations, RL–BP achieved a 0.58 mmHg (95% CI 0.37–0.79) reduction in SBP relative to clinicians' prescriptions (131.77 vs. 132.35 mmHg, p < 0.001). Fewer encounters showed uncontrolled HTN (SBP >140 mmHg) under RL–BP than under clinicians' prescriptions (16% vs. 27%, p < 0.001). The RL–CVD was consistent with clinicians’ prescriptions in 98.4% of encounters. In the remaining 946 encounters (1.6%) with discrepant recommendations from RL and clinicians’, the mean FRS reduced 3.53% (95% CI 2.94–4.12) under RL–CVD compared with clinicians’ prescriptions (13.65% vs. 17.18%, p < 0.001), with fewer encounters showing high FRS risk (> 20%; 25% vs. 31%, p < 0.01). Collectively, these results showed high concordance between the optimized RL algorithms and clinicians’ prescriptions for single-target management for patients with T2DM. However, there were more frequent discrepancies between RL–multimorbidity and clinicians. The RL–multimorbidity algorithm was consistent with clinicians' prescriptions in 71.1% of encounters. In the remaining 102,184 encounters (28.9%) with discrepant prescriptions, 16,436 (16.1%), 9800 (9.6%), and 48,283 (47.3%) encounters had uncontrolled A1c, uncontrolled HTN, and high FRS risk that was significantly lower than observed outcomes under clinicians’ prescriptions (20.4%, 20.5%, and 54.8%, respectively).
Table 3.
Performance of RL algorithms with comparison between RL and clinicians for glycemic control, hypertension control, and CVD prevention
| RL–glycemia | |||
| Encounters for which the algorithm’s recommendation differed from the observed clinician's prescription [n (%)] | 15,578 (13.9) | ||
| RL–glycemia | Clinician's prescription | p-Value | |
| A1c [mean (SE)] | 7.80 (0.01) | 8.09 (0.01) | < 0.001 |
| A1c >8% [n (%)] | 5421 (34.8) | 6617 (42.5) | < 0.001 |
| RL–BP | |||
| Encounters for which the algorithm’s recommendation differed from the observed clinician's prescription [n (%)] | 20,251 (17.1) | ||
| RL–BP | Clinician's prescription | p-Value | |
| SBP [mean (SE)] | 131.77 (0.06) | 132.35 (0.11) | < 0.001 |
| SBP >140 mmHg [n (%)] | 3256 (16.1) | 5390 (26.6) | < 0.001 |
| RL–CVD | |||
| Encounters for which the algorithm’s recommendation differed from observed clinician's prescription (N(%)) | 946 (1.6) | ||
| R–CVD | Clinician's prescription | p-Value | |
| FHS [mean (SE)] | 13.65 (0.26) | 17.18 (0.36) | < 0.001 |
| FHS >20% [n (%)] | 237 (25.1) | 299 (31.6) | < 0.001 |
| RL–multimorbidity | |||
| Encounters for which the algorithm’s recommendation differed from the observed clinician's prescription [n (%)] | 102,184 (28.9) | ||
| RL–multimorbidity | Clinician's prescription | p-Value | |
| A1c [mean (SE)] | 7.14 (0.003) | 7.19 (0.005) | < 0.001 |
| A1c >8% [n (%)] | 16,436 (16.08) | 20,879 (20.43) | < 0.001 |
| SBP [mean (SE)] | 129.40 (0.03) | 129.58 (0.05) | < 0.001 |
| SBP >140 mmHg [n (%)] | 9800 (9.59) | 20,957 (20.51) | < 0.001 |
| FHS [mean (SE)] | 21.89 (0.04) | 25.61 (0.05) | < 0.001 |
| FHS >20% [n (%)] | 48,283 (47.3) | 55,957 (54.8) | < 0.001 |
RL reinforcement learning, CVD cardiovascular disease, SE standard error, BP blood pressure, SBP systolic blood pressure
To understand when and how RL makes different prescriptions from clinicians, Table 4 compares consistent and discrepant encounters by patient demographics and clinical characteristics. The most significantly associated factor was severity at the time of the encounter. For RL–glycemia, encounters with higher A1c were more likely to have different recommendations (average A1c 8.1% for discrepant encounters vs. 7.5% for consistent encounters, p < 0.001). For RL–BP, encounters with higher SBP were more likely to have different recommendations (average SBP 132.85 vs. 131.00 mmHg, p < 0.001).
Table 4.
Comparison of RL and clinicians for glycemic control, BP, and CVD prevention.
| Features | T2DM [n = 15,578] | HTN [n = 20,251] | CVD [n = 946] | Multimorbidity [n = 102,184] | ||||
|---|---|---|---|---|---|---|---|---|
| Prescription consistency (%) | No (13.89) | Yes (86.11) | No (17.08) | Yes (82.82) | No (1.63) | Yes (98.37) | No (28.88) | Yes (71.12) |
| Age, years | 65.89 (13.77) | 64.25 (13.69) | 69.42 (12.54) | 68.79 (12.79) | 68.39 (11.65) | 68.87 (12.10) | 66.24 (13.36) | 65.87 (13.64) |
| Males, % | 47.21 | 45.34 | 43.39 | 43.65 | 54.50 | 46.41 | 45.59 | 44.89 |
| Ethnicity (%) | ||||||||
| Black | 33.53 | 34.61 | 31.15 | 32.45 | 16.27 | 26.85 | 33.40 | 33.68 |
| Native American | 0.51 | 0.44 | 0.28 | 0.23 | 0.53 | 0.32 | 0.39 | 0.37 |
| Asian | 4.47 | 4.26 | 4.04 | 3.88 | 3.57 | 4.14 | 4.54 | 4.32 |
| White | 53.91 | 52.07 | 57.80 | 56.62 | 72.88 | 61.49 | 53.30 | 53.18 |
| Smokers, % | 6.98 | 6.82 | 5.64 | 6.01 | 8.99 | 6.14 | 6.89 | 6.67 |
| SBP, mmHg | 127.59 (14.35) | 127.28 (13.73) | 132.85 (16.68) | 131.00 (14.99) | 125.72 (14.08) | 127.52 (13.63) | 131.10 (15.89) | 128.65 (14.30) |
| DBP, mmHg | 74.38 (8.68) | 74.13 (8.51) | 75.25 (10.12) | 74.36 (9.27) | 73.63 (8.26) | 73.47 (8.28) | 74.13 (9.52) | 74.02 (8.72) |
| BMI, kg/m2 | 31.84 (6.92) | 32.01 (7.19) | 32.33 (6.88) | 31.49 (6.81) | 29.98 (5.69) | 30.76 (6.63) | 32.06 (6.67) | 31.51 (6.94) |
| Triglycerides, mg/dL | 163.57 (104.98) | 157.51 (96.43) | 155.81 (86.83) | 150.20 (81.06) | 197.54 (163.01) | 159.31 (97.69) | 159.19 (94.16) | 154.27 (89.03) |
| Creatinine, mg/dL | 0.96 (0.38) | 0.99 (0.41) | 1.05 (0.44) | 1.07 (0.48) | 1.05 (0.43) | 1.02 (0.43) | 1.06 (0.45) | 1.02 (0.45) |
| Total-C, mg/dL | 172.25 (39.40) | 172.97 (38.85) | 173.76 (38.87) | 172.81 (38.75) | 180.62 (46.24) | 176.05 (42.67) | 170.21 (39.66) | 173.57 (39.70) |
| LDL-C, mg/dL | 90.81 (32.68) | 91.27 (32.80) | 93.00 (33.05) | 92.03 (32.70) | 93.38 (37.30) | 93.46 (35.92) | 89.77 (33.36) | 92.04 (33.45) |
| HDL-C, mg/dL | 49.59 (15.07) | 50.89 (15.56) | 50.20 (14.71) | 51.32 (15.31) | 49.77 (16.04) | 51.46 (14.86) | 49.29 (14.34) | 51.29 (15.34) |
| A1c, % | 8.11 (1.81) | 7.51 (1.62) | 6.95 (1.30) | 6.84 (1.25) | 6.85 (1.29) | 6.82 (1.24) | 7.09 (1.38) | 7.08 (1.43) |
Demographic characteristics of patients having encounters at which RL and clinicians prescribed consistently versus differently. Categorical variables are expressed as frequency (%), and continuous variables are expressed as the mean (SD) of biomarkers
RL reinforcement learning, BP blood pressure, CVD cardiovascular disease, T2DM type 2 diabetes mellitus, HTN hypertension, SBP systolic blood pressure, DBP diastolic blood pressure, BMI body mass index, C cholesterol, LDL low-density lipoprotein, HDL high-density lipoprotein, SD standard deviation
The efficacy of the RL prescriptive algorithms was consistently observed across T2DM patients, and sex, racial, and age subgroups (Tables 5, 6, 7). Specifically, African American T2DM patients, and T2DM patients aged older than 60 years, observed higher efficacies from the RL algorithms than clinicians’ prescriptions compared with the observed efficacies in White patients and patients aged 60 years and younger. For example, A1c under RL–glycemia for African American patients was 0.39% lower than under clinicians’ treatment. In contrast, A1c under RL–glycemia was 0.28% lower than under clinicians’ treatment for White patients. Patients aged 60 years and younger observed higher efficacy, with A1c under RL–glycemia 0.47% lower than that under clinicians’ treatment, than those older than 60 years of age, with A1c under RL–glycemia 0.19% lower than that under clinicians’ treatment.
Table 5.
Subgroup results of the glycemic control RL algorithm
| Subgroup | No. of encounters | RL benefit relative to clinician policy | ||
|---|---|---|---|---|
| A1c under RL | A1c under clinician | Benefit | ||
| Male | 7072 | 7.87 (0.01) | 8.20 (0.02) | − 0.33 (0.02) |
| Female | 8506 | 7.73 (0.01) | 8.00 (0.02) | − 0.27 (0.02) |
| Age > 60 years | 9548 | 7.63 (0.01) | 7.82 (0.02) | − 0.19 (0.01) |
| Age ≤ 60 years | 6030 | 8.06 (0.02) | 8.53 (0.03) | − 0.47 (0.02) |
| White ethnicity | 8427 | 7.54 (0.01) | 7.81 (0.02) | − 0.28 (0.02) |
| Black ethnicity | 5181 | 8.16 (0.02) | 8.55 (0.03) | − 0.39 (0.02) |
| Other ethnicity | 1970 | 7.94 (0.03) | 8.10 (0.04) | − 0.16 (0.04) |
| Smoker | 1026 | 8.08 (0.04) | 8.40 (0.06) | − 0.32 (0.05) |
| Non-smoker | 14552 | 7.78 (0.01) | 8.07 (0.01) | − 0.30 (0.01) |
RL reinforcement learning
Table 6.
Subgroup results of the BP control RL algorithm
| Subgroup | No. of encounters | RL benefit relative to clinician policy | ||
|---|---|---|---|---|
| SBP under RL | SBP under clinician | Benefit | ||
| Male | 8108 | 131.32 (0.09) | 132.45 (0.17) | − 1.13 (0.17) |
| Female | 12,143 | 132.07 (0.08) | 132.29 (0.14) | − 0.22 (0.14) |
| Age >60 years | 16,151 | 131.43 (0.07) | 132.34 (0.12) | − 0.90 (0.12) |
| Age ≤60 years | 4100 | 133.12 (0.13) | 132.43 (0.25) | 0.68 (0.24) |
| White ethnicity | 11,925 | 130.35 (0.07) | 131.22 (0.14) | − 0.87 (0.14) |
| Black ethnicity | 6536 | 134.19 (0.11) | 135.12 (0.20) | − 0.93 (0.19) |
| Other ethnicity | 1790 | 132.41 (0.23) | 129.79 (0.39) | 2.62 (0.38) |
| Smoker | 951 | 132.34 (0.31) | 132.55 (0.54) | − 0.21 (0.53) |
| Non-smoker | 19,300 | 131.74 (0.06) | 132.35 (0.11) | − 0.60 (0.11) |
BP blood pressure, RL reinforcement learning, SBP systolic blood pressure
Table 7.
Subgroup results of the multimorbidity control RL algorithm
| Subgroup | No. of encounters | RL benefit relative to clinician policy (standard of care) | ||||||
|---|---|---|---|---|---|---|---|---|
| A1c | SBP | Triglycerides | Total cholesterol | LDL cholesterol | HDL cholesterol | CVD risk | ||
| Male | 43,816 | − 0.09 (0.01) | − 0.32 (0.07) | − 5.27 (0.50) | − 0.10 (0.17) | 0.08 (0.14) | 0.71 (0.06) | − 5.09 (0.09) |
| Female | 58,368 | − 0.02 (0.01) | − 0.07 (0.06) | − 1.99 (0.33) | − 1.23 (0.16) | − 0.61 (0.14) | − 0.41 (0.06) | − 2.68 (0.05) |
| Age >60 years | 75,924 | 0.01 (0.00) | − 0.59 (0.05) | − 0.43 (0.29) | − 0.05 (0.13) | 0.27 (0.12) | − 0.24 (0.05) | − 5.70 (0.06) |
| Age ≤60 years | 26,260 | − 0.23 (0.01) | 1.02 (0.09) | − 11.97 (0.72) | − 2.75 (0.25) | − 1.99 (0.21) | 0.98 (0.08) | 2.03 (0.07) |
| White ethnicity | 60,029 | − 0.02 (0.00) | − 0.02 (0.06) | − 3.78 (0.37) | − 1.60 (0.15) | − 1.12 (0.13) | 0.16 (0.06) | − 4.04 (0.07) |
| Black ethnicity | 31,775 | − 0.12 (0.01) | − 0.92 (0.09) | 1.79 (0.47) | − 0.52 (0.22) | − 0.43 (0.19) | − 0.64 (0.08) | − 3.39 (0.08) |
| Other ethnicity | 10,380 | − 0.02 (0.01) | 1.17 (0.15) | − 17.02 (1.08) | 3.50 (0.39) | 4.70 (0.33) | 1.78 (0.13) | − 2.82 (0.14) |
| Smoker | 5747 | − 0.10 (0.02) | − 0.52 (0.20) | − 16.54 (1.71) | − 1.31 (0.55) | − 0.73 (0.46) | 2.14 (0.17) | − 10.41 (0.26) |
| Non-smoker | 96,437 | − 0.05 (0.00) | − 0.16 (0.05) | − 2.61 (0.28) | − 0.71 (0.12) | − 0.29 (0.10) | − 0.05 (0.05) | − 3.31 (0.05) |
RL reinforcement learning, SBP systolic blood pressure, LDL low-density lipoprotein, HDL high-density lipoprotein, CVD cardiovascular disease
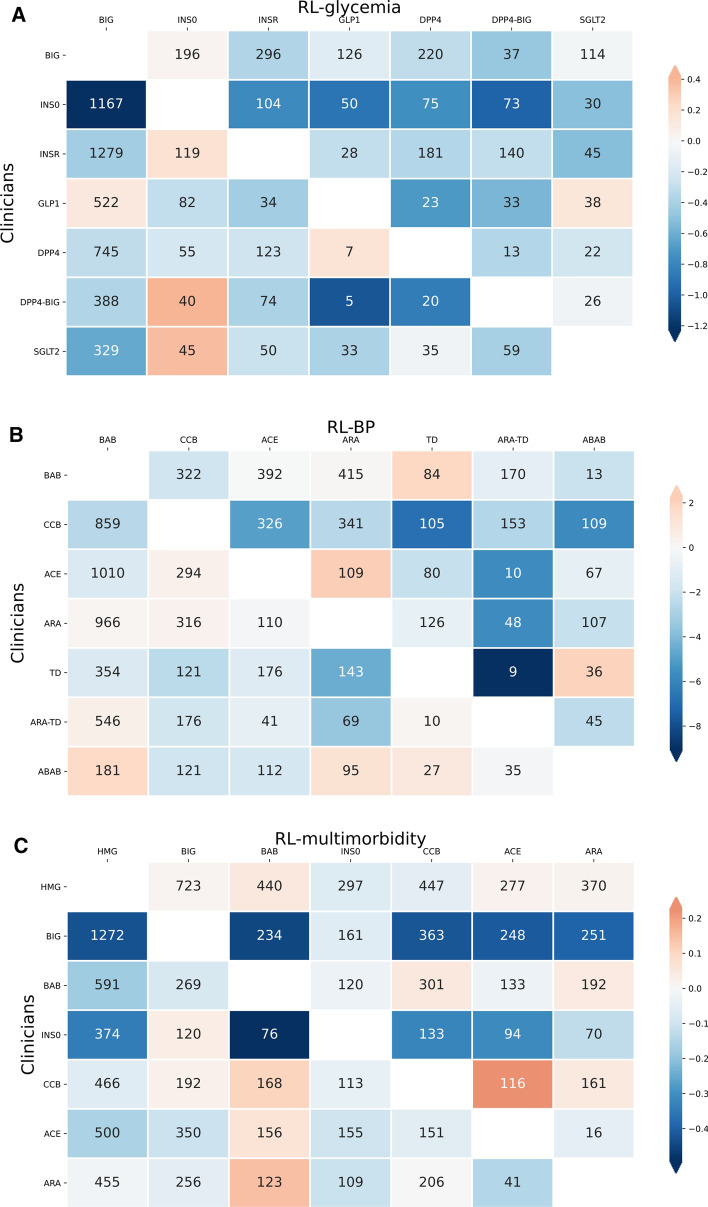
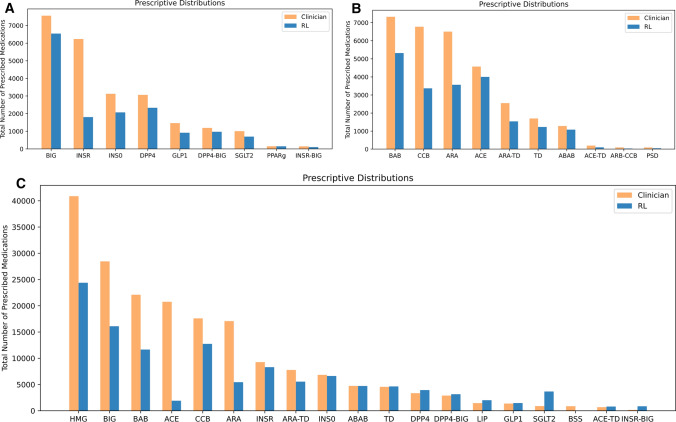
The patterns of different treatment recommendations, along with the resulting differences in health outcomes, for RL–glycemia, RL–BP, and RL–multimorbidity, are illustrated in Fig. 1. In the case of RL–glycemia, the most frequently observed discrepancy (1167 encounters) was that clinicians prescribed insulin monotherapy (INSO) while RL prescribed biguanide type (BIG). On these encounters, RL–glycemia achieved, on average, 1.22% lower A1c than clinicians. In the case of RL–BP, the most frequently observed discrepancy (1010 encounters) was that clinicians prescribed ACE inhibitors (ACE), while RL prescribed BABs. On these encounters, RL–BP achieved a 6.78 mmHg lower SBP. The most frequently observed discrepancy between RL–multimorbidity and clinicians’ prescription was biguanide type (BIG) prescribed by clinicians, and HMG-CoA reductase inhibitors (HMG) prescribed by RL–multimorbidity, observed in 1272 patient encounters. On these discrepant encounters, RL–multimorbidity achieved a 0.15% higher A1c but 2.42% lower CVD risk and 0.30 mmHg lower SBP. Overall, RL algorithms tended to prescribe fewer medications than clinicians (Fig. 2).
Fig. 1.
Patterns of the most frequent discrepant RL recommendations and clinicians’ prescriptions for (a) RL–glycemia, (b) RL–BP, and (c) RL–multimorbidity. Each cell and the numbers represent patients for whom RL (labels on the x axis) recommended a different regimen from the regimen given by clinicians (labels on the y axis). The color in each cell quantifies the improvement in health outcomes achieved by the RL recommendation relative to the clinician’s prescription, with blue indicating benefits of the RL recommendation and orange indicating worsening outcomes relative to the clinician’s prescription. (a) Indicates the mean A1c reduction (%) of RL–glycemia (labels on the x axis) compared with clinicians (labels on the y axis); (b) indicates the mean SBP decrease (mmHg) of RL–BP (labels on the x axis) compared with clinicians (labels on the y axis); and (c) indicates the mean difference of multimorbidity reward from RL–multimorbidity (labels on the x axis) compared with clinicians (labels on the y axis). RL–CVD was consistent with clinicians’ prescriptions for the vast majority of encounters, and thus was not shown in this figure. RL reinforcement learning, SBP systolic blood pressure
Fig. 2.
Prescription medication use by RL versus clinicians. Total number of drugs prescribed for (a) blood glucose control, (b) BP control, and (c) multimorbidity management. RL reinforcement learning
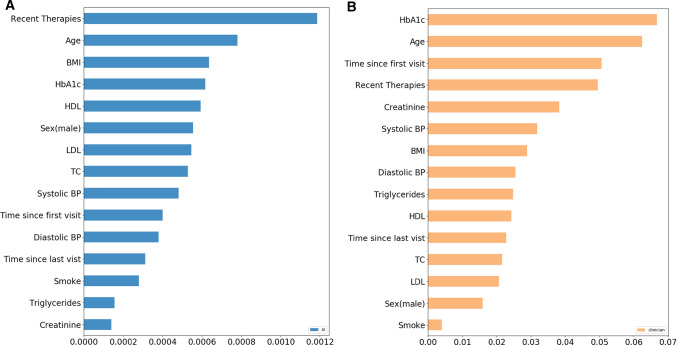
Figure 3 shows the importance of features associated with the RL–multimorbidity algorithm and clinicians’ prescriptions. In general, there was reasonable agreement between the feature importance estimates of RL–multimorbidity and those identified by the clinicians. A1c is the most important feature for clinicians, while RL–multimorbidity was most influenced by recent therapies, age, BMI, and A1c. One difference is the importance of creatinine in the clinicians’ prescriptions, but it was not as important for RL–multimorbidity. Another difference is the reduced role of the time since first encounter in RL–multimorbidity compared with clinicians’ prescriptions.
Fig. 3.
Feature importance of (a) RL–multimorbidity and (b) clinician prescription. RL reinforcement learning, BMI body mass index, HDL high-density lipoprotein, LDL low-density lipoprotein, TC total cholesterol, BP blood pressure
Discussion
To our best knowledge, this is the first RL-assisted prescriptive algorithm for personalized single and multimorbidity outcome management for patients with T2DM. Using an EHR database, the developed RL algorithm can efficiently recommend treatment regimens to optimize patient health outcomes incorporating their individual demographic and treatment history. Compared with other machine-learning methods, the RL approach has a particular advantage as it can efficiently learn complex dynamic drug–disease and drug–drug interactions in the presence of high temporal variation, uncertain outcomes, and long-term treatment effects [15, 19]. RL recommendations showed high levels of concordance with clinicians’ prescriptions for single outcome optimizations of glycemia, BP, and CVD risk control. This demonstrates the feasibility of using RL for T2DM management, and indicates that clinicians make near-optimal decisions with regard to single-outcome management.
RL–multimorbidity recommendations showed more frequent discrepancy with clinicians' prescriptions as well as the recommendations by single-outcome RL algorithms. This provides data-driven evidence that optimizing multimorbidity management is different from optimizing single outcomes in parallel. For example, in the 1272 patient encounters with the most frequently observed discrepancy between RL–multimorbidity and clinicians, their average A1c was 7.0%, SBP was 127.2 mmHg, and CVD risk was 12.6%. For these encounters, clinicians prescribed BIG to prioritize glycemic control, while RL–multimorbidity prescribed HMG for lipid-lowering. This indicates challenges and uncertainties of multimorbidity management for patients with borderline and balanced levels of severities in multiple chronic conditions [26, 27]. RL–multimorbidity showed overall improvements in managing the three outcomes simultaneously, significantly reducing the number of encounters with uncontrolled glycemia, uncontrolled HTN, and high FRS CVD risk.
Although both clinicians and RL–multimorbidity place high importance on similar factors, these factors are ranked differently. RL algorithms did not weigh features that were not included in the reward functions, such as creatinine, as much as clinicians who consider it an important renal function biomarker. This indicates a potential challenge of the RL algorithms using single-directed reward outcomes as the optimization goal. Ideally, a comprehensive reward function should incorporate domain knowledge and adverse events, such as hypoglycemia and kidney comorbidity, to achieve optimized outcomes while balancing the risks of adverse events [28].
Typical limitations with EHR data are their unobserved medication adherence, partially observed clinical data at each encounter, and uncontrolled time span between encounters [29]. However, the RL algorithms were designed to incorporate these uncertainties under real-world scenarios. In particular, if there were observable patient characteristics that were associated with higher non-adherence to a certain treatment leading to lower levels of efficacy, RL would be able to identify this and prescribe different treatments for patients with those characteristics.
Although our evaluation methodology controls for several confounding factors that could explain differences in treatment effects, we can only estimate counterfactual outcomes under RL recommendations for patients with discrepant prescriptions. In addition, the T2DM patient population from NYULH ambulatory care may not be representative of the United States T2DM population. To ultimately validate the efficacy of the RL algorithms, randomized clinical trials with patients randomly assigned to RL and clinician mechanism would be needed.
Conclusions
In this study, we demonstrated the feasibility of using RL prescriptive algorithms for patients with T2DM to manage their multimorbidity based on test data from an ambulatory care center. The RL–glycemia, RL–BP, and RL–CVD algorithms showed high concordance (83–98%) with clinicians’ prescriptions, while RL–multimorbidity showed relatively low concordance (71%) for multimorbidity management. For patient encounters in which the RL recommendations differed from the clinician prescriptions, RL prescriptions showed significantly improved health outcomes compared with clinicians’ prescriptions. Potentially, the algorithm can be integrated into EHR platforms to assist physicians for T2DM management with dynamic real-time suggestions of personalized treatment paths.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Hua Zheng, Ilya O. Ryzhov, Wei Xie, and Judy Zhong report no conflicts of interest. Judy Zhong is funded by NIA R01AG054467 and NIA R01AG065330.
Author Contributions
WX, JZ, and IOR initiated the study. WX, IOR, and HZ designed the data analyses, algorithm, and experiments. JZ provided the EHR data, clinical assessment, interpretation of subgroup efficacy, model performance and feature importance, and connections with clinicians’ workflow. HZ wrote the paper in conjunction with JZ All authors have read and approved the final manuscript, contributing edits where applicable. WX and JZ take full responsibility for the work, including the study design, access to data, and the decision to submit and publish the manuscript.
Declaration
Funding
JZ is funded by NIA R01AG054467 and NIA R01AG065330.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose
Footnotes
The original article has been updated: Due to co-corresponding author name inclusion.
Change history
2/25/2021
A Correction to this paper has been published: 10.1007/s40265-021-01484-3
Contributor Information
Wei Xie, Email: w.xie@northeastern.edu.
Judy Zhong, Email: judy.zhong@nyulangone.org.
References
- 1.Druss BG, Marcus SC, Olfson M, Tanielian T, Elinson L, Pincus HA. Comparing the national economic burden of five chronic conditions. Health Aff (Millwood). 2001;20(6):233–241. doi: 10.1377/hlthaff.20.6.233. [DOI] [PubMed] [Google Scholar]
- 2.Petrie JR, Guzik TJ, Touyz RM. Diabetes, hypertension, and cardiovascular disease: clinical insights and vascular mechanisms. Can J Cardiol. 2018;34(5):575–584. doi: 10.1016/j.cjca.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piette JD, Kerr EA. The impact of comorbid chronic conditions on diabetes care. Diabetes Care. 2006;29(3):725–731. doi: 10.2337/diacare.29.03.06.dc05-2078. [DOI] [PubMed] [Google Scholar]
- 4.Whitty CJM, MacEwen C, Goddard A, Alderson D, Marshall M, Calderwood C, et al. Rising to the challenge of multimorbidity. BMJ. 2020;368:l6964. doi: 10.1136/bmj.l6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett K, Mercer SW, Norbury M, Watt G, Wyke S, Guthrie B. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. Lancet. 2012;380(9836):37–43. doi: 10.1016/S0140-6736(12)60240-2. [DOI] [PubMed] [Google Scholar]
- 6.Moffat K, Mercer SW. Challenges of managing people with multimorbidity in today's healthcare systems. BMC Fam Pract. 2015;16:129. doi: 10.1186/s12875-015-0344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Spall HG, Toren A, Kiss A, Fowler RA. Eligibility criteria of randomized controlled trials published in high-impact general medical journals: a systematic sampling review. JAMA. 2007;297(11):1233–1240. doi: 10.1001/jama.297.11.1233. [DOI] [PubMed] [Google Scholar]
- 8.Smith SM, Soubhi H, Fortin M, Hudon C, O'Dowd T. Managing patients with multimorbidity: systematic review of interventions in primary care and community settings. BMJ. 2012;345:e5205. doi: 10.1136/bmj.e5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: A report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. Circulation. 2019;140(11):e596–e646. doi: 10.1161/CIR.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burt VL, Whelton P, Roccella EJ, Brown C, Cutler JA, Higgins M, et al. Prevalence of hypertension in the US adult population. Results from the Third National Health and Nutrition Examination Survey, 1988–1991. Hypertension. 1995;25(3):305–313. doi: 10.1161/01.hyp.25.3.305. [DOI] [PubMed] [Google Scholar]
- 11.Tejedor M, Woldaregay AZ, Godtliebsen F. Reinforcement learning application in diabetes blood glucose control: A systematic review. Artif Intell Med. 2020;104:101836. doi: 10.1016/j.artmed.2020.101836. [DOI] [PubMed] [Google Scholar]
- 12.Ernst D, Stan G, Goncalves J, Wehenkel L, editors. Clinical data based optimal STI strategies for HIV: a reinforcement learning approach. Proceedings of the 45th IEEE Conference on Decision and Control; 13–15 Dec 2006: San Diego, CA.
- 13.Zhao Y, Zeng D, Socinski MA, Kosorok MR. Reinforcement learning strategies for clinical trials in nonsmall cell lung cancer. Biometrics. 2011;67(4):1422–1433. doi: 10.1111/j.1541-0420.2011.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escandell-Montero P, Chermisi M, Martínez-Martínez JM, Gómez-Sanchis J, Barbieri C, Soria-Olivas E, et al. Optimization of anemia treatment in hemodialysis patients via reinforcement learning. Artif Intell Med. 2014;62(1):47–60. doi: 10.1016/j.artmed.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med. 2018;24(11):1716–1720. doi: 10.1038/s41591-018-0213-5. [DOI] [PubMed] [Google Scholar]
- 16.Bertsimas D, Kallus N, Weinstein AM, Zhuo YD. Personalized diabetes management using electronic medical records. Diabetes Care. 2017;40(2):210–217. doi: 10.2337/dc16-0826. [DOI] [PubMed] [Google Scholar]
- 17.Bertsimas D, Dunn J, Mundru N. Optimal prescriptive trees. INFORMS J. Optim. 2019;1(2):164–183. doi: 10.1287/ijoo.2018.0005. [DOI] [Google Scholar]
- 18.Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2(10):749–760. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutton RS, Barto AG. Reinforcement learning: an introduction. 2. Cambridge: The MIT Press; 2018. [Google Scholar]
- 20.Mnih V, Kavukcuoglu K, Silver D, Graves A, Antonoglou I, Wierstra D et al. Playing atari with deep reinforcement learning. INIPS Deep Learning Workshop; 2013.
- 21.Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, Bellemare MG, et al. Human-level control through deep reinforcement learning. Nature. 2015;518(7540):529–533. doi: 10.1038/nature14236. [DOI] [PubMed] [Google Scholar]
- 22.D'Agostino RB, Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, et al. General cardiovascular risk profile for use in primary care: the Framingham heart study. Circulation. 2008;117(6):743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 23.Imbens G, Rubin DB. Causal inference for statistics, social, and biomedical sciences: an introduction. Cambridge: Cambridge University Press; 2015. [Google Scholar]
- 24.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Proceedings of the 31st International Conference on Neural Information Processing Systems; Long Beach: Curran Associates Inc.; 2017. pp. 4768–77.
- 25.Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2(10):749–760. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Groot V, Beckerman H, Lankhorst GJ, Bouter LM. How to measure comorbidity: a critical review of available methods. J Clin Epidemiol. 2003;56(3):221–229. doi: 10.1016/S0895-4356(02)00585-1. [DOI] [PubMed] [Google Scholar]
- 27.Huntley AL, Johnson R, Purdy S, Valderas JM, Salisbury C. Measures of multimorbidity and morbidity burden for use in primary care and community settings: a systematic review and guide. Ann Fam Med. 2012;10(2):134–141. doi: 10.1370/afm.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mataric MJ. Reward functions for accelerated learning. Machine learning proceedings 1994. Amsterdam: Elsevier; 1994. pp. 181–189. [Google Scholar]
- 29.Bosworth HB, Granger BB, Mendys P, Brindis R, Burkholder R, Czajkowski SM, et al. Medication adherence: a call for action. Am Heart J. 2011;162(3):412–424. doi: 10.1016/j.ahj.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.