Abstract
Synthetic lethal screens are used to discover new combination treatments for cancer. In traditional high-throughput synthetic lethal screens, compounds are tested at a single dose, and hit selection is based on threshold activity values from the variance of the efficacy of the compounds tested. The limitation of the single-dose screening for synthetic lethal screens is that it does not allow for the robust detection of differential activities from compound collections with a broad range of potencies and efficacies. There is therefore a need to develop screening approaches that enable the identification of compounds with synthetic lethal effects based on changes in both potency and efficacy. Here we describe the implementation of a dose response–based synthetic lethal screen to find drugs that enhance or mitigate the cytotoxic effect of an immunotoxin protein (HA22). We developed a data analysis framework for the selection of compounds with enhancing or mitigating cytotoxic activities based on the use of dose-response parameters. The data analysis framework includes an ensemble ranking approach that allows the use of multiple dose-response parameters in a nonparametric fashion. Quantitative high-throughput screening (HTS) enables the identification of compounds with synthetic lethal activity not identified by single-dose HTS.
Keywords: qHTS, immunotoxin, synthetic lethal screen, ranking
Introduction
The identification of compounds that have a synthetic lethal effect in tumor cells is driving the search for combination therapies for the treatment of cancer.1–3 High-throughput screening (HTS) using cell proliferation assays has been implemented to identify compounds with differential activity in the presence of sublethal doses of a chemotherapeutic treatment or in paired isogenic cell lines with driver oncogenic mutations.4 In a traditional HTS campaign, compounds are tested at one concentration (e.g., 10 μM) to make the implementation of these large screens practical and economically feasible. The differential cytotoxic activity of a compound between two treatments or matched cell lines is then determined by calculating a ratio of normalized percent viability in each condition (e.g., chemotherapeutic treatment vs. no treatment or wild type vs. mutated gene). This analytical method is based on the differential efficacy of the compound, that is, the ability to distinguish the different inhibitory effect of compounds on cell viability in each screening arm. However, this method does not distinguish differential effects of compounds that are fully toxic in both conditions at the dose of compounds screened, even though their potencies (IC50) might be several fold different and, therefore, could potentially be used to selectively enhance killing at lower doses than those chosen in a screen.5,6 Quantitative HTS (qHTS) tests compounds in a dose-response format and fits the results to dose-response curves, which are then analyzed to assign a heuristic curve class (CC), summarizing the quality of the curve fit, IC50, maximum response (MAXR), and area under the curve (AUC) values for each compound in each assay condition.7,8 The CC also classifies the dose responses based on the nature of the asymptotes, efficacy, and goodness of fit (R2).9 However, CCs can be confounded by a variety of factors because of poor curve fits, mostly due to compounds with small efficacies, bell-shaped curves due to insolubility or other artifacts at the higher doses tested, and, to a lesser extent, outlying data points. In an ideal case, a compound would exhibit similar curves in the two treatments and only differ in their potencies (potency shift). More commonly, however, compounds will exhibit both potency (IC50) and efficacy shifts under each treatment condition. The AUC is used as an alternative parameter that can be calculated from a dose-response curve and implicitly considers both potency and efficacy.10 However, this parameter is still not an ideal solution because the AUC for a compound exhibiting a shift in potency but no shift in efficacy can be the same as (or similar to) the AUC for a compound exhibiting a shift in efficacy but not a shift in potency.5,6 Hit selection methods that combine the different dose-response curve parameters should therefore be developed and tested to ensure that relevant compounds are selected from chemical synthetic lethal screens. In practice, a combination of methods to help prioritize differentially active compounds, together with the manual inspection of the dose responses of the selected compounds at each key step of the selection process, is critical for the final selection of synthetic lethal compound candidates.
We implemented dose response–based qHTS to identify compounds that produce an enhanced or reduced cytotoxic effect in the presence of an immunotoxin (HA22)11,12 engineered to target CD22-expressing Nalm6 cells, which are used as a model of acute lymphoblastic leukemia (ALL). This engineered CD22-targeting toxin is currently in clinical development for the treatment of hairy cell leukemia and other B-cell malignancies, including ALL.13,14 The qHTS was implemented using a library of 466 compounds that have been clinically approved or are being developed for the treatment of cancer, so that compounds identified from the screen can potentially be efficiently developed into effective clinical combinations.15 This library of compounds includes both general cytotoxic (e.g., cisplatin, gemcitabine, doxorubicin) as well as targeted (e.g., kinase inhibitors targeting PI3K, mTOR, EGFR) compounds. The library exhibits redundancy for some target classes, such that there are several compounds with the same primary target. In addition to identifying new combinations for existing drugs, the biological annotation of the compounds in this collection allowed us to further investigate pathways involved in enhancement and resistance to the immunotoxin treatment.
Initial experiments indicated that the Nalm6 cells were very susceptible to the cytotoxic effect of the compounds in this oncology collection. Thus, it was difficult to distinguish between the cytotoxicity induced by the compound itself and the enhancement of immunotoxin cytotoxicity when the compounds were screened in single-dose format. We used the qHTS approach in conjunction with a combination of hit selection methods in an attempt to better distinguish differential cytotoxic effects of the compounds from the primary qHTS. Here we describe the data obtained from a cell viability synthetic lethal screen of the MIPE (Mechanism Interrogation PlatE) oncology collection and analytical methods that were used to identify compounds belonging to target classes that significantly enhanced or mitigated the cytotoxic effect of HA22.
Materials and Methods
A 1536-Well Cell Viability Assay for qHTS
Nalm6 cells were grown in phenol red–free RPMI with L-glutamine, 1× penicillin/streptomycin (Gibco, ThermoFisher Scientific, Fair Lawn, NJ). A library of 466 oncology small-molecule chemotherapeutics was tested using a cell viability assay in a 1536-well format as described previously.15 Briefly, 5 μL of a cell suspension (200 cells/μL) in media with 50 ng/mL HA22, 1 ng/mL HA22, or no immunotoxin added was dispensed to each well (1000 cells per well) of a white, solid, 1536-well microplate (catalog number 789173-F; Greiner Bio-One, Monroe, NC) with a Multidrop Combi Reagent dispenser and a small pin cassette (Thermo Scientific, Fisher Scientific, Fair Lawn, NJ). Then, 23 nL of compound solution in DMSO was added using a pintool dispenser (Kalypsys, San Diego, CA). Bortezomib (2 mM stock in DMSO, final 9 μM) was dispensed as positive control for cell death. Next, 23 nL of a plain DMSO solution was added to those wells that did not receive compound so that all wells in a plate had the same amount of DMSO in each of them. Cells were incubated with the compounds and HA22 (or phosphate-buffered saline [PBS]) for 48 h at 37 °C and 5% CO2. Then, 4 μL of CellTiterGlo solution (Promega, Madison, WI) was added to the cells, and after a 15-min incubation at 25 °C, luminescence was measured using a ViewLux reader (PerkinElmer, Waltham, MA).
Data Preprocessing and Curve Fitting
The raw relative luminescence units (RLUs) from the ViewLux plate reader were first normalized to the positive controls (0% viability, 9 μM bortezomib) and negative controls (100% viability, plain DMSO). Given a RLU value for a well, denoted as c, the normalized value was generated using 100(c − n)/(n − i) + 100, where n and i represent the median of the negative and positive control wells, respectively. Compounds were tested at 11 doses starting at 46 μM and using 3-fold dilutions. Percent viability data from each compound dose were then fitted to a four-parameter Hill curve using a custom algorithm.9 The curve fit analysis generates many parameters, including a calculated IC50; MAXR, which is the measured percentage viability at the highest concentration (46 μM) tested; and the AUC. The AUC was calculated using the trapezoidal rule over all measured responses.16 Finally, the qHTS analysis algorithm also assigns each dose response a CC (see Southall et al.17 for a detailed discussion on definition of curve classes) that allows for the rapid classification of inactive and active compounds, as well as to separate complete dose-response curves from partial ones. In this work, we consider active compounds those with CCs ±1.1, ±1.2, ±2.1, or ±2.2 (here, a negative value corresponds to cytotoxic compound and a positive value corresponds to a compound increasing cell proliferation), and inactive compounds are those with a curve class of 4. The rest of the compounds with CCs ±1.3, ±1.4, ±2.3, ±2.4, 3, and 5 were considered to have inconclusive activity and were labeled as “Other.” These are rather restrictive constraints, but its use does not affect the underlying methods employed as it simply defines the working set of compounds.
Analysis of Differential Activities
Each treatment data set was compared with a corresponding untreated arm screened the same day to minimize differences in cell responses due to cell culture conditions on different days. In the first analysis method, we identified “obviously” differentially active compounds as those that exhibited an inactive curve in the vehicle (PBS) treatment (CC 4) and an active curve in the HA22 treatment (CCs −1.1, −1.2, −2.1, and −2.2 for enhancers and CCs 1.1, 1.2, 2.1, and 2.2 for mitigators). For the second method, we considered differences or fold changes (i.e., ratio) of several individual curve parameters between treatment and vehicle, and then the differences or fold changes were scaled using the formula (x − mean)/standard deviation, using the values of each differential parameter for the set of 466 tested compounds as the reference population.18 A scaled value of 3 (enhancers) or −3 (mitigators) was used as a cutoff to select differentially active compounds. For the retest data, because the sample population was enriched for selected active hits, the data were scaled using the activity of the 466 compounds from the primary screen as the reference population, and the formula (x − median)/mean absolute deviation (MAD) was used to calculate the scaled values. This scaling mechanism allows us to compare different parameters (or rather their differences or ratios as the case may be) on the same scale. For this analysis, we used the three individual dose-response parameters, MAXR, log IC50, and AUC. Since log IC50 is calculated from dose-response fits (see above), compounds that had no activity and for which a dose response could not be fitted did not have a log IC50 and were discarded from the analysis. For log IC50 and MAXR, we evaluated the difference between the parameter value in PBS- and HA22-treated cells. For AUC, we calculated a ratio between HA22 treatment and vehicle and then log2 transformed the values of the ratios. Log2 ratio instead of differences for AUC was chosen because it appeared to provide better scaling for the differentiation between treatments. Compounds that enhanced the cytotoxic effect of HA22 had positive scaled values, and those that mitigated had negative scaled values. In a third analysis approach, we used an ensemble ranking method19,20 rather than a selection based on individual parameters. In this ensemble ranking method, we first computed fold changes or differences (as described above) between the control and treatment arms for multiple parameters, including log AC50, MAXR, and AUC. The compounds were first ranked by each fold change variable. Then a ranking for each compound was obtained by computing the product of its rank in each variable. This rank product approach21 is a convenient way to compute an ensemble ranking over a set of disparate ranks. Intuitively, a compound that is ranked highly by multiple parameters would be expected to have a high rank overall. A simple permutation test can also be performed to compute an empirical p-value (although a method to compute exact p-values based on an analytical form for the rank statistic has also been proposed22). The final ensemble rank is then used to order the data set. Finally, a fourth method to determine differential activity of the compounds was used that calculated the differences in percent viability between the two treatments (HA22 vs. vehicle) at each compound concentration screened, and then the differences were scaled as described above at each concentration using the differences in percent viability for the whole set of 466 compounds at each dose as the respective reference population. Scaled values of more than 3 (enhancers) and less than −3 (mitigators) were used as a cutoff to select those compounds with a differential effect at any one of the concentrations tested. For the retest data, because the sample population was enriched for selected active hits, scaling of delta mean percent activity per dose was calculated at each dose using the activity of the 466 compounds at each dose from the primary screen as reference population. We considered this approach since it is equivalent to hit selection from multiple single-point screens, run at different concentrations.
Target Enrichment Analysis
Given a selection of compounds, we identified the primary targets, based on manual curation of the literature, for these compounds and computed the enrichment for each target compared with background, using the Fisher exact test.23 For this test, the background was defined as all the targets annotated in the MIPE oncology collection.
Results
A cell viability assay was developed in a 1536-well microtiter plate format for Nalm6 cells using the CellTiterGlo reagent, which measures cellular metabolic adenosine triphosphate (ATP) levels, after a 48-h incubation with compound. A library of 466 oncology chemotherapeutic agents was tested at 11 doses (starting at 46 μM followed by 3-fold dilutions) using two sublethal doses of H22A immunotoxin: a dose of 1 ng/mL, which produced approximately an IC30 (low dose), and a dose of 50 ng/mL, which produced an IC80 (high dose). The data quality for the treatment and vehicle-treated plates was good, with average Z′ factors24 of 0.75 and 0.5 for the low and high H22A treatment, respectively (see Suppl. Fig. S1 for plots of median RLU and Z′ factors for each plate and each condition of the screen).
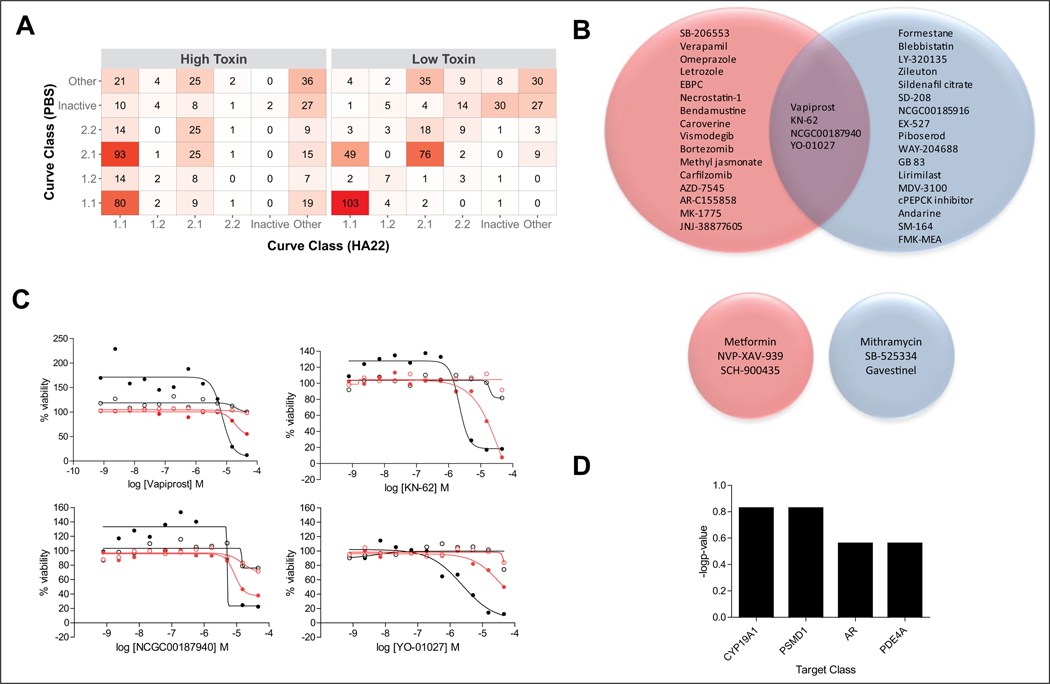
The results from the dose-response quantitative qHTS were analyzed to assign CCs as described in Materials and Methods. Supplemental Table S1 shows the number of compounds in each CC for each HA22 treatment screen and respective vehicle arm. Figure 1A displays a confusion table summarizing the concordance between the CC values for the HA22 versus control arms, in the high and low HA22 dose screens. Compounds that had differential cytotoxic activity between HA22 treated and untreated cells were first selected based on CC assignments. There were 20 and 21 enhancer compounds (CCs −1.1, −1.2, −2.1, and −2.2 for the treated arm and CC 4 for the untreated arm) for the high- and low-dose HA22 dose conditions, respectively. Four of those compounds were active in both dose conditions (Fig. 1B, top), including vapiprost, a prostanoid TP antagonist; KN-62, a calmodulin-dependent protein kinase II inhibitor; NCGC-00187940, a 15-lipoxygenase inhibitor; and YO-01027, a γ-secretase inhibitor. Dose responses of these four compounds in the different screening conditions are shown in Figure 1C. In general, these compounds have no activity in the untreated arm and are very efficacious in the HA22 treatment arm (complete killing at the highest dose tested), although they are weak in potency. There were three and three mitigating compounds (CCs 1.1, 1.2, 2.1, and 2.2 for HA22-treated arm and CC 4 for the untreated arm) for the high- and low-dose HA22 dose conditions, respectively. There was no overlap between these two sets of three compounds (Fig. 1B, bottom). The target redundancy for many of the compounds in the collection allowed us to evaluate whether there was target enrichment in the selected hits. Target analysis of the 37 unique compounds identified as HA22 enhancers show that aromatase inhibitors (CYP19A1), proteasome (PSDM1) inhibitors, androgen receptor (AR) antagonists, and phosphodiesterase (PDE4A) inhibitors were enriched, although not in a statistically significant manner (p-values ranging from 0.15–0.25) (Fig. 1D).
Figure 1.
Selection of compounds based on curve class (CC) analysis. (A) Correlation table of number of compounds in each CC for the synthetic lethal quantitative high-throughput screening (qHTS) for HA22 enhancers and mitigators. The higher the number of compounds in a CC correlation box, the darker the red color. Each CC bin includes both positive and negative curve classes. The “Other” bin includes CCs ±1.3, ±1.4, ±2.3, ±2.4, ±3, and 5. (B) Venn diagram of enhancer hits selected based on HA22-treated CCs −1.1, −2.1, −1.2, and −2.2 and vehicle-treated CC 4 (top Venn diagrams), as well as mitigator hits selected based on HA22-treated CCs 1.1, 2.1, 1.2, and 2.2 and vehicle-treated CC 4 (bottom Venn diagrams). Red circles correspond to high HA22 dose treatment arm, and blue circles correspond to low HA22 dose treatment arm. (C) Dose responses of the four enhancer compounds that overlap between low and high HA22 dose treatment. Solid black circles are high-dose HA22 treatment arm; open black circles are for the corresponding vehicle arm; solid red circles are low-dose HA22 treatment arm; open red circles are the corresponding vehicle arm. (D) Bar graph of the target classes enriched (−log p-value) for the 36 enhancer hits identified using the CC method. Complete list of hit compounds is included in supplemental material.
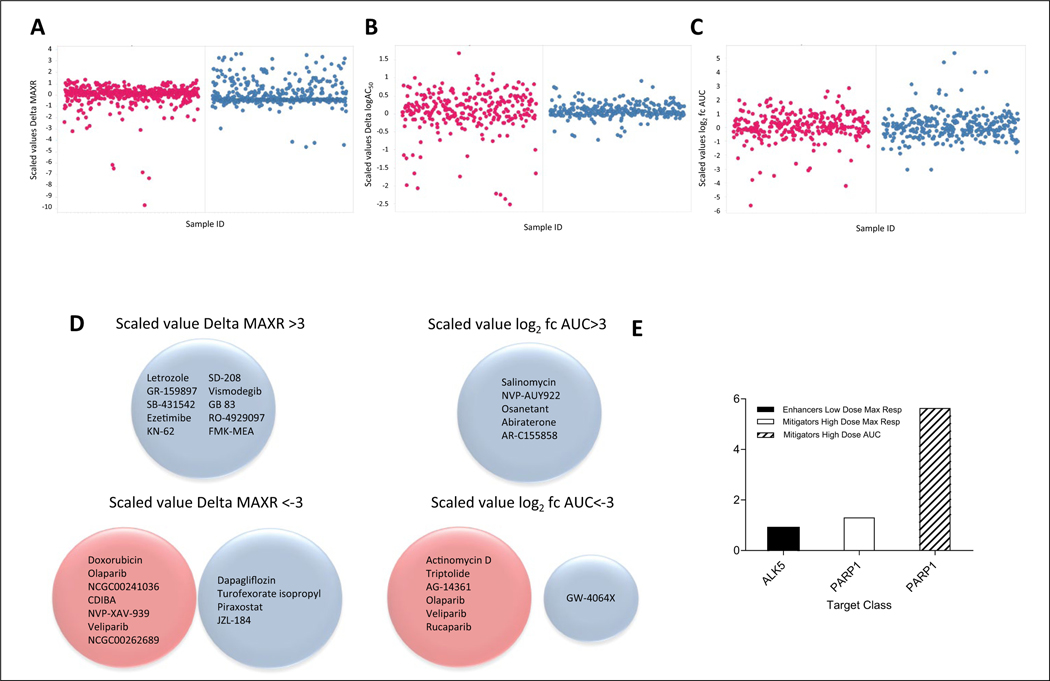
Additional analysis in search of compounds with differential activity in the presence of HA22 treatment was done using three dose-response curve parameters: MAXR, log IC50, and AUC. Because the log IC50 and AUC are obtained from dose response fitting, compounds for which dose responses cannot be fitted because of lack of activity will not have a log IC50 and AUC value. Furthermore, the algorithm at times generates IC50 values for compounds that are essentially inactive, and therefore these values are artifactual. Therefore, for the log IC50 and AUC analysis, we only included compounds that have robust dose responses (CCs −1.1, −1.2, –2.1, and −2.2 for enhancers or 1.1, 1.2, 2.1, and 2.2 for mitigators). To calculate differential activities, each of the differences (MAXR and log IC50) or ratio (AUC) between HA22-treated and control arms for each compound and each parameter was calculated, and then a scaled value was computed based on the total number of compounds for which there was a value for the parameter as described in Materials and Methods. The scaled values allowed us to use the same arbitrary cutoff of ±3 to identify differentially active compounds. Figure 2A–C shows the plots for the scaled values of the difference (MAXR and log IC50) or fold change AUC for the low- and high-dose HA22 treatment screens. Ten compounds had a scaled value >3 (enhancers of HA22 effect) by difference in MAXR, all in the low-dose HA22 treatment screen (Fig. 2A,D). Eleven compounds were also mitigators of the cytotoxic effect of the HA22 (scaled value <−3) by MAXR, seven from the high-dose HA22 treatment and four from the low-dose treatment screen, respectively, with no overlap between the two sets of compounds (Fig. 2A,D). No compounds met the scaled value >3 or <−3 criteria for differences in log IC50, at either the high or low dose of HA22 (Fig. 2B). However, there were two compounds with a 100-fold difference in potency, everolimus and osanetant, in the low-dose HA22 treatment. Five compounds enhanced the cytotoxic effect of the HA22 by AUC (scaled value log2 fold change [fc] AUC >3) in the low-dose HA22 treatment, and no compounds were enhancers in the high-dose treatment (Fig. 2C,D). Seven compounds mitigated the HA22 cytotoxic effect by fold change in AUC (scaled value log2 fc AUC <−3), six from to the high-dose HA22 treatment and one from the low-dose HA22 treatment (Fig. 2C,D). Target analysis (Fig. 2E) of the compounds with the HA22-enhancing cytotoxic effect by each of the parameters shows that those hits identified using the MAXR method had a statistically nonsignificant (p = 0.12) enrichment for ALK5 inhibitors. In contrast, poly(ADP-ribose) polymerase (PARP) inhibitors were significantly enriched in the compounds with mitigator activity, both by MAXR and AUC analysis, with p-values of 0.05 and 5.6e−6 for each method, respectively.
Figure 2.
Selection of compounds based on differential activity using parameters from the dose-response curve fit analysis. (A) Scatterplot of scaled values of differential (delta) maximum response (% viability at the maximum dose tested) for each compound. (B) Scatterplot of scaled values delta logIC50 for each compound. (C) Scatterplot of scaled values log2 fold change (fc) AUC for each compound. (D) Venn diagrams of hits selected based on the different parameters. In all plots and diagrams, red circles correspond to high HA22 dose treatment arm and blue circles correspond to low HA22 dose treatment arm. (E) Bar graph of the target classes enriched (−log p-value) for the 36 enhancer hits identified using the different curve class parameters. Complete list of hit compounds is included in supplemental material.
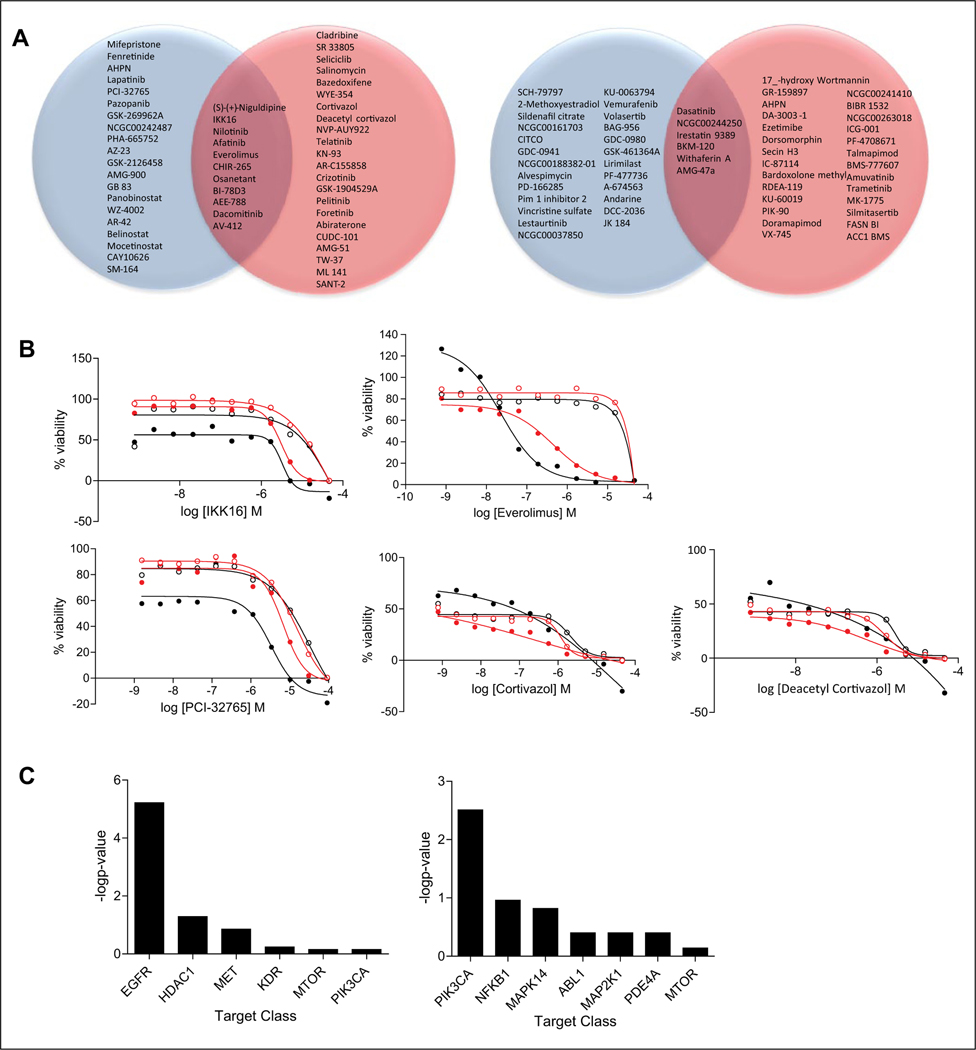
We also explored an ensemble rank product approach19,20 that included different dose-response parameters simultaneously as a way to sort compounds in terms of differential behavior. For this analysis, we computed fold changes (HA22 treatment vs. control) independently for AUC, MAXR, and AC50. The fold changes were then ranked (larger fold changes were assigned better ranks), and the rank product for each compound was computed. The compounds were then again ranked according to the rank product, and this analysis was performed for the high- and low-dose treatments separately. As for AC50-based analysis described above, while this approach is applicable to any type of curve fit, it is important to note that potency can be assigned even when the curve is of poor quality. In such cases, the potency may not be meaningful. Based on these considerations, we focused the ensemble rank analysis on the subset of compounds that were considered active with good-quality curves (CCs −1.1, −1.2, −2.1, and −2.2 for enhancers and 1.1, 1.2, 2.1, and 2.2 for mitigators) in both the treatment and control arms. This filter identified 308 and 331 compounds from the high- and low-dose treatments respectively (see supplemental material for lists of compounds). The ensemble ranking scheme implicitly identifies mitigators and enhancers of the HA22 response by placing enhancers at the top of the ranked list and mitigators at the bottom. Since this is a ranking procedure, one must decide how many compounds will be selected (as opposed to selecting a threshold). This decision is driven by various factors, including biological relevance, target enrichment, and number of compounds that can be practically pursued in further in vitro and in vivo assays. When considering the top 10% enhancers of each ranked list (high- and low-dose HA22 treatment), there are 11 compounds with enhancing HA22 cytotoxic activity in common (Fig. 3A, left Venn diagram): niguldipine (calcium channel blocker and α1-adrenergic receptor antagonist), IKK-16 (an IκB kinase inhibitor), nilotinib (an ABL1 inhibitor), afatinib (an epidermal growth factor receptor [EGFR] inhibitor), everolimus (mTOR inhibitor), CHIR-265 (BRAF inhibitor), osanetant (neurokinin 3 receptor antagonist), BI-78D3 (a MAPK inhibitor), AEE-788 (EGFR inhibitor), dacomitinib (EGFR inhibitor), and AV-412 (EGFR inhibitor). Figure 3B displays the dose-response curves for the three highest ranked enhancer compounds for the low-dose and high-dose treatments, respectively. The dose-response curves for all the 11 common compounds in the (high and low) treatment and control arms are displayed in Supplemental Figures S4 and S5. Based on the curves, the differential enhancing effect in the low-dose setting is somewhat smaller than in the high-dose setting, but in general, most compounds exhibit clear differential behavior with respect to control. Target enrichment analysis shows that there is a clear statistical enrichment (p = 0.01) for EGFR inhibitors for the 54 enhancer hits identified by this method (Fig. 3C, top). When considering the top 10% mitigators of each ranked list (high- and low-dose HA22 treatment), there are six compounds with mitigating HA22 cytotoxic activity in common (Fig. 3A, right Venn diagram). These compounds are dasatinib (Abl inhibitor), NCGC00244250 (TYK2 inhibitor), irestatin 9389 (ERN1 inhibitor), BKM-120 (PIK3 inhibitor), withaferin A (NFKβ inhibitor), and AMG-47a (LCK inhibitor). Target enrichment analysis shows that there is no clear statistical enrichment for any target class, although there is some enrichment for PI3K inhibitors for the 56 mitigator hits identified by this method (Fig. 3C, bottom). The current analysis procedure considers each parameter to be of equal importance, but this could easily be modified to give more weight to potency than efficacy, for example. To understand whether the ranking approach identified compounds that showed similar differential behavior in the high and low doses, we examined how many compounds were similarly ranked within the top 5%, 10%, and so on of the two dose treatments. The number of common compounds in the top 10% of the ranked lists from the two dose conditions is summarized in Supplemental Figure S6. It is clear that the different dose conditions lead to relatively unique ranked lists.
Figure 3.
Selection of compounds based on the ensemble ranking method. (A) Venn diagram of top 10% ranked enhancers (left Venn diagram) and mitigators (right Venn diagram) of HA22 cytotoxic effects. Red circles correspond to high-dose HA22 treatment arm, and blue circles correspond to low-dose HA22 treatment arm. (B) Dose responses for three top-ranked compounds by ensemble ranking method for the high- and low-dose HA22 treatment. Everolimus and IKK16 were selected as hits in both dose treatments. Solid black circles are high-dose HA22 treatment arm; open black circles are for the corresponding vehicle arm; solid red circles are low-dose HA22 treatment arm; open red circles are the corresponding vehicle arm. (C) Bar graph of the target classes enriched (−log p-value) for the 54 enhancer hits (top) and 56 mitigator hits (bottom) identified using the ensemble ranking method. Complete list of hit compounds is included in supplemental material.
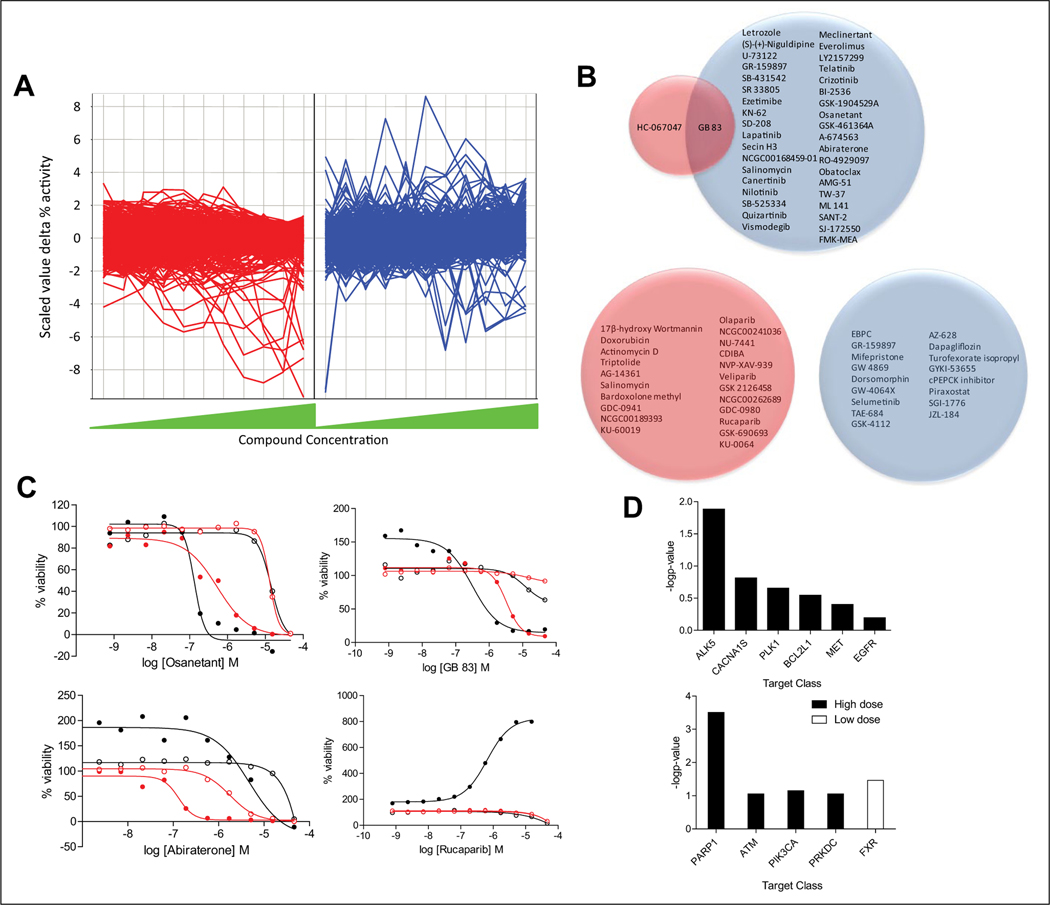
Finally, compounds with HA22 enhancing or mitigating cytotoxic effects were also selected by analyzing the results at each individual concentration of compound screened by calculating the differences in percent viability between the two treatments (HA22 vs. control) at each compound concentration and then transforming this difference in percent viability into a scaled value of percent viability difference at each compound concentration so that we could compare differential activity across the different compound concentrations. Scaled values of >3 for enhancers and <−3 for mitigators were used as cutoff to determine a significant differential effect for each compound at each dose (Fig. 4A). Thirty-eight compounds were found to be enhancers with a scaled value of >3 in at least one compound concentration in the low-dose HA22 treatment and two compounds in the high-dose treatment (Fig. 4A,B, top Venn diagram). One compound was common between the low and high HA22 treatment, GB83, a protease activated receptor 2 (PAR2) antagonist (Fig. 4B, top Venn diagram). Twenty-two compounds were found to be HA22 mitigators in the high-dose treatment and 17 in the low-dose treatment (Fig. 4A,B, bottom Venn diagram). There was no overlap between the two sets of compounds (Fig 4B, bottom Venn diagram). Three compounds showed an enhancement effect of scaled value of >3 at more than three compound concentrations, which included osanetant, abiraterone (dose responses shown in Fig. 4C), and everolimus (dose responses shown in Fig. 3B). Also shown in Figure 4C are dose responses for GB-83, the one enhancer compound that is common between the high- and low-dose HA22 treatment; finally, the dose responses for rucaparib, as a representative of the PARP1 inhibitors that mitigate HA22 cytotoxic effects, are shown as well. Target enrichment analysis within the 39 compounds identified to be HA22 cytotoxicity enhancers by this method included ALK5 inhibitors (p = 0.01) and, nonsignificantly (p > 0.05), Ca2+ channel blockers, PLK1 inhibitors, BCL2 inhibitors, Met inhibitors, and EGFR inhibitors (Fig. 4D, top). There was some target enrichment within the 22 compounds found to be mitigators in the high-dose treatment, particularly to PARP1 (p = 0.0003) and, less significantly, to ATM kinase and PIK3AC inhibitors (Fig. 4D, bottom). Farnesoid X receptor (FXR) antagonists were significantly enriched as mitigators in the low-dose HA22 treatment (p = 0.03) (Fig. 4D, bottom).
Figure 4.
Selection of compounds based on per individual compound concentration analysis. (A) Line plot of scaled values difference (delta) percent viability at each dose of compound tested. Each line corresponds to a compound. Plot on the left (red) and plot of the right (blue) correspond to the high- and low-dose HA22 treatment, respectively. (B) Venn diagrams of selected enhancer hits with scaled values of delta percent viability >3 (top Venn diagram) and mitigator hits with scaled values of delta percent viability <−3 (bottom Venn diagram) for at least one dose of compound screened. (C) Dose responses for two compounds with scaled values of delta percent viability >3 at three compound concentrations, osanetant and abiraterone (everolimus also meets these criteria, and dose responses are shown in Fig. 3B); GB-83, the one enhancer compound that is common between the high- and low-dose HA22 treatment using this selection method; and rucaparib, as a representative of the poly(ADP-ribose) polymerase 1 (PARP1) inhibitors that mitigate HA22 cytotoxic effects. (D) Target enrichment (−log p-value) for the 38 enhancers (top) and mitigators (bottom) selected by scaled values delta activity >3 (enhancers) or <−3 (mitigators) for at least one dose of compound screened. Complete list of hit compounds is included in supplemental material.
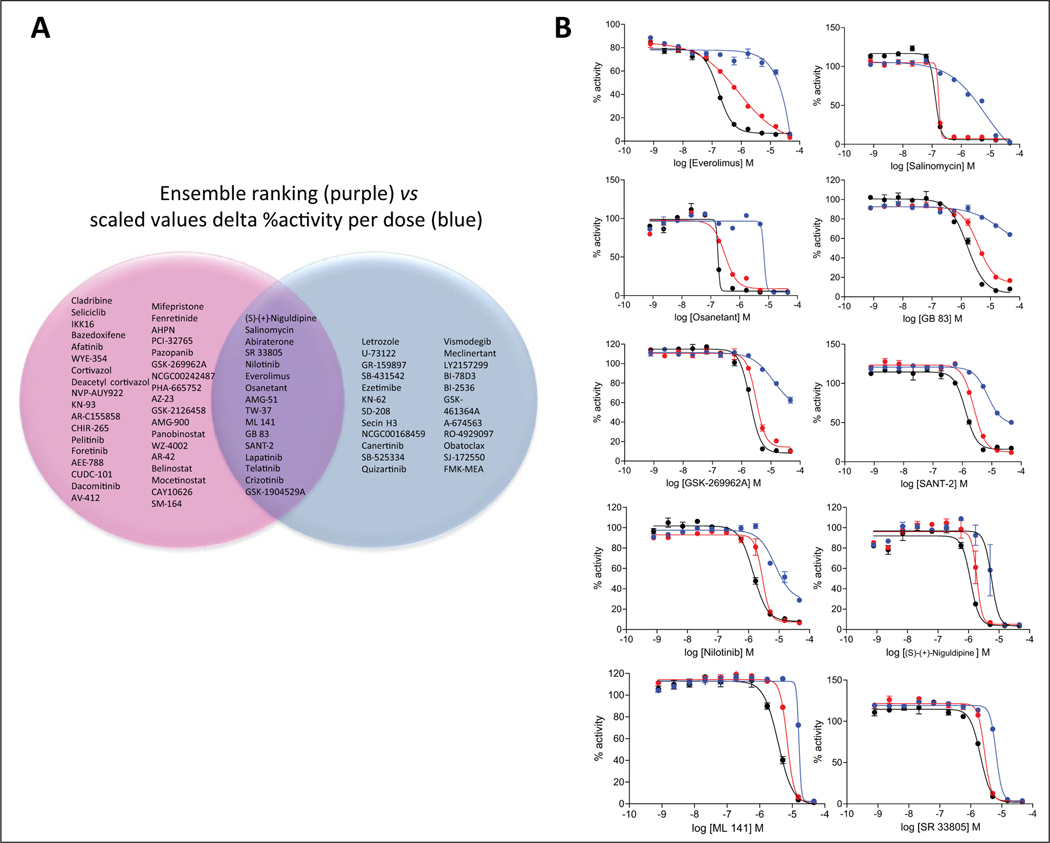
Figure 5A and Supplemental Figure S7 summarize the overlap between the lists of enhancer compounds selected by the different analysis methods described above, particularly comparing the scaled values for delta percent activity per dose versus the dose-response aggregate parameters. The 10 compounds identified using the delta MAXR method were all included in the 39 compounds identified by the delta percent activity per dose method. There was minimal overlap (only six compounds) between the 37 compounds identified using CCs and the 39 compounds identified by the delta percent activity per dose method. These six compounds were letrozole, an aromatase inhibitor; KN-62, a calmodulin-dependent protein kinase II inhibitor; SD-208, a TGF-bR1 (ALK5) inhibitor; vismodegib, a hedgehog antagonist; GB-83, a PAR2 antagonist; and FMK, a MAPKAP-K1 (RSK; p90Rsk) inhibitor. Four of the six enhancers identified using the fold change in AUC were included in the 39 compounds identified by the delta percent viability per dose method, including everolimus, salinomycin, osanetant, and abiraterone. Finally, 16 compounds (Fig. 5A) were common enhancers of HA22 by both the ensemble and differences in percent viability at individual concentrations methods. The 16 compounds included everolimus, salinomycin, osanetant, and abiraterone, as well as niguldipine and SR33805, both calcium channel blockers.
Figure 5.
(A) Venn diagram for enhancers of HA22 cytotoxic effect identified using the ensemble ranking method (purple) and by the scaled values delta percent activity per dose method (blue). (B) Dose responses for 10 of the 16 compounds that overlapped between the ensemble ranking and individual compound concentration analysis methods confirmed after retesting in triplicate. Blue, red, and black dose-response curves correspond to the phosphate-buffered saline (PBS), low HA22, and high HA22 treatment arms, respectively. Error bars correspond to SEM, n = 3.
One hundred unique HA22 enhancer compounds, including all the hits identified from the different scaled value >3 selection methods and the top 10% ranked from the ensemble ranking method, were retested using the same treatment conditions as in the primary screen, PBS, HA22 (low concentration), and HA22 (high concentration), in triplicate (data are included in supplemental material file qHTS data Excel spreadsheet in the tabs named “Hit Retest”). Supplemental Figure S8 shows that the correlation between the primary qHTS and the retest data at vehicle and each of the HA22 treatment conditions was reasonable, ranging from R2 = 0.56 to 0.73, depending on the activity parameter. The one exception is the MAXR at high-dose HA22, for which the R2 is 0.15, but this is to be expected because of the low signal due to decreased cell viability. Supplemental Figure S9 shows the scaled value plots for the differential activity using each of the parameters. Of the 100 compounds retested, 40 compounds confirmed as differentially active by at least one of the analysis methods used by the scaled value >3 criteria or the CC method (Table 1). ALK5, CACNA1S, and NOTCH1 were targets represented with more than one compound. A breakdown of the confirmed hits by each of the selection criteria in which the compounds were selected from was as follows: 4 of 20 by CC at high endotoxin, 2 of 21 by CC in low endotoxin, 9 of 10 by scaled value delta MAXR in low endotoxin, 2 of 10 by scaled value delta MAXR in high endotoxin, 3 of 6 by scaled value log fc AUC in low endotoxin, 0 of 1 by scaled value log fc AUC in high endotoxin, 22 of 38 by scaled value log delta percent activity in at least one compound dose in low endotoxin, and 0 of 2 by scaled value log delta percent activity in at least one compound dose in high endotoxin. Of the 16 compounds that were common enhancers of HA22 by both the ensemble and differences in percent viability at individual concentrations methods, 10 reproduced (62% confirmation rate) with a scaled value delta mean percent viability >3 in at least one dose of compound tested and by the differential AUC, AC50, and MAXR (three confirmed by scaled value delta mean percent viability >3 and AUC and AC50, four confirmed by scaled value delta mean percent viability >3 and MAXR, and three by scaled value delta mean percent viability >3) in either treatment condition, including everolimus, salinomycin, osanetant, niguldipine, ML 141, nilotinib, GSK-1904529A, GB-83, SANT-2, and SR-33805 (dose responses of triplicate retesting shown in Fig. 5B).
Table 1.
Forty Compounds Confirmed to Be Cytotoxic Enhancers of HA22 by at Least One of the Analysis Methods Described.
| Sample ID | Sample Name | Gene Symbol |
|---|---|---|
| NCGC00183285-04 | Nilotinib | ABL1 |
| NCGC00025230-01 | SB-431542 | ALK5 |
| NCGC00165898-02 | SD-208 | ALK5 |
| NCGC00186024-01 | SB-525334 | ALK5 |
| NCGC00249388-01 | LY2157299 | ALK5 |
| NCGC00159453-02 | Zileuton | ALOX5 |
| NCGC00263150-01 | Andarine | AR |
| NCGC00168109-03 | Secin H3 | ARHGEF1 |
| NCGC00025015-01 | (S)-(+)-niguldipine | CACNA1S |
| NCGC00025379-01 | SR 33805 | CACNA1S |
| NCGC00162398-03 | KN-62 | CAMKK2 |
| NCGC00263209-01 | ML 141 | CDC42 |
| NCGC00016083-14 | Verapamil | DRD2 |
| NCGC00263144-01 | WZ-4002 | EGFR |
| NCGC00182055-02 | Bazedoxifene | ER |
| NCGC00186046-01 | Caroverine hydrochloride | GLUR1 |
| NCGC00015917-04 | SB-206553 | HTR1A |
| NCGC00250378-01 | Piboserod hydrochloride | HTR4 |
| NCGC00253439-01 | GSK-1904529A | IGFR1 |
| NCGC00263104-01 | Foretinib | KDR |
| NCGC00263189-01 | JNJ-38877605 | MET |
| NCGC00242507-01 | Everolimus | MTOR |
| NCGC00025281-01 | Blebbistatin | MYH2 |
| NCGC00263019-01 | WAY-204688 | NFKB1 |
| NCGC00263162-01 | RO-4929097 | NOTCH1 |
| NCGC00263188-01 | YO-01027 | NOTCH1 |
| NCGC00095134-03 | Ezetimibe | NPC1L1 |
| NCGC00263107-01 | GB 83 | PAR2 |
| NCGC00263130-01 | cPEPCK inhibitor | PCK1 |
| NCGC00263118-01 | Lirimilast | PDE4A |
| NCGC00250391-01 | AZD-7545 | PDK2 |
| NCGC00025179-01 | Mifepristone | PGR |
| NCGC00025091-04 | U-73122 | PLCG1 |
| NCGC00263239-01 | FMK | RPS6KA1 |
| NCGC00242497-02 | Vismodegib | SHH |
| NCGC00263210-01 | SANT-2 | SMO |
| NCGC00025092-01 | GR-159897 | TACR2 |
| NCGC00263110-01 | Osanetant | TACR3 |
| NCGC00025325-01 | Vapiprost hydrochloride | TBXA2R |
| NCGC00168477-02 | Salinomycin | WNT1 |
Discussion
Although many HTS approaches have been described for combination screening of compounds,25,26 the challenge remains screening for combinations of compounds (in DMSO solution) with biological reagents (in aqueous solution). Here we describe a high-throughput synthetic lethal screening platform of small molecules in combination with a biological therapeutic and a data-processing framework that allows for the incorporation of both compound potency and efficacy to identify drugs that enhance or mitigate the effect of the biological therapeutic. The approach and analysis described here allow a protein drug (in this case an immunotoxin) to be screened with candidate small molecular weight chemical drugs. This is important because protein drugs are more fragile in handling and cannot always tolerate the same solvents used in a small-molecule HTS environment. In our synthetic lethal screening approach, compounds are screened in dose responses (46 μM to 0.8 nM) instead of the traditional one-dose screening so that enhancing and mitigating cytotoxic effects on the activity of the protein drug can be determined by changes in both potency and efficacy. Selection of compounds with enhancing and mitigating cytotoxic effects from dose-response synthetic lethal screen results required development of a new data analysis framework that incorporated features of the dose responses such us IC50, percent viability at the maximum concentration of compound tested, and AUC.
In this study, we investigated four analytical approaches to quantify the differential activity of compounds in each treatment arm from dose-response qHTS data. Compounds defined as “obviously” differential by using CC classification appear to be mostly weak in potency and do not show to be significantly enriched for target classes represented in the compound collection tested. Our analysis shows that comparing compounds based on individual dose curve parameters may be intuitive (e.g., potency or efficacy shifts), but it is not always a reliable approach to characterize differential activity behavior, unless all possible confounding variables are controlled. For example, identifying differential potencies requires that efficacy and slope be more or less equivalent between the two treatments. The use of the parameter AUC is attractive choice because it captures the entire dose response in a single number and should provide a measure of both the potency and efficacy of the compounds. However, based on these individual methods, there were no common compounds identified to have a significant enhancing or mitigating differential effect, and the target enrichment for each method did not produce statistically significant enriched targets, except for PARP inhibitors as mitigators using the AUC parameter.
We considered an ensemble ranking analysis method that provides a nonparametric approach to ranking compounds using multiple parameters. Here, parameters are curve-fit parameters (e.g., log AC50 and AUC) or curve-derived parameters (e.g., MAXR). This ensemble method avoids having to choose a specific parameter a priori, as well as selecting arbitrary thresholds. The downside is that the final ranking may not match user expectations since the rank of one parameter may offset the rank of another parameter in the final ensemble rank. Importantly, our analysis has shown that the rank method can be thrown off by curve parameters derived from poor-quality or nonsensical curve fits. As a result, we recommend that using the curve class as a filter to focus on good- to medium-quality curves leads to a more robust ranking analysis. We also note that although the AUC metric can be considered to “include” log AC50 and MAXR, inclusion of all three parameters in the ensemble rank is still meaningful. First, the maximum observed Pearson correlation (R2) between the three parameters was 0.13 across the various doses, and thus the parameters are largely orthogonal. Second, the AUC exhibits some degeneracy. Specifically, AUC cannot differentiate between a highly potent compound and a very toxic compound. Similarly, inactive compounds and compounds with poor potency can have similar AUC values. In the face of this degeneracy, we believe that inclusion of all three parameters in the ranking scheme is valid. We also explored the effect of individual parameters on the final ensemble rank, summarized in Supplemental Figure S10. Briefly, we generated the ensemble rank using log AC50, AUC, and MAXR. Then we computed a new ranking by excluding log AC50. This ranking was compared with the original by identifying the fraction of compounds in common between the rankings at a given percentage of the ranked list. For identical rankings, this results in a flat curve at Y = 1.0. Instead, Supplemental Figure S10 shows that this is not the case. For example, if we consider the top 25% of the original ranked list, only 70% of those compounds from the original ranking occur in the ranking where MAXR is excluded. Yet if log AC50 is excluded instead, we observe 77% of the compounds from the original ranking occurring in the new ranking. This suggests that compared with the ensemble ranking using all three parameters, loss of MAXR is more detrimental than log AC50. The ensemble ranked method identified compounds that had a clear differential activity between treatments by manual inspection of the dose-response curves, and the compounds identified were significantly enriched for several target classes. Compared with the compounds identified using the various thresholding methods previously described, we observed some overlaps. For example, using the AUC fold change approach, we observed that the six enhancers identified in the low-dose setting were found in the top 10% of the low-dose ranked list. But in contrast, if we considered the 10 enhancers identified using the shift in MAXR in the low-dose setting, none of them occur in the top 10% of the corresponding ranked list, and in fact, only three occur, at relatively high ranks. This is mainly driven by the strict constraint employed in the ranking analysis that required a compound to have a good-quality curve in both treatment and control conditions. But in addition, thresholding approaches based in single parameters, by definition, ignore other characteristics of the curve that are explicitly taken into account by the ensemble rank method.
Finally, analyzing the data using differences in percent viability at individual concentrations of compounds tested produced about 40 compounds with significant differential activity in at least one concentration of compound, and overall there was significant enrichment for compounds targeting a few target classes. Although this approach is not fundamentally different from analyzing differences in percent viability from single-dose screening, by doing dose-response qHTS, we incorporate the fact that enhancer and mitigator effects are compound and compound dose dependent. In addition, this method does not need a prerequisite for compounds with good-quality curves, because it does not use parameters obtained from dose-response curve fitting.
At the end, after retesting in triplicate 100 selected compounds from the primary qHTS, 40 compounds were confirmed to be cytotoxic enhancers of HA22 by either of the analysis methods described, corresponding to a ~9% hit rate of the MIPE oncology collection (Table 1). Ten of the 16 compounds were common enhancers of HA22 by both the ensemble and differences in percent viability at individual concentration methods that confirmed as actives. In our experience, this is a reasonable number of compounds to validate in further validation studies (unpublished data).
Does qHTS help in identifying differentially active compounds?
The preceding discussion has focused on the use of dose-response curves to identify differentially active compounds. Single-point screening is a common (and cheaper) paradigm for HTS, and we were also interested in investigating whether using dose-response screening data would allow us to identify differentially active molecules more robustly than single-dose screening. To evaluate how the compounds selected using a traditional single-dose screening compared with a dose-response screen, we retrospectively selected hits that were differentially active at 5 and 15 μM of compound, which are concentrations close to those normally used in HTS. No compounds with HA22-enhancing cytotoxic activity met the scaled value delta percent viability of >3 at either compound concentration for the high-dose HA22 treatment. Only eight and two compounds met the criteria of scaled value delta percent viability of >3 for the low-dose HA22 treatment at 5 μM and 15 μM concentrations, respectively, with no compounds overlapping between the two sets of compounds. Within the eight compounds selected at 5 μM, everolimus and osanetant, as well as (S)-(+)-niguldipine and GB-83, represented the target classes of MTOR inhibitors, NK antagonists, Ca2+ channel blockers, and PAR1 antagonists, respectively. However, representatives of other target classes found using the qHTS method were missed, including ALK5 inhibitors and PLK1 inhibitors. The two compounds found at 15 μM were SR 33805, a Ca2+ channel antagonist, and ML 141, a Cdc42 GTPase inhibitor. A similar analysis was done looking for mitigators at 5 μM and 15 μM. In this case, there was good overlap between hits found at 5 μM and 15 μM at each low- and high-dose HA22 treatment, but as seen with the qHTS dose analysis, there was no overlap between hits found at low- and high-dose HA22 treatment. The same eight compounds were found to mitigate the HA22 cytotoxic effect, and four of the PARP inhibitors were identified, as well as KU-0064, an ATM kinase inhibitor. Five compound mitigators were identified at 5 μM and 15 μM, with four overlapping between the two.
In conclusion, our multipronged analysis of dose-response HTS synthetic lethal data combined with target enrichment analysis enabled by the gene annotation and redundancy in our oncology collection highlights that the use of dose-response data provides more comprehensive functional results compared with single-point data. In addition, our results suggest that computed dose-response parameters such as CC, log IC50, and AUC might not individually be the best methods to identify hits from a synthetic lethal screen. Rather, the ensemble rank product approach represents a truly nonparametric approach that is sufficiently flexible to take into account multiple curve-derived parameters. Finally, methods that analyze percent viability of individual compound doses can also provide a good sampling of activities within target classes and produce compounds with obvious differentially active compounds by manual inspection of the dose-response curves, and they do not required preselecting compounds with good-quality dose responses. Although the approaches and results described here lend themselves to automated analyses, we stress that manual inspection of curves is a key step in the analytic workflow.
Supplementary Material
Acknowledgments
We thank Paul Shinn and Sam Michael, at the National Center for Advancing Translational Sciences, for help with the implementation of the screens.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the intramural programs of the National Cancer Institute and the National Center for Advancing Translational Sciences.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Data and Materials Available
Primary screen results are available as supplementary material. All cell lines used in this study are available for use, and all reagents, including the drugs explored, are commercially available.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Chan DA; Giaccia AJ Harnessing Synthetic Lethal Interactions in Anticancer Drug Discovery. Nat. Rev. Drug Discov. 2011, 10, 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prahallad A; Bernards R. Opportunities and Challenges Provided by Crosstalk between Signalling Pathways in Cancer. Oncogene 2016, 35, 1073–1079. [DOI] [PubMed] [Google Scholar]
- 3.Kaelin WG Jr. The Concept of Synthetic Lethality in the Context of Anticancer Therapy. Nat. Rev. Cancer 2005, 5, 689–698. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari E; Lucca C; Foiani M. A Lethal Combination for Cancer Cells: Synthetic Lethality Screenings for Drug Discovery. Eur. J. Cancer 2010, 46, 2889–2895. [DOI] [PubMed] [Google Scholar]
- 5.Jang IS; Neto EC; Guinney J; et al. Systematic Assessment of Analytical Methods for Drug Sensitivity Prediction from Cancer Cell Line Data. Pac. Symp. Biocomput. 2014, 19, 63–74. [PMC free article] [PubMed] [Google Scholar]
- 6.Fallahi-Sichani M; Honarnejad S; Heiser LM; et al. Metrics Other Than Potency Reveal Systematic Variation in Responses to Cancer Drugs. Nat. Chem. Biol. 2013, 9, 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inglese J; Auld DS; Jadhav A; et al. Quantitative High-Throughput Screening: A Titration-Based Approach That Efficiently Identifies Biological Activities in Large Chemical Libraries. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JH; Kang ZB; Ardayfio O; et al. Application of Titration-Based Screening for the Rapid Pilot Testing of High-Throughput Assays. J. Biomol. Screen. 2013, 19, 651–660. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y; Jadhav A; Southal N; et al. A Grid Algorithm for High Throughput Fitting of Dose-Response Curve Data. Curr. Chem. Genomics 2010, 4, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haibe-Kains B; El-Hachem N; Birkbak NJ; et al. Inconsistency in Large Pharmacogenomic Studies. Nature 2013, 504, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang S; Nagata S; Onda M; et al. HA22 (R490A) Is a Recombinant Immunotoxin with Increased Antitumor Activity without an Increase in Animal Toxicity. Clin. Cancer Res. 2005, 11, 1545–1550. [DOI] [PubMed] [Google Scholar]
- 12.Pastan I; Hassan R; Fitzgerald DJ; et al. Immunotoxin Therapy of Cancer. Nat. Rev. Cancer 2006, 6, 559–565. [DOI] [PubMed] [Google Scholar]
- 13.Kreitman RJ; Tallman MS; Robak T; et al. Phase I Trial of Anti-CD22 Recombinant Immunotoxin Moxetumomab Pasudotox (CAT-8015 or HA22) in Patients with Hairy Cell Leukemia. J. Clin. Oncol. 2012, 30, 1822–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wayne AS; Fitzgerald DJ; Kreitman RJ; et al. Immunotoxins for Leukemia. Blood 2014, 123, 2470–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathews LA; Keller JM; Goodwin BL; et al. A 1536-Well Quantitative High-Throughput Screen to Identify Compounds Targeting Cancer Stem Cells. J. Biomol. Screen. 2012, 17, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atkinson K. An Introduction to Numerical Analysis; John Wiley: New York, 1989; 2nd ed. [Google Scholar]
- 17.Southall N; A. J.; Huang R; et al. Enabling the Large Scale Analysis of Quantitative High Throughput Screening Data In Handbook of Drug Screening; Seethala R, Zhang L, Eds.; CRC Press: Boca Raton, FL, 2009; pp. 442–464. [Google Scholar]
- 18.Birmingham A; Selfors LM; Forster T; et al. Statistical Methods for Analysis of High-Throughput RNA Interference Screens. Nat. Methods 2009, 6, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bansal M; Yang J; Karan C; et al. A Community Computational Challenge to Predict the Activity of Pairs of Compounds. Nat. Biotech. 2014, 32, 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costello JC; Heiser LM; Georgii E; et al. A Community Effort to Assess and Improve Drug Sensitivity Prediction Algorithms. Nat. Biotech. 2014, 32, 1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitling R; Armengaud P; Amtmann A; et al. Rank Products: A Simple, Yet Powerful, New Method to Detect Differentially Regulated Genes in Replicated Microarray Experiments. FEBS Lett. 2004, 573, 83–92. [DOI] [PubMed] [Google Scholar]
- 22.Eisinga R; Breitling R; Heskes T. The Exact Probability Distribution of the Rank Product Statistics for Replicated Experiments. FEBS Lett. 2013, 587, 677–682. [DOI] [PubMed] [Google Scholar]
- 23.Fisher R. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar]
- 24.Zhang JH; Chung TD; Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 25.Borisy AA; Elliott PJ; Hurst NW; et al. Systematic Discovery of Multicomponent Therapeutics. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 7977–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathews Griner LA; Guha R; Shinn P; et al. High-Throughput Combinatorial Screening Identifies Drugs That Cooperate with Ibrutinib to Kill Activated B-Cell–Like Diffuse Large B-Cell Lymphoma Cells. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.