Abstract
SARS-CoV-2 has been efficient in ensuring that many countries are brought to a standstill. With repercussions ranging from rampant mortality, fear, paranoia, and economic recession, the virus has brought together countries to look at possible therapeutic countermeasures. With prophylactic interventions possibly months away from being particularly effective, a slew of measures and possibilities concerning the design of vaccines are being worked upon. We attempted a structure-based approach utilizing a combination of epitope prediction servers and Molecular dynamic (MD) simulations to develop a multi-epitope-based subunit vaccine that involves the two subunits of the spike glycoprotein of SARS-CoV-2 (S1 and S2) coupled with a substantially effective chimeric adjuvant to create stable vaccine constructs. The designed constructs were evaluated based on their docking with Toll-Like Receptor (TLR) 4. Our findings provide an epitope-based peptide fragment that can be a potential candidate for the development of a vaccine against SARS-CoV-2. Recent experimental studies based on determining immunodominant regions across the spike glycoprotein of SARS-CoV-2 indicate the presence of the predicted epitopes included in this study.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, vaccine design, spike protein, docking, MD simulations, adjuvant
1. Introduction
In December 2019, the town of Wuhan in Hubei, China, experienced an outbreak of a highly pathogenic virus prevalent with high transmissibility across humans and potentially responsible for varied symptoms associated with respiratory-based complications. Months after the outbreak, the disease has spread across the globe, infecting 14 million people and responsible for the deaths of another 5.9 lakh (at submission). It has all been implicated in a pathogen belonging to the genus Betacoronavirus of the Coronaviridae family and has been identified as the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). The WHO declared it a pandemic on 12th March 2020 allowing countries across the world to take appropriate measures to fend themselves from the disease and prevent its spread.
The disease transmission has left very few countries free from the repercussions of the disease with surmounting pressure on hospital settings and resources to contain this public health emergency. The scientific community is on an overdrive to look into prophylactic measures as the first line of defense against the disease. The development of rapid diagnostics to determine and isolate disease affected individuals along with repurposing and identifying existing FDA-approved drugs to provide substantial treatment are underway. Still, going by the looks of it and the unprecedented pace of development of therapeutics, it would require several months to cross over several clinical trials and finally be made available to the population.
The study of SARS-CoV-2 has been easier due to the manifestation of a similar homolog of the disease SARS-CoV more than a decade ago and comprises of similar structural counterparts that aid in the process of infection, including a spike glycoprotein, membrane glycoprotein, an envelope protein and a nucleocapsid protein (Morse et al., 2020; Wan et al., 2020). The genome sequence and its analysis were done promptly and made available (Lu et al., 2020; Wu et al., 2020). Several structures over SARS-Cov-2 S protein in interaction with human ACE-2 (hACE-2) have been made publicly available over the RCSB PDB (Shang et al., 2020; Walls et al., 2020; Wrapp et al., 2020; Yan et al., 2020), allowing the scientific community to proceed towards the development of probable therapeutic measures ranging from repurposing drugs (Li & De Clercq, 2020), design of small molecule inhibitors, identification of unique targets and elucidating molecular mechanisms of the viral protein machinery (Gordon et al., 2020) and how it establishes itself inside the host body. The interaction between the spike glycoprotein and the human ACE-2 receptor is important as it initiates the process of viral entry into human beings through contact with an infected individual. The identification of a probable site upstream of the receptor-binding motif (which includes the residues on S1 that interact with hACE-2), which undergoes cleavage and requires priming by TMPRSS2 before stable fusion of the viral membrane complex to the hACE2 (Hoffmann et al., 2020) and involves sufficient nonreversible conformational changes that put into motion the process of viral entry (Liu et al., 2020; Walls et al., 2020).
A vaccine can be a useful measure against this positive, single-stranded RNA virus. Several candidate vaccines are already underway since they are considered the only reasonable step in immunizing individuals worldwide and stopping the disease from steamrolling through countries. This paper investigates the utilization of the spike glycoprotein of SARS-CoV-2 as a potential immunogen for designing a multi-epitope peptide-based subunit vaccine with a chimeric adjuvant in tow. The spike glycoprotein is evidently an appropriate immunogen capable of eliciting neutralizing antibodies, which will inadvertently lead to the establishment and elicit an immune response to prevent viral entry and act as a prophylactic (Wrapp et al., 2020).
The work envisages the prediction of significant epitopes and the design of a multi-epitope-based peptide subunit vaccine coupled with a chimeric adjuvant. The immunogen considered involves the two domains of the spike glycoprotein involved in binding with hACE-2 followed by viral and cellular membrane fusion through S1 and S2, respectively. The cleavage site present between S1 and S2 post-fusion has not been considered due to previous apprehensions on the immune system's hypersensitivity with reference to the entire spike glycoprotein of SARS-CoV. The utilization of these epitopes and the vaccine construct across an experimental setting will help provide essential evaluation and aid in developing a vaccine to generate a robust immunological prophylactic response against SARS-CoV-2.
2. Methods
2.1. Sequence retrieval for SARS-CoV-2 spike glycoprotein
The SARS-CoV-2 spike glycoprotein sequence (PDB ID: 6VSB) (Wrapp et al., 2020) was checked for specific subunits and domains. S1 subunit (27–526 residues) comprising the Receptor Binding Domain (331–524 residues) along with the S2 subunit (663–1146) were identified from Wrapp et al. (Wrapp et al., 2020). The corresponding structure submitted over RCSB PDB (Wrapp et al., 2020) was utilized for visualization of the SARS-CoV-2 spike protein using UCSF Chimera (Pettersen et al., 2004).
2.2. Prediction of B-cell epitopes
The sequences of S1 and S2 domains were separately used to determine linear B cell epitopes through different servers. We used the Artificial Neural Network (ANN) approach-based server ABCPred (Saha & Raghava, 2006), Bcepred (Saha & Raghava, 2004), and the Immune Epitope Database and Analysis Resource (IEDB) based linear B cell epitope prediction tools (Chou & Fasman, 1978; Emini et al., 1985; Jespersen et al., 2017; Karplus & Schulz, 1985; Kolaskar & Tongaonkar, 1990; Parker et al., 1986) to predict probable epitope sequences across the query sequences. The latter two servers utilize physicochemical parameters like hydrophilicity, polarity, surface accessibility, and flexibility to predict a B cell epitope as has been previously evidenced. The predicted linear B cell sequences from ABCPred were matched with the probable epitopes predicted over the IEDB-based Bepipred 2.0 based linear prediction of epitopes for the different parameters inclusive of surface accessibility area, hydrophilicity, polarity and flexibility, and the results from the two servers were corroborated. These same parameters were then again checked with the predicted epitopes by Bcepred. Consensus sequences that emerged from all three were considered to be the potential epitopes in this regard. Since conformational or discontinuous B cell epitopes could be predicted through ElliPro (Ponomarenko et al., 2008) and Discotope (Kringelum et al., 2012), they were utilized for analysis of the consensus sequences arrived upon earlier. Consensus 16-mer epitopes were predicted using these servers.
2.3. Prediction of MHC II based helper T lymphocytes (CD4+)
The probable MHC II binding epitopes on S1 and S2 domains were predicted, and a consensus approach was utilized again to arrive at the conclusive sequences. The prediction of helper T lymphocytes was made using MHCPred (Hattotuwagama et al., 2004), SYFPEITHI (Rammensee et al., 1999), NetMHCIIpan 3.2 server (Jensen et al., 2018), and the IEDB server (Paul et al., 2016; Wang et al., 2010). A consensus selection of epitopes from the four different servers allowed us to improve upon and circumvent the limitations associated with the prediction of MHC II binders due to their polymorphic nature, peptide length variation, and determination of the appropriate peptide-binding core. Hence, looking into the limitations, these servers were determined to be the best amongst available servers that can be utilized in this study. Each of these prediction servers was compared with experimental datasets to assess their performance. SYFPEITHI predicts nonamer sequences based on the weighted contribution of each amino acid sequence present across a predicted epitope sequence. MHCPred allows the prediction of 9-mer epitopes based on multivariate statistical methods, and NetMHCIIpan 3.2 enables the determination of 15-mer epitope fragments but with limited allele-specific choices. The IEDB-based MHC II binding epitope sequence prediction server was also utilized because it employs a combination of methods ranging from ANN to SMM and ranks probable epitopes on the basis of a percentile score and an IC50 score. The sequences predicted over the different servers were either 15 mers or 9 mers depending on the server used. Based on the consensus selection across these platforms and overlapping regions of the predicted epitope sequences, uniform 15-mer epitopes were selected.
2.4. Prediction of MHC I based cytotoxic T lymphocytes (CD8+)
The prediction of Cytotoxic T Lymphocyte cells (TCTL) through MHC I binding servers involved the utilization of NetMHC 4.0 (Andreatta & Nielsen, 2016), MHC-NP (Giguère et al., 2013), NetCTL 1.2 (Larsen et al., 2007), and the IEDB-based T cell epitope tools (Moutaftsi et al., 2006). All these servers predict a nonamer epitope sequence using a default dataset along with probable interacting human leukocyte antigen alleles with SARS-CoV identified from the literature. Employing a consensus selection of the predictions from the four servers, we were able to list the appropriate TCTL epitopes with relative confidence. The NetMHC 4.0 utilizes an Artificial Neural Network to predict epitopes, NetCTL 1.2 server allows the identification of epitopes based on improved datasets with a sensitivity of 0.80 and specificity of 0.97 across the filtering threshold employed, MHC-NP employs a Machine Learning approach towards the prediction of naturally processed epitopes by the MHC, whereas IEDB-based prediction of T cell epitopes sorts epitopes based on the percentile score and low IC50 values across a combination of ANN and SMM approaches based on an appropriate peptide library. The MHC I alleles specific for a SARS-CoV-2 manifestation were based on the alleles confirmed during the outbreak of SARS-CoV at the beginning of the millennium.
2.5. Validation of predicted epitopes
The consensus sequences thus arrived upon can be considered as being capable of eliciting the necessary immune response. Additionally, each of the sequences was filtered based on their predicted antigenicity over Vaxijen (Doytchinova & Flower, 2007), allergenicity over AllerTop (Dimitrov et al., 2014) and Algpred (Saha & Raghava, 2006), toxicity over ToxinPred (Gupta et al., 2015), ability of eliciting Interferon-gamma over IFN-G (Dhanda et al., 2013) servers. Also, the epitope sequences were matched with human proteins to prevent cases of antibody response against a self-antigen with the Multiple Peptide Match Tool. Each of the epitope sequences was matched with their corresponding secondary structure over the cryoEM structure (PDB ID: 6VSB) (Wrapp et al., 2020) and listed as either alpha-helices, coils, strands or beta-sheets. This was done to allow for sequence arrangement for improved molecular modeling. Based on the conservation of the surface glycoprotein across the different strains deposited over the GISAID repository (https://www.gisaid.org/about-us/mission/) and the opinion that not much variability has been observed across these strains, we checked the coverage of the epitope sequences we have predicted with the sequences from the repository of surface glycoprotein and also the genome sequence deposited over NCBI from India (Sarkale et al., 2020).
2.6. Design of adjuvant
In a peptide-based subunit vaccine, the predicted B and T cell epitopes are not sufficient to elicit a strong immune response to generate the necessary prophylactic measures. Therefore, a suitable adjuvant must also be added to the vaccine design (Shi et al., 2019). Recently, several approaches have been made to utilize a chimeric adjuvant comprising two or more separately identified adjuvants (Albin et al., 2019; Pavot et al., 2014). We used a similar approach and carried out an in silico design comprising of three different adjuvants that have been used earlier in different vaccines or as agonists of TLR-4 receptors. A recent review of suitable adjuvants for different TLRs using this approach has been published (Gupta et al., 2020). It was backed up by experimental evidence of the identified adjuvants capable of eliciting an immune response on interaction with toll-like receptors (TLRs) separately and downstream immune signaling generating the necessary prophylactic measures that can be expected from a vaccine against SARS-CoV-2 (Fu et al., 2020). The generation of neutralization antibodies by the peptide sequences of the surface glycoprotein has already been evidenced in previous instances. Hence, their linking with a triagonist chimeric adjuvant supports their utilization as a suitable prophylactic measure. A suitable rigid linker was utilized to join the peptide sequence and their position was rearranged at the N-terminal to design several constructs of the vaccine. A single construct with adjuvants at the N and C-terminal of the vaccine was also considered.
2.7. Design and characterization of vaccine constructs
Appropriate linkers were utilized to join intra B cell epitopes, T-Helper, and T-Cytotoxic epitopes and also between them in the vaccine construct (Chen et al., 2013; Li et al., 2016). The designed construct comprises the B cell epitopes linked to the adjuvant through a rigid EAAAK linker (helix forming) toward the N-terminal, followed by a GPGPG linker connecting the THTL epitopes and an AAY linker for the TCTL epitopes toward the C-terminal. A DP linker was utilized to link the three adjuvants at the N terminal. The three adjuvants were rearranged each time, and the vaccine constructs were analyzed for physicochemical properties using ProtPARAM tool (Gasteiger et al., 2005), allergenicity (Dimitrov et al., 2014), antigenicity (Doytchinova & Flower, 2007), and scanned for probable stimulants of interferon gamma (Dhanda et al., 2013). The ToxinPred database (Gupta et al., 2015) was used to analyze each of the units of the vaccine construct and hence each of these constructs can be considered nontoxic.
2.8. Molecular Modeling of vaccine constructs
The main challenge associated with the molecular modeling of the vaccine constructs was that the adjuvant-specific region and the epitope-specific region matched with two different templates. Although in both cases there was sufficient sequence coverage that calls for homology modeling of the vaccine constructs, the multi-template alignment led to a modeled structure comprising mainly of strands, which would have made the modeled constructs unstable. An initial approach included modeling the two parts of the vaccine construct separately using a single template, but linking them through loops and energy minimizations of these constructs fell through. The ROBETTA server (Song et al., 2013), which allows for comparative modeling, brought the most promising model of the vaccine constructs with sufficient secondary structure coverage. Each of these constructs was then deposited over the GALAXY web server (Shin et al., 2014) for refinement. Based on select parameters on which each modeled construct was refined, the best structures were then carried forward for validation through Ramachandran Plot, Z-score over PROCHECK (Laskowski et al., 1993) and ProSA-web server (Wiederstein & Sippl, 2007). Additional validation was carried out using ERRAT-3D server (Colovos & Yeates, 1993). Each of the modeled vaccine constructs was also checked for sequence mismatches.
2.9. Stability of the modeled vaccine constructs
Each of these constructs was then assessed for its stability using the ProtParam server. The various markers for stability are made available through the Instability index (II) (Guruprasad et al., 1990), PEST hypothesis (Rechsteiner & Rogers, 1996), in vivo half-life, and the isoelectric point (Dice & Goldberg, 1975). The assessments based on Ramachandran Plot, ERRAT 3 D (Colovos & Yeates, 1993), WHATCHECK (Hooft et al., 1996), and the ProSA-web server (Wiederstein & Sippl, 2007) allowed us to select a single vaccine construct for a prolonged molecular dynamic simulation to determine the stability of the construct and possible use in in vivo settings.
2.10. Molecular dynamics simulations
MD simulations were performed to obtain elaborate insights on the dynamic stability of the constructs using the GROMACS suits and GROMOS 54A7 force field (Abraham et al., 2015; Schmid et al., 2011). Constructs were subjected to minimization in a cubical box with the SPC/E water model using the steepest descent followed by the conjugate gradient algorithm (Schmid et al., 2011). Counter-ions were added to neutralize the system. Temperature and pressure were maintained using the modified Berendsen thermostat and Parrinello-Rahman barostat at 300 K and 1 bar, respectively (Berendsen et al., 1984; Bussi et al., 2007). All the systems were run for 100 ns in the NPT ensemble before short equilibration of 2 ns in NVT and NPT ensembles. Long-range electrostatic interactions were evaluated using Particle Mesh Ewald (PME) algorithm with a cutoff of 1.4 nm, PME order 4 and Fourier spacing 0.16 (Essmann et al., 1995). Short-range interactions were evaluated up to 1.4 nm. Water hydrogens and other bonds were constrained by employing SETTLE and LINCS algorithm, respectively (Hess et al., 1997; Miyamoto & Kollman, 1992). Periodic boundary conditions were applied in all three (X, Y and Z) directions. Trajectories were analyzed and visualized using GROMACS inbuilt tools and VMD (Humphrey et al., 1996).
2.11. Molecular docking of vaccine construct with TLR-4
The rationale behind selecting TLR-4 (PDB ID: 3FXI_A) (Park et al., 2009) is the fact that in the case of SARS-CoV, HIV, Influenzae, and other RNA-based viruses, this Toll-like receptor has been experimentally evidenced to be implicated (Akira et al., 2006). TLR-4 structure was prepared for docking by removing all water molecules and bound ligands, adding polar hydrogen atoms and optimization at physiological pH 7.4 on UCSF Chimera. No other methods were employed on the TLR-4 structure prior to docking. The binding sites over TLR-4 were determined through available PDB IDs that showed binding of an adjuvant lipopolysaccharide with the Toll-like receptor and also through ElliPro (Ponomarenko et al., 2008) and Castp (Tian et al., 2018). It was carried out through Patchdock (Schneidman-Duhovny et al., 2005) and Firedock (Mashiach et al., 2008).
2.12. MM/GBSA based evaluation of docked pose
Post docking, the top pose was evaluated through Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) calculations over the HawkDock Server (Chen et al., 2016). The server allows the docked pose to be assessed on a per residue basis across Van der Waal potentials, electrostatic potentials, polar solvation free-energy, and solvation free energy through empirical models. The docked pose is minimized for 5000 steps, including 2000 cycles of steepest descents and 3000 cycles of conjugate gradient minimizations based on an implicit solvent model, the ff02 force field.
3. Results
3.1. Sequence retrieval for SARS-CoV-2 spike glycoprotein
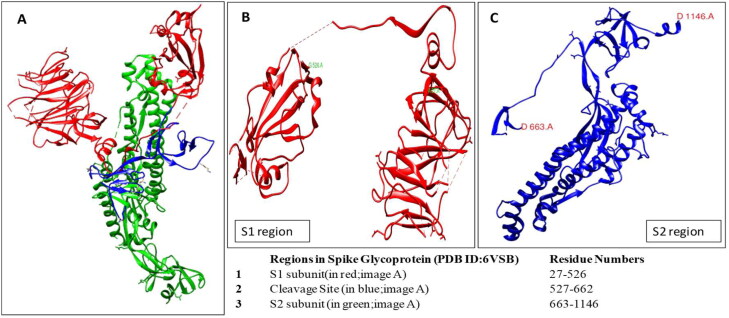
The SARS-CoV-2 surface glycoprotein sequence was retrieved from PDB ID: 6VSB in FASTA format. The UCSF Chimera visualization software (Pettersen et al., 2004) was utilized to edit the structure and include the S1 and the S2 domains as separate entities. The use of S1 and S2 domains separately without including the cleavage site, which falls in between the two domains (Figure 1) for the development of an immunogen is due to previously evidenced cases of increased immune hypersensitivity utilizing the entire length of spike glycoprotein as a vaccine immunogen (Jaume et al., 2012).
Figure 1.
The S1& S2 domains used in the investigation are reproduced from PDB ID: 6VSB.
3.2. Prediction of B cell, THTL, and TCTL epitopes
The selected epitopes were based on multiple servers which used different algorithms that lead each server to arrive at the predicted epitope. Based on the stringent filters applied in each case, the epitopes predicted over the different servers were manually curated to reach upon the consensus sequences in each case. All the epitope sequences specific for S1 and S2, separately predicted by the servers, are listed. In the case of B cell (Chou & Fasman, 1978; Jespersen et al., 2017; Saha & Raghava, 2004, 2006) and MHC II (Hattotuwagama et al., 2004; Jensen et al., 2018; Paul et al., 2016; Rammensee et al., 1999; Wang et al., 2010) epitopes, 15 mers epitope sequences were generated, while in the case of MHC I (Andreatta & Nielsen, 2016; Giguère et al., 2013; Larsen et al., 2007; Moutaftsi et al., 2006) epitope sequences, nonamer fragments were generated.
The predicted sequences were matched with the data generated by IEDB (Grifoni et al., 2020) on significant epitopes present on the surface glycoprotein, which did mention the need for analysis of the predicted sites and hence the utility of the stringent filters over the different servers followed by validation. The utilization of specific alleles that have been identified in the previous manifestation of the SARS-CoV has helped us sort the predicted T cell epitopes appropriately on the different servers, which otherwise would have made prediction biased (Greenbaum et al., 2011; Ng et al., 2004; Spínola, 2016; Weiskopf et al., 2013). The selected epitopes are listed in Table 1.
Table 1(A).
Predicted and selected B cell epitopes from S1 and S2.
| B cell epitopes | |||||||
|---|---|---|---|---|---|---|---|
| Sequence | Antigen | Allergen | IFN-G | Toxin | Mol Wt (Dalton) | pI | Length (amino acids) |
| YVGYLQPRTFLLKYNE | Positive | Negative | Positive | Negative | 2004.32 | 8.43 | 16 |
| EVRQIAPGQTGKIADY | Positive | Negative | Positive | Negative | 1745.95 | 6.17 | 16 |
| TGKIADYNYKLPDDFT | Positive | Negative | Positive | Negative | 1861.04 | 4.43 | 16 |
| YFKIYSKHTPINLVRD | Positive | Negative | Positive | Negative | 1994.32 | 9.53 | 16 |
| TVEKGIYQTSNFRVQP | Positive | Negative | Positive | Negative | 1867.09 | 8.26 | 16 |
| HVTYVPAQEKNFTTAP | Positive | Negative | Positive | Negative | 1803 | 6.75 | 16 |
3.3. Validation and filtering of selected epitopes
The various parameters against which the selected epitopes and adjuvants were filtered included antigenicity, allergenicity, toxicity, the capability of generating interferon-gamma response (Table 1A–C), and chances of being identified as self-antigen by the human immune system. Among the epitope sequences filtered, 100 percent sequence coverage was detected in all but single residue mutations in four of them. These residues were not considered in the vaccine constructs, and the aberrations were generally isolated to the S1 domain only (Figure 2B). Hence, sequence coverage based validation of the selected epitopes was also carried out. The list of epitopes considered for the design of the vaccines did not include any sequences that did not conform to the 100% sequence coverage. The percent identity matrix was utilized to indicate that the Indian sequence shows more than 90% identity with the sequence retrieved from PDB ID:6VSB, and the epitopes utilized show 100% sequence coverage across the sequence submitted from India of the spike glycoproteins (Sarkale et al., 2020) (Figure 2).
Table 1(B).
Predicted and selected THTL cell epitopes from S1 and S2.
| T HTL(MHCII, CD4) | |||||||
|---|---|---|---|---|---|---|---|
| Sequence | Antigen | Allergen | IFN-G | Toxin | Mol Wt (Dalton |
pI | Length (amino acids) |
| QSLLIVNNATNVVIK | positive | Negative | Positive | Negative | 1625.93 | 8.75 | 15 |
| AYYVGYLQPRTFLLK | positive | Negative | Positive | Negative | 1832.18 | 9.53 | 15 |
| REGVFVSNGTHWFVT | positive | Negative | Positive | Negative | 1735.92 | 6.75 | 15 |
| AIPTNFTISVTTEIL | positive | Negative | Positive | Negative | 1619.88 | 4 | 15 |
| GVVFLHVTYVPAQEK | positive | Negative | Positive | Negative | 1686.97 | 6.75 | 15 |
Table 1(C).
Predicted and selected TCTL cell epitopes from S1 and S2.
| T CTL (MHC I, CD8) | |||||||
|---|---|---|---|---|---|---|---|
| Sequence | Antigen | Allergen | IFN-G | Toxin | Mol Wt (Dalton |
pI | Length (amino acids) |
| VLPFNDGVY | positive | Negative | Positive | Negative | 1023.15 | 3.8 | 9 |
| LSETKCTLK | positive | Negative | Positive | Negative | 1022.22 | 8.2 | 9 |
| STQDLFLPF | positive | Negative | Positive | Negative | 1067.21 | 3.8 | 9 |
| YYVGYLQPRTF | positive | Negative | Positive | Negative | 1406.6 | 8.5 | 11 |
| FNATRFASV | positive | Negative | Positive | Negative | 1012.13 | 9.75 | 9 |
| SIAIPTNFTISVTTEILP | positive | Negative | Positive | Negative | 1917.23 | 4 | 18 |
| STECSNLLL | positive | Negative | Positive | Negative | 979.11 | 4 | 9 |
| LPPLLTDEM | positive | Negative | Positive | Negative | 1028.23 | 3.67 | 9 |
| AEIRASANL | positive | Negative | Positive | Negative | 944.06 | 6.05 | 9 |
Figure 2.
Percent Identity Matrix (PIM) of SARS-CoV-2 spike glycoprotein from GISAID (A) and (B) represents that apart from four of the epitopes selected, all have 100% sequence coverage across the submitted Indian sequence of SARS-CoV-2. The overall sequence similarity of the submitted Indian sequence across the seven other sequences from GISAID indicates the overall coverage sequence of the epitopes selected for vaccine design falls within acceptable parameters.
3.4. Design of adjuvant
The triagonist chimeric adjuvant has the propensity to elicit immune responses to allow for its use as a suitable adjuvant against SARS-CoV-2. RS09 (Shanmugam et al., 2012), TR-433 (Tandon et al., 2018), and an MTB based Hsp70 partial sequence (Dabaghian et al., 2015; Reed et al., 2016) were utilized in this endeavor. An appropriate rigid polar linker (DP) was utilized in this regard. Among the designed constructs, all of the rearrangements were validated based on their antigenicity score predicted by Vaxijen (Doytchinova & Flower, 2007). The adjuvants considered were all verified as TLR-4 agonists. The use of fragments of Mycobacterial hsp70 towards the generation of cytokines and natural killer cells, and also for antigen-specific CTL responses has been verified (Moyle, 2017; Multhoff et al., 2001; Wang et al., 2005). The initial constructs and the parameters across which they were assessed are mentioned in Table 1A and 1B, supplementary material. The use of lipopolysaccharide based adjuvants has been abrogated in this study due to constraints of utilizing modeling, docking, and simulation-based studies from their perspective.
3.5. Design and characterization of vaccine constructs
The arrangement and validation of antigenicity of the vaccine constructs based on the linkers and the adjuvants led to the terminal selection of five different vaccine constructs. These constructs were then subsequently characterized based on the ProtParam parameters, which determined the molecular weight and isoelectric point and predicted each to be a stable construct. Also, the constructs were found to be suitably antigenic, nontoxic, and non-allergens. Regions of the vaccine constructs were predicted to have sufficient B cell epitopes and capable of generating interferon-gamma response, and have 100% sequence coverage (Table 2 and Figure 3).
Table 2.
Validation of the five major vaccine constructs designed.
| Construct | Antigen | Allergen | IFN-G | Toxin | Length (aa) | Mol Wt (Dalton) |
pI |
|---|---|---|---|---|---|---|---|
| C1 | Positive | Negative | Positive | Negative | 460 | 50088.79 | 9.29 |
| C2 | Positive | Negative | Positive | Negative | 447 | 49165.7 | 9.25 |
| C3 | Positive | Negative | Positive | Negative | 468 | 51662.61 | 9.24 |
| C4 | Positive | Negative | Positive | Negative | 469 | 51775.77 | 9.24 |
| C5 | Positive | Negative | Positive | Negative | 474 | 52246.29 | 9.23 |
Figure 3.
Designed vaccine constructs from C_1 to C_5 with linkers and adjuvants. Vaccine construct arrangement A, B, and C refer to the three different adjuvants used. Designed Constructs based on rearrangement and numbered appropriately.
3.6. Molecular Modeling and validation of vaccine constructs
The vaccine constructs thus designed were made through comparative modeling over ROBETTA, and revealed a structure better than the other platforms utilized in this regard Figure 4.
Figure 4.
CM based modeling of designed vaccine constructs over ROBETTA server. The blue regions are the epitope sequences with the red indicating the adjuvants used and their positions.
Validation and refinement of the modeled structures revealed a stable construct in each case, which can be prominently detected by the predominant presence of defined secondary structures across the span of the construct interspersed by coils and helices. The modeled constructs were refined through the GALAXY server (Table 3) and validated through Ramachandran Plots and Z-score from ProSA-webserver (Table 4C).
Table 3.
Refinement based on GALAXY SERVER of modeled constructs.
| Construct | Rama | Poor rotamer | Mol probity | RMSD (Å) |
|---|---|---|---|---|
| C_5 | 96.6 | 0.5 | 1.779 | 0.36 |
| C_3 | 95.3 | 0.5 | 1.967 | 0.37 |
| C_4 | 96.6 | 0.5 | 1.774 | 0.36 |
| C_2 | 96.6 | 0 | 1.83 | 0.35 |
| C_1 | 96.1 | 0.3 | 1.792 | 0.35 |
Table 4(C).
Validation of modeled vaccine constructs.
| Ramachandran plot |
|||||
|---|---|---|---|---|---|
| Construct | Allowed + Favored regions (in percent) | Disallowed regions (in percent) | Z Score | ERRAT 3D | WHATCHECK |
| C1 | 91.5 + 7.3 + 0.8 | 0.5 | −4.02 | 88.1395 | Pass |
| C2 | 93.2 + 5.7 + 0.3 | 0.8 | −4.59 | 83.698 | Pass |
| C3 | 88.9 + 10.9 + 0.0 | 0.2 | −4.31 | 86.0045 | Pass |
| C4 | 92.1 + 6.9 + 0.2 | 0.7 | −4.62 | 90.4444 | Pass |
| C5 | 93.1 + 5.1 + 0.5 | 1.2 | −5.57 | 87.3085 | Pass |
3.7. Stability of the modeled vaccine constructs
Attempts were made to assess the stability of the designed constructs before moving on to docking and simulation studies with TLRs. Since it comprises three different adjuvants, an ensemble of different sequences interspersed across the spike glycoprotein, it becomes imperative that the constructs be evaluated. In this endeavor, we utilized the Expasy ProtParam Tool and based our assumptions on the PEST hypothesis and the Instability Index determinants, which include analysis of the residues that make any biomolecule have lower half-life or increased instability (includes Proline (P), Glutamic acid (E), Serine (S), Threonine (T), Methionine (M) and Glutamine (Q)) in the designed constructs and including Guruprasad’s basis (Guruprasad et al., 1990) that the relative abundance of Asparagine (N), Lysine (K) and Glycine (G) has been found in stable proteins (Table 4B). Moreover, basic or neutral isoelectric points also indicate a stable in vivo half-life for the modeled constructs. Since we cannot assess it entirely on these assumptions, we investigated the stability of the constructs through a 100 ns Molecular Dynamics Simulation. But due to constraints of computational time and effort, we assessed the stability of Construct_4 and 5 since they gave the best estimate of stability based on our assumptions as listed in Table 4(A)–(C). The above mentioned parameters went into this assumption coupled with the Ramachandran Plot, ERRAT-3D, and the Z-score. Figure 5A and Figure 3A, supplementary material, indicates the RMSD fluctuation of the stable vaccine constructs 4 and 5, which have been demarcated as the most stable constructs based on these parameters.
Table 4(B).
Assessment of stability based on PEST hypothesis and Guruprasad’s observations.
| Construct | P | E | S | T | Q | M | NKG |
|---|---|---|---|---|---|---|---|
| C1 | 6.5 | 5.4 | 4.8 | 9.8 | 3.9 | 0.4 | 19.5 |
| C2 | 7.2 | 5.4 | 4.5 | 10.1 | 3.8 | 0.2 | 19.4 |
| C3 | 7.1 | 5.3 | 4.9 | 9.6 | 3.8 | 0.4 | 19.2 |
| C4 | 7 | 5.3 | 4.9 | 9.6 | 3.8 | 0.4 | 19.2 |
| C5 | 7 | 5.5 | 4.9 | 9.5 | 3.8 | 0.4 | 19.2 |
Table 4(A).
The ProtParam based physicochemical based validation of vaccine constructs.
| Construct | Length (amino acids) | Mol Wt (Dalton) | Instability index | Half life (hrs) | pI |
|---|---|---|---|---|---|
| C1 | 460 | 50088.79 | 27.64 | 1.4 | 9.29 |
| C2 | 447 | 49165.7 | 26.89 | 4.4 | 9.25 |
| C3 | 468 | 51662.61 | 29.49 | 1.4 | 9.24 |
| C4 | 469 | 51775.77 | 28.12 | 4.4 | 9.24 |
| C5 | 474 | 52246.29 | 28.25 | 4.4 | 9.23 |
Figure 5.
(A) C-alpha RMSD of Vaccine Construct_4 for a 100 ns MD simulation; (B) RMSF plot comprising C-alpha atoms based on Vaccine Construct_4 from 0–30 ns; (C) RMSF plot comprising C-alpha atoms based on Vaccine Construct_4 from 30–100 ns, the peaks represent the regions where the loop is in abundance based on residue sequence (across residue index); (D) initial rearrangement is depicted through superimposed frames of Vaccine Construct_4 at 0 ns (in red) and at 30 ns (in blue); (E) rearrangement through superimposed frames at 30 ns (in red) and at 100 ns (in blue).
3.8. MD of vaccine construct
During the initial phase of MD simulation, subsequent conformational changes in the vaccine construct were observed, as illustrated in Figure 5A. During the rest of the simulation time average Root Mean Square Deviation (RMSD) of the C-alpha atoms remained constant around 9 Å (Figure 5A). This suggests that after the initial structural rearrangements, the vaccine construct achieved a stable three-dimensional structure, as evident in Figure 5A, and the superimposed structures obtained at 0 and 30 ns in Figure 5D, followed by another at 30 and 100 ns in Figure 5E. Further, to identify the regions of the vaccine construct that contribute to structural rearrangements, Root Mean Square Fluctuations (RMSF) of C-alpha atoms were calculated at 0–30 ns and another from 30–100 ns in Figure 5B and 5C, respectively.
The residue fluctuation is due to the low distribution of stable secondary structure rearrangements in the region between residues 153–176 and 412–424, accompanied by an abundance of loops, as observed from the RMSF plots. The abundance of loops confers its affinitive ability for its complementary receptor. An evaluation of the secondary structure features present in the vaccine construct was evaluated through a Dictionary of Secondary Structure Plot (DSSP) (Kabsch & Sander, 1983) in Figure 1, supplementary material.
The simulation indicates that Construct_4 conforms to the stability directives that are expected of it and can be reproduced under experimental conditions. Considering the MD, we extrapolated our results to include Construct 5 as a suitable vaccine construct that can be considered in the development of vaccines since they scored on a similar index across the parameters on which we assessed and went ahead with the simulation study of Construct_4. MD simulation of vaccine construct 5 was carried out in a similar manner (Figure 3, supplementary material).
3.9. Docking of the construct with TLR-4
Based on the MD simulation and the observed conformational changes in the modeled vaccine construct 4, molecular docking was carried out between TLR4 receptor and the vaccine construct 4 through Patchdock (Schneidman-Duhovny et al., 2005), which carries out docking of rigid molecules based on a protocol that employs molecular shape representation, surface patch mapping, followed by filtering and scoring. A semi-flexible docking protocol was employed involving the vaccine constructs post-simulation and the associated TLR-4 structure. The obtained results were refined and ranked using Firedock (Mashiach et al., 2008) based on a global energy value (in kJ/mol) that helps determine the binding affinity of the molecules being considered. The results indicate that the post-simulation Construct_4 does bind TLR-4 with a high binding affinity (Table 2, supplementary material) with a Global Energy Value (which is an analog of ranking based on binding affinity) of −24.18 kJ/mol. The interface between Construct_4 and TLR4 was analyzed post-docking through UCSF Chimera (Pettersen et al., 2004) and DimPlot (Laskowski & Swindells, 2011). These mainly include the non-covalent interactions that are determined through distance constraints between the two docked molecules and involve hydrogen bonds and hydrophobic interactions across the interface. Each of the residues has been specified that contribute to these interactions has been visualized as spheres and depicted on the basis of the corresponding non-covalent interaction across the protein-protein interface in Figure 6A. A similar docking was carried out between the modeled vaccine construct 4 (pre-MD simulation) with TLR-4 with a comparable affinity value (−9.10 kJ/mol). The docking with the simulated vaccine construct 4 implicates the following residues E187, K203, E255, Y311, N335, N336, Y414 and K417 involved in H-bonds across the interface with TLR-4. The docking with the initial vaccine construct 4 shows the interface residues involved in interacting with TLR4 includes S330, C371, S372, Y414, G328, S372, L375, Y391 and Y414 among others (Figure 4B, supplementary material). This can be attributed to the post-simulation transition that is represented in the structural rearrangement (Figure 5E). The change in residues forming hydrogen bonds across the interface indicates the role of polar residues like E187, K203, N335. This change in the residue pattern can conclusively indicate that the docking pose had changed significantly post the 100 ns simulation run (Figure 4A, supplementary material, and Figure 6A). The docking with the modeled vaccine construct 5 indicated residues like T258, G261, P262, G263, P264, G265, A266, I2667, T269 are involved in binding with TLR-4 only (Figure 2, supplementary material).
Figure 6.
(A) The interface of the Vaccine Construct 4 (Chain B: red) and TLR4 (Chain A: blue) are marked by the residues involved across the protein-protein interface through UCSF Chimera. The residues have been marked by green-colored spheres in between. (B) Each residue across the interface has been labeled and the corresponding interactions they take part in across the interface through DimPlot.
The docking can be conclusively used to assume that the binding of the vaccine construct to the TLR receptor may elicit signal cascades responsible for the desired immune response towards conferring a suitable prophylactic effect. The utilization of servers that allow simulation of immune response generated based on vaccine dosage was not done in this regard owing to the limitations of the predictions across experimental settings.
3.10. MM/Gbsa based evaluation of docked pose
Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) methods are utilized to determine binding free energies for macromolecules and have been used efficiently in the scoring of protein-protein interactions and suitably circumvent the computationally extensive free-energy calculation methods. Datasets employed to validate their role by comparing their scoring based on experimentally solved protein-protein interactions indicates the superiority of MM/GBSA based calculation models in determining correct docking poses. MM/GBSA has been evaluated to be better than the Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) when evaluating protein-protein interactions (Chen et al., 2016). The HawkDock Server (Chen et al., 2016) has been used explicitly in this purpose post-docking to evaluate the docked pose as being accurate. The residues that were implicated across the protein-protein interface on LigPlot of the top-scoring pose by FireDock have been assessed by a scoring function based on force-field parameters influenced through MM/GBSA calculations with similar residues being implicated. The overall binding free energy of the TLR-4 and vaccine construct was predicted to be −23.18 kcal/mol (Table 3, supplementary material).
4. Discussion
One of the most potent options that are being explored to curtail the spread of SARS-CoV-2 includes the design and development of appropriate vaccines. The entire process of vaccine development involves an extensive timeline ranging from experimental to clinical settings. In recent times, with advancement in molecular immunology and the development of several epitope mapping methods, several semi-empirical approaches at vaccine design have been introduced. Multi-epitope-peptide-based subunit vaccines based on predicted B and T cell epitopes utilizing similar bioinformatics tools has made remarkable strides. Despite the cons of low immunogenicity and multiple doses, the trade-off with eliciting neutralizing antibodies, humoral immune response, and relative safety when associated with attenuated or inactivated virus vaccines plays in favor of these approaches (Shang et al., 2020).
Looking into the probable limitations, strategies have been employed to design an epitope-based peptide vaccine utilizing the major subunits of the spike glycoprotein of SARS-CoV-2. In selecting the concerned immunogen, S1 and the S2 domains of the surface glycoprotein have been utilized separately, keeping in mind the limitations associated with using the entire surface glycoprotein (Jaume et al., 2012; Kam et al., 2007). The prediction of the three different types of epitopes (B cells, THTL and TCTL) by utilizing multiple servers in each case based on different algorithms and selection based on a consensus involving all the servers, including experimental basis behind alleles (HLA Class I and Class II), helps add sustenance to the concluded list of epitopes (Greenbaum et al., 2011; Grifoni et al., 2020; Ng et al., 2004; Spínola, 2016; Weiskopf et al., 2013). The predicted epitopes were found to be experimentally identified across a recent microarray study of mapped epitopes on the surface glycoprotein (Wang et al., 2020). Each of the predicted epitopes was verified based on described parameters and were found to be antigenic, non-allergenic, nontoxic, and share high coverage across the spike glycoprotein of SARS-CoV-2 sequences (Sarkale et al., 2020). The epitope sequences were also matched with a recently deposited SARS-CoV-2 sequence from Gujarat, India (QJC19491.1) and showed complete sequence coverage for all Indian sequences of the spike glycoprotein deposited to date. Based on the necessity of an immunostimulant, an adjuvant's role becomes important, and specifically, a chimeric adjuvant comprising TLR-4 agonists was designed and validated to have overlapping B cell epitope regions through the IEDB server. The adjuvant sequences have been experimentally evidenced to generate neutralizing antibodies, cytokines, and natural killer cells. A rigorous assessment of the implicated toll-like receptor in the case of SARS-CoV-2 has been carried out before deciding upon the use of TLR4 agonists which is based mainly on the association of TLR-4 with viral surface glycoprotein in SARS-CoV and Respiratory Syncytial Virus (RSV).The adjuvant and the predicted epitope sequences were appropriately joined by rigid linkers based on their arrangement. Even the chimeric adjuvant sequences were linked by short rigid linkers (XP)n to aid in ensuring stability. GPGPG and AAY linkers were used as intra HTL and CTL linkers, respectively. The separation between the vaccine components has been arrived upon by the role of EAAAK as a linker between adjuvant and the peptide epitope sequences (Chen et al., 2013; Li et al., 2016). The designed vaccine constructs based on the arrangement of the chimeric adjuvant were validated and concluded to be highly antigenic, non-allergenic, and nontoxic. This makes up for the limitations of subunit vaccines as mentioned above.
The stability of the vaccine constructs was assessed based on physico-chemical parameters that make it suitable for purification across experimental settings. The presence of cysteine bonds (without introducing any engineered mutations) points to the stability of the modeled constructs. The presence of defined secondary structure characteristics across the modeled construct, mainly strand and loop regions, has led us to verify the dynamic stability through a 100 ns MD simulation run. Moreover, the assessment of stability through amino acid composition (Guruprasad et al., 1990; Rechsteiner & Rogers, 1996), correlation with in vivo half-life periods, and calculated isoelectric points (Dice & Goldberg, 1975) help us identify three of the five modeled vaccine constructs to exhibit similar stability parameters. The utilization of the ProSA web server (Wiederstein & Sippl, 2007), SAVES server for Z-score and Ramchandran plot (Laskowski et al., 1993), ERRAT-3D (Colovos & Yeates, 1993) scores, helps validate the modeled constructs. Based on the arrangement of the adjuvant, CTL, HTL and B cell epitopes, five different vaccine constructs were considered and modeled as mentioned in Table 1A and 1B, supplementary material. The utilization of linkers interspersed between the different regions allows for a suitable stable vaccine construct, as indicated through Tables 3 and 4. Overlaps based on T cell and B cell epitopes were kept in mind to ensure continuation, as indicated in Table 1. The rationale behind the vaccine construct is primarily based on the thermo-stability of the designed protein sequence. Rearranging the vaccine constructs based on the different predicted epitopes would lead to a marked decrease in stability of the protein construct as is indicated in Tables 3 and 4. The vaccine construct would be ideally endocytosed and broken up into smaller peptides before presentation through antigen presentation systems that elicits a specific response against the peptides presented. Hence, rearranging the vaccine construct would lead to an unexpected change in generated immune response.
A semi-flexible docking of TLR-4 and Construct_4 was carried out. Although there are no experimental data to insinuate any such interaction but the structural features of the vaccine construct and the functional characteristics of TLR-4, an accepted dock model can be achieved in this scenario. Moreover, the N terminal comprises TLR-4 agonists which helps compound the interaction between the two proteins. Based on TLR-4 PDB structures (Park et al., 2009) and their corresponding binding sites followed by Ellipro (Ponomarenko et al., 2008) and Castp (Tian et al., 2018) predictions of binding sites over the receptor, docking was carried out with the vaccine construct over PatchDock (Schneidman-Duhovny et al., 2005) server, and the top pose based on binding affinity values was selected. The interacting residues at the protein-protein interface were mapped. The interaction occurs over the surfaces of TLR-4 which has a high electrostatic potential. The mapped non-covalent interactions are sufficient to indicate probable binding to the receptor surface through DimPlot and visualization through UCSF Chimera. The post docking MM/GBSA evaluation validates the docked pose and the involved residues across the protein-protein interaction interface.
The binding of B cell epitope sequences in the vaccine construct with the predicted B cell regions over TLR-4 will help in the generation of innate immune responses through plasma B cells producing antibodies and memory B cells that confer long-lasting prophylactic response against the virus. Moreover, recent MD studies have indicated that the trimer of spike protein is more stable as compared to the monomer, and hence may influence the stability of spike-based vaccines.
Since no experimental data is evidenced to verify the docking of the TLR4 and the vaccine construct, a theoretical determination of the protein-protein interaction between them may be carried out by running extensive MD simulations between both the proteins (tempered binding) and determining their association and dissociation profiles alongside residence time to assess the best docking pose of the two proteins in their energy minimized conditions. This may circumvent the limitations associated with the unavailability of experimental evidence to a certain extent, which could not be carried out due to computational constraints.
Supplementary Material
Acknowledgements
AJ acknowledges the Department of Biotechnology, Govt. of India for the Ramalingamswami Re-entry Fellowship-2019.
Authors' contributions
SS and DM conceived the idea of this study and designed the experiments together. DM performed the experiments. AJ, SS, DM and JP analyzed the data. DM, SS, AJ and JP contributed to drafting the manuscript.
Disclosure statement
Authors declare no conflict of interest.
References
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Akira, S., Uematsu, S., & Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell, 124(4), 783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Albin, T. J., Tom, J. K., Manna, S., Gilkes, A. P., Stetkevich, S. A., Katz, B. B., Supnet, M., Felgner, J., Jain, A., Nakajima, R., Jasinskas, A., Zlotnik, A., Pearlman, E., Davies, D. H., Felgner, P. L., Burkhardt, A. M., & Esser-Kahn, A. P. (2019). Linked toll-like receptor triagonists stimulate distinct, combination-dependent innate immune responses. ACS Central Science, 5(7), 1137–1145. 10.1021/acscentsci.8b00823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta, M., & Nielsen, M. (2016). Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics (Oxford, England), 32(4), 511–517. 10.1093/bioinformatics/btv639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J., Postma, J. V., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bussi, G., Donadio, D., & Parrinello, M. (2007). Canonical sampling through velocity rescaling. The Journal of Chemical Physics, 126(1), 014101. 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Chen, F., Liu, H., Sun, H., Pan, P., Li, Y., Li, D., & Hou, T. (2016). Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking . Physical Chemistry Chemical Physics, 18(32), 22129–22139. 10.1039/c6cp03670h [DOI] [PubMed] [Google Scholar]
- Chen, X., Zaro, J. L., & Shen, W.-C. (2013). Fusion protein linkers: Property, design and functionality. Advanced Drug Delivery Reviews, 65(10), 1357–1369. 10.1016/j.addr.2012.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, P. Y., & Fasman, G. D. (1978). Prediction of the secondary structure of proteins from their amino acid sequence. Advances in Enzymology and Related Areas of Molecular Biology, 47, 45–148. 10.1002/9780470122921.ch2 [DOI] [PubMed] [Google Scholar]
- Colovos, C., & Yeates, T. O. (1993). Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Science, 2(9), 1511–1519. 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabaghian, M., Latifi, A. M., Tebianian, M., Dabaghian, F., & Ebrahimi, S. M. (2015). A truncated C-terminal fragment of Mycobacterium tuberculosis HSP70 enhances cell-mediated immune response and longevity of the total IgG to influenza A virus M2e protein in mice. Antiviral Research, 120, 23–31. 10.1016/j.antiviral.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Dhanda, S. K., Vir, P., & Raghava, G. P. (2013). Designing of interferon-gamma inducing MHC class-II binders. Biology Direct, 8, 30. 10.1186/1745-6150-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice, J. F., & Goldberg, A. L. (1975). Relationship between in vivo degradative rates and isoelectric points of proteins. Proceedings of the National Academy of Sciences of the United States of America, 72(10), 3893–3897. 10.1073/pnas.72.10.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, I., Bangov, I., Flower, D. R., & Doytchinova, I. (2014). AllerTOP v.2-a server for in silico prediction of allergens. Journal of Molecular Modeling, 20(6), 2278. 10.1007/s00894-014-2278-5 [DOI] [PubMed] [Google Scholar]
- Doytchinova, I. A., & Flower, D. R. (2007). VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics, 8(1), 4. 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini, E. A., Hughes, J. V., Perlow, D., & Boger, J. (1985). Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. Journal of Virology, 55(3), 836–839. 10.1128/JVI.55.3.836-839.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fu, Y., Cheng, Y., & Wu, Y. (2020). Understanding SARS-CoV-2-mediated inflammatory responses: From mechanisms to potential therapeutic tools. Virologica Sinica, 35(3), 266–266. 10.1007/s12250-020-00207-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. In The proteomics protocols handbook (pp. 571–607). Springer. [Google Scholar]
- Giguère, S., Drouin, A., Lacoste, A., Marchand, M., Corbeil, J., & Laviolette, F. (2013). MHC-NP: Predicting peptides naturally processed by the MHC. Journal of Immunological Methods, 400–401, 30–36. 10.1016/j.jim.2013.10.003 [DOI] [PubMed] [Google Scholar]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., O'meara, M. J., Guo, J. Z., Swaney, D. L., Tummino, T. A., & Huttenhain, R. (2020). A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum, J., Sidney, J., Chung, J., Brander, C., Peters, B., & Sette, A. (2011). Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics, 63(6), 325–335. 10.1007/s00251-011-0513-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni, A., Sidney, J., Zhang, Y., Scheuermann, R. H., Peters, B., & Sette, A. (2020). A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host & Microbe, 27(4), 671–680.e2. 10.1016/j.chom.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N., Regar, H., Verma, V. K., Prusty, D., Mishra, A., & Prajapati, V. K. (2020). Receptor-ligand based molecular interaction to discover adjuvant for immune cell TLRs to develop next-generation vaccine. International Journal of Biological Macromolecules, 152, 535–545. 10.1016/j.ijbiomac.2020.02.297 [DOI] [PubMed] [Google Scholar]
- Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., & Raghava, G. P. (2015). Peptide toxicity prediction. In Computational peptidology (pp. 143–157). Springer. [DOI] [PubMed] [Google Scholar]
- Guruprasad, K., Reddy, B. B., & Pandit, M. W. (1990). Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Engineering, 4(2), 155–161. 10.1093/protein/4.2.155 [DOI] [PubMed] [Google Scholar]
- Hattotuwagama, C. K., Guan, P., Doytchinova, I. A., Zygouri, C., & Flower, D. R. (2004). Quantitative online prediction of peptide binding to the major histocompatibility complex. Journal of Molecular Graphics & Modelling, 22(3), 195–207. 10.1016/S1093-3263(03)00160-8 [DOI] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J., & Fraaije, J. G. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N.-H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft, R. W., Vriend, G., Sander, C., & Abola, E. E. (1996). Errors in protein structures. Nature, 381(6580), 272–272. 10.1038/381272a0 [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jaume, M., Yip, M., Kam, Y., Cheung, C., Kien, F., Roberts, A., Li, P., Dutry, I., Escriou, N., & Daeron, M. (2012). SARS CoV Subunit Vaccine: Antibodymediated Neutralisation and Enhancement. Hong Kong Med J, 18( Suppl 2), 31-6. [PubMed]
- Jensen, K. K., Andreatta, M., Marcatili, P., Buus, S., Greenbaum, J. A., Yan, Z., Sette, A., Peters, B., & Nielsen, M. (2018). Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology, 154(3), 394–406. 10.1111/imm.12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen, M. C., Peters, B., Nielsen, M., & Marcatili, P. (2017). BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Research, 45(W1), W24–W29. 10.1093/nar/gkx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W., & Sander, C. (1983). Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22(12), 2577–2637. 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- Kam, Y. W., Kien, F., Roberts, A., Cheung, Y. C., Lamirande, E. W., Vogel, L., Chu, S. L., Tse, J., Guarner, J., Zaki, S. R., Subbarao, K., Peiris, M., Nal, B., & Altmeyer, R. (2007). Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine, 25(4), 729–740. 10.1016/j.vaccine.2006.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, P., & Schulz, G. (1985). Prediction of chain flexibility in proteins. Naturwissenschaften, 72(4), 212–213. 10.1007/BF01195768 [DOI] [Google Scholar]
- Kolaskar, A., & Tongaonkar, P. C. (1990). A semi‐empirical method for prediction of antigenic determinants on protein antigens. FEBS Letters, 276(1–2), 172–174. 10.1016/0014-5793(90)80535-Q [DOI] [PubMed] [Google Scholar]
- Kringelum, J. V., Lundegaard, C., Lund, O., & Nielsen, M. (2012). Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Computational Biology, 8(12), e1002829. 10.1371/journal.pcbi.1002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, M. V., Lundegaard, C., Lamberth, K., Buus, S., Lund, O., & Nielsen, M. (2007). Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics, 8, 424. 10.1186/1471-2105-8-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. ACS Publications. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26(2), 283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- Li, G., & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- Li, G., Huang, Z., Zhang, C., Dong, B.-J., Guo, R.-H., Yue, H.-W., Yan, L.-T., & Xing, X.-H. (2016). Construction of a linker library with widely controllable flexibility for fusion protein design. Applied Microbiology and Biotechnology, 100(1), 215–225. 10.1007/s00253-015-6985-3 [DOI] [PubMed] [Google Scholar]
- Liu, W., Morse, J. S., Lalonde, T., & Xu, S. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Nature. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach, E., Schneidman-Duhovny, D., Andrusier, N., Nussinov, R., & Wolfson, H. J. (2008). FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Research, 36(Web Server issue), W229–W232. 10.1093/nar/gkn186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., & Kollman, P. (1992). SETTLE: An analytical version of the shake an RATTLE algorithms for molecular simulation. Journal of Computational Chemistry, 13(8), 952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Morse, J. S., Lalonde, T., Xu, S., & Liu, W. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem, 21(5), 730–738 [DOI] [PMC free article] [PubMed]
- Moutaftsi, M., Peters, B., Pasquetto, V., Tscharke, D. C., Sidney, J., Bui, H.-H., Grey, H., & Sette, A. (2006). A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nature Biotechnology, 24(7), 817–819. 10.1038/nbt1215 [DOI] [PubMed] [Google Scholar]
- Moyle, P. M. (2017). Biotechnology approaches to produce potent, self-adjuvanting antigen-adjuvant fusion protein subunit vaccines. Biotechnology Advances, 35(3), 375–389. 10.1016/j.biotechadv.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Multhoff, G., Pfister, K., Gehrmann, M., Hantschel, M., Gross, C., Hafner, M., & Hiddemann, W. (2001). A 14-mer Hsp70 peptide stimulates natural killer (NK) cell activity. Cell Stress & Chaperones, 6(4), 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M. H., Lau, K.-M., Li, L., Cheng, S.-H., Chan, W. Y., Hui, P. K., Zee, B., Leung, C.-B., & Sung, J. J. (2004). Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. The Journal of Infectious Diseases, 190(3), 515–518. 10.1086/421523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B. S., Song, D. H., Kim, H. M., Choi, B.-S., Lee, H., & Lee, J.-O. (2009). The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature, 458(7242), 1191–1195. 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- Parker, J., Guo, D., & Hodges, R. (1986). New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry, 25(19), 5425–5432. 10.1021/bi00367a013 [DOI] [PubMed] [Google Scholar]
- Paul, S., Sidney, J., Sette, A., & Peters, B. (2016). TepiTool: A pipeline for computational prediction of T cell epitope candidates. Current Protocols in Immunology, 114(1), 18.19.1–18.19.24. 10.1002/cpim.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavot, V., Rochereau, N., Rességuier, J., Gutjahr, A., Genin, C., Tiraby, G., Perouzel, E., Lioux, T., Vernejoul, F., Verrier, B., & Paul, S. (2014). Cutting edge: New chimeric NOD2/TLR2 adjuvant drastically increases vaccine immunogenicity. Journal of Immunology, 193(12), 5781–5785. 10.4049/jimmunol.1402184 [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Ponomarenko, J., Bui, H.-H., Li, W., Fusseder, N., Bourne, P. E., Sette, A., & Peters, B. (2008). ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics, 9, 514. 10.1186/1471-2105-9-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee, H.-G., Bachmann, J., Emmerich, N. P. N., Bachor, O. A., & Stevanović, S. (1999). SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics, 50(3–4), 213–219. 10.1007/s002510050595 [DOI] [PubMed] [Google Scholar]
- Rechsteiner, M., & Rogers, S. W. (1996). PEST sequences and regulation by proteolysis. Trends in Biochemical Sciences, 21(7), 267–271. 10.1016/S0968-0004(96)10031-1 [DOI] [PubMed] [Google Scholar]
- Reed, S. G., Hsu, F.-C., Carter, D., & Orr, M. T. (2016). The science of vaccine adjuvants: Advances in TLR4 ligand adjuvants. Current Opinion in Immunology, 41, 85–90. 10.1016/j.coi.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Saha, S., & Raghava, G. (2006). AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Research, 34(Web Server issue), W202–W209. 10.1093/nar/gkl343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., & Raghava, G. P. S. (2004). BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. International Conference on Artificial Immune Systems, Springer, pp. 197–204. [Google Scholar]
- Saha, S., & Raghava, G. P. S. (2006). Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins, 65(1), 40–48. 10.1002/prot.21078 [DOI] [PubMed] [Google Scholar]
- Sarkale, P., Patil, S., Yadav, P. D., Nyayanit, D. A., Sapkal, G., Baradkar, S., Lakra, R., Shete-Aich, A., Prasad, S., Basu, A., Dar, L., Vipat, V., Giri, S., Potdar, V., Choudhary, MLal., Praharaj, I., Jain, A., Malhotra, B., Gawande, P., … Abraham, P. (2020). Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian Journal of Medical Research, 151(2), 200-209. 10.4103/ijmr.IJMR_1029_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, N., Eichenberger, A., Choutko, A., Riniker, S., Winger, M., Mark, A., & van Gunsteren, W. (2011). Definition and testing of the GROMOS force-field versions 54A7 and 54B7. European Biophysics Journal, 40(7), 843–856. 10.1007/s00249-011-0700-9 [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny, D., Inbar, Y., Nussinov, R., & Wolfson, H. J. (2005). PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Research, 33(Web Server issue), W363–W367. 10.1093/nar/gki481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–228. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, W., Yang, Y., Rao, Y., & Rao, X. (2020). The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines, 5(1), 3. 10.1038/s41541-020-0170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam, A., Rajoria, S., George, A. L., Mittelman, A., Suriano, R., & Tiwari, R. K. (2012). Synthetic Toll like receptor-4 (TLR-4) agonist peptides as a novel class of adjuvants. PloS One, 7(2), e30839. 10.1371/journal.pone.0030839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, S., Zhu, H., Xia, X., Liang, Z., Ma, X., & Sun, B. (2019). Vaccine adjuvants: Understanding the structure and mechanism of adjuvanticity, Vaccine. Vaccine, 37(24), 3167–3178. 10.1016/j.vaccine.2019.04.055 [DOI] [PubMed] [Google Scholar]
- Shin, W.-H., Lee, G. R., Heo, L., Lee, H., & Seok, C. (2014). Prediction of protein structure and interaction by GALAXY protein modeling programs. Bio Design, 2, 1–11. [Google Scholar]
- Song, Y., DiMaio, F., Wang, R. Y.-R., Kim, D., Miles, C., Brunette, T., Thompson, J., & Baker, D. (2013). High-resolution comparative modeling with RosettaCM. Structure (London, England: 1993), 21(10), 1735–1742. 10.1016/j.str.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spínola, H. (2016). HLA loci and respiratory infectious diseases. Journal of Respiratory Research, 2, 56–66. [Google Scholar]
- Tandon, A., Pathak, M., Harioudh, M. K., Ahmad, S., Sayeed, M., Afshan, T., Siddiqi, M., Mitra, K., Bhattacharya, S. M., & Ghosh, J. K. (2018). A TLR4-derived non-cytotoxic, self-assembling peptide functions as a vaccine adjuvant in mice. Journal of Biological Chemistry, 293(51), 19874–19885. 10.1074/jbc.RA118.002768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, W., Chen, C., Lei, X., Zhao, J., & Liang, J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46(W1), W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 183(6), 1735. 10.1016/j.cell.2020.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94(7). 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Hou, X., Wu, X., Liang, T., Zhang, X., Wang, D., Teng, F., Dai, J., Duan, H., & Guo, S. (2020). SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Central Science. 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Sidney, J., Kim, Y., Sette, A., Lund, O., Nielsen, M., & Peters, B. (2010). Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics, 11, 568. 10.1186/1471-2105-11-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Whittall, T., McGowan, E., Younson, J., Kelly, C., Bergmeier, L. A., Singh, M., & Lehner, T. (2005). Identification of stimulating and inhibitory epitopes within the heat shock protein 70 molecule that modulate cytokine production and maturation of dendritic cells. Journal of Immunology (Baltimore, MD: 1950), 174(6), 3306–3316. 10.4049/jimmunol.174.6.3306 [DOI] [PubMed] [Google Scholar]
- Weiskopf, D., Angelo, M. A., de Azeredo, E. L., Sidney, J., Greenbaum, J. A., Fernando, A. N., Broadwater, A., Kolla, R. V., De Silva, A. D., de Silva, A. M., Mattia, K. A., Doranz, B. J., Grey, H. M., Shresta, S., Peters, B., & Sette, A. (2013). Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proceedings of the National Academy of Sciences of the United States of America, 110(22), E2046–E2053. 10.1073/pnas.1305227110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein, M., & Sippl, M. J. (2007). ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35(Web Server issue), W407–W410. 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.