Abstract
Aim:
Quxie capsule(QX), a TCM compound, had shown benefit on survival outcomes for metastatic colorectal cancer(mCRC) patients and could inhibit tumor growth through immune regulation. This study aimed to evaluate whether such effect is associated with gut microbiome modulation.
Method:
We conducted a randomized double-blinded placebo controlled clinical trial in Xiyuan Hospital, China Academy of Chinese Medical Sciences. All patients were randomly assigned into QX or placebo control group. Before and after 1-month interventions, we collected patients’ stool samples for microbiome analysis by 16s rRNA sequencing approaches, as well as blood samples to analyze T lymphocyte subsets by flow cytometry methods. Microbiome analysis among groups was done through bioinformation analysis platform. The study had been proved by the ethics committee of Xiyuan Hospital (2016XLA122-1) had been registered on Chinese Clinical Trial Registry (registration number: ChiCTR2000029599). All patients consented before enrollment.
Results:
We randomly assigned 40 patients and 34 were finally analyzed. Among them, 29% were female, with an average age of 63 years old, and 74% had liver or lung metastasis. Both CD4 T(TH) cell and CD8 T(TC) cell counts increased after QX treatment, while TH cells were significantly more in QX than in control group (737 vs 449, P = .024). Microbiome community analysis on Class level showed that the proportion of Actinobacteria declined in the control group, but significantly increased after QX treatments (0.83% vs 4.7%, P = .017). LEfSe analysis showed that after treatments, samples from QX group were highly related with Oscillibacter, Eubacterium, and Lachnospiraceae. RDA analysis showed that after QX interventions, stool samples and microbiome species had relevance with TC/TH cells counts but were not statistically significant. Heatmap analysis on Genus level revealed that after QX treatments, higher amounts of TH cells were significantly associated with less abundance of g_Bifidobacterium (coef. −0.76, P = .002), Collinsella (coef.−0.61, P = .02), Ruminiclostridium_9 (coef. −0.64, P = .01).
Conclusion:
QX capsule could enhance TH cells level among mCRC patients and increase the abundance of gut anticancer bacteria such as Actinobacteria as well as butyrate-producing bacteria such as Lachnospiraceae. These results indicated that QX capsule might have the property of dual effects of antitumor and immunity enhancement, both mediated by the microbiome.
Keywords: metastatic colorectal cancer, Traditional Chinese Medicine, gut microbiome, T cell immunity, randomized clinical trial
Introduction
Colorectal cancer (CRC) is fourth most frequently diagnosed cancer and second cause of cancer related death around the world.1 About 20% of CRC patients were diagnosed with stage IV disease (metastatic CRC, mCRC),2 and approximately 14% to 50% of early or local advanced CRC patients would develop into local recurrence or distant metastatic disease.3 With the advances of systematic treatments such as targeted therapy and immunotherapy, the survival outcome of mCRC has increased significantly during the past decade.4 However, real-world data showed that the overall survival time of mCRC was 18 to 20 month on average and 5-year survival rate was about 12%.5-7 A recent study in China revealed that the medical cost of mCRC treatment rose up from 2000 to 5000 dollars per cycle/patient during recent years.8 Due to economic burden or side effects, many Chinese cancer patients would utilize Traditional Chinese Medicine (TCM) for cancer treatment or symptom control.9
The Quxie (QX) capsule is a TCM herbal medicine which was developed from a traditional TCM formula, Yin-Yang Gongji Granule, and has been utilized to treat CRC patients in Xiyuan Hospital of the China Academy of Chinese Medical Sciences for more than 10 years. The composition of QX includes 20 different herbs such as Euodiae Fructus, Zingiberis Rhizoma, Cinnamomi Cortex and others. Based on TCM theory, QX capsule’s treatment effect is eliminating pathogenic factors and stimulating the body’s healthy Qi (Zheng Qi). A previous randomized controlled trial (RCT) had indicated that QX capsule along with conventional chemotherapy showed survival benefit relative to chemotherapy alone among mCRC patients (23 vs 14 months).10 Another RCT showed that QX capsule could not only prolong overall survival, but also improve mCRC patients’ quality of life.11
Basic research had been carried out to investigate the mechanism of QX capsule’s effect on treating mCRC. Previous studies found that QX capsule triggered apoptosis of human colon cancer HCT116 cells by elevating the level of the protein caspase-3, while increasing the cytokine IFN-γ level of mice. Chen et al found that Foxo1 protein expression was higher in tumor treated with QX, while knocking down Foxo1 in HCT116 cells partially blocked QX’s antiproliferative activity.12 In addition, QX capsule could regulate the Th1/Th2 and Th17/Treg cells immune balance of CT26 mouse through Foxo1 mediated upregulating T-bet protein and downregulating Foxp3.13 Based on these findings, QX capsule’s antitumor effect is related to cell apoptosis and the body’s immune response.
The gut microbiome, which has been found to be associated with body metabolism and the immune system, plays an important role in CRC development and treatment.14 Wang et al found that the gut microbiome of CRC patients was enriched in Escherichia/Shigella, Klebsiella, Streptococcus more than that of healthy volunteers, while Roseburia and other butyrate-producing bacteria related to Lachnospiraceae were less.15 Such changes in the gut microbiome community result in DNA damage to colonic epithelial cell (CEC) by genotoxin-producing bacteria such as Escherichia coli and contribute to CRC development.16 Meanwhile, lack of butyrate-producing bacteria decreases short chain fatty acids (SCFAs) secretion, which would impair inflammation suppression and anti-carcinogenic progress.17 SCFAs were also found to induce Foxp3+ T regulatory cell differentiation.18 As mentioned above, Foxp3 could mediate QX capsule inducing cell apoptosis and regulating immune balance for CRC. However, whether the gut microbiome participates in this process is still unknown.
In this study, we aimed to investigate QX capsule’s effect on mCRC patients’ gut microbiome and the association with T cell immunity changes. We hypothesized that QX capsule could moderate the gut microbiome and such mechanism have association with T cell immunity among mCRC patients.
Methods
We conducted a randomized double-blinded placebo controlled trial in Xiyuan Hospital of China Academy of Chinese Medical Sciences between April 2018 and March 2019. The study had been approved by the Ethics Committee of Xiyuan Hospital (2016XLA122-1) and had been registered on the Chinese Clinical Trial Registry (registration number: ChiCTR2000029599). All patients were consented by written documents before enrollment.
Population
We enrolled stage IV colorectal cancer patients with unresectable metastatic diseases. Other inclusion criteria included age between 18 to 80, having finished chemo, radio, targeted or immunotherapy for at least 1 month and estimated survival time longer than half a year. Exclusion criteria included intestinal obstruction, failure in orally taking medicine or other malabsorption diseases. Patients who had received antibiotic or probiotic treatments were also excluded from the study.
Randomization
Participants were randomized into the QX or control group with a 1:1 ratio. A third-party statistician used computer-generated numbers (SAS 9.2) sealed in opaque envelopes. All participants were sequentially randomized into different group after screening and consent. The sequential numbers were also printed on the external packing and bottles of all the medicine.
QX Capsule Preparation
QX capsule contains TCM herbs as following: Euodiae Fructus, Zingiberis Rhizoma, Cinnamomi Cortex, Aconiti Radix Cocta, Coptidis Rhizoma, Pinelliae Rhizoma, Citri Exocarpium Rubrum, Poria, Arecae Semen, Magnoliae Officinalis Cortex, Aurantii Fructus Immaturus, Acori Tatarinowii Rhizoma, Corydalis Rhizoma, Ginseng Radix Et Rhizoma, Aquilariae Lignum Resinatum, Platycodonis Radix, Crotonis Semen Pulveratum, and Gleditsiae Sinensis Fructus. All herbs were powdered and mixed into capsules by the Pharmaceutical Center of Xiyuan Hospital under the quality control of the institution (production date: May 1, 2017; expiration date: May 01, 2019). The placebo capsules which had exactly the same appearance and weight as the QX capsule were manufactured by the Beijing Institute of Food and Drug. All capsules were stored in a storage room under constant temperature and humidity.
Intervention
In the QX group, patients orally took QX capsule (6-10 capsules per time according to patients’ tolerance) twice a day, from day 1 to day 20, and stopped taking for 10 days (in total 30 days per cycle). In the control group, we gave patients placebo capsules. The intervention of QX or placebo capsule lasted for 1 cycle (1 month). Meanwhile, patients in both groups would also receive usual TCM herbal medicine for health promotion because it was unethical to provide mere placebo capsule for patients with advanced cancer diseases. The TCM herbal medicine prescriptions for all participants were similar (usually tonic herbs which stimulate healthy qi based on TCM theory) and did not overlap with the composition of QX capsule. We offered all patients 1 cycle of QX capsule as compensation. All interventions were blinded to researchers, physicians, and patients from the external packing to the appearance and weight of the capsule.
Patients’ Information and Sample Collection
We collected patients’ basic information including gender, age, as well as patients’ disease information including primary tumor sites, and metastatic organ. We collected patients’ blood samples (2 mL peripheral venous blood) before and after interventions for T lymphocyte subset analysis. Patients’ stool samples were collected before and after interventions. We used OMNIgene-GUT OME-200 tube to store the stool sample (DNA Genitek Inc, Ottawa, Canada). The tube could transport and store stabilized DNA at ambient temperature for 60 days.
T Lymphocyte Subset Analysis
T lymphocyte subset analysis was done by the hematology laboratory of Xiyuan Hospital by utilizing flow cytometry methods. Actual counts and ratio of CD4+ T cells (helper T cells, TH) and CD8+ T cells (Killer T cell, TC) of blood samples were presented in a paper report for each patient.
Microbiome Analysis
Microbial DNA was extracted from stool samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to manufacturer’s protocols. The final DNA concentration and purification were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system (GeneAmp 9700, ABI, USA). The PCR reactions were conducted using the following program: 3 minutes of denaturation at 95°C, 27 cycles of 30 seconds at 95°C, 30 seconds for annealing at 55°C, and 45 seconds for elongation at 72°C, and a final extension at 72°C for 10 minutes. PCR reactions were performed in triplicate 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase and 10 ng of template DNA. The resulting PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, USA) according to the manufacturer’s protocol. Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
Statistical Analysis
We used an analysis of variance or chi-square test or t-test to compare baseline variables between groups. Actual counts of TH and TC cells before and after intervention were compared by independent 2-sample t-test within QX or control group. All analyses mentioned above were performed using SPSS 21 (IBM Corp., USA). Microbiome analysis was done through Majorbio Cloud (Majorbio Bio-Pharm Technology Co. Ltd., Shanghai, China), an online bioinformation analysis platform. Alpha diversity on Operational Taxonomic Units (OTU) level was measured by Chao, Shannon and ACE indexes and compared before and after treatments by student t-test within QX or control group. Principal coordinates analysis (PCoA) was used to detect beta diversity between groups. Species difference analysis among groups was tested on the Phylum level as well as LEfSe multi-level. We then analyzed the counts of TH and TC cells as environmental factors to detect their association with samples from different groups and microbiome on the genus level by using RDA/CCA analysis and heatmap analysis. Differences were considered statistically significant if the p value was lower than 0.05. As an exploratory study, we defined the sample size as 40 in consideration of 5% of the participants who may drop out and lost to follow-up.
Results
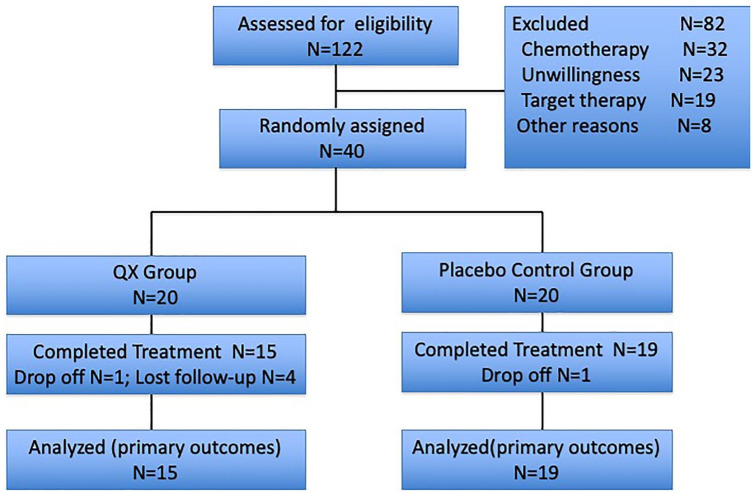
Between April 2018 and February 2019, we screened 122 mCRC patients. Of 40 patients who were randomly assigned, 2 patients dropped off (1 from the QX group who decided to quit the study right after randomized assignment, 1 from the placebo group who dropped off due to disease progression) and 4 patients lost to follow up (all from QX group). In total, 15 patients in the QX group and 19 patients in the control group completed all treatments and were finally analyzed (Figure 1).
Figure 1.
CONSORT diagram.
Participants Characteristics
The mean age of these 34 patients was 63 ± 15 years old. Among them, 29% were female, 44% patients had rectal cancer, followed by right colon (29.4%) and left colon (26.4%). There was no significant difference between the distribution of gender, age, and primary tumor site. There were 74% patients who had liver and lung metastasis; however, fewer patients with liver metastasis were inthe QX group than in the control group (P = .011; Table 1).
Table 1.
Patients (n = 34) Basic Information in Different Groups.
| Characteristics | QX group N (SD/%)* (n = 15) | Control group N (SD/%) (n = 19) | P value*** |
|---|---|---|---|
| Gender | |||
| Male | 12 (80) | 12 (63.2) | .29 |
| Female | 3 (20) | 7 (36.8) | |
| Age (year’ old) | 61 (19) | 64 (12) | .64 |
| Primary tumor site | |||
| Right colon | 4 (26.7) | 6 (31.6) | .27 |
| Left colon | 6 (40) | 3 (15.8) | |
| Rectum | 5 (33.3) | 10 (52.6) | |
| Metastasis site** | |||
| Lung (n = 13) | 8 (57.1) | 5 (27.8) | .09 |
| Liver (n = 12) | 2 (14.3) | 10 (58.8) | .011 |
| Other (n = 10) | 4 (28.6) | 6 (33.3) | .77 |
The proportions (%) were calculated with cases of certain characters and participants analyzed in each group.
Comparisons were done within patients who had lung, liver or other metastatic sites respectively because multiple metastatic sites might exist for 1 patient.
Statistical significance was set at P = .05.
T Lymphocyte Subset Analysis
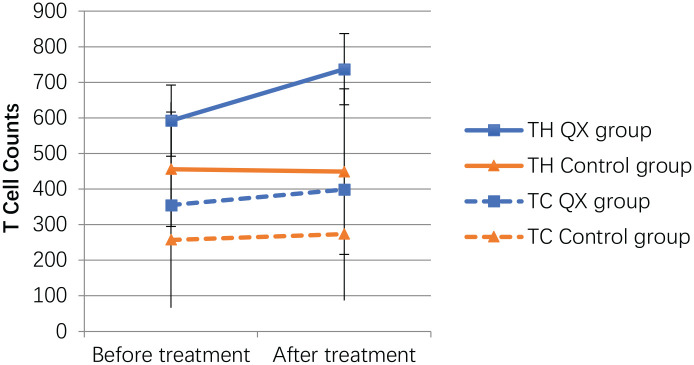
In the QX group, TH cells counts significantly increased from 592.3 ± 330.3 to 737 ± 376.9, however, they decreased from 455.8 ± 160.7 to 449.2 ± 232.6 in the control group. TH cells counts were significantly higher in the QX group than in the control group after treatment (P = .024, 95% CI −534.0, −41.6). TC cells counts also increased in the QX group, however, there was no significant difference in changes between groups (Figure 2).
Figure 2.
T lymphocytes subsets counts in different groups before and after treatments.
Gut Microbiome Analysis in Different Groups
High throughout pyrosequencing was performed with an Illumina MiSeq platform to generate 1 842 432 valid sequences from 68 stool samples of 34 patients. In total, 862 OTUs (Operational Taxonomic Units) were obtained.
Alpha Diversity and Enterotypes Analysis
Alpha analysis on OTU level showed that ACE/Sobs/Chao indexes increased after interventions in QX group, while they declined in the control group, however there were no statistically significant differences between groups (Table 2).
Table 2.
Student t-Test for Alpha Diversity Analysis on OTU Level Before and After Treatments in QX and Control Groups.
| ACE | Chao | Shannon | Simpson | Sobs | |
|---|---|---|---|---|---|
| QX | |||||
| Before | 236.4 ± 96.2 | 235.0 ± 98.3 | 3.1 ± 0.6 | 0.1 ± 0.09 | 207.0 ± 91.2 |
| After | 244.3 ± 103.1 | 247.7 ± 103.1 | 2.9 ± 0.8 | 0.2 ± 0.1 | 215.3 ± 91.1 |
| P value | .83 | .73 | .63 | .40 | .8 |
| Control | |||||
| Before | 242.2 ± 137.0 | 239.7 ± 144.7 | 3.0 ± 1.0 | 0.16 ± 0.16 | 206.0 ± 129.6 |
| After | 227.7 ± 142.9 | 227.3 ± 143.8 | 2.7 ± 0.9 | 0.18 ± 0.15 | 203.2 ± 130.6 |
| P value | .76 | .80 | .41 | .62 | .95 |
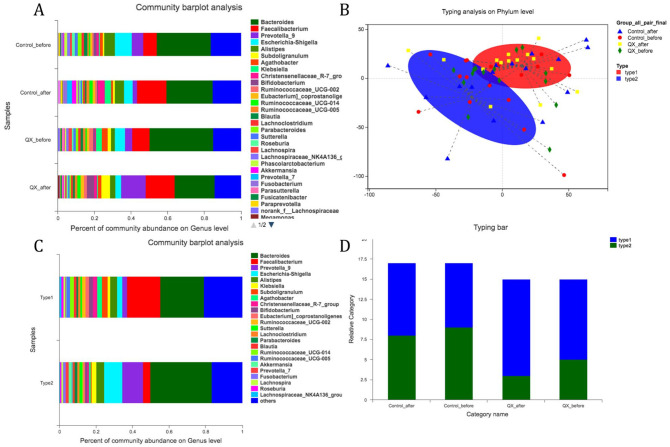
Community barplot analysis showed that the proportion of major bacteria on the Genus level such as Bacteroides and Escherichia-Shigella had declined in both the QX and control groups after treatment. In the control group, the proportion of Faecalibacterium increased after treatment, while in the QX group it was Prevotella_9 that increased after treatment. This indicated that different interventions might be associated with changes in enterotype of bacterial distribution (Figure 3A). Thus, we performed typing analysis of samples from different groups. Results showed that there were 2 specific enterotypes (ET) (Figure 3B), while type1 was ET-FB (Faecalibacterium/Bacteroides) type and type2 was ET-PB (Prevotella/Bacteroides)) type (Figure 3C). Distribution of ET-FB were higher among samples after QX treatment, but no significant variance was detected (Figure 3D).
Figure 3.
Community barplot analysis and enterotype analysis in QX and control group. (A) Community barplot analysis for different groups. (B) Micro-biome enterotype typing analysis on Phylum level for samples from both QX and control groups before or after treatments. (C) Community barplot analysis for enterotype 1 and enterotype 2 derived from typing analysis in Figure 2B. (D) Difference on the distribution of enterotype 1 and enterotype 2 in different groups.
Beta Diversity and Community Difference Analysis
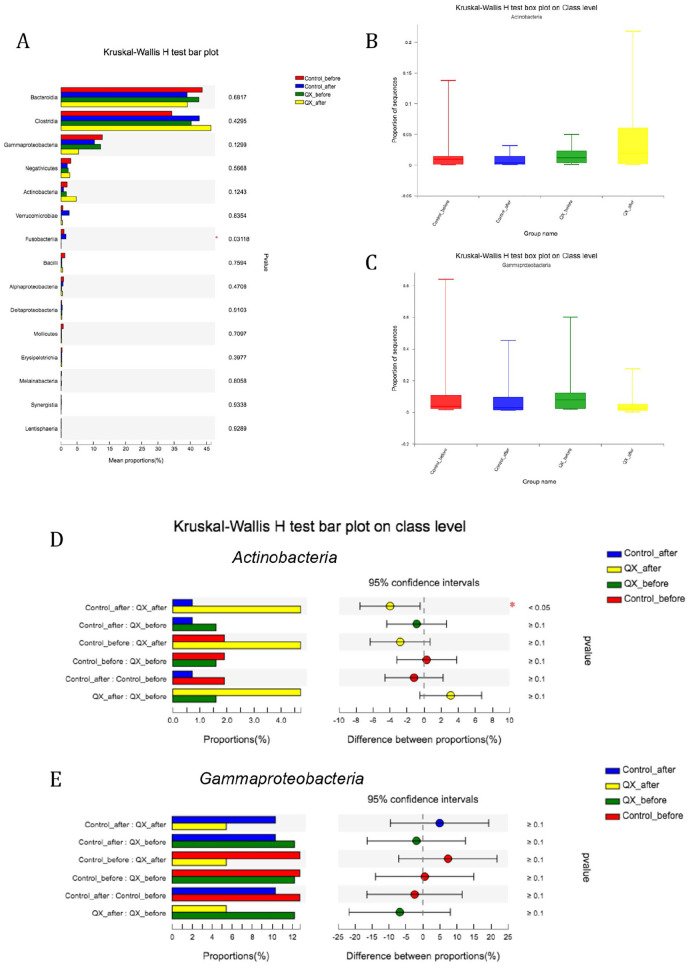
For beta analysis, PCoA on OTU level did not show significant differences on principal component 1 (PC1) before and after treatment in both the QX group and the control group (P = .43 and .34, respectively). Community difference analysis on Class level showed that both in the QX and control groups, the proportion of Clostridia increased after treatments (P = .43), while proportion of Bacteroidetes and Gammaproteobacteria decreased (P = .7 and .12, respectively, Figure 4A). Specifically, the proportion of Actinobacteria declined in the control group after treatments, but significantly increased after QX treatment (proportion after control treatment vs after QX treatments, 0.83% vs 4.7%, P = .017, Figure 4B and D). We also found the proportion of Gammaproteobacteria after treatment was higher in the control group than in QX group but not statistically significant (14.5% vs 5.4%, P = .25, Figure 4C and E).
Figure 4.
Difference of community proportions on class level among different groups. (A) Kruskal-Wallis H test bar plot among groups showed that the abundance of Actinobacteria increased in QX group and decreased in control group (P = .12). There was significant difference on the abundance of Fusobacterlia (P = .03), however the value is too minimum in QX group. (B) and (C) Box-plot showing the proportion of Actinobacteria and Gammaproteobacteria among QX and Control groups before and after treatments. (D) and (E) Comparisons between 2 groups for the proportion of Actinobacteria and Gammaproteobacteria. Statistical significance was set at P = .05.
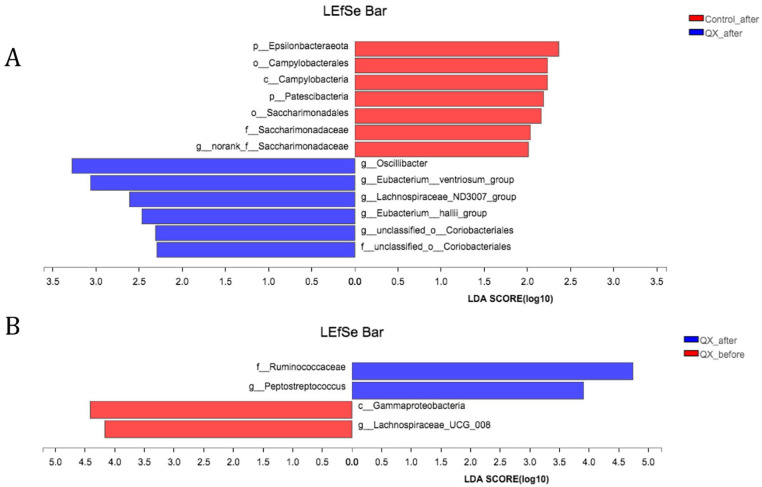
We then performed linear discriminant analysis of effect size (LEfSe) multi-level community difference analysis (from Genus to Phylum level) among groups. After treatments, samples from QX group were highly related with g_Oscillibacter, g_Eubacterium_ventriosum_group, g_Lachnospiraceae_ND3007_group, g_Eubacteriu_hallii_group, g_unclassified_o_coriobacteriales, while samples from control group were highly related with p_Epsilonbacteraeota, Campylobacteria, p_Patescibateria, Saccharimonadaceae (Figure 5A). Meanwhile, samples after QX treatments were related with f_Ruminococcaceae, g_Peptostreptococcus, compared with c_Gammaproteobacteria, and g_Lachnospiraceae_UCG_008 for samples before QX treatments (Figure 5B).
Figure 5.
LEfSe multi level species differences analysis between groups. (A) Samples after QX treatments and after control interventions. (B) Samples of QX group before and after treatments.
Environmental Factors Analysis of T Lymphocyte Subset and Microbiome in Different Groups
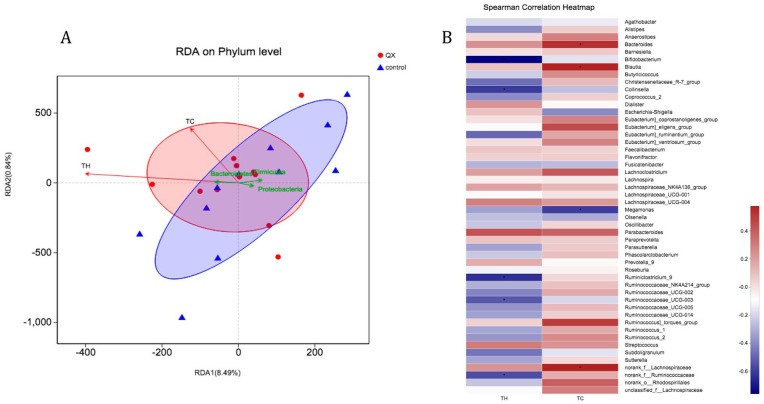
When taking TH and TC cells counts as environmental factors, RDA analysis showed that after QX and control interventions, TC and TH cells counts were associated with the distribution of stool samples and microbiome species from both groups but not statistically significantly (P = .42 and .11, respectively, Figure 6A). As seen in Figure 6A, TH and TC cell counts had higher relevance with stool samples from QX group than those from control group.
Figure 6.
Environmental factor analysis (A) and heatmap analysis (B) with TH and TC cells with samples and bacteria between QX and control group after interventions. (A) All samples were after 1-month intervention (red cycle or blue triangle). RDA analysis showed that TH cell and TC cells had higher relevance with samples after QX treatments than control group (Acute angle: positive correlation; obtuse angle: negative correlation; right angle: no correlation). Green arrows represent different bacteria. (B) Heatmap analysis between T cells and bacteria was done within QX group after treatments. Statistical significance was set at P = .05.
To further evaluate T cells’ association with certain bacteria, we performed heatmap analysis with samples after QX or control intervention. After QX treatment, on the Genus level, higher numbers of TH cells were significantly related with lower abundance of g_Bifidobacterium (coef. −0.76, P = .002), Collinsella (coef. −0.61, P = .02), Ruminiclostridium_9 (coef. −0.64, P = .01), and Ruminococcaceae_UCG-003 (coef. −0.54, P = .047). Meanwhile, higher amounts of TC cells were significantly related with lower abundance of Megamonas (coef. −0.60, P = .02), and with higher abundance of Bacteroides (coef. 0.54, P = .047), Blautia (coef. 0.58, P = .03) and norank_f_Lachnospiraceae (coef. 0.58, P = .03) (Figure 6B). There was no significant correlation between TH and TC cells with bacteria of samples after the control intervention.
Side Effects
Among 15 patients who received QX treatments, 2 of them reported diarrhea about 5 to 10 times per day. Such symptoms were relieved after reducing the dose of QX capsules. One patient in the control group reported hypertension and was hospitalized during the intervention.
Conclusion
In this study we found that compared with the control group, QX capsule could significantly increase numbers of T cells, especially TH cells of mCRC patients. After a 1-month intervention, fecal samples of both groups showed reduced abundance of Bacteroidetes and Escherichia-Shigella, as well as increased abundance of Firmicutes in the control group and Prevotella in the QX group. Specifically, the proportion of Actinobacteria was significantly higher after QX treatment than after control intervention, while the proportion of Gammaproteobacteria was relatively lower. Samples after QX treatment had higher relevance with TH and TC cell counts. Meanwhile, higher numbers of TH cell were significantly related with lower abundance of Actinobacteria on the Class level, and TC cells were positively related with Bacteroidetes and Blautia on the Genus level.
So far there are few studies about TCM herbal medicine’s effect on the interaction of the gut microbiome and CRC. Lv et al found that CT26 tumor-bearing mice fed with the TCM formula Gegen-Qinlian Decoction (GQD), had higher abundance of Erysipelotrichaceae, Lactobacillus, and Parasutterella in the gut microbiome. Furthermore, when using GQD together with anti-PD1 therapy, g_Bacteroides was enriched and the effect of anti-PD1 was enhanced, with higher levels of CD8 T cells and lower levels of CD4 T cells.19 Another TCM formula, CanMei formula, was found to upregulate the abundance of the genera Alistipes, Faecalibaculum, Lactobacillus, and Parabacterioides, while decreasing Desulfovibrionacea in colitis-associated colorectal tumor model mice, which might be associated with suppression of IL-17C and NF-κB to prevent CRC development.20 Yang et al found that Ganoderma Lingzhi, a TCM herb, could reduce Clostridium coccoides and Clostridium leptum, and increase Akkermansia muciniphila in rats fed high-fat diets and could reduce production of secondary bile acids, which is important to CRC.21 These findings provide important knowledge suggesting that different TCM herbal medicines could modulate certain gut bacteria and impact CRC development or treatment effect through immune, anti-inflammatory, or metabolic mechanisms, which is consistent with our results. However, to the best of our knowledge, our study is one of the first to investigate TCM herbs and the interaction of the microbiome and CRC in human beings.
Our study specifically found that QX could increase the abundance of Actinobacteria in stool samples of mCRC and such change was negatively related with numbers of TH cells. In 2017, Zhou et al reported an important finding that certain bacteria of the gut microbiome had anti-malignancy effects not only on primary tumor but also on remote metastases.22 Those bacteria mostly belonged to Actinobacteria on Phylum level and other phyla such as Proteobacteria and Firmicutes, however, the mechanism was unknown. It is known that Actinobacteria is an important bacterium which produces 50% to 60% of antibiotics in nature, for example streptomycin, oxytetracycline, tetracycline and gentamicin.23 In 2018, Ma et al published a paper in Science indicating that antibiotics could impact the gut microbiome and reduce the production of secondary bile acid while promoting primary bile acid (BA), thus regulating natural killer T (NKT) cells and inhibiting tumor growth of MYC liver cancer in a mouse model. They also found that CD8 T cells were not necessarily related with the microbiome-mediated BA-NKT antitumor pathway.24 Together, the antitumor effects of QX capsule might be related with the gut microbiome including Actinobacteria-mediated metabolism and natural killing immunity such as BA/NKT pathway, but this still needs further investigation.
Despite the negative relationship between Actinobacteria and TH cells which had been revealed in this study, we still observed that QX capsule could significantly increase TH cell counts among mCRC patients. Previous animal studies also supported such results. One of the possible explanations is that QX capsule might have dual effects of anti-tumor and body immunity enhancement, which is similar to TCM theory of eliminating pathogenic factors and stimulating body Zheng Qi synergistically. A previous vitro study revealed that SiJunZi decoction, a TCM formula stimulating spleen Qi, could induce the production of SFCAs which were associated Enterococcus, Sutterella, Butyricimonas, and Streptococcus.1 In our study, we also found that QX treatment was also positively related with Eubacterium, Lachnospiraceae, and Ruminococcaceae which were SFCA-producing and immunity-enhancing bacteria.25 In addition, SFCAs have been proved to upregulate TH cells in the tumor microenvironment18,26 and might explain QX capsule’s role on immunity enhancement.
We have certain limitations in this study. As mentioned above, QX capsule is a TCM compound which contains 20 different herbs and the active components are still not clear. Systemic pharmacological analysis is helpful to find out candidate herbal medicine and their effective components, which could target gut microbiome modulation, metabolism and immunity regulation for CRC. Since the human gut microbiome community is relatively stable under the ability of homeostasis, a 1-month intervention might be insufficient to observe long-term interaction between herbs and bacteria. However, mCRC patients’ conventional treatments such as systemic therapy or supportive care are more likely to be adjusted due to disease progression or other comorbidities. According to the inclusion and exclusion criteria of our study, a 1-month intervention phase is ethically appropriate for the study population. Although the current study was an RCT, results showed that there were significantly more patients with liver metastasis in control group than in QX group. This might add bias to the distribution of microbiome before and after intervention between 2 groups since patients with liver metastasis tend to have a more severe disease burden and worse survival outcomes.
From this RCT study, we found that QX capsules could enhance TH cells among mCRC patients and increase the abundance of gut anti-malignancy bacteria such as Actinobacteria. Although the abundance of Actinobacteria was negatively related with TH cells, other SFCA-producing and immunity-stimulating bacteria such as Lachnospiraceae were also enriched after QX treatments. These results indicated QX capsules might have dual effects of anti-tumor and stimulating TH cell immunity both mediated by the gut microbiome. Our study provided a better understanding on the mechanism of TCM compounds treating CRC and shed light on future study to detect active components of QX capsule which could target microbiome-immune response effects.
Acknowledgments
We thank all patients and their families who supported and participated our study. We also thank Dr Jun Mao from Memorial Sloan Kettering Cancer Center in the US for his guidance of the study design, as well as Dr Xu Yun and Zhang Tong who helped with patient recruitment.
Footnotes
Author Contributions: LS: manuscript drafting, study design, and statistical analysis. YunY: patient recruitment and observation. DC: study design. YufY: study design and administration support.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was funded by a grant from National Natural Science Foundation of China (General program 2015, No. 81573781).
Data Accessibility Statement: Some data generated or used during the study are available from the corresponding author by request.
ORCID iD: Lingyun Sun  https://orcid.org/0000-0002-7191-6177
https://orcid.org/0000-0002-7191-6177
References
- 1. Gao B, Wang R, Peng Y, et al. Effects of a homogeneous polysaccharide from Sijunzi decoction on human intestinal microbes and short chain fatty acids in vitro. J Ethnopharmacol. 2018;224:465-473. [DOI] [PubMed] [Google Scholar]
- 2. Fakih MG. Metastatic colorectal cancer: current state and future directions. J Clin Oncol. 2015;33:1809-1824. [DOI] [PubMed] [Google Scholar]
- 3. Joachim C, Macni J, Drame M, et al. Overall survival of colorectal cancer by stage at diagnosis: data from the Martinique Cancer Registry. Medicine (Baltimore). 2019;98: e16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dekker E, Tanis PJ, Vleugels JLA, et al. Colorectal cancer. Lancet. 2019;394:1467-1480. [DOI] [PubMed] [Google Scholar]
- 5. Abdel-Rahman O. A real-world, population-based study of the outcomes of patients with metastatic colorectal cancer to the peritoneum treated with or without cytoreductive surgery. Int J Colorectal Dis. 2020;35:719-725. [DOI] [PubMed] [Google Scholar]
- 6. Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahbari NN, Carr PR, Jansen L, et al. Time of metastasis and outcome in colorectal cancer. Ann Surg. 2019;269:494-502. [DOI] [PubMed] [Google Scholar]
- 8. Shen L, Li Q, Wang W, et al. Treatment patterns and direct medical costs of metastatic colorectal cancer patients: a retrospective study of electronic medical records from urban China. J Med Econ. 2020;23:456-463. [DOI] [PubMed] [Google Scholar]
- 9. Sun L, Mao JJ, Vertosick E, et al. Evaluating cancer patients’ expectations and barriers toward traditional Chinese medicine utilization in China: a patient-support group-based cross-sectional survey. Integr Cancer Ther. 2018;17:885-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang T, Yang YF, He B, et al. Efficacy and safety of quxie capsule () in metastatic colorectal cancer: a double-blind randomized placebo controlled trial. Chin J Integr Med. 2018; 24:171-177. [DOI] [PubMed] [Google Scholar]
- 11. Yang YF, Chen ZX, Xu Y. Randomized controlled study on effect of Quxie capsule on the median survival time and qualify of life in patients with advanced colorectal carcinoma. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2008;28:111-114. [PubMed] [Google Scholar]
- 12. Ning D, Yufei Y, Yinan L, et al. The mechanism study of Qingxue granule and Quxie capsule in inhibiting HCT-116 proliferation. Chin J Basic Med Tradit Chin Med. 2013;19:38-40. [Google Scholar]
- 13. Chen D, Yang Y, Yang P. Quxie capsule inhibits colon tumor growth partially through foxo1-mediated apoptosis and immune modulation. Integr Cancer Ther. 2019;18:1534735419846377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabit H, Cevik E, Tombuloglu H. Colorectal cancer: the epigenetic role of microbiome. World J Clin Cases. 2019;7:3683-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. 2019;11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shuwen H, Xi Y, Quan Q, et al. Relationship between intestinal microorganisms and T lymphocytes in colorectal cancer. Future Oncol. 2019;15:1655-1666. [DOI] [PubMed] [Google Scholar]
- 19. Lv J, Jia Y, Li J, et al. Gegen Qinlian decoction enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by remodelling the gut microbiota and the tumour microenvironment. Cell Death Dis. 2019;10:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H, Hui D, Li Y, et al. Canmei formula reduces colitis-associated colorectal carcinogenesis in mice by modulating the composition of gut microbiota. Front Oncol. 2019;9: 1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Y, Nirmagustina DE, Kumrungsee T, et al. Feeding of the water extract from Ganoderma lingzhi to rats modulates secondary bile acids, intestinal microflora, mucins, and propionate important to colon cancer. Biosci Biotechnol Biochem. 2017;81:1796-1804. [DOI] [PubMed] [Google Scholar]
- 22. Zhou YJ, Zhao DD, Liu H, et al. Cancer killers in the human gut microbiota: diverse phylogeny and broad spectra. Oncotarget. 2017;8:49574-49591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kothe E. Special focus: actinobacteria. J Basic Microbiol. 2018;58:719. [DOI] [PubMed] [Google Scholar]
- 24. Ma C, Han M, Heinrich B, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360:eaan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. dos Reis SA, da Conceição LL, Peluzio M. Intestinal microbiota and colorectal cancer: changes in the intestinal microenvironment and their relation to the disease. J Med Microbiol. 2019;68:1391-1407. [DOI] [PubMed] [Google Scholar]
- 26. Yuan C, Subramanian S. microRNA-mediated tumor-microbiota metabolic interactions in colorectal cancer. DNA Cell Biol. 2019;38:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]