Abstract
Chondroitin sulfate proteoglycan 4 (CSPG4) is a multifunctional transmembrane proteoglycan involved in spreading, migration and invasion of melanoma. In addition to the activating BRAF V600E mutation, CSPG4 was shown to promote MAPK signaling by mediating the growth-factor induced activation of receptor tyrosine kinases. However, it remains elusive which factors regulate CSPG4 expression. Therefore, the aim of the present study was to examine whether BRAF and MEK inhibitors have an effect on the expression of CSPG4. We exposed a panel of BRAF-mutant CSPG4-positive or -negative melanoma cell lines to BRAF and MEK inhibitors. Protein levels of CSPG4 were analyzed by flow cytometry (FACS), immunofluorescence microscopy (IF), and western blotting. CSPG4 mRNA levels were determined by quantitative PCR (qPCR). The prolonged exposure of cells to BRAF and MEK inhibitors resulted in markedly reduced levels of the CSPG4 protein in permanent resistant melanoma cells as well as decreased levels of its mRNA. We did not observe increasing levels of CSPG4 shedding into the culture supernatants. In addition, patient-derived matched tumor samples following therapy with kinase inhibitors showed decreased numbers of CSPG4-positive cells as compared to pre-therapy tumor samples. Our results indicate that BRAF and MEK inhibition downregulates CSPG4 expression until the cells have developed permanent resistance. Our findings provide the basis for further investigation of the role of CSPG4 in the development of drug-resistance in melanoma cells.
Keywords: melanoma, CSPG4, BRAF-mutated melanoma, MAPK pathway, drug resistance
Introduction
Malignant melanoma represents the most aggressive and deadliest form of skin cancer. It is responsible for 1.7% of newly diagnosed primary malignant cancers and leads to about 0.7% of all cancer-related deaths worldwide each year (1). Almost 50% of metastatic melanoma patients harbor a BRAF V600 mutation, among which the most common is a single nucleotide mutation resulting in the substitution of glutamic acid for valine (BRAF V600E) (2,3). BRAF V600E leads to a constitutive activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathway, as well as to insensitivity to negative feedback mechanisms (4). In recent years, the use of molecular inhibitors targeting mutant BRAF and its downstream effector MEK became a valid anti-melanoma therapeutic strategy. The first approved selective inhibitor of mutant BRAF was vemurafenib (PLX4032), which demonstrated highly beneficial early therapeutic effects (5). Unfortunately, a complete response is rarely observed due to either acquired or intrinsic resistance to the therapy (6,7). The combinational treatment with BRAF and MEK inhibitors resulted in an improved activity, but acquired resistance to the BRAF inhibitor remains the major obstacle to an effective cure of metastatic melanoma (8–10). Increasing evidence suggests that in addition to acquired mutations that confer drug resistance, phenotype plasticity is a key mechanism that allows for adaptation of melanoma cells to the drug and for drug resistance (11–14). Drug exposure - along with other unfavorable environmental conditions, such as hypoxia and nutrient starvation - leads to an early innate cell response in melanoma cells, resulting in multidrug resistance in so-called induced drug-tolerant cells (IDTCs) (15). The phenotype of IDTCs is reversible upon drug withdrawal. However, continuous exposure to the treatment eventually leads to transition of IDTCs into permanently resistant tumor cells.
The lack of optimal treatment options for patients with metastatic melanoma urged the cancer research community to look for novel strategies aiming at subverting eventual drug resistance and enhancing therapeutic efficacy. Targeting specific molecules, involved in multiple signaling pathways in melanoma cells and in several cellular functions important for cancer spreading, has the potential to improve clinical outcomes.
One such candidate target molecule is chondroitin sulfate proteoglycan 4 (CSPG4) (16,17). Structurally, CSPG4 is a single-pass type I transmembrane protein expressed as an either N-linked glycoprotein of approximately 280 kDa, or as an approximately 450 kDa chondroitin sulfate proteoglycan (18,19). The core protein of CSPG4 contains three major structural domains: A large, 2221-amino acid extracellular domain, a 25-amino acid hydrophobic transmembrane region, and a short 75-amino acid cytoplasmic tail (20). The extracellular region of CSPG4 contains the sequences for the modification with chondroitin sulfate (CS) chains that may confer different attributes of the cell (21–23). In the cytoplasmic domain of CSPG4, there are sites critical for CSPG4 function: The threonine phosphoacceptor sites phosphorylated by PKCα and ERK1/2, the PDZ domain binding motif that is the site for attachment of scaffold proteins, such as MUPP1, syntenin and GRIP1, and a proline-rich region (PRR) comprising a non-canonical SH3 protein interaction domain (16).
CSPG4 does not exhibit any known intrinsic catalytic activity (20). It functions as a scaffold protein, activating two major signaling pathways associated with oncogenic transformation, in particular the integrin-regulated FAK pathway and RTK signaling through the MAPK cascade, with downstream ERK1/2 (20). Activation of these pathways by CSPG4 leads to the regulation of a number of cellular functions that drive cytoskeletal reorganization, survival and chemoresistance, invasion, migration and proliferation, as well as epithelial to mesenchymal transition (EMT) in the radial growth phase of human melanomas (24,25).
Although CSPG4 received much attention as a potential therapeutic agent in recent years (16,17,26–31), there is little information concerning the mechanisms that regulate the expression of this proteoglycan (32). To the best of our knowledge, no study has investigated whether CSPG4 expression is altered by inhibiting signaling through the MAPK pathway.
In the present study, we demonstrated that a panel of induced-drug tolerant and drug-resistant melanoma cells exhibit lower levels of CSPG4 than the parental cells. Furthermore, exposure of the cells to the BRAF inhibitor PLX4032 resulted in markedly reduced levels of the CSPG4 protein in cell lysates, as well as decreased levels of its mRNA, with no increase in protein shedding into the culture supernatant. In addition, patient-derived tumor samples pre- and post-treatment with kinase inhibitors showed decreased numbers of CSPG4-positive cells after therapy. Our results indicate that BRAF and MEK inhibition can be regulatory factors of CSPG4 expression.
Materials and methods
Cell lines
The human melanoma cell line M14 was described previously (29) and the human melanoma cell lines WM9, WM35, WM164, WM1366, CJM, D24 and 451Lu were provided by Helmut Schaider's laboratory (University of Queensland, Australia). Cells were maintained in RPMI-1640 medium with 2 mM L-glutamine and 25 mM HEPES (Lonza Group, Ltd.), supplemented with either 10% FBS (M14, CJM and D24) or 5% FBS (WM9, WM35, WM164, WM1366 and 451Lu) and 1% penicillin-streptomycin (Gibco, Thermo Fischer Scientific, Inc.). Cells were cultured in a humidified atmosphere containing 5% CO2 and 95% ambient air at 37°C. The cell lines WM9, WM35, WM164, 451Lu and M14 were BRAF V600-mutated, CJM and WM1366 were NRAS Q61L-mutated, and D24 carried neither BRAF nor NRAS mutations. Prior to the experiments, all cell lines were tested negative for mycoplasma.
Antibodies
The mouse anti-CSPG4 monoclonal antibody clone 9.2.27 (#554275) was purchased from BD Biosciences. The donkey anti-mouse Alexa Fluor 488® (#A-21202) and goat anti-mouse Alexa Fluor 568® (#A-11004) secondary IgG antibodies were obtained from Life Technologies Corporation. Anti-CD271 (LNGFR) APC-conjugated human monoclonal antibody (#130-091-884) was purchased from MACS Miltenyi Biotec. The APC mouse IgG1, κ isotype control (FC) antibody (#400122) was obtained from BioLegend, Inc. The mouse monoclonal antibodies anti-Ki67 (sc-23900), as well as mAbs against CSPG4: Anti-NG2 clone G-9 (sc-166251) and anti-NG2 clone LHM 2 (sc-53389), were purchased from Santa Cruz Biotechnology, Inc. The primary anti-human CSPG4 antibody clone 132.38 (#ab50009) was purchased from Abcam. Control mouse IgG (#I8765) was obtained from Sigma-Aldrich (Merck KGaA). The mouse monoclonal antibodies anti-p44/42 MAPK (Erk1/2) clone L34F12 (#4696), anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) clone E10 (#9106), anti-Akt (pan) clone 40D4 (#2920), anti-phospho-Akt (Ser473) clone 587F11 (#4051) and anti-β-actin clone 8H10D10 (#3700) were purchased from Cell Signaling Technology, Inc. Anti-mouse IgG, HRP-linked secondary antibody (#7076) was obtained from Cell Signaling Technology, Inc.
Inhibitors and cell treatment
Inhibitors of mutant BRAF V600, vemurafenib (PLX4032) and dabrafenib (GSK2118436) and the MEK1/2 inhibitor trametinib (GSK1120212) were purchased from Selleckchem. Induced drug tolerant cells (IDTCs) were generated by exposure to PLX4032 (250 and 1,000 nM) for a minimum of 7 days. Drug resistant cells were obtained by exposure to PLX4032 (250 and 1,000 nM) for a minimum of 30 days. To confirm multidrug resistance of the cells, either additional MEK1/2 inhibitor GSK1120212 was applied (5 nM) or PLX4032 concentration was increased up to 10 µM. The media containing fresh drugs were replenished every third day for the period of the experiments.
In order to determine suboptimal doses of PLX4032 for each cell line, a CytoSelect™ MTT Cell Proliferation Assay (Cell Biolabs, Inc.) was performed according to the manufacturer's instructions. Briefly, cells were seeded in triplicates at a density of 6,000 per well in 96-well plates and subjected to the following concentrations of PLX4032: 0, 0.1, 1, 10, 100, 250, 500, 1,000, 5,000 and 10,000 nM for 72 h. Cells were then incubated with MTT reagent and solubilized. Absorbance was measured at 540 nm using a Spark® multimode microplate reader (Tecan Group Ltd.). Data are presented as percent of inhibition of PLX4032-exposed cells compared to the untreated cells.
Flow cytometry
Parental, induced-drug tolerant and drug resistant melanoma cells were harvested by scraping, washed with 1X PBS and dispensed into 5 ml polystyrene round-bottom FACS tubes. Cells were then incubated with Fixable Viability Dye eFluor® 506 (Affymetrix, eBioscience) according to the manufacturer's protocol. Next, the cells were washed with FACS buffer [0.5% BSA and 0.05% sodium azide (NaN3) in 1X PBS] and incubated with anti-CSPG4 antibody 9.2.27 (1:1,000) for 10 min at 4°C, washed with FACS buffer and incubated with donkey anti-mouse secondary IgG antibodies Alexa Fluor 488® (1:500) for 15 min at 4°C, protected from light. As an IgG control, cells were incubated with Alexa Fluor 488® secondary antibody only. For additional CD271 staining, the cells were incubated with anti-CD271-APC antibody (1:100) for 15 min at 4°C, protected from light. As a control, cells were incubated with the APC mouse IgG1, κ isotype antibody (1:100). Cells were washed and resuspended in FACS buffer. The samples were analyzed with a FACS Canto flow cytometer (BD Biosciences). FlowJo software version 10.6.1 (TreeStar Inc.) was used for analysis of the results.
Immunofluorescence
Melanoma cells were plated into 24-well cultivation plates with glass coverslips and were allowed to attach overnight. Cells were exposed to 1,000 nM PLX4032 for up to 7 days. At specific time points, indicated in the results, cells were washed with 1X PBS and fixed with 4% paraformaldehyde solution in 1X PBS for 30 min at room temperature (RT). The slides were washed with 1X PBS and the cells were blocked and permeabilized in blocking buffer (1X PBS containing 5% goat serum and 0.5% saponine) for 1 h at RT. Cells were incubated for 1 h at RT with anti-CSPG4 antibody 9.2.27, diluted 1:1,000 in blocking buffer, washed three times with 1X PBS and incubated for 1 h at RT with goat anti-mouse secondary IgG antibodies Alexa Fluor 568®, diluted 1:2,000 in blocking buffer. Finally, nuclei of the cells were counterstained with DAPI (1:10,000), slides were washed three times with 1X PBS and mounted on a glass slide using Fluoromount-G (Thermo Fisher Scientific, Inc.). A wide-field fluorescence microscope (Imager Z1/Zeiss) in combination with TissueFAXS software version 4.2.6245.1019 (TissueGnostic GmbH) was used to examine the slides.
RNA extraction and quantitative reverse transcription-PCR
Total RNA from non-treated and drug-exposed melanoma cells was isolated using an RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. cDNA was synthetized from 1 µg of total RNA using a High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The following primers were used (5′-3′): CSPG4_F, CCTCCTGCTGCAGCTCTACT and CSPG4_R, CTGAGGAGGCGTTCAGAAAC; GAPDH_F, ACGGATTTGGTCGTATTGGG and GAPDH_R, TGATTTTGGAGGGATCTCGC; hUBC_F, ATTTGGGTCGCAGTTCTTG and hUBC_R, TGCCTTGACATTCTCGATGGT. RT-qPCR analysis of CSPG4 and GAPDH or hUBC as an internal standards were performed on a 7900HT Fast-Real Time PCR System using the Power SYBR® Green PCR Master Mix according to manufacturer's instructions (Applied Biosystems, Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min, and the melting curve stage at 95°C for 15 sec, 60°C for 15 sec, and 95°C for 15 sec. The results were analyzed using the Sequence Detection Systems (SDS) software version 2.4 (Applied Biosystems, Thermo Fisher Scientific, Inc.) and relative gene expression levels were calculated as ΔCT. CSPG4 expression in melanoma cell lines was calculated as 100/ΔCT relative to GAPDH. Fold change expression of CSPG4 after treatment was calculated using the 2−∆∆Cq method (33).
Western blot analysis
Non-treated and drug-exposed melanoma cells were harvested by scraping and cell pellets were lysed in 1X RIPA buffer (Sigma-Aldrich, Merck KGaA) with 1X Protease/Phosphatase Inhibitor Cocktail (Cell Signaling Technology, Inc.). Lysates were incubated with Chondroitinase ABC (Sigma-Aldrich, Merck KGaA) at the working concentration 1 U/ml for 30 min at 37°C. Protein concentration in cell lysates was measured by Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.) and equal amounts of proteins were separated by SDS-PAGE (8% polyacrylamide gel) under reducing conditions and transferred onto polyvinylidene fluoride (PVDF) membranes (GE Healthcare Life Sciences, Thermo Fisher Scientific).
Equal volumes of supernatants of non-treated and drug-exposed melanoma cells were collected and concentrated eight times (from 400 to 50 µl) using a Vacuum Concentrator Centrifuge UNIVAPO 150 ECH (UniEquip GmbH). Next, 10 µl of concentrated supernatants were centrifuged at 14,000 × g for 30 min at 4°C to remove remaining aggregates. Five microliters of resulting supernatants were carefully collected and mixed 1:1 with ddH2O and with 4X reducing sample buffer. Samples were then separated by SDS-PAGE (6% polyacrylamide gel) under reducing conditions and transferred onto polyvinylidene fluoride (PVDF) membranes (GE Healthcare Life Sciences, Thermo Fisher Scientific, Inc.).
Membranes were blocked in 5% milk TBS-T for 1 h at RT and incubated with primary antibodies overnight at 4°C. The following dilutions of primary antibodies in 2% milk TBS-T were used: Anti-Ki67 (1:500), anti-NG2 clone G-9 (1:1,000), anti-NG2 clone LHM 2 (1:800), anti-Erk1/2 (1:2,000), anti-phospho-Erk1/2 (1:2,000), anti-Akt (1:3,000), anti-phospho-Akt (1:1,000) and anti-β-actin (1:1,000). Corresponding peroxidase-conjugated secondary mAbs were used (1:5,000). Blots were developed using the Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) and bands were visualized using the ChemiDoc Imaging System (Bio-Rad Laboratories, Inc.). The densitometric analysis of the intensity of the bands was performed using the ImageJ software (National Institutes of Health).
Immunohistochemistry
Formalin-fixed paraffin-embedded matched tumor samples from five patients before and after progression during a therapy with BRAF/MEK inhibitors from the archives of the Department of Dermatology and the Department of Pathology at the University Hospital St. Poelten, Karl Landsteiner University of Health Sciences were processed. The collection and storage of samples were performed according to local ethical guidelines. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Karl Landsteiner University (EC number: 1011/2019). The tissue was deparaffinized and stained using the BenchMark XT automated immune-staining platform (Ventana Medical System) and the ultraView Universal DAB detection system (Ventana Medical System). Antigen retrieval was performed before using prediluted, commercially available anti-CSPG4 antibodies (Abcam clone 132.38, dilution 1:500). Scoring of tissue slides was performed independently by 2 investigators. The percentage of positive cells and the intensity of staining were graded from 0 to 3+: 0, no staining; 1+, weak positive staining; 2+, moderate positive staining; 3+, strong positive staining. H&E staining was performed on 5-µm paraffin sections. The stained sections were examined using an Olympus BX53 microscope and photographed with an Olympus DP73 camera (Olympus Electronics).
Statistical analysis
Statistical analyses of CSPG4 expression in parental and PLX4032-exposed melanoma cells in western blotting and RT-qPCR as well as of ratios of pERK/ERK and pAKT/AKT in parental and PLX4032-exposed melanoma cells was carried out using the one-way ANOVA with Tukey's multiple comparison test. P-values <0.01 were regarded as significant (***P<0.0001, **P<0.001, *P<0.01). The Spearman's correlation test was used to determine the association between cell-based CSPG4 expression levels and CSPG4 ectodomain shedding levels. The results of the densitometric analyses of specific western blots were used for the calculation. P-value <0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism software version 4.03 (GraphPad Software, Inc.).
Results
Expression of CSPG4 in human melanoma cell lines
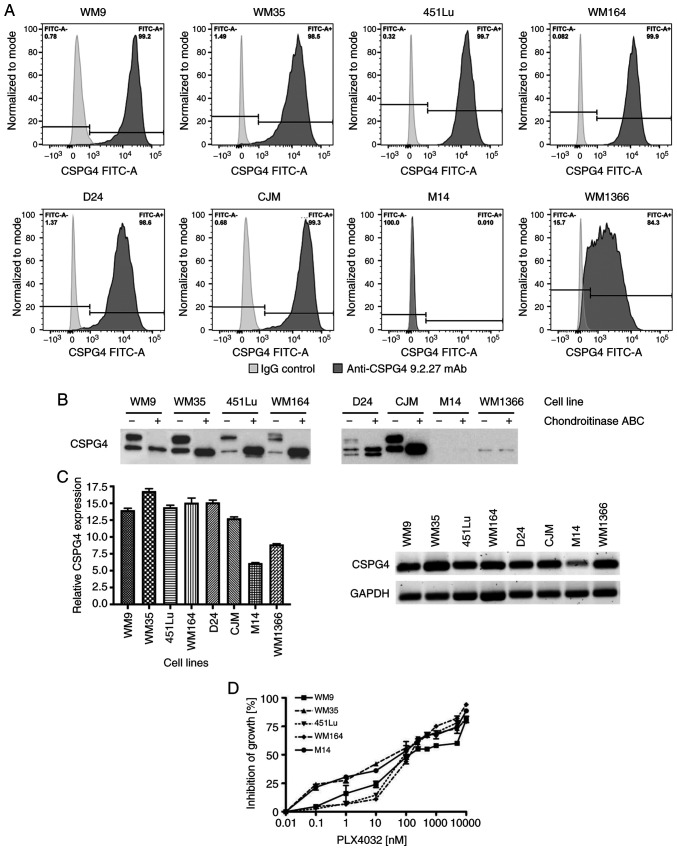
The protein and gene expression of CSPG4 was evaluated in a panel of melanoma cell lines: BRAF V600E-mutated (WM9, WM35, 451Lu, WM164 and M14), NRAS Q61L-mutated (CJM, WM1366) or carrying neither a BRAF nor an NRAS mutation (D24). CSPG4 protein expression on the cell surface was assessed by flow cytometry using the CSPG4-specific 9.2.27 mAb. Six out of eight cell lines tested showed a high percentage (>98%) of CSPG4-positive cells (WM9, WM35, 451Lu, WM164, D24 and CJM) (Fig. 1A). In the WM1366 cell line, a lower percentage of cells (~84%) was recognized by the anti-CSPG4 antibody. In contrast, CSPG4 was not detected on the surface of M14 cells (Fig. 1A). Western blot analysis (Fig. 1B) confirmed these results, by demonstrating expression of CSPG4 in all tested cell lines, except in M14. In WM9, WM35, 451Lu, WM164 and CJM cells, two major forms of CSPG4 were detected: A single, non-chondroitin sulfated glycoprotein of approximately 280 kDa (CSPG4-280), and a 450 kDa chondroitin sulfated proteoglycan (CSPG4-450). The D24 cell line demonstrated expression of an additional, lighter band of around 250 kDa, along with the 280 and 450 kDa forms of CSPG4. In WM1366 cells, CSPG4 was detected as a single protein of 250 kDa, which was not influenced by chondroitinase ABC treatment.
Figure 1.
CSPG4 expression in melanoma cell lines. (A) WM9, WM35, 451Lu, WM164, D24, CJM, M14 and WM1366 melanoma cells were subjected to flow cytometry to determine the percentage of CSPG4-positive cells using the anti-CSPG4 9.2.27 mAb. Donkey anti-mouse secondary antibody, Alexa Fluor 488®, was used as an IgG control. Data are representative of triplicates. (B) CSPG4 expression in WM9, WM35, 451Lu, WM164, D24, CJM, M14 and WM1366 cell lysates was evaluated by western blotting using the anti-CSPG4 mAb (clone LHM 2). Samples were untreated or treated with Chondroitinase ABC. In WM9, WM35, 451Lu, WM164 and CJM, CSPG4 was detected as a 280 kDa lower band and a 450 kDa upper band, in D24 as 250, 280 and 450 kDa bands, in WM1366 as 250 kDa band. (C) CSPG4 mRNA levels, normalized to the internal control (GAPDH), were analyzed by qPCR using specific primers. Bars represent mean ± SD from triplicates. Agarose gel electrophoresis confirmed the specific qPCR products. (D) Dose-response effect of PLX4032 on the growth of melanoma cell lines. The results are presented as percent inhibition of cell growth, compared to untreated cells. Bars represent mean ± SD from duplicates. PLX4032 at 0.01 nM corresponds to 0 nM PLX4032 (untreated cells), due to Log 10 scale representation of the x-axis. CSPG4, chondroitin sulfate proteoglycan 4.
In addition, CSPG4 mRNA levels were assessed in melanoma cells by qPCR. The presence of CSPG4 transcripts was confirmed in all cell lines exhibiting the expression of the proteoglycan (WM9, WM35, 451Lu, WM164, D24, CJM and WM1366) (Fig. 1C). Only very low amounts of CSPG4 mRNA were detected in the M14 cell line (Fig. 1C).
Differences in CSPG4 protein expression in parental, induced drug-tolerant and drug-resistant melanoma cells
Cell lines harboring the BRAF V600 mutation (WM9, WM35, 451Lu and WM164) with a high CSPG4 expression were used for the following experiments. The CSPG4-negative cell line M14 served as a control. To determine the most appropriate doses of PLX4032 on the different melanoma cell lines, dose-titration experiments were performed. Treatment of WM9, WM35, 451Lu, WM164 and M14 cell lines with increasing concentrations of PLX4032 resulted in a dose-dependent inhibition of all cell lines. The growth of all five cell lines exposed to PLX4032 in the concentration range of 250–1,000 nM was inhibited in 50–70% (Fig. 1D) and therefore, the concentrations of 250 and 1,000 nM PLX4032 were used for the study.
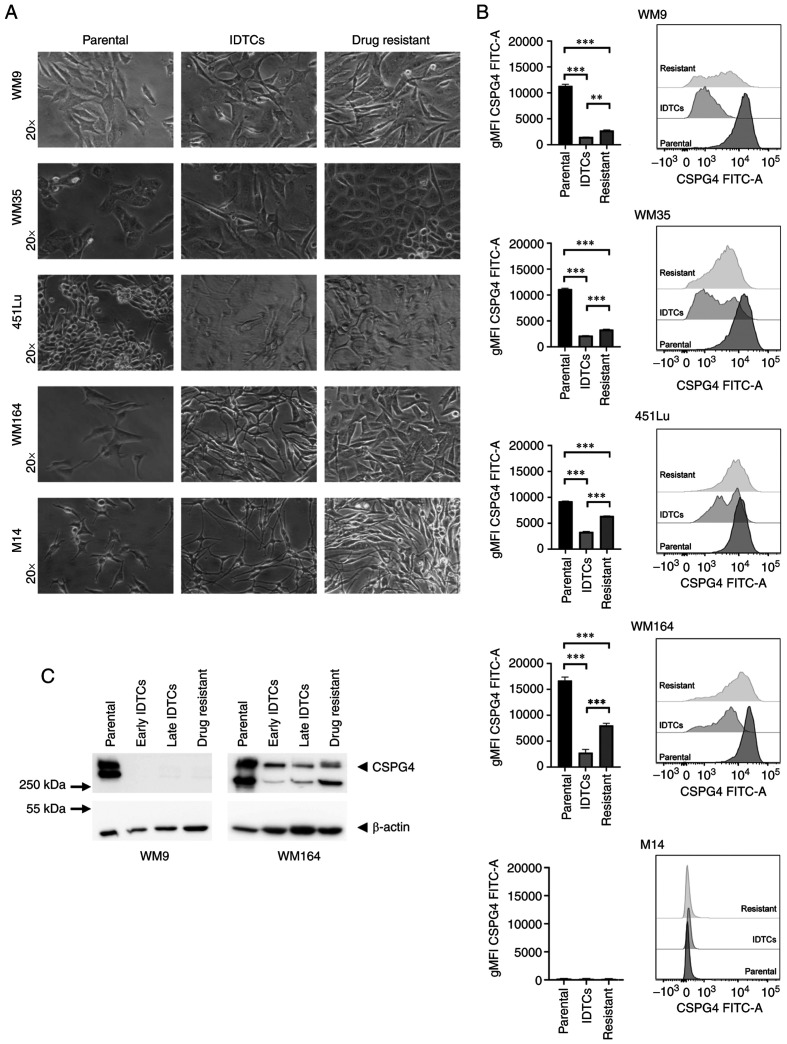
First, we generated induced drug-tolerant cells (IDTCs) and drug-resistant cells by exposing the parental cells to the BRAF inhibitor PLX4032. IDTCs demonstrated characteristic morphological changes, such as elongated shape and dendrite-like structures, as well as elevated CD271 expression (15). CD271 was used to verify the IDTC status in all experiments. Continuous exposure of IDTCs to PLX4032 resulted in the transformation into permanent drug-resistant cells that again proliferated (Fig. 2A). We next evaluated the expression of CSPG4 in different stages of BRAF inhibition by flow cytometry. Interestingly, the geometric mean intensity of the CSPG4 signal was significantly lower in IDTCs and drug-resistant cells, as compared to the parental cells for all analyzed cell lines (Fig. 2B). In 451Lu and WM164 resistant cells, the expression of CSPG4 was significantly higher than in IDTCs, but did not reach the level of the corresponding parental cells (Fig. 2B). WM9 and WM35 drug-resistant cells demonstrated a slightly but still significantly higher CSPG4 expression than IDTCs (Fig. 2B). M14 underwent the same morphological changes after exposure to PLX4032 as observed for the other cell lines. Expression of CSPG4 by M14 cells was not observed at any stage (Fig. 2A and B).
Figure 2.
Differences in CSPG4 expression in parental, induced drug-tolerant and drug-resistant melanoma cells. (A) WM9, WM35, 451Lu, WM164 and M14 cells were exposed to PLX4032 in order to generate induced drug-tolerant cells (IDTCs) and drug-resistant cells. The parental cells and corresponding IDTCs and resistant cells were examined by bright-field microscopy and representative photos were taken. (B) The expression of CSPG4 in parental, IDTCs and drug-resistant WM9, WM35, 451Lu, WM164 and M14 cells was evaluated by flow cytometry. Results are presented as geometric mean of fluorescence intensity (gMFI) (left panel) and representative histograms (right panel). Bars represent mean ± SD from triplicates. Statistical analysis was carried out using the one-way ANOVA with Tukey's multiple comparisons test. ***P<0.0001, **P<0.001. (C) Western blot analysis of CSPG4 (280 and 450 kDa) expression in WM9 and WM164 parental, early and late IDTCs as well as drug-resistant cells. β-actin (45 kDa) was used as a loading control. CSPG4, chondroitin sulfate proteoglycan 4; IDTCs, induced-drug tolerant cells.
These results obtained by flow cytometry were confirmed by western blotting. CSPG4 was not detected in IDTCs and drug-resistant WM9 cell lysates (Fig. 2C). In WM164 early and late IDTCs, the amounts of CSPG4-280 and CSPG4-450 were decreased, as compared to the parental cells. Interestingly, in drug-resistant cells the expression of CSPG4 was increased, mainly the 280 kDa component not decorated with chondroitin sulfate (Fig. 2C). Overall, the expression of this proteoglycan decreased upon BRAF inhibition in IDTCs and increased again in the corresponding resistant cell lines, but the exact time points of these events were cell line-specific.
CSPG4 protein expression over time upon BRAF inhibition
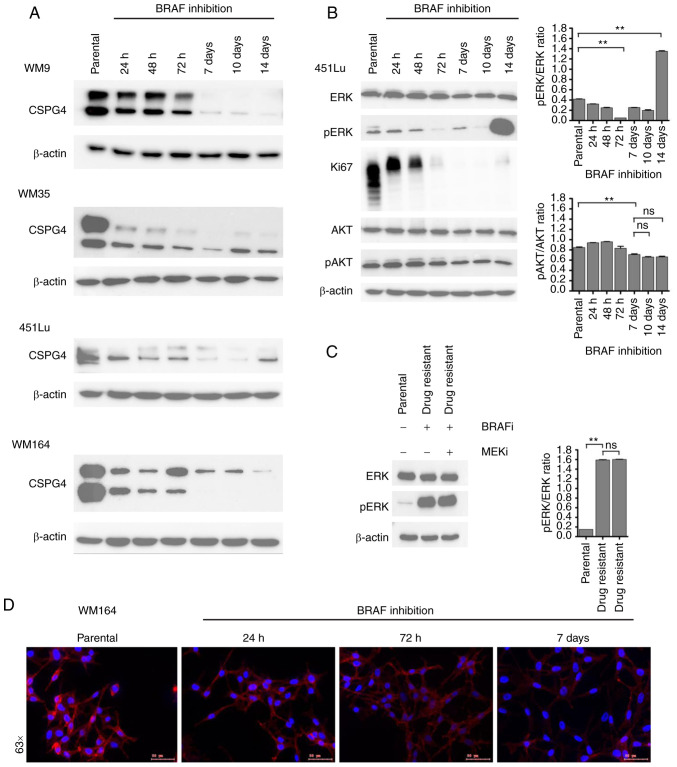
To narrow down the time frame when changes in CSPG4 expression upon BRAF inhibition occur, we performed time course experiments. We exposed four cell lines (WM9, WM35, 451Lu and WM164) to PLX4032 for 14 days. At specific time points, samples were collected and CSPG4 expression was analyzed by western blotting. On days 7, 10 and 14 in the WM9, WM35 and WM164 cell lines and on days 7 and 10 in the 451Lu cell line, cells presented an IDTC phenotype, as shown in Fig. 2A. Interestingly, each cell line exhibited certain dynamics of changes in CSPG4 expression (Fig. 3A). In WM9 cells, the CSPG4-280 and CSPG4-450 amounts were lower after 72 h of BRAF inhibition and from day 7 on they were detectable only in minute amounts. In the WM35 cell line, the expression of CSPG4-280 and CSPG4-450 decreased as soon as 24 h after drug exposure. Interestingly, the 280 kDa core protein was detected at each time point, while the chondroitin sulfate modified component decreased gradually over time. The amounts of CSPG4 in 451Lu cells were found to be lower already after 24 h. CSPG4-280 was detected until 72 h and continued to decrease until day 10, while CSPG4-450 was downregulated at each time point. On day 14, the 280 kDa core protein was detected again. The expression of the proteoglycan in WM164 cells was markedly lower after 24 h of BRAF inhibition and continued to decrease over time (Fig. 3A). Both CSPG4-280 and CSPG4-450 were detected only until the 72 h time point. From day 7 on, only CSPG4-450 was detectable, and its amounts decreased over time.
Figure 3.
Changes in CSPG4 expression over time upon BRAF inhibition. (A) WM9, WM35, 451Lu and WM164 cells were exposed to BRAF inhibitor (250 nM PLX4032) for 14 days. Samples were collected at indicated time points and CSPG4 (280 and 450 kDa) expression was analyzed in cell lysates by western blotting. β-actin (45 kDa) was used as a loading control. (B) Protein lysates of 451Lu parental and PLX4032-exposed cells were subjected to western blotting for expression levels of Ki67 (345/395 kDa), p44/42 MAPK (Erk1/2; 42/44 kDa), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204; 42/44 kDa), total AKT (60 kDa), phospho-AKT (Ser473; 60 kDa). β-actin (45 kDa) was used as a loading control. Histograms represent pERK and pAKT expression normalized to ERK and AKT, respectively (right panels). Statistical analysis was carried out using one-way ANOVA with Tukey's multiple comparisons test. **P<0.001; ns, not significant. (C) Lysates of 451Lu parental and PLX-resistant cells, exposed to BRAF and/or MEK inhibitor (250 nM PLX4032 and/or 5 nM GSK1120212, respectively) were subjected to western blotting to detect p44/42 MAPK (Erk1/2; 42/44 kDa) and phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204; 42/44 kDa). β-actin (45 kDa) was used as a loading control. Histogram represents pERK expression normalized to ERK (right panel). Statistical analysis was carried out using one-way ANOVA with Tukey's multiple comparisons test. **P<0.001; ns, not significant. (D) Immunofluorescent labelling of CSPG4 in WM164 parental and 1,000 nM PLX4032-exposed cells was performed using anti-CSPG4 9.2.27 mAb and goat anti-mouse IgG secondary antibody, Alexa Fluor 568®. CSPG4, chondroitin sulfate proteoglycan 4.
In addition to western blot analysis, immunofluorescence labelling of CSPG4 in WM164 parental and drug-exposed cells was performed (Fig. 3D). The cells showed a homogenous pattern of CSPG4 staining, with some punctate staining on the cell membrane, mainly in untreated cells. The immunofluorescence analysis confirmed lower CSPG4 levels in cells exposed to PLX4032, as compared to parental cells (Fig. 3D).
CSPG4 is known to be involved in the activation of two major cell signaling cascades, namely integrin/focal adhesion kinase (FAK) signaling and MAPK pathway. Therefore, we investigated whether - besides the fluctuations in CSPG4 expression - there were also changes in the activation of the pathway components, ERK and AKT. For this experiment, we chose 451Lu cells, as we previously observed a cycle of CSPG4 downregulation within 10 days of drug exposure and subsequent upregulation (after 14 days) (Fig. 3A). We evaluated the activity of ERK and AKT, as well as the expression of Ki67 in 451Lu cell lysates at specific time points of PLX4032 exposure by immunoblotting (Fig. 3B). The inhibition of cell proliferation was observed after 72 h and lasted until day 14, when a faint band of Ki67 was again detected, suggesting that the cells started to proliferate at this time point. Significant downregulation of ERK signaling was achieved after 72 h and on day 14 a reactivation of signaling was observed. After exposure of 451Lu cells to PLX4032 for 14 days, we observed that phosphorylation of ERK was still significantly high (Fig. 3C). The addition of the MEK inhibitor did not lead to downregulation of ERK signaling, suggesting the development of multidrug-resistance in the cells. Interestingly, after 7 days of treatment, significantly reduced AKT (Ser473) phosphorylation was detected and the similar phosphorylation level remained until day 14 (Fig. 3B).
Decreased levels of CSPG4 in WM9, WM35, 451Lu and WM164 melanoma cells after drug exposure are not due to ectodomain shedding
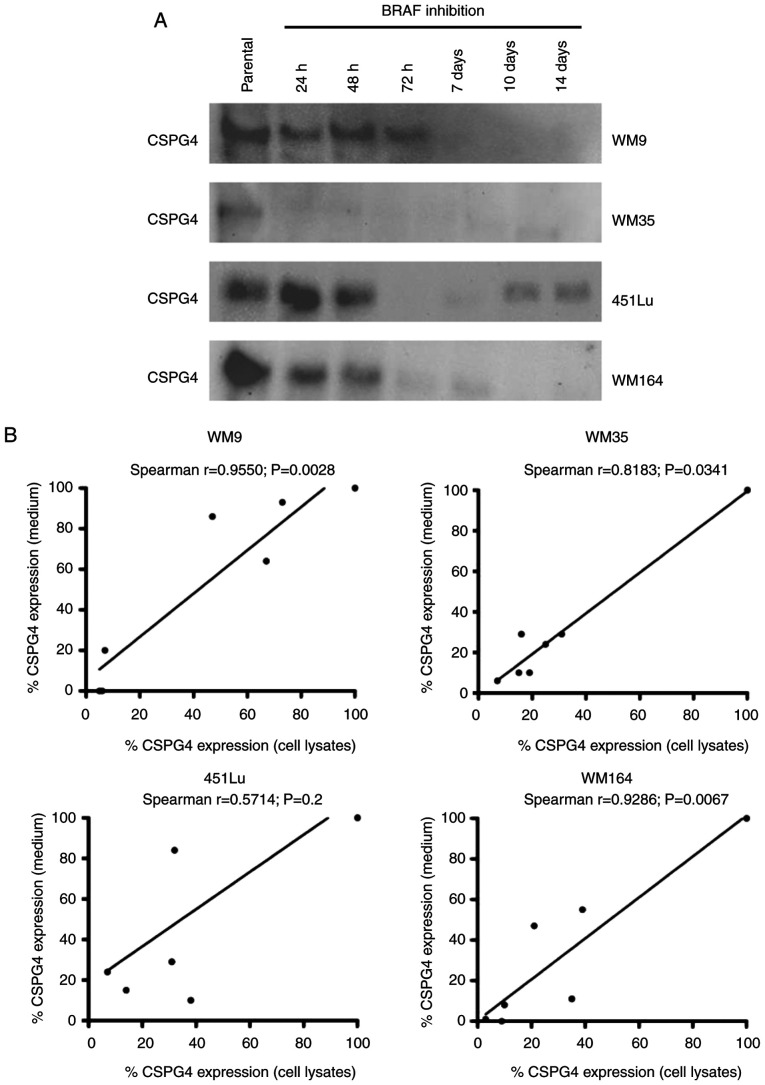
The extracellular domain of CSPG4 contains a number of putative proteolytic cleavage sites (34). Hence, in the next step, we investigated whether the observed low levels of CSPG4 in PLX4032-exposed cells were the result of protein shedding. Four cell lines (WM9, WM35, 451Lu and WM164) were exposed to PLX4032 for 14 days. Medium from cultured cells was collected at specific time points and analyzed for CSPG4 expression by western blotting (Fig. 4A). The analysis detected CSPG4 component of approximately 130 kDa. The highest amounts of shed protein were observed in all parental cells tested and decreased over time upon BRAF inhibition. The Spearman correlation test demonstrated a significant correlation between cell-based CSPG4 expression levels and CSPG4 ectodomain shedding levels for WM9 (Spearman r=0.9550), WM35 (Spearman r=0.8183) and WM164 (Spearman r=0.9286) cell lines (Fig. 4B). This indicates that increased synthesis of CSPG4 in these cell lines was associated with increased shedding of the CSPG4 ectodomain, suggesting that the inhibition of expression may occur on the genomic level.
Figure 4.
Shedding of CSPG4 ectodomain after exposure of melanoma cells to BRAF inhibitor. WM9, WM35, 451Lu and WM164 cells were exposed to 250 nM PLX4032 for 14 days. (A) Medium from cultured cells was collected at indicated time points and analyzed for CSPG4 (130 kDa) expression by western blotting. (B) Spearman's correlation analysis between cell-based CSPG4 expression levels with CSPG4 ectodomain shedding levels. The results are presented as the relative expression of CSPG4 in PLX4032-exposed cells compared to the CSPG4 expression in parental cells which was set to 100%. CSPG4, chondroitin sulfate proteoglycan 4.
Effect of BRAF inhibition on the CSPG4 gene level
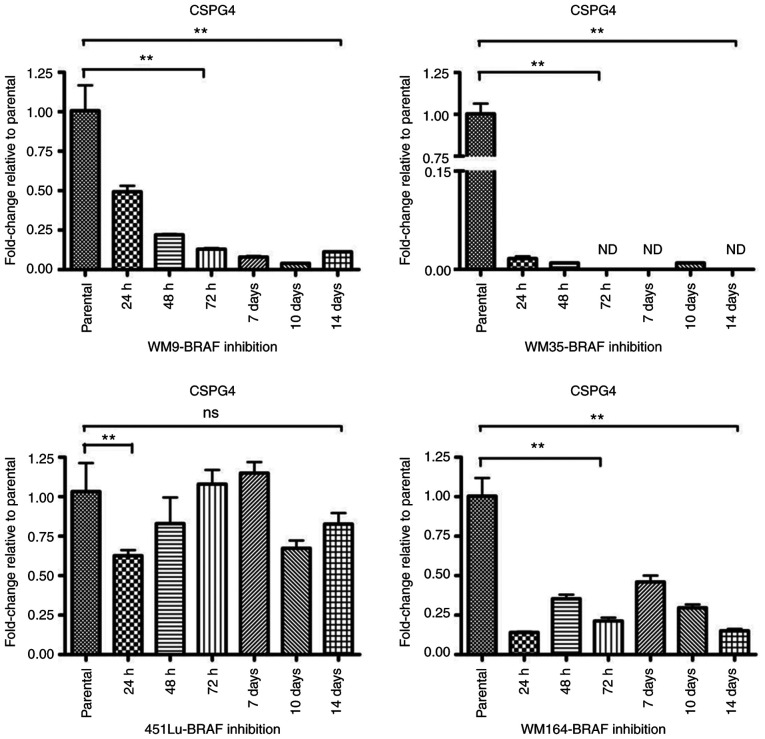
To investigate the gene expression of CSPG4 upon BRAF inhibition, RT-qPCR was performed. The relative messenger RNA levels of CSPG4 in PLX4032-exposed cells were decreased as compared to the parental cells (Fig. 5). As already observed at the protein level (Fig. 3A), changes in CSPG4 mRNA levels over time were specific for each cell line. In WM9 cells, CSPG4 mRNA levels decreased gradually over time (to 0.49±0.07 after 24 h, 0.22±0.01 after 48 h, 0.13±0.01 after 72 h, 0.08±0.01 after 7 days, 0.04±0.00 after 10 days) and started to increase slightly after day 14 (0.11±0.01). The most noteworthy CSPG4 downregulation was observed in the WM35 cell line. The PLX4032 treatment decreased the mRNA level of CSPG4 to 0.02±0.01 already after 24 h. The gene expression remained low for up to 14 days of treatment and was not detected (ND) on days 3, 7 and 14. CSPG4 mRNA levels in the 451Lu cell line were markedly reduced after 24 h to 0.63±0.06. No notable changes were observed after 3 and 7 days of PLX4032 treatment, as compared to parental cells. Reduced levels of CSPG4 mRNA were also detected after 48 h, 10 days and 14 days of PLX4032 exposure. In WM164 cells, CSPG4 mRNA levels fluctuated upon BRAF inhibition, but stayed lower than in parental cells, up to 14 days (Fig. 5).
Figure 5.
Downregulation of CSPG4 in melanoma cells after exposure to PLX4032. CSPG4 mRNA levels, normalized to the internal control: hUBC (WM9 and WM35) or GAPDH (451Lu and WM164) were analyzed by qPCR using specific primers. Bars represent mean ± SD from triplicates. Statistical analysis was carried out using one-way ANOVA with Tukey's multiple comparisons test. **P<0.001; ns, not significant; CSPG4, chondroitin sulfate proteoglycan 4.
CSPG4 expression is downregulated in vivo after tumor progression under BRAF/MEK inhibitor treatment
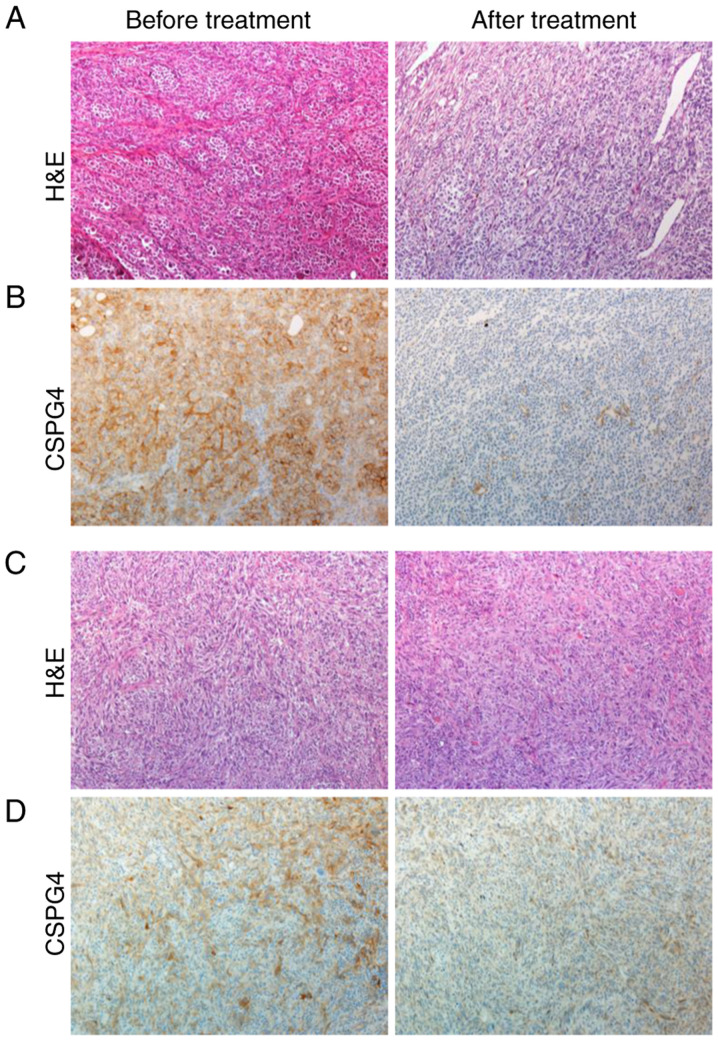
To support our in vitro data for the clinical relevance of CSPG4 expression before and after therapy with kinase inhibitors, patient-derived tumor samples were retrospectively analyzed by immunohistochemistry (Fig. 6). Four out of five pretreatment melanoma tissue samples showed a 2+ moderate positive staining for CSPG4 (Fig. 6B, left panel), and one pretreatment sample showed a 1+ weak positive staining for CSPG4 (Fig. 6D, left panel). In contrast, four out of five posttreatment melanoma samples stained negative for CSPG4, as shown in Fig. 6B (right panel), one sample stained 1+ for CSPG4 (Fig. 6D, right panel).
Figure 6.
CSPG4 expression in patient-derived matched pairs of melanoma tumor samples before and after therapy with kinase inhibitors. Tissue slides were prepared from biopsies of subcutaneous metastases of a melanoma patient before treatment (left panel) and after progression under therapy with dabrafenib and trametinib (right panel). (A and C) H&E staining, magnification ×100. (B and D) CSPG4 staining with anti-CSPG4 antibodies clone 132.38, magnification ×100. CSPG4, chondroitin sulfate proteoglycan 4; H&E, hematoxylin and eosin.
Discussion
Chondroitin sulfate proteoglycan 4 (CSPG4) is a multifunctional transmembrane proteoglycan involved in migration, spreading and invasion of melanoma (20). As reviewed by Ampofo et al, expression of CSPG4 is known to be influenced by inflammation and hypoxia and is regulated by methyltransferases, transcription factors and miRNAs (32). Identification of additional regulatory factors of CSPG4 expression may contribute to the understanding of CSPG4-mediated cellular functions and help design more effective treatments against melanoma. The present study demonstrated that BRAF inhibition is another important regulatory factor of CSPG4 expression.
We confirmed the presence of CSPG4 both at the protein and the gene level in a panel of seven out of eight melanoma cell lines. Regardless of the origin of the tumor cells (primary tumor or metastasis) and BRAF or NRAS mutation statuses, the majority of cell lines (WM9, WM35, 451Lu, WM164, D24 and CJM) displayed a uniform pattern of CSPG4 expression, as detected by anti-CSPG4 antibodies in western blotting. In line with other reports, within the same population of cells, CSPG4 was expressed both with a chondroitin sulfate (CS) chain and as an unmodified core protein (19,35). As anticipated, the M14 cell line, previously described as CSPG4-negative (30), did not show any CSPG4 expression.
The introduction of targeted therapies, such as BRAF and MEK inhibitors, has improved treatment options for metastatic melanoma patients. However, despite remarkable early therapeutic effects, a complete response is rarely observed due to acquired resistance (7).
Induced-drug tolerant cells (IDTCs), which constitute the step preceding the development of permanent drug resistance, are extremely important from a therapeutic perspective (15). Under hostile conditions (such as drug exposure, hypoxia or nutrient starvation), a fraction of tumor cells survives and over time can re-populate the tumor. Thus, targeting IDTCs in order to prevent or delay permanent drug resistance may enhance overall therapeutic efficacy. Since CSPG4 is a candidate target molecule, we examined whether its expression is altered in IDTCs and drug-resistant melanoma cells during exposure to kinase inhibitors. The amount of CSPG4 in IDTCs of all tested CSPG4-positive cell lines was markedly lower, as compared to parental cells. However, the presence of CSPG4 may not be required for a cell to become multidrug resistant since the CSPG4-negative cell line M14 underwent the same morphological and functional changes. After IDTCs escaped their slow cycling semiquiescent state and became permanently resistant, the expression of CSPG4 changed as well, in a cell line-specific manner. We demonstrated that different melanoma cell lines require different time spans to restore CSPG4 expression upon treatment with PLX4032, or that the proteoglycan in some cell lines is completely lost upon drug exposure. This effect did not change in a long-term experiment (up to 90 days) in all cell lines tested (data not shown). For a detailed understanding of CSPG4 expression dynamics, it is important to continuously follow its expression not only in short term cultures, but also in long-term experiments. Nevertheless, upregulated CSPG4 expression in permanent resistant melanoma cells could be a sign of the onset of additional resistance and survival mechanisms for cancer cells that will require further investigation (6).
Detailed analysis of cell lysates for CSPG4 expression upon BRAF inhibition within a time frame of up to 14 days revealed cell line-specific patterns of CSPG4 downregulation. In parental cells, CSPG4 was expressed both as a 280 kDa core protein and a 450 kDa chondroitin sulfate modified component. Upon BRAF inhibition, both forms gradually disappeared in WM9 cells, while in WM35 only the amount of the CS-decorated component decreased. In 451Lu cells, both CSPG4 forms decreased over time, and only the core protein appeared again. In WM164, both components initially decreased, but the CS-modified protein remained detectable for a much longer period of 14 days than the core protein. Previous studies showed that chondroitin sulfate decoration of the CSPG4 core protein may confer different attributes. First, CS chains may influence the CSPG4 distribution on the cell surface, focusing the core protein and proteoglycan into different microdomains in the plasma membrane (21). Second, CSPG4-linked chondroitin sulfate is critical for enhancement of matrix metalloproteinase (MMP) activity and thus extracellular matrix (ECM) degradation and increased tumor invasion (22). Finally, CS decoration facilitates an interaction of CSPG4 with fibronectin and α4β1-integrin, thereby modulating melanoma cell proliferation, adhesion, migration and invasion (20,23). Considering all these features, we speculate that the modifications of CSPG4 expression, and especially alterations in CS decoration, observed in our study, influence the overall behavior of the cell. As changes in CSPG4 upon BRAF inhibition are individual for different cell lines, it is likely that each cell line would exhibit different capabilities of CSPG4-dependent cellular functions.
CSPG4, as a membrane-spanning molecule, serves as a key proteoglycan that participates in signaling between the extracellular and intracellular compartments of the cell. In fact, CSPG4 was shown to promote MAPK signaling by mediating the growth-factor induced activation of receptor tyrosine kinases located on the cell surface. In addition, the cytoplasmic domain of CSPG4 contains a phosphoacceptor site at Thr2314, phosphorylated by ERK1/2, resulting in stimulation of cell proliferation (36). It was also described that the presence of CSPG4 is required for constitutive activation of the ERK1/2 pathway in melanoma cells possessing the BRAF V600E mutation (25). As demonstrated in 451Lu cells CSPG4 was downregulated upon BRAF inhibition. In addition, we observed the inhibition of cell proliferation and ERK1/2 phosphorylation at the same time points. In contrast, CS decoration seemed to play a role in AKT phosphorylation. The observed influence of the CS chain on AKT activation, independently from ERK1/2, is in line with studies showing that enhanced activation of FAK and ERK1/2 signaling by CSPG4 involves autonomous mechanisms (24).
The membrane proximal portion of extracellular domain of CSPG4 contains a number of putative proteolytic cleavage sites, leading to the release of CSPG4 from the cell surface (34). In our study, BRAF inhibition did not trigger shedding of CSPG4 in all tested cell lines. In fact, higher CSPG4 synthesis correlated significantly with greater CSPG4 release into the culture medium. This finding provided a rationale for investigating whether CSPG4 expression was regulated at the genomic level. Indeed, our results indicated markedly lower CSPG4 mRNA levels upon exposure to PLX4032 in all melanoma cell lines tested. Further studies are however required to fully describe the precise mechanism of CSPG4 downregulation. This process might involve DNA methylation, as it was shown that the promoter of CSPG4 contains many 5′CpG methylation sites (37). Another possibility is a regulatory mechanism involving a specific miRNA - miR129-2 (38). In addition to these mechanisms, CSPG4 can be regulated by several transcription factors, such as Sp1, Pax3 and Egr1 (32). The transcription factor Egr-1 is a crucial mediator in ERK-dependent signaling. In our investigation of the Egr-1 gene expression upon BRAF inhibition, no clear conclusion about a possible involvement of Egr-1 in CSPG4 downregulation could be made (data not shown).
CSPG4 has been found to be expressed in the majority (70% or greater) of superficial spreading and nodular human melanomas (20). It has not yet been examined whether its expression in tumors vary before and after treatment with kinase inhibitors. Our retrospective analysis of CSPG4 expression in melanoma tissue samples support the observations obtained in vitro, showing that CSPG4 is downregulated also in vivo after BRAF/MEK inhibition. The limited number of matched tumor samples did not permit to calculate a mathematical correlation of the CSPG4 expression between BRAF-mutant and drug-resistant melanoma samples. It would still be of great importance to perform a large, prospective, systemic quantitative study assessing CSPG4 expression before and after treatment with kinase inhibitors.
In conclusion, our results indicate that BRAF inhibition is a regulatory factor of CSPG4 expression in melanoma cells. Since CSPG4 plays a central role in coordinating cell proliferation, adhesion, migration and survival, downregulation of this proteoglycan may have a significant effect on the overall behavior of melanoma cells. The outcomes of this study provide the basis for further investigation of the underlying mechanisms and the role of CSPG4 in the development of drug-resistance in melanoma.
Acknowledgements
The authors would like to thank Helmut Schaider (Dermatology Research Centre, The University of Queensland Diamantina Institute, Translational Research Institute, The University of Queensland, Brisbane, Australia) for critical discussions throughout the project and for providing melanoma cell lines; Diana Mechtcheriakova and Anastasia Meshcheryakova (Department of Pathophysiology and Allergy Research, Center for Pathophysiology, Infectiology and Immunology, Medical University of Vienna, Austria) for technical assistance in qPCR experiments; Isabella Ellinger, Felicitas Mungenast and Martina Salzmann (Department of Pathophysiology and Allergy Research, Center for Pathophysiology, Infectiology and Immunology, Medical University Vienna, Austria) for support in immunofluorescence experiments; Johan Skaar, Rita Seeboeck (Department of Pathology, University Hospital St. Poelten, Karl Landsteiner University of Health Sciences, St. Poelten, Austria) and Vanessa Mayr (Department of Pathophysiology and Allergy Research, Center for Pathophysiology, Infectiology and Immunology, Medical University of Vienna, Austria) for technical assistance in the immunohistochemical analysis.
Glossary
Abbreviations
- AKT
protein kinase B
- CS
chondroitin sulfate
- CSPG4
chondroitin sulfate proteoglycan 4
- ECM
extracellular matrix
- ERK
extracellular signal-regulated kinase
- FAK
focal adhesion kinase
- GAG
glycosaminoglycan
- IDTCs
induced drug-tolerant cells
- mAb
monoclonal antibody
- MAPK
mitogen-activated protein kinase
- MEK
mitogen-activated protein kinase kinase
- PBS
phosphate-buffered saline
- RTK
receptor tyrosine kinase
Funding Statement
This project was funded by the NÖ Forschungs- und Bildungsges.m.b.H. (NFB), grant number: LSC15-007.
Funding
This project was funded by the NÖ Forschungs- und Bildungsges.m.b.H. (NFB), grant number: LSC15-007.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
KU and CH conceived the concept, designed the study, and analyzed and interpreted the data. TK and HB participated in designing the study and analyzing the data. KU and VV performed all experiments. MK contributed to the IHC analysis. KU, CH and HB wrote the manuscript. All authors read and revised the manuscript, and approved the final version. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated.
Ethics approval and consent to participate
All clinical samples were obtained from the collection of patients at the Department of Dermatology at the University Hospital St. Poelten, Karl Landsteiner University of Health Sciences. The collection and storage of samples were performed according to local ethical guidelines. Due to the retrospective nature of the study informed consent was waived by the Ethics Committee of the Karl Landsteiner University. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of the Karl Landsteiner University (EK number: 1011/2019).
Patient consent for publication
Not required according to local ethical guidelines.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A, Ugurel S. Melanoma. Lancet. 2018;392:971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 2.Spagnolo F, Ghiorzo P, Queirolo P. Overcoming resistance to BRAF inhibition in BRAF-mutated metastatic melanoma. Oncotarget. 2014;5:10206–10221. doi: 10.18632/oncotarget.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ascierto PA, Kirkwood JM, Grob JJ, Simeone E, Grimaldi AM, Maio M, Palmieri G, Testori A, Marincola FM, Mozzillo N. The role of BRAF V600 mutation in melanoma. J Transl Med. 2012;10:85. doi: 10.1186/1479-5876-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin M, Scotto D, Josephs DH, Mele S, Crescioli S, Bax HJ, Pellizzari G, Wynne MD, Nakamura M, Hoffmann RM, et al. BRAF inhibitors: Resistance and the promise of combination treatments for melanoma. Oncotarget. 2017;8:78174–78192. doi: 10.18632/oncotarget.19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1203421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson DB, Flaherty KT, Weber JS, Infante JR, Kim KB, Kefford RF, Hamid O, Schuchter L, Cebon J, Sharfman WH, et al. Combined BRAF (Dabrafenib) and MEK inhibition (Trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol. 2014;32:3697–3704. doi: 10.1200/JCO.2014.57.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, Chiarion Sileni V, Schachter J, Garbe C, Bondarenko I, et al. Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med. 2019;381:626–636. doi: 10.1056/NEJMoa1904059. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arozarena I, Wellbrock C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat Rev Cancer. 2019;19:377–391. doi: 10.1038/s41568-019-0154-4. [DOI] [PubMed] [Google Scholar]
- 14.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, Stuart DD. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menon DR, Das S, Krepler C, Vultur A, Rinner B, Schauer S, Kashofer K, Wagner K, Zhang G, Rad EB, et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015;34:4545. doi: 10.1038/onc.2014.432. [DOI] [PubMed] [Google Scholar]
- 16.Rolih V, Barutello G, Iussich S, De Maria R, Quaglino E, Buracco P, Cavallo F, Riccardo F. CSPG4: A prototype oncoantigen for translational immunotherapy studies. J Transl Med. 2017;15:151. doi: 10.1186/s12967-017-1250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilieva KM, Cheung A, Mele S, Chiaruttini G, Crescioli S, Griffin M, Nakamura M, Spicer JF, Tsoka S, Lacy KE, et al. Chondroitin sulfate proteoglycan 4 and its potential as an antibody immunotherapy target across different tumor types. Front Immunol. 2017;8:1911. doi: 10.3389/fimmu.2017.01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross AH, Cossu G, Herlyn M, Bell JR, Steplewski Z, Koprowski H. Isolation and chemical characterization of a melanoma-associated proteoglycan antigen. Arch Biochem Biophys. 1983;225:370–383. doi: 10.1016/0003-9861(83)90042-5. [DOI] [PubMed] [Google Scholar]
- 19.Campoli MR, Chang CC, Kageshita T, Wang X, McCarthy JB, Ferrone S. Human high molecular weight-melanoma-associated antigen (HMW-MAA): A melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit Rev Immunol. 2004;24:267–296. doi: 10.1615/CritRevImmunol.v24.i4.40. [DOI] [PubMed] [Google Scholar]
- 20.Price MA, Colvin Wanshura LE, Yang J, Carlson J, Xiang B, Li G, Ferrone S, Dudek AZ, Turley EA, McCarthy JB. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res. 2011;24:1148–1157. doi: 10.1111/j.1755-148X.2011.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stallcup WB, Dahlin-Huppe K. Chondroitin sulfate and cytoplasmic domain-dependent membrane targeting of the NG2 proteoglycan promotes retraction fiber formation and cell polarization. J Cell Sci. 2001;114:2315–2325. doi: 10.1242/jcs.114.12.2315. [DOI] [PubMed] [Google Scholar]
- 22.Iida J, Wilhelmson KL, Ng J, Lee P, Morrison C, Tam E, Overall CM, McCarthy JB. Cell surface chondroitin sulfate glycosaminoglycan in melanoma: Role in the activation of pro-MMP-2 (pro-gelatinase A) Biochem J. 2007;403:553–563. doi: 10.1042/BJ20061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida J, Meijne AM, Oegema TR, Jr, Yednock TA, Kovach NL, Furcht LT, McCarthy JB. A role of chondroitin sulfate glycosaminoglycan binding site in alpha4beta1 integrin-mediated melanoma cell adhesion. J Biol Chem. 1998;273:5955–5962. doi: 10.1074/jbc.273.10.5955. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Price MA, Neudauer CL, Wilson C, Ferrone S, Xia H, Iida J, Simpson MA, McCarthy JB. Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. J Cell Biol. 2004;165:881–891. doi: 10.1083/jcb.200403174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Price MA, Li GY, Bar-Eli M, Salgia R, Jagedeeswaran R, Carlson JH, Ferrone S, Turley EA, McCarthy JB. Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer Res. 2009;69:7538–7547. doi: 10.1158/0008-5472.CAN-08-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordaan S, Chetty S, Mungra N, Koopmans I, van Bommel PE, Helfrich W, Barth S. CSPG4: A target for selective delivery of human cytolytic fusion proteins and TRAIL. Biomedicines. 2017;5:37. doi: 10.3390/biomedicines5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann RM, Crescioli S, Mele S, Sachouli E, Cheung A, Chui CK, Andriollo P, Jackson PJM, Lacy KE, Spicer JF, et al. A novel antibody-drug conjugate (ADC) delivering a DNA mono-alkylating payload to chondroitin sulfate proteoglycan (CSPG4)-expressing melanoma. Cancers (Basel) 2020;12:1029. doi: 10.3390/cancers12041029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Geldres C, Ferrone S, Dotti G. Chondroitin sulfate proteoglycan 4 as a target for chimeric antigen receptor-based T-cell immunotherapy of solid tumors. Expert Opin Ther Targets. 2015;19:1339–1350. doi: 10.1517/14728222.2015.1068759. [DOI] [PubMed] [Google Scholar]
- 29.Hafner C, Breiteneder H, Ferrone S, Thallinger C, Wagner S, Schmidt WM, Jasinska J, Kundi M, Wolff K, Zielinski CC, et al. Suppression of human melanoma tumor growth in SCID mice by a human high molecular weight-melanoma associated antigen (HMW-MAA) specific monoclonal antibody. Int J Cancer. 2005;114:426–432. doi: 10.1002/ijc.20769. [DOI] [PubMed] [Google Scholar]
- 30.Pucciarelli D, Lengger N, Takacova M, Csaderova L, Bartosova M, Breiteneder H, Pastorekova S, Hafner C. Anti-chondroitin sulfate proteoglycan 4-specific antibodies modify the effects of vemurafenib on melanoma cells differentially in normoxia and hypoxia. Int J Oncol. 2015;47:81–90. doi: 10.3892/ijo.2015.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L, Favoino E, Wang Y, Ma Y, Deng X, Wang X. The CSPG4-specific monoclonal antibody enhances and prolongs the effects of the BRAF inhibitor in melanoma cells. Immunol Res. 2011;50:294–302. doi: 10.1007/s12026-011-8232-z. [DOI] [PubMed] [Google Scholar]
- 32.Ampofo E, Schmitt BM, Menger MD, Laschke MW. The regulatory mechanisms of NG2/CSPG4 expression. Cell Mol Biol Lett. 2017;22:4. doi: 10.1186/s11658-017-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Campoli M, Ferrone S, Wang X. Functional and clinical relevance of chondroitin sulfate proteoglycan 4. Adv Cancer Res. 2010;109:73–121. doi: 10.1016/B978-0-12-380890-5.00003-X. [DOI] [PubMed] [Google Scholar]
- 35.Yang J, Price MA, Wanshura LEC, He J, Yi M, Welch DR, Li G, Conner S, Sachs J, Turley EA, McCarthy JB. Chondroitin sulfate proteoglycan 4 enhanced melanoma motility and growth requires a cysteine in the core protein transmembrane domain. Melanoma Res. 2019;29:365–375. doi: 10.1097/CMR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makagiansar IT, Williams S, Mustelin T, Stallcup WB. Differential phosphorylation of NG2 proteoglycan by ERK and PKCalpha helps balance cell proliferation and migration. J Cell Biol. 2007;178:155–165. doi: 10.1083/jcb.200612084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo W, Wang X, Kageshita T, Wakasugi S, Karpf AR, Ferrone S. Regulation of high molecular weight-melanoma associated antigen (HMW-MAA) gene expression by promoter DNA methylation in human melanoma cells. Oncogene. 2006;25:2873–2884. doi: 10.1038/sj.onc.1209319. [DOI] [PubMed] [Google Scholar]
- 38.Yadavilli S, Scafidi J, Becher OJ, Saratsis AM, Hiner RL, Kambhampati M, Mariarita S, MacDonald TJ, Codispoti KE, Magge SN, et al. The emerging role of NG2 in pediatric diffuse intrinsic pontine glioma. Oncotarget. 2015;6:12141–12155. doi: 10.18632/oncotarget.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.