Abstract
Circular RNAs (circRNAs) are a class of novel endogenous transcripts with limited protein-coding abilities. CircRNAs have been demonstrated to function as critical regulators of tumor development and distant metastasis through binding to microRNAs (miRNAs) and interacting with RNA-binding proteins, thereby regulating transcription and translation. Emerging evidence has illustrated that certain circRNAs can serve as biomarkers for diagnosis and prognosis of cancer, and/or serve as potential therapeutic targets. Expression of functional circRNAs is commonly dysregulated in cancer and this is correlated with advanced Tumor-Node-Metastasis stage, lymph node status, distant metastasis, poor differentiation and shorter overall survival of cancer patients. Recently, an increasing number of studies have shown that circRNAs are closely associated with NSCLC. Functional experiments have revealed that circRNAs are intricately associated with the pathological progression of NSCLC. The present review provides an overview of the regulatory effect of circRNAs in the development and progression of NSCLC, taking into consideration various physiological and pathological processes, such as proliferation, apoptosis, invasion and migration, and their potential value as biomarkers and therapeutic targets.
Keywords: circRNAs, non-small cell lung cancer, carcinogenesis
1. Introduction
Lung cancer is the most common type of malignant tumor, and is the primary cause of cancer-related death worldwide (1). As the primary cause of mortality in men and women, the number of deaths from lung cancer was 135,720, which accounted for 22.5% of all cancer-related deaths in the United States in 2020 (2). Small cell lung cancer (SCLC) and non-SCLC (NSCLC), the two primary types of lung cancer, account for >85% of all lung cancer cases (3). Despite the progress in clinical management that has been made in the last few years, the 5-year overall survival (OS) rate of NSCLC is 15–21% (1,2). The primary cause of the low 5-year survival rate is that the majority of patients are diagnosed with advanced stage cancer with distant metastases at the first presentation (3). Additionally, cancer recurrence and drug resistance contribute to the high mortality rates of NSCLC (4). The poor prognosis of NSCLC can be attributed to the complicated and unclear molecular mechanisms underlying its development and progression (5,6). Therefore, it is vital to identify novel biomarkers or therapeutic targets to improve the prognosis of NSCLC.
Multiple factors may participate in the development and progression of NSCLC, including proliferation, autophagy, apoptosis, invasion, metastasis and drug resistance (7,8). One of the most fundamental characteristics of cancer cells is its ability to sustain chronic proliferation. However, proliferation and division of malignant cells is uncontrolled (9). Autophagy serves an important and complicated role in tumor development. Upregulation of autophagy in cancer therapy can promote the survival or death of tumor cells (10). Abnormal regulation of cell death, whether too little or too much, may contribute to several diseases. Aberrant initiation of apoptosis may lead to malignant transformation of NSCLC cells (11,12). Metastasis of NSCLC is a significant obstacle reducing the OS of NSCLC patients, and is considered a core step in the malignant progress of NSCLC (13). Although chemotherapy prolongs the OS of patients with NSCLC, tumor cells may acquire resistance, resulting in poor therapeutic effects, tumor metastasis and recurrence (7). Therefore, it is necessary to identify and determine the relationship between novel biomarkers and malignant behavior in NSCLC.
CircRNAs are a novel group of non-coding RNAs that do not possess 3′ and 5′ends, but instead form a closed-loop dissimilar to linear RNAs (14). CircRNAs were first detected in a virus by Sanger in 1976, and were initially deemed as irrelevant byproducts without any significant biological functions for a period of time (15). In the last decade, owing to the rapid advance of RNA-sequencing technologies, researchers have re-evaluated the crucial functions of circRNAs in the regulation of gene expression and in multiple diseases, such as carcinomas (16). Additionally, previous studies have indicated that circRNAs are conserved, stable and abundantly expressed in tissues and exosomes (17,18). According to reverse transcription quantitative (RT-qPCR) and reverse transcription-droplet digital (RT-ddPCR) qualification, 343 differentially-expressed circRNAs were identified between the plasma of patients with gastric cancer and healthy controls (19). CircRNAs are closely associated with tumorigenesis, development, proliferation, apoptosis, invasion and migration of various physiological and pathological processes in tumors (20). CircRNAs are extensively and stably expressed in the plasma and exosomes, indicating that they may serve as promising biomarkers in the prognosis and therapeutics of malignancies (21). The present review summarizes the relationship between circRNAs and the biological behaviors of NSCLC.
2. Classification and functions of circRNAs
Classification of circRNAs
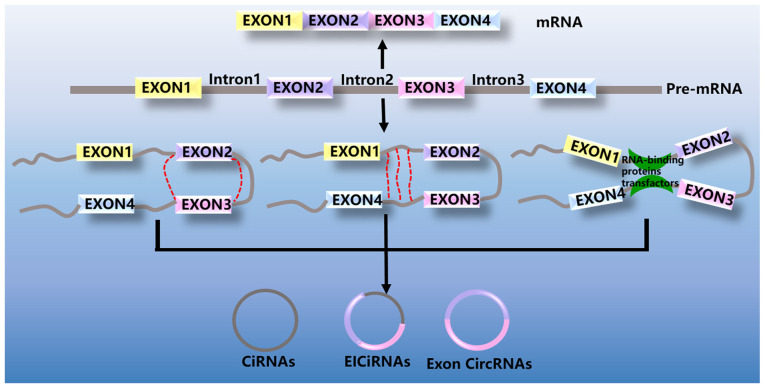
CircRNAs can be divided into four groups, exonic circRNAs (ecircRNAs), intronic circRNAs (ciRNAs), exon-intron circRNA (EIciRNAs) (Fig. 1) and tRNA intronic circRNAs (tricRNAs). EcircRNAs are generated from single or several exons. The majority of circRNAs are ecircRNAs, accounting for >80% of currently identified circRNAs. CircRNAs are primarily localized in the cytoplasm and may act as miRNA sponges, indirectly participating in the regulation of gene expression. CiRNAs are intron-derived circRNAs. CiRNAs are abundantly present in the nucleus and may modulate the expression of their parental genes. EIciRNAs contain both introns and exons that can regulate their parental genes in a cis manner. TricRNAs are derived from tRNA introns and can form stable circRNAs via pre-tRNA splicing (15,22).
Figure 1.
Biogenesis of circRNAs. CircRNA, circular RNA; ciRNAs, intronic circRNAs; EIciRNAs, exon-intron circRNA. CircRNA, circular RNA.
Functions of circRNAs
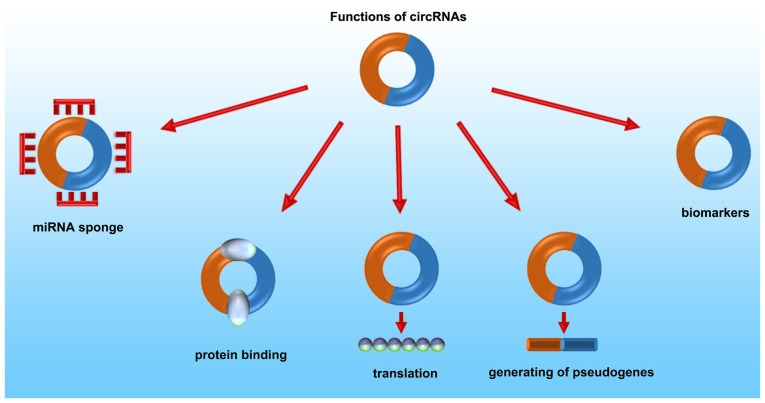
With the number of studies on circRNAs increasing, our understanding of the biological functions of circRNAs is ever growing. As shown in Fig. 2, an increasing number of studies have shown that circRNAs exhibit multiple functions, such as functioning as miRNA sponges, interacting with proteins, translation into proteins and regulation of transcription (23).
Figure 2.
Biological functions of circRNAs. CircRNA, circular RNA; miRNA, microRNA.
CircRNAs can sponge miRNAs
Recently, several studies have found that circRNA are primarily located in the cytoplasm. CircRNAs compete with miRNAs to regulate gene expression via miRNA response elements (MREs) (24). CircRNAs can increase the levels of the target genes of miRNAs, and circRNAs with this competitive function are termed competing endogenous (ce)RNAs. As a well-studied function of circRNAs, ceRNAs are widely involved in various circRNA-related diseases, particularly in cancer. For example, the expression of miR-7 is affected by ciRS-7 which possesses >70 selectively conserved binding sites for miR-7. When ciRS-7 efficiently binds to miR-7, the expression of miR-7 is attenuated and the activity of miR-7-target genes is increased. The ciRS-7/miR-7 axis participates in numerous diseases, such as breast cancer (25), cervical cancer (26), gastric carcinoma (27) and hepatocellular carcinoma (28,29). CircRNA zinc finger protein 609 (Circ-ZNF609) improves vascular endothelial dysfunction through upregulating the expression of myocyte enhancer factor 2A by serving as a ceRNA of miR-615-5p (30).
CircRNAs can bind with RNA-binding proteins (RBPs)
CircRNAs can also bind to proteins directly. Thus, they can act as protein sponges, similar in principal to their function as miRNA sponges. CircRNAs contain a high density of binding sites for RBPs, and they may affect the activity of related proteins through binding with them directly (31). A circular transcript from forkhead box O3 (Circ-Foxo3) is related with cell cycle progression. Circ-Foxo3 affects cell cycle progression via regulation of a G1/S transition through binding with cyclin-dependent kinase 2 (CDK2) and p21 (32). Circ-Foxo3 facilitates Foxo3 expression via interacting with the MDM proto-oncogene (MDM2) and p53, which leads to MDM2-induced p53 ubiquitination and subsequent degradation (33).
CircRNAs can be translated into proteins
Although most circRNAs serve as miRNA sponges and indirectly regulate the expression of mRNAs, emerging evidence has shown that certain circRNAs are translatable (34). CircRNAs may contain an open-reading frame (ORF), N6-methyladenosine modifications and/or internal ribosome entry site (IRES) elements. Hence, circRNAs can be translated into proteins accordingly (35,36). Moreover, circRNAs can translate into proteins via a rolling circle amplification mechanism in eukaryotic cells (37). Circ-ZNF609 contains an ORF and it can be translated into a protein via a splicing event (38). CircRNA F-box and WD repeat domain containing 7, expression of which is high in the brain, encodes F-box and WD repeat domains containing 7–185aa, and inhibits proliferation and cell cycle progression in cancer cells (39). A circular form of SNF2 histone linker PHD RING helicase, which contains an ORF driven by the IRES, is translated into SNF2 histone linker PHD RING helicase-146aa, which is a cancer suppressor in human glioblastoma (40).
CircRNAs can regulate transcription
CiRNAs are primarily located in the nucleus, and possess very little MRE activity for sponging miRNAs (41). Knockdown of ciRNAs decreases the expression of their parental genes. CiRNAs can regulate PolII transcription in a cis manner via an RNA-RNA interaction, and can mediate the expression of their parental genes (42,43). EIciRNAs interact with UI small nuclear ribonucleoprotein (snRNP) and PolII to regulate gene expression via RNA-RNA interactions. UI snRNP is indispensable for EIciRNA-mediated regulation of expression of parental genes (44). Ci-ankrd52, which accumulates at transcription sites, is an intron-derived circRNA that is produced from ANKRD52, and Ci-ankrd52 can interact with PolII to regulate transcription of the parent genes (45).
CircRNAs enable derivation of pseudogenes
Pseudogenes are non-functional residues that were formed during the evolution of a gene family, and they serve as essential markers in the field of evolutionary and comparative genomics (46). Pseudogenes may participate in cellular differentiation and cancer progression (47). There is an exon-exon junction in a reversed order in circRNA-derived pseudogenes. In both mice and humans, numerous circRNA-derived pseudogenes have been detected by a computational pipeline (CIRCpseudo) (48). In all mouse strains and in the rat reference genome, there are dozens of low-confidence circular SATB homeobox 1-derived pseudogenes. In the gorilla and chimp genomes, researchers identified the homologous sequences of human circular protein kinase, DNA-activated, catalytic submit-derived and circular calmodulin regulated spectrin associated protein 1-derived pseudogenes (49). Though reverse transcription may be involved in the non-colinear exon-exon junctions of pseudogenes, the mechanism of reverse transcription and translocation of circRNAs are still not distinct (23).
CircRNAs may serve as promising biomarkers
CircRNAs are stably expressed both intracellularly and in the plasma, due to their unique annular structure (14). RNA-seq analyses indicated that >1,000 circRNAs have been identified in human exosomes and may transfer biological activity to other cells (50). CircRNAs generated from cancer cells can enter into the blood circulation and can be detected easily, and may thus be used to distinguish between cancerous and healthy individuals (14). Exosomal circRNAs are thus potential biological markers of various types of cancer including NSCLC (17).
Roles of circRNAs in cancer
CircRNAs are extensively implicated in the pathological progression of multiple types of cancer, including gastric cancer, hepatocellular carcinoma, lung cancer, colorectal cancer and bladder cancer, amongst others (51). Moreover, these circRNAs exhibit dual roles; serving as oncogenes and tumor suppressors dependent on the type and potentially stage of cancer (52,53). In the following section, the roles of circRNAs in NSCLC are discussed in additional detail.
3. CircRNAs and NSCLC
Expression of circRNAs in NSCLC
With the development of next-generation sequencing technologies and advances in bioinformatics analysis, a large number of studies have shown that circRNAs are ectopically expressed in several types of tumors, including NSCLC. A total of 957 abnormally expressed circRNAs were identified by human circRNA microarray analysis in NSCLC tissues when compared with the adjacent normal tissue (54). In another study, 356 circRNAs were dysregulated in lung adenocarcinoma, including 204 upregulated circRNAs and 152 downregulated circRNAs (55). By utilizing circRNA chips, Mu et al (56) identified and annotated a total of 10,566 circRNAs in the peripheral whole blood of patients with lung adenocarcinoma. Amongst these, 78.14% of the circRNAs were exonic, and 3,009 circRNAs were upregulated, whereas 1,381 circRNAs were downregulated.
CircRNAs can be used as diagnostic biomarkers in NSCLC
CircRNAs produced by cancer cells can enter into the blood circulation and can be detected easily (57). Thus, they can be used to distinguish between patients with cancer from healthy individuals. Exosomal circRNAs are potential biological markers in a range of cancer types (14). With the development of circRNA research, a plethora of circRNAs may eventually be used as clinically diagnostic markers for the diagnosis of early-stage NSCLC (Table I).
Table I.
CircRNAs act as prognostic or diagnostic biomarkers of non-small cell lung cancer.
| ROC curve | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| First author, year | CircRNA name | circBase ID | Dysregulation | Correlation with clinical characteristics | Kaplan-Meier OS curves | AUC | Sensitivity | Specificity | (Refs.) |
| Zhang et al, 2018 | Circ_0014130 | Up | TNM stage, lymphatic metastasis | – | 0.878 | 0.87 | 0.848 | (58) | |
| Chen et al, 2019 | Circ_100146 | Up | Pathological classification, differentiation grade | – | 0.643 | 0.725 | 0.575 | (59) | |
| Qiu et al, 2018 | CircPRKCI | Up | T stage, TNM stage | Poor prognosis/independent factor (Multivariate analyses: HR=2.664, 95% CI: 1.327–5.347, P=0.006) | – | – | – | (60) | |
| Chen et al, 2018 | Circ_100395 | Down | Cancer metastasis, TNM stage | Poor prognosis | – | – | – | (63) | |
| Han et al, 2018 | CircBANP | Up | Cancer metastasis, TNM stage | Poor prognosis | – | – | – | (64) | |
| Zhang et al, 2018 and Su et al, 2018 | Circular RNAciRS-7/CDR1as | hsa_circ_0001946 | Up | TNM stage, lymphatic Metastasis, pathological classification | Shorter OS | (68,71) | |||
| Zhu et al, 2017 | Circ_0013958 | hsa_circ_0013958 | Up | TNM stage, lymphatic metastasis | – | 0.815 | 0.755 | 0.796 | (72) |
| Wang et al, 2019 | CircVANGL1 | Up | T stage, TNM stage | Poor prognosis | – | – | – | (85) | |
| Li et al, 2018 | CircPVT1 | Up | Distant metastasis | – | 0.803 | 0.825 | 0.675 | (86) | |
| Wang et al, 2019 | CircCRIM1 | Down | TNM stage, lymphatic metastasis | Poor prognosis | – | – | – | (94) | |
| SHI et al, 2020 | CircLARP4 | Down | – | Poor prognosis | – | – | – | (95) | |
CircRNA, circular RNA; TNM, Tumor-Node-Metastasis; ROC, receiver operating characteristic; AUC, area under the curve; OS, overall survival; HR, hazard ratio; CI, confidence interval.
The expression of hsa_circ_0014130 is associated with Tumor-Node-Metastasis (TNM) stage and lymphatic metastasis of NSCLC. Receiver operating characteristic (ROC) curves were used to determine the diagnostic potential of hsa_circ_0014130. The area under the ROC curve (AUC) was 0.878, the optimum critical value of hsa_circ_0014130 was 0.573, the sensitivity was 87% and specificity was 84.8%. Thus, hsa_circ_0014130 may serve as a biomarker for distinguishing NSCLC from normal tissues (58). CircRNA 100146 was shown to be augmented in 26 cases of NSCLC, and was associated with pathological stage and differentiation of lung cancer. ROC curve analysis indicated that the AUC was 0.643 (95% confidence interval: 0.521–0.764), the sensitivity was 72.5% and the specificity was 57.5%. Thus, circRNA 100146 may also be used as a diagnostic marker in NSCLC (59).
CircRNAs may serve as therapeutic targets in NSCLC
CircRNAs are stably expressed both intracellularly and in the plasma due to their annular structure. RNA-seq analyses indicated that >1,000 circRNAs are present in human exosomes and may transfer biological activity to other cells (50). Numerous circRNAs have been reported to be involved in the tumorigenesis and progression of NSCLC, and are being extensively assessed as potential therapeutic targets for the treatment of NSCLC (Table I).
Circular protein kinase C iota (CircPRKCI) is generated from exons 15 and 16 of the PRKCI gene (chr3:170013698-170015181) and is located at the 3q26.2 amplicon. CircPRKCI acts as a tumor promoting factor in lung adenocarcinoma (LAD), and circPRKCI is positively correlated with T stage and TNM stage in patients with LAD (60). Knockdown of circPRKCI led to a decrease in tumor size and tumor weight in nude mice. Patient-derived tumor xenografts (PDTXs) can be used as a translational model. Intratumoral injection of cholesterol-conjugated si-circPRKCI was used to clarify the therapeutic potential of circPRKCI. The findings showed that the growth of PDTX was decreased in the si-circPRKCI group. These results highlight the therapeutic potential of circPRKCI (60). At present, EGFR tyrosine kinase inhibitors (EGFR-TKIs) are widely used to treat NSCLC patients with EGFR-sensitive mutations. The combination of EGFR-TKIs (gefitinib) and knockdown of circPRKCI resulted in a more notable inhibitory effect than gefitinib or knockdown of circPRKCI alone. This suggests that a combination of EGFR-TKIs and attenuation of circPRKCI may exert a synergistic effect on reducing cancer progression (60).
Circular coiled-coil domain containing 66 (circCCDC66) is primarily located in the endoplasmic reticulum. CircCCDC66 is involved in several types of cancer and serves as a diagnostic and therapeutic biomarker (61). CircCCDC66 is highly expressed in LAD and in EGFR-resistant H1975 cells. EGFR is the primary target of TKIs for tyrosine kinase mutations, such as gefitinib and erlotinib in LAD chemotherapy. Knockdown of focal adhesion kinase (FAK) and hepatocyte growth factor reduces circRNA CCDC66 expression, separately. FAK was associated with metastasis and EMT. Meanwhile, administration of a FAK inhibitor, Y15, reduced metastasis. Conversely, nicotinic acetylcholine receptor α7 (nAchRα7) negatively regulates the expression of CCDC66β and circRNA CCDC66. The regulatory effect of nAchRα7 on circRNA CCDC66 is greater than that of FAK. Furthermore, knockdown of circRNA CCDC66 suppresses EMT and invasion, and augments cisplatin resistance in H23 cells. CircCCDC66 may thus serve as a novel therapeutic target for regulating EGFR-mediated tumorigenesis in NSCLC (62).
Prognostic potential of circRNAs in NSCLC
It is very important to evaluate the prognosis of patients with cancer. Aberrant expression of circRNAs has been reported to show extensive associations with clinical features of patients with NSCLC. Meanwhile, circRNAs are considered to possess valuable prognostic value as biomarkers in NSCLC.
According to a study with 69 cases of NSCLC, which used RT-qPCR qualification, hsa_circ_100395 expression was found to be lower in patients with advanced TNM stage. Additionally, Kaplan-Meier survival curve analysis showed that the survival rate of patients with lower expression of hsa_circ_100395 was lower (63). Circular BTG3 associated nuclear protein (Circ-BANP) was shown to be upregulated in lung cancer tissues and cell lines, and was higher in patients with stage III–IV cancer or in the metastatic tissue. Higher expression of circ-BANP was associated with reduced OS based on Kaplan-Meier curve analysis. Thus, circ-BANP may serve as an independent prognostic biomarker (64).
Based on the above studies, circRNAs may be used for the diagnosis, treatment and evaluation of prognosis of patients with NSCLC. In subsequent studies, increased attention should be paid to the molecular mechanisms by which circRNAs regulate cancer development/progression and in the clinical application of targeting circRNAs.
Functions of circRNAs in NSCLC
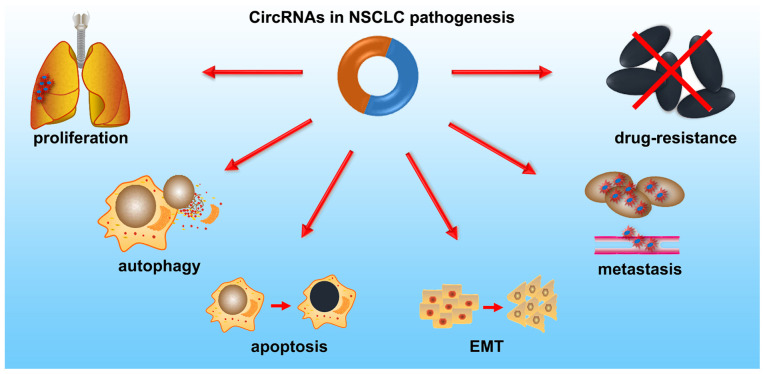
NSCLC pathogenesis is modulated by oncogenic or tumor suppressive circRNAs, via regulation of cell proliferation, autophagy, apoptosis, invasion, migration and EMT (Fig. 3). Additionally, circRNAs can act as independent prognostic biomarkers, and serve an important role in multidrug resistance (MDR) in NSCLC. The functions of circRNAs in NSCLC are discussed in the upcoming sections and are summarized in Table II.
Figure 3.
CircRNAs are extensively implicated in the pathogenesis of NSCLC. CircRNA, circular RNA; NSCLC, non-small cell lung cancer; EMT, epithelial-mesenchymal transition.
Table II.
Clinical value of circRNAs involved in the pathogenesis and progression of NSCLC.
| First author, year | CircRNA name | circBase ID | Dysregulation | Function | Target gene/pathway | Effect in NSCLC | (Refs.) |
|---|---|---|---|---|---|---|---|
| Chen et al, 2019 | Circ_100146 | – | Up | miRNA sponge | miR-615-5p↓, miR-361-3p↓, SF3B3↑ | Proliferation↑, invasion↑, apoptosis↓ | (59) |
| Qiu et al, 2018 | CircPRKCI | – | Up | miRNA sponge | miR-545↓, miR-589↓, E2F7↑ | Proliferation↑, migration↑, | (60) |
| Joseph et al, 2018 | CircCCDC66 | – | Up | – | – | Invasion↑, EMT↑, drug resistance (cisplatin) ↑ | (62) |
| Chen et al, 2018 | Circ_100395 | – | Down | miRNA sponge | miR-1228↓, TCF21↑ | Proliferation↑, migration↑, invasion↑ | (63) |
| Han et al, 2018 | CircBANP | – | Up | miRNA sponge | miR-503↓, LARP1↑ | Proliferation↑, migration↑, invasion↑ | (64) |
| Zhang et al, 2018 and Su et al, 2018 | Circular RNAciRS-7/CDR1as | hsa_circ_0001946 | Up | miRNA sponge | miR-7↓, EGFR↑, CCNE1↑, PIK3CD↑, NF-kB↑ | Proliferation↑, migration↑. invasion↑, apoptosis↓ | (68,71) |
| Zhu et al, 2017 | Circ_0013958 | hsa_circ_0013958 | Up | miRNA sponge | miR-134↓, CCND1↑ | Proliferation↑, invasion↑, apoptosis↓ | (72) |
| Chi et al, 2019 | CircPIP5K1A | hsa_circ_0014130 | Up | miRNA sponge | miR-600↓, HIF-1α↑ | Apoptosis↓, proliferation↑ | (74) |
| Nan et al, 2018 | CircNOL10 | hsa_circ_0000977 | Down | Protein binding, regulation of transcriptional | SCML1↑, HN polypeptide family | Apoptosis↑, proliferation↓, invasion↓ | (75) |
| Chen et al, 2020 | CircHIPK3 | hsa_circ_0000284 | Up | miRNA sponge | miR-124-3p↓, STAT3/PRKAA/AMPKa↑ | Proliferation↑, invasion↑, migration↑, autophagy↑ | (77) |
| Wang et al, 2019 | CircVANGL1 | – | Up | miRNA sponge | miR-195↓, Bax↑, Bcl-2↑ | Migration↑, invasion↑, apoptosis↓ | (85) |
| Li et al, 2018 | CircPVT1 | – | Up | miRNA sponge | miR-125b↓, E2F2↑ | Proliferation↑, migration↑, invasion↑, apoptosis↓ | (86) |
| Gao and Ye, 2020 | CircSOX4 | – | Up | miRNA sponge | miR-1270↓, PLAGL2↑ | Proliferation↑, invasion↑, migration↑ | (91) |
| Wang et al, 2018 | CircPTK2 | – | Down | miRNA sponge | miR-429↓, miR-200b-3p↓, TIF1γ↑ | Invasion↓, TGF-β-induced EMT↓ | (92) |
| Wang et al, 2019 | CircCRIM1 | hsa_circ_0002346 | Down | miRNA sponge | miR-93↓, miR-182↓, LIFR↑, MMP13↑, PI3K/AKT/ JAK1↑ | Invasion↓, metastasis↓ | (94) |
| Shi et al, 2020 | CircLARP4 | – | Down | Protein binding | SMAD7↑ | Invasion↓, migration↓ | (95) |
| Wan et al, 2016 | CircITCH | – | Down | miRNA sponge, regulation of parental genes | miR-7↓, miR-214↓, Wnt/β-catenin↓, ITCH↑ | Proliferation↓ | (96) |
| Huang et al, 2019 | Circ_0001946 | – | Down | miRNA sponge | miR-7-5p↓, miR-671-5p↓, miR-3156-5p↓, miR-1270↓, NER↑ | Proliferation↓, invasion↓, migration↓, drug resistance (cisplatin) ↓, apoptosis↑ | (100) |
| Li et al, 2019 | Circ_0002483 | – | Down | miRNA sponge | miR-182-5p↓, miR-520q-3p↓, miR-582-3p↓, miR-587↓, GRB2↑, FOXO1↑, FOXO3↑ | Proliferation↓, invasion↓, drug resistance (Taxol)↓ | (101) |
| Xiao et al, 2020 | Circ_103762 | – | Up | Protein binding | CHOP↓ | Proliferation↑, migration↑, invasion↑, drug resistance (MDR) ↑ | (102) |
CircRNA, circular RNA; miR/miRNA, microRNA; MDR, multi-drug resistance; NSCLC, non-small cell lung cancer; EMT, epithelial-mesenchymal transition.
CircRNAs in NSCLC proliferation and cell cycle progression
Cell cycle progression is an important factor in maintaining cell proliferation. The phenomenon of normal cells inhibiting division due to contact inhibition, is termed density-dependent inhibition of growth. When cells reach a finite density, they halt proliferation and the cell cycle arrests at the G0 phase of the cell cycle (65). Unrestricted cell proliferation and reduced apoptosis results in unlimited growth and distant metastasis of tumors. The proliferation of cancer cells represents a typical prognostic marker in the diagnosis of cancer (66). Abnormal expression of circRNAs leads to growth of NSCLC cells (67), suggesting that circRNAs serve a potential role in NSCLC treatments targeting unlimited proliferation.
CircRNAs modulate proliferation and cell cycle progression via sponging of miRNAs in NSCLC
CircRNAs serve their biological function in numerous ways. CircRNA-mediated sponging of miRNAs is the most-extensively studied circRNA mechanism.
CiRS-7, also known as cerebellar degeneration-related protein 1 antisense RNA (CDR1as), can absorb miRNAs, such as miR-7 and miR-671, and thus reduces the levels of CDR1 transcripts. CiRS-7 possesses over 70 binding sites with miR-7. Several reports have demonstrated that the CiRS-7/miR-7 axis contributes to several pathological processes, including NSCLC. CiRS-7 may increase cell viability and induce cell growth in NSCLC. CiRS-7 also significantly increases the expression of growth-related genes, including EGFR, cyclin E1 (CCNE1) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit δ (PIK3CD) (68). Lower expression levels of EGFR maintains cell cycle arrest, facilitating mitosis and preventing cell apoptosis (69). CCNE1 induces rapid progression of cells through the G1/S phase via activation of CDK2 (70). The effect of ciRS-7 on monitoring cell cycle progression is reversed by overexpression of miR-7. CiRS-7 also regulates cell proliferation, invasion, migration and apoptosis via targeting miR-7 to modulate nuclear factor-κB (NF-kB) (71).
Cyclin D1 (CCND1) is an important target of abnormally expressed circRNAs in NSCLC, such as circ_0013958 (72). Abnormal expression of circRNAs can regulate the cell cycle process and proliferation in NSCLC. The primary function of CCND1 is to promote cell proliferation. CCND1 can bind to and activate cyclin-dependent kinase CDK4, which is unique to the G1 phase (73). Circ_0013958 was identified as a sponge of miR-134, and circ_0013958 promotes the development of NSCLC via upregulation of oncogenic CCND1 (72).
A high-throughput microarray assay revealed that circular phosphatidylinositol-4-phophate 5-kinase type 1 alpha (circPIP5K1A) was significantly upregulated in NSCLC (58). CircPIP5K1A regulates the progression of NSCLC via activation of several signaling pathways. For example, circPIP5K1A promotes proliferation via a miR-600/hypoxia inducible factor-1α axis in NSCLC (74).
CircRNAs modulate proliferation and cell cycle progression via binding with RBPs and regulating transcription in NSCLC
Circular nucleolar protein 10 (circNOL10) is primarily expressed in the nucleus, and is generated from exons 6–12 of pre-NOL10 mRNA. The expression of circNOL10 is cooperatively regulated by pre-NOL10 methylation and by epithelial splicing regulatory protein 1, a splicing factor. CircNOL10 expression is low in lung cancer. CircNOL10 directly promotes the expression of sex comb on midleg-like 1 (SCML1) by suppression of ubiquitination, and also promotes the transcriptional regulatory effect of SCML1 on the humanin polypeptide family, ultimately inhibiting the progression of lung cancer (75).
CircRNAs participate in RNA splicing in NSCLC
The notable associations between the expression of circ-UBR5 and differentiation degree of NSCLC has been established. The differentiation of NSCLC is decreased following knockdown of circ-UBR5. Circ-UBR5 may thus be used to evaluate tumor differentiation, and as an indicator for the pathological grading of NSCLC. Circ-UBR5 binds to splicing regulatory factors, including KH domain containing RNA binding (QKI), NOVA alternative splicing regulator 1 and U1 snRNA. Circ-UBR5 additionally participates in differentiation via modulation of RNA splicing (76).
CircRNAs and NSCLC autophagy
Autophagy is a process of transporting damaged, denatured or aging proteins and organelles to lysosomes for digestion and degradation in cells. Autophagy serves an important and complicated role in tumor development. Upregulation of autophagy in cancer therapy can promote the survival or death of tumor cells (10). Abnormal activity of the mTOR signaling pathway, pathophysiological p53 expression and endoplasmic reticulum stress serve key roles in autophagy of NSCLC (12). Further studies have shown that autophagy is one of the most important pathogenic events in NSCLC development, leading to drug resistance, metastasis and poor prognosis (10).
CircRNAs modulate autophagy via sponging miRNAs in NSCLC
Circular homeodomain-interacting protein kinase 3 (circHIPK3) is derived from exon 2 of the HIPK3 gene, and circHIPK3 is primarily localized in the cytoplasm. Through an RFP-GFP-LC3B assay, Chen et al (77) reported that knockdown of circHIPK3 elevated autophagic flux in autophagy-induced cell lines (A549 and H838). CircHIPK3 may sponge miR-124-3p, a well-known tumor suppressor and autophagy regulator, and therefore indirectly regulate IL-6 receptor and STAT3. As a downstream factor of IL6R, STAT3 suppresses autophagy (78). Downregulation of circHIPK3 induces autophagy by modulation of miR-124-3p/STAT3/PRKAA/AMPKα signaling in NSCLC (77).
CircRNAs and NSCLC apoptosis
The unique morphology of cell death was first termed apoptosis by Kerr in 1972. Apoptosis is the process of programmed cell death which serves a crucial role in cell biology and life. The regulation of apoptosis must be strictly controlled (79). The imbalance in the expression ratio of pro-apoptotic proteins and anti-apoptotic proteins, such as the Bcl-2 protein family, p53 or inhibitor of apoptosis proteins are crucial for regulating cell death (80). The Caspases are generally divided into two groups. Caspase-1, 4, 5, 13 and 14, which primarily participate in the inflammatory process, and Caspase-2, 3, 6, 7, 8, 9 and 10, that either participate in initiation or execution of cell death (81). An increasing number of studies have illustrated that circRNAs regulate apoptosis in NSCLC, such as circ_0003645 (82) and circ_0074027 (83).
CircRNAs modulate apoptosis by sponging miRNAs in NSCLC
CircRNAs can affect the expression of apoptosis-related proteins by sponging miRNAs. The activation, expression and regulation of a series of proteins including the Caspase family of proteins, Bax and Bcl-2 family of proteins are involved in apoptosis. Circular VANGL planar cell polarity protein 1 (CircVANGL1) is generated from exons 3–4 of the VANGL1 gene. CircVANGL1 was reported as an oncogene in bladder cancer (84). Additionally, circVANGL1 was shown to reduce cell apoptotic rates in NSCLC. Silencing of circVANGL1 increased Bax expression and decreased Bcl-2 expression, and this effect was achieved by sponging of miR-195 in NSCLC (85).
Circular PVT1 (circPVT1) is generated from exon 3 of its host gene PVT1, and is flanked by two long introns (35,269 and 41,466 bp) on each side. In a total of 68 cases of NSCLC, the expression of circPVT1 was >2× higher than that in normal or paired paratumoral tissues (41/68 cases). Additionally, circPVT1 expression was significantly increased in 7 NSCLC cell lines compared with a human bronchial epithelial cell line (HBE cells). A luciferase assay showed that luciferase activity was promoted by c-Fos interacting with the circPVT1 promoter region. This results in upregulation of circPVT1 in NSCLC. C-Fos-induced circPVT1 modulates cell proliferation, invasion and migration, and induces cell apoptosis in NSCLC. CircPVT1 regulates carcinogenesis by downregulating miR-125b and upregulating E2F transcription factor 2 (E2F2) (86).
CircRNAs and NSCLC EMT, invasion and metastasis
Tumor metastasis refers to the process in which malignant tumor cells infiltrate into the surrounding tissues from their origin. The progression of tumor cell metastasis is divided into three stages: Adhesion, degradation and migration. Malignant tumor cells break through the basement membrane, move from its primary site (primary tumor) into lymphatic vessels, blood vessels or body cavities to ‘target’ tissues or organs, and form a distant secondary tumor with the same/similar histological type to that of the primary tumor (87). The EMT program is considered a key step and is closely involved in pathological states of tumor progression (88). EMT is considered as the driving factor of invasion and metastasis (89). Metastasis of NSCLC cells is a significant obstacle reducing the OS of NSCLC patients, and is considered a core step in the malignant progression of NSCLC (87). Thus, it is crucial that we improve our understanding of the mechanisms underlying metastasis. Moreover, it is widely accepted that circRNAs are related to the invasion and metastasis of NSCLC.
CircRNAs modulate EMT, invasion and metastasis through sponging miRNAs in NSCLC
Circular SRY-box transcription factor 4 (circ-SOX4) possesses a covalently closed cyclic structure, and has been shown to be upregulated in NSCLC. Reduced expression of Circ-SOX4 decreases the number of invasive and metastatic cells, and decreases the expression of the EMT related proteins, including N-cadherin, Vimentin, ZEB1, Slug, Twist, Snail, matrix metalloproteinase (MMP)2, MMP7 and MMP9. Overexpression of circ-SOX4 increases β-catenin expression in the cell nucleus and reduces its expression in the cell cytoplasm (increases translocation). Thus, circ-SOX4 results in activation of the Wnt pathway. c-MYC is upregulated upon Wnt pathway stimulation. Furthermore, c-MYC harbors two binding sites with circ-SOX4, and can increase its expression, highlighting the presence of a positive feedback loop between circ-SOX4 and c-MYC. Invasion, metastasis and EMT of NSCLC is promoted by circ-SOX4 via increased activity of the Wnt/β-catenin pathway, through increasing the expression of c-MYC (90). Gao and Ye (91) showed that circ-SOX4 was upregulated in LUAD Western blotting showed that expression of Wnt pathway related proteins and EMT representative proteins was increased when circ-SOX4 was overexpressed. These effects were altered via upregulated expression of miR-1270 and decreased expression of PLAG1 like zinc finger 2.
Three circRNAs, hsa_circ_0005273, hsa_circ_0008305, and hsa_circ_0003221, which are spliced from different exons of the pre-mRNA of PTK2, are all termed circular protein tyrosine kinase 2 (CircPTK2). In NSCLC, hsa_circ_0008305 expression is low in patients with distant metastasis. Hsa_circ_0008305 interacts with miR-429 and miR-200b-3p in NSCLC. The expression of tripartite motif containing 33 (TIF1γ) is reduced following overexpression of miR-429 and miR-200b-3p, as they can bind to the 3′-untralsted region (UTR) of TIF1γ. TIF1γ may mediate EMT and the TGF-β/Smad pathway by ubiquitinating Smad4. CircPTK2 participates in TGF-β-induced EMT and invasion. Mechanically, circPTK2 represses miR-429/miR-200b-3p expression and increases TIF1γ expression (92).
The process of tumor cell metastasis requires destruction of any physical barriers, such as the basement membrane and the extracellular matrix (ECM). MMPs are important proteases that degrade the ECM, and they serve an important role in tumor invasion and metastasis (93). Circular cysteine rich transmembrane BMP regulator 1 (circCRIM1), also known as hsa_circ_0002346, is an exon-related circRNA. LUAC patients with TNM stage II and III and lymph node metastasis exhibited lower circCRIM1 expression levels than patients with TNM stage I LUAD. Functional experiments revealed that circCRIM1 represses invasion and metastasis in LADC. CircCRIM1 serves as a miR-182/93 sponge. Resulting in upregulation of leukemia inhibitory factor receptor and increases MMP13 expression via activation of the PI3K/AKT/JAK1 signaling pathway (94).
CircRNAs modulate EMT, invasion and metastasis through binding with RBPs in NSCLC
Circular La ribonucleoprotein 4 (circLARP4), derived from the LARP4 gene, acts as a La-related RBP. In NSCLC, downregulated expression of circLARP4 is associated with a worse prognosis. Overexpression of circLARP4 suppresses the metastatic ability of SPCA1 cells. Additionally, the protein levels of the SMAD family member 7 (SMAD7) is upregulated following circLARP4 overexpression. Thus, circLARP4 negatively regulates invasion and metastasis of NSCLC by upregulating of SMAD7 (95).
CircRNAs modulate EMT, invasion and metastasis through sponging miRNAs and regulating expression of their parental genes
Dysregulation of the Wnt/β-catenin signaling pathway modulates EMT progression in a range of cancer types. CircRNAs modulate the Wnt/β-catenin pathway through different mechanisms. Circ-ITCH is located on chromosome 20q11.22. Circ-ITCH expression is significantly decreased in lung cancer tissues, and its expression is positively correlated with its parental gene, ITCH. Circ-ITCH inhibits the activity of the Wnt/β-catenin pathway. Western blot analysis showed that overexpression of circ-ITCH suppressed the protein expression levels of β-catenin. Subsequently, mRNA expression of c-Myc and CCND1, the two downstream binding partners of β-catenin, was reduced following circ-ITCH upregulation/overexpression. Thus, circ-ITCH interacts with miR-7 and miR-214, and participates in the progression of lung cancer (96).
CircRNAs and NSCLC drug resistance
Chemotherapy is a common method for treating cancer, including NSCLC. It has been found that cells that are resistant to certain chemotherapeutic drugs may also possess resistance to other structurally unrelated drugs via different mechanisms. This phenomenon of broad drug resistance is termed MDR (97). Whilst certain factors in tumor cells underlying the development of resistance to chemoradiation and targeted therapy have been characterized, the process and the underlying molecular mechanisms are still not completely understood. Recent studies have described the roles of circRNAs in drug resistant NSCLC (98,99).
CircRNAs modulate drug resistance through sponging miRNAs in NSCLC
One circRNA can interact with multiple miRNAs to moderate the nucleotide excision repair (NER) signaling pathway. Hsa_circ_0001946 is an exon-derived circRNA that is produced from CDR1 with a length of 1,485 nt. Hsa_circ_0001946 is located in chrX: 139865339-139866824. FISH analysis indicated that hsa_circ_0001946 is primarily present in the cytoplasm. Hsa_circ_0001946 functions as a tumor suppressor in NSCLC. Upregulation of hsa_circ_0001946 enhances the cisplatin sensitivity of A549 cells. Moreover, silencing of hsa_circ_0001946 activates the NER signaling pathway, which decreased cisplatin sensitivity of lung cancer. Hsa_circ_0001946 is implicated in regulation of the sensitivity of NSCLC cells to cisplatin via modulation of the NER signaling pathway. Hsa_circ_0001946 sponges four miRNAs (hsa-miR-7-5p, hsa-miR-671-5p hsa-miR-1270 and hsa-miR-3156-5p) to moderate NER signaling (100).
Circ_0002483 is located at chr8:141862969-141921766. Circ_0002483 is significantly downregulated in NSCLC tissue samples and in Taxol-resistant NSCLC cell lines. Lower levels of circ_0002483 is correlated with a poorer prognosis in patients with NSCLC. A Cell Counting Kit-8 assay showed that overexpression of circ_0002483 notably increased the sensitivity of NSCLC cells to Taxol. Using dual-luciferase reporter assays and an RNA immunoprecipitation assay, circ_0002483 was confirmed to competitively bind to miR-182-5p. Knockdown of miR-182-5p increases sensitivity to Taxol in A549 and H1299 cells. A luciferase assay indicated that miR-182-5p could bind to the 3′UTR of growth factor receptor bound protein 2 (GRB2), forkhead box O1 (FOXO1) and forkhead box O3 (FOXO3). Co-transfection of miR-182-5p and circ_0002483 restored GRB2, FOXO1 and FOXO3 expression and induced resistance to Taxol in NSCLC cells. These findings suggest that circ_0002483, serves as a miR-182-5p sponge, promotes GRB2, FOXO and FOXO3 expression and enhances the sensitivity of A549 and H1299 cells to the chemotherapeutic drug Taxol (101).
CircRNAs modulate drug resistance through binding with RBPs in NSCLC
CircRNA_103762 is significantly highly expressed in NSCLC tissues and cell lines, and its upregulated expression is closely correlated with shorter survival rates in patients with NSCLC. CircRNA_103762 is also upregulated in cisplatin-resistant H358/CDDP lung cancer cells. CircRNA_103762 represses the expression of DNA damage inducible transcript 3 and facilitates MDR in NSCLC (102).
CircRNAs modulate drug resistance through regulation of translation of their parental genes
Hsa_circ_0004350 and hsa_circ_0092857 are transcribed from eukaryotic translation initiation factor 3 subunit A (EIF3a). Hsa_circ_0004350 is located on chromosome 10:120.832.401–120.833.449 and hsa_circ_0092857 on chromosome 10:120.809.312-120.810.833, including three exons and two introns, and are differentially expressed in A549 and A549/DDP cells. Hsa_circ_0004350 and hsa_circ_0092857 are prominently associated with translation regulation based on analysis of data obtained from Metascape. Gene Ontology analysis showed that the overlapping RBPs of the two circEIF3as were regulators of translation, and they may exhibit functional synergy with their parental gene, EIF3a. Abnormal expression of hsa_circ_0004350 and hsa_circ_0092857 may impact the cisplatin resistance of lung cancer cells (103).
4. Conclusion
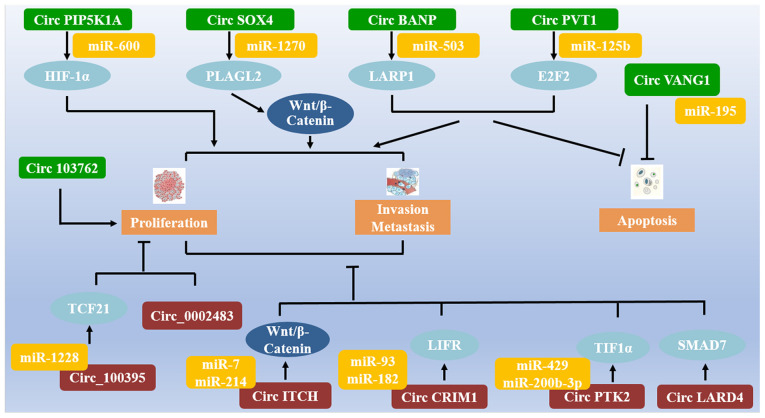
This review summarizes the findings of recent studies on circRNAs that may function as carcinogenic or tumor suppressor genes in NSCLC (Fig. 4). Several circRNAs participate in regulating the pathological progression of NSCLC. Compared with coding RNAs, miRNAs and lncRNAs, circRNA research is in its initial stages, and several problems still need to be addressed. To date several functions of circRNAs and their participation in the regulation of the progression of cancer have been identified, although considerably more remain undetermined. For NSCLC, in order to improve the prognosis and OS of patients, novel targeted therapeutic approaches are required. Further development of targeted circRNAs may become potential pivotal elements to improve our understanding of NSCLC. These mentioned circRNAs may serve as biomarkers of diagnosis and prediction in NSCLC. They may also serve as a means of non-invasive treatments. In future studies, additional attention should be paid to the role of circRNAs in the clinical diagnosis and treatment of NSCLC.
Figure 4.
Several circRNAs and their downstream targets in the regulation of the pathological progression of NSCLC. CircRNA, circular RNA; NSCLC, non-small cell lung cancer; miR, microRNA.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- NSCLC
non-small cell lung cancer
- circRNAs
circular RNAs
- miRNA
microRNA
- TNM
tumor node metastasis
- EMT
epithelial-mesenchymal transition
- SCLC
small cell lung cancer
- ceRNA
competing endogenous RNA
- ecircRNAs
exonic circRNAs
- ciRNAs
intronic circRNAs
- EIciRNAs
exon-intron circRNA
- tricRNAs
tRNA intronic circRNAs
- MRE
microRNA response element
Funding Statement
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81972522), Youth Talent Support Program of Liaoning Province (grant no. XLYC1907011), Key R&D Program of Liaoning Province (grant no. 2018225014) and Technological innovation fund of Shenyang Technology Division (grant nos. RC190008 and 19-112-4-023).
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81972522), Youth Talent Support Program of Liaoning Province (grant no. XLYC1907011), Key R&D Program of Liaoning Province (grant no. 2018225014) and Technological innovation fund of Shenyang Technology Division (grant nos. RC190008 and 19-112-4-023).
Availability of data and materials
Not applicable.
Authors' contributions
SL, YL, GQ, YL, XL, FM, NL and TX wrote the original manuscript. YW, BQ and SX reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Tsim S, O'Dowd CA, Milroy R, Davidson S. Staging of non-small cell lung cancer (NSCLC): A review. Respir Med. 2010;104:1767–1674. doi: 10.1016/j.rmed.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Giovannetti E, Toffalorio F, De Pas T, Peters GJ. Pharmacogenetics of conventional chemotherapy in non-small-cell lung cancer: A changing landscape? Pharmacogenomics. 2012;13:1073–1086. doi: 10.2217/pgs.12.91. [DOI] [PubMed] [Google Scholar]
- 5.Yu Y, He J. Molecular classification of non-small-cell lung cancer: Diagnosis, individualized treatment, and prognosis. Front Med. 2013;7:157–171. doi: 10.1007/s11684-013-0272-4. [DOI] [PubMed] [Google Scholar]
- 6.Minguet J, Smith KH, Bramlage P. Targeted therapies for treatment of non-small cell lung cancer-recent advances and future perspectives. Int J Cancer. 2016;138:2549–2561. doi: 10.1002/ijc.29915. [DOI] [PubMed] [Google Scholar]
- 7.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 8.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 9.Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle. 2009;8:532–535. doi: 10.4161/cc.8.4.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 11.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G, Pei F, Yang F, Li L, Amin AD, Liu S, Buchan JR, Cho WC. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. 2017;18:367. doi: 10.3390/ijms18020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlikos F, Harrington KJ, Syrigos KN. Key molecular mechanisms in lung cancer invasion and metastasis: A comprehensive review. Crit Rev Oncol Hematol. 2013;87:1–11. doi: 10.1016/j.critrevonc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Y, He D, Peng Z, Peng W, Shi W, Wang J, Li B, Zhang C, Duan C. Circular RNAs in cancer: An emerging key player. J Hematol Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 19.Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl) 2018;96:85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 20.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Xin Y. Circular RNAs: A new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21. doi: 10.1186/s13045-018-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z, Yang T, Xiao J. Circular RNAs: Promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 24.Bezzi M, Guarnerio J, Pandolfi PP. A circular twist on microRNA regulation. Cell Res. 2017;27:1401–1402. doi: 10.1038/cr.2017.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32:2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 26.Liu S, Zhang P, Chen Z, Liu M, Li X, Tang H. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 2013;587:2247–2253. doi: 10.1016/j.febslet.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 27.Kong D, Piao YS, Yamashita S, Oshima H, Oguma K, Fushida S, Fujimura T, Minamoto T, Seno H, Yamada Y, et al. Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene. 2012;31:3949–3960. doi: 10.1038/onc.2011.558. [DOI] [PubMed] [Google Scholar]
- 28.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
- 29.Sang M, Meng L, Sang Y, Liu S, Ding P, Ju Y, Liu F, Gu L, Lian Y, Li J, et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018;426:37–46. doi: 10.1016/j.canlet.2018.03.049. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, Liu B, Li XM, Yao J, Jiang Q, Yan B. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 36.Perriman R, Ares M., Jr Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4:1047–1054. doi: 10.1017/S135583829898061X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 41.Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 44.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M. Transcribed processed pseudogenes in the human genome: An intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 2005;33:2374–2383. doi: 10.1093/nar/gki531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, Asangani IA, Kothari V, Prensner JR, Lonigro RJ, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell. 2012;149:1622–1634. doi: 10.1016/j.cell.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2015;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L. CircRNA-derived pseudogenes. Cell Res. 2016;26:747–750. doi: 10.1038/cr.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei B, Tian Z, Fan W, Ni B. Circular RNA: A novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16:292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z, Ruan Y, Zhang H, Shen Y, Li T, Xiao B. Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci. 2019;110:3630–3638. doi: 10.1111/cas.14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Ge YZ, Xu L, Jia R. Circular RNA ITCH: A novel tumor suppressor in multiple cancers. Life Sci. 2020;254:117176. doi: 10.1016/j.lfs.2019.117176. [DOI] [PubMed] [Google Scholar]
- 54.Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, Di QG. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144:667–674. doi: 10.1007/s00432-017-2576-2. [DOI] [PubMed] [Google Scholar]
- 55.Li C, Zhang L, Meng G, Wang Q, Lv X, Zhang J, Li J. Circular RNAs: Pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145:2875–2889. doi: 10.1007/s00432-019-03045-4. [DOI] [PubMed] [Google Scholar]
- 56.Mu Y, Xie F, Huang Y, Yang D, Xu G, Wang C, Wu Q. Circular RNA expression profile in peripheral whole blood of lung adenocarcinoma by high: Throughput sequencing. Medicine (Baltimore) 2019;98:e17601. doi: 10.1097/MD.0000000000017601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8:2878. doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, Dai J, Zhang S, Yang Q, Yi Y, Jiang Y. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18:13. doi: 10.1186/s12943-019-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, Xu W, Zhang E, Wang J, Fang T, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 61.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joseph NA, Chiou SH, Lung Z, Yang CL, Lin TY, Chang HW, Sun HS, Gupta SK, Yen L, Wang SD, et al. The role of HGF-MET pathway and CCDC66 cirRNA expression in EGFR resistance and epithelial-to-mesenchymal transition of lung adenocarcinoma cells. J Hematol Oncol. 2018;11:74. doi: 10.1186/s13045-018-0557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17:2080–2090. doi: 10.1080/15384101.2018.1515553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, Cui J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503:2429–2435. doi: 10.1016/j.bbrc.2018.06.172. [DOI] [PubMed] [Google Scholar]
- 65.Pack LR, Daigh LH, Meyer T. Putting the brakes on the cell cycle: Mechanisms of cellular growth arrest. Curr Opin Cell Biol. 2019;60:106–113. doi: 10.1016/j.ceb.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 67.Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38:BSR20180570. doi: 10.1042/BSR20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther. 2018;11:3979–3987. doi: 10.2147/OTT.S158316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu XF, Liu ZC, Xie BF, Li ZM, Feng GK, Yang D, Zeng YX. EGFR tyrosine kinase inhibitor AG1478 inhibits cell proliferation and arrests cell cycle in nasopharyngeal carcinoma cells. Cancer Lett. 2001;169:27–32. doi: 10.1016/S0304-3835(01)00547-X. [DOI] [PubMed] [Google Scholar]
- 70.Nakayama K, Rahman MT, Rahman M, Nakamura K, Ishikawa M, Katagiri H, Sato E, Ishibashi T, Iida K, Ishikawa N, Kyo S. CCNE1 amplification is associated with aggressive potential in endometrioid endometrial carcinomas. Int J Oncol. 2016;48:506–516. doi: 10.3892/ijo.2015.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, Han S, Wu G. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 73.Donnellan R, Chetty R. Cyclin D1 and human neoplasia. Mol Pathol. 1998;51:1–7. doi: 10.1136/mp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chi Y, Luo Q, Song Y, Yang F, Wang Y, Jin M, Zhang D. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem. 2019;120:19019–19030. doi: 10.1002/jcb.29225. [DOI] [PubMed] [Google Scholar]
- 75.Nan A, Chen L, Zhang N, Jia Y, Li X, Zhou H, Ling Y, Wang Z, Yang C, Liu S, Jiang Y. Circular RNA circNOL10 inhibits lung cancer development by promoting SCLM1-mediated transcriptional regulation of the humanin polypeptide family. Adv Sci (Weinh) 2018;6:1800654. doi: 10.1002/advs.201800654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qin M, Wei G, Sun X. Circ-UBR5: An exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem Biophys Res Commun. 2019;503:1027–1034. doi: 10.1016/j.bbrc.2018.06.112. [DOI] [PubMed] [Google Scholar]
- 77.Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, Wang Z, Wang J, Kresty LA, Beer DG, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–671. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.You L, Wang Z, Li H, Shou J, Jing Z, Xie J, Sui X, Pan H, Han W. The role of STAT3 in autophagy. Autophagy. 2015;11:729–739. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Espinosa-Oliva AM, García-Revilla J, Alonso-Bellido IM, Burguillos MA. Brainiac caspases: Beyond the wall of apoptosis. Front Cell Neurosci. 2019;13:500. doi: 10.3389/fncel.2019.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D'Orazi G. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong RS. Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.An J, Shi H, Zhang N, Song S. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;511:921–925. doi: 10.1016/j.bbrc.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 83.Gao P, Wang Z, Hu Z, Jiao X, Yao Y. Circular RNA circ_0074027 indicates a poor prognosis for NSCLC patients and modulates cell proliferation, apoptosis, and invasion via miR-185-3p mediated BRD4/MADD activation. J Cell Biochem. 2020;121:2632–2642. doi: 10.1002/jcb.29484. [DOI] [PubMed] [Google Scholar]
- 84.Zeng Z, Zhou W, Duan L, Zhang J, Lu X, Jin L, Yu Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J Cell Physiol. 2019;234:3887–3896. doi: 10.1002/jcp.27162. [DOI] [PubMed] [Google Scholar]
- 85.Wang L, Ma H, Kong W, Liu B, Zhang X. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep. 2019;39:BSR20182433. doi: 10.1042/BSR20182433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Li X, Zhang Z, Jiang H, Li Q, Wang R, Pan H, Niu Y, Liu F, Gu H, Fan X, Gao J. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol Biochem. 2018;51:2324–2340. doi: 10.1159/000495876. [DOI] [PubMed] [Google Scholar]
- 87.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, Hengstschläger M, Dolznig H. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 88.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868:584–591. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 89.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 90.Wang L, Zheng C, Wu X, Zhang Y, Yan S, Ruan L, Dai H. Circ-SOX4 promotes non-small cell lung cancer progression by activating the Wnt/beta-catenin pathway. Mol Oncol. 2020 doi: 10.1002/1878-0261.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao N, Ye B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. 2020;20:2. doi: 10.1186/s12935-019-1065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Winer A, Adams S, Mignatti P. Matrix metalloproteinase inhibitors in cancer therapy: Turning past failures into future successes. Mol Cancer Ther. 2018;17:1147–1155. doi: 10.1158/1535-7163.MCT-17-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang L, Liang Y, Mao Q, Xia W, Chen B, Shen H, Xu L, Jiang F, Dong G. Circular RNA circCRIM1 inhibits invasion and metastasis in lung adenocarcinoma through the microRNA (miR)-182/miR-93-leukemia inhibitory factor receptor pathway. Cancer Sci. 2019;110:2960–2972. doi: 10.1111/cas.14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shi JQ, Wang B, Cao XQ, Wang YX, Cheng X, Jia CL, Wen T, Luo BJ, Liu ZD. Circular RNA_LARP4 inhibits the progression of non-small-cell lung cancer by regulating the expression of SMAD7. Eur Rev Med Pharmacol Sci. 2020;24:1863–1869. doi: 10.26355/eurrev_202002_20364. [DOI] [PubMed] [Google Scholar]
- 96.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhitomirsky B, Assaraf YG. Lysosomes as mediators of drug resistance in cancer. Drug Resist Updat. 2016;24:23–33. doi: 10.1016/j.drup.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 98.Fraipont F, Gazzeri S, Cho WC, Eymin B. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet. 2019;10:390. doi: 10.3389/fgene.2019.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong Y, Xu T, Zhong S, Wang B, Zhang H, Wang X, Wang P, Li G, Yang S. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984. doi: 10.1016/j.lfs.2019.116984. [DOI] [PubMed] [Google Scholar]
- 100.Huang MS, Liu JY, Xia XB, Liu YZ, Li X, Yin JY, Peng JB, Wu L, Zhang W, Zhou HH, Liu ZQ. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508. doi: 10.3389/fonc.2019.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li X, Yang B, Ren H, Xiao T, Zhang L, Li L, Li M, Wang X, Zhou H, Zhang W. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10:953. doi: 10.1038/s41419-019-2180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xiao G, Huang W, Zhan Y, Li J, Tong W. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP) J Clin Lab Anal. 2020;34:e23252. doi: 10.1002/jcla.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang MS, Yuan FQ, Gao Y, Liu JY, Chen YX, Wang CJ, He BM, Zhou HH, Liu ZQ. Circular RNA screening from EIF3a in lung cancer. Cancer Med. 2019;8:4159–4168. doi: 10.1002/cam4.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.