Abstract
The aim of the present study was to explore the effects of LINC00649 on the proliferation, migration and invasion of bladder cancer (BC) and identify possible mechanisms. Through TCGA database analysis of LINC00649 expression in bladder cancer and the association of LINC00649 with the BC patient prognosis, RT-qPCR was employed for detecting LINC00649 expression in 60 clinical tissue specimens and cell lines of bladder cancer. The lentivirus stable transfection or small interfering RNA was used to increase or decrease the LINC00649 expression level in T24 and UM-UC-3 cells. CCK8 and clone formation assay were utilized to observe the effects of LINC00649 on the proliferation and colony formation of BC cells. Transwell experiment was performed to detect the effects of LINC00649 on the migration and invasion of bladder cancer. Bioinformatics database was used to identify the possible downstream targets of LINC00649 while RT-qPCR, western blot analysis and dual luciferase reporter gene experiments were carried out to verify the possible molecular mechanism. The TCGA database analysis revealed a significantly high expression of LINC00649 in bladder cancer and an association of LINC00649 expression with overall survival rate of BC patients. As shown by RT-qPCR detection, LINC00649 expression was notably upregulated in BC tissues and BC cell lines. In addition, statistical analyses unveiled that highly expressed LINC00649 was clearly associated with poor overall survival of bladder cancer. Based on the in vitro cell experiment, upregulated LINC00649 considerately enhanced the proliferation, migration and invasion of BC cells, as opposed to those in T24 and UM-UC-3 cells by suppressing LINC00649. Mechanically, LINC00649 may promote the malignant progression of bladder cancer by regulating miR-15a-5p to promote the HMGA1 expression axis. Overall, LINC00649 upregulates HMGA1 expression by binding to miR-15a-5p to enhance the proliferation, migration and invasion of BC cells. Thus, LINC00649 is a potential biomarker and therapeutic target for bladder cancer.
Keywords: bladder cancer, LINC00649, miR-15a-5p, HMGA1, malignant progression
Introduction
Bladder cancer (BC) is an important malignancy in the urinary system, whose incidence ranks 11th among all malignant tumors, 7th among malignant tumors in male patients, and 10th among malignant tumors in female patients (1). Based on histopathology, BC is divided into urothelial carcinoma, squamous carcinoma, adenocarcinoma, and undifferentiated carcinoma, of which urothelial carcinoma is a major type, accounting for more than 90% of all malignant bladder tumors (2). In addition, according to the BC invasion and infiltration of the muscular layer, BC may be stratified into muscle invasive bladder cancer (MIBC) and non-muscle invasive bladder cancer (NMIBC), accounting for approximately 30 and 70%, respectively (3). MIBC has a higher degree of malignancy and a much higher probability of tumor recurrence and metastasis than NMIBC. High-grade MIBC is the major cause of mortality in BC patients (4). Mechanisms underlying the occurrence and development of BC remain unclear. Biological behaviors of BC cells are characterized by multicentricity, invasion, and susceptibility to recurrence. Some patients present with early stage NMIBC, progress to MIBC, and develop drug resistance post-treatment, with a high recurrence rate (5). Therefore, an in-depth study to clarify the biological process of BC and molecular regulatory network at various stages as early as possible can help patients diagnose tumors early, improve treatment effects as well as disease prognosis.
Non-coding RNAs (ncRNAs) are RNA transcripts, and the vast majority of ncRNAs are not translated into proteins (6). In spite of the lack of protein translation function, ncRNAs are vital participants in cellular activities. Based on different lengths, structures, and functions, ncRNAs are mainly divided into microRNA, rRNA, tRNA, lncRNA, and circular RNA, of which lncRNA is a transcript with more than 200 nt and no open reading frame (7). Although lncRNAs do not code proteins, they are expressed in many differentiated tissues or specific cancers. lncRNAs are regarded as noise produced during the transcription process and have no influence on biological effects. In addition, lncRNAs are considered irreplaceable in vital activities such as the regulation of the stability of telomeres, the control of nuclear structure and transcription, X inactivation, nucleosome generation, the regulation of mRNA splicing and maturity, and microRNA activity (8). Through epigenetic regulation of coding gene expression involved in cellular activities, lncRNAs regulate vital bioprocesses including cell autophagy, cell cycle, differentiation, proliferation, migration, invasion, and apoptosis (9). Therefore, lncRNAs are expected to become diagnostic markers and therapeutic targets for tumors. For example, lncRNA highly upregulated in liver cancer promotes the progression of liver cancer by binding to microRNA-186 to increase the expression of high mobility group AT-hook 2 (10), lncRNA LINC00958 inhibits the occurrence of pancreatic cancer by sponging microRNA-330-5p to downregulate the expression of paired box 8 (11), and lncRNA small nucleolar RNA host gene 1 (SNHG1) promotes the development of non-small cell lung cancer by spongy bonding to miR-145-5p to upregulate the expression of metadherin (12).
LINC00649 is an lncRNA abnormally highly expressed in BC through database screening; however, its biological roles and potential molecular mechanisms in BC remain unknown. This study investigated the expression of LINC00649 in BC tissues, adjacent tissues, normal cell lines, and BC cell lines, and analyzed the association of the expression with clinicopathological characteristics of BC patients. In addition, in vitro cell experiments were conducted to test the effects of LINC00649 on the proliferation, migration, and invasion of BC cells. Furthermore, bioinformatics and relevant experiments were used to explore the possible LINC00649 molecular mechanisms of action, thus providing a new direction and molecular target for the targeted therapy of BC.
Materials and methods
GEPIA database
A total of 404 BC tissues and 28 normal (control) tissues were obtained from the TCGA database to analyze mRNA expression profiles. GEPIA is a new interactive website for TCGA- and GTEx-based analysis of RNA sequence data (http://gepiacancer-pku.cn/index.html). LINC00649 expression in BC tissues and control tissues was analyzed, and the GEPIA database was used to calculate the correlation between LINC00649 expression and disease-free survival (DFS) and overall survival (OS) of BC patients.
Study subjects and sample collection
Approval for the study was obtained from the ethics committee of The First People's Hospital of Wenling (Zhejiang, China). The patients volunteered to participate in this study and signed a written informed consent form. All experiments were performed in accordance with the Declaration of Helsinki.
A total of 60 BC tissues and 60 adjacent tissues were collected from The First People's Hospital of Wenling. All tissue specimens were confirmed via pathological diagnosis by an experienced pathologist. Samples were frozen in liquid nitrogen (−196°C) immediately post-dissection and stored in a −80°C freezer. Age >55 years was considered the cut-off point as indicated in previous studies (13,14). Pathological classification and tumor staging were performed according to the cancer staging criteria of the Union for International Cancer Control. None of the patients had preoperative radiotherapy, chemotherapy history, or other tumor history and met the clinical and pathological data integrity. The patient sex, age, tumor staging, lymphatic metastasis, or distant metastasis was listed in Table I.
Table I.
Correlation between LINC00649 expression and clinicopathological factors.
| Expression of LINC00649 | ||||
|---|---|---|---|---|
| Factors | Cases | High | Low | P-value |
| Overall | 60 | 45 | 15 | |
| Sex | 0.9236 | |||
| Male | 43 | 32 | 11 | |
| Female | 17 | 13 | 4 | |
| Age | 0.5223 | |||
| ≤55 | 10 | 7 | 3 | |
| >55 | 50 | 38 | 12 | |
| Stage | ||||
| Primary tumor | 0.0056 | |||
| NMI (Ta+Tis+TI) | 30 | 17 | 13 | |
| MI (T2+T3+T4) | 30 | 28 | 2 | |
| Lymph node metastasis | 0.6325 | |||
| Absent (N0) | 20 | 15 | 5 | |
| Present (N1+N2+N3) | 40 | 30 | 10 | |
| Distant metastasis | 0.05869 | |||
| Absent (N0) | 54 | 40 | 14 | |
| Present (M1) | 6 | 5 | 1 | |
| Grade | 0.0969 | |||
| Low grade | 18 | 12 | 6 | |
| High grade | 42 | 33 | 9 | |
Cell culture
Normal control cell line SV-HUC-1 and human bladder cancer cell lines (5637, SW780, T24, and UM-UC-3) were purchased from the American Type Culture Collection (ATCC, Rockville). Cells were cultured in RPMI-1640 (Gibco) culture medium with 10% fetal bovine serum (FBS; Invitrogen) and placed in an incubator containing 5% CO2 at 37°C, followed by passage culturing at 80–90% of cell density.
Cell transfection
For LINC00649 overexpression, LINC00649 overexpression plasmids were constructed into the pSicoR lentiviral vector, and 293T cells were used for virus production. Lentiviruses were used to infect BC cells, and purinomycin was used for two weeks to produce LINC00649-overexpressed cell lines. Transfection efficiency was measured using RT-qPCR, and cells were collected for subsequent experiments. Then, 3,000 cells were seeded into a 6-well plate, culture medium was added, and transfection was performed at 70–80% of cell density at 37°C for 48 h, referencing the IFU of Lipofectamine 3000 (Invitrogen). GenePharma (Shanghai) designed and synthesized LINC00649 siRNA, miR-15a-5p mimics, HMGA1 overexpression plasmids, and corresponding negative control-transfected cells. The mixed solution was added to a cell culture plate or culture flask containing culture solution and shaken gently. After 24 h of culturing in a CO2 incubator at 37°C, subsequent experiments were conducted.
RNA extraction and real-time quantitative PCR
BC tissues and cells required for experiments were collected, and 1 ml TRIzol (Invitrogen) was added for cell lysis. Subsequently, chloroform (250 µl) was added and mixed for 30 sec, and centrifuged at 1,500 × g at 4°C for 15 min. The resultant aqueous phase was then drawn and added to isometric pre-cooling isopropanol. After centrifugation, precipitates were gently purged with 75% ethanol and dissolved in 30 µl DEPC water. The quantification of total RNA was performed with a NanoDrop 2000 (Thermo Fisher Scientific, Inc.) device and placed in a −80°C refrigerator for future use. Total RNA (1,000 ng) was reverse transcribed to cDNA using the PrimeScript RT reagent kit.
Quantitative PCR of miRNA was performed according to the IFU (Takara; code no. RR036A) of the miScript SYBR-Green PCR Kit. cDNA (500 ng) was used to conduct RT-qPCR using the ABI7900 device according to the IFU of SYBR®-Green Master Mix (Takara). GAPDH and U6 were used as an internal reference, respectively. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. The primer sequences used were as follows: LINC00649 forward: TCCCCACGTAAGGAGGGTAG, reverse: CAGCAACAGGCCTTGTCAAC; miR-15a-5p forward: CACAGAGTGTCGGAGGTGATTC, reverse: CCTGTAGTACGGGTATGTTGAGC; HMGA1 forward: TTTACAGAAGGAGCCCAGCG, reverse: AGGGAAGGGGGTACACAACT;GAPDH forward: GGAGCGAGATCCCTCCAAAAT, reverse: GGCTGTTGTCATACTTCTCATGG; U6 forward: CGCTTCGGCAGCACATATACTAAAATTGGAAC, reverse: GCTTCACGAATTTGCGTGTCATCCTTGC.
Cell proliferation experiment
EdU experiment: EdU kit (RiboBio, China) was used for the detection of cell proliferation. Then, 3,000 cells were seeded into 96-well plates at cells/well, and then 50 mM EdU solution was added to the culture medium. After 24 h, the cells were fixed in 4% formaldehyde and infiltrated using Triton X-100. Treated cells were incubated with the EDU reaction mixture and counterstained with DAPI. The staining results were observed under a fluorescence microscope and recorded according to the manufacturer. Images of five randomly selected fields of view were captured to calculate the number of EdU fusion cells.
For clone formation the transfected BC cells were digested using trypsin, re-suspended, and counted. The same number of BC cells was then calculated and plated into a 3.5-cm well. After 14 days of culturing using complete medium, crystal violet was used to stain visible colonies. Finally, stained colonies were photographed, counted, and compared among different groups.
Transwell experiment
Transwell experiments were performed using 24-well plates (Corning) with a membrane filter chamber at an aperture of 8 µm. For the invasion experiment, 100 µg Matrigel was precoated in the upper chamber. BC cells were seeded into the upper chamber at 4×104 cells/well using 100 µl FBS-free RPMI-1640 culture medium, while 500 µl RPMI-1640 culture medium with 10% FBS was added into the lower chamber. After 24 h of culturing in a cell incubator containing 5% CO2 at 37°C, cells in the upper chamber were removed using a cotton swab. Cells were fixed in methanol for 15 min, stained with crystal violet for 20 min, photographed in five random fields of view under a microscope (×200), and counted. For the migration experiment, steps were identical to those of the invasion experiment, except that no Matrigel was required.
Nucleoplasm isolation experiment
Cells were transferred to a 1.5-ml EP tube, with cytoplasmic lysate (RLA) added, incubated on ice for 20 min, and centrifuged at 1,500 × g at 4°C for 15 min to obtain cytoplasmic protein as the supernatant. After three repeated runs of washing and precipitation using RLA, the cells were incubated with nuclear lysate (RIPA), incubated on ice for 20 min, mixed for 30 sec every 5 min, and centrifuged at 11,000 × g at 4°C for 15 min to obtain nucleoproteins as the supernatant.
Luciferase assay
Bioinformatics website was used to predict the binding sites of miR-15a-5p to LINC00649 or HMGA1. The LINC00473 sequence was analyzed and predicted on the website (http://starbase.sysu.edu.cn/). Subsequently, 3×104 treated BC cells were seeded into 24-well plates. After 48 h of co-transfection with the corresponding plasmids (GenePharma Shanghai) and miR-15a-5p mimics or negative control according to the manufacturer, a Promega kit (Promega) was used to measure luciferase activity as per the IFU and recorded. The experiments were performed in triplicate, with the mean taken for statistical analysis.
Western blot analysis
The treated BC cells were collected. For protein extraction, the cells were added to the cell lysis solution (Beyotime) of protease inhibitor PMSF, placed on ice, centrifuged at 1,500 × g at 4°C for 15 min to collect the supernatant, and protein concentration was measured according to the BCA kit (Beyotime) manufacturer's instructions. After adding SDS-PAGE protein loading buffer, the cells were heated at 100°C to make albuminous degeneration. After loading, the transfer membrane was performed. The corresponding size of the PVDF membrane was cut as per the molecular weight for antigen blocking in 5% skim milk powder blocking buffer. Then the membrane was incubated with primary antibodies targeting HMGA1 (cat. no. ab202070; Abcam, Cambridge) or GAPDH (cat. no. ab128915; Abcam) overnight at 4°C. Next, the membranes were washed three times using TBS in Tween-20 and incubated with an HRP-linked goat antirabbit secondary antibody (cat. no. ab205718; Abcam) at room temperature for 2 h. The ECL Western Blotting Substrate Kit (cat. no. ab65623; Abcam) was used to detect protein signals.
Statistical analysis
Each experiment was performed in triplicate. Experimental data were reported as mean ± standard deviation and analyzed with GraphPad Prism 6 (GraphPad Software Inc.). If the data conformed to normal distribution, Student's t-test was used to compare the mean of measurement data between the two groups, and analysis of variance (ANOVA) was used to compare the difference between multiple groups with Tukey honestly significant difference (HSD) post hoc test. The correlation between LINC00649 and miR-15a-5p expression was assessed using the Pearson's correlation analysis. Kaplan-Meier analysis with log-rank tests were used for overall survival analysis. P-value <0.05 was considered statistically significant.
Results
LINC00649 is highly expressed in BC
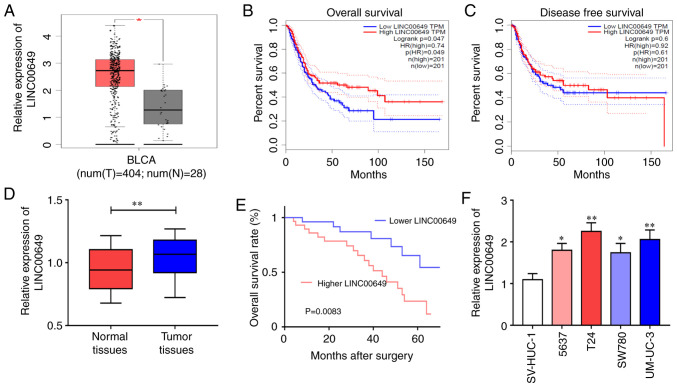
TCGA database analysis showed significantly higher expression of LINC00649 in BC tissues relative to control tissues (Fig. 1A). Further analysis revealed that LINC00649 was strongly associated with the OS of BC patients (HR=0.74, P=0.047) (Fig. 1B), but not significantly correlated with DFS (HR=0.61, P=0.6) (Fig. 1C). Subsequently, LINC00649 expressions in 60 BC tissues and control tissues were verified, which were consistent with the database results. LINC00649 expression was considerably increased in BC tissues (P<0.05) (Fig. 1D), and highly expressed LINC00649 indicated poor prognosis of BC patients (P<0.05) (Fig. 1E). The results were different from those of the TCGA database analysis. Further verification by RT-qPCR revealed significant upregulation of LINC00649 expression in the BC cell lines (P<0.05) (Fig. 1F). Moreover, the analysis of clinical data revealed an association between LINC00649 and muscle invasion of BC (P=0.0056), while no notable association with patient sex, age, tumor staging, lymphatic metastasis, or distant metastasis was observed (Table I). These results showed that LINC00649 may be important in BC.
Figure 1.
High LINC00649 expression in BC. (A) LINC00649 expression in BC tissues as analyzed by TCGA database. (B) Association of LINC00649 expression with the OS of BC patients as analyzed by TCGA database. (C) Correlation of LINC00649 expression with the DFS of BC patients as analyzed by TCGA database. (D) LINC00649 expression in BC tissues by RT-qPCR. (E) Analysis of the correlation between LINC00649 and the OS of BC patients. (F) LINC00649 expression in BC cells via RT-qPCR. *P<0.05; **P<0.01.
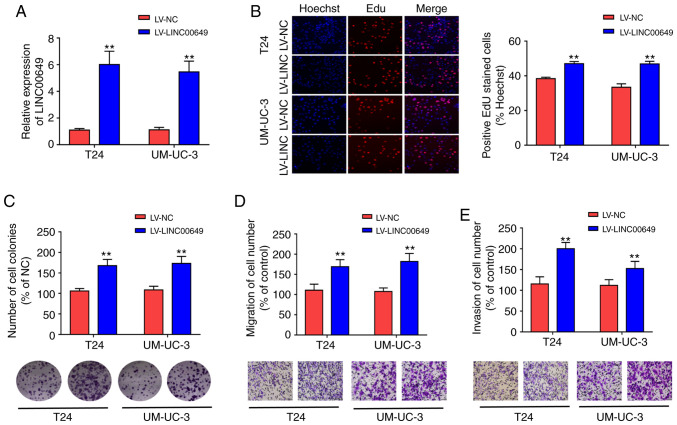
LINC00649 overexpression promotes the proliferation, migration, and invasion of BC cells
To explore the biological functions of LINC00649, lentivirus stable transfection was used to construct T24 and UM-UC-3 cell lines with highly expressed LINC00649, and RT-qPCR was performed to analyze LINC00649 expression, revealing that LV-LINC00649 enhanced LINC00649 expression (P<0.05) (Fig. 2A). Subsequently, EdU and clone formation experiments were used to assess the influence of LINC00649 on cell proliferation. As shown in Fig. 2B and C, highly expressed LINC00649 in T24 and UM-UC-3 cells promoted cell proliferation and clone formation. Transwell experiment revealed that the migration and invasion of T24 and UM-UC-3 cells in the LV-LINC00649 group were significantly enhanced relative to the LV-NC group (P<0.05) (Fig. 2D and E). These results indicated that LINC00649 overexpression notably enhanced the proliferation, migration, and invasion of BC cells.
Figure 2.
LINC00649 overexpression promoted the proliferation, migration and invasion of BC cells. (A) Stably highly expressed LINC00649 in T24 and UM-UC-3 cells as constructed by lentivirus stable transfection and transfection efficiency detected by RT-qPCR. (B) The influences of LINC00649 on T24 and UM-UC-3 cell proliferation by EdU experiment; magnification: ×200. (C) The influences of LINC00649 on the colony formation of T24 and UM-UC-3 cells via clone formation experiment. (D) The change of cell migration post LINC00649 overexpression in T24 and UM-UC-3 cells as detected by Transwell migration experiment; magnification, ×200. (E) Alteration of cell invasion post LINC00649 overexpression in T24 and UM-UC-3 cells as detected by Transwell invasion experiment; magnification, ×200. **P<0.01.
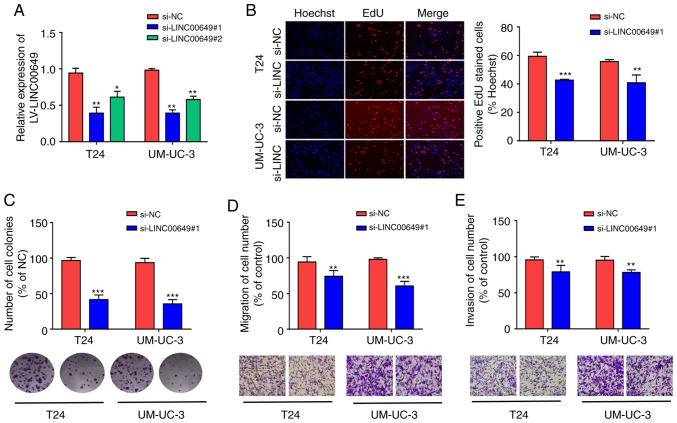
Inhibition of LINC00649 suppresses the proliferation, migration, and invasion of BC cells
LINC00649 expressions in T24 and UM-UC-3 were inhibited by siRNA. In addition, for transfection efficiency, RT-qPCR revealed that LINC00649 siRNA remarkably suppressed LINC00649 expression (P<0.05) (Fig. 3A). si-LINC00649#1 was selected for subsequent experiments owing to its higher interference efficiency. From the EdU experiment, si-LINC00649 notably inhibited the proliferation of T24 and UM-UC-3 cells (P<0.05) (Fig. 3B). Based on the results of the clone formation experiment, the inhibition of LINC00649 in T24 and UM-UC-3 cells suppressed cell colony formation. Furthermore, Transwell experiments indicated that the migration and invasion of T24 and UM-UC-3 cells in the si-LINC00649 group were notably weakened relative to the si-NC group (P<0.05) (Fig. 3D-E). These results suggest that the downregulation of LINC00649 significantly suppresses the proliferation, migration, and invasion of BC cells.
Figure 3.
Inhibition of LINC00649 suppressed the proliferation, migration and invasion of BC cells. (A) Downregulation of LINC00649 expression in T24 and UM-UC-3 cells by using siRNAs and transfection efficiency by RT-qPCR. (B) The influence of LINC00649 suppression on the proliferation of T24 and UM-UC-3 cells through EdU experiment; magnification, ×200. (C) The influences of LINC00649 inhibition on the colony formation of T24 and UM-UC-3 cells as detected by clone formation experiment. (D) The change of cell migration post-LINC00649 downregulation in T24 and UM-UC-3 cells by Transwell migration experiment; magnification, ×200. (E) The alteration of cell invasion post LINC00649 downregulation in T24 and UM-UC-3 cells via Transwell invasion experiment; magnification, ×200. *P<0.05; **P<0.01; ***P<0.001.
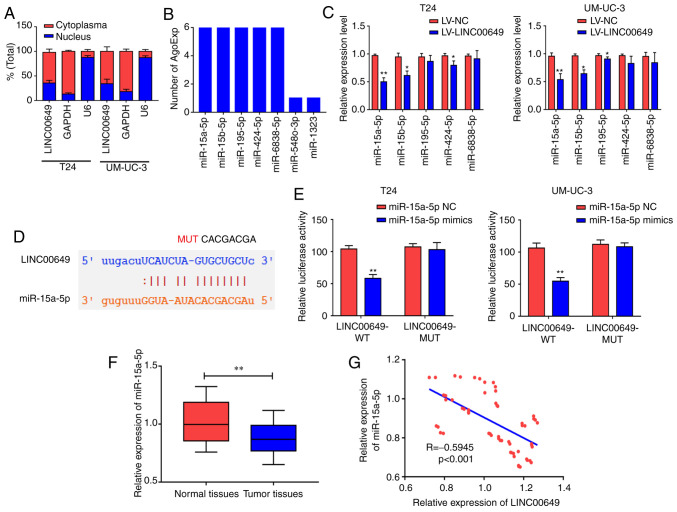
LINC00649 binds to miR-15a-5p
To investigate the possible molecular mechanisms of LINC00649 in BC, subcellular localization was performed via nucleoplasm isolation experiments. As shown in Fig. 4A, LINC00649 was mainly located in the cytoplasm of T24 and UM-UC-3 cells, suggesting that LINC00649 may participate in the regulation at the post-transcriptional level (P<0.05). Subsequently, the bioinformatics website was used to predict miRNAs that may be bound to LINC00649. The binding analysis found that miR-15a-5p, miR-15b-5p, miR-195-5p, miR-424-5p, and miR-6838-5p may be potential binding targets of LINC00649 (P<0.05) (Fig. 4B). LINC00649 was overexpressed in T24 and UM-UC-3 cells. In RT-qPCR, LINC00649 significantly suppressed the expression of miR-15a-5p, miR-15b-5p, and miR-424-5p, exerting the highest inhibition against miR-15a-5p; thus, miR-15a-5p was selected for subsequent studies (P<0.05) (Fig. 4C). Through predicted binding sites, LINC00649 wild-type plasmids (LINC00649 WT) and LINC00649 mutant plasmids (LINC00649 MUT) were designed and constructed (P<0.05) (Fig. 4D). For transfection efficiency, RT-qPCR revealed that miR-15a-5p mimics significantly promoted miR-15a-5p expression (P<0.05) (Fig. S1A). Their binding relationship was detected via a dual luciferase reporter gene experiment, which revealed that post-transfection with miR-15a-5p mimics in T24 and UM-UC-3 cells, luciferase activity was significantly decreased in LINC00649-WT 3′UTR group and was not notably changed in LINC00649-MUT 3′UTR group compared with the control group, indicating that miR-15a-5p bound to LINC00649 (P<0.05) (Fig. 4E). miR-15a-5p expression in BC and control tissues were analyzed by RT-qPCR, which revealed markedly low expression of miR-15a-5p in BC tissues (P<0.05) (Fig. 4F). Additionally, LINC00649 and miR-15a-5p expression levels were analyzed in BC tissues, and a negative correlation was observed between them (R=−0.5945, P<0.001) (Fig. 4G). These results showed that LINC00649 may bind to miR-15a-5p and suppress its expression in BC.
Figure 4.
LINC00649 bound to miR-15a-5p. (A) Subcellular localization of LINC00649 by nucleoplasm isolation experiment in T24 and UM-UC-3 cells. (B) Bioinformatics perdition of miRNAs that might be bound to LINC00649. (C) Expressions of miR-15a-5p, miR-15b-5p, miR-195-5p, miR-424-5p and miR-6838-5p via RT-qPCR post-overexpression of LINC00649 in T24 and UM-UC-3 cells. (D) The perdition of binding sites of LINC00649 to miR-15a-5p through bioinformatics website and the construction of LINC00649 wild-type plasmids and LINC00649 mutant plasmids. (E) miR-15a-5p bound to LINC00649 as indicated by the results of dual luciferase reporter gene. (F) miR-15a-5p expression in BC tissues via RT-qPCR. (G) Negative correlation of LINC00649 expression with miR-15a-5p expression in BC tissues. *P<0.05; **P<0.01.
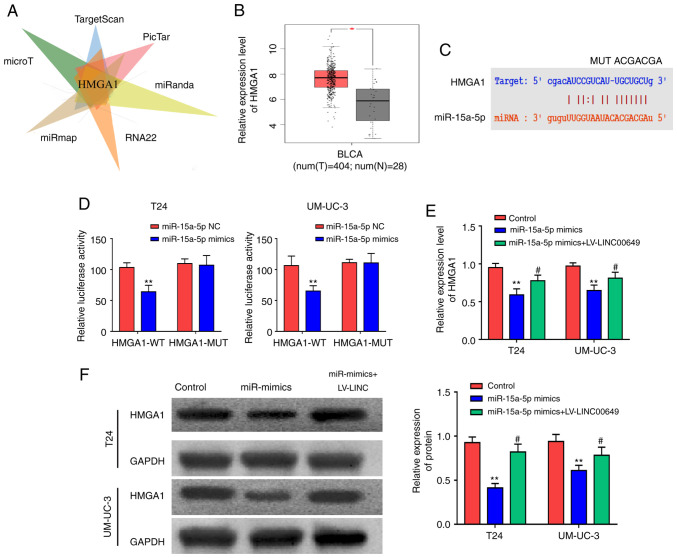
miR-15a-5p targeted HMGA1
Bioinformatics websites (microT, TargetScan, PicTar, miRanda, RNA22, and miRmap) were used to predict target genes that could bind to miR-15a-5p and found that HMGA1 was a potential target (P<0.05) (Fig. 5A). The analysis of the TCGA database revealed a significantly high expression of HMGA1 in BC tissues (P<0.05) (Fig. 5B). Using predicted binding sites, HMGA1 wild-type plasmids (HMGA1 WT) and HMGA1 mutant plasmids (HMGA1 MUT) were designed and constructed (P<0.05) (Fig. 5C). As shown by reporter gene results, following transfection of T24 and UM-UC-3 cells with miR-15a-5p mimics, luciferase was reduced in the HMGA1-WT 3′UTR group and was not notably different in the HMGA1-MUT 3′UTR group (P<0.05) (Fig. 5D), indicating that miR-15a-5p could bind to HMGA1. After miR-15a-5p and LINC00649 were highly expressed in both T24 and UM-UC-3 cells, RT-qPCR and western blot analysis were employed to examine HMGA1 expression and indicated that post miR-15a-5p overexpression, mRNA and protein expressions of HMGA1 were reduced, whereas LINC00649 overexpression partially reversed the inhibitory effect of miR-15a-5p on HMGA1 (P<0.05) (Fig. 5E and F). This indicated that LINC00649 enhanced HMGA1 expression by binding to miR-15a-5, thus exerting its role in BC.
Figure 5.
miR-15a-5p targeted HMGA1. (A) Possible targets of miR-15a-5p as predicted by bioinformatics website and HMGA1 identified as a potential target by taking the intersection point. (B) HMGA1 expression in BC tissues as analyzed by TCGA database. (C) The perdition of binding sites of HMGA1 to miR-15a-5p through bioinformatics website and the construction of HMGA1 wild-type plasmids and HMGA1 mutant plasmids. (D) HMGA1 targeted by miR-15a-5p as shown by the results of dual luciferase reporter gene. (E) HMGA1 expression via RT-qPCR post-overexpression of both miR-15a-5p and LINC00649 in T24 and UM-UC-3 cells. (F) Protein expression of HMGA1 through western blot post-overexpression of both miR-15a-5p and LINC00649 in T24 and UM-UC-3 cells. *P<0.05; **P<0.01; #P<0.05.
miR-15a-5p suppressed the proliferation, migration, and invasion of BC cells by inhibiting HMGA1
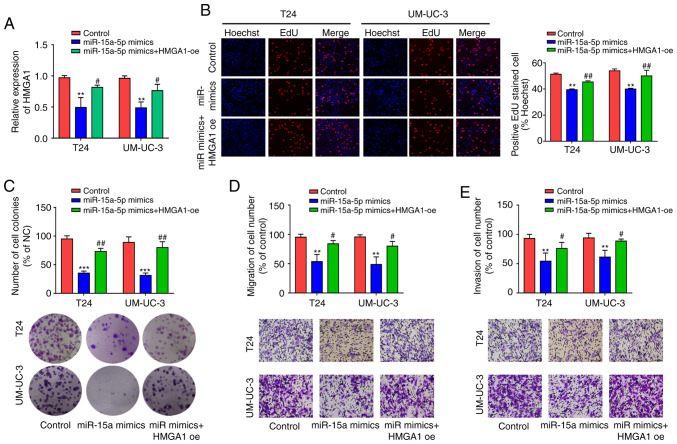
For transfection efficiency, RT-qPCR and western blot analysis revealed that HMGA1 OE markedly promoted HMGA1 mRNA and protein expressions (P<0.05) (Fig. S1B and C). After high expression of miR-15a-5p and HMGA1 in both T24 and UM-UC-3 cells, RT-qPCR was employed to detect HMGA1 expression and indicated that miR-15a-5p overexpression repressed HMGA1 expression, whereas HMGA1 overexpression showed a partial reversal effect on the inhibitory effect of miR-15a-5p (P<0.05) (Fig. 6A). Subsequently, the effects of miR-15a-5p and HMGA1 on cell proliferation, migration, and invasion were assessed in vitro. Results indicated that post miR-15a-5p overexpression in T24 and UM-UC-3 cells, cell proliferation, and colony formation were both markedly weakened, whereas HMGA1 overexpression reversed this inhibitory effect (P<0.05) (Fig. 6B and C). As such, miR-15a-5p overexpression in T24 and UM-UC-3 cells repressed cell migration and invasion, which was reversed by HMGA1 expression (P<0.05) (Fig. 6D and E). These results suggest that miR-15a-5p inhibits the proliferation, migration, and invasion of BC cells by inhibiting HMGA1.
Figure 6.
miR-15a-5p suppressed the proliferation, migration and invasion of BC cells by inhibiting HMGA1. (A) HMGA1 expression detection by RT-qPCR post-overexpression of both miR-15a-5p and HMGA1 in T24 and UM-UC-3 cells. (B) Cell proliferation detection by EdU experiment post overexpression of miR-15a-5p and HMGA1 in T24 and UM-UC-3 cells; magnification, ×200. (C) Cell clone formation by colony formation experiment post-overexpression of miR-15a-5p and HMGA1 in T24 and UM-UC-3 cells. (D) The change of cell migration via Transwell migration experiment post-overexpression of miR-15a-5p and HMGA1 in T24 and UM-UC-3 cells; magnification, ×200. (E) The change of cell invasion via Transwell invasion experiment post overexpression of miR-15a-5p and HMGA1 in T24 and UM-UC-3 cells; magnification, 200. **P<0.01; ***P<0.001; #P<0.05; ##P<0.01.
LINC00649 promotes the proliferation, migration, and invasion of BC cells by elevating HMGA1 expression
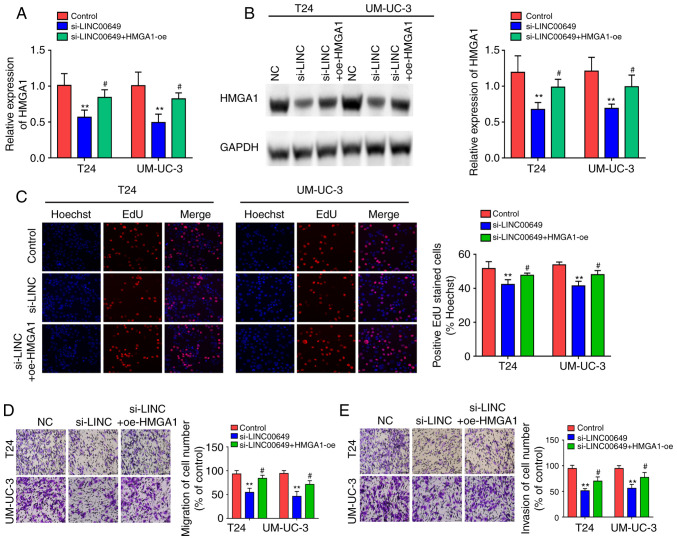
After co-transfection of LINC00649 siRNA and HMGA1 OE in both T24 and UM-UC-3 cells, RT-qPCR and western blotting were employed to examine HMGA1 expression and indicated that LINC00649 siRNA repressed HMGA1 expression, whereas HMGA1 overexpression showed a partial reversal effect on the inhibitory effect of si-LINC00649 (P<0.05) (Fig. 7A and B). Subsequently, the effects of LINC00649 and HMGA1 on cell proliferation, migration, and invasion were assessed in vitro. Results indicated that LINC00649 downregulation in T24 and UM-UC-3 cells markedly weakened cell proliferation, whereas HMGA1 overexpression reversed this inhibitory effect (P<0.05) (Fig. 7C). As such, LINC00649 downregulation in T24 and UM-UC-3 cells repressed cell migration and invasion, which was reversed by HMGA1 expression (P<0.05) (Fig. 7D and E). These results suggest that LINC00649 could promote the proliferation, migration, and invasion of BC cells by elevating HMGA1 expression.
Figure 7.
LINC00649 promoted the proliferation, migration and invasion of BC cells by elevating HMGA1 expression. (A and B) HMGA1 expression detection by RT-qPCR and western blot analysis after co-transfection of LINC00649 siRNA and HMGA1 OE in T24 and UM-UC-3 cells. (C) Cell proliferation detection by EdU experiment after co-transfection of LINC00649 siRNA and HMGA1 OE in T24 and UM-UC-3 cells; magnification, ×200. (D and E) The change of cell migration and invasion via Transwell experiment after co-transfection of LINC00649 siRNA and HMGA1 OE in T24 and UM-UC-3 cells; magnification, ×200. **P<0.01; #P<0.05.
Knockdown of LINC00649 inhibited the EMT of BC cells
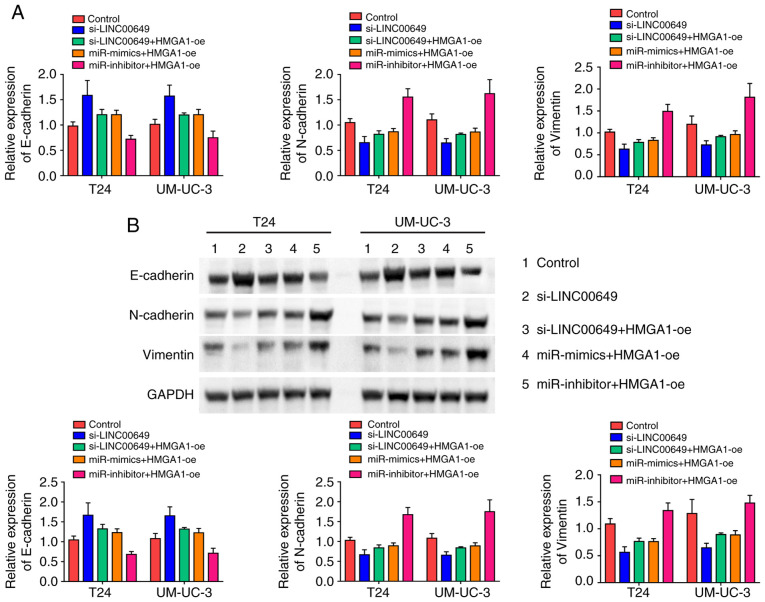
We determined whether the LINC00649/miR-15a-5p/HMGA1 regulatory axis regulated EMT in BC cells. The expressions of EMT markers were determined using RT-qPCR and western blot analysis. Knockdown of LINC00649 increased E-cadherin expression and decreased expression of N-cadherin and vimentin in BC cells, while the overexpression of HMGA1 promoted EMT. In addition, miR-15a-5p mimics increased E-cadherin expression and decreased N-cadherin and vimentin expressions in BC cells, while miR-15a-5p inhibitor showed an opposite effect (P<0.05) (Fig. 8A and B). These results indicated that the LINC00649/miR-15a-5p/HMGA1 regulatory axis promoted the EMT of BC cells.
Figure 8.
Knockdown of LINC00649 inhibited EMT of BC cells. (A and B) The expression of EMT markers were determined using RT-qPCR and Western blotting. Knockdown of LINC00649 increased the expression of E-cadherin and decreased the expression of N-cadherin and vimentin in BC cells, while miR-15a-5p could suppress EMT and HMGA1 showed the opposite effect.
Discussion
BC is the most prevalent malignant tumor in the urinary system, and its incidence is increasing worldwide (15). The occurrence and development of BC is a complex process involving multiple genes that are diversely regulated and may be associated with uncontrolled proto-oncogene activation, inactivation or inhibition of cancer suppressor genes, damage of self-repairing systems of DNA, extracellular matrix formation, and living habits of individual patients; however, specific mechanisms of action involved remain unclear (16). BC is currently mainly treated surgically. BC, especially MIBC, is characterized by a high recurrence rate, albeit with continuous improvement of surgical methods and techniques combined with comprehensive treatments such as radiotherapy, chemotherapy, and targeted drugs (17). Therefore, the exploration of molecular markers and new effective therapeutic targets for the early diagnosis of BC has become a hot spot in malignant bladder tumor research.
It has been suggested that lncRNAs may affect the occurrence and progression of multiple diseases, including malignant tumors, through the regulation of cell cycle, immune surveillance, cell differentiation, cell apoptosis, and other links (18–20). Although studies of BC-related lncRNAs started late, their progress has been rapid. lncRNA SNHG16 enhances the malignant development of BC by regulating the miR-98/STAT3/Wnt/β-catenin pathway axis (21), and lncRNA GAS5 contributes to the apoptosis of BC cells by inhibiting EZH2 transcription (13). However, further research on BC-related lncRNAs is required. Further in-depth studies on the search for highly sensitive and highly specific tumor markers, the influence of lncRNA on biological behaviors of BC cells, as well as specific mechanisms of action should also be considered.
Abnormally highly expressed lncRNA-LINC00649 in BC tissues was found through database analysis. LINC00649 was observed to be significantly highly expressed in BC tissues via RT-qPCR in 60 BC tissues and adjacent tissues. Results revealed that high expression of LINC00649 predicted poor prognosis of BC patients. Notably, we identified that the opposite also occurred between TCGA database and our results. Therefore, the study of more samples is required to verify the association between LINC00649 expression and BC patient OS. The in vitro cell experiments revealed that LINC00649 contributed to the proliferation, migration, and invasion of BC cells, indicating that it may act as a tumor promoter gene involved in the malignant progression of BC.
lncRNAs are characterized by sequence conservation and functional conservation, specific spatial secondary structure, strong tissue and cell expression specificity, and relative certainty of chromosomal localization, suggesting the vital biological functions of these genes that are widely expressed in human tissues and cells without protein-coding ability (22). Recent findings have shown that lncRNAs participate in and constitute important and complex gene expression regulatory networks in humans and may regulate gene expression via interaction with protein or DNA specificity. lncRNAs may regulate gene expression at multiple levels, such as transcriptional, post-transcriptional, and epigenetic regulation (23).
During a nucleoplasm isolation experiment, LINC00649 was mainly located in the BC cytoplasm, suggesting that it exerted crucial regulatory roles at the post-transcriptional level. The LINC00473 sequence was analyzed and predicted on the website (http://starbase.sysu.edu.cn/). We observed that LINC00473 had multiple miRNA binding sites and may act on multiple miRNAs, suggesting that it plays a role by binding to miRNA. miR-15a-5p is a potential target miRNA with a higher score and better correlation compared to other miRNAs. Further analysis showed that LINC00649 binds to miR-15a-5p and inhibits its expression, miR-15a-5p expression was notably downregulated in BC tissues, and LINC00649 expression was negatively correlated with miR-15a-5p expression. This indicated that LINC00649 promoted cancer by binding to miR-15a-5p.
Prediction using Bioinformatics website identified HMGA1 as a possible target of miR-15a-5p. HMGA1 has been identified in the human cervical cancer cell line HeLa with highly malignant proliferation. It is highly expressed in multiple bio-embryogeneses, but barely or only weakly expressed in most mature tissues. Additionally, it participates in many basic cellular processes, including cell cycle regulation, embryogenesis, tumor transformation, cell differentiation, apoptosis, metabolism, and DNA repair. Evidence indicates that HMGA1 is highly expressed in multiple human cancers such as cervical (24), breast (25), colorectal (26), and thyroid (27) cancer. The present study found that miR-15a-5p targeted HMGA1, and LINC00649 contributed to HMGA1 expression by binding to miR-15a-5p. Overexpression of miR-15a-5p repressed the proliferation, migration, and invasion of BC cells, which was partially reversed by simultaneous HMGA1 overexpression.
In conclusion, findings of the present showed that LINC00649 is abnormally highly expressed in BC, and that LINC00649 contributes to HMGA1 expression by binding to miR-15a-5p to enhance the proliferation, migration, and invasion of BC, thus enhancing its malignant progression. However, there were some limitations to the study. First, the role of LINC00649 in the regulation of HMGA1 expression was not directly assessed. Second, although it was demonstrated that LINC00649 is involved in the progression of BC by binding to miR-15a-5p to regulate HMGA1 expression, LINC00649 had multiple binding sites for miRNA. Thus, further studies are needed to determine whether LINC00649 participates in disease progression by binding to multiple miRNAs to regulate the expression of multiple tumor promoter genes. Finally, the biological effects and molecular mechanisms of LINC00649 were not verified in vivo.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data included in this study are available upon request by contact with the corresponding author.
Authors' contributions
XC and SC designed this study. XC conducted the experiments, and SC served as scientific advisor. XC and SC participated in writing or technical editing of the manuscript. Both authors have read and agreed to the published version of the manuscript.
Ethics approval and consent to participate
Approval for the study was obtained from the ethics committee of The First People's Hospital of Wenling (Zhejiang, China). The patients volunteered to participate in this study and signed a written informed consent form. All experiments were performed in accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Afonso J, Santos LL, Longatto-Filho A, Baltazar F. Competitive glucose metabolism as a target to boost bladder cancer immunotherapy. Nat Rev Urol. 2020;17:77–106. doi: 10.1038/s41585-019-0263-6. [DOI] [PubMed] [Google Scholar]
- 2.Alifrangis C, McGovern U, Freeman A, Powles T, Linch M. Molecular and histopathology directed therapy for advanced bladder cancer. Nat Rev Urol. 2019;16:465–483. doi: 10.1038/s41585-019-0208-0. [DOI] [PubMed] [Google Scholar]
- 3.Meng MV, Gschwend JE, Shore N, Grossfeld GD, Mostafid H, Black PC. Emerging immunotherapy options for bacillus calmette-guerin unresponsive nonmuscle invasive bladder cancer. J Urol. 2019;202:1111–1119. doi: 10.1097/JU.0000000000000297. [DOI] [PubMed] [Google Scholar]
- 4.Soria F, D'Andrea D, Pohar K, Shariat SF, Lotan Y. Diagnostic, prognostic and surveillance urinary markers in nonmuscle invasive bladder cancer: Any role in clinical practice? Curr Opin Urol. 2018;28:577–583. doi: 10.1097/MOU.0000000000000545. [DOI] [PubMed] [Google Scholar]
- 5.Dobruch J, Daneshmand S, Fisch M, Lotan Y, Noon AP, Resnick MJ, Shariat SF, Zlotta AR, Boorjian SA. Gender and bladder cancer: A collaborative review of etiology, biology, and outcomes. Eur Urol. 2016;69:300–310. doi: 10.1016/j.eururo.2015.08.037. [DOI] [PubMed] [Google Scholar]
- 6.Guo X, Hua Y. CCAT1: An oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol. 2017;143:555–562. doi: 10.1007/s00432-016-2268-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–1714. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Sallam T, Sandhu J, Tontonoz P. Long noncoding RNA discovery in cardiovascular disease: Decoding form to function. Circ Res. 2018;122:155–166. doi: 10.1161/CIRCRESAHA.117.311802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Chen F, Zhao M, Yang Z, Li J, Zhang S, Zhang W, Ye L, Zhang X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J Biol Chem. 2017;292:15395–15407. doi: 10.1074/jbc.M117.783738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, Tian YF, Peng CH, Shen BY, Chen YL, Wang YD. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019;446:49–61. doi: 10.1016/j.canlet.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32:3957–3967. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, Guo C, Wang L, Luo G, Huang C, Li Y, Liu D, Zeng F, Jiang G, Xiao X. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018;9:238. doi: 10.1038/s41419-018-0264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Xie D, Zhang H. Long noncoding RNA neuroblastoma-associated transcript 1 gene inhibits malignant cellular phenotypes of bladder cancer through miR-21/SOCS6 axis. Cell Death Dis. 2018;9:1042. doi: 10.1038/s41419-018-1090-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Soukup V, Capoun O, Cohen D, Hernandez V, Babjuk M, Burger M, Compérat E, Gontero P, Lam T, MacLennan S, et al. Prognostic Performance and Reproducibility of the 1973 and 2004/2016 World Health organization grading classification systems in non-muscle-invasive bladder cancer: A european association of urology non-muscle invasive bladder cancer guidelines panel systematic review. Eur Urol. 2017;72:801–813. doi: 10.1016/j.eururo.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: A global overview and recent trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Lobo N, Mount C, Omar K, Nair R, Thurairaja R, Khan MS. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol. 2017;14:565–574. doi: 10.1038/nrurol.2017.82. [DOI] [PubMed] [Google Scholar]
- 18.Lin C, Yang L. Long noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 2018;28:287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M, Li J, Zhuang C, Cai Z. Increased lncRNA ABHD11-AS1 represses the malignant phenotypes of bladder cancer. Oncotarget. 2017;8:28176–28186. doi: 10.18632/oncotarget.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Zhuang C, Liu Y, Li J, Dai F, Xia M, Zhan Y, Lin J, Chen Z, He A, et al. Tetracycline-inducible shRNA targeting antisense long non-coding RNA HIF1A-AS2 represses the malignant phenotypes of bladder cancer. Cancer Lett. 2016;376:155–164. doi: 10.1016/j.canlet.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 21.Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/β-catenin pathway axis. J Cell Biochem. 2018;119:9408–9418. doi: 10.1002/jcb.27257. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jandura A, Krause HM. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. 2017;33:665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Fu F, Wang T, Wu Z, Feng Y, Wang W, Zhou S, Ma X, Wang S. HMGA1 exacerbates tumor growth through regulating the cell cycle and accelerates migration/invasion via targeting miR-221/222 in cervical cancer. Cell Death Dis. 2018;9:594. doi: 10.1038/s41419-018-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanin R, Pegoraro S, Ros G, Ciani Y, Piazza S, Bossi F, Bulla R, Zennaro C, Tonon F, Lazarevic D, et al. HMGA1 promotes breast cancer angiogenesis supporting the stability, nuclear localization and transcriptional activity of FOXM1. J Exp Clin Cancer Res. 2019;38:313. doi: 10.1186/s13046-019-1307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei F, Zhang T, Deng SC, Wei JC, Yang P, Wang Q, Chen ZP, Li WL, Chen HC, Hu H, Cao J. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 2019;450:1–13. doi: 10.1016/j.canlet.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Zhong J, Liu C, Zhang QH, Chen L, Shen YY, Chen YJ, Zeng X, Zu XY, Cao RX. TGF-β1 induces HMGA1 expression: The role of HMGA1 in thyroid cancer proliferation and invasion. Int J Oncol. 2017;50:1567–1578. doi: 10.3892/ijo.2017.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in this study are available upon request by contact with the corresponding author.