Abstract
Over the past decade, long-read, single-molecule DNA sequencing technologies have emerged as powerful players in genomics. With the ability to generate reads tens to thousands of kilobases in length with an accuracy approaching that of short-read sequencing technologies, these platforms have proven their ability to resolve some of the most challenging regions of the human genome, detect previously inaccessible structural variants, and generate some of the first telomere-to-telomere assemblies of whole chromosomes. Long-read sequencing technologies will soon permit the routine assembly of diploid genomes, which will revolutionize genomics by revealing the full spectrum of human genetic variation, resolving some of the missing heritability, and leading to the discovery of novel mechanisms of disease.
INTRODUCTION
Studies of genetic variation and the discovery of the mutations underlying human disease are dependent on technological advances in molecular biology and conceptual advances in their application. Among such innovations, changes in sequencing platforms have often been regarded as revolutionary1. The DNA sequencing technology that has dominated genomics research for the past decade has undoubtedly been Illumina, a short-read, next-generation sequencing [G] platform that leverages a sequence-by-synthesis [G] approach to determine the order of nucleotides in a DNA strand (Figure 1a)2. Illumina’s DNA sequencing technology produces highly accurate (>99.9%) sequencing reads, which are inexpensive to generate on a massive scale (Table 1). These advantages have driven its ascent to become the current gold standard of clinical and research sequencing. Illumina next-generation sequencing has led to innumerable scientific discoveries over the past decade that have enhanced our understanding of evolution, adaptation, and disease through the discovery of pathogenic variants, including single-nucleotide variants [G], copy number variants [G], and insertions/deletions (indels [G])3–8. Importantly, the technology’s throughput has allowed it to serve as an assay for digital read-outs to investigate a myriad of biological phenomena, including chromatin accessibility, transcription factor occupancy, gene expression, and RNA binding among many other novel applications2.
Figure 1. Overview of short-read sequencing technologies.
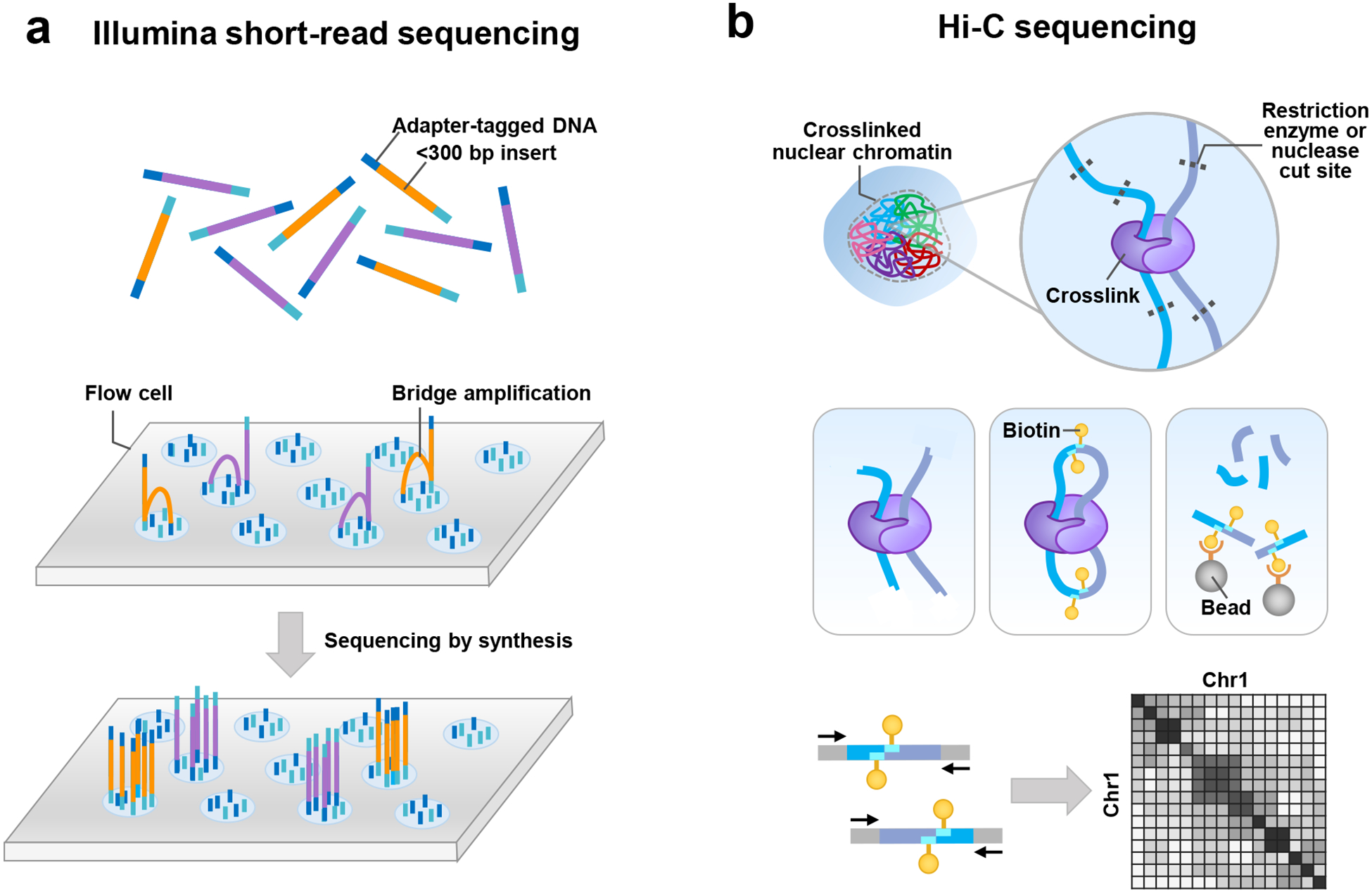
a) In short-read sequencing by Illumina, DNA fragments (yellow and purple) are ligated to adapters (blue and aqua) that contain unique molecular identifiers as well as sequences complementary to the oligonucleotides that are attached to the surface of a flow cell. The modified DNA is loaded onto a flow cell, and the adapters from the modified DNA hybridize to the oligonucleotides that coat the surface of the flow cell. Once the fragments have attached, cluster generation begins, where thousands of copies of each fragment are generated through a process known as bridge amplification. In this process, the strand folds over, and the adapter on the end of the molecule hybridizes to another oligonucleotide in the flow cell. A polymerase incorporates nucleotides to build double-stranded bridges of the DNA molecules, which are subsequently denatured to leave single-stranded DNA fragments tethered to the flow cell. This process is repeated over and over, generating several million dense clusters of double-stranded DNA. After bridge amplification, the reverse DNA strands are cleaved and washed away, leaving only the forward strands. Then, sequencing by synthesis begins, in which fluorescently labeled deoxyribonucleotide triphosphates (dNTPs) are incorporated into the newly synthesized DNA strand at each cycle. After incorporation, a laser excites the fluorophore on the strand, which emits a characteristic fluorescence emission signal that corresponds to the base. b) In Hi-C sequencing, nuclear chromatin is crosslinked with formaldehyde, which covalently bonds protein-DNA complexes in close proximity to each other. Crosslinked chromatin is digested with a restriction enzyme or nuclease, and single-stranded DNA overhangs are filled in and repaired with biotin-linked nucleotides before religating the DNA. Chemical crosslinks are reversed, proteins degraded, and the purified DNA is nonspecifically sheared (for example, by sonication). Biotin-labeled DNA is pulled down with streptavidin-conjugated beads and paired-end sequenced to reveal the junctions between two DNA loci (light and dark blue). Because the contact frequency between pairs of loci strongly correlates with distance, the majority of sequenced junctions encompass two loci from the same chromosome. As a result, Hi-C data can be used to provide linkage information between pairs of loci tens of megabases apart on a single chromosome (as shown in the contact map).
Table 1.
Data type, length, accuracy, throughput, and cost across long- and short-read technologies and platforms
| Sequencing technology | Platform | Data type | Read length (kb) | Read accuracy | Throughput per flow cell (Gb) | Estimated cost per Gb | Max throughput per year (Gb)* | ||
|---|---|---|---|---|---|---|---|---|---|
| N50 | Max | Mean | Max | ||||||
| PacBio | RS II† | CLR | 5–15 | >60 | 87–92% | 0.75–1.5 | 2 | $333–933a | 4,380 |
| Sequel | CLR | 25–50 | >100 | 5–10 | 20 | $98–195b | 17,520 | ||
| Sequel II | CLR | 30–60 | >200 | 50–100 | 160 | $13–26c | 93,440 | ||
| HiFi | 10–20 | >20 | >99% | 15–30 | 35 | $43–86c | 10,220 | ||
| ONT | MinION / GridION X5 | Long | 10–60 | >1000 | 87–98% | 2–20 | 30 | $50–500d | 21,900 (MinION) 109,500 (GridION X5) |
| Ultra-long | 100–200 | >1500 | 0.5–2 | 2.5 | $500–2000d | 913 (MinION) 4,563 (GridION X5) | |||
| PromethION | Long | 10–60 | >1000 | 50–100 | 180 | $21–42d | 3,153,600 | ||
| Illumina | NextSeq 550 | Single-end | 0.075–0.15 | 0.15 | >99.9% | 16–30 | >30 | $50–63e | >47,782 |
| Paired-end | 0.075–0.15 (x2) | 0.15 (x2) | 32–120 | >120 | $40–60e | >70,080 | |||
| NovaSeq 6000 | Single-end | 0.05–0.25 | 0.25 | 65–3000 | >3000 | $10–35f | >1,194,545 | ||
| Paired-end | 0.05–0.25 (x2) | 0.25 (x2) | |||||||
Assuming continuous, full-capacity sequencing on each instrument.
PacBio RS II support will end by 2021.
Current cost when sequencing with a SMRTbell Template Prep Kit 1.0 and SMRT Cell.
Current cost when sequencing with a SMRTbell Express Template Prep Kit 2.0 and SMRT Cell 1M.
Current cost when sequencing with a SMRTbell Express Template Prep Kit 2.0 and SMRT Cell 8M.
Current cost when sequencing with a Ligation or Rapid Sequencing Kit and an ONT R9.4.1 or R10.3 flow cell.
Current cost when sequencing with the NextSeq 500/550 Mid or High Output Kits v2.5.
Current cost when sequencing with the NovaSeq 6000 SP, S1, S2, or S4 Reagent Kits.
All estimates are listed in USD and exclude the cost for labor, instrumentation, maintenance, and computer resources.
However, application of short-read technologies to structural variant [G] detection and genome assembly more broadly has revealed a major shortcoming: limited read length. Reads <300 bases long, such as those typically produced by Illumina, are too short to detect >70% of human genome structural variation (>50 bp), with intermediate-size structural variation (<2 kb) especially underrepresented9. Moreover, entire swaths of our genome (>15%) remain inaccessible to assembly or variant discovery because of their repeat content or atypical GC content10. For example, even PCR-free, short-read genomic libraries show up to twofold reductions in sequence coverage when the GC composition exceeds 45%, limiting the ability to discover genetic variation in some of the most functionally important regions of our genome. These inaccessible parts of the genome include centromeres, telomeres, and acrocentric genomic regions, where massive arrays of tandem repeats predominate, as well as the 5% of our genome (and associated genes) mapping to large segmental duplications [G]11. Ironically, these regions also experience some of the highest mutation rates, both in the germline and soma3,12–14. As a result, some of the most mutable regions of our genome are typically understudied. These limitations have necessitated the development of methods that can resolve these more complex and dynamic regions of the genome.
One solution has been to develop short-read sequencing approaches that reconstruct the sequence of long DNA molecules. The key concept here is to use barcodes [G] to link short reads that originate from the same partitioned large molecule and to assemble the larger fragment into a DNA sequence. Linked-read [G]15–17, synthetic long-read18,19, and Hi-C20 technologies are all cost-effective methods that provide long-range information about the location of reads using only Illumina short reads. For example, Hi-C technology uses a proximity ligation approach to generate a genome-wide library from loci that were originally in close proximity to each other in the nucleus, with the majority of loci residing on the same chromosome (Figure 1b). Hi-C sequencing data can be used to provide long-range information between pairs of loci tens of megabases apart on the same chromosome, which has been shown to link contigs [G] in broken genome assemblies21, phase haplotypes22, and lead to the discovery of structural variation23. Although Hi-C outperforms simple short-read sequencing approaches for structural variant detection, the fundamental unit of assembly is still a short read, which greatly limits the ability to both detect and fully assemble structural variant regions, especially in larger repeats. For these applications, linked-read, synthetic long-read, and Hi-C approaches are generally inferior to strict long-read sequencing [G] approaches9.
In this Review, we focus on the two major long-read sequencing technologies, Pacific Biosciences (also known as single-molecule, real-time (SMRT) sequencing [G], or PacBio sequencing) and Oxford Nanopore Technologies (ONT). We compare them in terms of read accuracy, throughput, and cost to short-read technologies, such as Illumina. Additionally, we discuss the practical applications of these technologies in genomics, transcriptomics, and epigenomics and how they are enabling new biological insights. This Review does not provide a detailed assessment of the various software and algorithms related to genome assembly, which is an area of rapid development that has been discussed extensively elsewhere24–27. Instead, we focus on future directions, with a specific emphasis on studies of human disease and diversity, while recognizing that these technologies have had a huge impact more broadly across diverse species and phyla.
Long-read sequencing technologies
In contrast to short-read approaches, long-read technologies can generate long continuous sequences (ranging from 10 kb to >1 Mb in length) directly from native DNA, which, along with recent developments in throughput and accuracy, has substantially increased their utility and application28,29 (Figure 2). Both PacBio and ONT sequencing technologies produce reads that can readily traverse the most repetitive regions of the human genome, but underlying differences in their chemistry and sequence detection approaches influence their read lengths, base accuracies, and throughput.
Figure 2. Overview of long-read sequencing technologies.
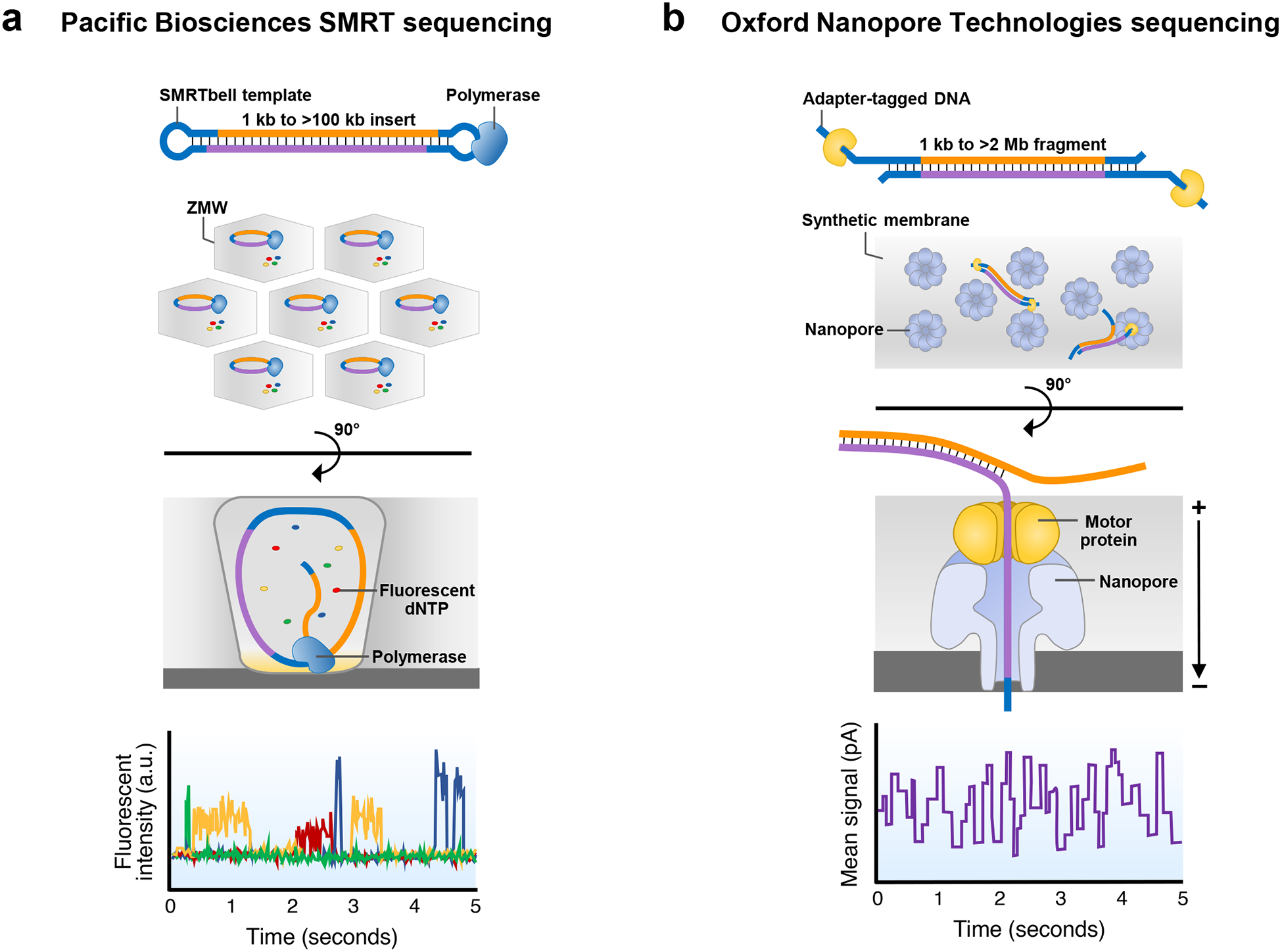
a) In single-molecule, real-time (SMRT) sequencing by Pacific Biosciences (PacBio), DNA (yellow for forward strand, dark blue for reverse strand) is fragmented and ligated to hairpin adapters (light blue) to form a topologically circular molecule known as a SMRTbell. Once the SMRTbell is generated, it is bound by a DNA polymerase and loaded onto a SMRT Cell for sequencing. Each SMRT Cell can contain up to eight million zero-mode waveguides (ZMWs), which are chambers that hold picoliter volumes. A light penetrates the lower 20–30 nm of each well, reducing the detection volume of the well to only 20 zeptoliters (10−21 liters). As the DNA mixture floods the ZMWs, the SMRTbell template and polymerase become immobilized on the bottom of the chamber. Fluorescently labeled dNTPs are added to begin the sequencing reaction. As the polymerase begins to synthesize the new strand of DNA, a fluorescent dNTP is briefly held in the detection volume, and a light pulse from the bottom of the well excites the fluorophore. Unincorporated dNTPs are not typically excited by this light but, in rare cases, can become excited if they diffuse into the excitation volume, thereby contributing to noise and error in PacBio sequencing. The light emitted from the excited fluorophore is detected by a camera, which records the wavelength and relative position of the incorporated base in the nascent strand. The phosphate-linked fluorophore is then cleaved from the nucleotide as part of the natural incorporation of the base into the new strand of DNA and released into the buffer, preventing fluorescent interference during the subsequent light pulse. The DNA sequence is determined by the changing fluorescence emissions that are recorded within each ZMW, with a different color corresponding to each DNA base (for example, green, T; yellow, C; red, G; blue, A). b) In Oxford Nanopore Technologies (ONT) sequencing, arbitrarily long DNA (orange for forward strand, purple for reverse strand) are tagged with sequencing adapters (light blue) preloaded with a motor protein on one or both ends. The DNA is combined with tethering proteins and loaded onto the flow cell for sequencing. The flow cell contains thousands of protein nanopores embedded in a synthetic membrane, and the tethering proteins bring the DNA molecules toward these nanopores. Then, the sequencing adapter inserts into the opening of the nanopore, and the motor protein begins to unwind the double-stranded DNA. An electric current is applied, which, in concert with the motor protein, drives the negatively charged DNA through the pore at a rate of about 450 bases per second. As the DNA moves through the pore, it causes characteristic disruptions to the current, known as a ‘squiggle’ [G]. Changes in current within the pore correspond to a particular k-mer (i.e., a string of DNA bases of length k) which is used to identify the DNA sequence.
Pacific Biosciences.
PacBio SMRT sequencing technology (Figure 2a) utilizes a topologically circular DNA molecule template, known as a SMRTbell [G], comprised of a double-stranded DNA insert with single-stranded hairpin adapters on either end. The DNA insert can range in length from 1 kb to over 100 kb, which allows long sequencing reads to be generated. Once the SMRTbell is assembled, it is bound by a DNA polymerase and loaded onto a SMRT Cell [G], which contains up to several million zero-mode waveguides (ZMWs) [G], for sequencing. During the sequencing reaction, the polymerase processes around the SMRTbell template and incorporates fluorescently labeled dNTPs into the nascent strand. After each incorporation, a laser excites the fluorophore and a camera records the emission. Then, the fluorophore is cleaved from the nucleotide before the next dNTP is incorporated. This process is repeated thousands of times to reveal the identity and sequence of each base in the SMRTbell template. PacBio technology typically generates reads tens of kilobases long, which greatly exceeds the <300 base read length obtained with Illumina sequencing30–33.
Oxford Nanopore Technologies.
ONT long-read sequencing technology (Figure 2b) employs linear DNA molecules rather than circular ones. These linear DNA molecules are typically one to several hundred kilobases in length but can be several megabases long34–37. ONT sequencing begins by first attaching a linear DNA molecule to a sequencing adapter, which is preloaded with a motor protein. The DNA mixture is loaded onto a flow cell [G], which contains hundreds to thousands of nanopores embedded in a synthetic membrane. The motor protein unwinds the double-stranded DNA and, together, with an electric current, drives the negatively charged DNA through the pore at a controlled rate. As the DNA translocates through the pore, it causes characteristic disruptions to the current, which are analyzed in real time to determine the sequence of the bases in the DNA strand. With ONT sequencing, reads over 1 Mb in length have been generated34, with the longest reported read close to 2.3 Mb in length when computationally stitched together from shorter reads37. Together, these achievements have pushed the genomics community into the realm of megabase-sized sequence reads for the first time.
Long-read sequencing data types
Based on new developments in sequencing chemistry and differences in DNA preparation, each of the long-read sequencing technologies can now produce different types of long reads that vary both in their length and their accuracy (Table 1). These diverse data types are, consequently, beginning to be used for specific applications. While long-read base accuracies have been reviewed elsewhere38–40, below we provide a limited meta-analysis of recently generated long-read datasets to illustrate the relative lengths and base accuracies of each of these data types (Figure 3, Supplementary Information).
Figure 3. PacBio and ONT long-read data types.
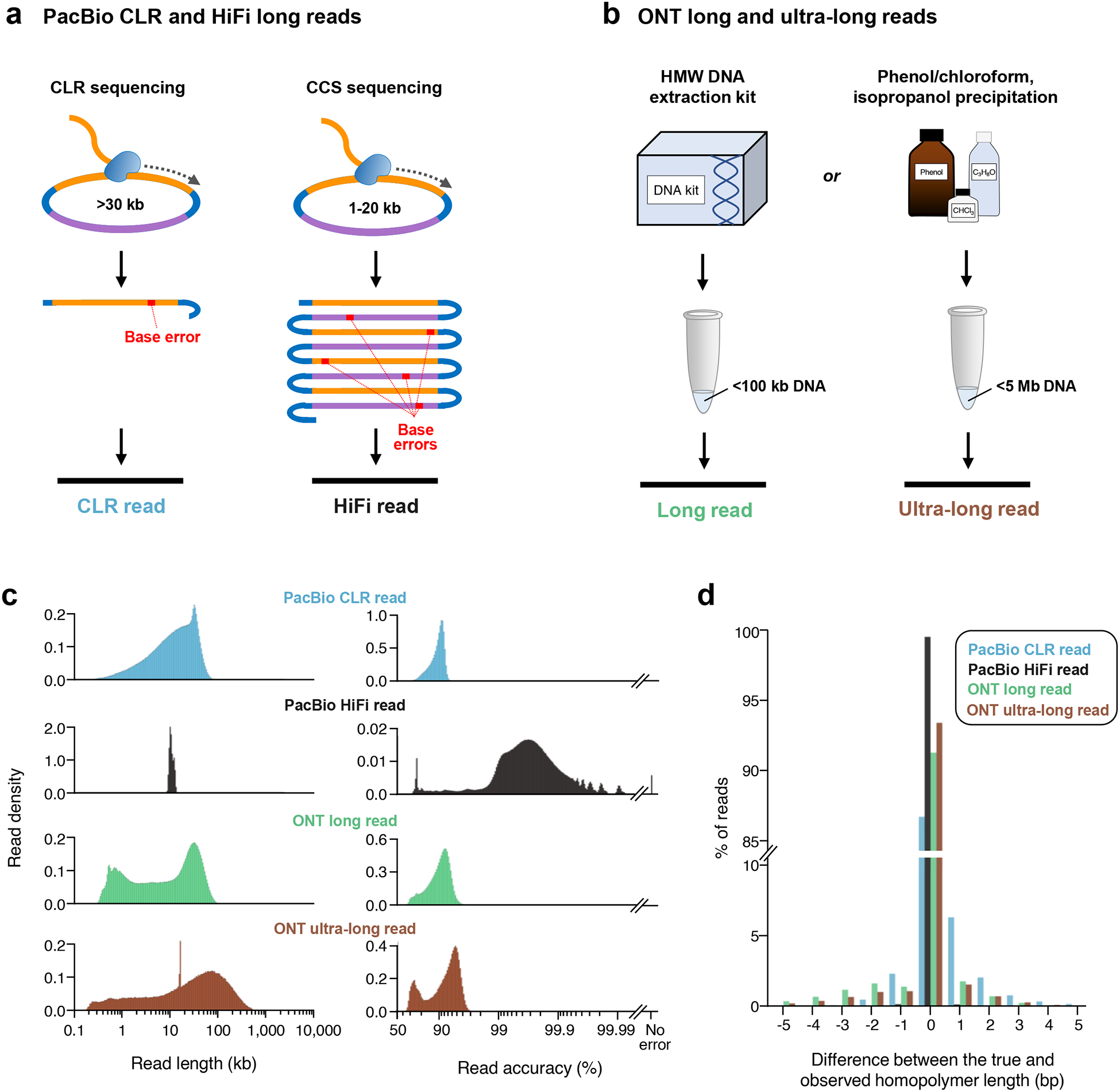
a) The PacBio platform can generate continuous long reads (CLR) or high-fidelity (HiFi) reads. CLR data is generated by sequencing a SMRTbell template containing a >30 kb DNA insert (yellow for forward strand, dark blue for reverse strand). Because of the large insert size, the polymerase often only completes a single pass through one strand of the template. A base is incorrectly called in about 1 out of every 10 bases, resulting in an error rate of 8–15% in the CLR. HiFi reads are generated by circular consensus sequencing (CCS) of a SMRTbell template containing a 10–30 kb DNA insert. The smaller insert size allows the polymerase to make several passes around the SMRTbell template. A consensus sequence is produced from the subreads, resulting in an error rate of ≤1% in the HiFi read. b) The ONT platform can generate long or ultra-long reads. To generate long and ultra-long ONT reads, high-molecular-weight (HMW) DNA is first extracted from cells or tissue. This extraction is commonly performed using either a commercially available DNA extraction kit, such as Qiagen’s Puregene kit or Genomic-tip 500/G kit, or via traditional methods, such as a phenol-chloroform extraction followed by either an ethanol or isopropanol precipitation. Kit-extracted DNA most often generates long (10–100 kb) reads, whereas high-molecular-weight DNA extracted by phenol-chloroform generates ultra-long (>100 kb) reads. c) Read length distributions and base accuracies of PacBio and ONT long-read data types differ. Shown are plots of the read length and accuracy distributions for: PacBio HG002 CLR data generated on the Sequel II platform; PacBio CHM13 HiFi data generated on the Sequel II platform; ONT CHM13 long-read data generated on the PromethION; and ONT ultra-long reads generated on the MinION and GridION. Read accuracy was estimated by aligning raw reads from each data type to GRCh38 and counting alignment differences as errors in the reads. Links to the publicly available datasets, a description of the methods used, and the code required to reproduce the analysis are provided in a Supplementary Note. A similar analysis was also performed in which raw reads were aligned to the T2T CHM13 assembly34, and differences in alignment between the reads and the highly curated ChrX were counted to estimate read accuracy. PacBio HiFi reads have a visibly higher read accuracy distribution when aligned to the CHM13 T2T assembly than GRCh38 because the high accuracy of the HiFi reads (>99%) is sufficient to detect differences between the two genome assemblies, which are interpreted as base errors. The other long-read data types are not accurate enough to detect differences between the two genome assemblies. Consequently, the accuracy distribution for these other data types are similar (Supplementary Figure 1a and Supplementary Note). d) Homopolymer accuracy differs between PacBio and ONT long-read data types. Shown is a plot of the homopolymer accuracy for the PacBio CLR, PacBio HiFi, ONT long, and ONT ultra-long datasets used in panel c. Homopolymer error was estimated by aligning raw reads from each data type to GRCh38 and comparing the observed homopolymer length in the reads to the homopolymer length. A similar analysis was performed where raw reads were aligned to the T2T CHM13 assembly34, and homopolymer error was estimated based on the comparison between the observed homopolymer length in the reads and the true homopolymer length in the highly curated ChrX assembly. In both cases, homopolymers ≥5 bases were assessed for accuracy (Supplementary Figure 1b and Supplementary Note).
PacBio continuous long reads.
Continuous long reads (CLR) are currently the most common PacBio data type. CLR are generated by first constructing standard SMRTbell template libraries with DNA inserts >30 kb in length (Figure 3a). Because of the large insert size in these molecules, the polymerase makes only one or a few passes around the template. In our meta-analysis, CLR subreads [G] ranged from 5–60+ kb in length with an estimated single-pass [G] subread accuracy of 85–92%. Homopolymer [G] runs of nucleotides were more error-prone than other sequence contexts, with only ~85% of homopolymers ≥5 bases long accurately identified (Figure 3c and 3d, Supplementary Figure 1b, Supplementary Note), consistent with those reported elsewhere31,41–44. Although the single-pass accuracy of CLR is low compared to Illumina short-read accuracy (which is >99.9%45), the error mode is remarkably stochastic in nature. As a result, errors can be corrected with polishing tools [G], such as Quiver46 and Arrow, which leverage CLR alignments, along with their underlying raw pulse information, to infer the true sequence of the regions based on sequence consensus with >99.9% accuracy at coverages exceeding 40-fold, which is easily obtained in a typical experiment46. Additional steps are typically employed to increase the accuracy and minimize residual indels, such as error-correction with Illumina data generated from the same individual (for example, with Pilon47, Racon48, Freebayes49,50, and NextPolish51); however, error-correction with short-read data is limited in repetitive regions (owing to ambiguous mappings) and regions with extreme GC content (owing to reduced coverage arising from biases in short-read sequencing). CLR data can be generated with the RS II, Sequel, and Sequel II systems. Whereas the RS II and Sequel platforms only generate up to 2 Gb and 20 Gb of data per flow cell, respectively, the more recent Sequel II system with 8 million (8M) ZMWs is capable of generating up to 160 Gb per flow cell in CLR mode (Table 1). Thus, it is now possible to obtain >40X sequencing coverage [G] of a human genome with only one or two Sequel II flow cells, resulting in >99.9% consensus sequence accuracy. Although still more expensive than Illumina, it is now feasible to contemplate population-scale sequencing of a few hundred samples and family-based sequencing for variant discovery and genome assembly based on Sequel II throughput cost reductions (Table 1)9,52.
PacBio high-fidelity reads.
High-fidelity (HiFi) sequence reads represent the most recent data type to be developed by PacBio. They are the first data type that is both long (>10 kb) and highly accurate (>99%). Here, smaller DNA inserts, 10–30 kb in length, are assembled into SMRTbell templates and subjected to sequencing via circular consensus sequencing (CCS) [G] (Figure 3a). Because of the relatively small size of the DNA insert, the polymerase is able to make several passes through the SMRTbell template, resulting in extremely long polymerase reads [G] (read N50 [G] > 150 kb) that each contain several subreads (both forward and reverse complement of the template). Owing to the improved efficiency of the DNA polymerase during CCS, the subread throughput of the HiFi protocol is improved over CLR (>200 Gb versus 100 Gb) but requires significantly longer movie times (30 hours) to generate datasets because accuracy is dependent on more passes. Subreads from a single polymerase read are then computationally combined via the CCS algorithm to create a HiFi consensus read, resulting in a total yield of 15–25 Gb of HiFi from a single SMRT Cell 8M. Thus, approximately three 8M SMRT Cells must be used to generate the 25-fold sequencing coverage of a human genome considered sufficient for de novo assembly52,53, equating to approximately two to three times the cost of CLR data (Table 1). It should be noted that each SMRT Cell 8M is run sequentially on the Sequel II and, therefore, takes several days to generate 25-fold sequencing coverage. Additionally, the process of converting subreads into HiFi reads via the CCS algorithm carries a significant computational investment and can require >10,000 CPU hours per SMRT Cell 8M of data52,53. However, recent improvements in the CCS algorithm have reduced this time to <2,000 CPU hours per SMRT Cell 8M of data (see https://github.com/PacificBiosciences/ccs#does-speed-impact-quality-and-yield). Typically, the CCS algorithm requires 3–4 subreads from the same molecule to eliminate the majority of stochastic errors and to achieve the minimum accuracy of 99%53. Once generated, our meta-analysis indicates that HiFi reads have a median accuracy >99.9%, with >99.5% of homopolymers ≥5 bases in length accurately resolved, consistent with those reported elsewhere53,54 (Figure 3c and 3d, Supplementary Figure 1, Supplementary Note). The high accuracy of PacBio HiFi sequence data has improved variant discovery, reduced time to assembly, and provided access to even more complex regions of repetitive DNA including the contiguous assembly of some human centromeres52,53,55. In fact, >50% of the regions previously inaccessible with Illumina short-read sequence data in GRCh37 are now accessible with HiFi reads53. Although especially useful for cDNA sequencing due to its comparatively high accuracy, it is generally thought that HiFi reads will ultimately replace CLR for most human genome sequencing applications. However, the cost (Table 1) and computational resources required to generate HiFi data currently limit its widespread adoption.
ONT long reads.
ONT read lengths can surpass PacBio by an order of magnitude or more by generating contiguous sequence hundreds to thousands of kilobases in length34–36, although, in practice, such reads represent a small proportion of the total read length distribution. These enormous read lengths are facilitated by the unique pore chemistry essential to ONT, which allows molecules to translocate through the nanopore regardless of their length. Various studies have shown that the main factor limiting ONT read lengths is the extraction and preparation of high-molecular-weight DNA34–36. These different methods of preparation underlie the two main types of ONT data: the standard long (10–100 kb) read and the specialized ultra-long (>100 kb) read (Figure 3b).
The most common type of read generated via ONT sequencing is the standard ONT long read [G]. In our meta-analysis, these reads were typically 10–100 kb in length and 87–98% accurate, on average, although a small portion of these have an accuracy as low as 69%. About 91% of homopolymers ≥5 bases in length were accurately called in raw ONT long reads, which is 3% higher than PacBio CLR reads but ~8% lower than PacBio HiFi reads (Figure 3c and 3d, Supplementary Figure 1, Supplementary Note). Our findings are consistent with previous reports34,36,56. It should be noted that ONT raw read accuracy is highly dependent on the base-calling algorithm used38,57, and recent improvements to these algorithms have increased raw read accuracy substantially in the past five years38. Additionally, several methods have been developed to improve the consensus read accuracy of ONT long reads, including INC-Seq58, HiFRe59, and 1D2 sequencing60, which can result in an ONT consensus read accuracy close to that of a PacBio HiFi read, at ~97–98%58,60.
Long-read data can be generated on any of the three standard ONT platforms: the MinION, GridION x5, and the PromethION. These three platforms differ in their flow cell capacity. The MinION, a pocket-sized device, can hold one flow cell, whereas the GridION X5 can hold up to five, and the PromethION generates data from up to 48 flow cells at a time. Importantly, the MinION and GridION X5 use the same type of flow cell, with 2,048 individual nanopores split into 512 channels, whereas the PromethION uses a different type of flow cell with 12,000 nanopores split into 3,000 channels. Because each channel can only sequence with one nanopore at a time, the MinION and GridION X5 are only able to sequence with 512 nanopores at a time per flow cell, while the PromethION is able to sequence ~5.9 times this amount (3,000 nanopores) at a time per flow cell. As a result, the PromethION provides nearly 6 times as much throughput per flow cell relative to the MinION or GridION X5, with 50–100 Gb of long-read data generated per PromethION flow cell36 compared to 2–20 Gb generated on each MinION and GridION X5 flow cell34,35,56. When accounting for the fact that the PromethION can sequence up to 48 flow cells simultaneously, the PromethION throughput far exceeds that of the PacBio Sequel II and the Illumina NovaSeq (Table 1).
For low-throughput applications, ONT also offers the Flongle (or flow cell dongle), which is an adapter compatible with the MinION and GridION X5 platforms. The Flongle uses a different type of flow cell that contains 126 nanopores in as many channels, allowing all 126 nanopores to be sequencing at one time. A clear advantage of the Flongle is that it allows for smaller, frequent, rapid tests to be carried out at a fraction of the cost of MinION or GridION X5 flow cells. Additionally, the portability of the Flongle and MinION allow them to be transported in standard overhead bins of airlines and readily moved into the field without the need for complex and unwieldy instrumentation. The Flongle has been used in diverse clinical and field applications to detect influenza in clinical respiratory samples61 and diagnose lower respiratory infections62. Additionally, the MinION has been used to track small bacterial and viral genomes, such as those during the 2015 Ebola outbreak63. Together, the portability and rapid sequencing speed of the Flongle and MinION make them ideal for genomic sequencing applications in the field and clinic.
ONT ultra-long reads.
Another type of read that can be generated on ONT sequencing platforms is the ONT ultra-long read [G]. These reads were first generated by Josh Quick35 (see https://lab.loman.net/2017/03/09/ultrareads-for-nanopore/) and are typically >100 kb in length34,35 but can be up to several megabases long37. Our meta-analysis shows that read accuracy is similar for ONT ultra-long reads and ONT long reads, with most reads averaging 87–98% accuracy and a small fraction having a base accuracy as low as 68% (Figure 3c, Supplementary Figure 1, Supplementary Note), consistent with previously published reports34,35. In addition, ultra-long reads have >93% of homopolymers ≥5 bases in length accurately called, similar to long reads (Figure 3d, Supplementary Figure 1, Supplementary Note). Although ultra-long reads shatter records with respect to read length, their throughput is much lower than that of standard long reads. Only 500 Mb to 2 Gb of ultra-long-read data are typically produced per flow cell on the MinION and GridION X5, with a maximum throughput of 2.5 Gb34,35. As a result, the generation of 20-fold ultra-long-read sequence data can take several weeks on a GridION X5 platform when running at full capacity, which is substantially longer than the time it takes to generate standard ONT long-read data on the same device (Table 1). Attempts to generate ultra-long-read data on the PromethION have been met with limited success36, which we speculate is because of the lack of compatible sequencing kits required to generate ultra-long reads. With improved kit compatibility, it is likely that ultra-long-read throughput will improve, increasing its utility for whole-genome applications.
PacBio and ONT long- and ultra-long-read sequencing data have begun to significantly impact several areas of human genetics research, including genome assembly9,30,33–36,64, variant discovery3,31,32,54, disease association29,65–68, and human genetic diversity69–71. New methods have evolved to apply the different long-read sequencing data types to each of these areas of research. In some cases, such as the complete assembly of human genomes, the different data types can be complementary.
Genome assembly with long reads
One of the first applications of long-read sequencing has been to improve the assembly of genomes, as read lengths are now sufficiently long to traverse most repeat structures of the genome. For diploid genomes, such as in humans, the challenge is now to achieve accurate haplotype resolution from telomere to telomere without guide from a reference.
De novo genome assembly.
De novo genome assembly is the process by which randomly sampled sequence fragments are reconstructed to determine the order of every base in a genome72. Stitched-together sequence fragments are referred to as contigs [G], and in the ideal case, there is one contig per chromosome. Short-read technology has been problematic for the de novo assembly of mammalian genomes and has typically resulted in hundreds of thousands of gaps, owing to repetitive sequences that cannot be traversed by short reads. Numerous studies have shown that long-read genome assemblies are superior in their contiguity by orders of magnitude when compared to previous short-read and Sanger-based sequencing approaches30,32,33,35,70,71 (Table 2). For example, in early 2015, there were 99 mammalian genome assemblies in GenBank with an average contig N50 [G] of only 41 kb, but none of them used long-read sequencing as the predominant data type27. As of early 2020, there are more than 800 genome assemblies available through GenBank that used either PacBio or ONT data with contig N50 lengths greater than 5 Mb, including some of the first human genomes: NA1287835, CHM1332, HX170, and AK171. This >100-fold increase in assembly contiguity has been driven not only by longer reads but also by the development of genome assembly tools optimized for long-read data (such as Canu73, HiCanu55, Peregrine74, FALCON75, Flye76, wtdbg2 (or RedBean)77, and Shasta36) as well as optical mapping [G] (for example, from Bionano Genomics)30,34,70,71,78 and electronic mapping [G] (for example, from Nabsys)79,80 tools that can improve assembly contiguity and accuracy. Importantly, it is now becoming tractable for individual laboratories (as opposed to large consortia) to sequence and assemble human genomes in a few weeks at levels of contiguity approximate to or exceeding that of the Human Genome Project31,36,81 (Figure 4a). For example, Shafin et al. generated 11 highly contiguous (median NG50 = 18.5 Mb) human genome assemblies with long-read ONT data with only three PromethION flow cells and six hours of compute time on a 28-core machine with >1 TB of RAM per genome. Similarly, Chin and Khalak assembled human genomes in less than 100 minutes (30 CPU hours; not including the one-time computational cost of generating the PacBio HiFi reads) with a contig N50 > 20 Mb with only PacBio HiFi data74. For comparison, an alignment of ~30X short-read Illumina data can take up to 100 CPU hours82,83.
Table 2.
Statistics of human genome assemblies generated with various data types and assembly algorithms
| Genome assembly | Data type [coverage; read N50 (kb)] | Assembler | Size (Mb) | # of contigs | Contig N50 (Mb) | Estimated cost | Ref. |
|---|---|---|---|---|---|---|---|
| hg1 | Multi-technology | GigAssembler, PHRAP | 2.69 | 149,821 | 0.082 | $300,000,000 | 72 |
| hg38 | Multi-technology | Multiple algorithms | 3.01 | 998 | 57.88 | not determined | 161 |
| YH | Illumina (56-fold; <0.075) | SOAPdenovo | 2.91 | 361,157 | 0.02 | $1,600a | 162 |
| CHM13 | PacBio CLR (77-fold; 17.5) | FALCON | 2.88 | 1,916 | 29.30 | $2,700b | 30 |
| PacBio HiFi (24-fold; 10.9) | FALCON | 3.00 | 2,116 | 31.92 | $4,100b | 52 | |
| Canu | 3.03 | 5,206 | 25.51 | ||||
| PacBio CLR (77-fold; 17.5) and ONT (50-fold; 70.4) | Canu | 2.94 | 590 | 72.00 | $55,000c | 34 | |
| HG002 | PacBio HiFi (28-fold; 13.5) | FALCON | 2.91 | 2,541 | 28.95 | $2,700b | 53 |
| PacBio HiFi (28-fold; 13.5) | Canu | 3.42 | 18,006 | 22.78 | |||
| ONT (47-fold; 48.7) | Shasta | 2.80 | 1,847 | 23.34 | $5,000d | 36 | |
| Flye | 2.82 | 1,627 | 31.25 | ||||
| Canu | 2.90 | 767 | 33.06 | ||||
| NA12878 | Illumina (103-fold; 0.101) | ALLPATHS-LG | 2.79 | 231,194 | 0.02 | $2,900a | 163 |
| ONT (29-fold; 10.6 5-fold; 99.8) | Flye | 2.82 | 782 | 18.18 | $4,000d | 76 | |
| Canu | 2.82 | 798 | 10.41 | 35 | |||
| NA12878 (phased) | PacBio HiFi (30-fold; 10.0) | Peregrine | 2.97 [H1] 2.97 [H2] | 9,334 [H1] 9,127 [H2] | 19.6 [H1] 18.7 [H2] | $4,100b | 22 |
| HG00733 | ONT (73-fold; 29.6) | Shasta | 2.78 | 2,150 | 24.43 | $6,000d | 36 |
| Flye | 2.81 | 1,852 | 28.76 | ||||
| Canu | 2.90 | 778 | 44.76 | ||||
| HG00733 (phased) | PacBio HiFi (33-fold; 13.4) and Strand-seq (5-fold) | Peregrine | 2.90 [H1] 2.91 [H2] | 2,618 [H1] 2,557 [H2] | 28.0 [H1] 29.2 [H2] | $9,000e | 91 |
Multi-technology: Clone-by-clone hierarchical sequencing with short and Sanger reads.
H1 and H2 refer to the first and second haplotype in the diploid genome assembly, respectively.
Current cost when generated on the NovaSeq using S4 flow cells and multiplexing.
Current cost when generated on the Sequel II.
Current cost when generated on the Sequel II (PacBio CLR) and GridION (ONT).
Current cost when generated on the PromethION.
Current cost when generated on the Sequel II (PacBio HiFi) and HiSeq 2500 (Illumina).
All estimates are listed in USD and exclude the cost for labor, instrumentation, maintenance, and computer resources.
Figure 4. Long-read data improves genome assembly.
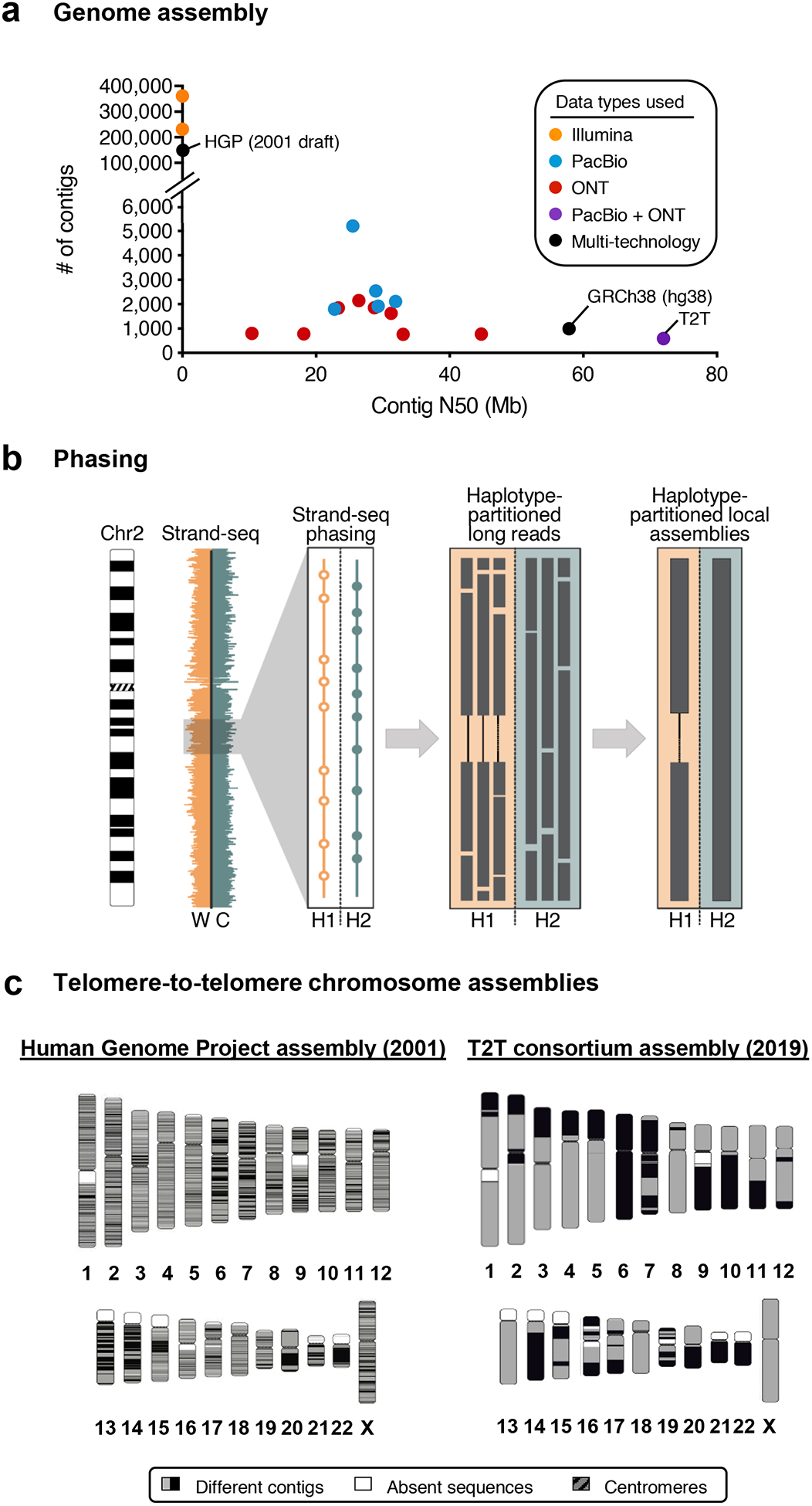
a) The plot shows the number of contigs and the contig N50 for 18 unphased human genome assemblies listed in Table 2. Genomes assembled from long-read data (PacBio or ONT) have fewer contigs and higher contig N50s compared to those assembled from short-read data (Illumina). Combining long-read data types (PacBio + ONT) produces a genome assembly with even fewer contigs and a higher contig N50, surpassing that of the reference genome (GRCh38, hg38) in contiguity. b) The schematic illustrates a genome assembly phasing approach known as Strand-seq164. In this approach, the template strand [i.e. the Watson (W, orange) or Crick (C, teal) strand)] is sequenced via short-read sequencing to generate template-specific short reads. When the W and C template strands are inherited from either parent, these templates-specific reads can be assigned to either parental homologue based on the direction they map to a genome assembly. For example, here, we show Strand-seq reads aligned to chromosome 2 and binned in 200 kb genomic stretches (orange and teal bars). Strand-seq reads containing a haplotype-specific SNP are able to partition long reads into haplotype 1 (H1, empty circles) or haplotype 2 (H2, filled circles). Haplotype-partitioned long reads permit the detection of structural variation, such as the deletion in H1 shown here, and can be assembled to generate haplotigs that span the region, thereby generating a phased genome assembly. c) Chromosome ideograms are shown that compare the 2001 Human Genome Project assembly (hg1)72 and the 2019 T2T assembly (CHM13 rel3 assembly). hg1 had >145,000 gaps and nearly 150,000 contigs, whereas the rel3 assembly has <1000 gaps and <1000 contigs (see Table 2 for additional statistics). Contigs are represented by alternating black and gray blocks, absent sequences are represented by white blocks, and centromeres are represented by red blocks.
Polishing and phasing.
Although speed is important, long-read genome assemblies have frequently been criticized for their reduced accuracy83. However, with proper correction and assessment, long-read assemblies can rival those generated by Illumina or Sanger sequencing84. Unpolished assemblies typically suffer from many small indel errors, which complicate gene annotation50. The majority of these errors can be resolved using polishing tools (such as Racon48, Nanopolish63,85,86, MarginPolish36, HELEN36, Quiver46, Arrow, and Medaka) and error-correction with short-read sequence data generated from the same individual47. Recent developments in base-calling algorithms and the generation of highly accurate long-read sequence data types such as HiFi are eliminating dependencies on short-read data polishing52,53,84. A major focus moving forward is the generation of high-quality, fully phased diploid genomes where both haplotypes are represented84. This procedure essentially converts a 3 Gb collapsed human genome into a 6 Gb genome that represents both maternal and paternal complements, which has the advantage of increasing overall sensitivity for variant discovery9. Fortunately, phased de novo genome assembly [G] is now becoming feasible with new strategies that take advantage of parental information to phase long reads (such as trio binning [G]87), computational methods that take advantage of the inherent phasing present in long-read data (such as FALCON-Unzip75), and methods that apply orthogonal technologies to phase single-nucleotide polymorphisms in long-read data (such as Strand-Seq9,88,89, Hi-C90, and, in the past, 10x Genomics9) (Figure 4b). The fundamental concept here is straightforward: by physically or genetically phasing an individual genome, the long-read data can be partitioned into two parental genome datasets that can be independently assembled. Such a procedure is particularly valuable for resolving structural variation and its haplotype architecture91 because structural differences between haplotypes have often led to hybrid representations or collapses in the assembly that do not reflect the true sequence and are, therefore, biologically meaningless92.
Telomere-to-telomere chromosome assemblies.
The ultimate genome assembly is a single contig per chromosome, where the order and orientation of the complete chromosome sequence is resolved from telomere to telomere (T2T). More than half of the remaining gaps in long-read genome assemblies correspond to regions of segmental duplications27,52,54,91 and can be readily identified by increased read depth. These collapses result from a failure to resolve highly identical sequences. However, these regions can be assembled with over 99.9% accuracy using approaches that partition the underlying long reads using a graph of paralogous sequence variants [G]93, such as SDA (Segmental Duplication Assembler)54. The human reference genome has been the gold standard for mammalian genomes since its first publication in 2001, and there has been considerable investment over the past two decades to improve its accuracy and continuity. Notwithstanding, even in its current iteration (GRCh38, or hg38), the number of contigs greatly exceeds the number of chromosomes (998 contigs versus 24 chromosomes) with most of the major gaps corresponding to large repetitive sequences present in centromeres, acrocentric DNA, and segmental duplications (Table 2). Application of both ONT and PacBio technologies to the essentially haploid CHM13 human genome [G] has shown that we are on the cusp of generating T2T genome assemblies. By using both these sequencing data types and improved assembly algorithms, the CHM13 human genome has been represented as 590 contigs, including a complete T2T assembly of the X chromosome34 (Figure 4c, Table 2). Key to this advance was the generation of high-coverage ultra-long ONT data, which allowed for greater contiguity than GRCh38 (81.3 Mb versus 57.9 Mb) and, for the first time, a reconstruction of the highly repetitive centromeric alpha-satellite array on the X chromosome. However, the T2T assembly process is far from automated, requiring considerable manual curation, and hundreds of collapsed repeats still remain to be resolved genome-wide. Nevertheless, efforts to automate centromere assembly (such as with CentroFlye94 and HiCanu55) are underway. Further developments, such as improved assembly tools that optimize the processing and assembly of PacBio HiFi sequence data or that couple it to ONT ultra-long-read data, will be required before T2T chromosome assemblies can be routinely generated for diploid genomes. Routine and accurate assembly of T2T human chromosomes from diploid genomes will likely take years, not just because specialized data types (that is, ultra-long-read sequence reads) are more expensive and take longer to generate, but because it will involve uncharted territories of the human genome. For many regions, including centromeric, acrocentric, and large regions of segmental duplication, the sequence has not been correctly assembled even once, so any computational assembly algorithm geared to such regions54,94 will require painstaking validation and assessment.
Understanding variation with long reads
Improved accuracy and continuity of genome assemblies necessarily enhances our understanding of more complex forms of genetic variation, and this, in turn, improves our understanding of mutation and evolutionary processes.
Large-scale structural variant detection and disease.
Long-read genome sequencing has substantially enhanced our understanding of the full spectrum of human genetic variation32,33,64. A comparison of the same individuals sequenced with Illumina short-read and PacBio long-read platforms, for example, showed that 47% of the deletions and nearly 78% of insertions were missed by Illumina whole-genome sequencing (WGS) [G] even after applying 11 different variant callers designed to detect insertions, deletions, inversions, and duplications in genomes9. Most of the gains in sensitivity involve intermediate-size variants ranging from 50 bp to a few kb in size. Similarly, an analysis of difficult-to-assay sequences from 748 human genes where mapping quality is low for some individual coding exomes reported remarkable improvements in sensitivity, including the discovery of potentially pathogenic variants associated with Alzheimer’s disease95. Similarly, there is evidence of increased sensitivity for the detection of indels less than 50 bp30,96, although this effect has been more difficult to quantify due to the predominant error types in long-read data. Accompanying this increase in sensitivity has been a spate of new structural variant callers (SMRT-SV33, MsPAC93, Phased-SV9, Sniffles97, and PBSV53) designed to discover, sequence and, in some cases, phase structural variants based on specific long-read sequence signatures and local assembly. These callers rely on the alignment of long-read data to a reference genome via specialized algorithms (such as BLASR98, NGMLR97, minimap299, MHAP100); however, as the speed and accuracy of generating fully phased and assembled human genomes increases, it is likely that many of these discovery tools will be supplanted by direct comparisons of assembled genomes for variant discovery30. Although there have been substantial gains in variant discovery, particular classes, including large copy number variants and inversions mapping within or near large segmental duplications, are still difficult to resolve solely with existing long-read technology9.
An immediate application of this increased sensitivity has been the discovery and sequencing of more complex forms of disease-causing variation56,101–108, including novel GGC repeat expansions associated with neuronal intranuclear inclusion disease and adults with leukoencephalopathy65,66,109; founder SINE-VNTR-Alu retrotransposon insertions responsible for X-linked Dystonia-Parkinsonism in the Philippines110; novel candidate mutations associated with schizophrenia and bipolar disorder111; pentanucleotide repeat expansions linked to familial and sporadic cases of benign adult myoclonic epilepsy in Japan and China103,109; and the discovery of large complex triplications and regions of segmental uniparental disomy [G] associated with Temple syndrome112. Here, too, specialized algorithms have been developed to detect and accurately predict short tandem repeat (STR) expansions as well as predict methylation status of the flanking regions from underlying long-read sequence data (for example, STRique)113. Expanding catalogs of sequence-resolved structural variation are identifying new lead variants associated with both expression quantitative trait loci (eQTLs) [G] and genome-wide association studies (GWAS) [G]31 and suggesting candidate loci for repeat-associated instability diseases114. Importantly, these discoveries are leading to new insights regarding disease mechanisms, such as the reported finding that TTTCA repeat expansions within introns associate with myoclonic epilepsy irrespective of the protein-coding gene in which they are found, potentially because of RNA-mediated toxicity linked to their transcription115. It is worth noting that the layers of genomic complexity and structural variation revealed only through high-quality sequencing often yield insights into multiple diverse diseases. For example, the GGC repeat-expansion associated with NOTCH2NLC and neuronal intranuclear inclusion disease maps to human-specific segmental duplications on chromosome 1q21 that have been recently implicated in cortical neurogenesis and expansion of the frontal cortex during human evolution116,117. The presence of these duplications was used to predict and discover recurrent rearrangements associated with developmental delay, microcephaly, and macrocephaly68,118,119 and later schizophrenia67 (Figure 5a). It should be noted that mapping-based approaches were used in these studies to discover and resolve the structure of the variants in question, not whole-genome assembly65,101. Yet, these discoveries were often preceded by high-quality assembly of the gene model or the locus of interest, which were missing from the original human genome but now can be assembled using whole-genome assembly methods54. Mapping-based approaches are largely ineffective without high-quality references to compare.
Figure 5. Long-read data provides insights into the biological relevance of structural variation and human evolution and diversity.
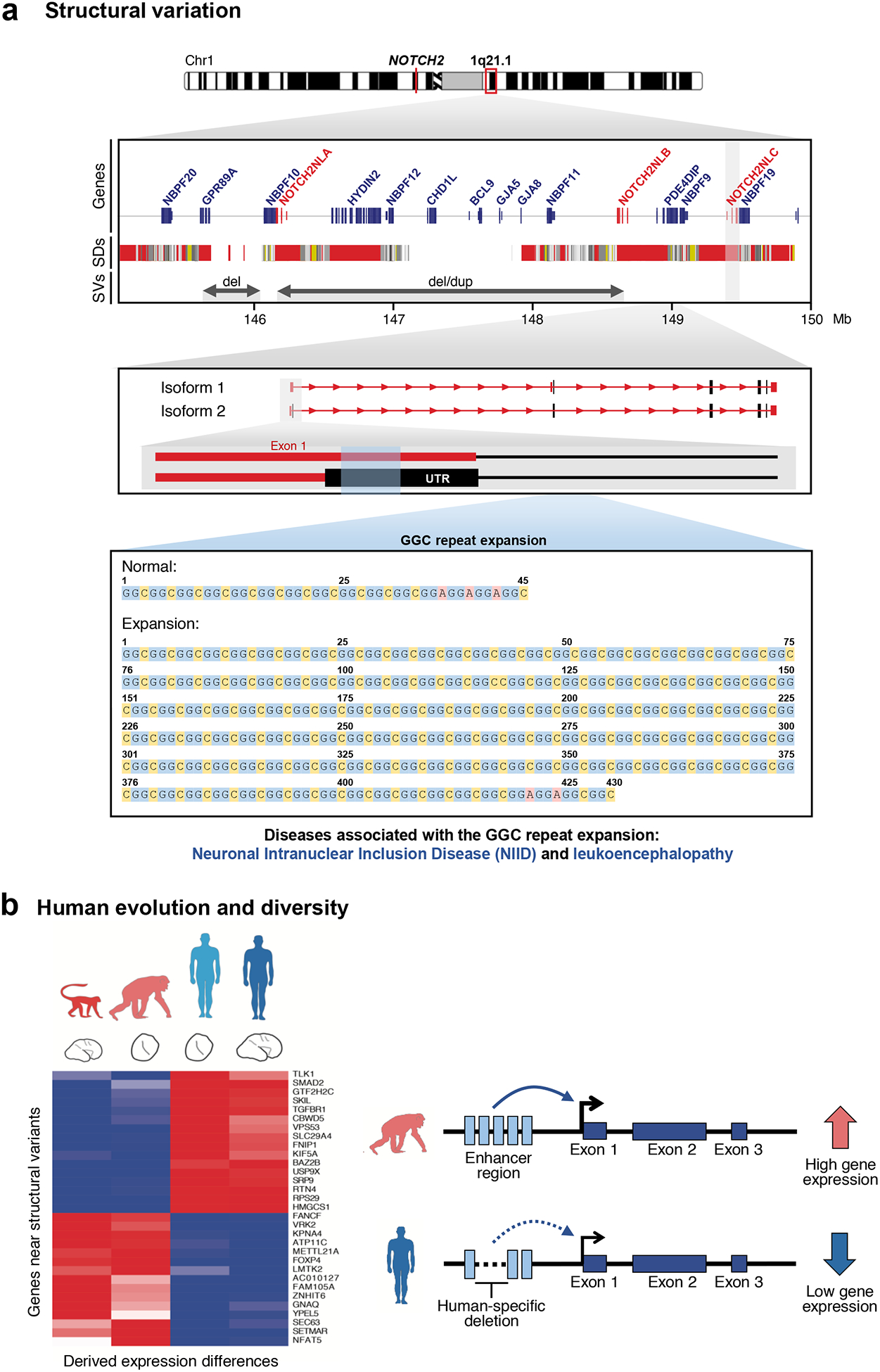
a) The NOTCH2NLA, B, and C genes are located within chromosome 1q21.1, a segmental duplication-rich region of the genome partially assembled by PacBio CLR sequencing of BAC clones116. The region was originally incorrectly assembled in the human reference genome116. Deletions and duplications mediated by the segmental duplication-rich region can cause thrombocytopenia-absent radius (TAR) syndrome165 as well as distal 1q21.1 deletion/duplication syndrome119,166. High-quality sequencing of the region allowed the breakpoints of these disease-causing rearrangements to be better defined and improved the annotation of human-specific NOTCH2NL duplicate genes116. Subsequent sequencing of patients affected with neuronal intranuclear inclusion disease (NIID) and leukoencephalopathy using long-read PacBio CLR and ONT sequencing recently identified a GGC repeat expansion in Exon 1 of NOTCH2NLC in affected patients66 (exons are in red; untranslated regions (UTRs) are in gray). Expansion of the repeat is associated with the production of anti-sense transcripts whose role is uncertain but may interfere with the expression and regulation of the gene family. Figure adapted from Ref. 66. SDs, segmental duplications; SVs, structural variants. b) Heatmap of differentially expressed genes located near structural variants in chimpanzee and human. Differences in macaque, chimpanzee, and human brain expression are shown for genes where a human-specific structural variant maps within 50 kbp of a transcription start and end. Structural changes, such as a deletion of an enhancer region as shown here, can cause changes in gene expression fundamental to brain development30.
Human genetic diversity and evolution.
Implicit in the sequencing and assembly of new human genomes and in increased structural variation discovery is an improved understanding of human genetic diversity and the mutational processes that have shaped our genomes31–36,53,64,70,71,78,81,90 (Figure 5b). For example, long-read sequencing of a modest diversity panel of 15 human genomes identified almost 100,000 structural variants—most of which were previously unknown31. Among these, variable number tandem repeats (VNTRs) [G] were shown to be the most non-randomly distributed with almost half mapping to the last 5 Mb of subtelomeric regions, possibly owing to increased rates of double-strand breaks in these regions31. Comparison of human and nonhuman primate genomes sequenced with PacBio have doubled the number of structural variants associated with brain expression differences specific to the human lineage30 and identified large-scale changes potentially important in the evolution of ape lineages120. Recent sequencing and assembly of large copy number polymorphisms have identified structural variants associated with both positive selection and introgression [G] that are largely specific to certain human populations69. For example, a 386 kb duplication polymorphism was fully sequenced and assembled that is effectively specific to individuals of Melanesian descent. Remarkably, the duplication, as well as the duplicated genes within, arose in the archaic Denisova lineage and was subsequently introgressed back into the human ancestor through interbreeding. The duplication shows multiple signatures of positive selection and is now present in 79% of Melanesians but virtually absent in other populations. The discovery and sequencing of such complex structural variants further improves genotyping even among short-read datasets making it feasible to enhance association studies31,32. For this reason, the NIH recently launched an initiative, the Human Pangenome Reference Sequence Project, to sequence and assemble more than 350 diverse human genomes using long-read sequencing platforms121.
Beyond DNA sequencing
In addition to genome assembly and variant discovery, long-read sequencing has been applied to molecules other than DNA, enabling the detection, for example, of full-length RNA isoforms122–124 as well as modifications of native RNA and DNA96,125–127.
Full-length RNA sequencing.
A major strength of long-read sequencing technology is the ability to determine the sequence of full-length RNA transcripts arising from genes. Both PacBio and ONT are able to resolve the sequence of full-length RNA molecules, either via cDNA sequencing (PacBio and ONT)128–131 or native RNA sequencing (ONT)122–124. Such sequence data improves gene annotation and simplifies downstream analysis by eliminating the need to reconstruct isoforms based on the error-prone assembly of short RNA-seq reads. The primary method used by PacBio to identify full-length RNA molecules is Iso-Seq129, which involves cDNA synthesis, PCR amplification, and SMRTbell ligation followed by CCS. The Iso-Seq method has been successfully used to capture novel isoforms54,70,71,129,132 and validate new gene models54 in diverse genomes69 (Figure 6a). Similar to the CCS mode of PacBio, ONT has developed rolling circular amplification of concatemerized sequences (known as R2C2) as a means to improve the accuracy of cDNA sequence133. In contrast to PacBio, which depends on cDNA synthesis, ONT sequencing technology can be applied to native RNA molecules to capture the full-length isoforms122. Native RNA-seq has the advantage that it ensures all RNA molecules are captured, including long transcripts often missed during cDNA synthesis owing to their length or complexity130. Furthermore, it avoids sequence biases frequently introduced during PCR amplification of cDNA134. Full-length poly(A) transcriptomes have been readily obtained using ONT native RNA-seq123,124. Additionally, native RNA-seq has revealed novel isoforms arising from disease-risk genes associated with psychiatric disorders135 and chronic lymphoid leukemia136, which may provide new targets for early disease detection in clinical settings and for pharmaceutical treatments.
Figure 6. Long-read platforms can be used to sequence RNA and detect nucleic acid modifications.
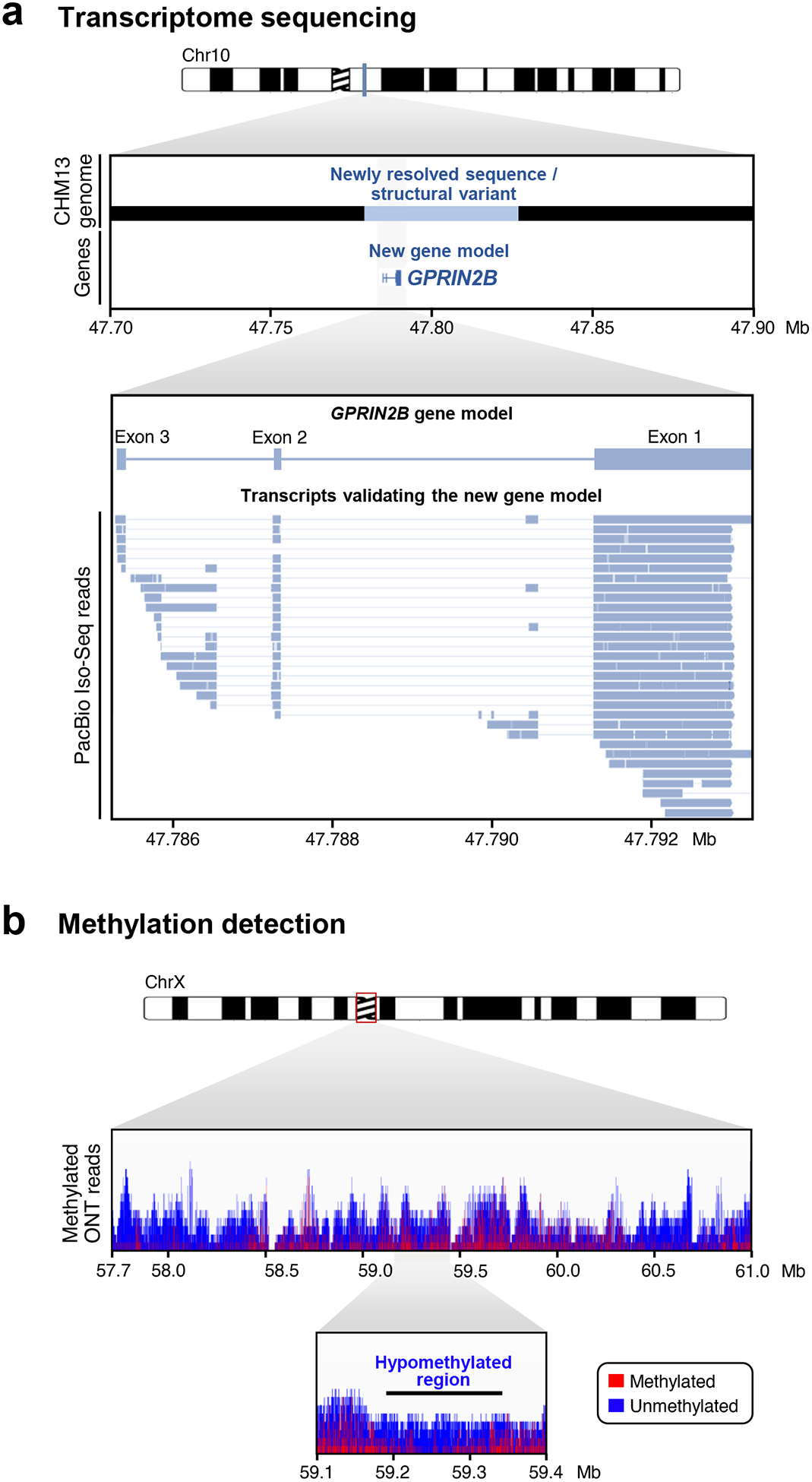
a) Long-read RNA sequencing can be used for full-length isoform discovery. A newly resolved sequence on chromosome 10 of the CHM13 genome (dark blue) revealed a previously undiscovered gene, GPRIN2B (light blue). Using PacBio Iso-Seq, full-length transcripts were identified that completely span GPRIN2B, validating the new gene model. Adapted from Ref. 54. b) The assembly of the entire X chromosome centromere reveals that the majority of the α-satellite repeat region is heavily methylated, except for a ~93 kb hypomethylated region, which was discovered via ONT long-read sequencing of native DNA molecules and subsequent analysis with the methylation detection tool, Nanopolish86. Adapted from Ref. 34.
DNA and RNA methylation detection.
Because both PacBio and ONT target native unamplified templates for sequencing, the DNA and RNA molecules retain base modifications, allowing epigenomic changes to be detected through polymerase kinetics96,125,126,137 or current changes, respectively86,126,127. Prior to the development of these technologies, the most common base modification that could be detected was methylated cytosine, using an indirect approach known as bisulfite sequencing. With bisulfite sequencing, DNA is treated with bisulfite, which converts cytosine to uracil but leaves modified cytosines unaffected. Short-read sequencing of the resulting DNA along with an untreated control allows for the identification of modified cytosines. However, it does not discriminate between cytosine modifications138 nor does it allow for the detection of other modified bases. Native DNA and RNA sequencing via PacBio and/or ONT presents substantial advantages over standard bisulfite-sequencing methods because it allows for a more diverse array of modifications to be identified, including 4-methylcytosine (4-mC), 5-methylcytosine (5-mC), 5-hydroxymethylcytosine (5-hmC), N6-methyladenine (6-mA), and 8-oxoguanine (8-oxoG)127,139–144. Additionally, direct sequencing of native molecules simplifies the process by eliminating the need to prepare bisulfite-treated samples that are sequenced separately from the untreated samples145. Similarly, long-read sequencing technologies greatly facilitate the detection of modified RNA bases by eliminating the use of highly specialized protocols to detect diverse types of modifications146–149. Thus, direct sequencing of native DNA and RNA molecules is expanding the fields of epigenomics and epitranscriptomics by allowing for the detection of previously unrecognized modifications on DNA and RNA concurrent with sequencing.
To detect modifications on DNA, PacBio depends on detecting changes in polymerase kinetics during SMRT sequencing96,125,126,137. Kinetic characteristics, such as the arrival time and duration between two successive base incorporations, yield information about polymerase or reverse transcriptase kinetics that facilitate base modification detection. Because various modifications affect polymerase kinetics differently, SMRT sequencing can identify these kinetic signatures at base-pair resolution but typically requires high sequence coverage (25- to 250-fold) to do so139. Targeted enrichment of select DNA loci via CRISPR-Cas9150,151 has shown promise for enabling the higher sequence coverage needed for accurate base modification detection. PacBio SMRT sequencing has led to the discovery of methylation profile differences in diseased and healthy individuals109 and has been used to identify novel hypermethylated regions in the genome152. For example, Ishiura and colleagues found that novel CGG repeat expansions associated with neural intranuclear inclusion disease were hypermethylated when compared to their unexpanded counterparts109. Additionally, Suzuki and colleagues uncovered novel LINE elements that were methylated in the human genome, which were previously missed with bisulfite sequencing152.
ONT sequencing is also able to detect modifications on native DNA and RNA molecules with high accuracy owing to the characteristic current disruption imparted by the modified base as it translocates through the nanopore86,126,127,142. Several computational tools have been developed to detect DNA and RNA modifications based on these characteristic disruptions: Nanopolish85,86, signalAlign127, DeepSignal153, mCaller154, DeepMod155, and Tombo156. These tools have been used to uncover methylation states in previously inaccessible regions of the genome and transcriptome, such as the chromosome X centromere34 (Figure 6b), as well as genes implicated in cancer157, leading to new biological insights. In particular, the finding that the human X chromosome centromere is methylated across the entire DXZ2 α-satellite repeat array except for a ~93 kb pocket of hypomethylation suggests differences in transcriptional regulation in these repeat-dense regions34. Additionally, the discovery that structural variants are differentially methylated in cancer cells is providing insight into the complex epigenetic characteristics of structurally variant regions implicated in cancer157. As more and more phased human genome assemblies become available, it may become possible to determine the methylation status of each allele, which could lead to important discoveries that lie at the root of allelic epigenetic variation.
CONCLUSIONS AND FUTURE PERSPECTIVES
Sequencing technology is the ‘microscope’ by which geneticists study genetic variation, and it is clear that long-read technologies have provided us with a new ‘lens and objective’ for understanding DNA and RNA variation, structure, and organization. Although the two predominant long-read technologies are competitive, some of the best results have been obtained when the sequencing platforms are used to complement one another. For example, the first T2T assembly of the human X chromosome leveraged both the accuracy of deep PacBio CLR data along with ultra-long ONT data to traverse centromeric regions. ONT sequencing generates the longest contiguous sequence reads and is the most portable, whereas PacBio produces some of the most accurate long-read data and is beginning to rival that of next-generation sequencing. Both technologies use native DNA as opposed to amplified products as templates for sequencing and, thus, provide access to more uniform and biologically meaningful data. Continued reductions in cost, improvements in accuracy, and increases in throughput will make these technologies more commonplace in the lab, field, and clinic. With the ability to now sequence, assemble, and phase human genomes at levels of continuity exceeding that of the first human genome project for a few thousand dollars, the field of human genetics has forever changed. We are now embarking on an era where all genetic variation in an individual will be completely discovered in the next few years. Hundreds and ultimately thousands of new human reference genomes will be produced. In addition, light sampling (~10- to 15-fold sequence coverage) of thousands of individuals provides an alternative strategy for improved variant discovery from a population perspective158,159. These advances will dramatically improve our understanding of human heritability, population diversity, mutational processes, and the genetic basis of disease. Notably, adoption of long-read technology will also change how we discover and catalog human variation. Variation will not be discovered by simply aligning reads to a single reference genome and inferring genetic differences but rather by sequencing and assembling complete haplotypes for which complex genetic variation is fully sequence resolved. The next steps will likely involve the development of graph-based reference genomes using new standards, such as Variant Graph Toolkit160. Functional data will be superimposed on these complete genomes, including epigenetic and transcriptomic differences that occur ultimately at the cellular and developmental level.
The wealth of additional information afforded by single-molecule, long-read sequencing compared with short-read sequencing promises a more comprehensive understanding of genetic, epigenetic, and transcriptomic variation and its relationship to human phenotype.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M.J. Chaisson (USC) and D. Porubsky for assistance with figures, K. Munson for technical assistance and commentarial insight, and T. Brown for assistance in editing this manuscript. This work was supported, in part, by grants from the U.S. National Institutes of Health (NIH HG010169 to E.E.E.) and National Institute of General Medical Sciences (NIGMS 1F32GM134558-01 to G.A.L.). M.R.V. was supported by a National Library of Medicine (NLM) Big Data Training Grant for Genomics and Neuroscience (5T32LM012419-04). E.E.E. is an investigator of the Howard Hughes Medical Institute.
GLOSSARY
- Next-generation sequencing
A sequencing method in which an entire genome is sequenced from fragmented DNA, producing short (<300 bp) sequencing reads at high speeds and low costs.
- Sequence-by-synthesis
A sequencing technology used primarily by Illumina, in which a DNA polymerase synthesizes a strand of DNA complementary to a template by incorporating a fluorescently labeled deoxynucleotide triphosphate (dNTP) that is imaged to identify the base and then cleaved before the process is repeated to determine the order and identity of each base in the DNA strand.
- Single-nucleotide variants
Instances in which a single base within a read or genome differs from the base found at the same position in other individuals or populations.
- Copy number variants
Instances in which a sequence of nucleotides within a genome vary in the number of copies among individuals or populations.
- Indels
Insertions or deletions of bases in sequence data or in DNA or RNA.
- Structural variant
A genetic variant >50 bp in length that includes insertions, deletions, inversions, or translocations of DNA segments, or copy number differences.
- Segmental duplications
Long DNA sequences (typically >1 kb in length) that have nearly identical sequences (90–100% identity) and exist in multiple genomic locations as a result of duplication events.
- Barcodes
A series of known bases added to a template molecule either through ligation or amplification. After sequencing, these barcodes can be used to identify which sample a particular read is derived from.
- Linked-read
A synthetic long-read DNA sequencing method wherein short-read sequencing is applied to long DNA molecules to “link” reads together from the same original long molecule.
- Contigs
Contiguous stretches of DNA sequence without gaps that have been assembled solely based on direct sequencing information.
- Long-read sequencing
A sequencing method used by PacBio and Oxford Nanopore Technologies (ONT), wherein native DNA or RNA molecules are sequenced in real time, often without the need for amplification, producing long (>10 kb) reads.
- Single-molecule, real-time (SMRT) sequencing
A DNA sequencing method used by Pacific Biosciences (PacBio), wherein a single DNA molecule is sequenced and derived in real time, with no pause after the detection of the bases.
- SMRTbell
A double-stranded DNA template used by PacBio wherein both DNA ends are capped with hairpin adapters, known as SMRTbell adapters. A SMRTbell template is topologically circular and structurally linear.
- SMRT Cell
A flow cell comprised of arrays of zero-mode waveguide (ZMW) nanostructures used during PacBio sequencing.
- Zero-mode waveguides (ZMWs)
A nanophotonic device that confines light to a small observation volume and is a part of the SMRT Cell used during PacBio sequencing.
- Flow cell
A disposable component of short- and long-read sequencing platforms that houses the chemistry to sequence DNA or RNA.
- Subreads
The sequence of nucleotides derived from a single pass as a DNA polymerase traverses a DNA molecule multiple times. A subread is trimmed to exclude any adapter sequence.
- Single-pass
A single pass is one single iteration through a molecule by the DNA polymerase, used in the SMRT sequencing approach from PacBio.
- Homopolymer
A sequence run of two or more identical bases.
- Polishing tools
Computational methods used to improve genome assembly accuracy, in which sequencing reads are compared to the assembly in order to derive a more accurate consensus sequence.
- Sequencing coverage
The average number of unique reads that align to, or ‘cover’, a known reference sequence or genome.
- Circular consensus sequencing (CCS)
A process used by PacBio in which multiple overlapping reads from a circular DNA molecule are aligned to each other and the most likely base at each position is determined.
- Polymerase reads
The sequence of nucleotides derived from one or more passes of the DNA polymerase around a SMRTbell template, including both adapters and inserts. Polymerase reads are trimmed to exclude any low-quality regions.
- Read N50
The sequence length of the shortest read at 50% of the total sequencing dataset sorted by read length. In other words, half of the sequencing dataset is in reads larger than or equal to the read N50 size.
- ONT long read
A read generated by sequencing of DNA or RNA molecules that is typically 10–100 kb long.
- ONT ultra-long read
A read generated by ONT sequencing DNA or RNA molecules that is >100 kb long.
- Contig
A continuous (or ‘contiguous’) sequence of DNA created by assembling overlapping sequencing reads.
- Contig N50
The sequence length of the shortest contig at 50% of the total genome length sorted by contig length. In other words, half of the genome sequence is contained in contigs larger than or equal to the contig N50 size.
- Optical mapping
A technique commonly used to scaffold sequence contigs that involves constructing ordered genomic maps from single molecules of DNA with a fluorescent readout.
- Electronic mapping
A technique commonly used to scaffold sequence contigs that involves constructing ordered genomic maps from single molecules of DNA with an electronic readout.
- Phased de novo genome assembly
A genome assembly in which the maternal and paternal haplotypes are resolved.
- Trio binning
A method in which short reads from two parental genomes are used to partition long reads from an offspring into haplotype-specific sets prior to assembly of each haplotype.
- Paralogous sequence variants
Paralogous sequence variants are genomic differences that appear between the different copies (or paralogs) of segmental duplications.
- CHM13 human genome
A complete hydatidiform mole (CHM) genome that has lost maternal genetic information and duplicated the paternal information. This genome is currently the focus of the T2T genome assembly efforts due to its essentially haploid nature and stable karyotype.
- Whole-genome sequencing (WGS)
Sequencing of the entire genome without using methods for sequencing selection.
- Uniparental disomy
Inheritance of two copies of a chromosome or segments of a chromosome from one parent, instead of one copy from each parent.
- Expression quantitative trait loci (eQTLs)
Loci that explain a fraction of the genetic variant of a gene expression phenotype.
- Genome-wide association studies (GWAS)
An approach used in genetics research to associate specific genetic variations with particular traits.
- Variable number tandem repeats (VNTRs)
A genomic sequence of at least six base pairs that is repeated in tandem within a genome and varies in repeat number among individuals.
- Introgression
The transfer of genetic information from one species to another as a result of hybridization between them and repeat backcrossing.
- Squiggle
The sequencing output exclusively used by ONT, wherein DNA translocation through the nanopore causes shifts in voltage that are directly correlated to the k-mer within the pore.
Footnotes
COMPETING INTERESTS
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc.
WEBLINKS
Arrow: https://github.com/PacificBiosciences/GenomicConsensus
Nanopolish: https://github.com/jts/nanopolish
REFERENCES
- 1.van Dijk EL, Jaszczyszyn Y, Naquin D & Thermes C The third revolution in sequencing technology. Trends Genet. 34, 666–681 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Shendure J et al. DNA sequencing at 40: past, present and future. Nature 550, 345–353 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Sudmant PH et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudmant PH et al. Global diversity, population stratification, and selection of human copy-number variation. Science 349, aab3761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42, 790–793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kircher M et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonson TS et al. Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Sudmant PH et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; The study compares multiple sequence and mapping technologies for the genomes of three parent–child trios and quantifies the amount of missing genetic variation. They develop a method, Phased-SV, that partitions long-read data based on phased single-nucleotide polymorphisms, which resolves the sequence of both structural haplotypes.
- 10.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey JA, Yavor AM, Massa HF, Trask BJ & Eichler EE Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11, 1005–1017 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkinson A, Chen Y & Eyre-Walker A The large-scale distribution of somatic mutations in cancer genomes. Hum. Mutat 33, 136–143 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Hills M, Jeyapalan JN, Foxon JL & Royle NJ Mutation mechanisms that underlie turnover of a human telomere-adjacent segmental duplication containing an unstable minisatellite. Genomics 89, 480–489 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Hastings PJ, Lupski JR, Rosenberg SM & Ira G Mechanisms of change in gene copy number. Nat. Rev. Genet 10, 551–564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng GXY et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nature Biotechnology 34, 303–311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F et al. Haplotype phasing of whole human genomes using bead-based barcode partitioning in a single tube. Nat. Biotechnol 35, 852–857 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Wang O et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 29, 798–808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R et al. Illumina synthetic long read sequencing allows recovery of missing sequences even in the “finished” C. elegans genome. Scientific Reports 5, 1–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters BA et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature 487, 190–195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman-Aiden E et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghurye J et al. Integrating Hi-C links with assembly graphs for chromosome-scale assembly. PLOS Computational Biology 15, e1007273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garg S et al. Efficient chromosome-scale haplotype-resolved assembly of human genomes. bioRxiv 810341 (2019) doi: 10.1101/810341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harewood L et al. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biology 18, 125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu J, Mohamadi H, Warren RL, Yang C & Birol I Innovations and challenges in detecting long read overlaps: an evaluation of the state-of-the-art. Bioinformatics 33, 1261–1270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung H, Winefield C, Bombarely A, Prentis P & Waterhouse P Tools and strategies for long-read sequencing and de novo assembly of plant genomes. Trends Plant Sci. 24, 700–724 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Sedlazeck FJ, Lee H, Darby CA & Schatz MC Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nature Reviews Genetics 19, 329–346 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Chaisson MJP, Wilson RK & Eichler EE Genetic variation and the de novo assembly of human genomes. Nature Reviews Genetics 16, 627–640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollard MO, Gurdasani D, Mentzer AJ, Porter T & Sandhu MS Long reads: their purpose and place. Hum Mol Genet 27, R234–R241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantere T, Kersten S & Hoischen A Long-Read Sequencing Emerging in Medical Genetics. Front Genet 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kronenberg ZN et al. High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Audano PA et al. Characterizing the major structural variant alleles of the human genome. Cell 176, 663–675.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article provides a large catalog of sequence-resolved structural variants based on long-read sequence analysis of a diverse panel of 15 genomes and identifies instances where the human reference has a minor allele for a structural variant. It also develops a machine-learning-based approach for genotyping sequence-resolved structural variants in Illumina whole-genome shotgun sequence data, which led to the discovery of eQTLs and new lead variants for GWAS.
- 32.Huddleston J et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaisson MJP et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes one of the first methods for sequencing and assembling structural variation from long-read sequence data. It shows that the majority of these variants are novel, and, thus, a large amount of human genetic variation is missed with short-read sequencing approaches.
- 34.Miga KH et al. Telomere-to-telomere assembly of a complete human X chromosome. bioRxiv 735928 (2019) doi: 10.1101/735928. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows that PacBio and ONT long reads are able to generate a de novo genome assembly superior in contiguity to all other genome assemblies (including hg38). Importantly, it reveals the first T2T sequence assembly of a human chromosome and shows that it is possible to resolve megabase-sized arrays of near-identical tandem repeats (that is, the centromere) with long and ultra-long reads.
- 35.Jain M et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article demonstrates that ultra-long ONT reads can be used for de novo human genome assembly. Additionally, this assembly resolved both haplotypes of the human major histocompatibility (MHC) locus for the first time.
- 36.Shafin K et al. Efficient de novo assembly of eleven human genomes using PromethION sequencing and a novel nanopore toolkit. bioRxiv 715722 (2019) doi: 10.1101/715722. [DOI] [Google Scholar]; This article describes the rapid assembly of 11 human genomes using long ONT reads, and it debuts a new assembler (Shasta) and polisher (HELEN). This article provides the methodological basis for scalability in human genome assembly using long reads.
- 37.Payne A, Holmes N, Rakyan V & Loose M BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35, 2193–2198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rang FJ, Kloosterman WP & de Ridder J From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biology 19, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ardui S, Ameur A, Vermeesch JR & Hestand MS Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res 46, 2159–2168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carneiro MO et al. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics 13, 375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eid J et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Korlach J Understanding Accuracy in SMRT® Sequencing. 9. [Google Scholar]
- 43.Rhoads A & Au KF PacBio Sequencing and Its Applications. Genomics Proteomics Bioinformatics 13, 278–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weirather JL et al. Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis. F1000Res 6, 100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fox EJ, Reid-Bayliss KS, Emond MJ & Loeb LA Accuracy of next generation sequencing platforms. Next Gener Seq Appl 1, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chin C-S et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Walker BJ et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaser R, Sović I, Nagarajan N & Šikić M Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrison E & Marth G Haplotype-based variant detection from short-read sequencing. (2012).
- 50.Gordon D et al. Long-read sequence assembly of the gorilla genome. Science 352, aae0344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Fan J, Sun Z & Liu S NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- 52.Vollger MR et al. Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. Ann. Hum. Genet (2019) doi: 10.1111/ahg.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenger AM et al. Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology 519025 (2019) doi: 10.1101/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article introduces PacBio HiFi reads as a new data type and reveals the power of highly accurate (>99%), long (>10 kb) reads for de novo genome assembly and structural variant detection.
- 54.Vollger MR et al. Long-read sequence and assembly of segmental duplications. Nature Methods 16, 88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article quantifies the extent to which segmental duplications remain unassembled in long-read genomes. Additionally, it describes a method to locally reconstruct segmental duplications by partitioning long-read sequence data using paralogous sequence variant graphs and locally assembling them.
- 55.Nurk S et al. HiCanu: accurate assembly of segmental duplications and allelic variants from high-fidelity long reads. bioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miao H et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 155, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wick RR, Judd LM & Holt KE Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biology 20, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li C et al. INC-Seq: accurate single molecule reads using nanopore sequencing. Gigascience 5, 34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson BD, Eisenstein M & Soh HT High-fidelity nanopore sequencing of ultra-short DNA targets. Anal. Chem 91, 6783–6789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.1D squared kit available in the store: boost accuracy, simple prep. Oxford Nanopore Technologies http://nanoporetech.com/about-us/news/1d-squared-kit-available-store-boost-accuracy-simple-prep (2017).
- 61.Lewandowski K et al. Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. Journal of Clinical Microbiology 58, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charalampous T et al. Rapid diagnosis of lower respiratory infection using nanopore-based clinical metagenomics. bioRxiv 387548 (2018) doi: 10.1101/387548. [DOI] [Google Scholar]
- 63.Quick J et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pendleton M et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12, 780–786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okubo M et al. GGC repeat expansion of NOTCH2NLC in adult patients with leukoencephalopathy. Ann. Neurol (2019) doi: 10.1002/ana.25586. [DOI] [PubMed] [Google Scholar]
- 66.Sone J et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet 51, 1215–1221 (2019). [DOI] [PubMed] [Google Scholar]; The authors show that PacBio CLR and ONT long reads can detect structural variation in clinically relevant disease-risk genes, which were previously missed with short-read whole-exome and whole-genome sequencing.
- 67.Stefansson H et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharp AJ et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet 38, 1038–1042 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Hsieh P et al. Adaptive archaic introgression of copy number variants and the discovery of previously unknown human genes. Science 366, eaax2083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe large structural variants, originating in Neanderthals or Denisovans, that show signs of adaptation and positive selection in the Melanesian population. In particular, the paper uses long reads to assemble a 386 kb duplication polymorphism that is present in 79% of Melanesians but generally absent from other populations, demonstrating the importance of developing new human reference genomes.
- 70.Shi L et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun 7, 12065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seo J-S et al. De novo assembly and phasing of a Korean human genome. Nature 538, 243–247 (2016). [DOI] [PubMed] [Google Scholar]
- 72.International Human Genome Project Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 73.Koren S et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chin C-S & Khalak A Human genome assembly in 100 minutes. bioRxiv 705616 (2019) doi: 10.1101/705616. [DOI] [Google Scholar]; This article describes a unique and fast genome assembly algorithm called Peregrine that uses PacBio HiFi data. This long-read assembler is able to assemble a human genome in <100 minutes or ~30 CPU hours.
- 75.Chin C-S et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolmogorov M, Yuan J, Lin Y & Pevzner PA Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Ruan J & Li H Fast and accurate long-read assembly with wtdbg2. bioRxiv 530972 (2019) doi: 10.1101/530972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steinberg KM et al. High-quality assembly of an individual of Yoruban descent. bioRxiv 067447 (2016) doi: 10.1101/067447. [DOI] [Google Scholar]
- 79.Oliver JS et al. High-definition electronic genome maps from single molecule data. bioRxiv 139840 (2017) doi: 10.1101/139840. [DOI] [Google Scholar]
- 80.Udall JA & Dawe RK Is it ordered correctly? Validating genome assemblies by optical mapping. Plant Cell 30, 7–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ameur A et al. De novo assembly of two Swedish genomes reveals missing segments from the human GRCh38 reference and improves variant aalling of population-scale sequencing data. Genes (Basel) 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio] (2013). [Google Scholar]
- 83.Watson M & Warr A Errors in long-read assemblies can critically affect protein prediction. Nat. Biotechnol 37, 124–126 (2019). [DOI] [PubMed] [Google Scholar]
- 84.Koren S, Phillippy AM, Simpson JT, Loman NJ & Loose M Reply to ‘Errors in long-read assemblies can critically affect protein prediction’. Nat. Biotechnol 37, 127–128 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Loman NJ, Quick J & Simpson JT A complete bacterial genome assembled de novo using only nanopore sequencing data. Nature Methods 12, 733–735 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Simpson JT et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]; The authors unveil a method to detect methylated cytosines in raw ONT reads based on characteristic signal disruptions in ONT data using a computational tool called Nanopolish. This tool has been used to map methylation within the centromere for the first time.
- 87.Koren S et al. De novo assembly of haplotype-resolved genomes with trio binning. Nature Biotechnology 36, 1174–1182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate a method to phase haplotypes for de novo genome assembly known as ‘trio binning’ in which reads from the parents are used to identity and partition reads from the child into haplotypes prior to sequence assembly.
- 88.Porubský D et al. Direct chromosome-length haplotyping by single-cell sequencing. Genome Res 26, 1565–1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patterson M et al. WhatsHap: weighted haplotype assembly for future-generation sequencing reads. Journal of Computational Biology 22, 498–509 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Kronenberg ZN et al. Extended haplotype phasing of de novo genome assemblies with FALCON-Phase. bioRxiv 327064 (2019) doi: 10.1101/327064. [DOI] [Google Scholar]
- 91.Porubsky D et al. A fully phased accurate assembly of an individual human genome. bioRxiv 855049 (2019) doi: 10.1101/855049. [DOI] [Google Scholar]
- 92.Eichler EE Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17, 661–669 (2001). [DOI] [PubMed] [Google Scholar]
- 93.Rodriguez OL, Ritz A, Sharp AJ & Bashir A MsPAC: A tool for haplotype-phased structural variant detection. Bioinformatics (2019) doi: 10.1093/bioinformatics/btz618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bzikadze AV & Pevzner PA centroFlye: Assembling centromeres with long error-prone reads. bioRxiv 772103 (2019) doi: 10.1101/772103. [DOI] [Google Scholar]
- 95.Ebbert MTW et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biology 20, 97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feng Z et al. Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLoS Comput. Biol 9, e1002935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sedlazeck FJ et al. Accurate detection of complex structural variations using single molecule sequencing. Nat Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chaisson MJ & Tesler G Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berlin K et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nature Biotechnology 33, 623–630 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Mizuguchi T et al. A 12-kb structural variation in progressive myoclonic epilepsy was newly identified by long-read whole-genome sequencing. J. Hum. Genet 64, 359–368 (2019). [DOI] [PubMed] [Google Scholar]
- 102.Merker JD et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet. Med 20, 159–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zeng S et al. Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with familial cortical myoclonic tremor with epilepsy. J. Med. Genet 56, 265–270 (2019). [DOI] [PubMed] [Google Scholar]
- 104.Reiner J et al. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet–Biedl Syndrome 9 (BBS9) deletion. NPJ Genom Med 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sato N et al. Spinocerebellar ataxia type 31 is associated with ‘inserted’ penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet 85, 544–557 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dutta UR et al. Breakpoint mapping of a novel de novo translocation t(X;20)(q11.1;p13) by positional cloning and long read sequencing. Genomics 111, 1108–1114 (2019). [DOI] [PubMed] [Google Scholar]
- 107.de Jong LC et al. Nanopore sequencing of full-length BRCA1 mRNA transcripts reveals co-occurrence of known exon skipping events. Breast Cancer Res. 19, 127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wenzel A et al. Single molecule real time sequencing in ADTKD- MUC1 allows complete assembly of the VNTR and exact positioning of causative mutations. Sci Rep 8, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ishiura H et al. Noncoding CGG repeat expansions in neuronal intranuclear inclusion disease, oculopharyngodistal myopathy and an overlapping disease. Nat. Genet 51, 1222–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Aneichyk T et al. Dissecting the causal mechanism of X-linked Dystonia-Parkinsonism by integrating genome and transcriptome assembly. Cell 172, 897–909.e21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Song JHT, Lowe CB & Kingsley DM Characterization of a human-specific tandem repeat associated with bipolar disorder and schizophrenia. Am. J. Hum. Genet 103, 421–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carvalho CMB et al. Interchromosomal template-switching as a novel molecular mechanism for imprinting perturbations associated with Temple syndrome. Genome Med 11, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Giesselmann P et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nature Biotechnology 37, 1478–1481 (2019). [DOI] [PubMed] [Google Scholar]
- 114.Sulovari A et al. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc. Natl. Acad. Sci. U.S.A (2019) doi: 10.1073/pnas.1912175116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lei XX et al. TTTCA repeat expansion causes familial cortical myoclonic tremor with epilepsy. Eur. J. Neurol 26, 513–518 (2019). [DOI] [PubMed] [Google Scholar]
- 116.Fiddes IT et al. Human-specific NOTCH2NL genes affect Notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suzuki IK et al. Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mefford HC et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. New England Journal of Medicine 359, 1685–1699 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brunetti-Pierri N et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet 40, 1466–1471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.He Y et al. Long-read assembly of the Chinese rhesus macaque genome and identification of ape-specific structural variants. Nat Commun 10, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Advancing the reference sequence of the human genome. Genome.gov https://www.genome.gov/news/news-release/NIH-funds-centers-for-advancing-sequence-of-human-genome-reference.
- 122.Garalde DR et al. Highly parallel direct RNA sequencing on an array of nanopores. Nature Methods 15, 201–206 (2018). [DOI] [PubMed] [Google Scholar]; The authors describe a method to sequence full-length native RNA molecules with ONT sequencing technologies, simplifying the process by removing the steps to convert RNA into cDNA prior to sequencing.
- 123.Workman RE et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. bioRxiv 459529 (2018) doi: 10.1101/459529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Soneson C et al. A comprehensive examination of Nanopore native RNA sequencing for characterization of complex transcriptomes. Nat Commun 10, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flusberg BA et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7, 461–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vilfan ID et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J Nanobiotechnology 11, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rand AC et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sharon D, Tilgner H, Grubert F & Snyder M A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol 31, 1009–1014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Au KF et al. Characterization of the human ESC transcriptome by hybrid sequencing. PNAS 110, E4821–E4830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows that full-length mRNA transcripts can be sequenced from end to end to identify novel gene isoforms using a PacBio method called Iso-Seq. This article also provides a catalog of the poly(A) transcriptome in human embryonic stem cells (hESCs) using a combination of Iso-Seq and short-read sequencing data.
- 130.Byrne A et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun 8, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oikonomopoulos S, Wang YC, Djambazian H, Badescu D & Ragoussis J Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations. Scientific Reports 6, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dougherty ML et al. Transcriptional fates of human-specific segmental duplications in brain. Genome Res. 28, 1566–1576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Volden R et al. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. PNAS 115, 9726–9731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aird D et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biology 12, R18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clark MB et al. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Mol Psychiatry 1–11 (2019) doi: 10.1038/s41380-019-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tang AD et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. bioRxiv 410183 (2018) doi: 10.1101/410183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Clark TA et al. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40, e29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang Y et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE 5, e8888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pacific Biosciences: Detecting DNA base modifications using single molecule, real-time sequencing. https://www.pacb.com/wp-content/uploads/2015/09/WP_Detecting_DNA_Base_Modifications_Using_SMRT_Sequencing.pdf (2015).
- 140.Frommer M et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A 89, 1827–1831 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.An N, Fleming AM, White HS & Burrows CJ Nanopore detection of 8-oxoguanine in the human telomere repeat sequence. ACS Nano 9, 4296–4307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu H et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leger A et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. bioRxiv 843136 (2019) doi: 10.1101/843136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lorenz DA, Sathe S, Einstein JM & Yeo GW Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base specific resolution. RNA rna.072785.119 (2019) doi: 10.1261/rna.072785.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li Y & Tollefsbol TO DNA methylation detection: Bisulfite genomic sequencing analysis. Methods Mol Biol 791, 11–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schaefer M, Pollex T, Hanna K & Lyko F RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37, e12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Levanon EY et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol 22, 1001–1005 (2004). [DOI] [PubMed] [Google Scholar]
- 148.Incarnato D et al. High-throughput single-base resolution mapping of RNA 2΄-O-methylated residues. Nucleic Acids Res 45, 1433–1441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bakin AV & Ofengand J Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol 77, 297–309 (1998). [DOI] [PubMed] [Google Scholar]
- 150.Tsai Y-C et al. Amplification-free, CRISPR-Cas9 targeted enrichment and SMRT sequencing of repeat-expansion disease causative genomic regions. bioRxiv 203919 (2017) doi: 10.1101/203919. [DOI] [Google Scholar]
- 151.Hafford-Tear NJ et al. CRISPR/Cas9-targeted enrichment and long-read sequencing of the Fuchs endothelial corneal dystrophy–associated TCF4 triplet repeat. Genetics in Medicine 21, 2092–2102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Suzuki Y et al. AgIn: measuring the landscape of CpG methylation of individual repetitive elements. Bioinformatics 32, 2911–2919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ni P et al. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics doi: 10.1093/bioinformatics/btz276. [DOI] [PubMed] [Google Scholar]
- 154.McIntyre ABR et al. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat Commun 10, 579 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu Q et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat Commun 10, 2449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stoiber M et al. De novo identification of DNA modifications enabled by genome-guided nanopore signal processing. bioRxiv 094672 (2017) doi: 10.1101/094672. [DOI] [Google Scholar]
- 157.Lee I et al. Simultaneous profiling of chromatin accessibility and methylation on human cell lines with nanopore sequencing. bioRxiv 504993 (2019) doi: 10.1101/504993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Beyter D et al. Long read sequencing of 1,817 Icelanders provides insight into the role of structural variants in human disease. bioRxiv 848366 (2019) doi: 10.1101/848366. [DOI] [PubMed] [Google Scholar]
- 159.National Institutes of Health (NIH) — All of Us. https://allofus.nih.gov/.
- 160.Garrison E et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nature Biotechnology 36, 875–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schneider VA et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27, 849–864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li R et al. Building the sequence map of the human pan-genome. Nat. Biotechnol 28, 57–63 (2010). [DOI] [PubMed] [Google Scholar]
- 163.Gnerre S et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. PNAS 108, 1513–1518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sanders AD, Falconer E, Hills M, Spierings DCJ & Lansdorp PM Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat Protoc 12, 1151–1176 (2017). [DOI] [PubMed] [Google Scholar]
- 165.Thrombocytopenia-absent radius syndrome: Background, pathophysiology, epidemiology. https://reference.medscape.com/article/959262-overview.
- 166.Rosenfeld JA et al. Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur J Hum Genet 20, 754–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


