Abstract
Schistosome parasites kill 250,000 people every year. Treatment of schistosomiasis relies on the drug praziquantel. Unfortunately, a scarcity of molecular tools has hindered the discovery of new drug targets. Here, we describe a large-scale RNA interference screen in adult Schistosoma mansoni examining the function of 2,216 genes. We discovered 250 genes with phenotypes affecting neuromuscular function, tissue integrity, stem cell maintenance, and parasite survival. Leveraging these data, we prioritize compounds with activity against the parasites and uncover a pair of protein kinases (TAO and STK25) that cooperate to maintain muscle-specific mRNA transcription. Loss of either of these kinases results in paralysis and worm death in a mammalian host. These studies may help expedite therapeutic development and invigorate studies of these neglected parasites.
One Sentence Summary:
An RNAi screen examines the function of 20% of S. mansoni genes uncovering new phenotypes and suggesting new therapeutic targets.
Studies of gene function in intra-mammalian schistosome parasites have been limited to relatively small numbers of genes(1). Therefore, we developed a large-scale RNAi screening platform on adult schistosomes that prioritized a list of 2,320 of the worm’s ~10,000 protein coding genes (Fig. S1A, Table S1). We generated dsRNAs, and treated adult male and female pairs with dsRNA over the course of a 30-day experiment (Fig. 1A). After filtering genes that either did not amplify by PCR, or failed to generate sufficient concentrations of dsRNA, a total of 2,216 genes were screened (Table S1).
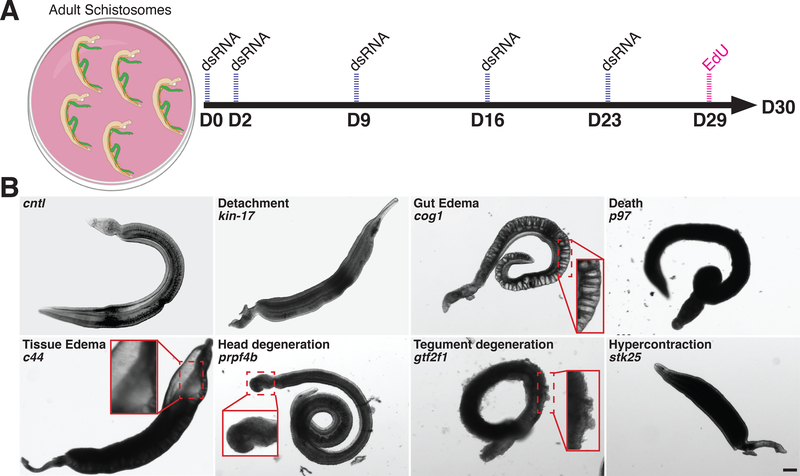
Fig. 1. Summary of RNAi phenotypes.
(A) RNAi treatment regime. Parasites were monitored for visible abnormalities and at D29 EdU was added to media to label proliferative cells. (B) Categories of RNAi phenotypes observed. kin-17 (Smp_023250), cog1 (Smp_132980), p97 (Smp_018240), c44 (Smp_136260), prpf4b (Smp_068960), gtf2f1 (Smp_088460), stk25 (Smp_096640). Scale bar: 100 μm
Schistosomes live in the veins surrounding the host intestines, and attach to the vascular endothelium to avoid being swept away in the blood and trapped in host organs. Under in vitro culture conditions healthy parasites attach to the substrate with a combination of their oral and ventral suckers (Movie S1). We thus reasoned that substrate attachment would be a useful quantitative metric to define RNAi treatments that affect parasite vitality and predict in vivo survival. Therefore, we monitored parasites for 30 days to identify substrate attachment and other visible defects.
Schistosomes possess adult somatic stem cells (neoblasts) that rejuvenate parasite tissues, including the intestine and tegument (skin)(2, 3). The parasites also contain large numbers of proliferative germline stem cells (GSCs) in their reproductive organs(2) which are essential for producing eggs; the central driver of parasite-induced pathology in vivo(4). Therefore, we monitored the maintenance of neoblasts and GSCs by labeling parasites with the thymidine analog EdU prior to the conclusion of the experiment (Fig. 1A). Due to the variable rate at which the reproductive organs of female worms degenerate during in vitro culture(5), stem cell proliferation was only monitored in male worms. For genes with RNAi phenotypes uncovered during our screen, we confirmed gene identity by sequencing, and mitigated potential off-target effects by designing an additional dsRNA targeting a non-overlapping gene region or by examining sequence identity of hits with other S. mansoni genes (Fig. S1). These studies identified 195 genes with fully-penetrant attachment phenotypes of which 121 possessed phenotypes in addition to attachment, including: tissue and intestinal edema (36), head (26) and/or tegument (78) degeneration, muscular hypercontraction (6), and death (36) (Fig. 1B, Table S2). RNAi of an additional 66 genes resulted in stem cell maintenance defects but caused no other visible phenotypes (e.g., attachment) suggesting an essential role for stem cell maintenance (Fig. S2, Table S3). We cannot rule out the possibility of false negatives among the genes with no phenotype and encourage greater scrutiny of such genes by alternate knockdown approaches or analysis of different phenotypic readouts.
Of the 66 genes required for stem cell survival, RNAi of over 90% (60/66) led to defects in the maintenance of both proliferative cells in the male testes and neoblasts (Fig. S2). However, for a minority of genes this maintenance defect appeared to be specific to either proliferative cells in the testes or the neoblasts (Fig. S2). Gene Ontology enrichment analysis identified genes important for protein translation, including gene products involved in ribosomal structure, tRNA aminoacylation, and rRNA processing as putative regulators of proliferative cell maintenance (Fig. S3A). Although this could reflect an enhanced sensitivity of actively proliferating cells to alterations in protein translation, studies have highlighted “non-housekeeping” roles for translational regulators in controlling stem cell function(6).
Of the 195 genes essential for parasite attachment, a large fraction share sequence similarity with other organisms, including other medically-relevant schistosome species (Table S4). Additionally, most genes with an attachment phenotype (172/195 genes; 88%) possess a close homolog from C. elegans or D. melanogaster that likewise has a loss-of-function phenotype (Table S5). Gene Ontology analyses of genes with attachment phenotypes further revealed that the dominant group of enriched genes were those encoding components necessary for protein turnover via the ubiquitin-proteasome system (UPS) (Fig. S3B). Proteolysis is important for larval and adult viability in vitro(7, 8) and our data identified that key components from virtually every arm of the UPS were required for adult parasite vitality during in vitro culture (Fig. S4A). Indeed, RNAi of nearly all of UPS components resulted in extensive tissue degeneration and in some cases adult parasite death (Fig. S4B–C).
To determine if any genes associated with attachment phenotypes encoded proteins targeted by existing pharmacological agents, we searched the literature and the ChEMBL database(9) (Table S6). This analysis uncovered 205 compounds potentially targeting 49 S. mansoni proteins. We selected 14 of these compounds (Table S7) and examined the activities of these compounds on worms cultured in vitro either by automated worm movement tracking or parasite attachment. More than half of the compounds tested (8/14) on worms at 10 μM reduced parasite movement >75% and half of the compounds (7/14) caused fully-penetrant substrate attachment defects by D7 post-treatment (Fig. 2A–B, Movie S2). Similarly, 7 of these compounds affected the movement of post-infective larvae (schistosomula), suggesting activity against multiple life-cycle stages (Table S8).
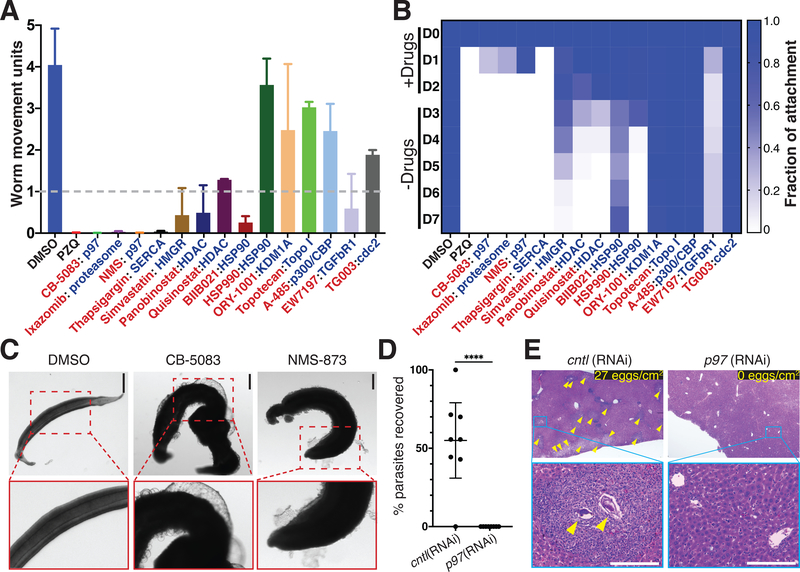
Fig. 2. Compounds prioritized from RNAi studies have effects on schistosomes in vitro.
(A) Compounds (red text) predicted to target schistosome proteins (blue text) essential for parasite attachment at 10 μM for their effects on worm motility. n = 12 worm pairs, n = 3 biological replicates; data are mean +/− SD. Dashed line, threshold for 75% reduction in motility (B) Heatmap showing time course measuring the fraction of male worms attached to the substrate following treatment with compounds for 72 hours as in A. n = 3 biological replicates (C) Effects of CB-5083 or NMS-873 (10 μM, 72 hrs) treatment on male parasites. (D) Percent recovery p97(RNAi) or control(RNAi) male parasites surgically transplanted into mice. n = 8 transplants for each group. ****, p < 0.0001, t-test; data are mean +/− 95% CI. (E) Hematoxylin and Eosin staining of livers from mice receiving control or p97(RNAi) parasites. Egg-induced granulomas (yellow arrows). (top right) counts of eggs/cm liver section are shown, counts from sections from n = 3 livers. Scale bars: C, E, 100 μm.
Among the compounds that emerged from these studies was simvastatin, an HMG-CoA reductase inhibitor, with known effects on parasites both in vitro and in vivo(10). The proteasome inhibitor ixazomib affected both schistosome movement (Fig. 2A) and attachment (Fig. 2B), mirroring a recent report using the proteasome inhibitor bortezomib (7). However, the most potent effects on adult parasites were observed with CB-5083 and NMS-873 that inhibit the UPS component p97 by either competing with ATP (11) or binding to an allosteric site(12), respectively (Fig. S5A). Similar to the death observed following long-term p97 RNAi treatment (Fig. 1B), both p97 inhibitors led to death in vitro (Movie S3). Despite their distinct biochemical mechanisms of action, we noted similar deformations in the structure of the parasite tegument following treatment with either CB-5083 or NMS-873, suggesting that these compounds have similar pharmacological effects on the parasite (Fig. 2C).
We then assessed if NMS-873 and CB-5083 affected UPS function by measuring the accumulation of ubiquitinated proteins using an antibody that recognizes K48 polyubiquitinated proteins marked for proteasome-mediated destruction(13). We observed the accumulation of polyubiquitinated proteins following RNAi of p97 and after treatment of schistosomes with either CB-5083 or NMS-873 (Fig. S5B). Consistent with CB-5083 or NMS-873 acting via p97 inhibition to blunt protein degradation, we found that p97 RNAi-treatment together with low concentrations of either drug led to additive increases in polyubiquitinated protein accumulation (Fig. S5C). We also observed accumulation of polyubiquitinated proteins following either RNAi of proteasome subunit beta type-2 or treatment with ixazomib (Fig. S5D). These effects on the degradation of ubiquitinated proteins appeared to be specific to inhibition of UPS function, rather than a non-specific effect due to reduced worm vitality (Fig. S5D).
We then depleted UPS components using RNAi and surgically transplanted these worms into the mesenteric veins of recipient mice (Fig. S6A) and measured parasite egg deposition in host tissues and parasite survival. Following hepatic portal perfusion, we recovered about 55% of transplanted control RNAi-treated worms (Figs. 2D, S6B) and these parasites established patent infections, depositing large number of eggs into the livers of recipient mice (Figs. 2E, S6C–D). In contrast, we failed to recover parasites following hepatic portal perfusion from mice receiving p97 (Fig. 2D) or proteasome subunit beta type-2 (Fig. S6B). As a consequence, the livers in these mice were devoid of eggs and we observed no signs of egg-induced granulomas (Figs. 2E, S6C–D). We also observed RNAi-treated parasites at various stages of infiltration by host immune cells (Fig. S6E–F). This suggests that these parasites are unable to remain in the portal vasculature and are cleared via the liver by the immune system. Taken together, these data highlight the essentially and druggability of the schistosome UPS.
Another group of potentially druggable targets to emerge from our RNAi screen were protein kinases, 19 of which led to defects in either parasite attachment or stem cell maintenance. RNAi of two STE20 serine-threonine kinases: tao and stk25, which are homologs of the human TAO1/2/3 and STK25/YSK1 protein kinases, respectively, led to rapid detachment from the substrate (Fig. S7A) and a concomitant posterior paralysis and hypercontraction of the body, such that the parasites took on a distinctive banana-shaped morphology (Figs. 3A, S7B, Movie S4). Aside from RNAi of stk25 and tao, this banana-shaped phenotype of worms was unique, only observed following RNAi of a CCM3/PDCD10 homolog (Smp_031950), a known heterodimerization partner with the mammalian STK25 kinase(14). We failed to observe death of either stk25- or tao- depleted parasites during in vitro culture. However, following surgical transplantation, we noted a reduction in the recovery of tao or stk25 RNAi-treated parasites from recipient mice and these recipient mice displayed few signs of egg-induced granuloma formation (Figs. 3B–C, S7C–D). Thus, both tao and stk25 appear to be essential for schistosome survival in vivo.
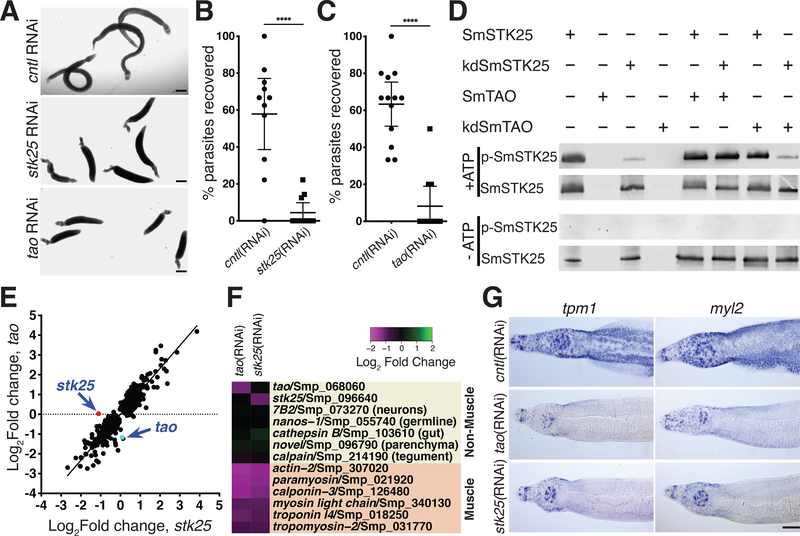
Fig. 3. The protein kinases SmSTK25 and SmTAO are essential to maintain muscular function.
(A) RNAi of stk25 or tao causes parasite hypercontraction. (B-C) Percent recovery stk25(RNAi), tao(RNAi), or control(RNAi) male parasites surgically transplanted into mice. n ≥ 11 transplants for each group. ****, p < 0.0001, t-test; data are mean +/− 95% CI. (D) Western blot to detect phospho-T173 (p-SmSTK25) or total SmSTK25 following an in vitro kinase reaction +/− ATP. kinase dead SmTAO(kdSmTAO). Representative of n = 2 experiments. (E) Scatter plot showing the relationship between the differentially expressed genes (p adj < 0.001, Benjamini-Hochberg corrected Wald test) following stk25 or tao RNAi-treatment (r = 0.9, p < 0.0001, Pearson correlation). (F) Heatmap showing expression of tissue-specific transcripts following RNAi of tao or stk25. (G). in situ hybridization for tropomyosin 1 (tpm1, Smp_340760) and a myosin light chain (myl2, Smp_132670) following RNAi at D13. Scale bars: A, 500 μm; G, 100 μm.
Given the unique and specific nature of the stk25 and tao associated “banana” phenotype we reasoned that these kinases may be acting in concert to mediate signaling processes in the worm. The Drosophila STK25 ortholog (GCK3) is a substrate of TAO and these proteins function in a signaling cascade essential for tracheal development(15). Likewise we observed that recombinant S. mansoni STK25 (SmSTK25) could serve as a substrate for the S. mansoni TAO (SmTAO) in an in vitro kinase assay (Fig. S8A–B). The human STK25 is activated by phosphorylation of a conserved threonine residue within its activation loop(16). By mass spectrometry we observed that this conserved threonine within the predicted SmSTK25 activation loop (T173) was phosphorylated following incubation of recombinant SmTAO with catalytically inactive SmSTK25 (Fig. S8C–D). Using an antibody that recognizes activation loop phosphorylation in STK25 orthologs(16), western blotting revealed SmSTK25 T173 autophosphorylation following an in vitro kinase reaction; this signal was abrogated in controls lacking ATP and when the SmSTK25 catalytic K48 residue was mutated to R (Figs. 3D, S8E). Consistent with our mass spectrometry results, we detected robust phosphorylation of T173 when recombinant SmTAO was incubated with kinase dead SmSTK25 (Fig. 3D), suggesting that SmTAO can phosphorylate a key residue for the activation of the mammalian STK25.
We reasoned that the schistosome TAO and STK25 might be acting in a signaling module to mediate critical processes in the parasite. To define these processes, we performed transcriptional profiling on RNAi-treated parasites just prior to observing detachment and hypercontraction (Day 6 and Day 9 for tao and stk25 RNAi treatments, respectively) (Fig. S9A–B). We found that expression of differentially regulated genes following RNAi of either tao or stk25 were correlated (Fig. 3E) and that more than half of these differentially regulated genes were common in both datasets (Fig. S9C, Table S9). Importantly, RNAi of either tao or stk25 was specific, not affecting expression of the other kinase gene of this pair (Fig. 3E–F).
Examining the tissue-specific expression of differentially-regulated genes on an adult schistosome single cell expression atlas using cells from schistosome somatic tissues(17), we found that roughly 40% (51/129) of the most down-regulated genes following tao and stk25 RNAi were specific markers of parasite muscle cells (Fig. S10A–C, Table S9). Indeed, nearly half of all mRNAs specifically-enriched in muscle cells (60/135) from this single cell atlas, including key muscle contractile proteins, were down-regulated following RNAi of both tao and stk25 (Figs. 3F, S10B, D, Table S10). Importantly, these transcriptional effects appeared to be largely specific to parasite muscles (Figs. 3F, S10B, Table S10).
In principle, loss of muscle-specific mRNAs could be due to either loss of muscle cells or down-regulation of muscle-specific mRNAs. We thus labeled F-actin in schistosome muscle fibers with phalloidin and performed in situ hybridization to detect muscle-specific mRNAs. RNAi-treated parasites exhibited a reduction in the expression of mRNAs encoding the contractile proteins Tropomyosin 1 and a Myosin Light Chain by in situ hybridization (Fig. 3G), but no qualitative defects in phalloidin staining of body wall muscles (Fig. S11). Thus, it appears that these kinases are required to maintain the transcription of many muscle-specific mRNAs in intact muscle cells. Interestingly, the heads of tao and stk25 RNAi parasites retained their capacity for movement (Movie S4) and partially maintained the expression of muscle-specific mRNAs (Fig. 3G). Thus, there appears to be a relationship between the maintenance of muscle-specific mRNA expression and locomotion.
Taken in their entirety, our data suggest that STK25 and TAO kinases cooperate in the schistosome to mediate signaling essential for sustaining the transcription of muscle-specific mRNAs. As a consequence, loss of either SmSTK25 or SmTAO activity results in muscular function defects compromising parasite survival in vivo. As whole-body knockouts of mouse STK25 are homozygous viable displaying no obvious deleterious phenotypes(18), SmSTK25 represents an attractive target for therapeutic intervention. Similar cross-examination of genes associated with other phenotypic classes (e.g., tissue edema) may provide insights into the specificity of the various phenotypes observed in this work.
Technological advances have paved the way for large-scale analyses of gene function in protozoan parasites(19–21), but, have been lacking in helminth parasites. Our systematic analysis of gene function in schistosomes allowed us to effectively prioritize targets and potential specific inhibitors with activities on worms. Future efforts should not only further explore the potency and selectivity of compounds our studies have uncovered (Fig. 2A–B, Table S6), but larger libraries of compounds with known molecular targets (22). However, to mitigate potential issues with host toxicity it is also worth exploring whether parasite-selective inhibitors of validated target proteins can be uncovered. Not only does this study enhance our understanding of schistosome biology, it provides a new lens to prioritize genes of interest in other medically- and agriculturally-important parasitic flatworms. Collectively, we anticipate this study will expedite the discovery of new anti-helminthics.
Supplementary Material
Acknowledgements:
We thank M. McConathy, C. Furrh, and G. Pagliuca for technical assistance, F. Hunter and N. Bosc for advice about retrieving data from ChEMBL, and M. Cobb and E. Ross for suggestions regarding kinase studies. Infected mice and B. glabrata snails were provided by the National Institute of Allergy and Infectious Diseases (NIAID) Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD, USA) through National Institutes of Health (NIH)-NIAID Contract HHSN272201700014I for distribution through BEI Resources.
Funding: The work was supported by the National Institutes of Health R01AI121037 (JJC), the Welch Foundation I-1948-20180324 (JJC), the Burroughs Wellcome Fund (JJC), and the Wellcome Trust 107475/Z/15/Z (JJC/KFH/MB) and 206194 (MB). CP was supported by Howard Hughes Medical Institute Gilliam Fellowship and National Science Foundation Graduate Research Fellowship SPA0001848.
Footnotes
Competing interests: The authors declare no competing interest.
Data and materials availability: Videos of RNAi attachment phenotypes can be accessed at www.collinslab.org/schistocyte or at https://doi.org/10.5061/dryad.zs7h44j4v. RNAseq analyses have been deposited in the NCBI Gene Expression Omnibus GSE146720.
References
- 1.Guidi A et al. , Application of RNAi to Genomic Drug Target Validation in Schistosomes. PLoS Negl Trop Dis 9, e0003801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins JJ III et al. , Adult somatic stem cells in the human parasite Schistosoma mansoni. Nature 494, 476–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins JJ III, Wendt GR, Iyer H, Newmark PA, Stem cell progeny contribute to the schistosome host-parasite interface. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearce EJ, MacDonald AS, The immunobiology of schistosomiasis. Nat Rev Immunol 2, 499–511 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Chen R, Collins JJ III, Systematically improved in vitro culture conditions reveal new insights into the reproductive biology of the human parasite Schistosoma mansoni. PLoS Biol 17, e3000254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buszczak M, Signer RA, Morrison SJ, Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159, 242–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibo-Verdugo B et al. , The Proteasome as a Drug Target in the Metazoan Pathogen, Schistosoma mansoni. ACS Infect Dis, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nabhan JF, El-Shehabi F, Patocka N, Ribeiro P, The 26S proteasome in Schistosoma mansoni: bioinformatics analysis, developmental expression, and RNA interference (RNAi) studies. Exp Parasitol 117, 337–347 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Mendez D et al. , ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 47, D930–D940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojo-Arreola L et al. , Chemical and genetic validation of the statin drug target to treat the helminth disease, schistosomiasis. PLoS One 9, e87594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson DJ et al. , Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 28, 653–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnaghi P et al. , Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat Chem Biol 9, 548–556 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Newton K et al. , Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Ceccarelli DF et al. , CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J Biol Chem 286, 25056–25064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poon CLC et al. , A Hippo-like Signaling Pathway Controls Tracheal Morphogenesis in Drosophila melanogaster. Dev Cell 47, 564–575 e565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preisinger C et al. , YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14–3-3zeta. J Cell Biol 164, 1009–1020 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendt GR et al. , A single-cell RNAseq atlas of the pathogenic stage of Schistosoma mansoni identifies a key regulator of blood feeding. bioRxiv, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amrutkar M et al. , Genetic Disruption of Protein Kinase STK25 Ameliorates Metabolic Defects in a Diet-Induced Type 2 Diabetes Model. Diabetes 64, 2791–2804 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alsford S et al. , High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res 21, 915–924 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bushell E et al. , Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 170, 260–272 e268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidik SM et al. , A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 166, 1423–1435 e1412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janes J et al. , The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc Natl Acad Sci U S A 115, 10750–10755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basch PF, Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J Parasitol 67, 179–185 (1981). [PubMed] [Google Scholar]
- 24.Collins JJ III et al. , Genome-Wide Analyses Reveal a Role for Peptide Hormones in Planarian Germline Development. PLoS Biol 8, e1000509 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins JJ III, King RS, Cogswell A, Williams DL, Newmark PA, An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl Trop Dis 5, e1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindelin J et al. , Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins JN, Collins JJ III, Tissue Degeneration following Loss of Schistosoma mansoni cbp1 Is Associated with Increased Stem Cell Proliferation and Parasite Death In Vivo. PLoS Pathog 12, e1005963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Helminth Genomes Consortium, Comparative genomics of the major parasitic worms. Nat Genet 51, 163–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamore SD et al. , Deconvoluting Kinase Inhibitor Induced Cardiotoxicity. Toxicol Sci 158, 213–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch JJ 3rd, Van Vleet TR, Mittelstadt SW, Blomme EAG, Potential functional and pathological side effects related to off-target pharmacological activity. J Pharmacol Toxicol Methods 87, 108–126 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Bowes J et al. , Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat Rev Drug Discov 11, 909–922 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Whatley KCL et al. , The repositioning of epigenetic probes/inhibitors identifies new anti-schistosomal lead compounds and chemotherapeutic targets. PLoS Negl Trop Dis 13, e0007693 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcellino C et al. , WormAssay: a novel computer application for whole-plate motion-based screening of macroscopic parasites. PLoS Negl Trop Dis 6, e1494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteland HL et al. , An Abies procera-derived tetracyclic triterpene containing a steroid-like nucleus core and a lactone side chain attenuates in vitro survival of both Fasciola hepatica and Schistosoma mansoni. Int J Parasitol Drugs Drug Resist 8, 465–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendt GR et al. , Flatworm-specific transcriptional regulators promote the specification of tegumental progenitors in Schistosoma mansoni. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trudgian DC, Mirzaei H, Cloud CPFP: a shotgun proteomics data analysis pipeline using cloud and high performance computing. J Proteome Res 11, 6282–6290 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Trudgian DC et al. , CPFP: a central proteomics facilities pipeline. Bioinformatics 26, 1131–1132 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Craig R, Beavis RC, TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Geer LY et al. , Open mass spectrometry search algorithm. Journal of proteome research 3, 958–964 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Elias JE, Gygi SP, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods 4, 207–214 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Trudgian DC, Singleton R, Cockman ME, Ratcliffe PJ, Kessler BM, ModLS: post-translational modification localization scoring with automatic specificity expansion. J. Proteomics Bioinform 5, 283–289 (2012). [Google Scholar]
- 43.Bolt BJ et al. , Using WormBase ParaSite: An Integrated Platform for Exploring Helminth Genomic Data. Methods Mol Biol 1757, 471–491 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Rozanski A et al. , PlanMine 3.0-improvements to a mineable resource of flatworm biology and biodiversity. Nucleic Acids Res 47, D812–D820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris TW et al. , WormBase: a modern Model Organism Information Resource. Nucleic Acids Res 48, D762–D767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurmond J et al. , FlyBase 2.0: the next generation. Nucleic Acids Res 47, D759–D765 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou HJ et al. , Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the p97 AAA ATPase (CB-5083). J Med Chem 58, 9480–9497 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Kupperman E et al. , Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res 70, 1970–1980 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Treiman M, Caspersen C, Christensen SB, A tool coming of age: thapsigargin as an inhibitor of sarco-endoplasmic reticulum Ca(2+)-ATPases. Trends Pharmacol Sci 19, 131–135 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Slater EE, MacDonald JS, Mechanism of action and biological profile of HMG CoA reductase inhibitors. A new therapeutic alternative. Drugs 36 Suppl 3, 72–82 (1988). [DOI] [PubMed] [Google Scholar]
- 51.Scuto A et al. , The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph- acute lymphoblastic leukemia cells. Blood 111, 5093–5100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arts J et al. , JNJ-26481585, a novel “second-generation” oral histone deacetylase inhibitor, shows broad-spectrum preclinical antitumoral activity. Clin Cancer Res 15, 6841–6851 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Lundgren K et al. , BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol Cancer Ther 8, 921–929 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Menezes DL et al. , The novel oral Hsp90 inhibitor NVP-HSP990 exhibits potent and broad-spectrum antitumor activities in vitro and in vivo. Mol Cancer Ther 11, 730–739 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Maes T et al. , ORY-1001, a Potent and Selective Covalent KDM1A Inhibitor, for the Treatment of Acute Leukemia. Cancer Cell 33, 495–511 e412 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Caceres G et al. , Determination of chemotherapeutic activity in vivo by luminescent imaging of luciferase-transfected human tumors. Anticancer Drugs 14, 569–574 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Lasko LM et al. , Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin CH et al. , Discovery of N-((4-([1,2,4]triazolo[1,5-a]pyridin-6-yl)-5-(6-methylpyridin-2-yl)-1H-imidazol-2 -yl)methyl)-2-fluoroaniline (EW-7197): a highly potent, selective, and orally bioavailable inhibitor of TGF-beta type I receptor kinase as cancer immunotherapeutic/antifibrotic agent. J Med Chem 57, 4213–4238 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Muraki M et al. , Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem 279, 24246–24254 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.