Abstract
Background
To develop an effective vaccine against a novel viral pathogen, it is important to understand the longitudinal antibody responses against its first infection. Here we performed a longitudinal study of antibody responses against SARS-CoV-2 in symptomatic patients.
Methods
Sequential blood samples were collected from 39 individuals at various timepoints between 0 and 154 days after onset. IgG or IgM titers to the receptor binding domain (RBD) of the S protein, the ectodomain of the S protein, and the N protein were determined by using an ELISA. Neutralizing antibody titers were measured by using a plaque reduction assay.
Findings
The IgG titers to the RBD of the S protein, the ectodomain of the S protein, and the N protein peaked at about 20 days after onset, gradually decreased thereafter, and were maintained for several months after onset. Extrapolation modeling analysis suggested that the IgG antibodies were maintained for this amount of time because the rate of reduction slowed after 30 days post-onset. IgM titers to the RBD decreased rapidly and disappeared in some individuals after 90 days post-onset. All patients, except one, possessed neutralizing antibodies against authentic SARS-CoV-2, which they retained at 90 days after onset. The highest antibody titers in patients with severe infections were higher than those in patients with mild or moderate infections, but the decrease in antibody titer in the severe infection cohort was more remarkable than that in the mild or moderate infection cohort.
Interpretation
Although the number of patients is limited, our results show that the antibody response against the first SARS-CoV-2 infection in symptomatic patients is typical of that observed in an acute viral infection.
Funding
The Japan Agency for Medical Research and Development and the National Institutes of Allergy and Infectious Diseases.
Research in context.
Evidence before this study
Immediately after the emergence of SARS-CoV-2, the antibody response in infected individuals against SARS-CoV-2 infection was unknown. We searched PubMed for articles up to August 2020, using the keywords “SARS-CoV-2″ and “antibody response”, “antibody” or “antibodies”. The published research consistently reported that antibodies against the S and N proteins and neutralizing antibodies are detectable in individuals infected with the virus. However, it was unclear whether the severity of disease affects the antibody response against SARS-CoV-2. Furthermore, the antibody titer against SARS-CoV-2 was reported to decline during the early phase especially in asymptomatic or mildly symptomatic individuals.
Added value of this study
We found that the IgG titers to the S and N proteins peaked at about 20 days after onset, gradually decreased thereafter, and were maintained for over 150 days after onset. Our modeling analysis indicated that the reduction rate of the IgG titers slowed down after 30 days post-onset. Neutralizing antibodies against authentic SARS-CoV-2 were also retained at 90 days after onset. The highest antibody titers in patients with severe infections were higher than those in patients with mild or moderate infections, but the decrease in antibody titer in the severe group was more remarkable than that in the other two groups, resulting in all patients having similar antibody titers after 60 days post-onset.
Implications of all the available evidence
The publication of a paper on the decline of antibodies in asymptomatic or mildly symptomatic individuals exposed to SARS-CoV-2 spurred discussions regarding the reinfection of recovered COVID-19 patients, vaccine efficacy, and building herd immunity. Our results show that the antibody response against the first SARS-CoV-2 infection is typical of that observed in an acute viral infection. Furthermore, a robust and rapid antibody response is likely triggered by a second exposure in individuals who were previously infected with SARS-CoV-2, leading to the suppression of virus growth in the respiratory organs due to the immune responses induced upon the second exposure. Individuals who recover from an initial symptomatic SARS-CoV-2 infection will, therefore, likely be refractory to subsequent exposure to this virus for several months after onset.
Alt-text: Unlabelled box
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China at the end of 2019 and spread throughout the world, resulting in a pandemic [1]. SARS-CoV-2 causes coronavirus disease-2019 (COVID-19), which can be asymptomatic or cause fever, cough, severe pneumonia, and death [2]. As of 12 September 2020, more than 28 million cases of COVID-19 including approximately 1 million deaths have occurred (https://covid19.who.int/).
IgG and IgM against the spike (S) and nucleocapsid (N) proteins of SARS-CoV-2 are elicited in most COVID-19 patients within 1–2 weeks after onset and contribute to viral clearance 3, 4, 5, 6, 7. The S protein is the surface glycoprotein and binds to the cellular viral receptor, angiotensin converting enzyme-2 (ACE-2), on host cells via its receptor binding domain (RBD) [8,9]. Accordingly, the S protein, especially the RBD, is a major target for neutralizing antibodies [10, 11, 12, 13, 14]. Antibodies against the S and N proteins and neutralizing antibodies are detectable in individuals infected with the virus [15,16]. It has been reported that the severity of disease affects the antibody titers [17,18], although one group reported no difference in antibody levels between severe and non-severe patients [19]. Therefore, it remains unclear whether the severity of disease affects the antibody response against SARS-CoV-2. Furthermore, some studies have reported that the antibody titer against SARS-CoV-2 declines during the early convalescent phase, especially in asymptomatic or mildly symptomatic individuals [4,15,17,20], whereas other groups have shown that the antibody levels are relatively stable for 2–5 months [21, 22, 23, 24, 25]. It has been suggested that humoral immunity in such individuals might not be long-lasting, and that in a worst case scenario, individuals with no symptoms or mild symptoms may be re-infected with SARS-CoV-2 [26,27]. To clarify these points, here we compared the longitudinal antibody response to SARS-CoV-2 infection in 39 COVID-19 patients who exhibited mild, moderate, or severe symptoms, at several timepoints from 0–154 days after onset.
Methods
2.1 Ethics and biosafety statements
Human samples were collected by following protocols approved by the Research Ethics Review Committee of the Institute of Medical Science, the University of Tokyo (approval number 2019–71–0201). Signed informed consent was obtained from all participants.
All experiments with SARS-CoV-2 were performed in biosafety level 3 (BSL3) laboratories at the University of Tokyo, which were approved for such use by the Ministry of Agriculture, Forestry, and Fisheries, Japan.
Cells
VeroE6 cells and Vero cells expressing human serine protease TMPRSS2 (Vero-TMPRSS2) [28] were maintained in DMEM containing 10% fetal calf serum (FCS), 1 mg/ml G418, 100 units/mL penicillin, 100 µg/mL streptomycin, and 5 μg/ml plasmocin prophylactic (Invivogen) and incubated at 37 °C under 5% CO2. We used these cells after clearance of mycoplasma.
2.3 Viruses
SARS-CoV-2 (UT-NCGM02/Human/2020/Tokyo) [29] virus was propagated in VeroE6 cells and titrated in Vero-TMPRSS2 cells.
2.4 Patient samples
COVID-19 patients (n = 39) were recruited to this study between January 2020 and April 2020 and gave signed, informed consent. They were then assigned to one of the following three groups based on their clinical status: mild (hospitalized, symptomatic, not requiring supplemental oxygen); moderate (hospitalized, symptomatic, requiring supplemental oxygen); or severe [hospitalized, symptomatic, requiring tracheal intubation or extracorporeal membrane oxygenation (ECMO) or who succumbed to their infection] (Table S1). Blood samples (serum or plasma) were obtained from each patient on the indicated days after onset for other assessments or antibody tests during hospitalization and after discharge. These samples were incubated at 56 °C for more than 30 min to inactivate the virus. The sample type (serum or plasma) did not affect the ELISA titers. HPCo-010′s case study and the antibody repertoire of some of these patients have been reported elsewhere (Ravichandran et al. 2020 submitted and [30,31]).
2.5 ELISA
Ninety-six-well Maxisorp microplates (Nunc) were incubated with 2 μg/ml of recombinant RBD [5,32] or ectodomain of the S protein, the N protein or with PBS at 4 °C overnight and were then incubated with 5% skim milk in PBS containing 0.05% tween-20 (PBS-T) for 1 h at room temperature. Serum or plasma samples were initially diluted 40-fold in PBS-T containing 5% skim milk, and subsequently serially 4-fold diluted (40- to 2,621,440-fold). The microplates were reacted for 1 h at room temperature with the diluted serum or plasma samples in duplicate, followed by peroxidase-conjugated goat anti-human IgG, Fcγ Fragment specific antibody (Jackson Immuno-Research) or anti-human IgM, Fcμ Fragment specific antibody (Jackson Immuno-Research). 1-Step Ultra TMB-Blotting Solution (Thermo fisher scientific) was then added to each well and incubated for 3 min at room temperature. The reaction was stopped by the addition of 2 M H2SO4 and the optical density at 450 nm (OD450) was immediately measured. The average OD450 values of two PBS-wells were subtracted from the average OD450 values of two RBD-, ectodomain-, or N protein-wells for background correction. A subtracted OD450 value of 0.1 or more was regarded as positive, and the maximum dilution to give a positive result was used as the ELISA titer.
2.6 Plaque reduction assay
Serum or plasma samples were initially diluted 10-fold in the DMEM containing 5% FCS, 10 mM HEPES, 100 μg/ml gentamicin sulfate, and 2.5 μg/ml amphotericin B and then serially 3-fold diluted in duplicate. The diluted serum or plasma samples were incubated with approximately 100–200 plaque-forming unit (PFU) of SARS-CoV-2 at room temperature for 1 h. The virus-sample mixtures were inoculated into Vero-TMPRSS2 cells and incubated for 1 h at 37 °C. After the mixture was removed, the cells were incubated with DMEM containing 5% FCS, 10 mM HEPES, 100 μg/ml gentamicin sulfate, 2.5 μg/ml amphotericin B, and 1% agar for 2 days at 37 °C. The maximum dilution to reduce the plaque number by more than 50% compared to the control (i.e., no serum or plasma present) was used as neutralization titer.
2.7 Modeling
A Bayesian hierarchical model was used to estimate the day of peak response for each measurement and to aid visualization. Measurement i, in individual, j, was modelled as yij = hj f(tij, αj, βj) exp(-λjtij), where tij is the time of the measurement, f(tij, αj, βj) is the cumulative gamma distribution function at time tij with shape αj and inverse scale βj, λj is the decay rate, and hj is the maximum response if λj = 0 [33]. Posterior distributions of each parameter were sampled for each individual. These distributions were in turn captured by overarching distributions in a standard hierarchical model. Concretely: hj ~ N(μh, σh), αj ~ N(μα, σα), βj ~ N(μβ, σβ) and λj ~ N(μλ, σλ), where N(μ, σ) denotes a normal distribution with mean μ, and standard deviation σ. Finally, the likelihood of each measurement was modelled as yij ~ N(0, σ), allowing the posterior distribution to be sampled using the NUTS MCMC algorithm implemented in Stan [34]. Analyses were conducted on data transformed to have unit standard deviation. Weakly informed priors, N(0, 5), were used throughout and six chains of 4000 iterations were run for each assay. Excellent parameter convergence was obtained. Titrations were transformed by yij = logn(mij/d) where mij is the titration, n = 4 for IgG, IgM, Ectodomain, n = 3 for PRNT50, and d is the starting dilution in the titration series. The half-life of each antibody was estimated by using a linear mixed model that included the log-titer as a dependent variable and days after peak as an independent variable with individual identifiers as random effects. Half-lives were then obtained as log42 (log32 for neutralization) divided by the coefficients of the days after peak.
2.8 Statistical analysis
Antibody titers were log-transformed before all statistical processes and are presented as geometric mean titers. Continuous values were compared between multiple independent groups by analysis of variance (ANOVA) with Tukey's pair-wise comparison as necessary. Paired comparisons within each group were performed by using the paired t-test. Values for clinical status between groups were compared by using Kruskal-Wallis test or Fisher's exact test as appropriate. P-values < 0.05 were considered statistically significant, and SPSS 25.0 (IBM) was used for all analyses.
2.9 Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
3.1 Clinical status of the COVID-19 patients in this study
We analyzed the longitudinal antibody response of 39 symptomatic COVID-19 patients against SARS-CoV-2 infection (Tables 1 and S1). The median age of the patients was 62 years (interquartile range [IQR], 49.5–69.5) and 27 (69.2%) were men (Table 1). The median time from onset to hospital admission was 6 (IQR, 4–8) days. Twenty-five of the patients (64.1%) had at least one comorbidity, such as hypertension (28.2%), diabetes (25.6%), dyslipidemia (12.8%), respiratory diseases (10.3%), and/or thyroid diseases (5.1%). At the end of the study period, 32 patients had been discharged and seven patients had died. The 39 symptomatic patients were divided into three groups based on their recorded clinical status: 13 were assigned to the mild group, 12 to the moderate group, and 14 to the severe group (see Methods for stratification criteria and Table S1). There was a significant difference in the median age of the mild (52 years), moderate (56.5 years), and severe groups (67 years) (p = 0.022) and in the percentage of diabetes in each group (7.7%, 16.7%, and 50.0%, respectively) (p = 0.041) (Table 1). Other items showed no statistical difference. Sequential blood samples were collected from all 39 individuals at various timepoints between 0 and 154 days after onset (Table S2).
Table 1.
Clinical status of mild, moderate, and severe patients.
| Mild (n = 13) | Moderate(n = 12) | Severe (n = 14) | Total (n = 39) | P value | ||
|---|---|---|---|---|---|---|
| Age(Median, IQR) | 52 (48–64) | 56.5 (46.5–69.5) | 67 (62–75) | 62 (49.5–69.5) | 0.022† | |
| Gender (%) | Male | 7 (53.8%) | 9 (75.0%) | 11 (78.6%) | 27 (69.2%) | 0.386* |
| Female | 6 (46.2%) | 3 (25.0%) | 3 (21.4%) | 12 (30.8%) | ||
| Days between onset and hospitalization (Median, IQR) | 5 (5–7) | 5 (3.5–9) | 6 (4–9) | 6 (4–8) | 0.801† | |
| Comorbidity (%) | Any | 5 (38.4%) | 9 (75.0%) | 11 (78.6%) | 25 (64.1%) | 0.071* |
| Hypertension | 2 (15.4%) | 5 (41.7%) | 4 (28.6%) | 11 (28.2%) | 0.406* | |
| Diabetes | 1 (7.7%) | 2 (16.7%) | 7 (50.0%) | 10 (25.6%) | 0.041* | |
| Dyslipidemia | 2 (15.4%) | 2 (16.7%) | 1 (7.1%) | 5 (12.8%) | 0.716* | |
| Respiratory disease | 2 (15.4%) | 2 (16.7%) | 0 (0%) | 4 (10.3%) | 0.363* | |
| Thyroid disease | 1 (7.7%) | 1 (8.3%) | 0 (0%) | 2 (5.1%) | 0.528* | |
| Other | 1 (7.7%) | 3 (25%) | 2 (14.3%) | 6 (15.4%) | – | |
| Outcome (%) | Discharged | 13 (100%) | 12 (100%) | 6 (42.9%) | 31 (79.5%) | – |
| Still hospitalized | 0 (0%) | 0 (0%) | 2 (14.3%) | 2 (5.1%) | – | |
| Dead | 0 (0%) | 0 (0%) | 6 (42.9%) | 6 (15.4%) | – |
P values were calculated by using.
Fisher's exact test or the †Kruskal-Wallis test.
3.2 Antibody responses against SARS-CoV-2 infection
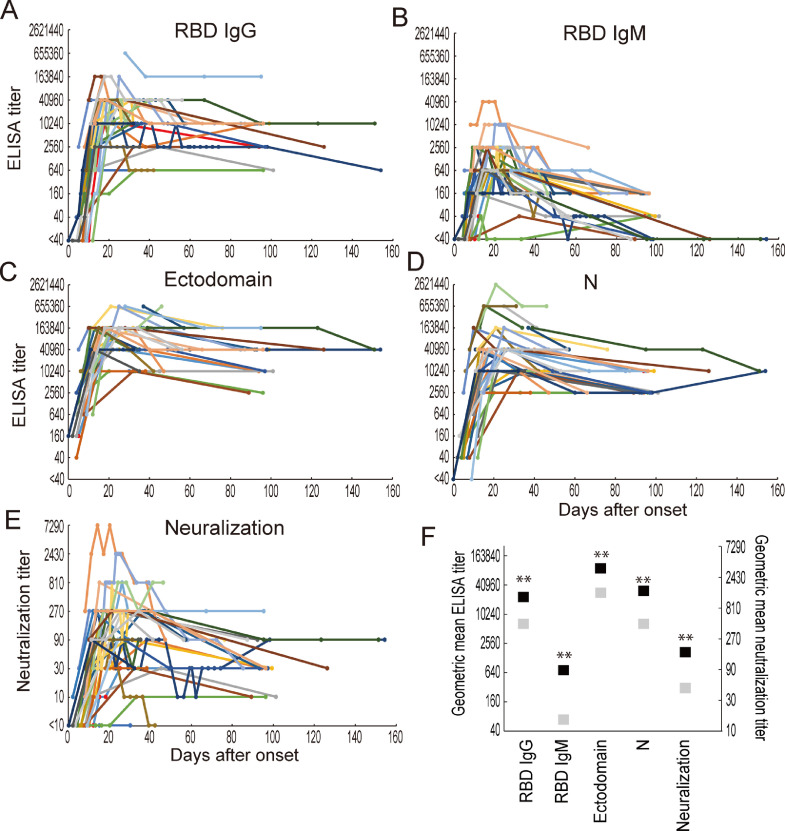
To evaluate the antibody response of each patient after onset, we performed ELISAs and neutralization assays using the RBD or ectodomain of the S protein, or the N protein and authentic SARS-CoV-2, respectively. The longitudinal changes in antibody titers of the 39 patients are shown in Figs. 1 and S1–S3. All patients possessed detectable IgG against all antigens tested and detectable IgM against the RBD (Figs. 1A, 1B, and S1–S3). Although the IgG response to all antigens tested was elicited within 14 days of onset, the IgG response to the ectodomain was detected earlier than that to the RBD and the N protein (Figs. 1B–1D, and S1–S3). The IgG titers to the RBD, ectodomain, and N protein peaked at about 20 days after onset, decreased thereafter, and were maintained for several months after onset. The IgM response against the RBD was not induced before that of IgG (Fig. 1A and 1B), as reported previously [6,35]. The IgM to the RBD decreased rapidly, resulting in some individuals becoming anti-RBD IgM-negative after 90 days post-onset. In the neutralization tests, all patients, except for HPCo-023, possessed neutralization antibodies against authentic SARS-CoV-2 (Figs. 1E and S1–S3). Although the neutralization titer showed a similar trend to that of the IgM response to the RBD, it was maintained after 90 days post-onset. HPCo-023, from whom no neutralizing antibody was detected, was discharged from the hospital without his condition worsening, indicating that factors other than neutralization antibodies contribute to virus clearance. To better understand the decline in antibody titers over time, we calculated the geometric mean titers at the highest titer found over the entire study period (Fig. 1F, black square) and after 60 days post-onset (Fig. 1F, gray square). The geometric mean highest IgG and IgM titers against the RBD, ectodomain, and N protein and neutralization titers after 60 days post-onset decreased significantly from the peak titers. The reduction in the IgM titer against the RBD was 1/10.8, whereas that in the IgG titer against the RBD, ectodomain, and N protein ranged from 1/3.18–1/4.67, suggesting that IgG antibody levels tend not to decrease as rapidly as IgM antibody levels.
Fig. 1.
Longitudinal change in antibody titers of 39 COVID-19 patients. Blood samples were obtained from 39 COVID-19 patients at several timepoints between 0 and 154 days after onset. IgG titers against the RBD (A), the ectodomain of the S protein (C), and the N protein (D); IgM titers against the RBD (B); and neutralization titers (E) were determined by performing an ELISA and a neutralization test using a wild-type virus. The maximum dilution of blood samples that gave a positive result in the ELISA or reduced the plaque number by more than 50% was selected as the ELISA or neutralization titer. Lines of the same color indicate data from the same patient. (F) Geometric mean antibody titers at the highest titer over the entire study period (black) and at least 60 days post-onset (gray). **, p<0.01 (paired t-test).
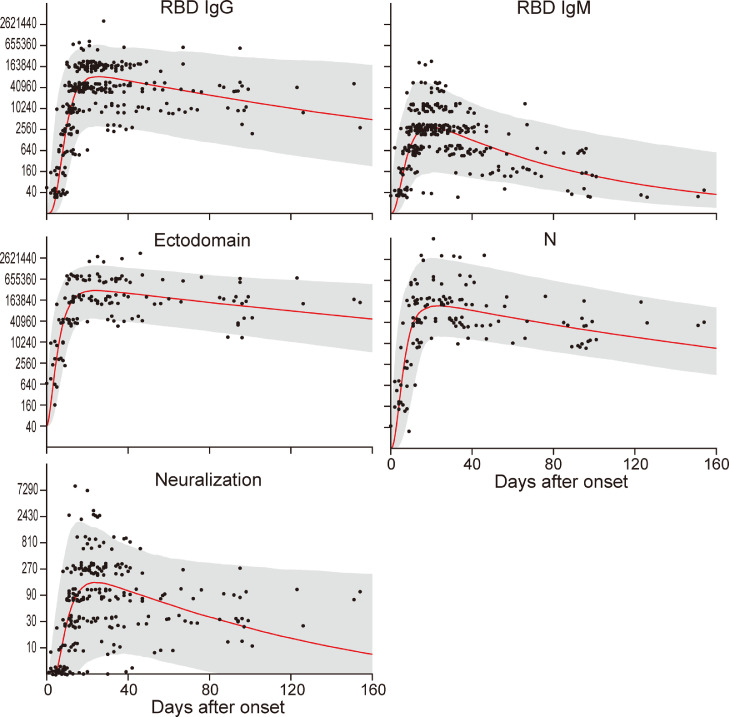
To understand the overall trend of the antibody responses in these 39 patients, we visualized the group level responses from a hierarchical model (Fig. 2). The peak RBD IgG response estimated from the posterior distribution of parameters occurred on day 22.8 [interquartile range (IQR): 19.5–26.0]. For RBD IgM, these values were 18.8 (15.3–21.5) and similarly for the ectodomain, 22.5 (19.3–25.8) and the N protein, 20.3 (17.0–23.8). Neutralizing antibody titers reached a peak 23.8 days after onset (IQR: 19.3–29.0). Although the IgM titer against the RBD and the neutralization titer were predicted to drop below the limit of detection within 6 months, the IgG titers against the RBD, ectodomain, and N protein were predicted to be retained for a long time. We next calculated the half-lives of the antibody titers by using data after the peak antibody titers or after 30 days post-onset (Table 2). The half-lives of the RBD-IgG, RBD-IgM, ectodomain, N protein, and neutralization titers after each peak titer were 32.9, 19.3, 49.3, 35.5, and 39.6 days, respectively, whereas those more than 30 days post-onset were 57.9, 26.6, 60.6, 43.4, and 71.8 days, respectively (Table 2), indicating that the half-lives of the antibody titers are extended at later timepoints. These data demonstrate that the rate of antibody decline slows down at later timepoints post-infection. Taken together, our findings show that the antibody titers reach their maximum values at approximately 20 days after onset and gradually decrease but the IgG antibodies are maintained for a long time because the rate of reduction slows down after 30 days post-onset. Therefore, such antibodies might play protective roles against subsequent SARS-CoV-2 infection.
Fig. 2.
Modeling the longitudinal response in 39 patients. The gray area indicates the 95% high density interval of the posterior distribution from the hierarchical model across all patients. The red line indicates the median level in the posterior distribution. Jitter in the y-dimension has been added to aid visualization of the data points.
Table 2.
Antibody titer half-lives.
| Half-life (day) of antibody titer (95%CI) |
|||
|---|---|---|---|
| Severity | After peak titer | After 30 days post-onset* | |
| RBD-IgG | Mild | 56.6 (33.1–194.3) | 2804 (67.7– −71.2) |
| Moderate | 34.5 (24.9–55.9) | 101.3 (32.1– −88.0) | |
| Severe | 27.7 (22.8–35.4) | 42.8 (31.8–65.2) | |
| Combined | 32.9 (27.7–40.7) | 57.9 (40.5–95.0) | |
| RBD-IgM | Mild | 23.3 (18.1–32.7) | 43.4 (23.3–315.1) |
| Moderate | 22.9 (18.2–31.1) | 40.2 (19.0– −334.0) | |
| Severe | 16.6 (14.5–19.3) | 22.7 (18.0–30.7) | |
| Combined | 19.3 (17.3–21.9) | 26.6 (21.3–35.5) | |
| Ectodomain | Mild | 41.5 (28.4–76.8) | 50.0 (24.4– −951.5) |
| Moderate | 63.5 (38.0–192.7) | 130.8 (29.0– −52.1) | |
| Severe | 47.0 (33.1–81.5 | 66.9 (33.9–2194) | |
| Combined | 49.3 (38.3–69.3) | 60.6 (36.4–180.0) | |
| N | Mild | 37.5 (25.2–73.2) | 29.2 (18.4–70.7) |
| Moderate | 42.1 (27.5–89.8) | 1412 (33.7– −35.7) | |
| Severe | 30.4 (23.0–44.9) | 48.5 (28.9–151.1) | |
| Combined | 35.5 (28.6–46.9) | 43.4 (29.4–83.1) | |
| Neutralization | Mild | 39.3 (23.2–126.2) | 28.8 (17.3–84.6) |
| Moderate | 36.8 (22.8–95.6) | −556.8 (33.2– −29.7) | |
| Severe | 40.7 (28.2–73.1) | 107.8 (43.6– −228.9) | |
| Combined | 39.6 (30.0–58.1) | 71.8 (39.8–369.8) | |
Negative half-lives theoretically correspond to doubling times as calculated by using the statistical model. However, they should be interpreted as “infinite” if such a reversal after peak-out is biologically unlikely.
3.3 Comparisons of antibody responses in patients with mild, moderate, or severe disease
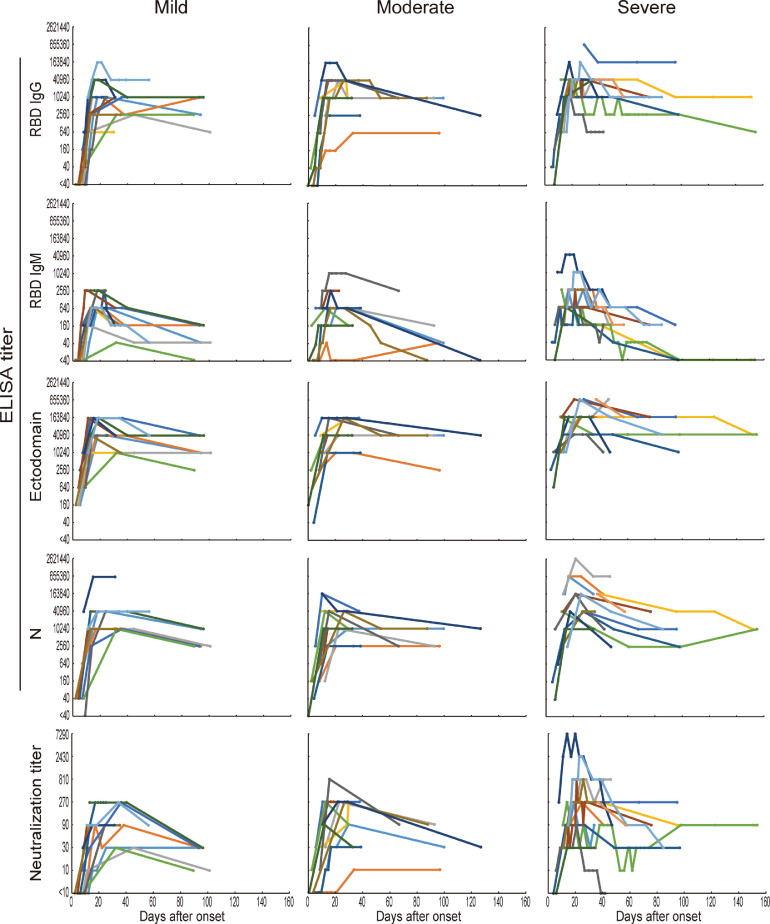
Because severity is thought to be one of the factors that affects the antibody response against SARS-CoV-2 infection [4,17,20], we compared the longitudinal changes in antibody titers of patients with mild, moderate, or severe infections. The IgG and IgM responses against the RBD, ectodomain, or N protein were similar between the mild, moderate, and severe groups (Fig. 3). The half-lives of the RBD-IgG, RBD-IgM, ectodomain, N protein, and neutralization titers at least 30 days post-onset were longer than those after the peak titer, except for the antibody titer against the N protein in the mild group (Table 2).
Fig. 3.
Changes in antibody titers of patients with mild, moderate, or severe infections. The changes in antibody titers against the RBD, ectodomain, and N protein shown in Fig. 1 were divided into three groups (mild, moderate, and severe) according to disease severity. Lines of the same color in each group indicate data from the same patient.
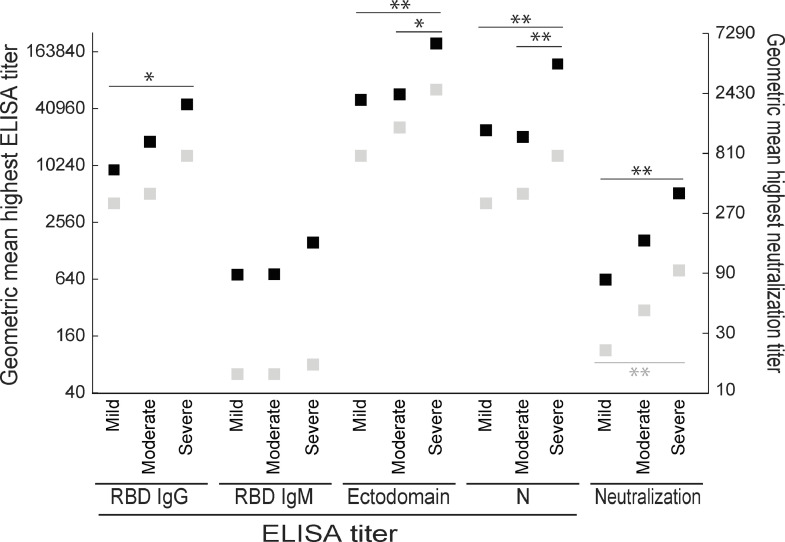
Next, we compared the geometric mean antibody titers at the highest titers over the entire study period and at least 60 days post-onset among the mild, moderate, and severe groups. Over the entire study period, the IgG titer to the RBD and the neutralization titer in the severe patient group were significantly higher than those in the mild patient group, whereas the antibody titers to the ectodomain and N protein in the severe group were significantly higher than those in the mild and moderate groups (Fig. 4). After 60 days post-onset, although the neutralization titer in the severe group was significantly higher than that in the mild group, the other antibody titers tested were not significantly different between the two groups. The IgM titer to the RBD was similar among the three groups at both points. Taken together with our longitudinal analysis of the three groups, these results show that the highest antibody titers in severe patients are higher than those in mild (or moderate) patients, but the decrease in antibody titer in the severe infection cohort is more remarkable than that in the mild or moderate infection cohorts, resulting in all patients having similar antibody titers after 60 days post-onset (Fig. 4).
Fig. 4.
Geometric mean highest antibody titers over the entire study period and after 60 days post-onset. The geometric mean titers of RBD IgG, RBD IgM, ectodomain, N proteins, and neutralization in the mild, moderate, and severe groups at the highest titers over the entire study period (black squares) and after 60 days post-onset (gray squares) were calculated. * and **, p<0.05 and p<0.01, respectively (one-way ANOVA followed by Tukey's test).
Discussion
Here we compared the longitudinal changes in antibody titers against SARS-CoV-2 among 39 COVID-19 patients, who exhibited mild, moderate, or severe symptoms, until several months post-onset. Antibodies against the RBD, ectodomain, and N protein of SARS-CoV-2 were detectable at approximately 10 days after onset and reached a peak at approximately 20 days after onset. The peak IgG titers against the RBD, ectodomain, and N protein, and the neutralization titer in the severe group tended to be higher than those in the other groups. However, the decrease in antibody titers was greater in the severe group over time, such that there were ultimately no differences in these antibody titers among the three groups, except for the neutralizing titers between the severe and mild groups. Similar reductions in antibody titers (i.e., within 30–60 days of onset) have been reported in COVID-19 patients [4,15,17,20] and there is concern that antibodies against SARS-CoV-2 may not be long-lasting. However, in our study, antibody titers in all 39 patients were maintained for several months post-onset, suggesting that antibodies against SARS-CoV-2 do not drop below the detection limit in this timeframe. Although the sample size of our study is small, similar antibody responses in other COVID-19 patient cohorts have been reported recently [16,21, 22, 23, 24, 25]. Such antibody behavior has been observed in the immune response to primary infections with other acute viral pathogens [36,37]. Since it is unclear what the long-term antibody titer changes will be (i.e., more than six months after onset) and what the antibody response in asymptomatic infected individuals will be, follow-up assessments of recovered individuals for more than a year should be considered.
Although the small sample size in our study is a limitation, the statistical power is sufficient to detect biologically meaningful differences because the variations in antibody titer among cases within each group were relatively small. We can assume a SD of 1.0 log-titer based on the observed statistics, and a paired t-test with the 18 samples can detect the difference of 0.8 log-titer and ANOVA with the 39 samples can detect the difference of 1.1 log-titer as a statistical significance with the power of >80%. This means that the titer ratio of 2.7 to 4.6 between groups can be detected in each comparison. Therefore, the sample size in this study was sufficient to detect differences.
We found that the antibody response against the first exposure to the newly emerged SARS-CoV-2 was consistent in the COVID-19 patients of the mild, moderate, and severe groups. Although levels of neutralizing antibodies tend to decrease as time passes, one study reported that 804 recovered patients showed no clinical reinfections within three months of hospital discharge [38] and only one asymptomatic reinfection has been reported [39]. These findings suggest that a robust and rapid antibody response relative to the initial SARS-CoV-2 infection is likely triggered by the second exposure to SARS-CoV-2, meaning that the virus growth in the respiratory organs could be suppressed by the immune response and no clear symptoms might appear. However, as yet, we know little about the immune correlates of protection against SARS-CoV-2, which will be important for long-term follow-up of infected patients and clinical studies of COVID-19 vaccines. Yet, we cannot rule out the possibility that acquired non-neutralizing antibodies against the S protein may enhance the pathogenicity of the virus via antibody-dependent enhancement (ADE) activity. ADE has been observed in vitro and in vivo in dengue patients and its animal models 40, 41, 42 and ADE in vitro or by serum/antibody transfer in an animal model has been reported for Ebola and Zika viruses [43,44]. Numerous studies involving COVID-19 patients and animal models have been conducted all over the world, and the induction of neutralizing antibodies by infection and vaccination and the efficacy of monoclonal antibody, serum, or plasma therapies have been reported [12,29,45]. But no studies describing an increase in pathogenicity due to ADE have been reported yet, which suggests that ADE may be limited in human SARS-CoV-2 infection. Nevertheless, we should consider looking closely at ADE in individuals who have recovered from COVID-19 or received a COVID-19 vaccine.
Data sharing
All data used and obtained in this study are available in the manuscript and supporting information.
Author contributions
S. Yamayoshi and Y.K. designed the study. S. Yamayoshi, A.Y., and M.I. performed the experiments. S.C., M.O., J.M., and T.H. assisted with the experiments. O.A., M.N., I.N., M.K., K. Mitamura, K.Y., K. Maeda, H.K., T.O., R.B., K.F., H. Nagai, S. Yamamoto, M.S., E.A., J.O., S.H., T.S., Y.M., H. Nakajima., H.M., N.O., N.S., and H.Y. collected the clinical samples and provided clinical data. K.I.H coordinated the clinical sample collection. S. Yamayoshi, M.N., D.P., and Y.K. analyzed the data. S. Yamayoshi, M.N., D.P., and Y.K. wrote the manuscript. All authors reviewed and approved the manuscript.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
We thank Maki Kiso, Calvin Duong, Shu Yanagimoto, Takayuki Kurosawa, Noriko Kinoshita, Takahide Kikuchi, Daisuke Taniyama, Kazuto Ito, Taketomo Maruki, Shigehiro Hagiwara, Daisuke Arai, Hidenori Takahashi, Genta Nagao, Shunichiro Konishi, Arinobu Tojo, Tokiko Nagamura-Inoue, Etsuko Nagai, Hiromi Isshiki, Mika Kogayu, and Tomoko Sato for assistance of experiments. We also thank Susan Watson for editing the manuscript.
Funding
This work was supported by a Research Program on Emerging and Re-emerging Infectious Diseases (JP19fk0108113 and JP19fk0108166), a Project Promoting Support for Drug Discovery (JP20nk0101612, JP20nk0101614, and JP20nk0101603), the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) (JP19fm0108006), and the Japan Program for Infectious Diseases Research and Infrastructure (JP20wm0125002 and 20fk0108272) from the Japan Agency for Medical Research and Development (AMED), and the National Institutes of Allergy and Infectious Diseases funded Center for Research on Influenza Pathogenesis (CRIP; HHSN272201400008C).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eclinm.2021.100734.
Contributor Information
Seiya Yamayoshi, Email: yamayo@ims.u-tokyo.ac.jp.
Yoshihiro Kawaoka, Email: yoshihiro.kawaoka@wisc.edu.
Appendix. Supplementary materials
References
- 1.Zhu N., Zhang D., Wang W. A Novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 Novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To K.K., Tsang O.T., Leung W.S. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.H. Patterns of IgG and IgM antibody response in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amanat F., Stadlbauer D., Strohmeier S. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long Q.X., Liu B.Z., Deng H.J. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 7.Okba N.M.A., Muller M.A., Li W. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan M., Liu H., Wu N.C. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369(6507):1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouwer P.J.M., Caniels T.G., van der Straten K. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zost S.J., Gilchuk P., Case J.B. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584(7821):443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan J., Xing S., Ding L. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 2020;32(3) doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zost S.J., Gilchuk P., Chen R.E. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med. 2020;26(9):1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seow J., Graham C., Merrick B., et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv 2020: 2020.07.09.20148429.
- 16.Li K., Huang B., Wu M. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun. 2020;11(1):6044. doi: 10.1038/s41467-020-19943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long Q.X., Tang X.J., Shi Q.L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y., Zuiani A., Fischinger S. Quick COVID-19 healers sustain anti-SARS-CoV-2 antibody production. Cell. 2020 doi: 10.1016/j.cell.2020.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang Y., Liu T., Wei Y. Scoring systems for predicting mortality for severe patients with COVID-19. EClinicalMedicine. 2020;24 doi: 10.1016/j.eclinm.2020.100426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383(11):1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford K.H.D., Dingens A.S., Eguia R. Dynamics of neutralizing antibody titers in the months after severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wajnberg A., Amanat F., Firpo A. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020:eabd7728. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seow J., Graham C., Merrick B. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nature Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isho B., Abe K.T., Zuo M. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Science Immunol. 2020;5(52):eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Long Q.-.X., Deng H.-.J. Longitudinal Dynamics of the Neutralizing Antibody Response to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. Clinical Infect Diseases. 2020 doi: 10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quadeer A.A. Immunodominant epitopes based serological assay for detecting SARS-CoV-2 exposure: promises and challenges. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckerle I., Meyer B. SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet. 2020;396(10250):514–515. doi: 10.1016/S0140-6736(20)31482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuyama S., Nao N., Shirato K. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai M., Iwatsuki-Horimoto K., Hatta M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117(28):16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito M., Adachi E., Yamayoshi S. Gargle lavage as a safe and sensitive alternative to swab samples to diagnose COVID-19: a case report in Japan. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azekawa S., Namkoong H., Mitamura K., Kawaoka Y., Saito F. Co-infection with SARS-CoV-2 and influenza a virus. IDCases. 2020;20:e00775. doi: 10.1016/j.idcr.2020.e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stadlbauer D., Amanat F., Chromikova V. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol. 2020;57(1):e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salje H., Cummings D.A.T., Rodriguez-Barraquer I. Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature. 2018;557(7707):719–723. doi: 10.1038/s41586-018-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter B., Gelman A., Hoffman M.D. Stan: a Probabilistic Programming Language. J Stat Softw. 2017;76(1):1–29. doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun B., Feng Y., Mo X. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;9(1):940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puschnik A., Lau L., Cromwell E.A., Balmaseda A., Zompi S., Harris E. Correlation between dengue-specific neutralizing antibodies and serum avidity in primary and secondary dengue virus 3 natural infections in humans. PLoS Negl Trop Dis. 2013;7(6):e2274. doi: 10.1371/journal.pntd.0002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sande C.J., Mutunga M.N., Okiro E.A., Medley G.F., Cane P.A., Nokes D.J. Kinetics of the neutralizing antibody response to respiratory syncytial virus infections in a birth cohort. J Med Virol. 2013;85(11):2020–2025. doi: 10.1002/jmv.23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mumoli N., Vitale J., Mazzone A. Clinical immunity in discharged medical patients with COVID-19. Int J Infect Dis. 2020;99:229–230. doi: 10.1016/j.ijid.2020.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.To K.K., Hung I.F., Ip J.D. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huisman W., Martina B.E., Rimmelzwaan G.F., Gruters R.A., Osterhaus A.D. Vaccine-induced enhancement of viral infections. Vaccine. 2009;27(4):505–512. doi: 10.1016/j.vaccine.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halstead S.B., Mahalingam S., Marovich M.A., Ubol S., Mosser D.M. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis. 2010;10(10):712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katzelnick L.C., Gresh L., Halloran M.E. Antibody-dependent enhancement of severe dengue disease in humans. Science. 2017;358(6365):929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuzmina N.A., Younan P., Gilchuk P. Antibody-dependent enhancement of ebola virus infection by human antibodies isolated from survivors. Cell Rep. 2018;24(7):1802–1815. doi: 10.1016/j.celrep.2018.07.035. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bardina S.V., Bunduc P., Tripathi S. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science. 2017;356(6334):175–180. doi: 10.1126/science.aal4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers T.F., Zhao F., Huang D. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.