Abstract
In this study, the complete mitogenome sequence of the broad-nosed pipefish Syngnathus typhle (Linnaeus, 1758) (Syngnathiformes: Syngnathidae) has been sequenced by next-generation sequencing method. The assembled mitogenome consisting of 16,488bp, includes 13 protein coding genes, 22 transfer RNAs and 2 ribosomal RNAs genes. The overall base composition\ of \the broad-nosed pipefish is 27.72% for A, 27.81% for C, 16.53% for G, 27.94% for T and show 87% identities to Syngnathus schlegeli (GenBank: AP012318). The complete mitogenome of the broad-nosed pipefish, provides essential and important DNA molecular data for further phylogeography and evolutionary analysis for seahorse family.
Keywords: Broad-nosed pipefish, mitogenome, next-generation sequencing
The broad-nosed pipefish Syngnathus typhle (Linnaeus, 1758) occurs in Eastern Atlantic: Norway, Baltic Sea and the British Isles to Morocco. Usually, it is found along the coasts and estuaries at a temperature range from 8° to 24 °C (Dawson 1990). Spawns in summer (Muus & Nielsen 1999), the male carries the eggs in a brood pouch that is found under the tail (Breder & Rosen 1966). Its maximum length reached 35 cm in TL. The snout is compressed and taller than the eye diameter, and anterior trunk rings not fused ventrally.
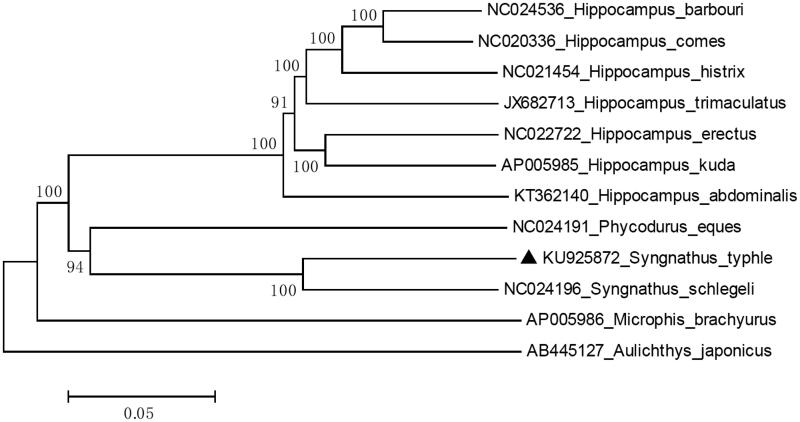
Samples of the broad-nosed pipefish were live transported from Shanghai Changfeng Ocean World Aquarium (belonging to Merlin Entertainment Group, United Kingdom), and the muscle was preserved in pure alcohol. The specimens were stored in Fish Specimens Museum in Shanghai Ocean University, the accession number is SHOU20150044003. Then, their genomic DNA was extracted from muscle by using Genomic DNA Purification Kit (GeneMark, Taichung, Taiwan). The methods for genomic DNA extraction, library construction and next-generation sequencing were followed by previous publication (Shen et al. 2014). The raw next-generation sequencing reads were de novo assembled by commercial software (Geneious V8, Auckland, New Zealand) to produce a single, circular form of complete mitogenome with about an average 338.4 X coverage (13,496 out of 9,637,558, 0.0014%). The complete mitochondrial genome of the broad-nosed pipefish was 16,488 bp in size (GenBank: KU925872), includes 13 protein-coding genes, 22 transfer RNAs and two ribosomal RNAs genes. The overall base composition of the broad-nosed pipefish is 27.72% for A, 27.81% for C, 16.53% for G, 27.94% for T and show 87% identities to Syngnathus schlegeli (GenBank: AP012318) (Figure 1).
Figure 1.
Neighbour-joining (NJ) tree of 12 species complete mitochondrial genome sequence. The phylogenetic relationships of broad-nosed pipefish show 87% identities to Syngnathus schlegeli using Aulichthys japonicus as an outgroup.
The protein coding and tRNA genes of the broad-nosed pipefish mitogenome were predicted by using DOGMA (Wyman et al. 2004), ARWEN (Laslett & Canback 2008) and Mito Annotator (Iwasaki et al. 2013) tools. Some ambiguous annotation sites are manual inspected. All protein-coding genes were encoded on H-strand with exception of protein-coding genes of ND6. All tRNA genes were encoded on H-strand with exception of tRNA-Glu, tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr and tRNA-Ser. All the 13 mitochondrial protein-coding genes share the start codon ATG, except for COI (GTG start codon). The stop codon TAA is present in COI, ATP8, ND3, ND4L, ND5 and ND6; TAG is present in ND1; and incomplete stop codon ‘TA–’ is found in COIII and ATP6, and ‘T––’ is found in COII, ND2, ND4 and Cytb.
The longest one is ND5 gene (1836 bp) in all protein-coding genes, whereas the shortest is ATP8 gene (168 bp). The two ribosomal RNA genes, 12S rRNA gene (936 bp) and 16S rRNA gene (1660 bp), are located between tRNA-Phe and tRNA-Leu and separated by tRNA-Val. We expect that the present result would elucidate the further phylogenetic approach among different species of Syngnathidae.
To validate the phylogenetic position of S. typhle, we used MEGA6 software (Tamura et al. 2013) to construct a Neighbor-joining (NJ) (with 1000 bootstrap replicates) containing complete mitogenomes of 12 species (Figure 1). In conclusion, the complete mitogenome of S. typhle provides essential and important DNA molecular data for phylogenetic and evolutionary analysis for marine seahorse.
Acknowledgments
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by a grant from Shanghai Collaborative Innovation Centre for Aquatic Animal Genetics and Breeding (ZF1206).
References
- Breder CM, Rosen DE.. 1966. Modes of reproduction in fishes. Neptune City, NJ: T.F.H. Publications. [Google Scholar]
- Dawson CE. 1990. Syngnathidae In: Quero JC, Hureau JC, Karrer C, Post A, Saldanha L, editors. Check-list of the fishes of the eastern tropical Atlantic (CLOFETA). Lisbon: JNICT; Paris: SEI; UNESCO, Vol. 2 p. 658–664. [Google Scholar]
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al.. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline . Mol Biol Evol. 30:2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B.. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175. [DOI] [PubMed] [Google Scholar]
- Muus BJ, Nielsen JG.. 1999. Sea fish. Scandinavian fishing year book. Denmark: Hedehusene; p. 340. [Google Scholar]
- Shen KN, Yen TC, Chen CH, Li HY, Chen PL, Hsiao CD.. 2014. Next generation sequencing yields the complete mitochondrial genome of the flathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA. [Epub ahead of print]. doi: 10.3109/19401736.2014.963799. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0 . Mol Biol Evol. 30:2725–2729. [\PMC][ 10.1093/molbev/mst197] [24132122]. [DOI] [PMC free article] [PubMed] [Google Scholar]