Pandemic progress in Kenya
By the end of July 2020, Kenya had reported only 341 deaths and ∼20,000 cases of COVID-19. This is in marked contrast to the tens of thousands of deaths reported in many higher-income countries. The true extent of COVID-19 in the community was unknown and likely to be higher than reports indicated. Uyoga et al. found an overall seroprevalence among blood donors of 4.3%, peaking in 35- to 44-year-old individuals (see the Perspective by Maeda and Nkengasong). The low mortality can be partly explained by the steep demographics in Kenya, where less than 4% of the population is 65 or older. These circumstances combine to result in Kenyan hospitals not currently being overwhelmed by patients with respiratory distress. However, the imposition of a strict lockdown in this country has shifted the disease burden to maternal and child deaths as a result of disruption to essential medical services.
Science, this issue p. 79; see also p. 27
By May 2020, 1 in 20 Kenyan adults had SARS-CoV-2 antibodies, when fewer than 100 COVID-19 deaths had been reported nationally.
Abstract
The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Africa is poorly described. The first case of SARS-CoV-2 in Kenya was reported on 12 March 2020, and an overwhelming number of cases and deaths were expected, but by 31 July 2020, there were only 20,636 cases and 341 deaths. However, the extent of SARS-CoV-2 exposure in the community remains unknown. We determined the prevalence of anti–SARS-CoV-2 immunoglobulin G among blood donors in Kenya in April–June 2020. Crude seroprevalence was 5.6% (174 of 3098). Population-weighted, test-performance-adjusted national seroprevalence was 4.3% (95% confidence interval, 2.9 to 5.8%) and was highest in urban counties Mombasa (8.0%), Nairobi (7.3%), and Kisumu (5.5%). SARS-CoV-2 exposure is more extensive than indicated by case-based surveillance, and these results will help guide the pandemic response in Kenya and across Africa.
Africa accounts for 17% of the global population (1) but by late July 2020 accounted for only 5% of the global COVID-19 cases and 3% of global COVID-19 deaths reported (2). This disparity has been attributed to limited capacity for diagnosis, timely implementation of stringent containment measures, a younger population structure, and a predominance of asymptomatic and mild infections (3, 4). The first case of COVID-19 in Kenya was detected on 12 March 2020. Within 1 week, the government instituted containment measures to limit the spread of the virus (5). By 31 July national surveillance recorded 20,636 cases and 341 deaths (6). This increase in cases is notably slower than the epidemic in Wuhan, Europe, or the United States. Recently, it has been suggested that “the virus is spreading… …with an attenuated outcome in Africa” [(7), p. 626], but there are few data available to confirm or refute this assertion.
In countries affected early in the pandemic, serological surveillance was used to define cumulative incidence. For example, at the release of lockdown in Wuhan, 9.6% of staff resuming work were found to have antibodies to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (8). At the end of the epidemic wave in Spain, seropositivity was 5.0% in a random population sample of 60,897 (9). As the epidemic curve declined in Geneva, seroprevalence rose over 3 weeks from 4.8 to 10.9% (10). Currently, there are few estimates of SARS-CoV-2 seroprevalence in Africa in the literature (11).
Movement restrictions, in response to COVID-19, have limited the conduct of fieldwork for population-based serosurveys. Several countries have monitored seroprevalence in blood transfusion donors (12, 13) or expectant mothers attending antenatal clinics (14). Here, we report the results of a pragmatic national serosurvey using residual blood samples from transfusion donors across Kenya and a highly sensitive and specific assay for anti–SARS-CoV-2 spike immunoglobulin G (IgG).
We validated a widely used enzyme-linked immunosorbent assay (ELISA) for SARS-CoV-2 IgG (15) with 910 serum samples from the prepandemic period and 174 sera from polymerase chain reaction (PCR)–defined SARS-CoV-2 cases, and a well-characterized five-sera panel from the National Institute of Biological Standards and Control (NIBSC) in the UK. For either receptor-binding domain (RBD) or whole spike, specificity was higher when using a ratio of the sample optical density (OD)/negative control OD than when using the raw sample OD plus 3 standard deviations to define seropositivity (table S1). By using OD ratios, both RBD and spike ELISAs correctly classified 901 of 910 prepandemic samples as seronegative (table S1). However, the spike ELISA detected more seropositives (166 of 179 compared with 145 of 179 for RBD ELISA) among sera from SARS-CoV-2 PCR-positive individuals (fig. S2, A and B). On the basis of these data, we defined anti–SARS-CoV-2 IgG seropositivity as an OD ratio >2 and selected the spike ELISA for this study. The sensitivity and specificity, at this threshold, were 92.7% [95% confidence interval (CI), 87.9 to 96.1%] and 99.0% [95% CI, 98.1 to 99.5%], respectively (figs. S3, A and B, S5, and S6; and table S1). As previously noted (15), the RBD and whole-spike ELISA responses were highly correlated (fig. S3C), with very little interassay variation (fig. S4).
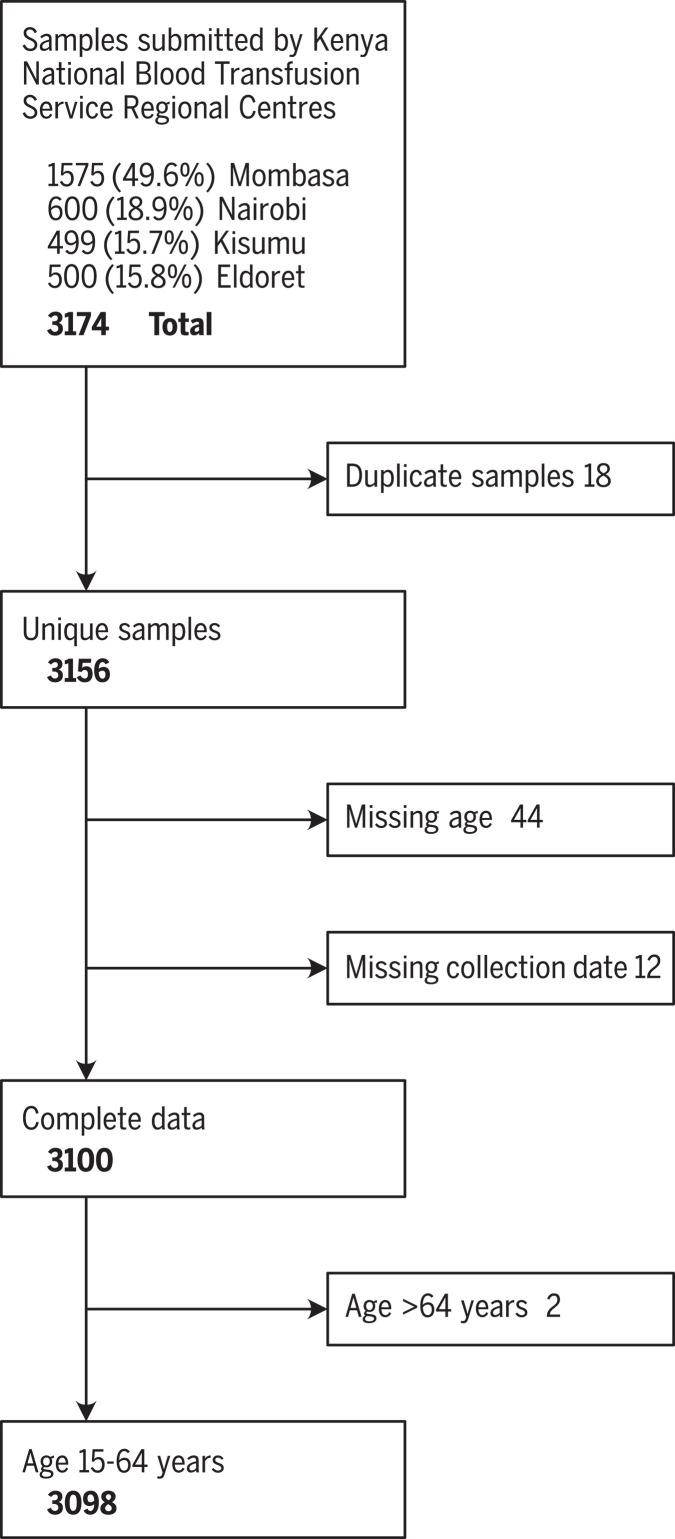
A total of 3174 blood transfusion samples were collected from four Kenya National Blood Transfusion Service (KNBTS) regional blood transfusion centers that are supported by several satellites and hospitals between 30 April and 16 June 2020, from individuals aged 15 to 66 years. Approximately half of the samples were drawn in Mombasa; the remainder were evenly distributed between Nairobi, Kisumu, and Eldoret (Fig. 1 and table S2). We excluded 18 duplicate samples, 56 records missing data on age or collection date, and two records from individuals aged ≥65 years. Policy in Kenya is to avoid blood donation from individuals >65 years, and we excluded these other data points as potentially unreliable. These exclusions left 3098 samples for further analysis (Fig. 1).
Fig. 1. Participant flow diagram for SARS-CoV-2 seroprevalence study of blood donors in Kenya.
Exclusion criteria for the selection of samples with complete data.
Of the 3098 samples, 174 were positive for anti–SARS-CoV-2 spike IgG, giving a crude seroprevalence of 5.6% (95% CI, 4.8 to 6.5%). Crude seroprevalence varied by age (P = 0.046), ranging between 3.4 to 7.0% among adults 15 to 54 years; all 71 donors aged 55 to 64 years were seronegative (Table 1). Crude seroprevalence did not vary by sex (P = 0.50) but did vary geographically, from 1.9% in the Rift Valley region to 10.0% in the Western region (P = 0.002) (Table 1).
Table 1. Crude, population-weighted, and test performance–adjusted SARS-CoV-2 anti-spike protein IgG seroprevalence by participant characteristics and regions.
Prevalence estimates were calculated by using multilevel regression and poststratification (MLRP) to account for differences in the sample population and the national population and subsequently adjusted for assay sensitivity and specificity.
| All samples |
Seropositive samples |
Crude seroprevalence |
Kenya population (2019 Census) |
Bayesian population-weighted seroprevalence* |
Bayesian population-weighted, test-adjusted seroprevalence* |
||||
| % | (95% CI) | % | (95% CI) | % | (95% CI) | ||||
| Age | |||||||||
| 15 to 24 years | 808 | 49 | 6.1 | 4.5 to 7.9 | 9,733,174 | 5.1 | 3.7 to 6.9 | 4.4 | 2.7 to 6.4 |
| 25 to 34 years | 1242 | 66 | 5.3 | 4.1 to 6.7 | 7,424,967 | 4.9 | 3.6 to 6.4 | 4.2 | 2.8 to 6.0 |
| 35 to 44 years | 714 | 50 | 7.0 | 5.2 to 9.1 | 4,909,191 | 5.9 | 4.3 to 8.1 | 5.2 | 3.3 to 7.7 |
| 45 to 54 years | 263 | 9 | 3.4 | 1.6 to 6.4 | 3,094,771 | 3.8 | 1.9 to 6.0 | 3.0 | 1.1 to 5.4 |
| 55 to 64 years | 71 | 0 | 0 | 1,988,062 | 3.4 | 0.7 to 6.2 | 2.9 | 0.7 to 5.7 | |
| Sex | |||||||||
| Male | 2540 | 146 | 5.7 | 4.9 to 6.7 | 13,388,243 | 4.4 | 2.9 to 6.2 | 3.6 | 1.9 to 5.8 |
| Female | 558 | 28 | 5.0 | 3.4 to 7.2 | 13,761,922 | 5.5 | 4.4 to 6.8 | 4.8 | 3.5 to 6.4 |
| Regions | |||||||||
| Central | 105 | 7 | 6.7 | 2.7 to 13.2 | 3,452,213 | 5.6 | 2.9 to 10.0 | 4.9 | 1.9 to 9.7 |
| Mombasa | 550 | 51 | 9.3 | 7.0 to 12.0 | 792,072 | 8.3 | 6.1 to 10.9 | 7.8 | 5.4 to 10.8 |
| Other Coast | 973 | 39 | 4.0 | 2.9 to 5.4 | 1,671,097 | 3.7 | 2.6 v 5.1 | 2.9 | 1.6 to 4.6 |
| Eastern/N. Eastern | 242 | 11 | 4.5 | 2.3 to 8.0 | 5,176,080 | 4.3 | 2.5 to 7.0 | 3.5 | 1.4 to 6.6 |
| Nairobi | 235 | 21 | 8.9 | 5.6 to 13.3 | 3,002,314 | 7.6 | 4.9 to 11.2 | 7.1 | 4.2 to 11.2 |
| Nyanza | 442 | 30 | 6.8 | 4.6 to 9.5 | 3,363,813 | 6.0 | 4.2 to 8.4 | 5.2 | 3.1 to 7.9 |
| Rift Valley | 481 | 8 | 1.7 | 0.7 to 3.3 | 7,035,581 | 2.1 | 1.1 to 3.6 | 1.5 | 0.4 to 3.1 |
| Western | 70 | 7 | 10.0 | 4.1 to 19.5 | 2,656,995 | 7.0 | 3.5 to 13.1 | 6.3 | 2.5 to 13.1 |
| Total | 3,098 | 174 | 5.6 | 4.8 to 6.5 | 27,150,165 | 4.9 | 3.9 to 6.2 | 4.3 | 2.9 to 5.8 |
*Reweighted prevalence estimates based on demographic data from the 2019 Kenya Population and Housing Census
Compared with the 2019 Kenya Population and Housing Census, our participants were more commonly male (82.0% in our study versus 49.3% in the census), had more persons aged 25 to 34 years (40.1 versus 27.3%), and more residents of coastal counties (49.2 versus 9.1%) (Table 2). We therefore adjusted the prevalence estimate for the demographics of the sample using poststratification, and for the sensitivity and specificity of the test.
Table 2. General characteristics of the study population compared with the national population of Kenya.
N is the number of individuals in each stratum.
| Blood transfusion samples | Kenya National Census 2019 | ||||
| N | % | N | % | ||
| Age | 15 to 24 years | 808 | 26.1 | 9,733,174 | 35.8 |
| 25 to 34 years | 1,242 | 40.1 | 7,424,967 | 27.3 | |
| 35 to 44 years | 714 | 23.0 | 4,909,191 | 18.1 | |
| 45 to 54 years | 263 | 8.5 | 3,094,771 | 11.4 | |
| 55 to 64 years | 71 | 2.3 | 1,988,062 | 7.3 | |
| Sex | Male | 2540 | 82.0 | 13,388,243 | 49.3 |
| Female | 558 | 18.0 | 13,761,922 | 50.7 | |
| Regions | Central | 105 | 3.4 | 3,452,213 | 12.7 |
| Mombasa | 550 | 17.8 | 792,072 | 2.9 | |
| Other Coast | 973 | 31.4 | 1,671,097 | 6.2 | |
| Eastern/Northeastern | 242 | 7.8 | 5,176,080 | 19.1 | |
| Nairobi | 235 | 7.6 | 3,002,314 | 11.1 | |
| Nyanza | 442 | 14.3 | 3,363,813 | 12.4 | |
| Rift Valley | 481 | 15.5 | 7,035,581 | 25.9 | |
| Western | 70 | 2.3 | 2,656,995 | 9.8 | |
| Total | Kenya 15 to 64 years | 3098 | 27,150,165 | ||
The Bayesian population-weighted and test-adjusted seroprevalence for Kenya was 4.3% (95% CI, 2.9 to 5.8%) (Table 1), and the posterior sensitivity and specificity estimates were 92.4% (95% CI, 88.0 to 95.6%) and 98.9 (95% CI, 98.2 to 99.5%), respectively. Seroprevalence was higher (4.2 to 5.2%) in the younger age groups (15 to 44 years) and declined in the older age groups (45 to 64 years) but was similar for both sexes. Seroprevalence was highest for those living in Mombasa, Nairobi, and the Western region, although the number of observations for the Western region was small. The directly standardized seroprevalence estimates are presented in table S3. Seroprevalence was also calculated for counties that had at least 120 donors sampled. The three largest urban counties of Mombasa, Nairobi, and Kisumu had SARS-CoV-2 seroprevalence of 8.0% (95% CI, 5.5 to 11.1%), 7.3% (95% CI, 4.2 to 11.4%), and 5.5% (95% CI, 2.8 to 9.6%), respectively (table S4).
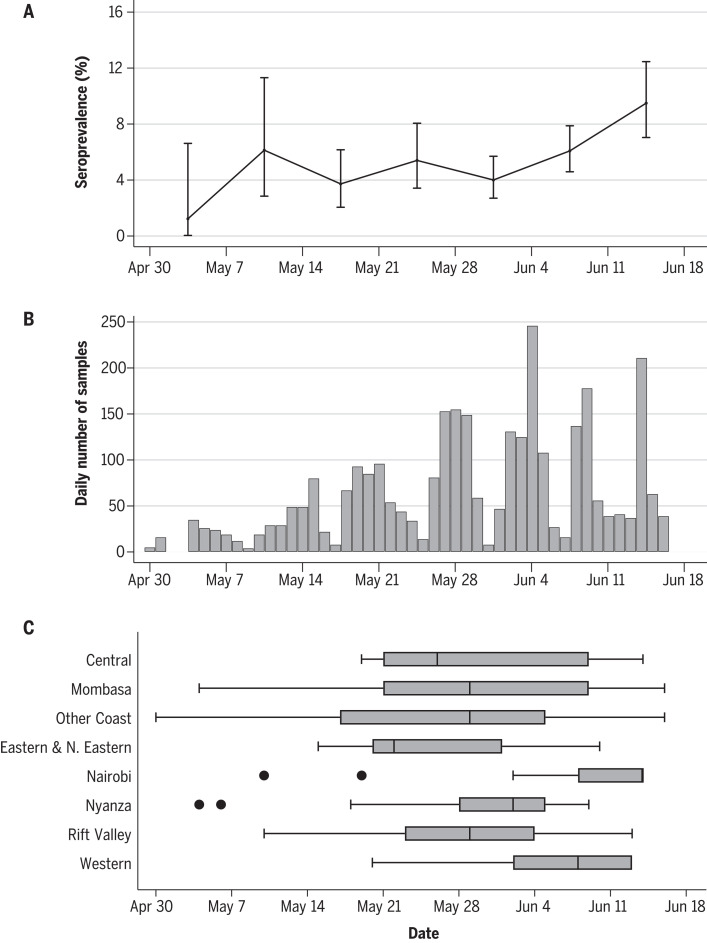
The frequency of blood donor sampling and crude seroprevalence estimates increased with time over the 7-week study period (Fig. 2). The median sample date was 30 May 2020, and the midpoint of the study was 24 May 2020. We did not adjust for sample date because the period of sampling varied for residents of different counties (Fig. 2C); instead, we show the variation in crude prevalence over time (Fig. 2A).
Fig. 2. Timeline of sampling for SARS-CoV-2 seroprevalence in blood donors in Kenya.
(A to C) Against the timeline of the sampling period, (A) is the weekly crude seroprevalence and 95% confidence interval, (B) is the daily frequency of samples collected, and (C) is the temporal distribution of samples by region. Shown are the proportion, counts, and regional distribution of donors during the study period.
Voluntary nonremunerated donors (VNRDs), who donate blood at community-based “blood drives,” made up only 7.6% (236 of 3098) of our sample of donors; the remainder were family replacement donors (FRDs) who provide a unit of blood in compensation for a transfusion received by a sick relative. The two groups did not differ significantly by age (P = 0.15) or sex (P = 0.51) (table S5). Crude seroprevalence was 8.5% (20 of 236) for VNRDs and 5.4% (154 of 2862) for FRDs. The median sample date for VNRDs (14 June 2020) was 2 weeks later than that for FRDs (29 May 2020).
Population exposure across Kenya, with a population-weighted test-adjusted seroprevalence of 4.3%, is considerably higher than was previously thought, on the basis of the cases and deaths reported to date. Seroprevalence was particularly high in the three urban counties: Mombasa (8.0%), Nairobi (7.3%), and Kisumu (5.5%). Consistent with other studies, seroprevalence did not vary significantly by sex (9, 10, 16); however, it peaked in 35- to 44-year-olds and was lowest for those ≥45 years, which is also consistent with existing reports in which seroprevalence was found to be lower in older adults (9, 10).
SARS-CoV-2 seroprevalence in our study is comparable with estimates from large population-based serosurveys in China, Switzerland, Spain, and the United States after the initial epidemic peak and after many tens of thousands of deaths (9, 10, 17, 18). Our results are also comparable with those of other surveys of blood donors in Brazil (13), Italy (12), and many parts of England (19). Kenya has an estimated population of 53 million in 2020, and 57% of the population is aged 15 to 64 years. If the transfusion donor seroprevalence of 4.3% was applied to all 15- to 64-year-olds, it would suggest approximately 1.3 million infections. However, by the median sample date, 30 May 2020, only 2093 cases had been detected (of which approximately 90% were asymptomatic), and there were 71 deaths among all ages (6). Although it is difficult to extrapolate our data directly to the whole population, they do strongly suggest that the infection is more widespread in Kenya than the current PCR test results suggest and indicate a need for more systematic testing. The current PCR testing strategy targets symptomatic individuals, health care workers, contacts of confirmed cases, international travelers, cross-border truck drivers, and residents of areas identified as “hotspots.”
What are the potential explanations for the divergence in the ratio of observed cases or deaths to serologically defined infections inferred from transfusion donors in Kenya, compared with many high-income countries? (i) The seroprevalence could be overestimated because of bias in the selection or behavior of blood transfusion donors. (ii) Cases could be underascertained by national public health surveillance, although it seems unlikely that reporting of deaths and severe cases could be reduced by several orders of magnitude, and hospitals in Kenya were not overwhelmed by admissions with respiratory illness. (iii) The steep demographic age-pyramid results in a smaller vulnerable age group. In Kenya, only 3.9% of the population is aged 65 years or greater, which is substantially less than, for example, 23.3% found in Italy; again, this would only explain a moderate reduction in severe cases or deaths (4). (iv) There may be alternative mechanisms of immunity to SARS-CoV-2, including cell-mediated immunity (20, 21), perhaps as a result of human coronavirus (HCoV)–elicited immunity (22, 23). Despite our prior work showing that HCoVs circulate in Kenya (24), we did not identify evidence of cross-reactive antibodies to endemic coronaviruses in our validation study.
Although blood donors are not representative of the Kenyan population as a whole, we adjusted for demographic bias in the sample structure by standardization against the age, sex, and regional distribution of the Kenyan population. A substantial proportion (43%) of the population of Kenya is outside the age range (15 to 64 years) sampled in this study, and the seroprevalence in children <15 years and adults >65 years is often lower (9, 10); our estimate for blood donors may be higher than the estimate for the population as a whole. Blood donors also differ from the general population in their risk of exposure to SARS-CoV-2. For example, potential donors are excluded from giving blood if they have been ill during the past 6 months, so the sample may underestimate the population prevalence of SARS-CoV-2 antibodies; however, people who are shielding at home are unlikely to be captured in our sample, leading to an overestimate of seroprevalence. Our exploration of the two distinct populations of blood donors, FRDs and VNRDs, suggests variation in the seroprevalence by donor group, but 92% (2862 of 3098) of our sample came from the group with lower seroprevalence, and exclusion of VNRDs reduced little the crude seroprevalence in our study, from 5.6 to 5.4%. Against these considerations, other countries have relied on blood transfusion donors for an early estimate of seroprevalence, but later estimates from random population samples have not been substantially different (25, 26).
A key strength of this study is the rigorous validation that included testing positive and negative control samples from the target population, as well as reference plasma from the UK NIBSC as part of a World Health Organization (WHO)–coordinated effort on SARS-CoV-2 seroepidemiology. In addition, we adopted a conservative seropositivity threshold to optimize assay specificity and sensitivity for our setting.
The pandemic response in countries with limited health care capacity has been driven by the aggressive implementation of control measures to limit transmission. Unfortunately, this strategy has been accompanied by enormous collateral costs, particularly in Africa. Modeled estimates of the disruptions of essential medical services, such as immunization and antenatal care, suggest an additional ~253,500 child deaths and 12,200 maternal deaths over 6 months in low- and middle-income countries (27). In the absence of social protection, the economic effects of lockdown are debilitating, so it is important to obtain an early measure of the trajectory of the epidemic.
Our study provides a national and regional estimate of population exposure to SARS-CoV-2 in an African country. The 4.3% prevalence in blood transfusion donors is in sharp contrast with the reported COVID-19 cases and deaths and supports the impression that disease may be attenuated in Africa (7).
Acknowledgments
We thank F. Krammer for providing the plasmids used to generate the RBD, spike protein, and CR3022 monoclonal antibody used in this work. Development of SARS-CoV-2 reagents was partially supported by the NIAID Centres of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C. The COVID-19 convalescent plasma panel (NIBSC 20/118) and research reagent for SARS-CoV-2 Ab (NIBSC 20/130) were obtained from the NIBSC, UK. We thank the blood donors and KNBTS staff who supported this work. We also thank the WHO SOLIDARITY II network for sharing of protocols and for facilitating the development and distribution of control reagents. This paper has been published with the permission of the director, Kenya Medical Research Institute. This study was approved by the Scientific and Ethics Review Unit (SERU) of the Kenya Medical Research Institute (Protocol SSC 3426). Before the blood draw, donors gave individual consent for the use of their samples for research. Ethical approval was obtained for collection, storage, and further use for the sample sets used in the validation assays (SERU numbers 1433, 3149, and 3426). Funding: This project was funded by the Wellcome Trust (grants 220991/Z/20/Z and 203077/Z/16/Z), the Bill and Melinda Gates Foundation (INV-017547), and the Foreign Commonwealth and Development Office (FCDO) through the East Africa Research Fund (EARF/ITT/039). S.U. is funded by DELTAS Africa Initiative [DEL-15-003], L.I.O.-O. is funded by a Wellcome Trust Intermediate Fellowship (107568/Z/15/Z), A.A. is funded by a DFID/MRC/NIHR/Wellcome Trust Joint Global Health Trials Award (MR/R006083/1), J.A.G.S. is funded by a Wellcome Trust Senior Research Fellowship (214320) and the NIHR Health Protection Research Unit in Immunisation, I.M.O.A. is funded by the United Kingdom’s Medical Research Council and Department For International Development through an African Research Leader Fellowship (MR/S005293/1) and by the NIHR-MPRU at UCL (grant 2268427 LSHTM). G.M.W. is supported by a fellowship from the Oak Foundation. C.N.A. is funded by the DELTAS Africa Initiative [DEL-15-003], and the Department for International Development and Wellcome (220985/Z/20/Z). Author contributions: Conceptualization and methodology: S.U., I.M.O.A., A.A., J.A.G.S., E.W.K., K.E.G., A.E., S.V., R.A., M.M., P.A., K.Ka., W.N., and G.M.W. Investigation: S.U., H.K.K., J.N., J.T., P.W., C.R., C.Y., K.Ki., E.Od., T.R., I.O., S.K., Z.N.M., J.N.G., D.M., C.N.A., E.Ot., L.N., L.I.O.-O., and G.M.W. Formal analysis: J.A.G.S., M.O., and C.B. Validation: T.L., D.W., H.K.K., J.N., J.T., L.I.O.-O., and G.M.W. Resources and funding acquisition: S.U., T.L., P.B., and G.M.W. Supervision: E.B., B.T., and P.B. Writing, original draft preparation: S.U., I.M.O.A., A.A., J.A.G.S., and G.M.W. Writing, review and editing: all authors. Competing interests: R.A., M.M., K.K., and P.A. are from the Ministry of Health, Government of Kenya. All other authors declare no competing interests. Data and materials availability: Deidentified data has been published on the Havard dataverse server (28). This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.”
Supplementary Materials
science.sciencemag.org/content/371/6524/79/suppl/DC1
Materials and Methods
Figs. S1 to S6
Tables S1 to S5
MDAR Reproducibility Checklist
References and Notes
- 1.United Nations Department of Economic and Social Affairs Population Division, “World Urbanization Prospects: The 2018 Revision,” custom data acquired via (2018); https://population.un.org/wup/DataQuery.
- 2.Africa Centres for Diseases Control and Prevention, “Coronavirus Disease 2019 (COVID-19)” (2020); https://africacdc.org/covid-19[accessed 21 July 2020].
- 3.Diop B. Z., Ngom M., Pougué Biyong C., Pougué Biyong J. N., The relatively young and rural population may limit the spread and severity of COVID-19 in Africa: A modelling study. BMJ Glob. Health 5, e002699 (2020). 10.1136/bmjgh-2020-002699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowd J. B., Andriano L., Brazel D. M., Rotondi V., Block P., Ding X., Liu Y., Mills M. C., Demographic science aids in understanding the spread and fatality rates of COVID-19. Proc. Natl. Acad. Sci. U.S.A. 117, 9696–9698 (2020). 10.1073/pnas.2004911117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ministry of Health Kenya, “Press Statement on the update of the coronavirus in the country and response measures” (2020); www.health.go.ke/wp-content/uploads/2020/03/Coronavirus-Press-Statement-March-17-2020.pdf.
- 6.Ministry of Health Kenya, “COVID-19 Situation Reports (SITREP)” (2020); www.health.go.ke/#1591180376422-52af4c1e-256b.
- 7.Mbow M., Lell B., Jochems S. P., Cisse B., Mboup S., Dewals B. G., Jaye A., Dieye A., Yazdanbakhsh M., COVID-19 in Africa: Dampening the storm? Science 369, 624–626 (2020). 10.1126/science.abd3902 [DOI] [PubMed] [Google Scholar]
- 8.Wu X., Fu B., Chen L., Feng Y., Serological tests facilitate identification of asymptomatic SARS-CoV-2 infection in Wuhan, China. J. Med. Virol. 92, 1795–1796 (2020). 10.1002/jmv.25904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M. A., Pérez-Olmeda M., Sanmartín J. L., Fernández-García A., Cruz I., Fernández de Larrea N., Molina M., Rodríguez-Cabrera F., Martín M., Merino-Amador P., León Paniagua J., Muñoz-Montalvo J. F., Blanco F., Yotti R., ENE-COVID Study Group , Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A nationwide, population-based seroepidemiological study. Lancet 396, 535–544 (2020). 10.1016/S0140-6736(20)31483-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stringhini S., Wisniak A., Piumatti G., Azman A. S., Lauer S. A., Baysson H., De Ridder D., Petrovic D., Schrempft S., Marcus K., Yerly S., Arm Vernez I., Keiser O., Hurst S., Posfay-Barbe K. M., Trono D., Pittet D., Gétaz L., Chappuis F., Eckerle I., Vuilleumier N., Meyer B., Flahault A., Kaiser L., Guessous I., Seroprevalence of anti–SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 396, 313–319 (2020). 10.1016/S0140-6736(20)31304-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chibwana M. G., Jere K. C., Kamng’ona R., Mandolo J., Katunga-Phiri V., Tembo D., Mitole N., Musasa S., Sichone S., Lakudzala A., Sibale L., Matambo P., Kadwala I., Byrne R. L., Mbewe A., Morton B., Phiri C., Mallewa J., Mwandumba H. C., Adams E. R., Gordon S. B., Jambo K. C., High SARS-CoV-2 seroprevalence in Health Care Workers but relatively low numbers of deaths in urban Malawi. Wellcome Open Res. 5, 199 (2020). 10.12688/wellcomeopenres.16188.132766597 [DOI] [Google Scholar]
- 12.L. Valenti et al., SARS-CoV-2 seroprevalence trends in healthy blood donors during the COVID-19 Milan outbreak. medRxiv 2020.2005.2011.20098442 [Preprint] 31 May 2020. 10.1101/2020.05.11.20098442 [DOI] [PMC free article] [PubMed]
- 13.Amorim Filho L., Szwarcwald C. L., Mateos S. O. G., Leon A. C. M. P., Medronho R. A., Veloso V. G., Lopes J. I. F., Porto L. C. M. S., Chieppe A., Werneck G. L.; Grupo Hemorio de Pesquisa em Covid-19 , Seroprevalence of anti–SARS-CoV-2 among blood donors in Rio de Janeiro, Brazil. Rev. Saude Publica 54, 69 (2020). 10.11606/s1518-8787.2020054002643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannery D. D., Gouma S., Dhudasia M. B., Mukhopadhyay S., Pfeifer M. R., Woodford E. C., Gerber J. S., Arevalo C. P., Bolton M. J., Weirick M. E., Goodwin E. C., Anderson E. M., Greenplate A. R., Kim J., Han N., Pattekar A., Dougherty J., Kuthuru O., Mathew D., Baxter A. E., Vella L. A., Weaver J., Verma A., Leite R., Morris J. S., Rader D. J., Elovitz M. A., Wherry E. J., Puopolo K. M., Hensley S. E., SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci. Immunol. 5, eabd5709 (2020). 10.1126/sciimmunol.abd5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T. H. O., Chromikova V., McMahon M., Jiang K., Arunkumar G. A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D. S., Lugo L. A., Kojic E. M., Stoever J., Liu S. T. H., Cunningham-Rundles C., Felgner P. L., Moran T., García-Sastre A., Caplivski D., Cheng A. C., Kedzierska K., Vapalahti O., Hepojoki J. M., Simon V., Krammer F., A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 26, 1033–1036 (2020). 10.1038/s41591-020-0913-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang A. T., Garcia-Carreras B., Hitchings M. D. T., Yang B., Katzelnick L. C., Rattigan S. M., Borgert B. A., Moreno C. A., Solomon B. D., Trimmer-Smith L., Etienne V., Rodriguez-Barraquer I., Lessler J., Salje H., Burke D. S., Wesolowski A., Cummings D. A. T., A systematic review of antibody mediated immunity to coronaviruses: Kinetics, correlates of protection, and association with severity. Nat. Commun. 11, 4704 (2020). 10.1038/s41467-020-18450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X., Sun J., Nie S., Li H., Kong Y., Liang M., Hou J., Huang X., Li D., Ma T., Peng J., Gao S., Shao Y., Zhu H., Lau J. Y.-N., Wang G., Xie C., Jiang L., Huang A., Yang Z., Zhang K., Hou F. F., Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. 26, 1193–1195 (2020). 10.1038/s41591-020-0949-6 [DOI] [PubMed] [Google Scholar]
- 18.F. P. Havers et al, Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23-May 12, 2020. JAMA Intern Med, 10.1001/jamainternmed.2020.4130 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Public Health England, “Sero-prevalence epidemiology, England. in Weekly Coronavirus Disease 2019 (COVID-19) Surveillance Report 2020 Week 28” (2020); https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/899301/Weekly_COVID19_Surveillance_Report_week_28.pdf.
- 20.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181, 1489–1501.e15 (2020). 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Bert N., Tan A. T., Kunasegaran K., Tham C. Y. L., Hafezi M., Chia A., Chng M. H. Y., Lin M., Tan N., Linster M., Chia W. N., Chen M. I.-C., Wang L.-F., Ooi E. E., Kalimuddin S., Tambyah P. A., Low J. G.-H., Tan Y.-J., Bertoletti A., SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020). 10.1038/s41586-020-2550-z [DOI] [PubMed] [Google Scholar]
- 22.Sette A., Crotty S., Pre-existing immunity to SARS-CoV-2: The knowns and unknowns. Nat. Rev. Immunol. 20, 457–458 (2020). 10.1038/s41577-020-0389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.K. W. Ng et al., Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans. bioRxiv 2020.2005.2014.095414 [Preprint] 23 July 2020. 10.1101/2020.05.14.095414 [DOI]
- 24.Otieno G. P., Murunga N., Agoti C. N., Gallagher K. E., Awori J. O., Nokes D. J., Surveillance of endemic human coronaviruses (HCoV-NL63, OC43 and 229E) associated with childhood pneumonia in Kilifi, Kenya. Wellcome Open Res. 5, 150 (2020). 10.12688/wellcomeopenres.16037.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.H. Ward et al., Antibody prevalence for SARS-CoV-2 following the peak of the pandemic in England: REACT2 study in 100,000 adults. medRxiv 2020.2008.2012.20173690v20173692 [Preprint] 21 August 2020. 10.1101/2020.08.12.20173690 [DOI]
- 26.Public Health England, National COVID-19 surveillance report week 40 (2020); https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/923668/Weekly_COVID19_Surveillance_Report_week_40.pdf.
- 27.Roberton T., Carter E. D., Chou V. B., Stegmuller A. R., Jackson B. D., Tam Y., Sawadogo-Lewis T., Walker N., Early estimates of the indirect effects of the COVID-19 pandemic on maternal and child mortality in low-income and middle-income countries: A modelling study. Lancet Glob. Health 8, e901–e908 (2020). 10.1016/S2214-109X(20)30229-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Uyoga et al, Replication Data for: Seroprevalence of anti–SARS-CoV-2 IgG antibodies in Kenyan blood donors. Harvard Database (2020); 10.7910/DVN/RENVC9. 10.7910/DVN/RENVC9 [DOI] [PMC free article] [PubMed]
- 29.K. Ministry of Health, “Policy guidelines on blood transfusion in Kenya. The National Blood Transfusion Service of Kenya” (2001); https://nbtskenya.or.ke/wp-content/uploads/2019/02/Policy-Guidelines-on-Blood-Transfusion-in-Kenya.pdfaccessed 20 July 2020.
- 30.A. Gelman, J. Hill, Data analysis using regression and multilevel/hierarchical models, in Analytical Methods for Social Research, A. M. Alvarez, N. L. Beck, L. L. Wu, Eds. (Cambridge Univ. Press, 2007). [Google Scholar]
- 31.Gelman A., Carpenter C., Bayesian analysis of tests with unknown specificity and sensitivity. J. R. Statistic. Soc. C 69, 1269–1283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelman A., Prior distributions for variance parameters in hierarchical models. Bayesian Anal. 1, 515–534 (2006). 10.1214/06-BA117A [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- S. Uyoga et al, Replication Data for: Seroprevalence of anti–SARS-CoV-2 IgG antibodies in Kenyan blood donors. Harvard Database (2020); 10.7910/DVN/RENVC9. 10.7910/DVN/RENVC9 [DOI] [PMC free article] [PubMed]
Supplementary Materials
science.sciencemag.org/content/371/6524/79/suppl/DC1
Materials and Methods
Figs. S1 to S6
Tables S1 to S5
MDAR Reproducibility Checklist