Abstract
Breast cancer (BC) remains one of the major causes of cancer deaths in women. Over half of all BCs carry genetic defects in the gene encoding p53, a powerful tumor suppressor. P53 is known as the ‘guardian of the genome’ because it is essential for regulating cell division and preventing tumor formation. Ral-interacting protein (RLIP) is a modular protein capable of participating in many cellular functions. Blocking this stress-responsive protein, which is overexpressed during malignancy, enables BC cells to overcome the deleterious effects of p53 loss more effectively. In the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas9) system, a single-guide RNA (sgRNA) recognizes a specific DNA sequence and directs the endonuclease Cas9 to make a double-strand break, which enables editing of targeted genes. Here, we harnessed CRISPR/Cas9 technology to target the RLIP gene in BC cells. We screened sgRNAs using a reporter system and lentivirally delivered them, along with Cas9, to BC cells for validation. We then assessed the survival, proliferation, and tumorigenicity of BC cells in vitro and the growth of tumors in vivo after CRISPR-mediated knockdown of RLIP. Doxycycline-inducible expression of Cas9 in BC cells transduced with lentiviral vectors encoding the sgRNAs disrupted the RLIP gene, leading to inhibition of BC cell proliferation both in vitro and in vivo, with resected tumors showing reduced levels of the survival and proliferation markers Ki67, RLIP, pAkt, and survivin, the cell cycle protein CDK4, and the mesenchymal marker vimentin, as well as elevated levels of the differentiation protein E-cadherin and pro-apoptotic protein Bim. Inducible Cas9/sgRNA-transduced BC cells without doxycycline treatment did not exhibit altered cell survival or proliferation in vitro or in vivo. Our study provides proof-of-concept that the CRISPR/Cas9 system can be utilized to target RLIP in vitro and in vivo.
This study provides proof-of-concept that the CRISPR/Cas9 system can be harnessed to target an oncogene (RLIP) to inhibit tumor growth both in vitro and in vivo.
Introduction
Globally, breast cancer (BC) is the most frequently diagnosed cancer and the most common cause of death from cancer in women, with an estimated 1.4 million new cases and ~458,000 deaths/year. Its incidence is highest in the USA and Northern Europe, intermediate in Southeastern Europe and South America, and lowest in Asia. BC accounts for 29% of all female cancers in the USA, with ~230,000 women diagnosed and ~40,000 dying each year. Overall the 5-year survival rate of women in the USA has improved significantly between 1960 and 2007, from 63 to 92% for white women and from 46 to 83% for African–American women (1–5). Screening mammography has enabled the diagnosis of localized disease at earlier stages, which has contributed substantially to improved survival rates, as 10-year overall survival rates are 88, 66, and 7%, respectively, for women with localized cancer (62% of cases), regional metastases (31% of cases), and distant metastases (6% of cases) at diagnosis (2). Furthermore, the morbidity of local therapy has improved as lumpectomy and radiation for localized or limited regional disease have become the standard for early-stage disease, and the refinement of techniques for radiation post-mastectomy may yield further improvement (3). The role of genetic heterogeneity on the natural history, prognosis, and response to therapy of BC has increased the importance of tissue procurement for biomarker studies to optimize selection of therapy (2,4,5).
BC cells deploy numerous resistance mechanisms that reduce the efficacy of chemotherapy (6,7), radiation (8,9), endocrine therapy (10), Her2-targeted therapy (11), and kinase inhibitor therapy (12). ATP-binding cassette (ABC) and non-ABC drug transporters mediate drug resistance by catalyzing the efflux of cytotoxic drugs and their metabolites (6,7). Peptide hormones such as EGF, TGF and WNT mediate pleiotropic resistance to a broad spectrum of anticancer therapies by protecting cells from apoptosis and promoting the survival of cancer stem cells through the regulation of TP53 (13), MYC (10), PI3KCA, and AKT (14). Clearly, more effective targeted drugs are needed to overcome the inherent and acquired resistance mechanisms utilized by BC cells to elude therapeutic interventions.
Over half of all BCs carry genetic defects in the p53 gene, a powerful tumor suppressor. P53 is known as the ‘guardian of the genome’ because it is essential for regulating cell division and preventing tumor formation (13). Loss of this gene increases the risk of developing BC, and the resultant tumors are highly resistant to treatment. However, blocking the stress-responsive, non-ABC transporter protein Ral-interacting protein (RLIP), which is existentially required for cancer formation, enables cells to overcome the deleterious effects of p53 loss more effectively, suggesting that RLIP could be an important therapeutic target in BC (15).
RLIP is a 76 kDa, multi-domain splice variant protein encoded by the Ral-binding protein 1 (RALBP1) gene in humans (16,17). RLIP, a downstream effector of the small GTPases RalA and RalB, was cloned as a Ral-binding protein and predicted to be an effector involved in the regulation of membrane plasticity, cell movement, and endocytosis (16–18). Through a series of biochemical, molecular, cellular, and animal studies, we have established that RLIP is the quantitatively dominant transporter of glutathione-electrophile conjugates (GS-Es) and the rate-regulating enzyme of the mercapturic acid pathway (MAP) (19,20). Studies in knockout mice, as well as mouse embryonic fibroblasts (MEFs), show up to 80% loss of GS-E transport function upon loss of RLIP (9,21). Other investigators have shown that RLIP is a Ral-regulated component of clathrin-dependent endocytosis that is bound to the AP2 clathrin adaptor protein and other endocytic components, including POB1 (17,18), and the endocytosis of EGF and insulin by mouse embryonic fibroblasts is reduced by 80% by RLIP knockout. The ability of RLIP mutants to reconstitute clathrin-dependent endocytosis in RLIP knockout mouse embryonic fibroblasts parallels their GS-E transport activity, indicating that the function of RLIP as an ATP-dependent transporter of GS-E in MAP is coupled with its function as a rate-determining ATPase in endocytosis (21). More remarkably, RLIP knockout mice are highly resistant to carcinogenesis caused by benzo[a]pyrene, the most potent known chemical carcinogen (21). These observations have led us to consider an existential role of RLIP in cancer and to hypothesize that RLIP depletion should have anticancer activity in BC.
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas9)-based genetic screens are a powerful new tool in biology. The discovery and engineering of the CRISPR/Cas9 system over the past several years has revolutionized genome editing techniques and functional genetics studies (22). Directed by an RNA chimera (single-guide RNA, or sgRNA), which comprises a direct sequence of 20 nucleotides and an RNA scaffold, the Cas9 endonuclease can generate double-strand breaks in a targeted DNA locus (23). In mammalian cells, double-strand breaks are repaired through non-homologous end-joining, which often leads to small insertions or deletions (indels) (23). Thus, the CRISPR/Cas9 system provides a simple way to abolish the expression of selected proteins by shifting the reading frame or disrupting splicing sites. Recently, inducible CRISPR/Cas9-based genome editing strategies have been developed. For example, some studies have used knock-in techniques to introduce doxycycline-inducible Cas9 into DNA loci. This inducible CRISPR/Cas9 system dramatically decreases off-target effects by limiting exposure of the genome to the Cas9/sgRNA complex (24). By simply altering the sequence of the sgRNA, one can reprogram Cas9 to target different sites in the genome with relative ease, but the on-target activity and off-target effects of individual sgRNAs can vary widely. Doxycycline-controlled tetracycline (Tet) systems provide a powerful and commonly used method for functional studies on the consequences of gene overexpression/downregulation (25,26). Thus, we endeavored to disrupt the RLIP gene using inducible Cas9 and sgRNAs that specifically recognize RLIP sequences. Here, we identified sgRNAs that specifically target RLIP and evaluated the effects of Cas9 and RLIP sgRNA on the survival, proliferation, and tumorigenicity of the cultured BC cells in vitro and on BC tumor growth in vivo. Overall, our findings validate RLIP as an attractive target in BC.
Materials and methods
Reagents
Horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies, doxycycline hydrochloride, and 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were purchased from Sigma–Aldrich (St. Louis, MO). Antibodies against pAKT (S473), CD31, Ki67, CDK4, Bcl2, survivin, Bim, vimentin, and E-cadherin were purchased from Santa Cruz Biotechnology (Columbus, OH) and Cell Signaling Technologies (Danvers, MA). CellTiter-Glo was procured from Promega (Madison, WI). Tet-free fetal bovine serum (US origin) was purchased from Access Cell Culture (Vista, CA). The terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) fluorescence detection kit was purchased from Promega. The avidin–biotin complex detection kit was procured from Vector (Burlingame, CA). The universal Mycoplasma detection kit was purchased from American Type Culture Collection (ATCC; Manassas, VA). The anti-RLIP antibodies were obtained as described previously (27).
Cell lines and culture
Untransformed human breast epithelial (MCF10a) and human BC (MCF7 and MDA-MB231) cell lines were purchased from ATCC. Cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (P/S). The Integrative Genomics Core (Beckman Research Institute of City of Hope, Duarte, CA) analyzed 15 different human short tandem repeats to authenticate the cell lines and test for interspecies contamination. Cells were also tested for Mycoplasma once every 3 months.
CRISPR/Cas9-RLIP sgRNA methodology
We designed two sgRNAs, sgRLIP-Blast-3p and sgRLIP-Blast-5p, to target 78 bp downstream of the 5′ splicing join point of exon 3 of RLIP (CTTGAAAACATGGATTCCCT) and 1015 bp downstream of the 3′ splicing join point of intron 2–3 (ATTTGAGGTCCTATTCAGTA), respectively. These sgRNAs were cloned into the viral vectors pX458 (Addgene: 48138), using BbsI digestion, and pLX-U6-sgR-BsmbI-Blast (constructed by Dr Chih-Hong Lou, Gene Editing and Viral Vector Core [GEVVC], City of Hope), using BsmbI digestion. All sgRNAs were tested for their cutting efficiency in HEK293T cells using Surveyor Mismatch Cleavage Assays. Then, pLX-U6-sgRLIP-Blast-5p, pLX-U6-sgRLIP-Blast-3p, and a Tet-inducible Cas9 vector, pCW-CAS9 (Addgene: #50661), were used for lentiviral production in HEK293T cells. Briefly, HEK293T cells were transfected with one of the lentiviral vector plasmids (pLX-U6-sgRLIP-Blast-5p, pLX-U6-sgRLIP-Blast-3p, or pCW-CAS9) and three helper plasmids (pCgp, containing the gag/pol gene; pCMV-Rev2, containing the rev gene; and pCMV-G, containing the VSV-G gene) using the calcium phosphate transfection method. The viral particles were harvested 72 h later, followed by PEG precipitation. The MCF7 and MDA-MB231 BC cells were transduced with pCW-CAS9 for 48 h, followed by puromycin (1 µg/ml) screening for 7 days. The puromycin-resistant cells were then transduced with both pLX-U6-sgRLIP-Blast-5p and pLX-U6-sgRLIP-Blast-3p viruses for 48 h, followed by blasticidin (5 µg/ml) screening for 10 days. After screening, multiple inducible Cas9/RLIP sgRNA (Cas/sg RLIP) clones were picked from each cell line, and 6–8 per line were provided by Dr Chih-Hong Lou (GEVVC).
MTT cell viability assay
MCF7 and MDA-MB231 Cas/sg RLIP clones (five clones per cell line) were seeded in 96-well, flat-bottomed microtiter plates (5000 cells per well) for 24 h. Each clone was seeded in two vertical rows (one for control and another for doxycycline treatment) in four different plates. After a 24-h incubation at 37°C, cells were treated with either saline or doxycycline (final concentration 0.5 µg/ml) for 24 (plate #1), 48 (plate #2), 72 (plate #3), or 96 h (plate #4) at 37°C in a CO2 incubator. Next, 10 µl MTT (5 mg/ml stock) was added, and after a 2-h incubation at 37°C, the medium was removed and the pellet was washed with saline. The resulting formazan crystals were dissolved in 100 µl of DMSO with gentle shaking for 2 h at room temperature, and the absorbance of each well was measured at 570 nm using a microplate reader (xMark™ Spectrophotometer, Bio-Rad, Hercules, CA). The percentage of surviving cells was calculated using background-corrected absorbance. The data shown represent the mean and standard deviation of eight replicate wells for each time point in three independent experiments.
Colony formation assay
Cell survival was evaluated using a standard colony formation assay. Aliquots of 5,000 or 10,000 MCF7 and MDA-MB231 Cas/sg RLIP clones (five per cell line, control and treated with doxycycline at a final concentration 0.5 µg/ml for 48 h) were added to 60 mm Petri dishes containing 4 ml culture medium. After 7 days, adherent colonies were fixed, stained with 0.5% methylene blue for 30 min, and counted using the Innotech Alpha Imager HP (9).
Detection of apoptosis by TUNEL assay
We assessed whether levels of RLIP in cultured BC cells could be depleted by Cas/sg RLIP followed by doxycycline treatment and whether RLIP uptake correlates with apoptosis, as measured by the TUNEL assay. MCF7 and MDA-MB231 Cas/sg RLIP clones (five per cell line, control and treated with doxycycline at a final concentration 0.5 µg/ml for 48 h) were grown on sterile coverslips in 12-well tissue culture-treated plates. The TUNEL assay was performed using a fluorescence detection kit (Promega), according to the protocol provided by the manufacturer.
Quantitative real-time PCR
qPCR was performed using RLIP primers and SYBR Green master mix on an ABI-7500 fast real-time PCR system. Briefly, 1 µg of total RNA from MCF7 and MDA-MB231 Cas/sg RLIP clones (five per cell line, control and treated with doxycycline at a final concentration of 0.5 µg/ml for 24 and 48 h) was used to synthesize cDNA by reverse transcription using an RT kit (Applied Biosystems). Data were analyzed using the 2−ΔΔCt method (28).
Western blotting
MCF7 and MDA-MB231 Cas/sg RLIP clones (0.5 × 106 cells/ml) grown in Dulbecco's modified Eagle's medium supplemented with 10% Tet-free fetal bovine serum were seeded in a six-well plate. The cells were treated for 0, 24, 48, 72, and 96 h with doxycycline (final concentration 0.5 µg/ml). Cell pellets were lysed using an SDS lysis buffer containing protease and phosphatase inhibitors (Roche; Indianapolis, IN) and briefly sonicated to dissociate the cell membranes. After centrifugation, the supernatant was collected. 50 µg of total protein from control and doxycycline-treated cells was separated on SDS-PAGE gels at 200 V for 1 h. The protein was then transferred to nitrocellulose membranes at 100 V for 90 min at 4°C. Blots were probed with primary antibodies overnight at 4°C. The following day, the blots were re-probed with anti-rabbit secondary antibodies for 1 h at room temperature. The membranes were developed using SuperSignal West Pico Chemiluminescent Substrate. Differences in the expression of the desired proteins were determined by densitometric scanning of the immuno-reactive bands. Nitrocellulose membranes were stripped in 62.5 mM Tris–HCl (pH 6.8) buffer containing 2% SDS and 100 mM β-mercaptoethanol for 30 min at 50°C. Stripped blots were washed five times in TBST, blocked, and re-probed with an alternative antibody. Equal loading of proteins was confirmed by stripping and re-probing the membranes with β-actin antibodies.
Selective disruption of RLIP by CRISPR/Cas9 induces tumor regression in a mouse model of BC
We implemented an inducible CRISPR/Cas9 strategy to investigate the effects of RLIP knockdown in suppression of tumor growth in nude mice xenograft model of BC. The inducible CRISPR/Cas9 platform enables the introduction of double-strand breaks within the gene of interest with unprecedented precision and efficiency. Animal studies were conducted in compliance with IACUC guidelines for the care and use of laboratory animals. Ten-week-old female nude (athymic nu/nu) mice (n = 30, Charles River Laboratories, Wilmington, MA) were housed in vented caging systems on a 12 h light/12 h dark cycle with uniform temperature and humidity. They were fed a standard diet of irradiated feed and allowed water ad libitum. About 2 × 106 MDA-MB231 cells carrying inducible Cas9 and control or RLIP sgRNA (Cas9/sg control and Cas9/sg RLIP, respectively) were implanted subcutaneously into the flanks of the mice. When the cross-sectional areas of the tumors reached ~30 mm2, the mice were stratified into five cohorts (5 mice in Groups #1–4; and 10 mice in Group #5): Group #1 mice received parental MDA-MB231 cells; Group #2 mice received MDA-MB231 cells transduced with a lentivirus expressing Cas9/sg control; Group #3 mice received MDA-MB231 cells transduced with Cas9/control sgRNA, followed by doxycycline treatment; Group #4 mice received MDA-MB231 cells transduced with a lentivirus expressing Cas9/sg RLIP; and Group #5 mice received MDA-MB231 cells transduced with lentivirus expressing Cas9/sg RLIP, followed by doxycycline treatment. Doxycycline was administered in the drinking water (2 mg/ml with 1% sucrose added to increase palatability) of Groups #3 and #5 for 2 weeks. As doxycycline has limited stability in water, the supply was changed every other day. Bodyweights and tumor size measurements (by caliper) were taken twice a week. The mice were monitored every other day and euthanized once there was a significant difference in the tumor sizes of mice in different groups. Upon euthanasia, the primary tumors were excised, and their weights and sizes were measured and compared between groups using unpaired Student’s t-tests. Tumors from half of the mice from each group were paraformaldehyde-fixed and paraffin-embedded for immunohistochemical analysis. Tumors from the remaining mice were snap-frozen in liquid nitrogen and stored at −80°C for molecular analysis.
Histopathological examination of tumor tissues for differentiation, proliferative, and angiogenic markers
Tumor tissues from nude mice bearing Cas9/sg control and Cas9/sg RLIP MDA-MB231 tumors, without and with doxycycline treatment, were fixed in buffered formalin for 12 h. About 5 µm-thick paraffin-embedded tissue sections were prepared and stained with hematoxylin and eosin (H&E) to assess hyperplasia. Sections were also analyzed for the expression of proteins involved in the epithelial–mesenchymal transition, such as E-cadherin and vimentin; CD31 to visualize blood vessels; and Ki67 and RLIP to assess cell proliferation using a Universal ABC detection kit (Vector). Immuno-reactivity is evident as a dark brown stain, whereas non-reactive areas display only the background color. Photomicrographs at 40× magnification were acquired using an Olympus DP72 microscope. Percent staining was determined by measuring positive immuno-reactivity per unit area. The intensity of antigen staining was quantified by digital image analysis using DP2-BSW software. Bars represent mean ± SE (n = 5); *P < 0.003 compared with controls.
Statistical analysis
The data are expressed as the mean ± SD and were evaluated using two-tailed, unpaired Student’s t-tests. Changes in tumor size and body weight over the experiment period were visualized by scatter plot. The differences between control and treatment groups were evaluated for statistical significance by analysis of variance followed by multiple comparison tests. Differences were considered statistically significant when the P-value was <0.05.
Ethics statement
No human subjects were involved in the present study. All animal studies were conducted according to a protocol approved by the City of Hope Animal Care and Ethics Committee (IACUC protocol #12024). Any mice showing signs of distress, pain, or suffering due to tumor burden were humanely euthanized.
Results
RLIP protein expression in human BC cell lines
RLIP protein levels are higher in various cancer cell lines and tissues than in their normal counterparts (27,29–32). In our recent studies, we investigated the protein expression of RLIP in immortalized non-tumorigenic mammary epithelial cells (MCF10a) and BC cells (MCF7, T47D, SKBR3, TMD231, and MDA-MB231). As expected, the protein expression levels of RLIP were higher in all BC cell lines than in MCF10a cells (33,34). This observation suggests that RLIP may be an attractive target for inhibiting BC cell growth.
Selective and efficient Cas9/sgRNA-directed disruption of RLIP in cancer cells
The CRISPR/Cas9 system has emerged as a novel and effective tool for gene knockout studies (22–24,35).
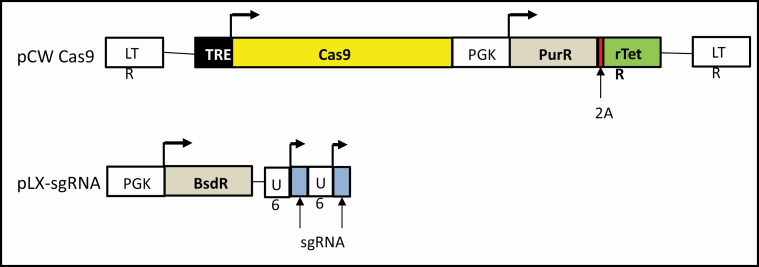
RLIP (human gene RalBP1, ral-binding protein, 18p11.22), a major and multi-specific transporter of MAP, is a protein that transports anticancer drugs out of the cancer cells, is upregulated in multiple cancers and is known to induce apoptotic- and drug-resistance. To target RLIP in cancer cells using the CRISPR/Cas9 system, we first screened for sgRNAs that specifically target RLIP. We used Tet-inducible Cas9 and selected sgRNAs to knockdown the RLIP gene in MCF7 and MDA-MB231 BC cell lines. The sgRNA sequences are presented in Methods section. The RLIP gene was knocked out to assess the role of MAP transporter in protecting BC cells from signaling events that impair their survival. These studies were conducted in consultation with Dr Chih-Hong Lou (GEVVC of City of Hope) (Figure 1).
Figure 1.
Target gene knockdown by CRISPR/Cas9. Schematic representation of a sensitive and efficient inducible CRISPR/Cas9 system: The pCW-CAS9 and pLX-sgRNA constructs were used for inducible CRISPR/Cas9-dependent RLIP gene knockdown.
Targeting RLIP with CRISPR/Cas9 inhibits cancer cell survival, proliferation, and tumorigenicity in vitro
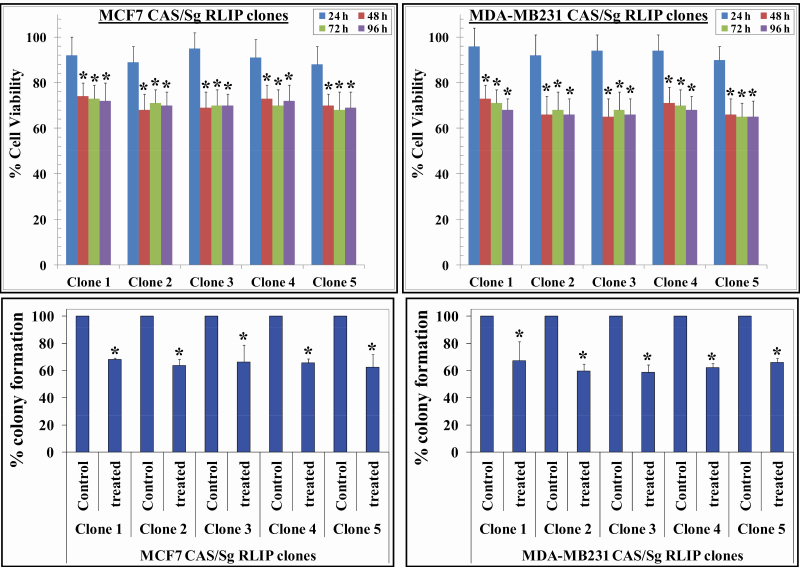
To validate the function of the selected sgRNAs at the endogenous targets, we transduced BC cells with lentiviral vectors encoding Cas9 and the corresponding sgRNA and determined whether such disruption of RLIP could affect BC cell survival, proliferation, and tumorigenicity in vitro. The transduced cells were subjected to cell survival and colony formation assays. Expression of Cas9 and RLIP sgRNA in MCF7 cells led to 32 ± 5% and 38 ± 4% reductions in survival and colony formation, respectively, suggesting that disrupting RLIP using Cas9 and sgRNA inhibits the survival and tumorigenicity of MCF7 BC cells (Figure 2). Similar Cas9 and RLIP sgRNA-induced reductions in survival and colony formation were also observed in MDA-MB231 cells, indicating inhibition of survival and tumorigenicity (Figure 2).
Figure 2.
RLIP knockdown activated by doxycycline inhibits cell survival and tumorigenicity. MCF7 and MDA-MB231 Cas9/sg RLIP clones were treated with doxycycline (final concentration 0.5 µg/ml) for 24–96 h, and MTT assays were used to assess cell viability. Values are presented as mean ± SD from three independent experiments with eight replicates each (n = 24). Bars represent the percentage of surviving cells after doxycycline (doxy) treatment at each time point for all five clones from both cell lines (*P < 0.05 compared with control) (upper panels). Colony formation assays were performed and colonies were counted using an Innotech Alpha Imager HP. The bars represent the percent colony formation (*P < 0.03 compared with control) (lower panels).
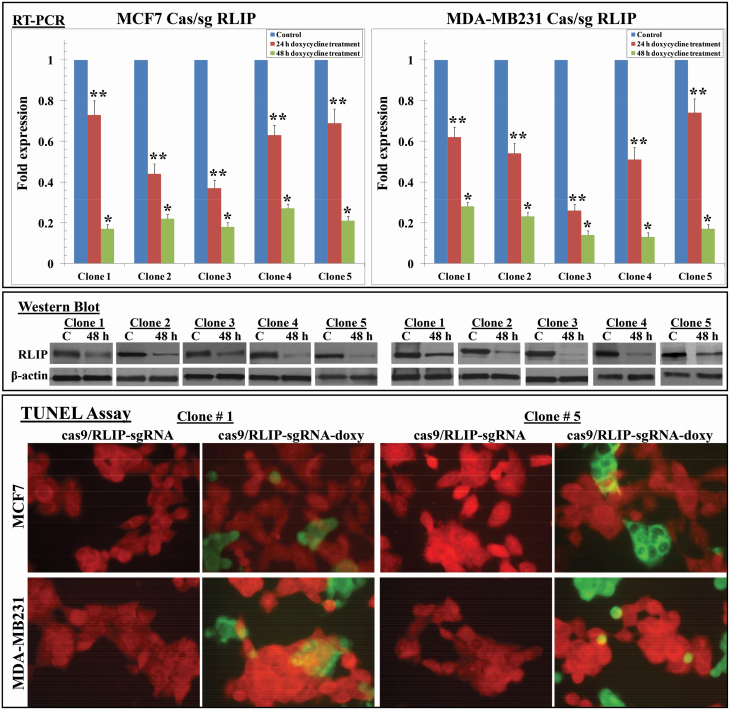
We also evaluated the effects of Cas9 and RLIP sgRNA on RLIP mRNA using qPCR. After lentiviral delivery of Cas9 and RLIP sgRNA, live cells were counted, and 5,000 cells were seeded into 96-well plates for the MTT assay and 50,000 were seeded into six-well plates for the qPCR assay using RLIP primers. The MTT assays performed after 48 h of treatment with doxycycline (0.5 µg/ml) showed that the average proportions of viable MCF7 and MDA-MB231 cells expressing Cas9 and RLIP sgRNA, relative to control cells, were 0.68 and 0.65, respectively, suggesting that RLIP disruption using Cas9 and sgRNA inhibits cell proliferation or survival. Because depletion of RLIP triggers apoptosis, MCF7 and MDA-MB231 Cas9/sg RLIP clones (five per cell line) were randomly selected for qPCR. The level of RLIP mRNA expression in MCF7 and MDA-MB231 clones treated with doxycycline for 48 h was decreased to 70–85% of that in control (untreated) clones (Figure 3 upper panels). Western blot analysis of the detergent-extracted crude membrane fractions of these clones revealed an approximately 50–80% reduction in RLIP expression in both MCF7 and MDA-MB231 clones after 48 h doxycycline treatment (Figure 3 middle panels). The mechanism of cytotoxicity was further assessed using an immunohistochemical TUNEL assay to measure apoptosis. TUNEL assays detect the appearance of DNA fragmentation, a late event in apoptosis. The TUNEL assay showed no detectable apoptosis in the cells that were not treated with doxycycline. Apoptosis was, however, seen in cells treated with doxycycline (Figure 3 lower panels). After demonstrating the efficiency of this inducible genome editing system in cell culture, we tested our system in a xenograft model of BC and evaluated whether targeting RLIP could suppress tumor growth in vivo.
Figure 3.
Effect of Cas/sg RLIP on RLIP gene and protein expression. qPCR using RLIP primers (upper panels), Western blot analysis using antibodies against RLIP (middle panels), and TUNEL assays (lower panels) were performed using MCF7 and MDA-MB231 Cas9/sg RLIP clones (without [control] or with doxycycline treatment, final concentration 0.5 µg/ml) and normalized against β-actin. In qPCR values are means with 95% confidence intervals (n = 4 for each clone); *P < 0.01 and **P < 0.03 compared with respective controls. Data were analyzed using the 2−ΔΔCt method. Effect of Cas9/sg RLIP on apoptosis. MCF7 and MDA-MB231 Cas9/sg RLIP clones without or with doxycycline treatment (0.5 µg/ml, 48 h) were grown on coverslips. TUNEL assays using a Promega fluorescence detection kit were performed using a Zeiss LSM 510 META laser scanning fluorescence microscope with 520 and >620 nm filters. Photographs presented were taken at identical exposure with 400x magnification. Apoptotic cells showed green fluorescence and characteristics of cell shrinkage.
Targeting RLIP with CRISPR/Cas9 blocks tumor growth in vivo
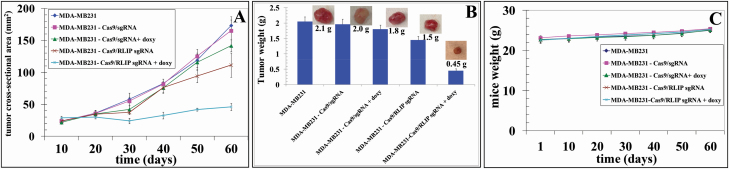
Human cancer cells grown as subcutaneous xenografts in immuno-deficient mice represent one of the most frequently used in vivo models for drug target validation and preclinical drug testing in translational cancer research (36). The xenografted cancer cells are often engineered for inducible expression/suppression of a gene of interest to enable a more precise assessment of its function(s). Inducible gene expression/suppression based on doxycycline-controlled Tet systems provides a powerful and commonly used method for functional studies on the consequences of gene overexpression/downregulation (37,38). To confirm the results of our in vitro studies, we used a subcutaneous xenograft model based on MDA-MB231 BC cells engineered for doxycycline-inducible knockdown of RLIP expression. MDA-MB231 BC cells carrying control or RLIP sgRNAs were injected subcutaneously into athymic nude mice. Palpable growth (i.e., ~30 mm2 cross-sectional area) was seen in all animals 15 days after cell implantation. Palpable tumor-bearing animals in Groups #3 and #5 were treated with doxycycline in drinking water (2 mg/ml) for 14 days, and tumors were analyzed for the expression of RLIP as well as anti- and pro-apoptotic proteins. We found that RLIP sgRNA dramatically inhibited tumor growth and tumor weights in the doxycycline-treated group (Figure 4 and Supplementary Figure 1, available at Carcinogenesis Online), and significantly lower RLIP expression was also observed in RLIP sgRNA-containing tumors of doxycycline-treated mice, compared with tumors isolated from mice injected with control cells or that did not receive doxycycline (Figure 5). Tumors grew more slowly in mice that received MDA-MB231-Cas9/sg RLIP xenografts + doxycycline (Group #5) than in mice in the other groups (Figure 4A). The average wet weights of the tumors at day 60 was also lower for the Cas9/sg RLIP sgRNA + doxycycline group (0.45 g) than for other groups (Group #1: 2.1 g, Group #2: 2.0 g, Group #3: 1.8 g, and Group #4: 1.5 g, respectively; Figure 4B). The average weight gain among mice in the five groups was similar, demonstrating that all treatments were safe and produced no toxicity (Figure 4C). No macroscopic evidence of metastasis to other organs was evident for any experimental groups. The deletion efficiency in the tumor cells was probably similar to what was observed in the cell culture experiments due to the existence of non-tumor cells in the tumor tissues. Moreover, the tumors isolated from mice from Groups #1–4 did not show any reduction in RLIP protein expression, suggesting that Cas9-mediated RLIP deletion was tightly controlled (Figure 5). These results demonstrate that the selective disruption of RLIP by Cas9/sg RLIP induced a significant reduction in tumor size compared with Cas9/sg control.
Figure 4.
Effect of CRISPR/Cas9-mediated RLIP knockdown on the size of subcutaneously implanted human BC (MDA-MB231) xenograft tumors in nude mice. Thirty 10-week-old mice were divided into five groups: (1) MDA-MB231, (2) MDA-MB231-Cas9/sg control, (3) MDA-MB231-Cas9/sg control + doxycycline, (4) MDA-MB231-Cas9/sg RLIP, and (5) MDA-MB231-Cas9/sg RLIP + doxycycline. Tumors grew more slowly in mice that received MDA-MB231-Cas9/sg RLIP + doxycycline (Group #5) than in mice in the other groups (panel A). The average wet weights of the tumors at day 60 was also lower for the Cas9/sg RLIP + doxycycline group (0.45 g) than for the other groups (Group #1: 2.1 g, Group #2: 2.0 g, Group #3: 1.8 g, and Group #4: 1.5 g, respectively; panel B). The Cas9/sg RLIP + doxycycline-treated mice did not exhibit any signs of overt toxicity, such as impaired movement or posture, indigestion, or areas of redness or swelling. The initial and final body weights of the controls and treated mice did not differ significantly (panel C).
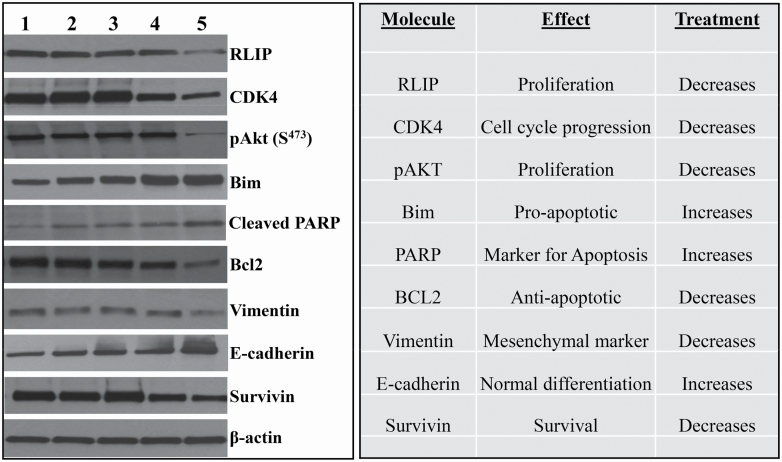
Figure 5.
CRISPR/Cas9-mediated RLIP knockdown regulates the levels of signaling proteins in human BC (MDA-MB231) xenograft tumors in nude mice. Western blot analyses of signaling proteins in MDA-MB231 human BC tumor tissue lysates from (1) MDA-MB231, (2) MDA-MB231-Cas9/sg control, (3) MDA-MB231-Cas9/sg control + doxycycline, (4) MDA-MB231-Cas9/sg RLIP, and (5) MDA-MB231-Cas9/sg RLIP +doxycycline groups. β-actin was used as a loading control.
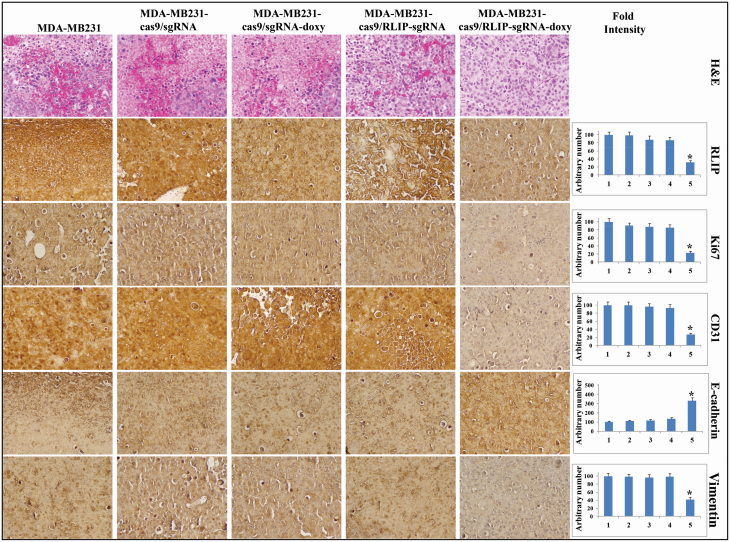
Tumor tissue lysates from mice in all five groups were assessed for proliferation, apoptosis, and cell cycle markers by Western blot analysis (Figure 5) and immunohistochemistry (Figure 6). The Western blot analyses revealed that tumors from mice treated with Cas9/sg RLIP + doxycycline had lower levels of the proliferation and survival proteins pAKT, survivin, CDK4, and Bcl2, as well as the mesenchymal marker vimentin, and higher levels of Bim, cleaved PARP, and the normal differentiation marker E-cadherin, compared tumors isolated from mice injected with control cells (Figure 5). Immunohistochemical staining of Cas9/sg RLIP + doxycycline tumor tissue sections showed remarkable disappearance of the proliferative marker Ki67, RLIP, and the angiogenesis marker CD31. E-cadherin, a marker of differentiation that signifies reversal of epithelial–mesenchymal transition, were remarkably higher, whereas levels of the vimentin were lower, in Cas9/sg RLIP + doxycycline tumor tissue compared with control tissue (Figure 6). These results established that targeting RLIP promotes anticancer activity in BC.
Figure 6.
Immunohistochemical analyses revealing that CRISPR/Cas9-mediated RLIP knockdown inhibits proliferation and angiogenesis markers in BC (MDA-MB231) xenograft tumors in mice. Sectioned tumor tissues from nude mice bearing Cas9/sg control and Cas9/sg RLIP MDA-MB231 tumors, without or with doxycycline treatment, were used for histopathologic analyses. Presented are H&E-stained sections and immunohistochemical analyses for the expression of RLIP, Ki67, CD31, E-cadherin, and vimentin. The difference between expression in sections from MDA-MB231 Cas9/sg RLIP tumor-bearing mice treated with doxycycline and sections from mice in the control groups was statistically significant (P < 0.02, determined by a two-tailed Student’s t-test). Photomicrographs at 40x magnification were acquired using an Olympus DP72 microscope. Percent staining was determined by measuring positive immunoreactivity per unit area. The intensity of antigen staining was quantified by digital image analysis using Pro Plus software. Bar represent mean ± SE (n = 5).
Discussion
Despite significant progress in our knowledge concerning the risk factors and genomic heterogeneity of BC, this malignancy remains a leading cause of cancer-related deaths in women worldwide (39). Metastatic spread to distant organs is the primary cause of morbidity and mortality in patients with BC (40). Women with BC may experience metastasis to the lungs, liver, and brain, but bone is the preferred site of metastatic spread for each subtype of the disease (43–71%) compared with the other sites (8–47%) (41). Present studies demonstrate the sensitivity of human BC cells to RLIP depletion or inhibition in vitro and in vivo.
The human RALBP1 gene encodes a 76 kDa splice variant protein, RLIP, which is a stress-protective MAP transporter protein that also plays a key role in regulating clathrin-dependent endocytosis as a Ral effector (17,18,21). Targeted inhibition or depletion of RLIP causes regression of human xenografts of lung, colon, prostate, and kidney cancer without inducing toxicity in nude mouse models (27,29–31,42). The present studies were performed to determine whether targeted depletion of RLIP using CRISPR/Cas9 inhibits breast tumor growth. Queries of the GEO, EGA, and TCGA databases revealed that RLIP expression correlates with overall or relapse-free survival in subsets of BC patients and is associated with significant differences in the frequencies of mutations and copy number alterations in several top genetically altered genes in BC, including PIK3CA, TP53, and MYC (43,44). Previous studies demonstrated that RLIP was expressed in human BC tissues, as well as BC cell lines (32–34). Knockdown of RLIP resulted in apoptotic death of MCF7 and SKBR3 cells, and xenograft studies in nude mice showed regression of BC upon targeted depletion of RLIP, as well as treatment with anti-RLIP antibodies that recognized an epitope of RLIP on the cell surface. Signaling studies showed that RLIP depletion inhibited endocytosis and signaling to Akt, MYC, STAT3/JAK2, CDK4, and ERK1/2 (21,33,34).
The CRISPR/Cas9 system is a powerful genome-editing tool that has been widely used for biomedical research (45). Programmable nucleases enable the targeting of specific genomic sequences, and we now show that cells containing specific cancer-causing sequences can be selectively controlled both in vitro and in vivo using CRISPR/Cas9. The results from our in vitro and in vivo studies suggest for the first time that RLIP has strong potential as a target for the treatment of metastatic BC. Furthermore, we observed that compared with tumor sections from control mice, sections from mice that received cells expressing RLIP sgRNA and Tet-inducible Cas9, followed by doxycycline treatment, exhibited lower RLIP, Ki67, CD31, and vimentin expression in immunohistological studies (Figure 6), as well as lower RLIP, pAKT, vimentin, survivin, Bcl2, and CDK4 expression and higher cleaved PARP, Bim, and E-cadherin expression in Western blot analyses (Figure 5). Loss of E-cadherin expression with a concomitant increase in the protein levels of mesenchymal markers (e.g., vimentin) is a biochemical hallmark of epithelial–mesenchymal transition, a normal physiological process that is also implicated in cancer metastasis (46–50). The expression of the targeted protein, RLIP, as well as its mRNA levels was also decreased to 50–85% after 48 h doxycycline treatment in both MCF7 and MDA-MB231 clones. These findings were mirrored in the results of TUNEL assays that detect the last step in apoptosis (Figure 3). These results indicated that our inducible multiplex system worked efficiently in vitro and in vivo.
It is important to note that the editing efficiency of CRISPR on the oncogene was not 100% and that the delivery efficiency of Cas9 and the sgRNA to cancer cells was also not 100%, which prevented this strategy from causing complete tumor remission. Furthermore, our successful application of CRISPR/Cas9 to target the cancer driver RLIP in vitro and in vivo could be utilized to investigate the functional and regulatory pathways of other oncogenic drivers. The present data demonstrate that targeting RLIP using CRISPR/Cas9 can control tumor growth in vivo, and our results also suggest that RLIP inhibition is probably the mechanism of action.
Significance
RLIP is an anti-apoptotic, non-ABC, GS-E transporter involved in receptor-ligand endocytosis, as well as in multi-drug transport and resistance. Partial inhibition of RLIP causes apoptosis in multiple cancer cell lines. Doxycycline-inducible expression of Cas9 in BC cells transduced with lentiviral vectors encoding RLIP sgRNAs disrupted the RLIP gene, leading to inhibition of BC cell proliferation both in vitro and in vivo. These findings indicate that RLIP should be a good target for cancer cell killing, as its downregulation promotes apoptosis through both drug-dependent and drug-independent effects.
Future perspectives
Drug agents for targeting RLIP have not been fully developed. The ability of RLIP to transport a wide variety of antineoplastic agents renders it a multi-drug transporter. Other multi-drug transporters have not proven to be ‘druggable,’ primarily because of toxicity. Whether this will be an issue for RLIP remains to be determined. In summary, the development of RLIP-targeting therapies for BC appear justified based on the demonstrated sensitivity of BC to RLIP knockdown, a lack of significant toxicity in laboratory studies in mice, and a reasonably well-defined mechanism of action that involves multiple key cancer-related signaling pathways. Our studies indicate that selecting patients for this treatment based on RLIP expression may not be required, but the presence of genetic alterations in related signaling molecules may be useful as predictive biomarkers to select patients who are probably to benefit from treatment.
Supplementary Material
Acknowledgements
We thank the personnel in the Animal Research Center and Pathology Cores for their invaluable assistance. The authors are grateful to Dr Chih-Hong Lou and Dr Jiing-Kuan Yee, Director of the Gene Editing and Viral Vector Core (GEVVC) at City of Hope, for technical assistance with CRISPR/Cas9 methodology. We sincerely thank Dr Ravi Salgia, MD, PhD, Professor and Chair, Department of Medical Oncology at City of Hope, for providing research space and support.
Glossary
Abbreviations
- BC
breast cancer
- CRISPR/Cas
clustered regularly interspaced short palindromic repeats/CRISPR-associated protein
- GS-E
glutathione-electrophile conjugate
- MAP
mercapturic acid pathway
- RLIP
ral-interacting protein
- sgRNA
single-guide RNA
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
Ethics approval and consent to participate
No human subjects were involved in the present study. All animal studies were conducted according to a protocol approved by the City of Hope Animal Care and Ethics Committee (IACUC protocol # 12024).
Consent for publication: Not Applicable
Availability of data and material
All data generated and analyzed during the current study are available from the corresponding author upon request.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported in part by grants from the United States Department of Defense (W81XWH-16-1-0641) and the National Cancer Institute of the National Institutes of Health (P30CA33572). Funding from the Beckman Research Institute of City of Hope is also acknowledged.
Conflict of Interest Statement: None declared.
Authors’ contributions
J.S. contributed to data collection. S.C. Contributed to data collection and analysis. D.H. Contributed to reviewing/editing the manuscript. S.A. Contributed to discussion and review/editing the manuscript. R.S. Contributed to data interpretation. S.S.S. Contributed to data collection and wrote the manuscript.
References
- 1. DeSantis, C.E. et al. (2016) Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA. Cancer J. Clin., 66, 31–42. [DOI] [PubMed] [Google Scholar]
- 2. Polyak, K. (2011) Heterogeneity in breast cancer. J. Clin. Invest., 121, 3786–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fisher, C.M. et al. (2014) Frontiers in radiotherapy for early-stage invasive breast cancer. J. Clin. Oncol., 32, 2894–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carlson, J.J. et al. (2013) The impact of the Oncotype Dx breast cancer assay in clinical practice: a systematic review and meta-analysis. Breast Cancer Res. Treat., 141, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diallo-Danebrock, R. et al. (2007) Protein expression profiling in high-risk breast cancer patients treated with high-dose or conventional dose-dense chemotherapy. Clin. Cancer Res., 13(2 Pt 1), 488–497. [DOI] [PubMed] [Google Scholar]
- 6. Awasthi, Y.C. et al. (2007) The non-ABC drug transporter RLIP76 (RALBP-1) plays a major role in the mechanisms of drug resistance. Curr. Drug Metab., 8, 315–323. [DOI] [PubMed] [Google Scholar]
- 7. Giai, M. et al. (1991) Chemoresistance in breast tumors. Eur. J. Gynaecol. Oncol., 12, 359–373. [PubMed] [Google Scholar]
- 8. Rodman, S.N. et al. (2016) Enhancement of radiation response in breast cancer stem cells by inhibition of thioredoxin- and glutathione-dependent metabolism. Radiat. Res., 186, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singhal, J. et al. (2008) RLIP76 in defense of radiation poisoning. Int. J. Radiat. Oncol. Biol. Phys., 72, 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Green, A.R. et al. (2016) MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br. J. Cancer, 114, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lluch, A. et al. (2014) Emerging EGFR antagonists for breast cancer. Expert Opin. Emerg. Drugs, 19, 165–181. [DOI] [PubMed] [Google Scholar]
- 12. Singhal, S.S. et al. (2010) Rlip76 transports sunitinib and sorafenib and mediates drug resistance in kidney cancer. Int. J. Cancer, 126, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muller, P.A. et al. (2013) p53 mutations in cancer. Nat. Cell Biol., 15, 2–8. [DOI] [PubMed] [Google Scholar]
- 14. Mohammed, M.K. et al. (2016) Wnt/β-catenin signaling plays an ever-expanding role in stem cell self-renewal, tumorigenesis and cancer chemoresistance. Genes Dis., 3, 11–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Awasthi, S. et al. (2018) Rlip depletion prevents spontaneous neoplasia in TP53 null mice. Proc. Natl. Acad. Sci. U. S. A., 115, 3918–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cantor, S.B. et al. (1995) Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol., 15, 4578–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jullien-Flores, V. et al. (1995) Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem., 270, 22473–22477. [DOI] [PubMed] [Google Scholar]
- 18. Morinaka, K. et al. (1999) Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene, 18, 5915–5922. [DOI] [PubMed] [Google Scholar]
- 19. Awasthi, S. et al. (1994) Adenosine triphosphate-dependent transport of doxorubicin, daunomycin, and vinblastine in human tissues by a mechanism distinct from the P-glycoprotein. J. Clin. Invest., 93, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Awasthi, S. et al. (2000) Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry, 39, 9327–9334. [DOI] [PubMed] [Google Scholar]
- 21. Singhal, S.S. et al. (2011) Glutathione-conjugate transport by RLIP76 is required for clathrin-dependent endocytosis and chemical carcinogenesis. Mol. Cancer Ther., 10, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hsu, P.D. et al. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157, 1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mali, P. et al. (2013) RNA-guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dow, L.E. et al. (2015) Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol., 33, U390–U398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aubrey, B.J. et al. (2015) An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep., 10, 1422–1432. [DOI] [PubMed] [Google Scholar]
- 26. Cawthorne, C. et al. (2007) Comparison of doxycycline delivery methods for Tet-inducible gene expression in a subcutaneous xenograft model. J. Biomol. Tech., 18, 120–123. [PMC free article] [PubMed] [Google Scholar]
- 27. Singhal, S.S. et al. (2007) Regression of lung and colon cancer xenografts by depleting or inhibiting RLIP76 (Ral-binding protein 1). Cancer Res., 67, 4382–4389. [DOI] [PubMed] [Google Scholar]
- 28. Singhal, S.S. et al. (2013) RLIP76 protein knockdown attenuates obesity due to a high-fat diet. J. Biol. Chem., 288, 23394–23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singhal, S.S. et al. (2006) Regression of melanoma in a murine model by RLIP76 depletion. Cancer Res., 66, 2354–2360. [DOI] [PubMed] [Google Scholar]
- 30. Singhal, S.S. et al. (2009) Regression of prostate cancer xenografts by RLIP76 depletion. Biochem. Pharmacol., 77, 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singhal, S.S. et al. (2009) RLIP76: a target for kidney cancer therapy. Cancer Res., 69, 4244–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang, C.Z. et al. (2015) RLIP76 expression as a prognostic marker of breast cancer. Eur. Rev. Med. Pharmacol. Sci., 19, 2105–2111. [PubMed] [Google Scholar]
- 33. Singhal, J. et al. (2017) 2’-Hydroxyflavanone: a novel strategy for targeting breast cancer. Oncotarget, 8, 75025–75037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singhal, J. et al. (2018) 2’-Hydroxyflavanone inhibits in vitro and in vivo growth of breast cancer cells by targeting RLIP76. Mol. Carcinog., 57, 1751–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shalem, O. et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sausville, E.A. et al. (2006) Contributions of human tumor xenografts to anticancer drug development. Cancer Res., 66, 3351–335 4, discussion 3354. [DOI] [PubMed] [Google Scholar]
- 37. Berens, C. et al. (2003) Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem., 270, 3109–3121. [DOI] [PubMed] [Google Scholar]
- 38. Wiznerowicz, M. et al. (2006) Tuning silence: conditional systems for RNA interference. Nat. Methods, 3, 682–688. [DOI] [PubMed] [Google Scholar]
- 39. van Zitteren, M. et al. (2011) Genome-based prediction of breast cancer risk in the general population: a modeling study based on meta-analyses of genetic associations. Cancer Epidemiol. Biomarkers Prev., 20, 9–22. [DOI] [PubMed] [Google Scholar]
- 40. Weigelt, B. et al. (2005) Breast cancer metastasis: markers and models. Nat. Rev. Cancer, 5, 591–602. [DOI] [PubMed] [Google Scholar]
- 41. Kennecke, H. et al. (2010) Metastatic behavior of breast cancer subtypes. J. Clin. Oncol., 28, 3271–3277. [DOI] [PubMed] [Google Scholar]
- 42. Awasthi, S. et al. (2008) RLIP76 and Cancer. Clin. Cancer Res., 14, 4372–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pereira, B. et al. (2016) The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun., 7, 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao, J. et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cao, J. et al. (2016) An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res., 44, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tomaskovic-Crook, E. et al. (2009) Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res., 11, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singhal, J. et al. (2019) RLIP inhibition suppresses breast-to-lung metastasis. Cancer Lett., 447, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singhal, S.S. et al. (2019) Synergistic efficacy of RLIP inhibition and 2’-hydroxyflavanone against DMBA-induced mammary carcinogenesis in SENCAR mice. Mol. Carcinog., 58, 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krishna, B.M. et al. (2019) Notch signaling in breast cancer: from pathway analysis to therapy. Cancer Lett., 461, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Singhal, S.S. et al. (2019) RLIP: an existential requirement for breast carcinogenesis. Biochim. Biophys. Acta. Rev. Cancer, 1871, 281–288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed during the current study are available from the corresponding author upon request.