Abstract
The pectinolytic genus Dickeya (formerly Erwinia chrysanthemi) comprises numerous pathogenic species which cause diseases in various crops and ornamental plants across the globe. Their pathogenicity is governed by complex multi-factorial processes of adaptive virulence gene regulation. Extracellular polysaccharides and lipopolysaccharides present on bacterial envelope surface play a significant role in the virulence of phytopathogenic bacteria. However, very little is known about the genomic location, diversity, and organization of the polysaccharide and lipopolysaccharide biosynthetic gene clusters in Dickeya. In the present study, we report the diversity and structural organization of the group 4 capsule (G4C)/O-antigen capsule, putative O-antigen lipopolysaccharide, enterobacterial common antigen, and core lipopolysaccharide biosynthesis clusters from 54 Dickeya strains. The presence of these clusters suggests that Dickeya has both capsule and lipopolysaccharide carrying O-antigen to their external surface. These gene clusters are key regulatory components in the composition and structure of the outer surface of Dickeya. The O-antigen capsule/group 4 capsule (G4C) coding region shows a variation in gene content and organization. Based on nucleotide sequence homology in these Dickeya strains, two distinct groups, G4C group I and G4C group II, exist. However, comparatively less variation is observed in the putative O-antigen lipopolysaccharide cluster in Dickeya spp. except for in Dickeya zeae. Also, enterobacterial common antigen and core lipopolysaccharide biosynthesis clusters are present mostly as conserved genomic regions. The variation in the O-antigen capsule and putative O-antigen lipopolysaccharide coding region in relation to their phylogeny suggests a role of multiple horizontal gene transfer (HGT) events. These multiple HGT processes might have been manifested into the current heterogeneity of O-antigen capsules and O-antigen lipopolysaccharides in Dickeya strains during its evolution.
Introduction
The genus “Dickeya” (D.) is a newly formed group of six pectinolytic phytopathogenic species formerly classified as Erwinia chrysanthemi [1]. Tuber soft rot and stem rot (Blackleg) caused by these phytopathogenic enterobacterales, Dickeya spp. are prevalent in the European countries [2]. Dickeya species such as D. dadantii and D. solani are regarded as two of the top ten bacterial plant pathogens of economic and scientific significance [3]. Several species of Dickeya cause soft rot in many economically important crops, with the most prominent ones affecting potato, maize, tomato, Chrysanthemum spp., banana, and Dianthus spp. [1, 4]. The pathogenicity and the host range of these pectinolytic Dickeya depend on multi-factorial processes including adhesion, penetration, interaction with virulence factors like flagella, lipopolysaccharide, exopolysaccharide (EPS) and other effector proteins, and their multi-layer regulation [5].
Capsular polysaccharides (CPS) and lipopolysaccharides (LPS) are the major bacterial surface polysaccharides that continuously evolve to protect bacterial pathogens against bacteriophages [6, 7]. CPS are high molecular weight acidic polysaccharides that differ depending on the mechanism of their synthesis and assembly [8, 9]. Bacterial LPS are the major outer membrane surface components present in most Gram-negative bacteria. LPS typically consists of three parts: (a) a hydrophobic domain known as lipid A (or endotoxin) (b) a nonrepeating "core" oligosaccharide and (c) a distal polysaccharide chain (or O-antigen lipopolysaccharide). Lipid A, the most conserved hydrophobic part of LPS, consists of glucosamine disaccharide units with β (1→ 6) linkages. These sugars are linked to acylated fatty acid chains in the bacterial membrane. The core-oligosaccharide of LPS is divided into two main parts—the inner core and the outer core. The inner core consists of 3-deoxy-D-mannooctulosonic acid (Kdo) and heptose sugars, whereas the outer core consists of hexose sugars like D- glucose, D-mannose, D-galactose, etc. The O-antigen component is the most diverse part of LPS and provides a high degree of variability with specificity. Enteropathogenic and enterohaemorrhagic E. coli, a Gram-negative enterobacterium, forms a group 4 capsule (G4C)/ O-antigen capsule [10]. Compared to LPS, G4Cs are not attached to the bacterial surface via lipid but rather with specific surface proteins [11]. O-antigen modifications influence different pathogen infection stages, including colonization (adherence), and play a role in bypassing the host defense mechanism by masquerading host molecules in both plants and animals [12, 13]. Both LPS and CPS also contribute to antibacterial defense by mitigating the effects of antimicrobial peptides [14]. These complex polysaccharides are key virulence factors in plant-pathogen interactions [15, 16]. Therefore, it is of utmost importance to determine their role in Dickeya host-pathogen interactions.
Bacterial soft-rot primary symptoms include pectin degradation of the middle lamella and primary plant cell wall caused by secreted pectinases [17]. These infected plant tissues appear as wet macerated foul-smelling rot. These symptoms are common to many different types of phytopathogenic bacteria. Therefore, focus on key virulence determinants and their genomic counterparts specific to the genus Dickeya is essential. LPS and CPS are key virulence determinants of many phytopathogenic bacteria, and their different components play a crucial role during pathogenesis [18–21]. Proper identification and classification of these virulence factors will guide the diagnosis and disease prevention in a systematic way.
In addition to the previous six classified Dickeya species, three more species are reported. Dickeya solani is a recent addition to the genus Dickeya as a taxonomically established species [22]. Additionally, two new species, "Dickeya aquatica” and “Dickeya fangzhongdai," were also reported [23, 24]. Phylogenetic classification based on multi-locus sequence analysis (MLSA) corroborates with whole genome-based average nucleotide identity (ANI) for species resolution [25]. Accounting for 54 strains of the genus Dickeya, we performed species delineation using criteria of RNA-polymerase beta subunit (rpoB) gene identity, whole genome-based species criteria of ANI, and whole genome based digital DNA-DNA hybridization (dDDH) method [26]. The rpoB gene is a promising phylogenetic marker as it is a single-copy gene in the bacterial genome and relatively immune to HGT [27]. Our study reflects on the classification of different species of Dickeya based on these species criteria and their co-occurrence with specific complex polysaccharide biosynthesis clusters. In this study, we provide comprehensive analyses of G4C or O-antigen capsule cluster diversity within the genus Dickeya. We also summarize the variation and organization of the conserved genomic islands that regulate the biosynthesis of G4C (OAg-capsule), O-antigen lipopolysaccharides (OAg-LPS), Enterobacterial Common Antigen (ECA), and other core lipopolysaccharide biosynthesis cluster in these strains along with their evolution among different species of Dickeya.
Material and methods
Bacterial strains data
A total of 54 genomes of Dickeya species are included in these analyses (Table 1). All the genomes were downloaded from the National Center for Biotechnology Information (NCBI) ftp server (ftp://ftp.ncbi.nlm.nih.gov/genomes, downloaded in March 2019) using their accession number as mentioned in Table 1.
Table 1. Group 4 capsule (G4C) coding gene cluster of different species in Dickeya.
| SL. No | Dickeya Strains | G4C/O-antigen capsule cluster | NCBI Accession | ||
|---|---|---|---|---|---|
| Size (Kb) | No. Of ORF | GC content (%) | |||
| G4C Group I | |||||
| 1 | D. dianthicola GBBC 2039 | 22.3 | 20 | 52.5 | AOOM01000000 |
| 2 | D. dianthicola NCPPB 453 | 28.9 | 25 | 48.4 | AOOB01000000 |
| 3 | D. dianthicola DE440 | 28.9 | 25 | 48.4 | PJJB00000000 |
| 4 | D. dianthicola IPO 980 | 28.9 | 25 | 48.4 | AOOS01000000 |
| 5 | D. dianthicola ME23 | 28.9 | 25 | 48.4 | CP031560 |
| 6 | D. dianthicola RNS04.9 | 28.9 | 25 | 48.4 | CP017638 |
| 7 | D. dianthicola SS70 | 28.9 | 25 | 48.4 | QESZ00000000 |
| 8 | D. dianthicola WV516 | 28.9 | 25 | 48.4 | PJJC00000000 |
| 9 | D. dianthicola NCPPB 3534 | 28.9 | 25 | 48.4 | AOOK01000000 |
| 10 | D. dadantii NCPPB 898 | 31.3 | 27 | 48.9 | AOOE01000000 |
| 11 | D. dadantii DSM 18020 | 31.3 | 27 | 48.9 | CP023467 |
| 12 | D. dadantii NCPPB 3537 | 31.3 | 28 | 48.9 | AOOL01000000 |
| 13 | D. chrysanthemi NCPPB 516 | 31 | 27 | 47.6 | AOOC00000000 |
| 14 | D. sp. NCPPB 3274 | 36.1 | 31 | 50.2 | AOOH01000000 |
| 15 | D. dadantii 3937 | 32.1 | 26 | 48.1 | CP002038 |
| * | D. paradisiaca NCPPB 2511 | 6.8 | 5 | 51.9 | AONV01000000 |
| G4C Group II | |||||
| 1 | D. sp. 2B12 | 34.7 | 30 | 53.1 | JSYG00000000 |
| 2 | D. sp. FVG1-MFV-O17 | 34.7 | 30 | 53.1 | RJLR00000000 |
| 3 | D. sp. FVG10-MFV-A16 | 34.8 | 30 | 53.1 | RJLS00000000 |
| 4 | D. zeae EC1 | 34.8 | 30 | 50.8 | CP006929 |
| 5 | D. zeae DZ2Q | 34.8 | 30 | 50.9 | APMV00000000 |
| 6 | D. zeae Ech586 | 34.8 | 30 | 51.4 | CP001836 |
| 7 | D. zeae MS1 | 34.9 | 30 | 51.2 | APWM00000000 |
| 8 | D. zeae NCPPB 3531 | 34.7 | 30 | 51.2 | AOOI01000000 |
| 9 | D. zeae ZJU1202 | 34.8 | 30 | 50.8 | AJVN00000000 |
| 10 | D. fangzhongdai B16 | 36.7 | 31 | 55.4 | JXBN00000000 |
| 11 | D. fangzhongdai S1 | 36.7 | 31 | 55.4 | JXBO00000000 |
| 12 | D. sp. MK7 | 36.7 | 31 | 55.4 | AOOO01000000 |
| 13 | D. sp. Secpp 1600 | 36.7 | 31 | 55.3 | CP023484 |
| 14 | D. fangzhongdai DSM 101947 | 37.8 | 32 | 55.2 | CP025003 |
| 15 | D. fangzhongdai M005 | 37.6 | 32 | 55.1 | JSXD00000000 |
| 16 | D. fangzhongdai ND14b | 37.6 | 32 | 55.1 | CP009460 |
| 17 | D. fangzhongdai PA1 | 37.8 | 32 | 55.2 | CP020872 |
| 18 | D. dadantii subsp. dieffenbachiae NCPPB 2976 | 37.4 | 32 | 55.3 | AOOG01000000 |
| 19 | D. zeae MK19 | 35.6 | 31 | 51.2 | AOOR01000000 |
| 20 | D. zeae MS2 | 35.6 | 31 | 51.2 | CP025799 |
| 21 | D. zeae NCPPB 2538 | 35.6 | 31 | 51.2 | AOOF01000000 |
| 22 | D. zeae NCPPB 3532 | 35.6 | 31 | 51.2 | AONW01000000 |
| 23 | D. solani IPO 2222 | 35.9 | 31 | 54.8 | CP015137 |
| 24 | D. solani D s0432-1 | 35.9 | 31 | 54.8 | CP017453 |
| 25 | D. solani F012 | 35.9 | 31 | 54.8 | PDVN00000000 |
| 26 | D. solani GBBC 2040 | 35.9 | 31 | 54.8 | AONX01000000 |
| 27 | D. solani IFB0223 | 35.9 | 31 | 54.8 | CP024710 |
| 28 | D. solani IFB 0099 | 35.9 | 31 | 54.8 | JXRS01000000 |
| 29 | D. solani IFB_0158 | 35.9 | 31 | 54.8 | PENA00000000 |
| 30 | D. solani IFB_0221 | 35.9 | 31 | 54.8 | PEMZ00000000 |
| 31 | D. solani MK10 | 35.9 | 31 | 54.8 | AOOP01000000 |
| 32 | D. solani MK16 | 35.9 | 31 | 54.8 | AOOQ01000000 |
| 33 | D. solani PPO 9019 | 35.9 | 31 | 54.8 | CP017454 |
| 34 | D. solani PPO 9134 | 35.9 | 31 | 54.8 | JWLT00000000 |
| 35 | D. solani RNS 07.7.3B | 35.9 | 31 | 54.8 | JWLR00000000 |
| 36 | D. solani RNS 08.23.3.1.A | 35.9 | 31 | 54.8 | CP016928 |
| 37 | D. chrysanthemi NCPPB 3533 | 33.5 | 28 | 53.1 | AOOJ00000000 |
| 38 | D. solani RNS 05.1.2A | 36.6 | 31 | 54.0 | JWMJ00000000 |
Identification and annotation of G4C (OAg-capsule) and OAg-LPS biosynthesis gene clusters
An NCBI blastp homology tool was used to find the OAg-capsule and OAg-LPS gene cluster within the genus Dickeya. Briefly, we used wza, wzb, wzc and rfbB, rfbA, rfbC protein sequences of Dickeya solani type strain IPO 2222T (NCBI Accession no. CP015137.1) for blastp query (S1 File) to identify OAg-capsule cluster in the non-redundant protein sequence (nr) database of NCBI in genus Dickeya (taxid:204037). Similarly, for identification of OAg-LPS biosynthesis gene clusters, we used wzm and wzx protein sequence of Pseudomonas aeroginosa PAO1 (NCBI Accession AAG08836.1 and AAG06541.1 respectively) (S1 File). We used blastp, E-value cut-off of 2e-06 for the study. We could identify these clusters in 16 out of 54 Dickeya strains (Table 1), for which a complete annotated genome was available. For further investigation, we downloaded genomes of 54 Dickeya strains (Table 1) from the NCBI genome database as the nucleotide (.fna) file and created the nucleotide blast database using standalone NCBI blast+ facility [28]. We took conserved flanking genes of OAg-capsule, OAg-LPS gene cluster, and other gene clusters described in the result section and performed tblastn and blastn search against the nucleotide blast database. The results of tblatn and blastn helped to identify the full gene cluster with precise coordinates of nucleotides. We extracted the sequences of these clusters from their genomes and used the RAST annotation server (http://rast.nmpdr.org/) to annotate them [29]. After annotation, we manually verified each Open Reading Frame (ORF) for their homology to genes from the NCBI database using blastp and further manually curated them for analysis. We generated the annotated OAg-capsule and OAg-LPS clusters using EasyFig software [30] and enhanced their appearance using Adobe illustrator CS6 software.
Taxonomic and phylogenetic analysis of Dickeya strains
Phylogenetic analysis based on the complete rpoB gene was performed by clustalW alignment and MEGA X software [26, 31]. Similarly, taxonomic classification based on the complete rpoB gene was obtained by NCBI BLASTn with a ≤ 97.7% sequence identity cut-off was used as a species selection criterion [26]. The entire rpoB gene sequence identity was determined by the Unipro-Ugene toolkit [32]. Whole genome-based species criteria like Average Nucleotide Identity (ANI) and digital DNA-DNA hybridization (dDDH) of ≥ 94.4% and ≥ 70% were used respectively based on the standard recommendations [33–35]. Whole genome-based ANI were calculated using Jspecies software, and ANIb values were obtained [35]. Similarly, dDDH recommended values were obtained using the Genome-to-Genome Distance Calculator web server (https://ggdc.dsmz.de/) [36]. All the sequences for the identified clusters are made available at figshare repository with following link “https://figshare.com/s/91c9625e09e09bda5df3”.
Results
Identification of polysaccharide biosynthesis clusters in Dickeya
In the revolutionary era of genomics, we have access to tremendous genomic sequence data from the public database, such as NCBI (NCBI Resource Coordinators, 2016). We retrieved 73 complete and draft whole-genome sequences of members of the genus Dickeya from NCBI, as of March 5th, 2019. We further limited our selection to 54 strains, in which we were able to obtain full sequences of the group 4 capsule (G4C) coding region from whole-genome sequences (Table 1).
We identified conserved polysaccharide biosynthesis gene clusters in Dickeya and successfully characterized five such conserved genomic regions, including putative O-antigen clusters, with exceptions to certain species/strains. In this study, we further provide comprehensive comparisons of diverse group 4 capsule (G4C)/OAg-capsules, putative O-antigen lipopolysaccharide (OAg-LPS) coding clusters, enterobacterial common antigen (ECA) gene clusters, core-lipopolysaccharide biosynthesis clusters, and an additional alternative O-antigen biosynthesis lipopolysaccharide region in these strains.
Identification of Group 4 Capsule (G4C) or OAg-capsule coding region in Dickeya
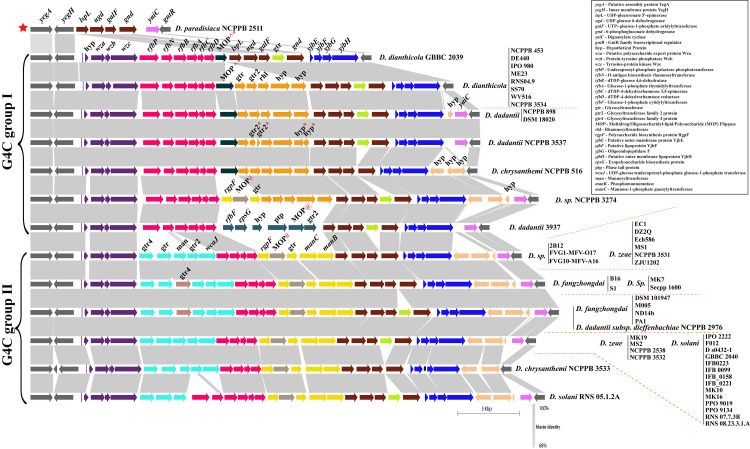
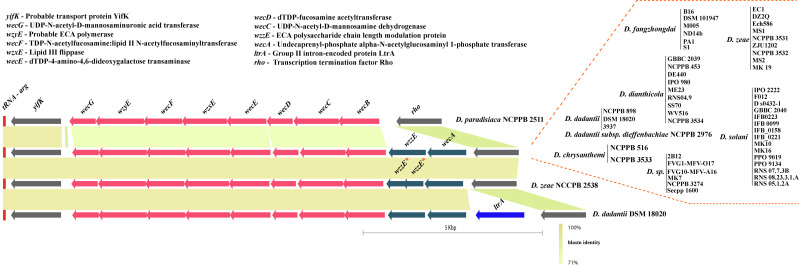
The group 4 capsule (G4C) polysaccharides are frequently identical to the cognate lipopolysaccharide O side chain. Therefore, it is often referred to as the O-antigen capsule (OAg-capsule) in E. coli [37]. Biosynthesis of various polysaccharides, including O-antigen, are often linked to the 39 bp conserved JUMPstart (for “just upstream of many polysaccharides starts”) element present at the beginning of these clusters [38]. Here, we found conserved JUMPstart sequence homology at the beginning of the cluster (marked as the blue bar, Fig 1) in the genus Dickeya, except for strain Dickeya paradisiaca NCPPB 2511. Sequence homology and alignment to the 39 bp JUMPstart sequence in Dickeya is shown in S1 Fig. These G4C cluster genes are flanked by membrane protein YegA and transport protein YegH (yegA-yegH) on the left and transcriptional regulator GntR (gntR) on the right as shown in grey color (Fig 1). All 54 studied genomes of Dickeya strains contain this conserved locus in their genomes.
Fig 1. Group 4 capsule or O-antigen capsule coding biosynthesis gene clusters of Dickeya.
Conserved flanking genes are shown in grey on both ends. Gene abbreviations are mentioned in the right corner box. JUMPstart sequence homology was marked with blue bar after initial flanking gene yegH. Identical sequence homology for ORFs or clusters of ORFs are shown by using the same color. Gene names and abbreviations are shown in the figure. Blastn identity is shown by a grey color scale among the clusters. Blastn identity is shown by a grey color scale among the clusters. O-antigen diversity is grouped into two major G4C groups, group I and group II, however an exception to this is D. paradisiaca NCPPB 2511 marked as red star at the beginning of the figure Respective strain names are mentioned for each cluster at the right end with red dotted boundaries. Different strains of the same species are listed as a vertical bar. @, #, and ^ denote different homologs of the corresponding gene. * denotes a pseudogene. All gene clusters are drawn to scale.
Based on their sequence identities and variation, we studied these clusters individually and grouped them as represented in Fig 1. There are two major types of G4C cluster groups, i.e., G4C group I and G4C group II as marked in Fig 1 (Table 1). These groups are distributed among different species of Dickeya and exhibit diversity. The average GC content of G4C clusters is significantly different compared to their average genomic GC content (Table 1). This feature indicates the possible role of HGT events to shape their G4C cluster organization. These G4C clusters also vary in size as well as numbers of Open Reading Frames (ORFs), as listed in Table 1.
The beginning of both group I and group II G4C clusters contain three ORFs including wza, wzb, and wzc genes that are respectively similar to yccZ-etp-etk genes of the cps/wca operon responsible for the production of colanic acid (CA)-EPS synthesis (Fig 1, violet color) [37, 39]. Another yjbEFGH operon (marked in blue in Fig 1) was also consistently present in both the G4C group strains and probably has a role in exopolysaccharide biosynthesis [37, 40]. This operon is also paralogous to the ymcDCBA operon known for O-antigen biosynthesis in E. coli [37, 40]. However, the role of the yjbEFGH operon is uncertain in Dickeya but is speculated to be part of O-antigen capsule biosynthesis.
Structure, variation, and diversity of G4C (O-antigen capsule) group I coding region in Dickeya
Fifteen Dickeya strains contain group I G4C cluster (Table 1). The G4C group I cluster is characterized by the presence of both a set of six genes, i.e., rfbP, rfbN, rfbB, rfbA, rfbC, rfbD and a set of five genes i.e., gtr, gtr2, rhl, and two hyps clustered within the conserved flanking genes (Fig 1). Four genes from these clusters, rfbB, rfbA, rfbC, and rfbD, are known to be involved in dTDP-rhamnose biosynthesis rfbBACD operon, which is an important precursor for the biosynthesis of various cell-wall polysaccharides, EPS and LPS [41, 42]. This rfbBACD operon is present in all G4C group I and II strains, likely coding for O-antigen (Fig 1). However, five genes, encoding for two glycosyltransferases, a rhamnosyltransferase, and two hypothetical proteins, are uniquely present in G4C group I members except for D. dianthicola GBBC 2039 and D. dadantii 3937 (shown in orange color, Fig 1). In D. dadantii NCPPB 3537, glycosyltransferase and the hypothetical proteins from the five gene clusters are present as pseudogenes (marked with *, in Fig 1). Furthermore, a multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase coding ORF is present in all the group I members, just after the rfbD gene. In D. sp. NCPPB 3274 and D. dadantii 3937, different homologs of MOP flippase are present as a cluster of three and six genes, respectively (Fig 1). The cluster of three genes in D. sp. NCPPB 3274 contains rgpF, MOP# flippase, and a glycosyltransferase (Fig 1). Similarly, the six genes with distinct MOP@ flippase in D. dadantii 3937 encodes for rgbF, epsG, hypothetical protein (hyp), phage-tail protein (ptp), MOP@ flippase (MOP@) and glycosyltransferase family 2 protein (gtr2).
Interestingly, we found that D. paradisiaca NCPPB 2511 contains smallest G4C cluster of 6.8 Kbp (Kilobasepair) and consists of only five ORFs which encode for lspL, ugd, galF, gnd, and yaiC genes. Four out of these five genes namely lspL, ugd, galF, and gnd, are present in both of the G4C clusters group I and II, as shown in brown color in Fig 1 and Table 1. Our phylogenetic distribution study (Fig 4) also indicates D. paradisiaca NCPPB 2511 to be evolutionary ancestral strain. Owing to presence of its genes in both the G4C group I and II cluster we have denoted as star marked individual cluster (Fig 1).
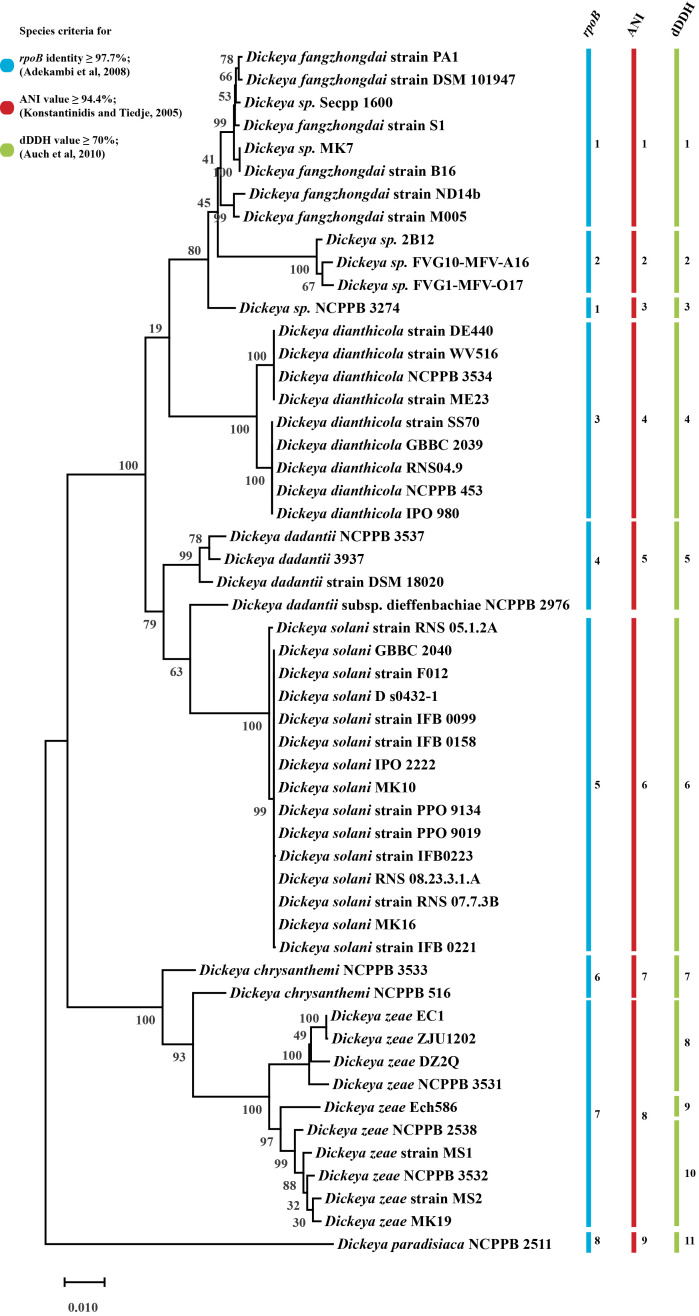
Fig 4. Maximum-likelihood phylogenetic tree of Dickeya based on the complete rpoB sequence and its species delineation.
The phylogenetic tree was obtained by clustalW alignment with a complete deletion method and 500 bootstrap values. Individual color bars and respective species group numbers have been assigned to different species criteria of rpoB, ANI, and dDDH.
In this study, we found that D. dianthicola strains and the major D. dadantii species exclusively contains G4C group I clusters. However, one of D. chrysanthemi strain, D. chrysanthemi NCPPB 516 is also part of the G4C group I cluster. The most extended LPS group I cluster of 36.1 Kb with 31 ORFs was present in strain D. sp. NCPPB 3274, (Table 1).
Structure and variation of G4C (O-antigen capsule) group II coding region in Dickeya
A total of 38 out of 54 strains contain G4C group II cluster homology (Table 1 and Fig 1). Our findings suggest that the genus Dickeya, D. fangzhongdai and D. solani species exclusively belong to G4C group II clusters. The smallest G4C group II cluster is 33.5 Kb, with 28 predicted ORFs in strain D. chrysanthemi NCPPB 3533. D. chrysanthemi is present in both G4C groups I and II with its single representative strains in each of the group. The largest group II G4C cluster is represented by D. fangzhongdai species and D. dadantii subsp. Dieffenbachiae NCPPB 2976 with a length of 37.8 Kb encoding 32 ORFs. G4C group II clusters appear differently from the G4C group I members in that they have two distinct five gene clusters shown in the sky-blue and yellow-colored region in Fig 1. The first five gene cluster consists of gtr4, gtr, man, gtr2 and wcaJ genes, while the second five gene cluster is represented by rgpF, MOP#, gtr, manC, and manB respectively (Fig 1). A slight variation of an additional gtr4 gene (shown in light brown color) in the first five gene cluster was present in species D. fangzhongdai and a few other strains (Fig 1).
The MOP# flippase gene, colored in olive, is present in all members with G4C group II clusters and in one G4C group I member, D. sp. NCPPB 3274. The MOP genes were present in three different gene sequences, and out of these, MOP^ is characteristic of G4C group I while MOP# is present in G4C group II containing Dickeya species except for D. sp. NCPPB 3274. In D. sp. NCPPB 3274, MOP# is present with the arrangement of three genes (rgpF, MOP#, gtr). The third MOP@ is present in D. dadantii 3937 of G4C group I. An additional glycosyltransferase (gtr), shown in bright green, was also present in different G4C group I and II cluster strains (Fig 1). D. solani RNS 05.1.2A contains rfbP and rfbN genes followed by the rfbBACD operon, similar to the G4C cluster group I members.
Despite finding distinct MOP flippase genes in these G4C clusters, the most prevalent O-antigen processing gene, wzx (flippase), wzy (polymerase), or wzm/wzt transport system (ABC transporters) [43] are not present within this locus.
Identification of putative O-antigen lipopolysaccharide (OAg-LPS) biosynthesis cluster
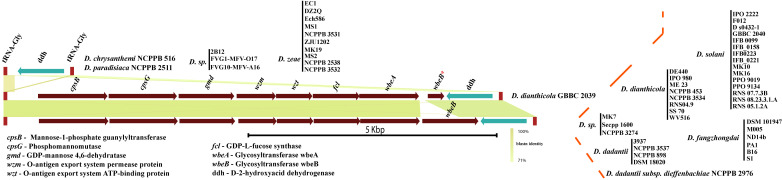
We further investigated the presence of wzx and wzm homologs in the genomes of Dickeya. Our findings suggest the presence of a putative O-antigen lipopolysaccharide (OAg-LPS) biosynthesis cluster in Dickeya. This particular locus contains an essential O-antigen transport system (wzm-wzt transport genes) with a cluster of eight genes—cpsB, cpsG, gmd, wzm, wzt, fcl, wbeA, and wbeB (Fig 2). This locus is flanked by the tRNA-Gly gene on the left and a D-2-hydroxy-acid dehydrogenase (ddh) gene on the right, followed by another tRNA-Gly gene (Fig 2). D. dianthicola GBBC 2039 contains wbeB as a pseudogene in this cluster. The most critical wzm-wzt genes are part of this conserved cluster, contributing to O-antigen transport in the periplasm. This small but quintessential putative O-antigen cluster contains colanic acid biosynthesis genes cpsB and cpsG. It also contains the GDP-L-fucose biosynthesis genes gmd and fcl, which are part of the colanic acid (CA) biosynthesis cluster in E. coli [44]. This locus is conserved in all 54 strains. Interestingly, a total of 15 strains, shown in Fig 2, do not contain LPS coding genes and end abruptly with only the ddh flanking ORF between two tRNA-Gly genes (Fig 2). This arrangement also occurs in all D. zeae species members.
Fig 2. Putative O-antigen lipopolysaccharide biosynthesis gene cluster in genus Dickeya.
Gene abbreviations are mentioned in the bottom-left corner. Identical colors show ORFs or clusters of ORFs with identical sequence homology. Gene names and abbreviations are shown in the figure. Blastn identity is shown with a green color scale among the clusters. Respective strain names are mentioned for each cluster at the right end with red dotted boundaries. Different strains of the same species are listed as a vertical bar. * denotes a pseudogene. All gene clusters are drawn to scale.
Since the O-antigen transport system is essential for all the species of Dickeya, we searched for it across the genomes of the remaining strains and did not find this putative O-antigen cluster at this locus. Instead, we found these transport genes along with LPS biosynthetic cluster at different locus, as described in the next section.
Complementary putative O-antigen lipopolysaccharide (OAg-LPS) cluster identification and characterization in Dickeya
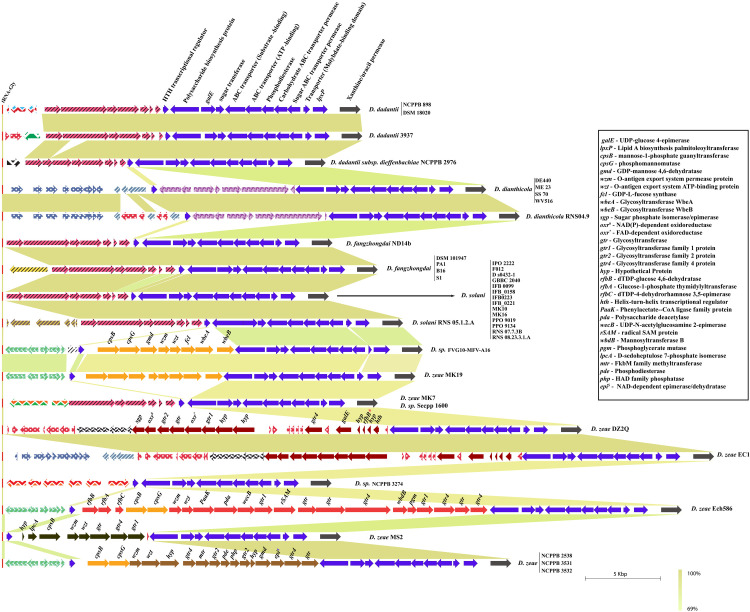
The alternative putative O-antigen lipopolysaccharide (OAg-LPS) locus is found to be in proximity of genes involved in the lipid A biosynthesis gene, lpxP. Moreover, this locus is flanked by a tRNA-Gly gene on the left and xanthine/uracil permease gene on the right (Fig 3). A cluster of ten ORFs, including the lpxP gene on the right end of this locus, is related to polysaccharide biosynthesis and marked with a navy blue color. The annotation of these 10 ORFs reveal a polysaccharide biosynthesis protein, galE, sugar transferase, ABC transporters (both substrate binding and ATP-binding), phosphodiesterase, carbohydrate ABC transporter permease, molybdate binding transporter protein, and lpxP gene. The role of this cluster of genes is not yet known and requires further experimental evidence.
Fig 3. Complementary alternative O-antigen biosynthesis gene cluster in Dickeya.
LPS biosynthesis genes are shown with solid color fill. However, non-LPS coding genes are marked with different color patterns based on their gene annotations. Identical gene homology clusters are shown with identical color or color patterns in this figure. Gene names and abbreviations are shown in the figure. * denotes a pseudogene. All clusters are drawn to scale with the blastn identity color scale.
However, the missing putative OAg-LPS cluster genes, as described in the previous section, are mostly present in the remaining 15 strains. As shown in Fig 3, ORFs marked with different color patterns do not show any relation to LPS biosynthesis genes according to their in silico functional annotation. ORFs in different color patterns are also marked based on identical nucleotide homology and clustering.
Out of these 15 Dickeya strains which lack putative OAg-LPS cluster, we identified a complete alternative OAg-LPS cluster in nine of them (Fig 3). D. sp. FVG10-MFV-A16 and D. zeae MK19 contain an identical arrangement of eight ORFs (cpsB, cpsG, gmd, wzm, wzt, fcl, wbeA, and wbeB) in a putative OAg-LPS cluster, just upstream of a polysaccharide biosynthesis protein (shown in orange color, Fig 3).
In D. zeae DZ2Q and D. zeae EC1, eight gene clusters (sgp, oxr#, gtr2, gtr, oxr*, gtr1, two hyps) are related to LPS biosynthesis, as shown in brown (Fig 3). Few other genes like gtr4, galE, rfbB*, two hyp, and hth related to LPS biosynthesis are also present and marked in brown for both the strains. The rfbB* gene is present as a pseudogene in both strains, D. zeae DZ2Q and D. zeae EC1. These strains lack wzm-wzt transporters at this locus. This ambiguity remains open for further investigation and discussion.
D. zeae NCPPB 2538, D. zeae NCPPB 3531, and D. zeae NCPPB 3532 have identical cluster arrangement, which contains 16 LPS biosynthesis related ORFs in addition to a 10 ORFs cluster at the end of this locus. D. zeae Ech586 and D. zeae MS2 strains contain entirely different clusters containing distinct wzm-wzt homologs. Individual ORFs and clusters of ORFs annotated with abbreviations and specific color are shown in Fig 3. In addition to the G4C group I cluster shown in Fig 1, D.zeae Ech586 contains a duplicate set of rfbB, rfbA, and rfbC at an alternative locus (Fig 3).
Phylogenetic distribution of O-antigen capsule (G4C) coding region across the genus Dickeya
Phylogenetic species groups were determined with whole genome-based species criteria of Average Nucleotide Identity (ANI) and digital DNA-DNA hybridization (dDDH) methods. Our study encompassing 54 Dickeya strains suggests the presence of nine species groups, based on ANI species criteria (≥ 94.4% identity) and 11 species groups, based on dDDH criteria (≥ 70% identity) (S2 Fig). Members of D. zeae are divided into three distinct species groups based on dDDH classification. We also used RNA polymerase beta subunit (rpoB) gene, found as single copy gene in bacterial genome and relatively immune to HGT, as a phylogenetic marker and a species delineation cut-off of ≥ 97.7% sequence identity, as interpreted in Fig 4 [26, 27] (53 strains are used for this analysis, data available in S1 Table). It shows eight distinct rpoB species grouping of 53 Dickeya strains and has similarity to the findings with ANI, except for strain D. sp. NCPPB 3274 (Fig 4). We compared the different G4C clusters present with each strain and their phylogenetic distance with each other (S3 Fig).
In our study of the variation of G4C (O-antigen capsule) group I and II coding region in Dickeya, we found that the G4C group I cluster is predominantly present in D. dianthicola, and D. dadantii strains and the G4C group II cluster is present in D. fangzhongadi, D. zeae, and D. solani, while D. sp. and D. chrysanthemi are distributed in both G4C group I and II clusters. These observations are not consistent with variation of putative O-antigen LPS biosynthesis cluster, and an alternative O-antigen LPS biosynthesis clusters in those species which may contribute to more variation of O-antigen of individual species.
Enterobacterial Common Antigen (ECA) biosynthesis cluster in Dickeya
The ECA biosynthesis cluster, or wec cluster, in Dickeya is flanked by conserved genes yifK on the left and rho on the right end. Additionally, a tRNA-Arg gene is present just before the yifK flanking gene. This locus is highly conserved with its constituent genomic organization. Fiftyone out of 54 studied strains contain an identical set of ten ORFs, namely wecG, wzyE, wecF, wzxE, wecE, wecD, wecC, wecB, wzzE, and wecA, between these flanking genes (Fig 5). However, D. paradisiaca NCPPB 2511 lacks wzzE and wecA at this locus. D. zeae NCPPB 2538 contains wzzE as a pseudogene in their cluster (Fig 5). D. dadantii DSM 18020 contains an additional ORF coding for the ltrA gene in this locus.
Fig 5. Enterobacterial Common Antigen (ECA) biosynthesis clusters in Dickeya.
* denotes a pseudogene. Gene abbreviations are mentioned at the top of the figure. All clusters are drawn to scale.
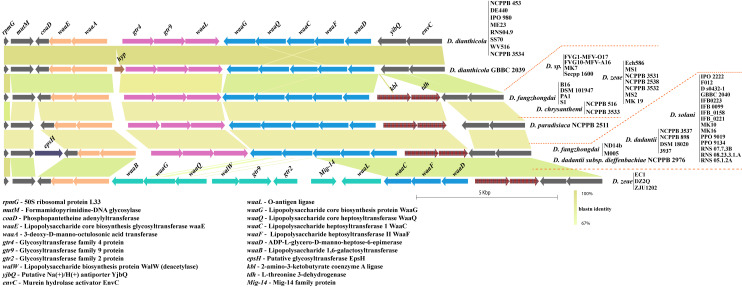
Core lipopolysaccharide biosynthesis waa cluster diversity in Dickeya
The waa gene cluster involved in core lipopolysaccharide biosynthesis in Klebsiella pneumoniae is present, between the coaD and kbl flanking genes [45]. A substantial similarity of this conserved locus is found in several species of Dickeya. However, in case of D. dianthicola, this locus is flanked by coaD and yjbQ. The yjbQ gene is the second downstream gene to kbl in other Dickeya strains (Fig 6). The majority of Dickeya members contain an array of 10 genes, i.e., waaE, waaA, gtr4, gtr9, waaL, waaG, waaQ, waaC, waaF, and waaD that constitutes the waa gene cluster (Fig 6). One additional epsH gene is also present before the coaD flanking gene in several strains of D. solani, D. dadantii, and D. fangzhongdai (Fig 6). D. zeae EC1, D. zeae DZ2Q, and D. zeae ZJU1202 contain different homologs of waaB, waaG, waaQ, waaL, and other genes walW, gtr9, gtr2, and a Mig-14, shown in green (Fig 6).
Fig 6. Core lipopolysaccharide biosynthesis (waa) cluster diversity in Dickeya.
Identical sequence homology for ORFs or clusters of ORFs are shown with matching color. Gene abbreviations are mentioned at the bottom of the figure. All clusters are drawn to scale.
Discussion
The genus Dickeya is a significant threat to herbaceous plants and vegetables. In Gram-negative bacteria, complex polysaccharides form an essential component of the bacterial outer surface. The surface polysaccharides enable bacteria to colonize the host and cause disease by evading plant immunity [46]. It is estimated that more than one capsule system is present in 40% of the total analyzed bacterial lineages [47]. The present study describes the genetic relatedness of the complete lipopolysaccharide O-antigen and G4C biosynthesis systems in eight distinct species of Dickeya encompassing 54 strains.
LPSs are recognized as one of the crucial pathogen-associated molecular patterns (PAMPs) and potent elicitors of PAMP triggered immunity (PTI) [48]. Long-chain O-antigen enables Xylella fastidiosa to delay host innate immune recognition and allows for effective establishment into the host [49]. The localized induced response is a common phenomenon and is observed in pepper leaves after treatment with different LPS components isolated from Xanthomonas campestris [50]. These responses include the production of phenolic compounds, alteration in the expression of pathogenesis related (PR) proteins, and prevention of HR responses caused by avirulent bacteria [51, 52]. The effect of different components of LPS of genus Dickeya on plant responses is not yet completely understood. Additionally, the revelation of LPS and its genomic counterparts as an important virulence factor remain mostly unidentified in the genus Dickeya.
The O-antigen capsule biosynthesis clusters in Dickeya are categorized into two different G4C groups as they are distinct from each other in their genetic makeup. This suggests a parallel co-evolution of the two G4C groups through genetic recombination among these strains or mutations in their genomes. The uniform structure of O-polysaccharides (a homopolymer of 6-deoxy-d-altrose: →2)-β-d-6dAltp-(1→) isolated from Dickeya solani strains from different geographic regions has been reported [53]. This observation is in agreement with our finding of a highly clonal arrangement of a putative G4C group II cluster and O-antigen LPS in D. solani, as shown in Figs 1 and 2. Other polysaccharide biosynthesis clusters are also consistently conserved for D. solani (Figs 3, 5, and 6). However, the novel differences between G4C groups I and II and other polysaccharide biosynthesis clusters might produce distinct O-antigen capsules but remain to be experimentally validated for the genus Dickeya. Enteropathogenic Escherichia coli is resistant to human α-defensin 5 and is linked to the additive effects of both G4C and lipopolysaccharide O-antigen [54]. Subsequently, both G4C and O-antigen LPS may contribute to virulence, pathogenicity, and better survival of Dickeya in plants, therefore altering host-pathogen interactions.
Many enterobacteria produce colanic acid (CA) or M-antigen as an exopolysaccharide (a negatively charged polymer of glucose, galactose, fucose, and glucuronic acid) [55]. However, LPSs act as a potent stimulator of innate or natural immunity in diverse eukaryotic species ranging from insects to humans [56]. This study also suggests that the modification of lipopolysaccharides with colanic acid might be possible in Dickeya, as LPS and colanic acid biosynthesis genes are related in origin and intermingled at these clusters. This combination of colanic acid, LPS core biosynthesis cluster, and waaL are responsible for the presence of M-antigen, which is a modified form of O-antigen in E. coli [55]. However, while this may be possible in Dickeya, it would be presumptive to conclude that such M-antigen is present and contributes to their diversity of surface molecules.
A recent study shows that genetic exchanges in bacteria encoding capsules are frequent and lead to changes in genome dynamics [46, 57]. Variation at the LPS biosynthesis cluster in phytopathogenic Gram-negative bacteria, Xanthomonas oryzae pv. oryzae (cause of bacterial blight of rice) is driven by multiple HGT events [58]. Continuing this observation of the O-antigen capsule/G4C coding genomic region in Dickeya indicates multiple HGT events and are responsible for the structure of the current complex polysaccharide biosynthesis cluster diversity. The presence of several t-RNA coding genes at the sites of complex polysaccharide biosynthesis clusters in Dickeya suggests that tRNA genes are favorable sites for recombination/integration of these LPS/Capsule coding pathogenicity clusters. These properties of G4C capsule and LPS biosynthesis clusters, preferentially linked to tRNA sites, could be prevalent in phytopathogenic bacteria and needs further investigation with a more diverse group of bacterial populations. Moreover, these clusters meet the classical features of pathogenicity islands in several cases [59, 60].
ECA and core lipopolysaccharide biosynthesis clusters (waa) are highly predictable and consistent in their genetic organization with less variation. However, the Dickeya genus, being a member of Enterobacteriaceae, is expected to have ECA and waa gene clusters with less or no variation (Figs 5 and 6). A recent transcriptomics and comparative genomics study using 100 genomes of Pectobacterium and Dickeya spp. identified the structural organization of G4C, wec, and waa gene clusters in Pectobacterium carotovorum subsp. brasiliense strain PBR 1692 [61]. Their experimental findings suggest further investigation is needed in these species, including Dickeya. We present a thorough comparative analysis of LPS biosynthesis and O-antigen capsule biosynthesis clusters present in Dickeya. In D. zeae, the heterogeneity of O-antigen capsule and waa gene clusters might be one of the several factors affecting their virulence differentially, as reported by Ming Hu and co-workers [62]. Apart from pathogens, the capsular proteins found in many non-pathogens may increase their ability to survive in constantly changing environmental conditions suggesting its role in niche specialization [63]. In turn, it requires extensive experimental evidence to ascertain the role of the different genetic organization of the O-antigen capsule coding regions to the differences observed in host-pathogen interactions. Overall, this study provides a better insight into how the genetic variations and organization of polysaccharide biosynthetic clusters may lead to changes in bacterial surface structure and therefore pathogenicity of Dickeya. The functional study of these genetic variations will lead to better understanding of their impact on pathogenicity.
Supporting information
(TIF)
Whole genome ANI species cut-off of 94.4% and above is marked with red color boxes while dDDH species cut-off of 70% and above were marked with green color boxes in this figure.
(TIF)
G4C coding clusters of 54 Dickeya strains were aligned to their phylogenetic tree based on rpoB and also classified with different color bar of ANI and dDDH. Clusters are drawn to scale and rpoB, ANI and dDDH species cut-off is mentioned with dot colored legends.
(TIF)
Sequence identity matrix with species cut-off of 97.7% or above identity is marked in blue color for the specified strains.
(XLSX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files. All the sequences for the identified clusters are made available at figshare: https://figshare.com/s/91c9625e09e09bda5df3.
Funding Statement
AR was supported by Department of Biotechnology, India (http://dbtindia.gov.in/), Ramalingaswami Re-entry Fellowship no. BT/HRD/35/02/2006 and currently, he is supported by Stakman Endowment fund 1750-11031-20088-END00E0822-5669146 from Department of Plant Pathology, University of Minnesota-Twin Cities, US. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Samson R, Legendre JB, Christen R, Fischer-Le Saux M, Achouak W, Gardan L. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dick. Int J Syst Evol Microbiol. 2005. [DOI] [PubMed] [Google Scholar]
- 2.Toth IK, van der Wolf JM, Saddler G, Lojkowska E, Hélias V, Pirhonen M, et al. Dickeya species: An emerging problem for potato production in Europe. Plant Pathology. 2011. 10.1111/j.1365-3059.2011.02552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology. 2012. 10.1111/j.1364-3703.2012.00804.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkholder WR, Mcfadden LA, Dimock EW. A bacterial blight ol Chrysanthemums. Phytopathology. 1953;43: 522–526 pp. [Google Scholar]
- 5.Reverchon S, Nasser W. Dickeya ecology, environment sensing and regulation of virulence programme. Environ Microbiol Rep. 2013. 10.1111/1758-2229.12073 [DOI] [PubMed] [Google Scholar]
- 6.Buttimer C, McAuliffe O, Ross RP, Hill C, O’Mahony J, Coffey A. Bacteriophages and bacterial plant diseases. Frontiers in Microbiology. 2017. 10.3389/fmicb.2017.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabanova AP, Shneider MM, Korzhenkov AA, Bugaeva EN, Miroshnikov KK, Zdorovenko EL, et al. Host specificity of the dickeya bacteriophage PP35 is directed by a tail spike interaction with bacterial O-antigen, enabling the infection of the alternative non-pathogenic bacterial host. Front Microbiol. 2019. 10.3389/fmicb.2018.03288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitfield C, Roberts IS. Structure, assembly, and regulation of expression of capsules in Escherichia coli. Molecular Microbiology. 1999. 10.1046/j.1365-2958.1999.01276.x [DOI] [PubMed] [Google Scholar]
- 9.Whitfield C. Biosynthesis and Assembly of Capsular Polysaccharides in Escherichia coli. Annu Rev Biochem. 2006. 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- 10.Goldman RC, White D, Orskov F, Orskov I, Rick PD, Lewis MS, et al. A surface polysaccharide of Escherichia coli O111 contains O-antigen and inhibits agglutination of cells by O-antiserum. J Bacteriol. 1982. 10.1128/JB.151.3.1210-1221.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahn A, Beis K, Naismith JH, Whitfield C. A novel outer membrane protein, wzi, is involved in the surface assembly of the Escherichia coli K30 group 1 capsule. J Bacteriol. 2003. 10.1128/jb.185.19.5882-5890.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerouge I, Vanderleyden J. O-antigen structural variation: Mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiology Reviews. 2002. 10.1111/j.1574-6976.2002.tb00597.x [DOI] [PubMed] [Google Scholar]
- 13.Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MAG. Masquerading microbial pathogens: Capsular polysaccharides mimic host-tissue molecules. FEMS Microbiology Reviews. 2014. 10.1111/1574-6976.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miajlovic H, Smith SG. Bacterial self-defense: How Escherichia coli evades serum killing. FEMS Microbiology Letters. 2014. 10.1111/1574-6968.12419 [DOI] [PubMed] [Google Scholar]
- 15.Peyraud R, Dubiella U, Barbacci A, Genin S, Raffaele S, Roby D. Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant J. 2017. 10.1111/tpj.13429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudolph KWE, Gross M, Ebrahim-Nesbat F, Nöllenburg M, Zomorodian A, Wydra K, et al. The role of extracellular polysaccharides as virulence factors for phytopathogenic pseudomonads and xanthomonads. 1994. 10.1007/978-94-011-0746-4_25 [DOI] [Google Scholar]
- 17.Charkowski AO. The Changing Face of Bacterial Soft-Rot Diseases. Annu Rev Phytopathol. 2018. 10.1146/annurev-phyto-080417-045906 [DOI] [PubMed] [Google Scholar]
- 18.Bedini E, De Castro C, Erbs G, Mangoni L, Dow JM, Newman MA, et al. Structure-dependent modulation of a pathogen response in plants by synthetic O-antigen polysaccharides. J Am Chem Soc. 2005. 10.1021/ja0452166 [DOI] [PubMed] [Google Scholar]
- 19.Erbs G, Molinaro A, Dow JM, Newman MA. Lipopolysaccharides and plant innate immunity. Subcell Biochem. 2010. 10.1007/978-90-481-9078-2_17 [DOI] [PubMed] [Google Scholar]
- 20.Li CH, Wang KC, Hong YH, Chu TH, Chu YJ, Chou IC, et al. Roles of different forms of lipopolysaccharides in ralstonia solanacearum pathogenesis. Mol Plant-Microbe Interact. 2014. 10.1094/MPMI-08-13-0248-R [DOI] [PubMed] [Google Scholar]
- 21.Denny TP. Involvement of Bacterial Polysaccharides in Plant Pathogenesis. Annu Rev Phytopathol. 1995. 10.1146/annurev.py.33.090195.001133 [DOI] [PubMed] [Google Scholar]
- 22.van der Wolf JM, Nijhuis EH, Kowalewska MJ, Saddler GS, Parkinson N, Elphinstone JG, et al. Dickeya solani sp. nov., a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int J Syst Evol Microbiol. 2014. 10.1099/ijs.0.052944-0 [DOI] [PubMed] [Google Scholar]
- 23.Tian Y, Zhao Y, Yuan X, Yi J, Fan J, Xu Z, et al. Dickeya fangzhongdai sp. nov., a plant pathogenic bacterium isolated from pear trees (Pyrus pyrifolia). Int J Syst Evol Microbiol. 2016. 10.1099/ijsem.0.001060 [DOI] [PubMed] [Google Scholar]
- 24.Parkinson N, DeVos P, Pirhonen M, Elphinstone J. Dickeya aquatica sp. nov., isolated from waterways. Int J Syst Evol Microbiol. 2014. 10.1099/ijs.0.058693-0 [DOI] [PubMed] [Google Scholar]
- 25.Marrero G, Schneider KL, Jenkins DM, Alvarez AM. Phylogeny and classification of Dickeya based on multi-locus sequence analysis. Int J Syst Evol Microbiol. 2013. [DOI] [PubMed] [Google Scholar]
- 26.Adékambi T, Shinnick TM, Raoult D, Drancourt M. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Microbiol. 2008. 10.1099/ijs.0.65440-0 [DOI] [PubMed] [Google Scholar]
- 27.Ogier JC, Pagès S, Galan M, Barret M, Gaudriault S. RpoB, a promising marker for analyzing the diversity of bacterial communities by amplicon sequencing. BMC Microbiol. 2019. 10.1186/s12866-019-1546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okonechnikov K, Golosova O, Fursov M, Varlamov A, Vaskin Y, Efremov I, et al. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012. 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- 33.Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010. 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peleg A, Shifrin Y, Ilan O, Nadler-Yona C, Nov S, Koby S, et al. identification of an Escherichia coli operon required for formation of the O-antigen capsule. J Bacteriol. 2005. 10.1128/JB.187.15.5259-5266.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobbs M, Reeves PR. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Molecular Microbiology. 1994. 10.1111/j.1365-2958.1994.tb01071.x [DOI] [PubMed] [Google Scholar]
- 39.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996. 10.1128/jb.178.16.4885-4893.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrières L, Aslam SN, Cooper RM, Clarke DJ. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology. 2007. 10.1099/mic.0.2006/002907-0 [DOI] [PubMed] [Google Scholar]
- 41.Rahim R, Burrows LL, Monteiro MA, Perry MB, Lam JS. Involvement of the rml locus in core oligosaccharide and O polysaccharide assembly in Pseudomonas aeruginosa. Microbiology. 2000. 10.1099/00221287-146-11-2803 [DOI] [PubMed] [Google Scholar]
- 42.Boels IC, Beerthuyzen MM, Kosters MHW, Van Kaauwen MPW, Kleerebezem M, De Vos WM. Identification and Functional Characterization of the Lactococcus lactis rfb Operon, Required for dTDP-Rhamnose Biosynthesis. J Bacteriol. 2004. 10.1128/jb.186.5.1239-1248.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samuel G, Reeves P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydrate Research. 2003. 10.1016/j.carres.2003.07.009 [DOI] [PubMed] [Google Scholar]
- 44.Andrianopoulos K, Wang L, Reeves PR. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998. 10.1128/JB.180.4.998-1001.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regué M, Climent N, Abitiu N, Coderch N, Merino S, Izquierdo L, et al. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J Bacteriol. 2001. 10.1128/JB.183.12.3564-3573.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Haeze W, Holsters M. Surface polysaccharides enable bacteria to evade plant immunity. Trends in Microbiology. 2004. 10.1016/j.tim.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 47.Rendueles O, de Sousa JAM, Bernheim A, Touchon M, Rocha EPC. Genetic exchanges are more frequent in bacteria encoding capsules. PLoS Genet. 2018. 10.1371/journal.pgen.1007862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman MA, Dow MJ, Molinaro A, Parrilli M. Invited review: Priming, induction, and modulation of plant defense responses by bacterial lipopolysaccharides. Journal of Endotoxin Research. 2007. 10.1177/0968051907079399 [DOI] [PubMed] [Google Scholar]
- 49.Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P, et al. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat Commun. 2018. 10.1038/s41467-018-02861-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman MA. The activity of lipid A and core components of bacterial lipopolysaccharides in the prevention of the hypersensitive response in pepper. Mol Plant-Microbe Interact. 1997. 10.1094/MPMI.1997.10.7.926 [DOI] [PubMed] [Google Scholar]
- 51.Dow M, Newman M, von Roepenack E. The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu Rev Phytopathol. 2000. 10.1146/annurev.phyto.38.1.241 [DOI] [PubMed] [Google Scholar]
- 52.Newman MA, Dow JM, Daniels MJ. Bacterial lipopolysaccharides and plant-pathogen interactions. European Journal of Plant Pathology. 2001. 10.1023/A:1008738817551 [DOI] [Google Scholar]
- 53.Ossowska K, Czerwicka M, Sledz W, Zoledowska S, Motyka A, Golanowska M, et al. The uniform structure of O-polysaccharides isolated from Dickeya solani strains of different origin. Carbohydr Res. 2017. 10.1016/j.carres.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 54.Thomassin JL, Lee MJ, Brannon JR, Sheppard DC, Gruenheid S, Le Moual H. Both group 4 capsule and lipopolysaccharide O-antigen contribute to enteropathogenic Escherichia coli resistance to human α-defensin 5. PLoS One. 2013. 10.1371/journal.pone.0082475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meredith TC, Mamat U, Kaczynski Z, Lindner B, Holst O, Woodard RW. Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J Biol Chem. 2007. 10.1074/jbc.M611034200 [DOI] [PubMed] [Google Scholar]
- 56.Alexander C, Rietschel ET. Bacterial lipopolysaccharides and innate immunity. Journal of Endotoxin Research. 2001. 10.1179/096805101101532675 [DOI] [PubMed] [Google Scholar]
- 57.Holt K.E., Lassalle F., Wyres K.L. et al. Diversity and evolution of surface polysaccharide synthesis loci in Enterobacteriales. ISME J. 2020. 10.1038/s41396-020-0628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patil PB, Sonti R V. Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice. BMC Microbiol. 2004. 10.1186/1471-2180-4-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt H, Hensel M. Pathogenicity Islands in Bacterial Pathogenesis. Clinical Microbiology Reviews. 2004. 10.1128/cmr.17.1.14-56.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gal-Mor O, Finlay BB. Pathogenicity islands: A molecular toolbox for bacterial virulence. Cellular Microbiology. 2006. 10.1111/j.1462-5822.2006.00794.x [DOI] [PubMed] [Google Scholar]
- 61.Bellieny-Rabelo D, Tanui CK, Miguel N, Kwenda S, Shyntum DY, Moleleki LN. Transcriptome and Comparative Genomics Analyses Reveal New Functional Insights on Key Determinants of Pathogenesis and Interbacterial Competition in Pectobacterium and Dickeya spp. Appl Environ Microbiol. 2019. 10.1128/AEM.02050-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu M, Li J, Chen R, Li W, Feng L, Shi L, et al. Dickeya zeae strains isolated from rice, banana, and clivia rot plants show great virulence differentials. BMC Microbiol. 2018. 10.1186/s12866-018-1300-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rendueles O, Garcia-Garcerà M, Néron B, Touchon M, Rocha EPC. Abundance and co-occurrence of extracellular capsules increase environmental breadth: Implications for the emergence of pathogens. PLoS Pathog. 2017. 10.1371/journal.ppat.1006525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Whole genome ANI species cut-off of 94.4% and above is marked with red color boxes while dDDH species cut-off of 70% and above were marked with green color boxes in this figure.
(TIF)
G4C coding clusters of 54 Dickeya strains were aligned to their phylogenetic tree based on rpoB and also classified with different color bar of ANI and dDDH. Clusters are drawn to scale and rpoB, ANI and dDDH species cut-off is mentioned with dot colored legends.
(TIF)
Sequence identity matrix with species cut-off of 97.7% or above identity is marked in blue color for the specified strains.
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All the sequences for the identified clusters are made available at figshare: https://figshare.com/s/91c9625e09e09bda5df3.