Abstract
The mitochondrion is critical for the survival of apicomplexan parasites. Several major anti-parasitic drugs, such as atovaquone and endochin-like quinolones, act through inhibition of the mitochondrial electron transport chain at the coenzyme Q:cytochrome c oxidoreductase complex (Complex III). Despite being an important drug target, the protein composition of Complex III of apicomplexan parasites has not been elucidated. Here, we undertake a mass spectrometry-based proteomic analysis of Complex III in the apicomplexan Toxoplasma gondii. Along with canonical subunits that are conserved across eukaryotic evolution, we identify several novel or highly divergent Complex III components that are conserved within the apicomplexan lineage. We demonstrate that one such subunit, which we term TgQCR11, is critical for parasite proliferation, mitochondrial oxygen consumption and Complex III activity, and establish that loss of this protein leads to defects in Complex III integrity. We conclude that the protein composition of Complex III in apicomplexans differs from that of the mammalian hosts that these parasites infect.
Author summary
Apicomplexan parasites cause numerous diseases in humans and animals, including malaria (Plasmodium species) and toxoplasmosis (Toxoplasma gondii). The coenzyme Q:cytochrome c oxidoreductase protein complex (Complex III) performs a central role in the mitochondrial electron transport chain of many eukaryotes. Despite being the target of several major anti-apicomplexan drugs, the protein composition of Complex III in apicomplexans was previously unknown. Our work identifies novel proteins in Complex III of apicomplexans, one of which is critical for complex function and integrity. Our study highlights divergent features of Complex III in apicomplexans, and provides a broader understanding of Complex III evolution in eukaryotes. Our study also provides important insights into what sets this major drug target apart from the equivalent complex in host species.
Introduction
Apicomplexans are a large phylum of intracellular, protozoan parasites that include the causative agents of malaria (Plasmodium species) and toxoplasmosis (Toxoplasma gondii). These parasites impose major economic and health burdens on human societies, and, in the absence of effective vaccines [1,2], there is a heavy reliance on drugs to treat disease. The parasite coenzyme Q:cytochrome c oxidoreductase complex (Complex III of the mitochondrial electron transport chain, ETC) represents one of the major drug targets in these parasites [3,4]. Numerous inhibitors of Complex III, including atovaquone and endochin-like quinolones, are in clinical use or in preclinical development against apicomplexans [5–7].
The ETC consists of a series of protein complexes that are embedded in the inner mitochondrial membrane. Electrons derived from the oxidation of mitochondrial substrates are donated via the action of dehydrogenases to a mobile electron carrier in the inner membrane called coenzyme Q (CoQ). CoQ exchanges electrons with Complex III at the Qo and Qi sites of the cytochrome b protein of Complex III, in a process called the Q cycle [8]. At the Qo site, electrons from reduced CoQ are donated to a heme moiety on cytochrome b, from where they are either donated on to cytochrome c, a mobile carrier protein in the mitochondrial intermembrane space, or donated back to CoQ at the Qi site. The transfer of electrons to cytochrome c occurs via an iron-sulfur cluster and a heme prosthetic group in the Rieske and cytochrome c1 proteins of Complex III, respectively. Electrons from cytochrome c are transported on to the cytochrome c oxidase complex (Complex IV), which facilitates electron transfer to the terminal electron acceptor, oxygen [9,10]. Electron transport through Complexes III and IV is coupled to the translocation of protons from the mitochondrial matrix into the intermembrane space, thereby generating a proton motive force across the inner membrane. This proton gradient is used for several important mitochondrial processes, including protein and solute import, and driving the activity of ATP synthase (Complex V) to generate ATP [11,12].
It is becoming increasingly apparent that the ETC of myzozoans–a eukaryotic lineage that includes apicomplexan parasites and their closest free living relatives, two phyla of marine algae called chromerids and dinoflagellates–differs considerably from that of other eukaryotes, including the animal hosts that apicomplexans infect [13–15]. For instance, Complexes IV and V from these organisms contain many subunits that lack homologs outside the myzozoan lineage [16–18]. Uncovering such diversity in a canonical mitochondrial process is of interest from both an evolutionary and a therapeutic standpoint. Evolutionarily, these studies provide insights into the diversification of mitochondria since eukaryote lineages diverged from their common ancestor >1.5 billion years ago [19]. Therapeutically, the discovery of novel proteins in a critical pathway such as the ETC highlights differences between parasites and their mammalian hosts that could open avenues for drug development.
The mechanism of action of Complex III inhibitors has been studied extensively, with most exploiting structural differences between the Qo and/or Qi sites of Complex III in parasites and host species to selectively target parasites [5–7]. Despite this, many important features of Complex III in apicomplexan parasites, including its protein composition, have not yet been elucidated. Here, we undertake a proteomic analysis of Complex III in T. gondii. Along with canonical subunits that are conserved in host organisms, we identified two highly divergent and two novel, apicomplexan-specific Complex III components. We demonstrate that one of the apicomplexan-specific subunits, which we term TgQCR11, is critical for parasite proliferation and ETC function through maintaining Complex III integrity. We conclude that Complex III of apicomplexans contains highly divergent and novel protein subunits, and differs considerably from the equivalent complex in animal hosts.
Results
T. gondii Complex III contains both canonical and novel protein subunits
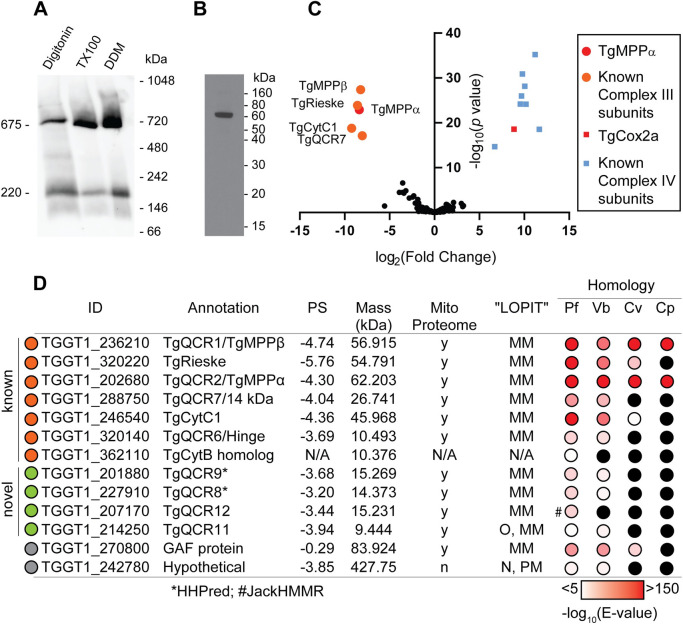
The putative mitochondrial processing peptidase alpha subunit (TgMPPα; www.toxodb.org gene ID TGGT1_202680) was shown previously to localize to the mitochondrion of T. gondii [20]. In many other eukaryotes, the MPPα protein functions in both the cleavage of mitochondrial-targeting presequences from mitochondrial matrix proteins as they are imported into the mitochondrion, and as one of the so-called Core proteins of Complex III [21]. To facilitate further characterization of TgMPPα, we introduced a TEV-HA tag into the 3’ end of the open reading frame of the native TgMPPα locus in T. gondii parasites (S1A and S1B Fig). To determine whether TgMPPα exists as part of a protein complex, we extracted proteins from TgMPPα-TEV-HA parasites using 1% (w/v) digitonin, 1% (v/v) Triton X-100 or 1% (w/v) n-Dodecyl β-D-maltoside (DDM) detergents, and separated solubilized protein complexes by blue native (BN)-PAGE. Western blotting with anti-HA antibodies revealed that the primary TgMPPα-TEV-HA complex is ~675 kDa in mass when solubilized in any of the three detergents (Fig 1A), though a fainter secondary complex at ~220 kDa was also observed. By contrast, the molecular mass of TgMPPα-TEV-HA when separated by SDS-PAGE was ~60 kDa (Fig 1B). We conclude that TgMPPα is a component of a ~675 kDa protein complex.
Fig 1. TgMPPα is part of a ~675 kDa protein complex and co-purifies with known components of Complex III.
(A) Western blot of proteins extracted from TgMPPα-TEV-HA parasites in 1% (w/v) digitonin, 1% (v/v) TX100 or 1% (w/v) DDM-containing lysis buffer, separated by BN-PAGE, and detected with anti-HA antibodies. (B) Western blot of proteins extracted from TgMPPα-TEV-HA parasites, separated by SDS-PAGE, and detected with anti-HA antibodies. (C) Volcano plot showing the log2 fold change vs -log10 p values of proteins purified from TgMPPα-TEV-HA vs TgCox2a-TEV-HA parasites using anti-HA immunoprecipitations and detected by mass spectrometry. To enable statistical comparisons, only proteins detected in all three independent experiments for both parasite lines are depicted. Proteins enriched in the TgMPPα-TEV-HA samples (p < 0.05; log2 fold change > 5) are labelled and shown in orange circles, with TgMPPα shown in red. Similarly, proteins previously identified as T. gondii Complex IV subunits [17] are shown in blue squares, with TgCox2a shown in red. (D) Table summarising the characteristics of proteins identified in proteomic analysis of the TgMPPα-TEV-HA complex (modelled after [18]). Proteins with homology to Complex III proteins are shown in orange circles, novel or divergent Complex III proteins are depicted by green circles, and proteins identified in the initial proteomic analysis but excluded in subsequent analyses are depicted in gray circles. Protein IDs were obtained from ToxoDB and proposed annotations are listed. The phenotype score (PS) of each gene predicts its importance for parasite proliferation, with scores < -2 typically found in genes that are important for proliferation [26]. Detection in the mitochondrial proteome (Mito Proteome, [17]) and its predicted cellular localisation (“LOPIT” [15]) are indicated (y = yes, n = no, N/A = not available, MM = mitochondrial membranes, O = outlier, N = nucleus, PM = plasma membrane). Homology indicates the tBLASTn expected value (E-value) between each T. gondii protein sequence and its closest match in Plasmodium falciparum (Pf), Vitrella brassicaformis (Vb), Chromera velia (Cv) or Cryptosporidium parvum (Cp) using EuPathDB searches. Black circles indicate a close match could not be identified. *, homology detected using HHPRED; #, homology detected using iterative JackHMMER searches.
To identify the proteins that comprise the TgMPPα-containing complex, we immunoprecipitated TgMPPα-TEV-HA and associated proteins, then subjected samples to mass spectrometry-based proteomic analyses. As a control for these experiments, we purified the unrelated cytochrome c oxidase complex (Complex IV of the ETC). To purify Complex IV, we introduced a TEV-HA tag into the 3’ end of the cytochrome c oxidase subunit 2a (TgCox2a; TGGT1_226590) open reading frame (S1C and S1D Fig), immunoprecipitated TgCox2a-TEV-HA and associated proteins, and performed separate mass spectrometry-based proteomic analyses. Using this approach, we identified five proteins–including TgMPPα–that were enriched in the TgMPPα-TEV-HA immunoprecipitation compared to the TgCox2a-TEV-HA immunoprecipitation across three independent experiments (Fig 1C and 1D and S1 Table). All five proteins are annotated as canonical components of Complex III, including the Core/mitochondrial processing peptidase protein TgMPPβ (TGGT1_236210), the iron-sulfur cluster protein TgRieske (TGGT1_320220), the so-called ‘14 kDa’ protein TgQCR7 (TGGT1_288750), and the cytochrome c1 heme protein TgCytC1 (TGGT1_246540) (S2 Fig). Two other canonical Complex III proteins–the ‘hinge’ protein TgQCR6 (TGGT1_320140) and the cytochrome b protein (TgCytB; TGGT1_362110)–were highly enriched in the TgMPPα immunoprecipitation but excluded from the quantitative analysis in Fig 1C because they were absent from at least one replicate of the TgCox2a control (Fig 1D and S1 Table). Note that the gene ID for TgCytB (TGGT1_362110) encodes a truncated protein, and may represent one of several fragmented TgCytB pseudogenes encoded by genetic material that occurs as ‘junk’ DNA in the nuclear genome of T. gondii, rather than the actual mitochondrial genome-encoded TgCytB gene [22,23].
Given that two canonical Complex III proteins were excluded from the initial analysis, we reasoned that a more comprehensive assessment of the data might reveal additional subunits that were likewise excluded. We shortlisted six additional proteins that were highly enriched in all three TgMPPα replicates relative to TgCox2a (either absent from all TgCox2a samples or > 100-fold more abundant in the TgMPPα purification than in the TgCox2a purification) (Fig 1D and S1 Table). Of these six proteins, five were annotated as hypothetical proteins (TGGT1_201880, TGGT1_227910, TGGT1_207170, TGGT1_214250, and TGGT1_242780) and one was annotated as a GAF domain-containing protein (TGGT1_270800). We utilized Localization of Organelle Proteins by Isotope Tagging (“LOPIT”) cellular localization data [15] coupled with our previously published mitochondrial proteome [17] to assess whether these proteins are likely to be mitochondrial. All the proteins were identified in both the mitochondrial proteome and the mitochondrial membrane fraction of the “LOPIT” dataset, with the exception of TGGT1_242780, which we excluded from further consideration (Fig 1D, gray).
We employed a phylogenomic approach to further assess the candidate Complex III proteins. The chromerid Vitrella brassicaformis contains the full suite of ETC complexes including Complex III, while Chromera velia lacks Complex III but retains the other ETC complexes [14]. We reasoned that if a protein has a homolog in V. brassicaformis but not in C. velia, this would support the hypothesis that it is a Complex III subunit. In addition, the apicomplexan Cryptosporidium parvum contains a highly reduced mitochondrion (termed mitosome) that lacks Complex III [24]. We further hypothesized that ‘true’ Complex III proteins were likely to be absent from C. parvum. To this end, we performed tBLASTn searches of EuPathDB using the identified proteins as queries to identify homologs in Plasmodium falciparum, V. brassicaformis, C. velia and C. parvum (Fig 1D). Since TgMPPα and TgMPPβ function together as a mitochondrial peptidase protein complex, separate from their function in Complex III, it is unsurprising that these proteins are found in all four species (Fig 1D). In contrast, most other canonical Complex III proteins have homologs in P. falciparum and V. brassicaformis but not in C. velia and C. parvum. An interesting exception is the TgRieske protein, which has a homolog in C. velia (Fig 1D). The CvRieske protein lacks the long N-terminus typical of other Rieske proteins, but retains the iron-sulfur cluster-containing core, implying this protein may have a secondary function in C. velia. A potential TgCytC1 homolog was also identified in C. velia (Fig 1D); however, this matched only to the N-terminus of TgCytC1, and lacked key resides including those that mediate heme binding. The CvCytC1 is likely not a functional CytC1 protein.
Of the additional proteins that we shortlisted, one has homologs in both chromerids (TGGT1_270800) and was therefore excluded from further analysis, three have homologs in V. brassicaformis but not C. velia (TGGT1_201880, TGGT1_227910, TGGT1_214250), and one appeared to be restricted to T. gondii (TGGT1_207170) (Fig 1D). Further analysis of TGGT1_207170 using iterative JackHMMER homology searches identified a homolog of this protein in P. falciparum and numerous other apicomplexans, but not in C. parvum, chromerids or dinoflagellates (Fig 1D). These data suggest that TGGT1_207170 may be an apicomplexan-specific protein.
To investigate whether the four shortlisted proteins (Fig 1D, green) have similarity to known Complex III proteins, we queried each protein against the Protein Data Bank (PDB) using HHPRED, a profile hidden Markov model search tool designed to identify homologous proteins with limited sequence similarity [25]. This analysis predicted that TGGT1_227910 has homology to the yeast Complex III protein QCR8 (percent probability 97.59 and E-value 0.00012) and TGGT1_201880 has homology to yeast QCR9 (percent probability 99.78 and E-value 4.9e-19). We therefore termed these two divergent Complex III subunits TgQCR8 and TgQCR9, respectively (Figs 1D and S2 and S3 and S4). The remaining two proteins were not matched to proteins from PDB with any confidence (percent probabilities <40 and E-values >20). In total, our proteomic analysis identified T. gondii homologs of nine out of the ten proteins from the well-studied Complex III of budding yeast, with no homolog to the yeast QCR10 protein apparent in T. gondii (S2 Fig). In addition, we identified two proteins that were restricted to the myzozoan or apicomplexan lineages. We termed these proteins TgQCR11 (TGGT1_214250) and TgQCR12 (TGGT1_207170), delineating them from the 10 “QCR” proteins from yeast (Figs 1D and S5 and S6). All the candidate Complex III proteins that were tested in a genome-wide CRISPR screen were predicted to be important for growth of the tachyzoite stage of T. gondii [26] (Fig 1D).
To begin to characterize the candidate Complex III proteins experimentally, we introduced FLAG epitope tags into the 3’ end of the open reading frames of the TgQCR8, TgQCR9, TgQCR11, and TgQCR12 loci in an existing TgMPPα-HA background strain [20] (S7 Fig). We then undertook western blot analysis and immunofluorescence assays to analyze the expression and cellular localization of these proteins. All four FLAG-tagged proteins were found to be between 15–20 kDa in mass and to localize to the mitochondrion (Fig 2). Interestingly, the relative abundance of these four proteins appeared to differ, with TgQCR11-FLAG and TgQCR8-FLAG the most abundant and TgQCR12-FLAG the least (S8A Fig).
Fig 2. Candidate Complex III subunits localize to the mitochondrion of T. gondii.
(Left) Western blot and (Right) immunofluorescence assay analysis of (A) TgMPPα-HA/TgQCR11-FLAG, (B) TgMPPα-HA/TgQCR8-FLAG, (C) TgMPPα-HA/TgQCR9-FLAG, or (D) TgMPPα-HA/TgQCR12-FLAG parasites. Western blots were detected with anti-FLAG antibodies, and immunofluorescence assays were detected with anti-FLAG (green) and the mitochondrial marker anti-TgTom40 (red) antibodies. Scale bars represent 2 μm.
We next sought to determine whether TgQCR11-FLAG, TgQCR8-FLAG, TgQCR9-FLAG and TgQCR12-FLAG exist in protein complexes. To do this, we extracted proteins from the four parasite lines and from the TgMPPα-TEV-HA parasite line using 1% (w/v) DDM, and separated these proteins by BN-PAGE. Western blotting using anti-FLAG antibodies revealed that all four proteins exist in a protein complex of ~675 kDa, approximately the same mass as the TgMPPα-TEV-HA complex when an adjacent lane run on the same gel was probed with anti-HA antibodies (Fig 3A). The relative abundances of these proteins in the ~675 kDa complex reflected their relative abundances in SDS-PAGE western blots (S8B Fig).
Fig 3. Candidate Complex III subunits are part of a ~675 kDa protein complex and interact with TgMPPα.
(A) Western blot of proteins extracted from TgMPPα-TEV-HA (left) or TgMPPα-HA/TgQCR11-FLAG, TgMPPα-HA/TgQCR8-FLAG, TgMPPα-HA/TgQCR9-FLAG and TgMPPα-HA/TgQCR12-FLAG parasites in 1% (w/v) DDM, separated by BN-PAGE, and detected with anti-HA or anti-FLAG antibodies. Images were obtained from a single membrane with lanes cut as indicated and probed with different concentration of antibodies. (B-E) Western blots of proteins extracted from (B) TgMPPα-HA/TgQCR11-FLAG, (C) TgMPPα-HA/TgQCR8-FLAG, (D) TgMPPα-HA/TgQCR9-FLAG or (E) TgMPPα-HA/TgQCR12-FLAG parasites, and subjected to immunoprecipitation using anti-HA (anti-HA IP) or anti-FLAG (anti-FLAG IP) antibody-coupled beads. Extracts include samples before immunoprecipitation (Total), samples that did not bind to the anti-HA or anti-FLAG beads (Unbound), and samples that bound to the anti-HA or anti-FLAG beads (Bound). Samples were separated by SDS-PAGE, and probed with anti-HA antibodies to detect TgMPPα-HA, anti-FLAG to detect TgQCRs, and anti-TgTom40 as a control to detect an unrelated mitochondrial protein.
As a direct test for whether TgQCR11-FLAG, TgQCR8-FLAG, TgQCR9-FLAG and TgQCR12-FLAG proteins are part of the same protein complex as TgMPPα-HA, we performed co-immunoprecipitation experiments. Immunoprecipitation of TgMPPα-HA with anti-HA antibodies co-purified each of the four FLAG-tagged proteins, but not the unrelated mitochondrial protein TgTom40 (Fig 3B–3E). Likewise, immunoprecipitation of TgQCR11-FLAG (Fig 3B), TgQCR8-FLAG (Fig 3C), TgQCR9-FLAG (Fig 3D) and TgQCR12-FLAG (Fig 3E) with anti-FLAG antibodies co-purified TgMPPα-HA but not TgTom40. In all instances, we identified a small proportion of TgMPPα-HA in the unbound fraction of the anti-FLAG immunoprecipitations (Fig 3B–3E). This could represent the ~220 kDa complex observed for TgMPPα-HA in BN-PAGE that is absent from the BN-PAGE analyses of the other proteins (Fig 3A). Together, these data indicate that TgMPPα-HA and the four TgQCR proteins exist in the same, ~675 kDa protein complex.
TgQCR11 is important for T. gondii proliferation and mitochondrial oxygen consumption
The existence of TgQCR11-FLAG, TgQCR8-FLAG, TgQCR9-FLAG and TgQCR12-FLAG in a complex with the core protein TgMPPα-HA suggests that these proteins are components of Complex III in T. gondii. To elucidate the importance and role of one of the apicomplexan-specific Complex III proteins, we undertook a functional characterization of TgQCR11. We first added a FLAG tag to the 3’ end of the TgQCR11 gene (S9A and S9B Fig) and then replaced the native promoter of TgQCR11 with an anhydrotetracycline (ATc)-regulated promoter (S9C and S9D Fig). We termed the resultant ATc-regulated TgQCR11 strain ‘rTgQCR11-FLAG’. We also added an HA tag to the 3’ end of the TgMPPα gene in this strain (S9E and S9F Fig), resulting in a strain we termed ‘rTgQCR11-FLAG/TgMPPα-HA’.
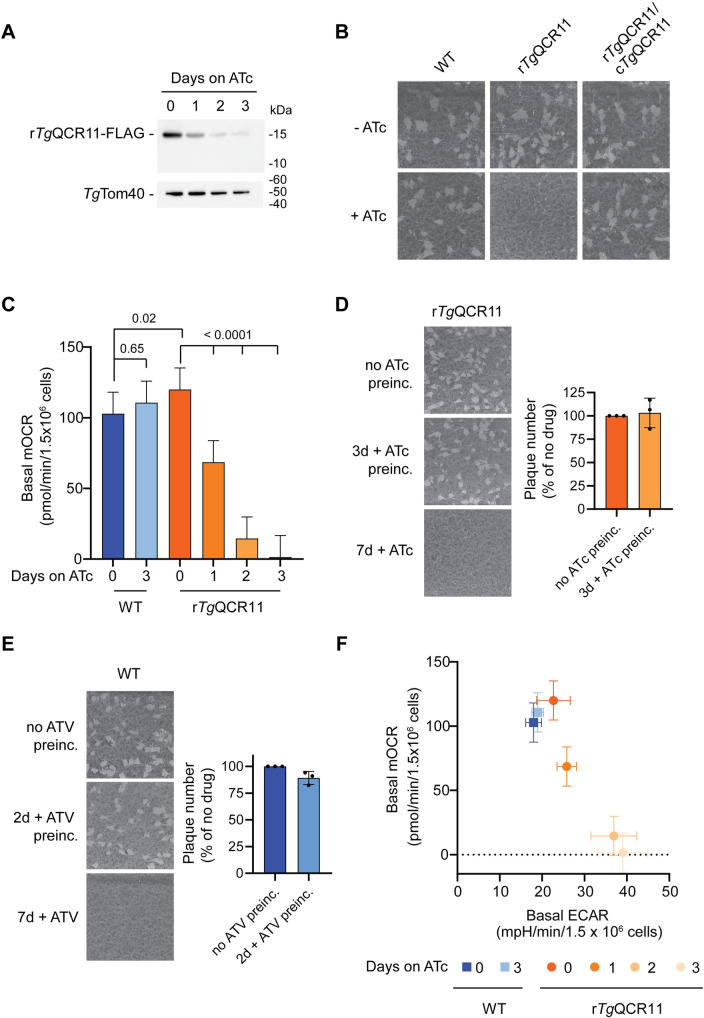
To examine the extent of TgQCR11 knockdown upon the addition of ATc, we cultured rTgQCR11-FLAG/TgMPPα-HA parasites in the absence of ATc or in the presence of ATc for 1–3 days, then separated proteins by SDS-PAGE and performed western blotting. TgQCR11-FLAG expression levels decreased substantially upon the addition of ATc, with only a small amount of protein detectable after 3 days in ATc (Fig 4A). To investigate the impact of TgQCR11-FLAG knockdown on parasite proliferation, we grew wild type (WT) and rTgQCR11-FLAG/TgMPPα-HA parasites in the absence or presence of ATc for 8 days and compared plaque sizes. Plaque sizes in rTgQCR11 but not WT parasites was severely impaired in the presence of ATc (Fig 4B). To determine whether this proliferation defect was specifically due to loss of TgQCR11, we complemented the rTgQCR11-FLAG strain with an additional copy of TgQCR11 expressed from the constitutively expressed α-tubulin promoter. Complementation with constitutively expressed TgQCR11 restored plaque formation in rTgQCR11 parasites cultured in the presence of ATc (Fig 4B), indicating that the proliferation defect we observed upon TgQCR11 knockdown was specifically due to loss of TgQCR11.
Fig 4. The apicomplexan-specific Complex III subunit TgQCR11 is important for parasite proliferation and mitochondrial oxygen consumption.
(A) Western blot of proteins extracted from rTgQCR11-FLAG/TgMPPα-HA parasites grown in the absence of ATc, or in the presence of ATc for 1–3 days, separated by SDS-PAGE, and detected using anti-FLAG and anti-TgTom40 antibodies (loading control). (B) Plaque assays measuring growth of WT, rTgQCR11-FLAG/TgMPPα-HA and complemented cTgQCR11-Ty1/rTgQCR11-FLAG/TgMPPα-HA parasites cultured in the absence (top) or presence (bottom) of ATc for 8 days. Assays are from a single experiment and are representative of 3 independent experiments. (C) Basal mitochondrial oxygen consumption rates (mOCR) of WT parasites grown in the absence of ATc or in the presence of ATc for 3 days (blue), and rTgQCR11-FLAG/TgMPPα-HA parasites grown in the absence of ATc or in the presence of ATc for 1–3 days (orange). A linear mixed-effects model was fitted to the data and values depict the least squares mean ± 95% CI of three independent experiments. ANOVA followed by Tukey’s multiple pairwise comparisons test was performed, with relevant p values shown. (D) Plaque assays of rTgQCR11-FLAG parasites grown in the absence of ATc (no ATc preinc; top) for 7 days, pre-incubated in ATc for 3 days before washing out and growing for a further 7 days in the absence of ATc (3d + ATc preinc; middle), or grown in the presence of ATc for all 7 days (7d + ATc; bottom). Quantifications of plaque number as a percent of the no ATc control are depicted to the right of the plaque assays, with bars representing the mean ± standard deviation of 3 independent experiments. (E) Plaque assays of WT parasites grown in the absence of atovaquone (no ATV preinc; top), pre-incubated in ATV for 2 days before washing out and growing for 7 days in the absence of ATV (2 days + ATV preinc; middle), or grown in the presence of ATV for all 7 days (7d + ATV; bottom). Quantifications of plaque number as a percent of the no ATV control are depicted to the right of the plaque assays, with bars representing the mean ± standard deviation of 3 independent experiments. (F) Basal mOCR versus basal extracellular acidification rate (ECAR) of WT parasites grown in the absence of ATc or in the presence of ATc for 3 days (blue), and rTgQCR11-FLAG/TgMPPα-HA parasites grown in the absence of ATc or in the presence of ATc for 1–3 days (orange). Data depict the mean mOCR and ECAR values ± 95% CI of the linear mixed-effects model (n = 3).
Oxygen acts as the final electron acceptor in the ETC. If TgQCR11 is an important component of Complex III, we hypothesized that knockdown of TgQCR11 would lead to defects in oxygen consumption in the parasite. To test this, we utilized a previously established assay to measure oxygen consumption in extracellular parasites using a Seahorse XFe96 extracellular flux analyzer [17]. We grew WT and rTgQCR11-FLAG/TgMPPα-HA parasites in the absence of ATc or in the presence of ATc for 1–3 days then measured their basal mitochondrial oxygen consumption rate (mOCR). While the presence of ATc did not impair basal mOCR in WT parasites, the basal mOCR of rTgQCR11-FLAG parasites was substantially depleted upon TgQCR11 knockdown (Fig 4C), indicating that TgQCR11 is important for mitochondrial oxygen consumption.
We wondered whether the severe deficiency in mOCR observed upon TgQCR11 knockdown was due to a specific defect in the ETC, or whether it was caused by a more general defect in mitochondrial function or parasite viability. To test whether TgQCR11 knockdown causes any gross defects in mitochondrial morphology, we grew rTgQCR11/TgMPPα-HA parasites in the absence or presence of ATc and performed immunofluorescence assays to visualize both the inner (TgMPPα-HA) and outer (TgTom40) mitochondrial membranes. This revealed no observable defects in mitochondrial morphology upon TgQCR11 knockdown (S10A Fig).
Next, we tested whether parasites remain viable following knockdown of TgQCR11. To do this, we pre-incubated rTgQCR11 parasites in the presence of ATc for 3 days (a time point when TgQCR11 is knocked down substantially and mOCR is minimal; Fig 4A and 4C), then washed out the ATc and allowed parasites to proliferate in the absence of ATc. After 7 days proliferation, we compared plaque number to parasites that had not been pre-incubated in ATc. Plaque size and number were similar between the ATc-pre-incubated and non-pre-incubated parasites when grown in the absence of ATc, while parasites grown in the presence of ATc for all 7 days underwent minimal proliferation (Fig 4D). These results indicate that TgQCR11 knockdown is reversible and that rTgQCR11 parasites grown in ATc for 3 days have similar viability to parasites grown in the absence of ATc.
We were curious whether treating parasites with the Complex III inhibitor atovaquone might phenocopy TgQCR11 knockdown in terms of parasite viability. To test this, we grew WT parasites in the presence of atovaquone (1 μM, 50 times the IC50 reported by [27]) for 2 days, then washed out the drug and grew parasites for a further 7 days (Fig 4E). While parasites grown in the presence of atovaquone for all 7 days did not form visible plaques, parasites that had been pre-incubated in atovaquone for 2 days showed a similar plaque number to the no drug control (Fig 4E). These results mirror those obtained in the genetic experiments and, together, these data imply that parasites remain viable for at least two days without a functional ETC.
To observe what happens to other aspects of parasite metabolism upon TgQCR11 knockdown, we measured the extracellular acidification rate (ECAR) of WT and rTgQCR11 parasites grown in the absence or presence of ATc. We have previously used ECAR as a general indication of parasite metabolic activity [17,28]. ECAR levels of WT and rTgQCR11 parasites grown in the absence of ATc or WT parasites grown in the presence of ATc were not significantly different (Figs 4F and S10B). By contrast, growth of rTgQCR11 parasites in the presence of ATc for 2 days resulted in a significant increase in ECAR (Figs 4F and S10B), indicating that parasites remained metabolically active upon TgQCR11 knockdown. The small but significant increase in ECAR upon TgQCR11 knockdown may indicate that the parasite compensates for loss of ETC activity by upregulating other aspects of cellular metabolism (e.g. glycolysis), though this needs to be studied further.
Together, these data indicate that the defects observed in mitochondrial oxygen consumption upon TgQCR11 knockdown were not due to general defects in mitochondrial morphology, parasite viability or cellular metabolism. We therefore conclude that TgQCR11 has an important and specific role in the ETC of T. gondii parasites, consistent with its association with Complex III.
TgQCR11 is important for the function and integrity of Complex III
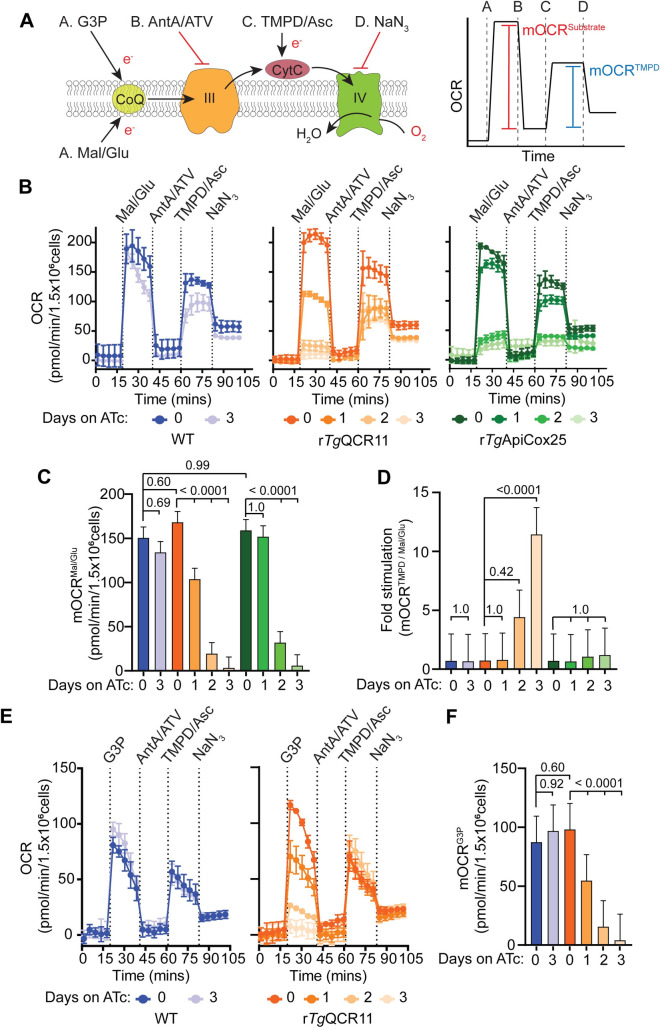
To establish whether TgQCR11 is important for Complex III function in T. gondii, we sought to undertake a more direct interrogation of the functionality of various ETC components upon TgQCR11 knockdown. To do this, we established an XFe96 flux analyzer-based assay that enabled us to measure substrate-dependent mOCR using digitonin-permeabilized extracellular parasites. Permeabilizing the plasma membrane of parasites with digitonin allows the passage of substrates with polar functional groups into parasites, where they can feed electrons into the ETC either directly or via mitochondrial metabolism. Supplying different combinations of substrates and inhibitors during the assay enables an assessment of the functionality of different ETC complexes (Fig 5A; [29]).
Fig 5. Loss of TgQCR11 leads to a specific defect in Complex III function.
(A) Schematic diagram of the assay measuring OCR in digitonin-permeabilized parasites, with inset (right) depicting a mock oxygen consumption rate (OCR) versus time graph to illustrate the typical response of WT parasites. Parasites are starved for 1 hour in base media to deplete endogenous energy sources, then permeabilized with 0.002% (w/v) digitonin before being subjected to the following injections of substrates or inhibitors: Port A, malate and glutamate (Mal/Glu) or glycerol 3-phopshate (G3P); Port B, antimycin A and atovaquone (AntA/ATV); Port C, TMPD and ascorbate (TMPD/Asc); Port D, sodium azide (NaN3). The mitochondrial OCR (mOCR) elicited by a substrate (red line, mOCRsubstrate) and the mOCR elicited by TMPD/Asc (blue line, mOCRTMPD) are then calculated from these data. CytC, cytochrome c; CoQ, coenzyme Q; III, Complex III; IV, Complex IV; e-, electrons. (B) Representative traces depicting OCR over time when supplying Mal/Glu (10 mM) as an energy source. WT (blue), rTgQCR11-FLAG/TgMPPα-HA (orange) and rTgApiCox25-HA (green) parasites were grown in the absence of ATc or in the presence of ATc for 1–3 days. Data represent the mean ± SD of three technical replicates, and are representative of three independent experiments. (C) Mal/Glu elicited mOCR (mOCRMal/Glu) of WT (blue), rTgQCR11-FLAG/TgMPPα-HA (orange) and rTgApiCox25-HA (green) parasites that were grown in the absence of ATc or in the presence of ATc for 1–3 days. A linear mixed-effects model was fitted to the data and values depict the least squares mean ± 95% CI from three independent experiments. ANOVA followed by Tukey’s multiple pairwise comparisons test was performed, with relevant p values shown. (D) Fold stimulation of mOCR by TMPD relative to Mal/Glu in WT (blue), rTgQCR11-FLAG/TgMPPα-HA (orange) and rTgApiCox25-HA (green) parasites that had been grown in the absence of ATc or in the presence of ATc for 1–3 days (mean ± 95% CI of the linear mixed-effects model; n = 3). ANOVA followed by Tukey’s multiple pairwise comparisons test was performed, with relevant p values shown. (E) Representative traces depicting OCR over time when supplying the TCA cycle-independent substrate G3P (10 mM) as an energy source. WT (blue) and rTgQCR11-FLAG/TgMPPα-HA (orange) parasites were grown in the absence of ATc or in the presence of ATc for 1–3 days. Data represent the mean ± SD of three technical replicates, and are representative of three independent experiments. (F) G3P elicited mOCR (mOCRG3P) of WT (blue) and rTgQCR11-FLAG/TgMPPα-HA (orange) parasites that were grown in the absence of ATc or in the presence of ATc for 1–3 days (mean ± 95% CI of the linear mixed-effects model; n = 3). ANOVA followed by Tukey’s multiple pairwise comparisons test was performed, with relevant p values shown.
We grew WT, rTgQCR11 and rTgApiCox25 (an ATc-regulatable strain that enables knockdown of the Complex IV protein TgApiCox25; [17]) parasites in the absence of ATc or in the presence of ATc for 1–3 days. Parasites were starved for 1 hour in base medium to deplete endogenous substrates, and then permeabilized with 0.002% (w/v) digitonin. The five readings taken before injection of substrate show that permeabilized parasites have very low OCR (Fig 5B), indicating that the 1 hour starvation successfully depleted endogenous substrates. Injection of the tricarboxylic acid (TCA) cycle substrates malate and glutamate caused an almost instantaneous increase in OCR in WT, rTgQCR11 and rTgApiCox25 parasites that were cultured in the absence of ATc (Fig 5B). As this OCR could be abolished by subsequent injection of the Complex III inhibitors antimycin A and atovaquone (Fig 5B), these data indicate that the ETC is functional in these parasites. The malate/glutamate-elicited mOCR of WT parasites grown for 3 days in the presence of ATc was not significantly different to WT parasites grown in the absence of ATc (Fig 5C), indicating that ATc itself does not impact ETC function. By contrast, knockdown of TgQCR11 and TgApiCox25 caused significant decreases in malate/glutamate-dependent mOCR (Fig 5B and 5C).
We next asked whether defects in malate/glutamate-dependent mOCR upon TgQCR11 or TgApiCox25 knockdown occurred upstream or downstream of cytochrome c. To test this, we injected the cytochrome c substrate N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD). Reduced TMPD donates electrons directly to cytochrome c, downstream of Complex III, and should therefore rescue mOCR in parasites with an ETC defect upstream of cytochrome c (Fig 5A). By contrast, a Complex IV defect should not be rescued by TMPD since Complex IV is downstream of cytochrome c. Injection of TMPD caused an increase in mOCR in rTgQCR11 parasites cultured in the presence of ATc (Fig 5B). By contrast, rTgApiCox25 parasites cultured for 2 or 3 days on ATc had very little TMPD-dependent mOCR (Fig 5B). Calculating the fold stimulation of mOCR by TMPD relative to malate/glutamate-dependent mOCR indicates that rTgQCR11 parasites grown for 3 days on ATc have 10-fold greater stimulation of mOCR by TMPD relative to malate (Fig 5D). This demonstrates that Complex IV activity is largely retained upon TgQCR11 knockdown and implies that the defect in ETC activity upon loss of this protein occurs upstream of cytochrome c. By contrast, rTgApiCox25 parasites grown for 3 days on ATc have little stimulation of mOCR by TMPD relative to malate/glutamate, indicating that loss of TgApiCox25 leads to a defect in Complex IV activity (Fig 5D).
The preceding data indicate that loss of TgQCR11 leads to defects in the ETC upstream of cytochrome c. Since malate/glutamate can feed into the ETC via the TCA cycle [13,30], it is conceivable that defects we observe in mOCR upon TgQCR11 knockdown could be the result of defects in the TCA cycle rather than a selective defect in Complex III. To address this, we measured mOCR using the substrate glycerol 3-phosphate, which donates electrons to CoQ independently of the TCA cycle (Fig 5A; [13,30]). Knockdown of TgQCR11 caused a similar decrease in glycerol 3-phosphate-dependent mOCR compared to malate/glutamate-dependent mOCR (Fig 5E and 5F). Since mOCR elicited by two different substrates, one of which is independent of the TCA cycle, was impaired by knockdown of TgQCR11, we conclude that loss of TgQCR11 likely leads to a selective defect in Complex III activity.
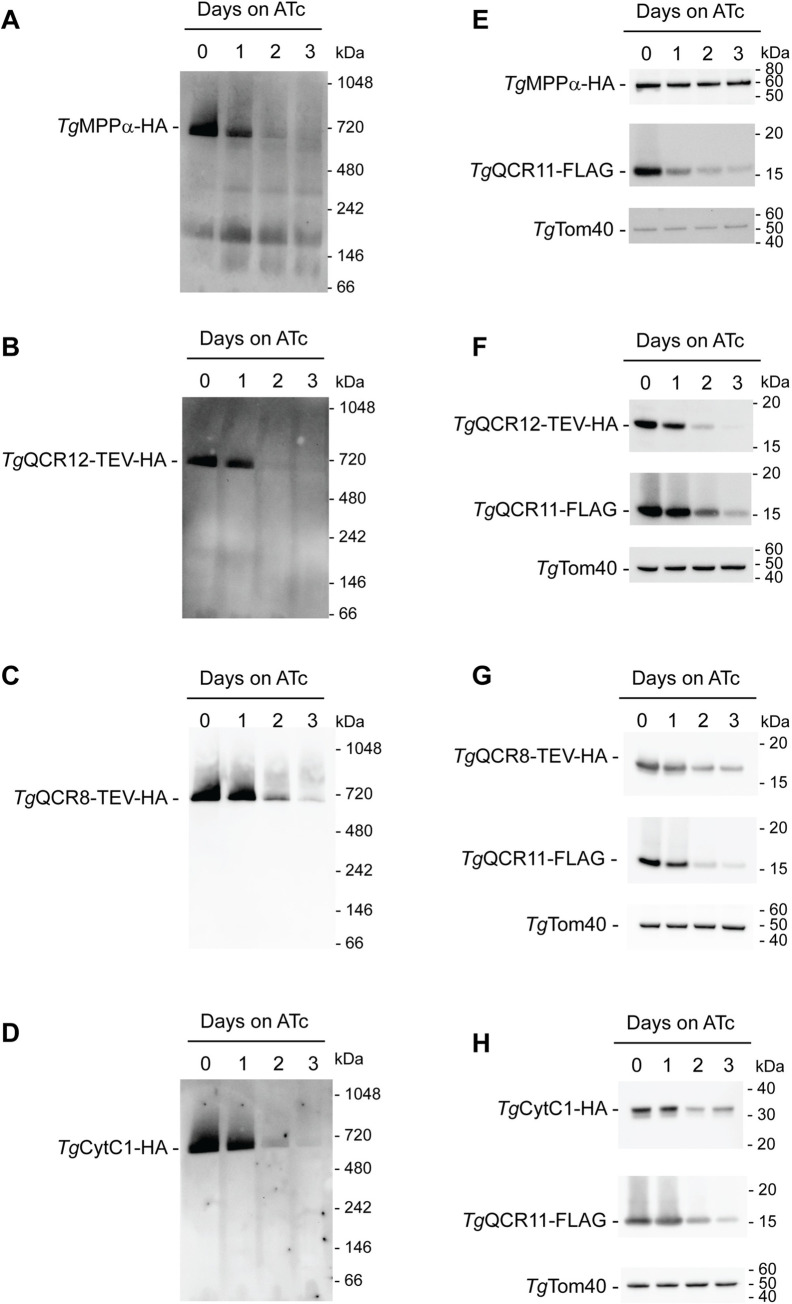
Given the severe defect in mOCR observed upon TgQCR11 knockdown, we wondered whether the integrity of Complex III is compromised by loss of TgQCR11. To assess this, we set out to measure the effects of TgQCR11 knockdown on several Complex III proteins, including TgMPPα, the apicomplexan specific subunit TgQCR12, the supernumerary subunit TgQCR8, and the catalytic subunit TgCytC1. We integrated TEV-HA tags into the 3’ ends of the TgQCR8 and TgQCR12 open reading frames (S11 Fig), or an HA tag into the 3’ end of the TgCytC1 open reading frame in the rTgQCR11-FLAG parasite line. We performed co-immunoprecipitation analysis and verified that TgCytC1 and TgQCR11 are part of the same protein complex (S12 Fig). We then grew rTgQCR11-FLAG/TgMPPα-HA, rTgQCR11-FLAG/TgQCR8-TEV-HA, rTgQCR11-FLAG/TgQCR12-TEV-HA and rTgQCR11-FLAG/TgCytC1-HA parasites in the absence of ATc or in the presence of ATc for 1–3 days and assessed Complex III integrity by BN-PAGE. Strikingly, the ~675 kDa complex of all four proteins was depleted upon TgQCR11 knockdown (Figs 6A–6D and S13), suggesting that Complex III assembly and/or stability may be impaired upon loss of TgQCR11. We wondered whether the abundance of Complex III proteins may also be affected by TgQCR11 knockdown, and assessed this by SDS-PAGE and western blotting. Interestingly, while the abundance of TgMPPα remained consistent (Figs 6E and S13), the abundance of TgQCR12, TgQCR8 and TgCytC1 decreased by day 2–3 on ATc (Figs 6F–6H and S13), suggesting that knockdown of TgQCR11 decreases the abundance of some Complex III proteins but not others. It is possible that TgMPPα abundance remains unchanged because, in addition to being a component of Complex III, it exists in a lower mass, ~220 kDa complex that our BN-PAGE analysis indicated is retained upon TgQCR11 knockdown (Fig 6A). This ~220 kDa complex could represent the mitochondrial processing peptidase in these parasites. Together, these data indicate that TgQCR11 is a novel, apicomplexan-specific subunit of Complex III in T. gondii that is critical for Complex III function by maintaining the integrity of this protein complex.
Fig 6. TgQCR11 is important for Complex III integrity.
(A-D) Western blots of proteins extracted from (A) rTgQCR11-FLAG/TgMPPα-HA, (B) rTgQCR11-FLAG/TgQCR12-TEV-HA, (C) rTgQCR11-FLAG/TgQCR8-TEV-HA and (D) rTgQCR11-FLAG/TgCytC1-HA parasites grown in the absence of ATc or in the presence of ATc for 1–3 days. Samples were prepared in 1% (w/v) DDM, separated by BN-PAGE, and detected with anti-HA antibodies. (E-H) Western blots of proteins extracted from (E) rTgQCR11-FLAG/TgMPPα-HA, (F) rTgQCR11-FLAG/TgQCR12-TEV-HA, (G) rTgQCR11-FLAG/TgQCR8-TEV-HA and (H) rTgQCR11-FLAG/TgCytC1-HA parasites that had been grown in the absence of ATc or in the presence of ATc for 1–3 days. Samples were separated by SDS-PAGE, and probed with anti-HA, anti-FLAG and anti-TgTom40 (loading control) antibodies. Western blots shown are representative of at least two independent experiments, with matched BN-PAGE and SDS-PAGE samples prepared from the same experiment. Quantifications of protein abundances are depicted in S13 Fig.
Discussion
In this study, we characterized Complex III of the mitochondrial ETC in T. gondii parasites, which we demonstrate exists as a ~675 kDa complex comprising 11 protein subunits (Fig 1). Our data are consistent with an independent, parallel study by MacLean and colleagues, who identified the same 11 subunits in a broader proteomic analysis of mitochondrial respiratory chain complexes in T. gondii [31]. The number of subunits we identified in T. gondii Complex III is similar to that reported for the bovine [32–34], yeast [35] and plant [21,36] complexes (11, 10 and 10 subunits, respectively), and the mass is similar to the fully-assembled yeast complex (670 kDa [37]). The overall architecture of Complex III in eukaryotes is broadly conserved as a homodimer, with one copy of each subunit per monomer [32–35,38], and this may also be the case for T. gondii. The sum of the predicted masses of the 11 T. gondii Complex III subunits is ~350 kDa (assuming TgCytB mass to be ~41 kDa; [22]), giving a mass of ~700 kDa for the Complex III homodimer, which is in the ballpark of the ~675 kDa we observed by BN-PAGE. A caveat to this conclusion is that we observed different abundances in some Complex III subunits (S8 Fig), raising the possibility that not all subunits exist in a strict 1:1 stoichiometry per monomer.
In our proteomic analysis of T. gondii Complex III, we identified numerous canonical subunits, including the three electron transporting subunits (TgRieske, TgCytB and TgCytC1), the two core proteins (TgMPPα and TgMPPβ), and two known additional (or so-called ‘supernumerary’) subunits, the ‘hinge’ protein TgQCR6 and ‘14 kDa’ protein TgQCR7. Our analysis also identified two highly divergent supernumerary subunits, TgQCR8 and TgQCR9, which were only identifiable using the HHPRED search tool. This indicates that, like several supernumerary proteins in Complexes IV and V of T. gondii [16–18], the ~1.5 billion years of evolution since the common ancestor of apicomplexans and other eukaryotes has resulted in considerable divergence in the sequences of these ETC proteins.
Intriguingly, our analysis also identified two novel Complex III subunits (TgQCR11 and TgQCR12) that are restricted to organisms closely related to T. gondii. TgQCR11 has homologs in other myzozoans–a lineage which comprises apicomplexans, as well as their closest free-living relatives, the chromerids and dinoflagellates–whereas TgQCR12 homologs are restricted to apicomplexans. These observations fit with an emerging narrative that mitochondria of myzozoans have evolved numerous proteins and functions that are unique amongst eukaryotes [15,17,39,40]. The reasons for these novelties are unclear, but it is conceivable that the evolution of proteins like TgQCR11 and TgQCR12 were necessitated by evolutionary pressures for greater ETC efficiency and/or improved mitochondrial energy generation in the marine (and later parasitic) niches in which these organisms evolved. We cannot rule out the possibility that TgQCR11 and TgQCR12 homologs do exist in other lineages of eukaryotes, but have diverged to the extent that they can no longer be detected through even the most sophisticated homology-based searches. In future, obtaining a Complex III structure from apicomplexans will help reconcile these possibilities and shed further light on how the highly diverged and novel subunits identified in this study contribute to Complex III function.
In other eukaryotes such as yeast, the supernumerary subunits of Complex III contribute to complex assembly and/or stability [41,42]. For example, yeast QCR9 (which is homologous to TgQCR9) is important for maintaining the integrity of Complex III [37,43,44], and yeast QCR10 (for which we identified no homolog in T. gondii) appears to have a role in stabilising the catalytic Rieske subunit in the complex [45]. Supernumerary subunits have also been proposed to contribute to the assembly of so-called “supercomplexes” between Complex III and other respiratory complexes [46,47]. For example, the yeast Complex III subunits Cor1 (the homolog of TgMPPβ) and QCR8 (the homolog of TgQCR8) interact with Complex IV within these supercomplexes [46,47]. We demonstrate that loss of TgQCR11 leads to severe defects in parasite proliferation and mitochondrial oxygen consumption (Fig 4), decreased abundance of other Complex III subunits, and the disappearance of the ~675 kDa Complex III (Fig 6). These observations are all consistent with TgQCR11 playing a critical role in maintaining Complex III integrity by mediating the assembly and/or the stability of this complex, much like supernumerary Complex III subunits from yeast. The exact role of TgQCR11 in these processes, however, remains elusive. Future functional analyses of TgQCR11, including an examination of its position within the structure of Complex III, may address its actual role in the complex. Much like our findings with TgQCR11, the parallel study by MacLean and colleagues found that knockdown of TgQCR8, TgQCR9 and TgQCR12 led to defects in the assembly of the TgRieske subunit into the complex [31], suggesting that the various novel and divergent components are all required for integrity of Complex III in T. gondii.
In this study, we have developed a powerful suite of assays to probe different stages and complexes of the ETC in T. gondii parasites. These assays rely on feeding permeabilized parasites with specific ETC substrates and inhibitors at set times during the assays. We used these assays to demonstrate that knockdown of TgQCR11 caused a specific defect in Complex III function (Fig 5), since 1) the mOCR elicited by two independent substrate combinations (malate/glutamate and glycerol 3-phosphate) was decreased upon TgQCR11 knockdown, and 2) the fold stimulation of TMPD-dependent mOCR compared to malate/glutamate-dependent mOCR was high. This is consistent with TgQCR11 functioning downstream of CoQ yet upstream of cytochrome c (i.e. in Complex III). By contrast, the Complex IV protein TgApiCox25 [17] had a low-fold simulation of TMPD-dependent mOCR compared to malate/glutamate-dependent mOCR, consistent with Complex IV functioning downstream of cytochrome c (i.e. in Complex IV). In future, these assays will enable an in-depth characterization of the function of ETC proteins and complexes, and provide a detailed understanding of the contribution of mitochondrial (and broader parasite) biosynthetic and metabolic pathways to the ETC and energy generation in these parasites. We also note that these approaches lend themselves to drug screening approaches for pin-pointing the target of ETC inhibitors in these parasites.
We observed that parasites remained viable after knockdown of TgQCR11 (Fig 4D), and that this was phenocopied by 2 day treatment with the Complex III inhibitor atovaquone (Fig 4E). These findings suggest that, at least in the short term and at the inhibitor concentration tested, parasites can survive but do not thrive in the absence of mitochondrial respiration. Precisely how parasites achieve this state remains uncertain, though the measurable increases in ECAR observed upon TgQCR11 knockdown (Figs 4F and S10B) suggests that parasites may respond to the impairment of Complex III by upregulating other metabolic pathways (such as glycolysis). Our earlier study of Complex IV demonstrated that parasites remain viable after loss of a key protein in this complex (TgApiCox25) [17], suggesting that parasite persistence is not specific to impairment of Complex III but is a general phenomenon when targeting the ETC. Determining the mechanism and duration of parasite persistence following the impairment of mitochondrial respiration has potentially important implications for therapeutics that target Complex III. Several clinical studies reported that 12–26% of patients experienced relapse of T. gondii infection after 1 year, and up to 75% after 6 years, following atovaquone therapy [48–50]. While the inability of atovaquone to eliminate T. gondii infection could be due to many factors, including the conversion of disease-causing tachyzoites to latent bradyzoite-containing tissue cysts [5], it is conceivable that the short-term persistence following ETC impairment may enable parasites to survive initial treatment until bradyzoite differentiation and/or atovaquone resistance arises.
Our work highlights the divergence of mitochondrial ETC Complex III composition in apicomplexan parasites, providing important insights into what sets this major drug target apart from the equivalent complex in host species. Future studies can now build on our findings to reveal how novel subunits of this complex contribute to Complex III function and druggability.
Materials and methods
Host cell and parasite culture
T. gondii tachyzoites were cultured in human foreskin fibroblasts (HFF), as previously described [28,51]. In paired anhydrotetracycline (ATc) knockdown experiments, ATc (0.5 μg/ml) or ethanol (vehicle control) was added to the media on required days. Plaque assays were performed as described previously [51], with 500 parasites added per flask and incubated for 7–8 days before staining with crystal violet. In the atovaquone washout experiments, 500 parasites were added per flask and allowed to invade host cells for 4 hours before addition of atovaquone (1 μM) to the flask. After 2 days incubation in the presence of atovaquone, the medium was removed, flasks were washed twice with atovaquone-free medium, and parasites allowed to grow for a further 7 days to allow plaques to develop before staining with crystal violet.
Genetic modifications of T. gondii
TATiΔku80 strain parasites [52] were used as the parental cell line in this study. All genetically modified parasites were cloned by flow cytometry before being characterized.
To incorporate a 3’ hemagglutinin tag containing a tobacco etch virus protease cleavage site (TEV-HA tag) into the loci of TgMPPα and TgCox2a, we generated a vector expressing a single guide RNA (sgRNA) targeting the region around the stop codon of TgMPPα and used an existing sgRNA-expression plasmid to target TgCox2a [17]. To generate the TgMPPα-targeting vector, we modified the pSAG1::Cas9-U6::sgUPRT plasmid (Addgene plasmid # 54467; [53]) using Q5 mutagenesis (New England Biolabs) as described previously [53]. For site-directed mutagenesis, we used the primers MPPα 3’rep CRISPR fwd and the Universal Reverse primer (S2 Table). We also amplified a TEV-HA tag containing 50 bp of flanking sequence either side of the TgMPPα or TgCox2a stop codon, using the primers MPPα tag fwd and MPPα tag rvs or Cox2a 3’ rep fwd and Cox2a 3’ rep rvs, with a TEV-HA tag template synthesized as a gBlock (IDT; S2 Table). The sgRNA expressing vectors, which also expressed GFP-tagged Cas9, were co-transfected into TATiΔku80 strain parasites along with the TEV-HA tags, with transfections performed as described previously [51]. GFP-positive parasites were selected by flow cytometry and cloned 3 days following transfection, then screened for successful integration using the primers MPPα 3’ screen fwd and MPPα 3’ screen rvs or Cox2a 3’ screen fwd and Cox2a 3’ screen rvs (S2 Table and S1 Fig).
To introduce 3’ FLAG epitope tags into the loci of TgQCR8, TgQCR9, TgQCR11 and TgQCR12, we generated vectors expressing a sgRNA targeting the region around their stop codons. To do this, we modified the pSAG1::Cas9-U6::sgUPRT vector using Q5 mutagenesis with gene specific 3’rep CRISPR fwd primers and the Universal Reverse primer (S2 Table). We also amplified a FLAG tag containing 50 bp of flanking sequence either side of the stop codon of each gene, using gene specific fwd and rvs primers, with FLAG tag template synthesized as a gBlock (IDT; S2 Table). We co-transfected the plasmid and PCR product into TgMPPα-HA strain parasites [20] and also into TATi/Δku80 strain parasites for TgQCR11, selected GFP positive parasites by flow cytometry 3 days post-transfection, then screened for successful integration using gene specific screening fwd and rvs primers (S2 Table and S7E and S7F and S9A and S9B Figs).
To introduce an ATc-regulated promoter into the TgQCR11 locus, we generated a vector expressing a sgRNA targeting the region around the start codon of TgQCR11. To do this, we modified the vector pSAG1::Cas9-U6::sgUPRT using Q5 mutagenesis with the primers QCR11 5’ CRISPR fwd and the universal reverse primer (S2 Table). We also PCR amplified the ATc-regulated promoter plus a ‘spacer’ region consisting of part of the T. gondii DHFR open reading frame and 3’ UTR using the pPR2-HA3 vector [54] as template and the primers QCR11 pro rep fwd and QCR11 pro rep rvs (S2 Table), which each contain 50 bp of sequence specific for the TgQCR11 locus. We co-transfected the plasmid and the ATc-regulatable promoter into TgQCR11-FLAG strain parasites, selected GFP positive parasites by flow cytometry 3 days post-transfection, then screened for successful integration of the ATc-regulatable promoter using the primers QCR11 5’ screen fwd and QCR11 5’ screen rvs (S2 Table and S9C and S9D Fig).
To generate a cell line where TgMPPα is HA tagged in the rTgQCR11-FLAG strain, we introduced a 3’ HA tag into the locus of TgMPPα using the vector described earlier that expresses a sgRNA targeting the region around the stop codon of TgMPPα. We also amplified a HA tag containing 50 bp of flanking sequence either side of the TgMPPα stop codon, using the primers MPPα tag fwd and MPPα tag rvs, with HA tag template synthesized as a gBlock (IDT; S2 Table). We co-transfected the plasmid and PCR product into rTgQCR11-FLAG strain parasites, selected GFP positive parasites by flow cytometry 3 days post-transfection, then screened for successful integration using the primers MPPα 3’ screen fwd and MPPα 3’ screen rvs (S2 Table and S9E and S9F Fig).
To generate a vector that constitutively expressed TgQCR11 for complementing the rTgQCR11 mutant, we ordered a gene block encoding the TgQCR11 open reading frame plus a Ty1 tag (IDT; S2 Table) and performed PCR using the primers QCR11 comp fwd and universal Ty1 rvs (S2 Table). We digested the resulting PCR product with BglII and XmaI and ligated this into the BglII and XmaI sites of the vector pUDTG. The pUDTG vector was generated by Yi Xue (ANU) by digesting the vector pUgCTH3 [55] with ApaI and HindIII, and ligating the excised UPRT flank into the equivalent sites of the vector pDTG (a kind gift from Chris Tonkin, Walter and Eliza Hall Institute). The resulting “TgQCR11 in pUDT-Ty1” vector contains a pyrimethamine-resistance DHFR cassette, a UPRT flanking sequence for integration into the non-essential UPRT locus of T. gondii, and fuses a C-terminal Ty1 epitope tag to the complementing TgQCR11 protein. This vector was linearized in the UPRT flanking sequence with MfeI, transfected into rTgQCR11-FLAG/TgMPPα-HA parasites, and selected on pyrimethamine as described [51].
To generate cell lines expressing either TgQCR8-TEV-HA or TgQCR12-TEV-HA in the rTgQCR11-FLAG parasite strain, we introduced 3’ TEV-HA tags into the loci of TgQCR8 and TgQCR12. We replicated the strategy for introducing FLAG tags into these loci that is described above, but instead co-transfected the sgRNA-expressing vectors with TEV-HA tags. We amplified TEV-HA tags containing 50 bp of flanking sequence either side of the stop codons using gene specific fwd and rvs primers, with TEV-HA tag template synthesized as a gBlock (IDT; S2 Table). Following transfection, we selected GFP positive parasites by flow cytometry, then screened for successful integration using gene specific screening fwd and rvs primers (S2 Table and S11 Fig).
SDS-PAGE, BN-PAGE and immunoblotting
Sodium dodecylsulfate (SDS)-polyacrylamide electrophoresis (PAGE), blue native (BN)-PAGE and immunoblotting were performed as described previously [20,56]. SDS-PAGE samples were separated on a 12% (v/v) acrylamide gel and BN-PAGE samples were separated on a 4–16% (v/v) acrylamide gel. Primary antibodies used included mouse anti-FLAG (1:100–1:2,000 dilution; Sigma clone M2), rat anti-HA (1:2000 dilution; Sigma clone 3F10), and rabbit anti-Tom40 (1:2,000 dilution; [20]). The secondary antibodies used were horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Abcam, catalog number ab6789), goat anti-rabbit IgG (Abcam, catalog number ab97051) and goat anti-rat IgG (Abcam, catalog number ab97057). For probing for mouse antibodies on immunoprecipitation western blots, HRP-conjugated anti-mouse TrueBlot ULTRA antibodies (Rockland, catalog number 18-18817-33) were used at 1:2,500 dilution. Blots were imaged using X-ray film or using a ChemiDoc MP imaging system (BioRad).
Immunoprecipitation and mass spectrometry
Immunoprecipitations were performed as described previously [20], except that parasite samples were solubilized in 1% (v/v) Triton X-100. HA-tagged proteins were purified using anti-HA affinity matrix (Sigma; rat anti-HA clone 3F10 antibodies) and FLAG-tagged proteins were purified using anti-FLAG M2 affinity gel (Sigma; mouse anti-FLAG clone M2 antibodies). For mass spectrometry sample preparation, parasite samples were solubilized in 1% (w/v) DDM, and processed as described previously [17]. Briefly, anti-HA affinity matrix bound with HA-tagged protein complexes were frozen at −80°C for 1 hr, then eluted at room temperature in 0.2 M glycine containing 1% (w/v) DDM (pH 2.3). Samples were neutralized in ammonium bicarbonate, then extracted in chloroform:methanol as described [57]. After extraction, the pellets were dried and stored at −80°C before mass spectrometry analysis.
Mass spectrometry was conducted as previously described [17]. Briefly, samples were resuspended in 8 M Urea, 50 mM Tris pH 8.3 followed by reduction and alkylation. Solubilized proteins were submitted to trypsin digestion overnight and the resulting peptides purified using the C18 stage tips procedure. Peptides were reconstituted in 0.1% formic acid and 2% acetonitrile, loaded onto a trap column and washed for 6 min before switching the precolumn in line with the analytical column. The separation of peptides was performed as previously described (11). Data were collected on a Q Exactive HF Orbitrap mass spectrometer (Thermo-Fisher Scientific) in Data Dependent Acquisition mode using m/z 350–1500 as MS scan range at 60 000 resolution, HCD MS/MS spectra were collected for the 10 most intense ions per MS scan at 15 000 resolution with a normalized collision energy of 28% and an isolation window of 1.4 m/z. Dynamic exclusion parameters were set as follows: exclude isotope on, duration 30 s and peptide match preferred. Other instrument parameters for the Orbitrap were MS maximum injection time 30 ms with AGC target 3 × 106, MSMS for a maximum injection time of 110 ms with AGT target of 1.1 × 104. The raw data were uploaded into Peaks Studio 10.5 (Bioinformatics Solution Inc., Waterloo, Canada) and processed with de novo peptide sequencing and Peaks DB using the ToxoDB (https://toxodb.org/toxo/) database together with common contaminants (cRAP). For peptides identification, the default settings were with precursor-ion and product-ion tolerances set to 10 ppm and 0.02 Da, respectively. Semispecific trypsin digest with a maximum of 1 missed cleavage was employed and peptides were searched with carbamidomethylation of cysteine set as fixed modification. To limit false-positive peptide identification, the false discovery rate (FDR) applied to peptide-spectrum match (PSM) was set to 1%, and at least 1 unique peptide per protein was used.
Immunofluorescence assays and microscopy
Immunofluorescence assays were performed as described previously [56]. Primary antibodies used were mouse anti-FLAG (1:500 dilution; Sigma clone M2), rat anti-HA (1:500 dilution; Sigma clone 3F10), and rabbit anti-Tom40 (1:2,000 dilution; [20]). Secondary antibodies used were goat anti-mouse Alexa Fluor 488 (1:500 dilution; Thermo Fisher Scientific, catalog number A-11029), goat anti-rat Alexa Fluor 488 (1:500 dilution; Thermo Fisher Scientific, catalog number A-11006), and goat anti-rabbit Alexa Fluor 546 (1:500 dilution; Thermo Fisher Scientific, catalog number A-11035). Images were acquired on a DeltaVision Elite deconvolution microscope (GE Healthcare) fitted with a 100X UPlanSApo oil immersion objective lens (NA 1.40). Images were deconvolved and adjusted for contrast and brightness using SoftWoRx Suite 2.0 software, and subsequently processed using Adobe Illustrator.
Seahorse XFe96 extracellular flux analysis
Experiments measuring the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of intact extracellular parasites were conducted as described previously [17,28]. We also developed experiments to assess the OCR in digitonin-permeabilized parasites utilizing specific ETC substrates. Briefly, parasites were harvested as for the XFe96 assays on intact parasites. Parasites were then washed once in base medium (Agilent Technologies), resuspended in base medium to 1.5 × 107 cells/mL and starved for 1 hour at 37°C to deplete endogenous ETC substrates. 1.5 × 106 parasites were added to wells of Cell-Tak-coated Seahorse XFe96 cell culture plates and adhered to the bottom by centrifugation (800 × g, 3 min). Base medium was removed and replaced with 175 μL mitochondrial assay solution (MAS) buffer (220 mM mannitol, 70 mM sucrose, 10 mM KH2PO4, 5 mM MgCl2, 0.2% w/v fatty acid-free bovine serum albumin, 1 mM EGTA, and 2 mM HEPES-KOH pH 7.4) containing 0.002% (w/v) digitonin. ETC substrates and inhibitors were loaded into Seahorse XFe96 sensor cartridge ports A-D, and injected into wells during the experiment. OCR measurements were obtained every 3 min for five repeats before and after injection of compounds (prepared in MAS buffer; concentrations given are final concentrations following injection). Port A: FCCP (1 μM) plus substrates. The substrates used were malate plus glutamate (10 mM each) or sn-glycerol 3-phosphate bis(cyclohexylammonium) salt (G3P; 25 mM). Port B: antimycin A and atovaquone (10 μM and 1 μM, respectively). Port C: N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride (TMPD; 0.2 mM) mixed with ascorbic acid (3.3 mM). Port D: sodium azide (NaN3; 10 mM). Substrate-elicited mOCR was calculated by subtracting the non-mitochondrial OCR (the values following antimycin A and atovaquone injection via Port B) from the OCR value obtained after substrate injection (Port A). Likewise, TMPD-elicited mOCR was calculated by subtracting the non-mitochondrial OCR from the OCR value obtained after TMPD injection (Port C). A minimum of 4 background wells were used in each plate, and 3 technical replicates were used for each condition.
Data analyses
Data from the Seahorse flux analysis were exported from the Seahorse Wave Desktop software (Agilent Technologies). A linear mixed effects model was applied to the data as described previously [17], setting the error between plates (between experiments) and wells (within experiments) as random effects, and the mOCR or ECAR values between cell lines and days on drug (ATc) as fixed effects. Analysis of the least square means of the values was performed in the R software environment. Statistical differences between these values were tested through ANOVA (linear mixed effects), with a post hoc Tukey test.
Data from the TgMPPα-TEV-HA and TgCox2a-TEV-HA mass spectrometry-based proteomic experiment were analyzed in the R software environment using the EdgeR package as described previously [17,58]. In the analysis performed to produce the volcano plot, only proteins identified in both data sets and each biological replicate were included, while subsequent analyses also considered proteins that were absent from one or more replicates of the TgCox2a-TEV-HA control.
Bioinformatic analyses
Amino acid sequences of T. gondii Complex III proteins were accessed from ToxoDB (www.toxodb.org). Initial identification of homologs in the apicomplexan parasites P. falciparum and C. parvum, and the chromerids V. brassicaformis and C. velia, was performed through Basic Local Alignment Search Tool (tBLASTn) searches of the EuPathDB Transcripts database (www.eupathdb.org., [59]). Where simple BLAST searches did not identify homologs, additional homology searches were performed using the iterative search tool JackHMMER (www.ebi.ac.uk/Tools/hmmer/search/jackhmmer) and the profile hidden Markov model based search tool HHPRED (https://toolkit.tuebingen.mpg.de/tools/hhpred, [25]). To identify homologs of TgQCR8 and TgQCR9 in yeast, humans, Arabidopsis and the parasitic dinoflagellate Perkinsus marinus, we performed JackHMMER and NCBI iterative PSI-BLAST [60] searches. We identified dinoflagellate homologs of TgQCR8, TgQCR9 and TgQCR11 in BLAST searches of Symbiodinium spp. genomes available at http://reefgenomics.org [61]. Transmembrane domain predictions were performed using TMHMM [62] and TMPred (https://embnet.vital-it.ch/software/TMPRED_form.html). Multiple protein sequence alignments of QCR8 (S3 Fig), QCR9 (S4 Fig), TgQCR11 (S5 Fig) and TgQCR12 (S6 Fig) were performed using Clustal Omega [63], and the graphical output was generated in BoxShade (https://embnet.vital-it.ch/software/BOX_form.html). All accession numbers are included in the supplementary figure legends.
Supporting information
(A) Diagram depicting the 3’ replacement strategy to generate TEV-HA-tagged TgMPPα. A sgRNA was designed to target the T. gondii genome near the stop codon of TgMPPα, and cause a double stranded break. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding a TEV-HA epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgMPPα stop codon. The homologous repair pathway of the parasite mediates integration of the PCR product into the TgMPPα locus. Forward and reverse primers were used to screen parasite clones for successful integration of the TEV-HA tag at the target site, yielding a 274 bp product in the native locus and a 414 bp product in the modified locus. (B) PCR screening analysis using genomic DNA extracted from putative TgMPPα-TEV-HA parasites (clones 1–6). Clones 1–4 and 6 yielded PCR products that indicated that these clones had been successfully modified. Genomic DNA extracted from wild type (WT) parasites was used as a control. (C) Diagram depicting the 3’ replacement strategy to generate TEV-HA-tagged TgCox2a. A sgRNA was designed to target near the stop codon of TgCox2a. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding a TEV-HA epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgCox2a stop codon. Forward and reverse primers were used to screen parasite clones for successful integration of the TEV-HA tag at the target site, yielding a 260 bp product in the native locus and a 400 bp product in the modified locus. (D) PCR screening analysis using genomic DNA extracted from putative TgCox2a-TEV-HA parasites (clones 1–6). All 6 clones yielded PCR products that indicated that these clones had been successfully modified. Genomic DNA extracted from wild type (WT) parasites was used as a control.
(TIF)
The yeast Complex III dimer structure (PDB: 3CX5) was imported into the EzMol program [64] and subunits were coloured as indicated, with one monomer of each subunit shown as surface display and the other as ribbons. No homolog of yeast QCR10 was detected in T. gondii (black), and no homologs of the T. gondii proteins TgQCR11 and TgQCR12 (purple) were found in yeast.
(TIF)
Alignment of QCR8 homologs from Toxoplasma gondii (TgQCR8; TGME49_214250), Plasmodium falciparum (PfQCR8; PF3D7_0306000), P. berghei (PbQCR8; PBANKA_0404400), Theileria equi (TeQCR8; BEWA_031210), Babesia bovis (BbQCR8; BBOV_IV004300), Vitrella brassicaformis (VbQCR8; Vbra_14054), Symbiodinium microadriaticum (SaQCR8; Smic7304), Arabidopsis thaliana (AtQCR8; NP_196156), Homo sapiens (HsQCR8; NP_055217) and Saccharomyces cerevisiae (ScQCR8; NP_012369). Dark shading indicates amino acid identity in ≥70% of the sequences, and light shading indicates amino acid similarity in ≥70% of the sequences. The positions of predicted transmembrane domains in TgQCR8 (TMPred prediction) and ScQCR8 (TMHMM prediction) are indicated by red boxes.
(TIF)
Alignment of QCR9 homologs from T. gondii (TgQCR9; TGME49_201880), P. falciparum (PfQCR9; PF3D7_0622600), P. berghei (PbQCR9; PBANKA_1121500), T. equi (TeQCR9; BEWA_007140), B. bovis (BbQCR9; BBOV_III007050), V. brassicaformis (VbQCR9; Vbra_943), Symbiodinium kawagutii (SkQCR9; Skav217368), Arabidopsis thaliana (AtQCR9; NP_190841), Homo sapiens (HsQCR9; NP_037519) and Saccharomyces cerevisiae (ScQCR9; NP_011699). Dark shading indicates amino acid identity in ≥70% of the sequences, and light shading indicates amino acid similarity in ≥70% of the sequences. The positions of predicted transmembrane domains in TgQCR9 (TMPred prediction) and ScQCR9 (TMHMM prediction) are indicated by red boxes.
(TIF)
Alignment of QCR11 homologs from T. gondii (TgQCR11; TGME49_214250), P. falciparum (PfQCR11; PF3D7_0722700), P. berghei (PbQCR11; PBANKA_0620200), B. bovis (BbQCR11; BBOV_IV004900), T. equi (TeQCR11; BEWA_032020), V. brassicaformis (VbQCR11; Vbra_12339), Perkinsum marinus (PmQCR11; XP_002780203), and S. kawagutii (SkQCR11; Skav223196). Dark shading indicates amino acid identity in >70% of the sequences, and light shading indicates amino acid similarity in >70% of the sequences. The position of predicted transmembrane domains in TgQCR11 (TMHMM prediction) is indicated by a red box.
(TIF)
Alignment of QCR12 homologs from T. gondii (TgQCR12; TGME49_207170), P. berghei (PbQCR12; PBANKA_1341100), P. falciparum (PfQCR12; PF3D7_1326000), T. equi (TeQCR12; BEWA_021660), and B. bovis (BbQCR12; BBOV_III005260). Dark shading indicates amino acid identity in ≥80% of the sequences, and light shading indicates amino acid similarity in ≥80% of the sequences. The position of predicted transmembrane domains in TgQCR12 (TMHMM prediction) is indicated by a red box.
(TIF)
Diagrams depict the 3’ replacement strategy to FLAG-tag target genes. sgRNAs were designed to target the T. gondii genome near the stop codon of target genes. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into TgMPPα-HA T. gondii parasites with a PCR product encoding a FLAG epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the stop codon. Genomic DNA extracted from wild type (WT) parasites was used as a control in PCRs. (A) Forward and reverse primers were used to screen parasite clones for integration of the FLAG tag at the TgQCR8 locus, yielding a 241 bp product in the native locus and a 338 bp product in the modified locus. (B) PCR screening using genomic DNA extracted from putative TgQCR8-FLAG parasites (clones 1–6). Clones 1, 4 and 6 yielded PCR products that indicated that these clones had been successfully modified. (C) Forward and reverse primers were used to screen parasite clones for integration of the FLAG tag at the TgQCR9 locus, yielding a 288 bp product in the native locus and a 378 bp product in the modified locus. (D) PCR screening using genomic DNA extracted from putative TgQCR9-FLAG parasites (clones 1–10). Clone 8 yielded a PCR product that indicated it had been successfully modified. (E) Forward and reverse primers were used to screen parasite clones for integration of the FLAG tag at the TgQCR11 locus, yielding a 1.3 kb product in the native locus and a 1.4 kb product in the modified locus. (F) PCR screening using genomic DNA extracted from putative TgQCR11-FLAG parasites (clones 1–5). Clone 4 yielded a PCR product that indicated it had been successfully modified. (G) Forward and reverse primers were used to screen parasite clones for integration of the FLAG tag at the TgQCR12 locus, yielding a 290 bp product in the native locus and a 398 bp product in the modified locus. (H) PCR screening using genomic DNA extracted from putative TgQCR12-FLAG parasites (clones 1–4). Clones 1–3 yielded PCR products that indicated that these clones had been successfully modified.
(TIF)
(A) Western blot of proteins extracted from TgMPPα-HA/TgQCR11-FLAG, TgMPPα-HA/TgQCR9-FLAG, TgMPPα-HA/TgQCR8-FLAG and TgMPPα-HA/TgQCR12-FLAG parasites, separated by SDS-PAGE, and detected with anti-FLAG and anti-Tom40 (loading control) antibodies. (B) Western blot of proteins extracted from TgMPPα-TEV-HA, TgMPPα-HA/TgQCR11-FLAG, TgMPPα-HA/TgQCR9-FLAG, TgMPPα-HA/TgQCR8-FLAG and TgMPPα-HA/TgQCR12-FLAG parasites, separated by BN-PAGE, and detected with anti-FLAG antibodies.
(TIF)
(A) Diagram depicting the 3’ replacement strategy to FLAG-tag TgQCR11. A sgRNA was designed to target the T. gondii genome near the stop codon of TgQCR11. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding a FLAG epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgQCR11 stop codon. Forward and reverse primers were used to screen parasite clones for integration of the FLAG tag at the TgQCR11 locus, yielding a 1.3 kb product in the native locus and a 1.4 kb product in the modified locus. (B) PCR screening using genomic DNA extracted from putative TgQCR11-FLAG parasites (clones 1–12). Clone 6 yielded a PCR product that indicated it had been successfully modified. Genomic DNA extracted from wild type (WT) parasites was used as a control. (C) Diagram depicting the promoter replacement strategy to generate ATc-regulated TgQCR11. A sgRNA was designed to target the T. gondii genome near the start codon of TgQCR11. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding the ATc regulated ‘t7s4’ promoter, which contains 7 copies of the Tet operon and a Sag4 minimal promoter, flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgQCR11 start codon. The PCR product also contain a ‘spacer’ region that separates the regulatable promoter from the native promoter of the TgQCR11 gene to enable sufficient regulation. Forward and reverse primers were used to screen parasite clones for successful integration of the regulatable promoter at the TgQCR11 locus, yielding a 1.4 kb product in the native, FLAG-tagged locus and a 3.3 kb product in the modified locus. (D) PCR screening using genomic DNA extracted from putative rTgQCR11-FLAG parasites (clones 1–9). Clones 1, 2, 4–7 yielded PCR products that indicated that these clones had been successfully modified. Genomic DNA extracted from wild type (WT) parasites was used as a control. (E) Diagram depicting the 3’ replacement strategy to HA-tag TgMPPα. A sgRNA was designed to target the T. gondii genome near the stop codon of TgMPPα. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into rTgQCR11-FLAG T. gondii parasites with a PCR product encoding a HA epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgMPPα stop codon. Forward and reverse primers were used to screen parasite clones for integration of the HA tag at the TgMPPα locus, yielding a 274 bp product in the native locus and a 387 bp product in the modified locus. (F) PCR screening using genomic DNA extracted from putative rTgQCR11/TgMPPα-HA parasites (clones 1–3). Clones 2 and 3 yielded PCR products that indicated that these clones had been successfully modified. Genomic DNA extracted from wild type (WT) parasites was used as a control.
(TIF)
(A) Immunofluorescence assays assessing mitochondrial morphology in rTgQCR11-FLAG/TgMPPα-HA parasites grown in the absence of ATc (top) or in the presence of ATc for 3 days (bottom). The outer mitochondrial membrane was labelled using antibodies against TgTom40 (red), and the inner mitochondrial membrane was labelled using anti-HA antibodies to detect TgMPPα-HA. Images are representative of 100 four-cell vacuoles examined in 2 independent experiments; scale bar represents 2 μm. (B) Basal extracellular acidification rate (ECAR) of WT parasites grown in the absence of ATc or in the presence of ATc for 3 days (blue), and rTgQCR11-FLAG/TgMPPα-HA parasites grown in the absence of ATc or in the presence of ATc for 1–3 days (orange). A linear mixed-effects model was fitted to the data and values depict the least squares mean ± 95% CI of three independent experiments. ANOVA followed by Tukey’s multiple pairwise comparisons test was performed, with relevant p values shown.
(TIF)
Diagrams depict the 3’ replacement strategy to TEV-HA-tag target genes. sgRNAs were designed to target the T. gondii genome near the stop codon of target genes. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into rTgQCR11-FLAG T. gondii parasites with a PCR product encoding a TEV-HA epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the stop codon. Genomic DNA extracted from wild type (WT) parasites was used as a control in PCRs. (A) Forward and reverse primers were used to screen parasite clones for integration of the TEV-HA tag at the TgQCR8 locus, yielding a 241 bp product in the native locus and a 377 bp product in the modified locus. (B) PCR screening using genomic DNA extracted from putative rTgQCR11-FLAG/TgQCR8-TEV-HA parasites (clones 1–12). Clone 9 yielded PCR products that indicated it had been successfully modified. (C) Forward and reverse primers were used to screen parasite clones for integration of the TEV-HA tag at the TgQCR12 locus, yielding a 290 bp product in the native locus and a 425 bp product in the modified locus. (D) PCR screening using genomic DNA extracted from putative rTgQCR11-FLAG/TgQCR12-TEV-HA parasites (clones 1–6). Clones 1–2 and 4–6 yielded PCR products that indicated that they had been successfully modified.
(TIF)
Western blots of proteins extracted from rTgQCR11-FLAG/TgCytC1-HA parasites, and subjected to immunoprecipitation using anti-HA (anti-HA IP) or anti-FLAG (anti-FLAG IP) antibody-coupled beads. Extracts include samples before immunoprecipitation (Total), samples that did not bind to the anti-HA or anti-FLAG beads (Unbound), and samples that bound to the anti-HA or anti-FLAG beads (Bound). Samples were separated by SDS-PAGE, and probed with anti-HA antibodies to detect TgCytC1-HA, anti-FLAG to detect TgQCR11, and anti-TgTom40 as a control to detect an unrelated mitochondrial protein.
(TIF)
(A-D) Quantification of BN-PAGE western blots from Fig 6. (A) rTgQCR11-FLAG/TgMPPα-HA, (B) rTgQCR11-FLAG/TgQCR12-TEV-HA, (C) rTgQCR11-FLAG/TgQCR8-TEV-HA and (D) rTgQCR11-FLAG/TgCytC1-HA parasites were grown in the absence of ATc or in the presence of ATc for 1–3 days. Band intensities were normalized to the matched SDS-PAGE TgTom40 control and expressed as a percent of the day zero control. Columns represent the mean ± SD of at least 2 independent experiments, with individual values depicted. (E-H) Quantification of SDS-PAGE western blots from Fig 6. (E) rTgQCR11-FLAG/TgMPPα-HA, (F) rTgQCR11-FLAG/TgQCR12-TEV-HA, (G) rTgQCR11-FLAG/TgQCR8-TEV-HA and (H) rTgQCR11-FLAG/TgCytC1-HA parasites were grown in the absence of ATc or in the presence of ATc for 1–3 days. Band intensities were normalized relative to the TgTom40 control and expressed as a percent of the day zero control. Columns represent the mean ± SD of at least 2 independent experiments, with individual values depicted. rTgQCR11-FLAG is shown in gray tones and HA-tagged proteins are shown in red tones.
(TIF)
Tab 1: List of proteins identified in the mass spectrometry-based proteomic analysis of proteins purified from the TgMPPα-TEV-HA and TgCox2a-TEV-HA immunoprecipitations. Included are the ToxoDB (www.toxodb.org) accession number, the name assigned to the protein in this manuscript if applicable, the total precursor intensity (area) for each biological replicate, the average total precursor intensity (area) of the three replicates, the fold change (average TgMPPα/average TgCox2a), and the predicted mass of the identified protein. Proteins are highlighted based on whether they were highly enriched in TgMPPα-TEV-HA (orange) or TgCox2a-TEV-HA (blue) pulldown. Tab 2: A list of the log fold change (logFC) and p values calculated for each protein identified in all replicates of the TgMPPα-TEV-HA and TgCox2a-TEV-HA immunoprecipitations following EdgeR analysis.
(XLSX)
(PDF)
Acknowledgments
We thank Harpreet Vohra and Michael Devoy (ANU) for assistance with flow cytometry, Michael Devoy for assistance with establishing the XFe96 assays, Teresa Neeman from the ANU Statistical Consulting Unit for assistance with data analysis, Chris Tonkin (WEHI) and Yi Xue (ANU) for sharing vectors, the ANU Toxo lab for comments on the manuscript, and the 2019 ANU Cell Biology course for contributing to the generation of parasite strains. We are grateful to EuPathDB for providing numerous datasets and search tools.
Data Availability
All relevant data are within the manuscript and its Supporting Information files with the exception of the mass spectrometry proteomics data. The mass spectrometry proteomics data have been deposited to the ProteomeXchange consortium via the Pride partner repository with the dataset identified PXD018781 and 10.6019/PXD018781.
Funding Statement
This work was supported by a Research School of Biology innovation grant to ER and GGvD, a National Health and Medical Research Council Ideas grant (GNT1182369) to GGvD, and an Australian Government Research Training Program Scholarship to JAH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang JL, Zhang NZ, Li TT, He JJ, Elsheikha HM, Zhu XQ. Advances in the Development of Anti-Toxoplasma gondii Vaccines: Challenges, Opportunities, and Perspectives. Trends Parasitol. 2019;35(3):239–53. Epub 2019/02/06. 10.1016/j.pt.2019.01.005 . [DOI] [PubMed] [Google Scholar]
- 2.King A. Building a better malaria vaccine. Nature. 2019;575(7784):S51–s4. Epub 2019/11/30. 10.1038/d41586-019-03639-5 . [DOI] [PubMed] [Google Scholar]
- 3.Goodman CD, Buchanan HD, McFadden GI. Is the Mitochondrion a Good Malaria Drug Target? Trends Parasitol. 2017;33(3):185–93. Epub 2016/10/30. 10.1016/j.pt.2016.10.002 . [DOI] [PubMed] [Google Scholar]
- 4.Mather MW, Henry KW, Vaidya AB. Mitochondrial drug targets in apicomplexan parasites. Curr Drug Targets. 2007;8(1):49–60. Epub 2007/02/03. 10.2174/138945007779315632 . [DOI] [PubMed] [Google Scholar]
- 5.Alday PH, Doggett JS. Drugs in development for toxoplasmosis: advances, challenges, and current status. Drug Des Devel Ther. 2017;11:273–93. Epub 2017/02/10. 10.2147/DDDT.S60973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doggett JS, Nilsen A, Forquer I, Wegmann KW, Jones-Brando L, Yolken RH, et al. Endochin-like quinolones are highly efficacious against acute and latent experimental toxoplasmosis. Proc Natl Acad Sci U S A. 2012;109(39):15936–41. 10.1073/pnas.1208069109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999;33(4):704–11. 10.1046/j.1365-2958.1999.01515.x . [DOI] [PubMed] [Google Scholar]
- 8.Mitchell P. The protonmotive Q cycle: a general formulation. FEBS Lett. 1975;59(2):137–9. Epub 1975/11/15. 10.1016/0014-5793(75)80359-0 . [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115(4):1936–89. Epub 2015/01/21. 10.1021/cr500266a . [DOI] [PubMed] [Google Scholar]
- 10.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272(5265):1136–44. 10.1126/science.272.5265.1136 . [DOI] [PubMed] [Google Scholar]
- 11.Santo-Domingo J, Demaurex N. The renaissance of mitochondrial pH. Journal of General Physiology. 2012;139(6):415–23. 10.1085/jgp.201110767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–8. Epub 1961/07/08. 10.1038/191144a0 . [DOI] [PubMed] [Google Scholar]
- 13.Hayward JA, van Dooren GG. Same same, but different: Uncovering unique features of the mitochondrial respiratory chain of apicomplexans. Mol Biochem Parasitol. 2019;232:111204 Epub 2019/08/06. 10.1016/j.molbiopara.2019.111204 . [DOI] [PubMed] [Google Scholar]
- 14.Flegontov P, Michalek J, Janouskovec J, Lai DH, Jirku M, Hajduskova E, et al. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol Biol Evol. 2015;32(5):1115–31. 10.1093/molbev/msv021 . [DOI] [PubMed] [Google Scholar]
- 15.Barylyuk K, Koreny L, Ke H, Butterworth S, Crook OM, Lassadi I, et al. A comprehensive subcellular atlas of the Toxoplasma proteome via hyperLOPIT provides spatial context for protein functions. Cell Host Microbe. 2020;28(5):752–766.e9. Epub 2020/10/13. 10.1016/j.chom.2020.09.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salunke R, Mourier T, Banerjee M, Pain A, Shanmugam D. Highly diverged novel subunit composition of apicomplexan F-type ATP synthase identified from Toxoplasma gondii. PLoS Biol. 2018;16(7):e2006128 Epub 2018/07/14. 10.1371/journal.pbio.2006128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidi A, Muellner-Wong LS, Rajendran E, Tjhin ET, Dagley LF, Aw VY, et al. Elucidating the mitochondrial proteome of Toxoplasma gondii reveals the presence of a divergent cytochrome c oxidase. Elife. 2018;7:e38131 Epub 2018/09/12. 10.7554/eLife.38131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huet D, Rajendran E, van Dooren GG, Lourido S. Identification of cryptic subunits from an apicomplexan ATP synthase. eLife. 2018;7:e38097 Epub 2018/09/12. 10.7554/eLife.38097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roger AJ, Munoz-Gomez SA, Kamikawa R. The Origin and Diversification of Mitochondria. Curr Biol. 2017;27(21):R1177–R92. Epub 2017/11/08. 10.1016/j.cub.2017.09.015 . [DOI] [PubMed] [Google Scholar]
- 20.van Dooren GG, Yeoh LM, Striepen B, McFadden GI. The Import of Proteins into the Mitochondrion of Toxoplasma gondii. J Biol Chem. 2016;291(37):19335–50. 10.1074/jbc.M116.725069 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser E, Eriksson A, Sjoling S. Bifunctional role of the bc1 complex in plants. Mitochondrial bc1 complex catalyses both electron transport and protein processing. FEBS Lett. 1994;346(1):83–7. Epub 1994/06/06. 10.1016/0014-5793(94)00312-2 . [DOI] [PubMed] [Google Scholar]
- 22.McFadden DC, Tomavo S, Berry EA, Boothroyd JC. Characterization of cytochrome b from Toxoplasma gondii and Q(o) domain mutations as a mechanism of atovaquone-resistance. Mol Biochem Parasitol. 2000;108(1):1–12. Epub 2000/05/10. 10.1016/s0166-6851(00)00184-5 . [DOI] [PubMed] [Google Scholar]
- 23.Ossorio PN, Sibley LD, Boothroyd JC. Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome of Toxoplasma gondii. J Mol Biol. 1991;222(3):525–36. Epub 1991/12/05. 10.1016/0022-2836(91)90494-q . [DOI] [PubMed] [Google Scholar]
- 24.Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: a story of gains, losses and retentions. Trends Parasitol. 2008;24(10):468–78. 10.1016/j.pt.2008.07.004 . [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430(15):2237–43. Epub 2017/12/21. 10.1016/j.jmb.2017.12.007 . [DOI] [PubMed] [Google Scholar]
- 26.Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, et al. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell. 2016;166(6):1423–35.e12. Epub 2016/09/07. 10.1016/j.cell.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfefferkorn ER, Borotz SE, Nothnagel RF. Mutants of Toxoplasma gondii resistant to atovaquone (566C80) or decoquinate. J Parasitol. 1993;79(4):559–64. Epub 1993/08/01. . [PubMed] [Google Scholar]
- 28.Tjhin ET, Hayward JA, McFadden GI, van Dooren GG. Characterization of the apicoplast-localized enzyme TgUroD in Toxoplasma gondii reveals a key role of the apicoplast in heme biosynthesis. J Biol Chem. 2020;295(6):1539–50. Epub 2020/01/09. 10.1074/jbc.RA119.011605 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salabei JK, Gibb AA, Hill BG. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat Protoc. 2014;9(2):421–38. 10.1038/nprot.2014.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vercesi AE, Rodrigues CO, Uyemura SA, Zhong L, Moreno SN. Respiration and oxidative phosphorylation in the apicomplexan parasite Toxoplasma gondii. J Biol Chem. 1998;273(47):31040–7. 10.1074/jbc.273.47.31040 . [DOI] [PubMed] [Google Scholar]
- 31.MacLean AEB H. R.; Silva M. F.; Ding S.; Hirst J.; Sheiner L. Complexome profile of Toxoplasma gondii mitochondria identifies a divergent cytochrome bc1 complex. BioRxiv preprint. 2020. 10.1101/2020.08.17.252163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281(5373):64–71. Epub 1998/07/04. 10.1126/science.281.5373.64 . [DOI] [PubMed] [Google Scholar]
- 33.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277(5322):60–6. Epub 1997/07/04. 10.1126/science.277.5322.60 . [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392(6677):677–84. Epub 1998/05/16. 10.1038/33612 . [DOI] [PubMed] [Google Scholar]
- 35.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8(6):669–84. Epub 2000/06/30. 10.1016/s0969-2126(00)00152-0 . [DOI] [PubMed] [Google Scholar]
- 36.Meyer EH, Taylor NL, Millar AH. Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J Proteome Res. 2008;7(2):786–94. Epub 2008/01/15. 10.1021/pr700595p . [DOI] [PubMed] [Google Scholar]
- 37.Zara V, Conte L, Trumpower BL. Identification and characterization of cytochrome bc(1) subcomplexes in mitochondria from yeast with single and double deletions of genes encoding cytochrome bc(1) subunits. Febs j. 2007;274(17):4526–39. Epub 2007/08/08. 10.1111/j.1742-4658.2007.05982.x . [DOI] [PubMed] [Google Scholar]
- 38.Xia D, Esser L, Tang WK, Zhou F, Zhou Y, Yu L, et al. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. Biochim Biophys Acta. 2013;1827(11–12):1278–94. Epub 2012/12/04. 10.1016/j.bbabio.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danne JC, Gornik SG, Macrae JI, McConville MJ, Waller RF. Alveolate mitochondrial metabolic evolution: dinoflagellates force reassessment of the role of parasitism as a driver of change in apicomplexans. Mol Biol Evol. 2013;30(1):123–39. 10.1093/molbev/mss205 . [DOI] [PubMed] [Google Scholar]
- 40.Jacot D, Waller RF, Soldati-Favre D, MacPherson DA, MacRae JI. Apicomplexan Energy Metabolism: Carbon Source Promiscuity and the Quiescence Hyperbole. Trends Parasitol. 2016;32(1):56–70. Epub 2015/10/17. 10.1016/j.pt.2015.09.001 . [DOI] [PubMed] [Google Scholar]
- 41.Zara V, Conte L, Trumpower BL. Biogenesis of the yeast cytochrome bc1 complex. Biochim Biophys Acta. 2009;1793(1):89–96. Epub 2008/05/27. 10.1016/j.bbamcr.2008.04.011 . [DOI] [PubMed] [Google Scholar]
- 42.Conte A, Papa B, Ferramosca A, Zara V. The dimerization of the yeast cytochrome bc1 complex is an early event and is independent of Rip1. Biochim Biophys Acta. 2015;1853(5):987–95. Epub 2015/02/17. 10.1016/j.bbamcr.2015.02.006 . [DOI] [PubMed] [Google Scholar]
- 43.Phillips JD, Graham LA, Trumpower BL. Subunit 9 of the Saccharomyces cerevisiae cytochrome bc1 complex is required for insertion of EPR-detectable iron-sulfur cluster into the Rieske iron-sulfur protein. J Biol Chem. 1993;268(16):11727–36. Epub 1993/06/05. . [PubMed] [Google Scholar]
- 44.Phillips JD, Schmitt ME, Brown TA, Beckmann JD, Trumpower BL. Isolation and characterization of QCR9, a nuclear gene encoding the 7.3-kDa subunit 9 of the Saccharomyces cerevisiae ubiquinol-cytochrome c oxidoreductase complex. An intron-containing gene with a conserved sequence occurring in the intron of COX4. J Biol Chem. 1990;265(34):20813–21. Epub 1990/12/05. . [PubMed] [Google Scholar]
- 45.Brandt U, Uribe S, Schägger H, Trumpower BL. Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J Biol Chem. 1994;269(17):12947–53. Epub 1994/04/29. . [PubMed] [Google Scholar]
- 46.Hartley AM, Lukoyanova N, Zhang Y, Cabrera-Orefice A, Arnold S, Meunier B, et al. Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1. Nat Struct Mol Biol. 2019;26(1):78–83. Epub 2019/01/02. 10.1038/s41594-018-0172-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathore S, Berndtsson J, Marin-Buera L, Conrad J, Carroni M, Brzezinski P, et al. Cryo-EM structure of the yeast respiratory supercomplex. Nat Struct Mol Biol. 2019;26(1):50–7. Epub 2019/01/02. 10.1038/s41594-018-0169-7 . [DOI] [PubMed] [Google Scholar]
- 48.Pearson PA, Piracha AR, Sen HA, Jaffe GJ. Atovaquone for the treatment of toxoplasma retinochoroiditis in immunocompetent patients. Ophthalmology. 1999;106(1):148–53. Epub 1999/01/26. 10.1016/S0161-6420(99)90021-0 . [DOI] [PubMed] [Google Scholar]
- 49.Katlama C, Mouthon B, Gourdon D, Lapierre D, Rousseau F. Atovaquone as long-term suppressive therapy for toxoplasmic encephalitis in patients with AIDS and multiple drug intolerance. Atovaquone Expanded Access Group. Aids. 1996;10(10):1107–12. Epub 1996/09/01. . [PubMed] [Google Scholar]
- 50.Winterhalter S, Severing K, Stammen J, Maier AK, Godehardt E, Joussen AM. Does atovaquone prolong the disease-free interval of toxoplasmic retinochoroiditis? Graefes Arch Clin Exp Ophthalmol. 2010;248(8):1187–92. Epub 2010/05/04. 10.1007/s00417-010-1379-9 . [DOI] [PubMed] [Google Scholar]
- 51.Jacot DL S.; Markus M.; Sheiner L.; Soldati-Favre D.; Striepen B. Genetic manipulation of Toxoplasma gondii. In: Weiss LMK, K., editor. Toxoplasma gondii the model apicomplexan—perspectives and methods Third edition ed London: Academic Press; 2020. p. 897–934. [Google Scholar]
- 52.Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, et al. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7(12):e1002392 10.1371/journal.ppat.1002392 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen B, Brown KM, Lee TD, Sibley LD. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio. 2014;5(3):e01114–14. 10.1128/mBio.01114-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katris NJ, van Dooren GG, McMillan PJ, Hanssen E, Tilley L, Waller RF. The apical complex provides a regulated gateway for secretion of invasion factors in Toxoplasma. PLoS Pathog. 2014;10(4):e1004074 10.1371/journal.ppat.1004074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajendran E, Hapuarachchi SV, Miller CM, Fairweather SJ, Cai Y, Smith NC, et al. Cationic amino acid transporters play key roles in the survival and transmission of apicomplexan parasites. Nat Commun. 2017;8:14455 10.1038/ncomms14455 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci U S A. 2008;105(36):13574–9. Epub 2008/09/02. 10.1073/pnas.0803862105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pankow S, Bamberger C, Calzolari D, Bamberger A, Yates JR, 3rd. Deep interactome profiling of membrane proteins by co-interacting protein identification technology. Nat Protoc. 2016;11(12):2515–28. 10.1038/nprot.2016.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–32. 10.1093/biostatistics/kxm030 . [DOI] [PubMed] [Google Scholar]
- 59.Aurrecoechea C, Barreto A, Basenko EY, Brestelli J, Brunk BP, Cade S, et al. EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017;45(D1):D581–d91. Epub 2016/12/03. 10.1093/nar/gkw1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. 10.1093/nar/25.17.3389 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liew YJ, Aranda M, Voolstra CR. Reefgenomics.Org—a repository for marine genomics data. Database (Oxford). 2016;2016 Epub 2016/12/28. 10.1093/database/baw152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. Epub 2001/01/12. 10.1006/jmbi.2000.4315 . [DOI] [PubMed] [Google Scholar]
- 63.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–45. Epub 2017/09/09. 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reynolds CR, Islam SA, Sternberg MJE. EzMol: A Web Server Wizard for the Rapid Visualization and Image Production of Protein and Nucleic Acid Structures. J Mol Biol. 2018;430(15):2244–8. Epub 2018/02/03. 10.1016/j.jmb.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]