Abstract
Monocytes are progenitors to macrophages and a sub-class of dendritic cells (MoDCs), but they also act as circulating sensors responding to environmental changes and disease. Technological advances have defined the production of classical monocytes in the bone marrow through identification of lineage determining transcription factors and proposed alternative routes of differentiation. Monocytes released into the circulation can be recruited to tissues by specific chemoattractants, responding to sequential niche-specific signals that determine their differentiation into terminal effector cells. Of interest, new aspects of monocyte biology in circulation are being revealed, exemplified here by discussing the influence of cancer on the systemic alteration of monocyte subset abundance and transcriptional profiles. These changes can act to enhance the metastatic spread of primary cancers and may offer therapeutic opportunities.
Monocytes: key players in the innate immune system.
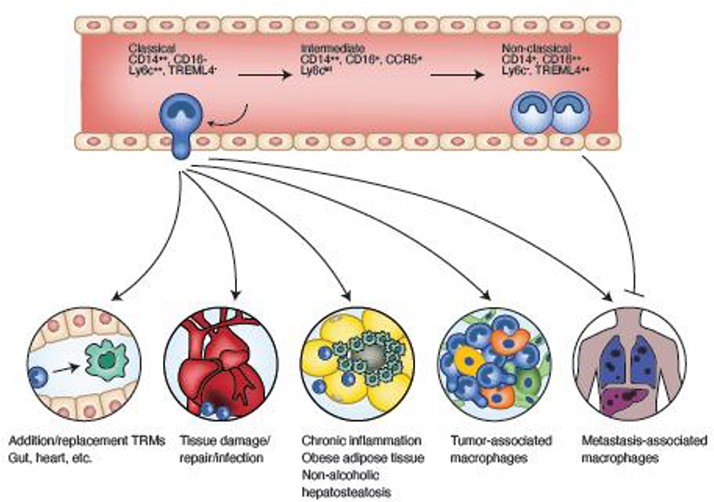
There are two major populations of monocytes in mouse and humans that have been termed classical and non-classical-- defined by specific markers (Table 1) as well as by an intermediate transitional population between classical and non-classical ones. During homeostasis, monocytes survey the vasculature or are recruited to continuously replenish tissue-resident macrophages (TEMs), for example in the gut. In response to inflammation, infection, or tissue damage, immune cell recruitment results in the development of effector macrophages repelling pathogens and mediating tissue repair, (Figure 1, Table 1) as well as a sub-class of dendritic cells (MoDCs) [1]. Traditionally, monocytes have been primarily viewed as “conduits” to processes involving macrophages or DCs, and as being beneficial to the mammalian host. However, the frequency, activity, and fate of monocyte populations has also been linked to many diseases, including autoimmunity, chronic inflammation, cardiovascular diseases, and cancer [2,3]. In these diseases, and others involving monocyte trafficking, environmental cues acting in the bone, blood, and tissues, dynamically modify the transcriptional landscape of monocytes, resulting in phenotypes that can potentiate disease severity (Figure 1, Table 1). Using the processes of monocyte development, here we discuss mechanisms of monocyte ontogeny, transcriptional regulation, and the relevance to tissue homeostasis. We also use cancer as an example of a disease state that can influence monocyte biology. We posit that a better understanding of this regulation is important, as it might contribute to advancing monocyte targeted therapies in cancer and in turn, be potentially applicable to other progressive diseases.
Key Table, Table 1.
Classical and Non-Classical Monocytes in Humans and Mice
| “Classical” “Inflammatory” | “Non-Classical” “Patrolling” | |
|---|---|---|
| Human surface antigen | CD14++ CD16− CCR2+ CSF1R | CD14+ CD16+ CSF1R |
| Mouse surface antigen | Ly6c++ Treml4− CCR2+ CSF1R | Ly6c− Treml4++ CSF1R |
| Recruitment | Recruited from blood through CCR2 release. | Migrate in response to CX3CL1 |
| Transcription factors | PU1, JUN, FOS, IRF8, KLF4 | PU1, NR4A1, KLF2 |
| Genes up-regulated | VCAN, CD163, CD63 S100A12, S100A8, | FCGR3A, IFITM1–3, CDKN1C, MTSS1 |
| Pathways up-regulated | Immune response, defense response, inflammatory response, chemotaxis, TLR, lysosome | Immune system process, leucocyte migration, cytoskeleton rearrangement |
| Metabolic profile | Carbohydrate Nucleotide production ROS production | Oxidative phosphorylation |
| Cytokines | TNFα, IL-1β, IL-6 |
Figure 1. Monocyte differentiation in blood and tissues.
Classical monocytes differentiate into non-classical monocytes through an intermediary stage. Markers are shown for human (top) and mouse (bottom). Classical monocytes extravasate into tissues to replace resident macrophages, participate in wound responses, contribute to pathogenic tissue inflammation, and promote tumor progression and metastasis. Non-classical monocytes are retained in vessels where they perform surveillance and monitor pathogenic insults. They can also extravasate at sites of metastasis and inhibit tumor cell establishment through a NK cell mediate mechanism.
Mammalian Monocyte Ontogeny
Monocyte development in the Bone
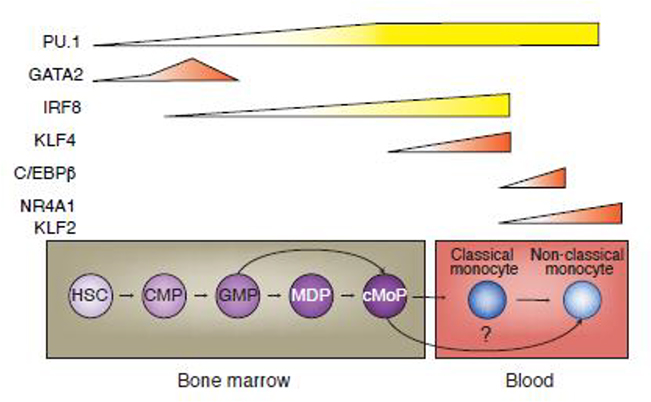
Postnatal monocytes are formed primarily in the bone marrow (BM) from the differentiation of hematopoietic stem cells (HSCs) into the common myeloid progenitor (CMP) cells, and subsequently, into granulocytic myeloid progenitor (GMP) cells; these in turn become bipotent macrophage dendritic cell progenitors (MDP), finally leading to the unipotent common monocyte progenitor (cMoP) stage (Figure 2) [4–6]. Though originally identified in mouse, these progenitors are also found in humans and are transcriptionally similar between both species [7,8]. Two recent studies demonstrated that cMoPs can directly arise from GMPs; thus, two developmental pathways – one excluding MDPs– might exist (Figure 2) [9,10]. The development of monocytes in the BM via their progenitors requires de novo gain of lineage determining transcription factors (LDTFs). The primary LDTF in monocytes is PU.1 (SPI1) whose expression increases throughout the development of monocyte progenitors [11]. The combinatorial effect of PU.1 upregulation, in addition to the collaborative binding and repression of other key LDTFs drive the progenitor fate towards a monocytic lineage (Figure 2) [11–19].
Figure 2. Differentiation of circulating monocytes from bone marrow progenitor cells.
Differentiation from early progenitors through progressively restricted ones that are then released into the circulation as classical monocytes (mice and humans) is regulated by the sequential expression of transcription factors (shown at the top). Alternative pathways to non-classical monocytes have also been proposed as indicated by the arrow. HSCs; hematopoietic stem cells, CMP; common myeloid progenitor cells, GMP; granulocytic myeloid progenitor cells, MDP; bipotent macrophage dendritic cell progenitor, cMoP; unipotent common monocyte progenitor.
Early in lineage commitment, the collaborative binding of PU.1 with C/EBPα is essential for HSC commitment to, and subsequent formation of CMPs [12]. Increasing amounts of PU.1 then steer CMP transition towards the GMP. This is accompanied by a large decrease in Gata2 gene expression, which is inhibited by PU.1 at the GMP stage [11,13]. The accessibility of these respective transcription factor (TF) binding motifs are similarly changed [14,15]. To ensure CMP progression into MDPs, high PU.1 amounts potentiate Interferon regulatory factor 8 (IRF8) TF expression, and collaborative binding of these two TFs temporarily blocks the binding of CCAAT - enhancer binding protein α (C/EBPα), ensuring monocyte/DC fate by inhibiting transit into the neutrophil lineage [16,17]. It is argued that it is at the CMP stage that commitment to a monocyte fate is determined via this combination of TFs [11,18]. MDPs then split into the DC progenitor cells or cMoPs. The amounts of IRF8 continue to rise during the transition of cMoPs into Ly6chi monocytes wherein PU.1 and IRF8 collaboratively bind to the Kruppel-like factor-4 (KLF4) enhancer that direct leads to Klf4 expression [19]. KLF4 is an essential factor for monocyte development as deletion of mouse Klf4 leads to the absence of Ly6chi monocytes, phenocopying Irf8−/− mice [19]. Noteworthy, monocyte differentiation in Irf8−/− mice can be partially rescued by KLF4 overexpression [20]. While much of this work has been conducted in mouse models, the recent use of CRISPR-Cas9 to modify gene expression in human induced pluripotent stem cells (iPS) offers a new mode to study TFs in monocyte development. For example, IRF8−/− iPS cells show no defect in developing into hematopoietic progenitor cells, but exhibit reduced capacity to generate monocytes, indicating that IRF8 is an important TF in human monocyte development [21]. However, a caveat of these studies is that human iPS-derived macrophages are similar to yolk sac derived macrophages in that they do not require TF C-MYB, typically reported to be essential for HSC development [22]. Thus, these iPS derived “monocytes” may in fact be equivalent to tissue-resident progenitors or perhaps, fetal liver monocytes [23].
Monocyte fate and transcriptional regulation in the blood
Ly6chi monocytes produced in the BM are subsequently released into circulation (Figure 2). Seminal work revealed the existence of two populations of human monocytes found in peripheral blood [24]: CD14Hi CD16− (classical) and CD14Lo CD16+ (nonclassical) populations (other markers shown in Table 1), at approximately, a 9 to 1 ratio [25]. Likewise, the presence of two circulating monocyte populations with distinct functions was confirmed in mice, although in this case, the populations were approximately equal in frequency and the high protein expression of TRM-like transcript protein-4, TLT (TREML4) was assigned as a robust marker for the non-classical population [1,25,26]. Microarray analysis and recent single cell sequencing suggest that these mouse monocyte populations, Ly6chi and Ly6clo, correspond to the human classical and nonclassical populations, respectively [27,28]. There is a third subset that on fluorescence activated cell sorting (FACS) plots, lies as an intermediary between classical and non-classical monocytes through intermediate CD14 and CD16 expression in humans and Ly6c and CCR2 in mice [29,30]. This diffuse population causes problems of gating on FACS that explain some of the functional discrepancies that have been noted in the two major subsets between mouse and human [31]. Tracking experiments of human BM monocytes –into the blood– that were labeled with a pulse of deuterium-labeled glucose to indicate A synthesis have shown that the non-classical population is derived from classical monocytes via this transitional intermediate population, replicating findings in mice that used BrdU incorporation into proliferating monocytes in the BM [32,33] (Figure 1). During homeostasis, the two established Ly6c+ and Ly6c− populations have coherent transcriptomes, while the intermediate population shows greater transcriptional diversity; this suggests that there may be various stages of differentiation in this transient population, although this remains to be robustly assessed [34]. However, in other studies, transcriptional diversity has also been reported within the Ly6C+ population, suggesting pre-determination of alternative fates in response to external stimuli, as discussed below [35].
PU.1 motifs and protein expression are increased across all monocyte subpopulations in mice but the accessibility of binding motifs for JUN (AP1) and KLF TFs are enriched in classical and non-classical monocytes, respectively [36]. The expression of the corresponding TFs that recognize these motifs correlates with monocyte identity; thus, FOS (binding to AP1 motif) and KLF2 (binds KLF motif) TFs are highly expressed in classical and non-classical monocytes, respectively [36,37]. This transition also requires C/EBPβ binding to the Nuclear receptor subfamily 4 group A member 1 (Nr4a1) promoter, stimulating the expression of this orphan nuclear receptor [34]. Moreover, Nr4a1 has been shown to be essential for mouse non-classical monocyte development, acting as a survival factor [38]. The specificity and interaction between these two TFs in non-classical monocytes is exemplified by the preferential loss of non-classical monocytes in the periphery and BM in mice in which Nr4a1 was genetically ablated (Nr4a1−/−) [38]. KLF2 binding to the Nr4a1 super-enhancer region E2 was also responsible for inducing Nr4a1 expression in mice [37]. Of note, deletion of the E2 region resulted in decreased Nr4a1 expression in classical and non-classical monocytes, but not in tissue-resident macrophages in which the gene was also highly expressed at steady state; this result is interesting as it might suggest that the E2 super enhancer is monocyte-specific, although this remains to be assessed [37]. Further, human monocytes showed histone acetylation marks at the E2 region –an indication of enhancer activation– suggesting that the transcriptional regulation of N4ra1/NR4A1 might be conserved between mouse and human [37]. However, the transition between classical and non-classical monocytes necessitating NR4A1 may be more complex. In mice, Muramyl dipeptide (MDP), a component of bacterial peptidoglycan, is a NOD2 receptor agonist and based on kinetic evaluation experiments, is thought to prompt the transition of monocytes from classical to non-classical monocytes. Specifically, intravenous administration of MDP into wildtype (WT) and Nr4a1−/− mice increased the total number of non-classical monocytes in blood, although to a much lesser degree in the latter [39]. Moreover, comparison of a variety of surface markers after MDP treatment showed that the their expression in newly arisen Ly6clo monocytes in Nr4a1−/− mice were similar to those of WT Ly6clo monocytes, with the exception of C-X-C 3 motif chemokine receptor 1 (Cx3cr1) and C-C-chemokine receptor type 2 (Ccr2) expression [39]. Collectively, these data suggest that there may be a potential alternative mechanism, other than Nr4a1, that might elicit the transition of classical monocytes to non-classical monocytes, at least in mice, and in the context of bacterial (or bacteria-simulated) infection.
In homeostatic conditions, classical and non-classical monocytes both contribute to maintaining tissue integrity. Non-classical monocytes patrol the vasculature, clearing dying endothelial cells, and protecting vessel health [40]. These cells do not appear to extravasate and thus, might be called endothelial macrophages. In the liver and spleen of mice, they occupy vascular niches that locally convert classical to non-classical monocytes via Notch signaling, upregulating expression of the TFs Nr4a1 and POU domain class 2 (Pou2f2), and significantly lowering Schlafen family member 5 (Slfn5) expression, at least in culture [41]. In disease states, non-classical monocytes can function to prevent tumor metastasis [42] (Figure 1) or to protect endothelial cells from apoptosis in atherosclerosis [43], either intrinsically, or indirectly via recruitment and crosstalk with other cell types. Alternatively, classical monocytes can migrate out of the vasculature to replenish tissue resident macrophages (TRM) in certain tissues, and to act as inflammatory mediators during acute and chronic inflammation, for example, in ischemic damage to the kidney or in non-alcoholic hepatosteatosis (Figure 1). Indeed, throughout their journey, classical monocytes encounter a variety of signals and cell types which result in dynamic changes to their transcriptomic and genomic architecture (Figure 2, 3).
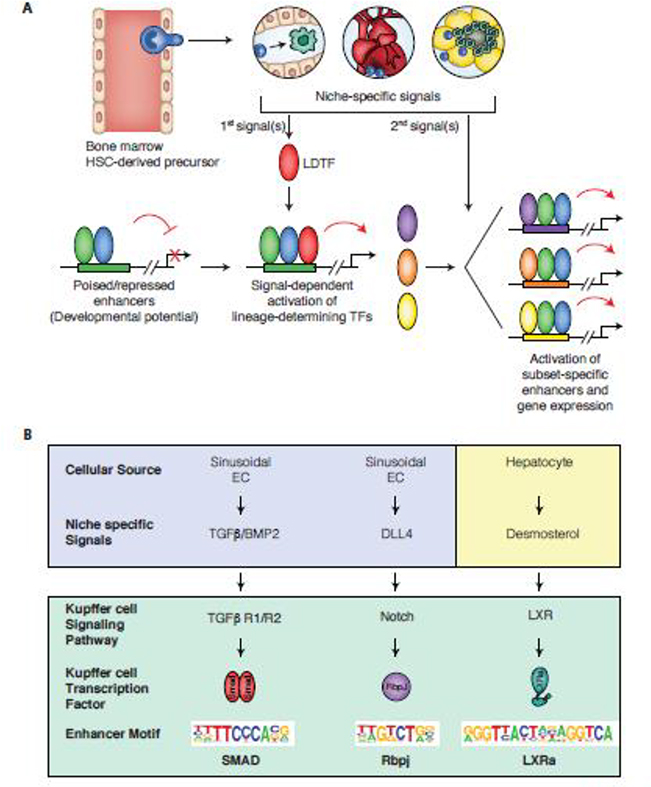
Figure 3. Molecular mechanisms by which niche-specific signals promote monocyte differentiation into different macrophage phenotypes (mice and humans).
Panel A. Niche specific signals induce the expression of lineage determining factors (LDTFs) that collaborate with core macrophage LDTFs to drive the selection of new enhancers. Pre-existing and new enhancers are further influenced by the actions of broadly expressed signal-dependent transcription factors that mediate the effects of internal and external signals.
Panel B. Sequence motifs enriched in Kupffer cell specific enhancers can enable inference of the transcription factors and upstream signaling pathways that drive differentiation from progenitor cells. For each motif, corresponding transcription factors, upstream signaling pathways, and sources of liver-specific signals are indicated.
Transcriptional imprinting of monocytes in homeostasis
TRMs are largely derived from embryonic sources and are capable of long-term self-renewal. However, in several tissues, including the gut [44], skin [45], and heart [46], in some cases, classical monocytes can contribute to a portion of the steady pool of TRMs, completely replacing them [44]. Thus, TRMs in many tissues but not all, e.g. the brain macrophages, microglia [47,48], are the product from two embryonic sources. In this context, transcriptional imprinting can drive the tissue phenotype. In mice, detailed dissection of the gut has revealed that TGFβR signaling in recruited classical monocytes is indispensable for the terminal differentiation of monocytes to intestinal macrophages, inducing the expression of several intestinal macrophage-specific genes [49]. Of note, gut macrophages are also characterized by high expression of TF Runx3, a mediator of TGFβ signaling, as compared to other TRMs [50]. Other reported factors that drive the gut macrophage phenotype include IL-10 [51,52], aryl hydrocarbon receptor ligands [53], microbiota byproducts [54], as well as a state of hypo-responsiveness to foreign ligands.
Numerous tracing and genetic ablation studies in mice have solidified the notion that classical monocytes replace existing TRMs to various extents in a tissue-specific manner [48,55]. Yet, little is known about the molecular mechanism orchestrating monocyte acquisition of the resulting tissue-specific phenotypes, potentially due to technical difficulties in isolating cells, and acquiring sufficient cell numbers for analysis, particularly in a homeostatic setting. Although artificial, niche depletion studies offer a modality to understand how monocytes acquire the molecular properties of TRMs under non-inflammatory conditions. For example, in a mouse model in which liver Kupffer cells (KC) were ablated, circulating Ly6chi monocytes rapidly colonized the liver and differentiated into KC that were capable of self-renewing [56]. Recently, two novel studies detailed certain specific molecular processes underlying the conversion of monocytes to KCs. By undertaking detailed kinetic analyses of the changes in monocyte transcriptional signatures as these differentiated into KCs and comparing these with the transcriptomes of hepatocytes, Liver Sinusoidal Endothelial Cells (LSECs), and pericytes over the same time period, the authors predicted likely cellular interactions that might imprint KC identity in mice [57] (Fig 3). By comparing changes in transcriptional signatures in these cell types in conjunction with analysis of enhancer landscapes, another study arrived at the same conclusion [58]. In addition, using the transcriptomes of different liver cell types, the study revealed that Notch ligand DLL4 and TGFβ/BMP family members expressed in liver sinusoidal endothelial cells LSECs sequentially drove the election and function of enhancers regulating KC-specific gene expression (Figure 3B). Further, hepatic stellate cells also expressed the monocyte and macrophage growth factor, Colony stimulating factor 1 (CSF1);this is relevant as it might serve as a niche signal that is important for ensuring the maintenance of KC identity, although this remains to be further tested [57]. Moreover, upon recruitment to the mouse liver, monocytes encountered Delta-Like 4 (DLL4), expressed on LSECs, and DLL4-mediated activation of Notch signaling converted recombination signal binding protein for immunoglobulin kappa J region (RBPJ) from a transcriptional repressor to an activator, thus activating pre-existing but poised enhancers in monocytes, and rapidly inducing the KC LDTF, Nr1h3 (encoding LXRα) [50,59]. Furthermore, LXR activity, also induced by desmosterol, allowed the selection of new enhancers comprising the repertoire of mature KC; and TGFβ/BMP-SMAD signaling also promoted and/or maintained KC-specific enhancer and gene expression [50,59]. While the liver is closed to monocyte colonization under such homeostatic conditions, collectively, these studies demonstrate how transcriptomic and epigenetic-led hypothesis generation can be used to predict key molecular pathways coordinating monocyte-to-TRM differentiation. Importantly this type of analysis should lead to the identification of signaling pathways that result in an improved understanding of tissue architecture at homeostasis, and thus, a better appreciation of what happens during disease. Indeed, a recent study also in mice, employed similar depletion methods in combination with adoptive transfer of monocytes followed by in-depth transcriptomic interrogation to qualitatively advance the understanding of the factors involved in establishing TRM identity in recruited monocytes in the gut [60]. Specifically, during monocyte-macrophage differentiation in the intestines, the expression of transcription factors E3-Sumo Ligase (Egr1, 2, and 3) were upregulated while expression of C/EBP and E2F family members were significantly downregulated along with TFs involved in regulating the immune response, such as Bach1, Bcl6, Irf9, Nfatc3, NfKb1, consistent with the hyporesponsive phenotype of intestinal macrophages [61]. Additionally, consistent with past reports, the expression of Spi1 (PU.1) was also downregulated in these intestinal macrophages. Also, in mice, TFs that were required for imprinting the identity of other TRMs, including Gata6 and LXRα, were increased in differentiated monocytes [50,59]. Thus, the use of additional mouse knockout studies and Chipseq to assess the expression of these transcription factors can greatly assist in further resolving the interplay that occurs between niche-specific signals, and the exact sequence of TF activation that is necessary to confer TRM identity onto recruited monocytes.
The monocyte studies reported here have been performed during homeostasis, but disease states affect both monocyte production and phenotypes [62]. These changes may work to amplify pre-existing heterogeneity within the monocyte population or have global population effects such as the induction of monocyte egress following depletion in response to LPS stimuli in humans [63]. For example, in mice, inflammation can trigger pre-existing sub-monocytic populations to become microbicidal macrophages and GM-CSF-dependent MoDCs. This divergence is controlled by Sfpi1, as haploinsufficiency in this TF has resulted in reduced MoDCs and monocyte numbers, but in turn, generated iNOS+ macrophages more efficiently, as shown from lineage tracking [35]. As an example, we now discuss the effects of cancer – as a disease state – on modulating monocyte biology.
Transcriptional imprinting of monocytes in cancer
There are at least three major contexts in which monocyte conditioning may occur in the context of cancer: within the BM or spleen, the circulation itself, or on arrival to the tumor microenvironment (TME). In mouse and human cancers, there is often monocytosis and granulopoiesis that leads to enlarged spleens [64]. However, the exact mechanism behind the elevation in total circulating monocyte numbers is unknown. Global profiling studies of mouse monocytes indicate relative uniformity in the transcriptional profiles of classical monocytes and thus, suggest limited cellular heterogeneity. However, at least a minor Tie2+ sub-population of monocytes has been described that gives rise to Tie2+ Tumor associated Macrophages (TAMs) that enhance angiogenesis [65]. Another population of blood cells that come from the BM in mice and humans has been called myeloid derived suppressor cells (MDSCs) because of their cytotoxic CD8+ T cell suppressive activities. These cells are thought to be immature, and two types have been reported, monocytic (M-MDSC) and granulocytic (G-MDSC) [66]. M-MDSC might be derived from the monocyte lineage but unfortunately, the markers used do not differentiate them from true monocytes, nor in many cases, immature neutrophils. A recent detailed analysis in a wide range of human pathologies including many cancer types, showed elevated G-MDSCs but not M-MDSCs in the blood of patients [67]. However, M-MDSCS derived from monocytes can form a population in tumors and these cells have cytotoxic CD8+ T cell suppressor activity [66,68]. The systemic effect of cancer influencing the production and increased number of different types of monocytes has been likened to emergency hematopoiesis relative to healthy individuals [66]. During emergency hematopoiesis, but not in cancer, novel monocytic sub-populations have been described: these include neutrophil-like monocytes, trained monocytes, and SatM monocytes. These subpopulations have been recently reviewed elsewhere [66,69].
True monocytes isolated by FACS sorting from the serum of patients with epithelial cancers are altered in their relative subset abundance, with an increase in non-classical monocytes relative to healthy individuals; moreover, both classical and non-classical monocyte populations are transcriptionally distinct from those of healthy individuals [70–72]. The mechanisms behind these alterations are unknown and it is not even known whether monocytes may have recirculated from the TME; nevertheless, there is similarity in the transcriptional signatures, cytokine profiles, and functions of monocytes isolated from the serum of patients with severe inflammatory response syndrome (SIRS) compared to those isolated from certain cancer patients [73–75]. This suggests that the conditioning of circulating monocytes might be due to inflammatory mediators, rather than tumor-specific signals. However, this might not always be the case. For example, monocytes in human breast cancer can have distinct transcriptional signatures compared to those found in certain inflammatory diseases (e.g. Lyme disease) and artificial intelligence-derived gene expression signatures might be predictive of the presence of cancer; these monocyte signatures might also be distinguished in the blood of cancer patients compared with those harboring other inflammatory diseases [71].
In addition to inflammatory factors, tumor cell-derived exosomes can play a role in modulating monocytic function [76,77]. Specifically, exosomes isolated from both human and mouse melanomas [77], can alter monocyte fate in mice in a manner that is dependent on the invasiveness of the original tumor. Thus, injection of exosomes isolated from highly metastatic melanoma tumors have resulted in pro-tumoral Ly6c+ monocytes being recruited to the pre-metastatic niche. Conversely, in mice, exosomes isolated from less invasive non-metastatic cancers have led to the expansion of anti-tumoral Ly6clo monocytes, and their recruitment to the bone and lungs; here, these monocytes exerted protective effects via NK cell recruitment/activation, as well as by inducing macrophage killing of metastatic cells [77]. On a transcriptional level, this appeared to be due to the presence of pigment epithelium-derived factor-expressing exosomes which increased Nr4a1 gene expression in circulating monocytes [77]. In mouse mammary cancer models, anthracycline chemotherapy was associated with expansion of Ly6chi monocytes recruited to the lung and increased rates of metastasis relative to untreated cancer-bearing mice. This was reported to be due to the release of extracellular vesicles by anthracycline-treated tumor cells; these extracellular vesicles in turn, increased CCL2 lung endothelial cell expression and elevated CCR2 expression on Ly6chi monocytes [78]. In addition, human monocytes previously exposed to pancreatic tumor-derived exosomes presented elevated amounts of phosphorylated signal transducer of transcription 1 alpha/beta (STAT1) relative to exosomes from healthy volunteers resulting in altered CD14 HLA-DR expression, and driving an immunosuppressive phenotype [79].
In mouse models, monocytes are recruited to the tumor microenvironment via a number of different chemotactic factors [80]. The recruitment and retention of Ly6chi monocytes at both primary and bone and lung metastatic sites is under the regulation of CCR2-CCL2 axis [63,81–84] (Figure 4). Using human xenograft metastatic models, classical human monocytes have also been shown to migrate into tumor sites in a CCR2-dependent manner [82]. These monocytes then differentiate into TAMs (primary) or metastasis (secondary) associated macrophages (MAMs) that play trophic roles in the tumor [82,85]. In the primary tumor, this CCL2-mediated recruitment [84] appears to be dominant, but is entirely genetically redundant, with factors such as Vascular derived endothelial growth factor A (VEGFA)) or CSF1 acting as powerful alternative monocyte recruitment factors [86,87]. Ablation of Csf1 using the null mutation osteopetrotic (Csf1op/op) in a mouse model of mammary cancer (caused by the Polyoma Middle T oncoprotein expression) results in few TAMs and a significant inhibition of tumor progression and metastasis relative to WT mice [87]. Moreover, in metastasis mouse models of breast cancer, inhibition of classical monocytic recruitment can reduce metastatic seeding and persistent tumor growth [82,85]. In contrast, in these models, nonclassical monocytes have been documented to be initially protective through enhancement of NK cell activity. However, as the disease progresses, the pro-tumoral effect of classical monocytes appears to win with classical monocytes becoming dominant [42,85]. In addition, CCL2 has been reported to be the primary driver of classical monocyte recruitment in a mouse model of breast cancer bone metastasis; these recruited monocytes –but not resident macrophages– can promote tumor outgrowth [88]. The chemokine CCL8 which is a factor released by TAMs in response to tumor derived factors such as TNFα to recruit monocytes, can also act through CCR2 to recruit these cells, as recently shown in vitro for human monocytes [71]. Additionally, the chemokine CCL5 can affect monocyte production within the BM as well as the subsequent recruitment of these cells to Breast Phyllodes tumors [89,90]. Lastly, increased amounts of CCL4 have also been reported to correlate with TAM infiltrates in colorectal tumors [91].
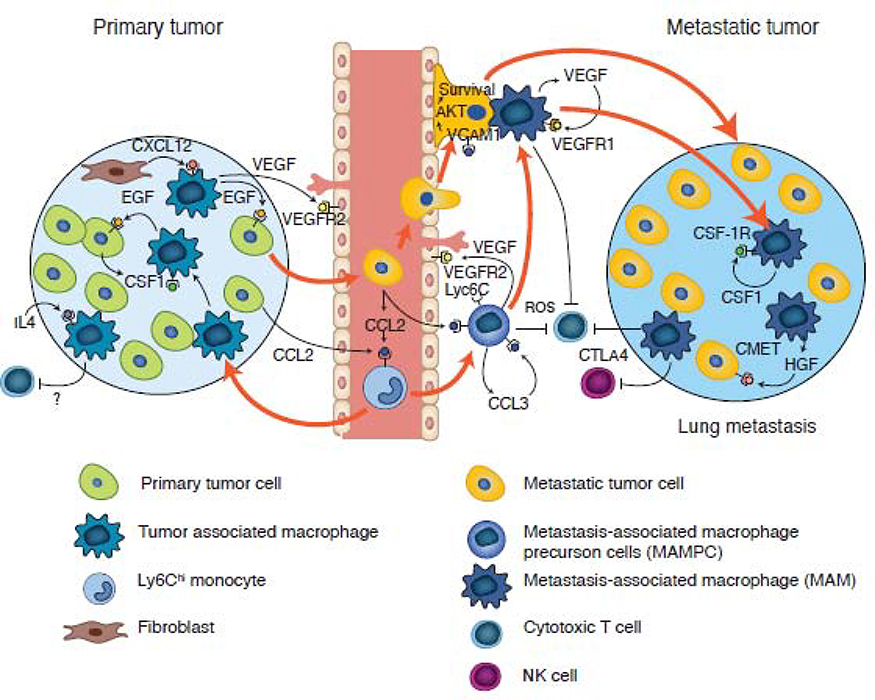
Figure 4. Origin of tumor- and metastasis-associated macrophages in mouse models of breast cancer.
Classical monocytes are recruited to both primary and metastatic sites via a CCL2-CCR2 driven chemokine signaling pathway. In the primary tumor, these recruited monocytes undergo sequential development into TAMs, which are first involved in a streaming behavior that drives tumor cell invasion via a CSF1-Epidermal growth factor (EGF) paracrine loop, before being recruited to vessels by fibroblast expressed CXCL12. On the vessel, these TAMs enhance tumor cell intravasation by expressing VEGFA. At metastatic sites, the circulating tumor cells recruit classical monocytes expressing VEGFA (causing vessel permeability) that upon arrival, promotes the extravasation of the latter. Subsequent signaling pathways (as indicated), result in monocyte differentiation through a progenitor stage (MAMPC) --that is immuno-suppressive via production of reactive oxygen species (ROS)-- into a MAM that confers survival signals to tumor cells via vascular cell adhesion protein 1 (VCAM1) engagement. These MAMs further differentiate, supporting metastatic tumor growth via the depicted signaling pathways; this process occurs in part via inhibition of cytotoxic CD8+ T cells and NK cell effector functions.
Detailed studies in mouse models of lung metastasis indicate that monocytes are not just conduits to MAMs; upon their arrival at the metastatic site, they express VEGFA that increases vessel permeability which in turn enhances metastatic cell seeding. Indeed, this activity can be recapitulated in vitro where classical monocytes can stimulate tumor cell transendothelial cell migration in a process mimicking extravasation [82]. Furthermore, monocytes, along with tumor cell derived VEGFA, can drive further differentiation of these progenitors into MAMs, promoting tumor cell survival and persistent growth [92]. Moreover, classical WT monocytes transferred into Ccr2−/− and Flt1−/− (VEGFR1) genetic knockout mice rescued the reduced metastasis phenotype produced by the effect of the null mutation, definitively revealing the monocyte origin of this metastasis-enhancing activity [82,92].
It is extremely difficult to define when a monocyte becomes a macrophage, but clearly, tissue engagement immediately changes their transcriptional profile. In mouse models of breast cancer lung metastatic disease, tumor-cell derived CCL2 drives an initial differentiation of the newly recruited Ly6c+ monocyte, through an autocrine CCL3-CCR1 mediated mechanism in the monocyte, to a precursor Ly6c++ CD11bhi cell [63]. This latter phenotype is immunosuppressive to cytotoxic CD8+ T cells through the production of reactive oxygen species (ROS) production, as shown by ex-vivo inhibitor experiments in cytotoxic CD8+ T-cell assays, but also enhances tumor cell viability in vivo [63]. Moreover, lineage tracing experiments have indicated that this intermediary cell always differentiates into a MAM that in turn, has a different transcriptional profile and different mechanism of immunosuppression via Cytotoxic T-lymphocyte protein 4 (CTLA4) checkpoint inhibitors, than the monocyte or its first progenitor; this has suggested that the sequential differentiation of the recruited monocyte to a mature macrophage can be influenced by the TME (Figure 4) [68]. These precursor cells can accumulate in the TME, as evidenced from observations that their rate of recruitment exceeds their differentiation into MAMs, thus acting as a bone fide pool of M-MDSCs in this tissue [93]. Their subsequent differentiation into MAMs can alter their transcriptional profile further, which is in turn different from that of TAMs in the primary tissue [30,93]. Supporting these data, TAMs in human breast and endometrial cancers have highly divergent transcriptional signatures relative to the circulating monocytes in the host, and relative to each other; this suggests that the individual TME has a significant impact on TAM differentiation [71].
From another angle, little is known about the transcriptional regulation of monocyte phenotypic and functional changes in cancers. In the model of bone metastasis, monocyte differentiation is driven by IL4 –and has been hypothesized to act through STAT6 [88]. However, to our knowledge, no systematic analysis has been performed on the outcomes of transcriptional regulation in monocytes when neoplasias are present. This lack of data reflects the lack of specify of genetic means of ablation in monocytes alone. Thus, the emergence of monocyte restricted cre-recombinases such as Ms4a3 cre in genetic mouse models might enable the feasibility of such important experiments [10].
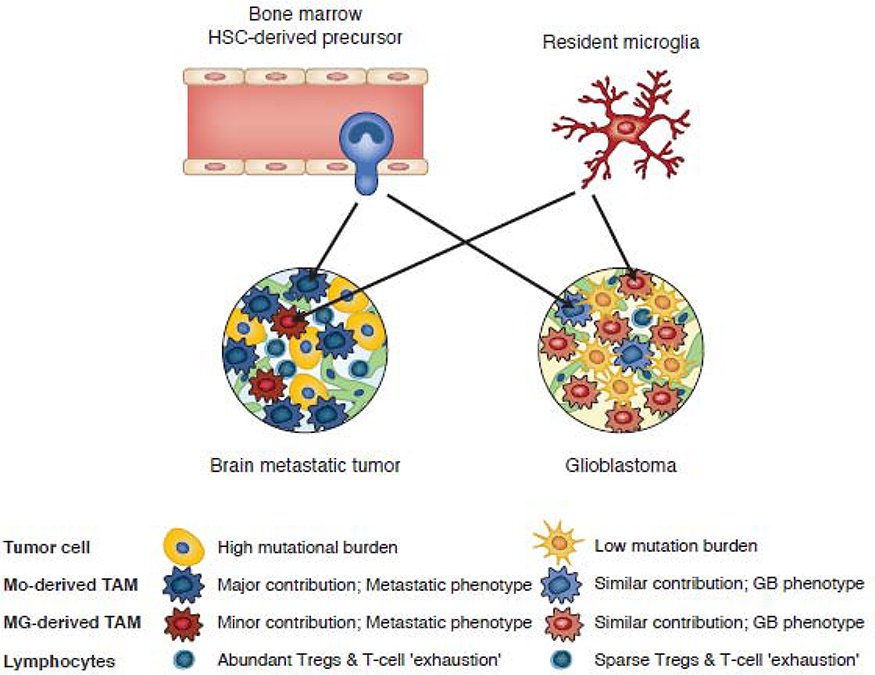
Technical advances in single cell RNA sequencing and high dimensional flow cytometry are being increasingly used to define the cell populations in primary and metastatic human tumors based on transcripts or protein markers at single cell resolution [94–96]. Indeed, single cell RNA-seq analysis of the immune cell populations of 8 primary human breast cancers have enabled the derivation of an immune cell atlas providing transcriptional profiles of monocytes, macrophages, dendritic cells, B cells, and various T cell subsets [95]. In contrast to the above studies, mouse models of breast cancer in which all TAMs and MAMs have been recruited from monocytes, two recent studies performed in-depth analysis of the immune cell populations of primary and metastatic human brain tumors (Figure 5) [96,97]. Both studies found that primary glioblastomas were occupied by both TRM, microglia-derived cells, and monocyte-derived cells, whereas in metastatic brain tumors, most TAMs were monocyte-derived (Figure 5). In addition, the glioblastoma TME imposed a distinct pattern of gene regulation on microglia and monocyte-derived macrophages relative to that found in metastatic tumors. In concert with these differences, metastatic tumors exhibited a more diverse immune cell population than primary glioblastomas, including a higher frequency of regulatory T cells (Figure 5) [96,97]. Collectively, these studies establish tumor-specific roles in the transcriptional imprinting of recruited monocyte-derived and resident macrophages in mouse and human tumors in vivo and provide important resources for further investigation.
Figure 5.
Proportions and tumor specific phenotypes of monocyte-derived and microglia-derived tumor associated macrophages in metastatic brain tumors and glioblastoma.
Concluding remarks
Monocytes are highly adaptable cells that are influenced by the plasma milieu and are subsequently transformed by signals encountered upon entry into a tissue niche. Substantial progress has been made in identifying environmental factors and downstream pathways that drive monocyte differentiation into a few of the many types of tissue macrophages. Importantly, we remain far from understanding the complete set of internal and external signals that are sufficient to establish any particular macrophage phenotype (see outstanding questions). Achieving this goal is made more challenging by the increasing appreciation of myeloid cell complexity, as revealed by single cell technologies. Nevertheless, the power of these new technologies needs to be tempered with further biological understanding and association studies with specific defined markers. For example, a report having defined 4 monocyte subsets in human blood [30] has now come under intense security as the Mono4 population in this report expressed NK cell markers, leading to the conclusion that there might only be three major monocyte populations with a mis-ascribed NK cell population [98]. We posit that it is obvious that while in transition from one population to another, individual cells in the transitional phase will all have slightly different transcriptomes. Thus, there can be endless discussions about diversity but what is most important is that a consensus map be achieved regarding the monocyte populations during homeostasis [98]. This baseline might inform an outstanding question on whether the developmental fates can be influenced by changing environments to give functionally different monocytic populations, or whether there are only whole-scale population shifts. The dramatic shifts in monocytic populations and transcriptomes in cancer suggest the latter. Therefore, another outstanding question is whether these shifts are simply markers of the systemic environment or whether they have functional outcomes influencing the disease state. Given the roles of monocytes in mouse models enhancing metastasis for example, we would argue the latter, but this will be a challenge to answer in humans.
Outstanding questions.
What is the mechanism behind the systemic alterations in monocyte abundance and their transcriptomes in different cancers? Can these alterations be reversed and can this reversal influence disease progression? Profound systemic changes could be an epi-phenomenon or be causal in disease. If the latter case, then reversal of changes may be therapeutic.
Are monocytes involved in reciprocal interactions with disease sites to which they are recruited and thus influence disease progression? If the tissue environment turns recruited monocytes into effector cells at specific sites, this opens the possibility of tissue-specific therapeutic interventions.
When does a monocyte become a macrophage, what controls these steps, and do these intermediate states have functions in vivo? Fate mapping is essential for studying monocyte differentiation into a macrophage in tissues. How many intermediates states are there and what are their functions?
Are tissue macrophage functions independent of an embryonic or bone marrow derived origin? Origin can determine function, offering the possibility of therapeutic discrimination.
Are monocytes and tissue resident macrophage precursors transcriptionally similar, and are they controlled in the adult vs the neonate by the same tissue environment cues?
If the progenitors are different, they might be independently targeted therapeutically.
This analysis raises a key question on how the signaling pathways that have been defined for the recruitment and differentiation of monocytes interact with LDTFs, and how these interact with the overlaid differentiation factors within the blood and tissue in cancer; this is only beginning to be understood in homeostasis. This question is relevant as it might be possible to derive interventions that can define a switch in cell fate or phenotype for therapeutic purposes. Thus, insights into intra- and inter-cellular crosstalk may better inform the roles of macrophages in homeostasis, immunity, and cancer, and are likely to be strengthened by emerging technologies such as spatial transcriptomics, where single cell transcriptomes can be revealed within the context of two- or three-dimensional tissue sections [e.g., [99]]. These methods may also enable improved identification of potential ligand receptor pairs in adjacent cells. In addition, the increasing ability to perform deep epigenetic analyses of relatively small populations of cells is enabling the inference of TFs and upstream signaling pathways that can regulate cell identity from dynamic enhancer landscapes in both normal [58] and pathogenic [100] tissue environments. A better understanding of the mechanisms controlling context-specific monocyte and TRM phenotypes will be essential for the rational development of methods that can beneficially alter their functions in disease.
Highlights.
Mammalian monocyte development in the bone marrow from hematopoietic stem cells is driven by evolving and dynamic genomic enhancer landscapes and sequential binding of Lineage determining transcription factors
Monocyte fates are highly diverse; their plasticity and niche-specific regulation are beginning to be revealed in contexts of homeostasis, inflammation, and cancer.
Murine models of liver injury reveal that the interactions of monocytes with liver sinusoidal cells can lead to sequential alterations of genomic enhancer landscapes that ultimately drive Kupffer cell-specific expression of enhancers and genes.
Murine models of genetic fate mapping, combined with studies of human cancers using single-cell transcriptome and proteome analysis, as well as bioinformatic-inferred algorithms mapping trajectories of cellular differentiation, demonstrate that the tumor-associated macrophage (TAM) phenotype is driven by tumor-specific features, regardless of their origin from monocytes or yolk-sac derived tissue resident monocytes.
Acknowledgements
This review was written with funding from CRUK C17950/A26783, Wellcome Trust (101067/Z/13/Z) and MRC Centre grant MR/N022556/1 to JWP, and AR was funded by a CRUK Clinical Research Fellowship (C157/A20919 – 1).
Funding supporting work related to this review by CKG was provided by NIH grant DK091183. C.Z.H. is supported by the Cancer Research Institute Irvington Postdoctoral Fellowship Program.
Footnotes
Conflicts of Interest
JWP is a co-founder, board member and consultant to “Macomics” an immuno-oncology company. AR, CKG and CZH declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geissmann F et al. (2003) Blood Monocytes Consist of Two Principal Subsets with Distinct Migratory Properties. Immunity 19, 71–82 [DOI] [PubMed] [Google Scholar]
- 2.Stansfield BK (2015) Clinical significance of monocyte heterogeneity. Clinical and Translational Medicine 4, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joyce JA and Pollard JW (2009) Microenvironmental regulation of metastasis. Nat Rev Cancer 9, 239–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hettinger J et al. (2013) Origin of monocytes and macrophages in a committed progenitor. Nat Immunol 14, 821–830 [DOI] [PubMed] [Google Scholar]
- 5.Fogg DK (2006) A Clonogenic Bone Marrow Progenitor Specific for Macrophages and Dendritic Cells. Science 311, 83–87 [DOI] [PubMed] [Google Scholar]
- 6.Akashi K et al. (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 [DOI] [PubMed] [Google Scholar]
- 7.Breton G et al. (2015) Defining human dendritic cell progenitors by multiparametric flow cytometry. Nature Protocols 10, 1407–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamura S et al. (2017) Identification of a Human Clonogenic Progenitor with Strict Monocyte Differentiation Potential: A Counterpart of Mouse cMoPs. Immunity 46, 835–848.e4 [DOI] [PubMed] [Google Scholar]
- 9.Yáñez A et al. (2017) Granulocyte-Monocyte Progenitors and Monocyte-Dendritic Cell Progenitors Independently Produce Functionally Distinct Monocytes. Immunity 47, 890–902.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z et al. (2019) Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell 178, 1509–1525.e19 [DOI] [PubMed] [Google Scholar]
- 11.Notta F et al. (2016) Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116–aab2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara-Astiaso D et al. (2014) Chromatin state dynamics during blood formation. Science 345, 943–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh JC et al. (2002) Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 17, 665–676 [DOI] [PubMed] [Google Scholar]
- 14.Corces MR et al. (2016) Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nature Genetics 48, 1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro JD et al. (2018) Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 173, 1–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schönheit J et al. (2013) PU.1 Level-Directed Chromatin Structure Remodeling at the Irf8 Gene Drives Dendritic Cell Commitment. CellReports 3, 1617–1628 [DOI] [PubMed] [Google Scholar]
- 17.Kurotaki D et al. (2014) IRF8 inhibits C/EBPα activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nature Communications 5, 3764–15 [DOI] [PubMed] [Google Scholar]
- 18.Naik SH et al. (2013) Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496, 229–232 [DOI] [PubMed] [Google Scholar]
- 19.Alder JK et al. (2008) Kruppel-Like Factor 4 Is Essential for Inflammatory Monocyte Differentiation In Vivo. The Journal of Immunology 180, 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurotaki D et al. (2013) Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood 121, 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sontag S et al. (2017) Modelling IRF8 Deficient Human Hematopoiesis and Dendritic Cell Development with Engineered iPS Cells. Stem Cells 35, 898–908 [DOI] [PubMed] [Google Scholar]
- 22.Mucenski ML et al. (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65, 677–689 [DOI] [PubMed] [Google Scholar]
- 23.Lee CZW et al. (2018) Studying tissue macrophages in vitro: are iPSC-derived cells the answer? Nat Rev Immunol 18, 716–725 [DOI] [PubMed] [Google Scholar]
- 24.Passlick B et al. (1989) Identification and Characterization of a Novel Monocyte Subpopulation in Human Peripheral Blood. Blood 74, 2527–2534 [PubMed] [Google Scholar]
- 25.Guilliams M et al. (2018) Developmental and Functional Heterogeneity of Monocytes. Immunity 49, 595–613 [DOI] [PubMed] [Google Scholar]
- 26.Olingy CE et al. (2019) Monocyte heterogeneity and functions in cancer. Journal of Leukocyte Biology 106, 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingersoll MA et al. (2010) Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115, e10–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zilionis R et al. (2019) Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 50, 1317–1334.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zawada AM et al. (2011) SuperSAGE evidence for CD14++CD16+ monocytes as a third monocyte subset. Blood 118, e50–e61 [DOI] [PubMed] [Google Scholar]
- 30.Villani A-C et al. (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cros J et al. (2010) Human CD14dim Monocytes Patrol and Sense Nucleic Acids and Viruses via TLR7 and TLR8 Receptors. Immunity 33, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel AA et al. (2017) The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 214, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yona S et al. (2013) Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 38, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mildner A et al. (2017) Genomic Characterization of Murine Monocytes Reveals C/EBPβ Transcription Factor Dependence of Ly6C- Cells. Immunity 46, 849–862.e7 [DOI] [PubMed] [Google Scholar]
- 35.Menezes S et al. (2016) The Heterogeneity of Ly6Chi Monocytes Controls Their Differentiation into iNOS+ Macrophages or Monocyte-Derived Dendritic Cells. Immunity 45, 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidl C et al. (2014) Transcription and enhancer profiling in human monocyte subsets. Blood 123, e90–e99 [DOI] [PubMed] [Google Scholar]
- 37.Thomas GD et al. (2016) Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6C low Monocytes while Preserving Macrophage Gene Function. Immunity 45, 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanna RN et al. (2011) The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C− monocytes. Nat Immunol 12, 778–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lessard A-J et al. (2017) Triggering of NOD2 Receptor Converts Inflammatory Ly6Chigh into Ly6Clow Monocytes with Patrolling Properties. Cell Reports 20, 1830–1843 [DOI] [PubMed] [Google Scholar]
- 40.Auffray C et al. (2007) Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666–670 [DOI] [PubMed] [Google Scholar]
- 41.Gamrekelashvili J et al. (2017) Regulation of monocyte cell fate by blood vessels mediated by Notch signalling. Nature Communications 7, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanna RN et al. (2015) Patrolling monocytes control tumor metastasis to the lung. Science 350, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quintar A et al. (2017) Endothelial Protective Monocyte Patrolling in Large Arteries Intensified by Western Diet and Atherosclerosis. Circ. Res. 120, 1789–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bain CC et al. (2014) Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 15, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamoutounour S et al. (2013) Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 [DOI] [PubMed] [Google Scholar]
- 46.Epelman S et al. (2014) Origin and Functions of Tissue Macrophages. Immunity 41, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginhoux F et al. (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulz C et al. (2012) A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 [DOI] [PubMed] [Google Scholar]
- 49.Schridde A et al. (2017) Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol 10, 1387–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavin Y et al. (2014) Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell 159, 1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi T et al. (2012) IL-10 regulates Il12b expression via histone deacetylation: implications for intestinal macrophage homeostasis. J. Immunol. 189, 1792–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zigmond E et al. (2014) Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 [DOI] [PubMed] [Google Scholar]
- 53.Chng SH et al. (2016) Ablating the aryl hydrocarbon receptor (AhR) in CD11c+ cells perturbs intestinal epithelium development and intestinal immunity. Sci Rep 6, 23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang PV et al. (2014) The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U.S.A. 111, 2247–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bain CC et al. (2016) Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nature Communications 7, ncomms11852–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott CL et al. (2016) Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nature Communications 7, 10321–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonnardel J et al. (2019) Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakai M et al. (2019) Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 51, 655–670.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mass E et al. (2016) Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238–aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gross-Vered M et al. (2019) Defining Monocyte Differentiation into Colonic and Ileal Macrophages. 46, 849–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lasitschka F et al. (2017) Human monocytes downregulate innate response receptors following exposure to the microbial metabolite n-butyrate. Immunity, Inflammation and Disease 5, 480–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapellos TS et al. (2019) Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front. Immunol. 10, 2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kitamura T et al. (2015) CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med 212, 1043–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casbon A-J et al. (2015) Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proc Natl Acad Sci USA 112, E566–E575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Palma M and Naldini L (2009) Tie2-expressing monocytes (TEMs): novel targets and vehicles of anticancer therapy? Biochim Biophys Acta 1796, 5–10 [DOI] [PubMed] [Google Scholar]
- 66.Canè S et al. (2019) The Endless Saga of Monocyte Diversity . Front. Immunol. 10, 1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cassetta L et al. (2020) Differential expansion of circulating human MDSC subsets in patients with cancer, infection and inflammation. J Immunother Cancer 8, e001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kitamura T et al. (2018) Monocytes Differentiate to Immune Suppressive Precursors of Metastasis-Associated Macrophages in Mouse Models of Metastatic Breast Cancer. Front. Immunol. 8, 69–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guilliams M et al. (2018) Developmental and Functional Heterogeneity of Monocytes. Immunity 49, 595–613 [DOI] [PubMed] [Google Scholar]
- 70.Chittezhath M et al. (2014) Molecular Profiling Reveals a Tumor-Promoting Phenotype of Monocytes and Macrophages in Human Cancer Progression. Immunity 41, 815–829 [DOI] [PubMed] [Google Scholar]
- 71.Cassetta L et al. (2019) Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell 35, 588–602.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamm A et al. (2016) Tumour-educated circulating monocytes are powerful candidate biomarkers for diagnosis and disease follow-up of colorectal cancer. Gut 65, 990–1000 [DOI] [PubMed] [Google Scholar]
- 73.Bergenfelz C et al. (2015) Systemic Monocytic-MDSCs Are Generated from Monocytes and Correlate with Disease Progression in Breast Cancer Patients. PLoS ONE 10, e0127028–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lapko N et al. (2017) Long-term Monocyte Dysfunction after Sepsis in Humanized Mice Is Related to Persisted Activation of Macrophage-Colony Stimulation Factor (M-CSF) and Demethylation of PU.1, and It Can Be Reversed by Blocking M-CSF In Vitro or by Transplanting Naïve Autologous Stem Cells In Vivo. Front. Immunol. 8, 463–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shalova IN et al. (2015) Human Monocytes Undergo Functional Re-programming during Sepsis Mediated by Hypoxia-Inducible Factor-1&alpha. Immunity 42, 484–498 [DOI] [PubMed] [Google Scholar]
- 76.Song X et al. (2016) Cancer Cell-derived Exosomes Induce Mitogen-activated Protein Kinase-dependent Monocyte Survival by Transport of Functional Receptor Tyrosine Kinases. J. Biol. Chem. 291, 8453–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plebanek MP et al. (2017) Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nature Communications 8(1)1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keklikoglou I et al. (2019) Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol 21, 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Javeed N et al. (2017) Immunosuppressive CD14HLA-DR monocytes are elevated in pancreatic cancer and ``primed´´ by tumor-derived exosomes. OncoImmunology 6, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mantovani A et al. (2004) Chemokines in the recruitment and shaping of the leukocyte infiltrate of tumors. Seminars in Cancer Biology 14, 155–160 [DOI] [PubMed] [Google Scholar]
- 81.Ma R-Y et al. Monocyte-derived macrophages promote breast cancer bone metastasis outgrowth. J Exp Med DOI: DOI 10.1084/jem.20191820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian B-Z et al. (2011) CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Franklin RA et al. (2014) The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arwert EN et al. (2018) A Unidirectional Transition from Migratory to Perivascular Macrophage Is Required for Tumor Cell Intravasation. Cell Reports 23, 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qian B et al. (2009) A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS ONE 4, e6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin EY et al. (2007) Vascular endothelial growth factor restores delayed tumor progression in tumors depleted of macrophages. Molecular Oncology 1, 288–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin EY et al. (2001) Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med 193, 727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma R-Y et al. (2020) Monocyte-derived macrophages promote breast cancer bone metastasis outgrowth. J Exp Med 217, 2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nie Y et al. (2019) Breast Phyllodes Tumors Recruit and Repolarize Tumor-Associated Macrophages via Secreting CCL5 to Promote Malignant Progression, Which Can Be Inhibited by CCR5 Inhibition Therapy. Clinical Cancer Research 25, 3873–3886 [DOI] [PubMed] [Google Scholar]
- 90.Ban Y et al. (2017) Targeting Autocrine CCL5-CCR5 Axis Reprograms Immunosuppressive Myeloid Cells and Reinvigorates Antitumor Immunity. Cancer Res 77, 2857–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.la Fuente López De, M. et al. (2018) The relationship between chemokines CCL2, CCL3, and CCL4 with the tumor microenvironment and tumor-associated macrophage markers in colorectal cancer. Tumor Biol. 40, 1–12 [DOI] [PubMed] [Google Scholar]
- 92.Qian B-Z et al. (2015) FLT1 signaling in metastasis-associated macrophages activates an inflammatory signature that promotes breast cancer metastasis. J Exp Med 212, 1433–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ojalvo LS et al. (2009) High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. The American Journal of Pathology 174, 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neftel C et al. (2019) An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Azizi E et al. (2018) Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Friebel E et al. (2020) Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell 181, 1–17 [DOI] [PubMed] [Google Scholar]
- 97.Klemm F et al. (2020) Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 181, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Günther P et al. (2019) A rule-based data-informed cellular consensus map of the human mononuclear phagocyte cell space. bioRxiv 10.1101/658179 [DOI] [Google Scholar]
- 99.Eng C-HL et al. (2019) Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature Publishing Group 568, 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seidman JS et al. (2020) Niche-Specific Reprogramming of Epigenetic Landscapes Drives Myeloid Cell Diversity in Nonalcoholic Steatohepatitis. Immunity 52, 1057–1074.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]