Abstract
The new coronavirus infection (COVID-19) is a major public health concern, with a high burden and risk for infection among patients and healthcare workers. Saliva droplets containing SARS-COV-2 are a major vector for COVID-19 infection, making saliva a promising alternative for COVID-19 testing using nasopharyngeal swab samples. To diagnose COVID-19 patients in the field, a point-of-care test (POCT) using saliva was conceptualized. We have developed a simple method for extracting RNA from saliva samples using semi-alkaline proteinase, a sputum homogenizer typically used for preparing samples for tuberculosis testing, and a subsequent simple heating step with no need for centrifugation or RNA extraction. Further, we newly developed a triplex reverse transcription loop-mediated isothermal amplification approach (RT-LAMP) which utilizes colorimetric readout using a heat block, with results evaluated with the unaided eye. In 44 clinical patients suspected of having COVID-19 infection, the test took 45 min, and resulted in a diagnostic sensitivity of 82.6% (19/23) and diagnostic specificity of 100% (21/21), compared to the reference standard. The limit of detection was 250 copies/reaction (25,000 copies/mL). Our newly developed POCT approach achieved simple RNA extraction and constant RT-LAMP detection. This POCT has the potential to be used for simple inspection stations in a field setting, helping reduce the risk of infection by simplifying and accelerating testing for COVID-19.
Abbreviations: ATCC, American Type Culture Collection; CFI, colori fluorescent indicator; HNB, hydroxynaphthol blue; LOD, limit of detection; PBS, phosphate-buffered saline; POCT, point-of-care test; RT-LAMP, reverse transcription loop-mediated isothermal amplification; RT-qPCR, reverse transcription quantitative real-time polymerase chain reaction; SAP, semi-alkaline protease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SS mixture, saliva-SAP mixture
Keywords: COVID-19; LAMP; Point-of-care test; POCT; SARS-CoV-2, semi alkaline proteinase
1. Introduction
With progressing globalization, the worldwide spread of infectious diseases has accelerated. The new coronavirus infection (COVID-19) is a major public health concern [1], with a high burden and risk for infection among patients and healthcare workers. A reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) testing for the diagnosis of COVID-19 has increased the use of saliva as an alternative to nasopharyngeal swab sampling [[2], [3], [4], [5], [6]].
Angiotensin-converting enzyme II (ACE2), the main receptor for SARS-CoV-2 entry into human cells, is highly expressed on oral cavity mucosa, especially tongue epithelial cells [3,7]. In Japan, many COVID-19 cases have been found in metropolitan entertainment districts, where food, drink, and conversation are frequent. Saliva droplets containing SARS-CoV-2 are a major vector for COVID-19 infection, making saliva a promising candidate for COVID-19 testing [2,4,8]. While the viral load of SARS-CoV-2 in saliva samples is lower than in nasopharyngeal swab samples [3,9], several researchers have reported comparable or better diagnostic sensitivity with saliva testing [2,[4], [5], [6],8].
The combined use of the direct-to-test addition by heating and RT-qPCR using a nasopharyngeal swab sample has proven successful due to reduced inhibitory effects [10]. However, the saliva sample must undergo high-speed centrifugation at 20,000 g for 5−30 min following purification with a commercial RNA extraction kit to remove potential inhibitors [[2], [3], [4], [5],8,9]. This tedious RNA extraction process requires a well-equipped laboratory and is a major hurdle for performing sensitive point-of-care testing (POCT) for COVID-19 using saliva.
Saliva is easier to sample and carries less risk of contamination to the collector than nasopharyngeal swabs [6]. For immunochromatographic assays, the sensitivity is relatively high for nasopharyngeal swabs with low carryover of sample-derived inhibitors, but decreases for saliva with high inhibitors [4]. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) is a more attractive option for POCT with robustness to inhibitors in the sample over immunochromatographic assays. Also, it has broader utility when a portable real-time detector is used with an in-house battery or combined use with a cost-effective heat block and colorimetric reagent [[11], [12], [13], [14]].
We have developed a simple method for extracting RNA from saliva samples using semi-alkaline proteinase (SAP), a sputum homogenizer typically used for preparing samples for tuberculosis testing, and a subsequent simple heating step with no need for centrifugation or RNA extraction. Further, we newly developed a triplex RT-LAMP approach which utilizes colorimetric readout usable of a heat block, with results evaluated with the unaided eye. Of note, colorimetric readout is more unambiguous than turbidimetric readout with the unaided eye, and more practical in the field than fluorescent LAMP analysis without color change dye. Here, we preliminarily evaluated the POCT performance of this approach with 44 clinical saliva samples.
2. Materials and methods
2.1. Design of LAMP primers
We used Primer Explorer V5 (primerexplorer.jp/lampv5e/index.html) to design three primer sets targeting the ORF 1ab, S, and ORF 7a regions to simultaneously detect SARS-CoV-2 with high sensitivity and robustness. To identify highly conserved nucleotide sequences with low similarity to SARS-CoV-1 sequences, we performed multiple sequence alignment using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo) of 490 whole-genome SARS-Cov-2 sequences from the DDBJ/EMBL/GenBank databases (www.ncbi.nlm.nih.gov/nucleotide/) submitted by researchers from five continents. The details of each primer and in-house reagent are shown in Appendix Table A1, Table A2, Table A3. Artificially synthesized SARS-CoV-2 sequences (Eurofins, Tokyo, Japan) corresponding to the three target regions and RNA extracted from healthy human saliva were used as positive and negative controls, respectively, for the triplex RT-LAMP format.
2.2. Reference method for SARS-CoV-2 detection from saliva
Saliva samples were collected from a total of 165 patients suspected of COVID19 infection at Kyoto University Hospital and Kyoto City Hospital between May and July 2020. Saliva samples were collected into a disposable sterilized plastic container through the drooling technique. Forty-five and 120 samples were determined to be positive and negative, respectively, according to the reference method shown below [15]. By random sampling, 44 of these samples were used for this study. The saliva samples were mixed thoroughly with 3 strength-volume of SAP (semi-alkaline protease, Suputazyme; Kyokuto Pharmaceutical Industrial, Tokyo, Japan) manually or using a vortex mixer for 15 s. After incubation for 15 min at room temperature to dissolve the saliva components, each of the saliva-SAP mixtures (SS mixtures) was added to a 1.5-mL microcentrifuge tube. Subsequent procedures were completed without interruption, or else the SS mixture was stored at −80 °C until used. For RNA extraction, 140 μL of each SS mixture was eluted into 60 μL using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). RT-qPCR using the N2 primers and probe was performed with 5 μL of the extracted RNA in 20 μL of reaction mixture in the LightCycler 480 System II (Roche, Basel, Switzerland) according to the protocol recommended by the National Institute of Infectious Diseases, Japan [15]. Ten-fold serial dilutions (5 to 5 × 106 copies/reaction) of the synthesized RNA containing the target sequence [15] were used to calculate the viral loads in clinical samples.
2.3. POCT for SARS-CoV-2 detection from saliva
In parallel, for POCT, the remaining SS mixture was heated at 95 °C for 5 min, and then, if visible large amount of the droplet had adhered on the 1.5-mL microcentrifuge tube lid, the tube was swung manually 1–2 times to quickly remove the aqueous droplet, as the prevention measure of contamination. Using 50 μL of RT-LAMP reaction mixtures (including 10 μL of template), the RT-LAMP reaction was monitored in real-time using a LightCycler 480 System II (Roche) at 63 °C for 30 min. The threshold time of positivity was set at 25 min, and time of positivity was automatically calculated. Amplification was also judged by endpoint readout with the unaided eye, which was interpreted as positive when the color change was observed from violet to sky blue.
2.4. Determination of limit of detection (LOD)
The LOD of triplex RT-LAMP was defined as the lowest concentration at which 95% of positive samples were detected using a 2-fold series of phosphate-buffered saline (PBS) dilutions with 20 replicates of 50,000, 25,000, 12,500, and 10,000 copies/mL of heat-inactivated SARS-CoV-2 (ATCC VR-1986HK; American Type Culture Collection, Manassas, VA, USA) (Table 1 ). The SARS-CoV-2 dilutions by PBS were heated at 95 °C for 5 min. After removal of the aqueous droplet on the tube lid by swinging the tube manually 1–2 times, 10 μL of the template was applied to 50 μL of RT-LAMP reaction mixture, as described above.
Table 1.
LOD determination of the triplex RT-LAMP with the heat-inactivated SARS-CoV-2, ATCC VR-1986HK in phosphate-buffered saline.
| Copies/mL | 50,000 | 25,000 | 12,500 | 10,000 |
|---|---|---|---|---|
| Copies/reaction | 500 | 250 | 125 | 100 |
| Positive/tested (%) | 20/20 (100%) | 20/20 (100%) | 14/20 (70%) | 9/20 (45%) |
ATCC, American Type Culture Collection.
LOD, Limit of detection.
RT-LAMP, Reverse transcription loop-mediated isothermal amplification detection.
SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2.
3. Results and discussion
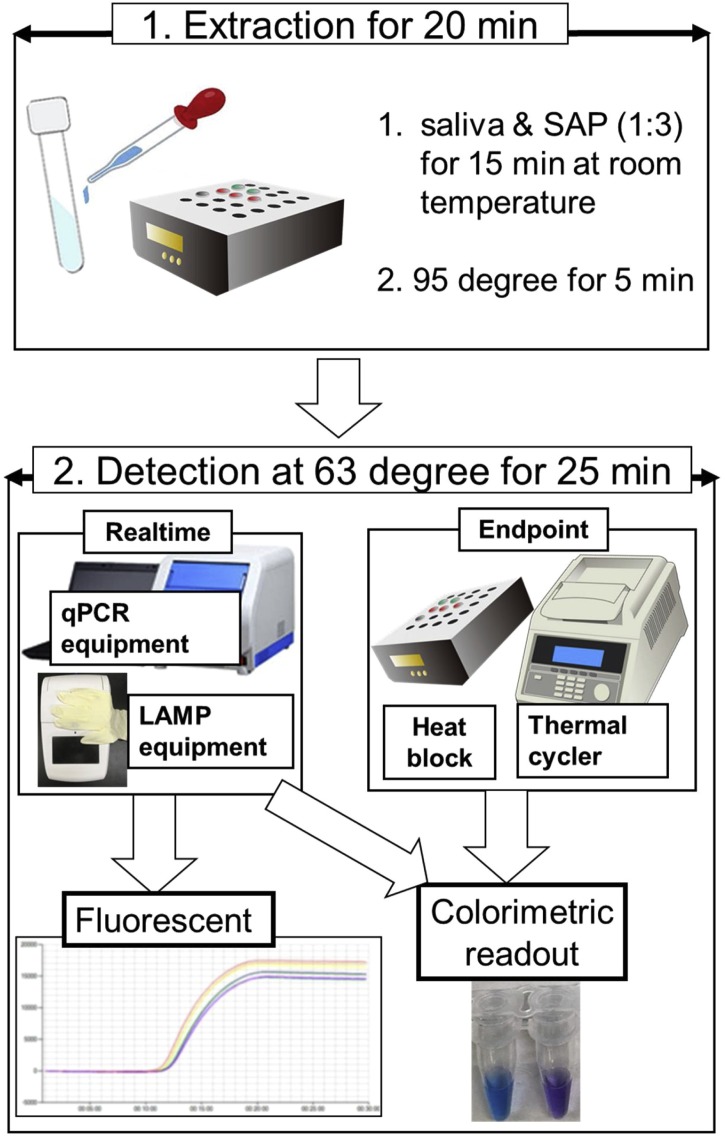
In 44 clinical patients suspected of having COVID-19 infection, 23 and 21 saliva samples resulted positive and negative by the reference method. The diagnostic sensitivity and specificity were 82.6% (19/23; 95% confidence interval [CI], 61.2%–95.1%) and 100% (21/21, 95% CI, 83.8%–100%) for the new POCT, which took 45 min from starting crude RNA extraction to final judgement (Fig. 1 , Table 2 ). Colorimetric endpoint readout with the unaided eye achieved robust judgement from violet to sky blue (Fig. 1). The results of the fluorescent real-time RT-LAMP detection by using the qPCR equipment, LightCycler 480 System II and endpoint judgement by the colorimetric readout with the unaided eye were constantly matched each other.
Fig. 1.
Concept of POCT using a combination of simple RNA extraction and LAMP equipment.
Table 2.
Results of POCT and reference standard in 44 clinical patients.
| Sample ID | Time of positivity by POCT (min:s) | Copies/reaction tube by reference standard | Copies/mL of saliva by reference standard |
|---|---|---|---|
| N = 23 | RT-qPCR positive | ||
| AR+26 | Negative | 13 | 1100 |
| AR+23 | Negative | 86 | 7400 |
| AR+24 | 23:32 | 170 | 15,000 |
| AR+03 | Negative | 180 | 15,000 |
| AR+11 | Negative | 230 | 20,000 |
| AR+22 | 12:05 | 380 | 33,000 |
| AR+20 | 14:35 | 590 | 51,000 |
| AR+04 | 14:34 | 1300 | 110,000 |
| AR+16 | 13:37 | 2500 | 210,000 |
| AR+21 | 12:11 | 3800 | 330,000 |
| AR+17 | 16:12 | 8000 | 690,000 |
| AR+06 | 13:11 | 9300 | 800,000 |
| AR+25 | 12:11 | 16,000 | 1,400,000 |
| AR+18 | 14:43 | 24,000 | 2,100,000 |
| AR+07 | 12:56 | 34,000 | 2,900,000 |
| AR+08 | 11:59 | 45,000 | 3,900,000 |
| AR+14 | 11:36 | 120,000 | 10,000,000 |
| AR+09 | 10:58 | 130,000 | 11,000,000 |
| AR+10 | 10:17 | 190,000 | 16,000,000 |
| AR+15 | 09:49 | 530,000 | 45,000,000 |
| AR+27 | 09:02 | 1,600,000 | 140,000,000 |
| AR+13 | 09:06 | 1,700,000 | 150,000,000 |
| AR+12 | 13:51 | 3,600,000 | 310,000,000 |
| N = 21 | |||
| Negative | RT-qPCR negative | ||
POCT, Point-of-care test.
RT-qPCR, Reverse transcription quantitative real-time polymerase chain reaction.
Vortexing or manual mixing was used to mix the saliva sample with the SAP solution. If a large amount of visible droplets adhered to the 1.5-mL microcentrifuge tube lid after heating at 95 °C for 5 min, the droplets were removed by swinging the 1.5-ml microcentrifuge tube 1–2 times. Neither of these manual operations affected the detection by POCT. Our newly developed POCT is capable of both simple endpoint judgement using a heat block and more rapid real-time judgement using a qPCR equipment or real-time LAMP equipment, as shown in Fig. 1.
As shown in Table 2, 18 clinical samples exceeding 380 copies/reaction (>33,000 copies/mL of saliva) according to reference standard measurement were positive on POCT between 9 min 02 s and 16 min 12 s from the start of amplification. Regarding the 3 samples showing low copy numbers of 170–230 copies/reaction (15,000–20,000 copies/mL of saliva) on RT-qPCR, 1 was ultimately positive at 23 min 32 s of amplification, while the other 2 were false negatives on POCT. This discrepant result is not surprising. This is because, as shown in Table 1, the LOD determined by 20 repetitions was 250 copies/reaction (25,000 copies/mL), but the frequency was 70% (14/20) positive at 125 copies/reaction (12,500 copies/mL). Samples with copy numbers in this range are on the LOD borderline between positive and negative, so we speculate that the results were unstable. Therefore, these findings were interpreted to indicate borderline samples near the LOD of POCT. Two other samples showing low copy numbers of 13–86 copies/reaction (1,100−7,400 copies/mL of saliva) were also false negatives, likely due to their low copy numbers being below the detection power of POCT.
4. Conclusions
Our newly developed POCT approach achieved simple RNA extraction and constant RT-LAMP without the need for centrifugation, or nucleic acid extraction kits. While this POCT does not require expensive equipment and performs comparably to an existing reference standard, of course, requiring further improvement for higher sensitivity and larger scale evaluation with clinical samples. There is a high need for POCT using saliva, due to a low risk of contamination and easiness of sample collection, but the immunochromatography is not a realistic option because of the high inhibitory effects in saliva [4]. This POCT using saliva thus has the potential to be used for simple inspection stations in a field setting, helping reduce the risk of infection by simplifying and accelerating testing for COVID-19.
Declaration of Competing Interest
The authors report no declarations of interest.
Ethics approval
The Ethics Committee of Kyoto University Graduate School and the Faculty of Medicine approved this study (R2379) and waived the need for obtaining informed consent from each patient.
CRediT authorship contribution statement
Wataru Yamazaki: Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing - original draft, Writing - review & editing. Yasufumi Matsumura: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Validation, Visualization, Writing - original draft, Writing - review & editing. Uraiwan Thongchankaew-Seo: Data curation, Investigation. Yasuko Yamazaki: Data curation, Investigation. Miki Nagao: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing - review & editing.
Acknowledgement
This research was supported by AMED under Grant Number JP20he0622031.
Appendix A
Table A1.
Primers used for RT-LAMP assay.
| Primer | Sequences (5′-3′) | Location* | Target |
|---|---|---|---|
| F3−1ab | TGCTAGATTTTACTTTTACACCAG | 4522−4545 | ORF1ab |
| B3−1ab | CAGGTGTTTTAGAAGAAGAAGT | 4757−4736 | |
| FIP-1ab | CCAAGTGGCATTGTAACAAGAGTTT-TAAAACAACTGTAGCGTCAC | 4617−4593, 4546−4565 | |
| BIP-1ab | TGCTCGGTATATGAGATCTCTCAA-CGCTGTAACAGCATCAGG | 4651−4674, 4720−4674 | |
| FLP-1ab | CATTTAGATCGTTAAGTGTGTTGA | 4592−4569 | |
| BLP-1ab | AGTGCCAGCTACAGTTTCT | 4675−4693 | |
| F3-S | TCTTTCACACGTGGTGTT | 21653−21670 | Spike glycoprotein |
| B3-S | GTACCAAAAATCCAGCCTC | 21885−21867 | |
| FIP-S | CATGGAACCAAGTAACATTGGAAAA-CCTGACAAAGTTTTCAGATCC | 21761−21737, 21677−21697 | |
| BIP-S | CTCTGGGACCAATGGTACTAAGAG-GACTTCTCAGTGGAAGCA | 21772−21795, 21855−21838 | |
| FLP-S | GGTAAGAACAAGTCCTGAGTTGAAT | 21732−21708 | |
| BLP-S | GTTTGATAACCCTGTCCTACCATT | 21796−21819 | |
| F3−7ab | GCACTGACTTGCTTTAGC | 27556−27573 | ORF7ab |
| B3−7ab | TGAAAGTTCAATCATTCTGTCTT | 27770−27748 | |
| FIP-7ab | AGGTGAAACTGATCTGGCACG-AATTTGCTTTTGCTTGTCCTG | 27645−27625, 27578−27598 | |
| BIP-7ab | AGACAAGAGGAAGTTCAAGAACTTT-TGAGTGTGAAGCAAAGTGT | 27658−27682, 27742−27724 | |
| FLP-7ab | GATAGACGTGTTTTACGCCGT | 27619−27599 | |
| BLP-7ab | CTCCAATTTTTCTTATTGTTGCGGC | 27686−27710 | |
RT-LAMP, reverse transcription loop-mediated isothermal amplification.
Corresponding to the GenBank accession No. NC_045512.2.
Table A2.
Composition of the RT-LAMP reaction mixture including the homemade master mix.
| RT-LAMP reaction mix | Triplex | |
|---|---|---|
| 4x LAMP Master Mix | 12.5 | μL |
| Distilled water | 11.5 | μL |
| 1x Primer mix for 1ab | 2.6 | μL |
| 1x Primer mix for S | 2.6 | μL |
| 1x Primer mix for 7a | 2.6 | μL |
| CFI | 2 | μL |
| Bst 2.0 DNA polymerase (8 units/μL) | 6 | μL |
| AMV Reverse Transcriptase for Genie III (20 units/μL) | 0.2 | μL |
| RNA template | 10 | μL |
| Total | 50 | μL |
| Homemade 4x LAMP master mix | For 500 | For 1000 | ||
|---|---|---|---|---|
| 5 M Betaine | 4 | mL | 8 | mL |
| 1 M Tris-HCl (pH8.8) | 0.5 | mL | 1 | mL |
| 3 M Potassium chloride (KCl) | 0.08 | mL | 0.16 | mL |
| 5.6 M (NH4)2SO4 | 0.04 | mL | 0.08 | mL |
| 1 M MgSO4 | 0.2 | mL | 0.4 | mL |
| 100 mM dATP | 0.35 | mL | 0.7 | mL |
| 100 mM dCTP | 0.35 | mL | 0.7 | mL |
| 100 mM dGTP | 0.35 | mL | 0.7 | mL |
| 100 mM dTTP | 0.35 | mL | 0.7 | mL |
| Tween20 | 0.05 | mL | 0.1 | mL |
| Total (for 500/1000 samples) | 6.27 | mL | 12.54 | mL |
| CFI | For 500 | |
|---|---|---|
| Distilled water | 0.6965 | mL |
| 10 mM HNB* | 0.3 | mL |
| Gel green (×10,000) | 0.0035 | mL |
| Total (for 500 samples) | 1.0 | mL |
| * Distilled water:HNB powder =1 mL:6 mg | ||
| Primer mix | For 500 | |
|---|---|---|
| FIP (Inner F) (100 μM) | 0.4 | mL |
| BIP (Inner B) (100 μM) | 0.4 | mL |
| LF (Loop F) (100 μM) | 0.2 | mL |
| LB (Loop B) (100 μM) | 0.2 | mL |
| F3 (Outer F) (100 μM) | 0.05 | mL |
| B3 (Outer B) (100 μM) | 0.05 | mL |
| Total (for 500 samples) | 1.3 | mL |
HNB, hydroxynaphthol blue; CFI, colori fluorescent indicator; RT-LAMP, reverse transcription loop-mediated isothermal amplification.
Table A3.
Manufacturers' list for RT-LAMP reaction mix.
| LAMP reagents | Manufacturers | Locations |
|---|---|---|
| 4x LAMP Master Mix | ||
| Tris-HCl | Wako Pure Chemical Industries | Osaka, Japan |
| Potassium chloride (KCl) | Sigma–Aldrich | St. Louis, MO, USA |
| (NH4)2SO4 | Sigma–Aldrich | St. Louis, MO, USA |
| Tween20 | Sigma-Aldrich | St. Louis, MO, USA |
| MgSO4 | New England Biolabs | Ipswich, MA, USA |
| Illustra dNTP set 100 mM, solutions 4 × 25 μmol | GE Healthcare | Little Chalfont, UK |
| 5M Betain Solution 5 mL | Sigma–Aldrich | St. Louis, MO, USA |
| CFI | ||
| HNB | MP Biomedicals | Aurora, OH, USA |
| Gel green (x10,000) | Biotium | Hayward, CA, USA |
| Bst 2.0 DNA polymerase (8 units/μL) | New England Biolabs | Ipswich, MA, USA |
| AMV Reverse Transcriptase, for Genie III (20 units/μL) | Nippon Gene | Tokyo, Japan |
| Primer (Recommended cartridge grade purification) | Hokkaido System Science | Sapporo, Japan |
HNB, hydroxynaphthol blue; CFI, colori fluorescent indicator; RT-LAMP, reverse transcription loop-mediated isothermal amplification.
References
- 1.World Health Organization (WHO). Coronavirus disease 2019 (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed 17 November 2020).
- 2.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T., et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 2020;81:e145–e147. doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., et al. Clinical Evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J. Clin. Microbiol. 2020;58:e01438–20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To K.K., Tsang O.T., Yip C.C., Chan K.H., Wu T.C., Chan J.M., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogels C.B., Watkins A.E., Harden C.A., Brackney D., Shafer J., Wang J., et al. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. 2021;2:1–18. doi: 10.1016/j.medj.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H., Zhong L., Deng J., Peng J., Dan H., Zeng X., et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58:e00776–20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., et al. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel) 2020;2020(9):182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckel F., Küsters F., Drossel B., Konert M., Mattes H., Schopf S. VariplexTM test system fails to reliably detect SARS-CoV-2 directly from respiratory samples without RNA extraction. Eur. J. Clin. Microbiol. Infect. Dis. 2020;2020(17):1–5. doi: 10.1007/s10096-020-03983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashida K., Kajino K., Hachaambwa L., Namangala B., Sugimoto C. Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for human African trypanosomiasis. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.J.I., Kim B.T., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]