Abstract
Background
Confirmatory testing of SARS-CoV-2 results is essential to reduce false positives, but comes at a cost of significant extra workload for laboratories and increased turnaround time. A balance must be sought. We analysed our confirmatory testing pathway to produce a more refined approach in preparation for rising case numbers.
Methods
Over a 10-week low prevalence period we performed confirmatory testing on all newly positive results. Turnaround time was measured and results were analysed to identify a threshold that could be applied as a cut-off for future confirmatory testing and reduce overall workload for the laboratory.
Results
Between 22/06/20 and 31/08/20 confirmatory testing was performed on 108 newly positive samples, identifying 32 false positive results (30 %). Turnaround time doubled, increasing by an extra 17 h. There was a highly statistically significant difference between initial Relative Light Unit (RLU) of results that confirmed compared to those that did not, 1176 vs 721 (P < 0.00001). RLU = 1000 was identified as a suitable threshold for confirmatory testing in our laboratory: with RLU ≥ 1000, 55/56 (98 %) confirmed as positive, whereas with RLU < 1000 only 12/38 (32 %) confirmed.
Conclusions
False positive SARS-CoV-2 tests can be identified by confirmatory testing, yet this may significantly delay results. Establishing a threshold for confirmatory testing streamlines this process to focus only on samples where it is most required. We advise all laboratories to follow a similar process to identify thresholds that trigger confirmatory testing for their own assays, increasing accuracy while maintaining efficiency for when case numbers are high.
Keywords: SARS-CoV-2, COVID-19, Confirmatory testing, False positive, Laboratory diagnosis
1. Introduction
As coronavirus disease (COVID-19) rates fell after the first peak in the UK, we became suspicious of potential false positive results in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) screening tests in our hospital. We identified patients who had negative admission swabs and no COVID-19 symptoms, yet had a subsequent single positive inpatient screening test, followed by further negative tests. A diagnosis of COVID-19 seemed highly improbable, yet each case prompted major infection control operations to isolate and contact trace within the hospital.
Our previously published work [1] shows how false positive results can occur in SARS-CoV-2 testing. Assuming our estimate of 99.9 % specificity [1], without confirmatory testing, the current count of 300,000 tests per day in the UK [2] could lead to over 100,000 false positive results a year. Confirmatory testing is crucial to prevent this. Erroneously positive results have significant consequences: nosocomial infection caused by placement on COVID-19 wards, delayed or cancelled hospital procedures, wasted infection control and contact tracing resources and inaccurate national reporting [3]. On 22/06/2020 our laboratory began confirmatory testing on all samples with a newly positive SARS-CoV-2 result.
The major drawbacks to confirmatory testing are the extra time taken to release a confirmed result and the additional workload it puts on already stretched laboratories. While relatively straightforward in a low prevalence setting, it quickly becomes impractical as case numbers rise. In preparation for the second wave of COVID-19, we aimed to streamline the process in our laboratory by analysing our results to identify those most and least likely to require confirmation. Here we report our findings, expand on our previously published work [1] (which included just the first month’s data), and outline methods that other laboratories can follow to improve their confirmatory testing pathways.
2. Methods
Over a 10-week period of low prevalence, from 22/06/20 to 31/08/20, we performed confirmatory testing on all newly positive SARS-CoV-2 results. Full methodology of our confirmatory testing pathway is outlined in our previous paper [1]. In brief, our laboratory used two SARS-CoV-2 testing platforms: an in-house real time reverse transcriptase polymerase chain reaction (RT-PCR) and the Aptima SARS-CoV-2 assay (Hologic Panther System) [4]. For confirmatory testing all newly positive samples were re-extracted and re-tested on the same and alternative platform, giving three results. We interpreted the overall result as confirmed if 3/3 or 2/3 tests were positive, or not confirmed (false positive) if 1/3 tests were positive, i.e. only the initial result was positive. If there was insufficient sample for three tests, we interpreted the overall result as confirmed if 2/2 were positive or indeterminate if only 1/2 was positive. Newly positive results were defined as those from patients for whom we had no previous positive SARS-CoV-2 results in our laboratory.
The vast majority of our samples were run on the Aptima SARS-CoV-2 assay due to its high throughput [5] and so analysis was focused on samples tested initially on this platform. The output of this assay is the Relative Light Unit (RLU). We used Excel and GraphPad prism to analyse the RLU of tests that confirmed as positive and those that did not. Turnaround time (TAT) was measured using our electronic laboratory tracking system.
3. Results
Over the 10-week period we performed 56,463 tests for SARS-CoV-2 RNA. There were 203 positive results, of which 108 were the first positive result for that patient. Of these, 69/108 (64 %) confirmed as positive, 32/108 (30 %) did not confirm (false positive) and 7/108 (6%) were indeterminate, Table 1 .
Table 1.
Confirmatory testing outcomes.
| Total samples | Confirmed | Did not confirm | Indeterminate | |
|---|---|---|---|---|
| In house PCR | 12 | 2 | 5 | 5 |
| Hologic Aptima SARS-CoV-2 Assay | 96 | 67 | 27 | 2 |
| Total | 108 | 69 | 32 | 7 |
TAT was measured between 11/07/20 and 31/08/20. There were 71 confirmatory tests in this period with a median additional TAT of 16h47, inter quartile range (IQR) 12h05-24h40, representing a doubling of TAT for these samples. The confirmatory testing pathway required a dedicated biomedical scientist each day.
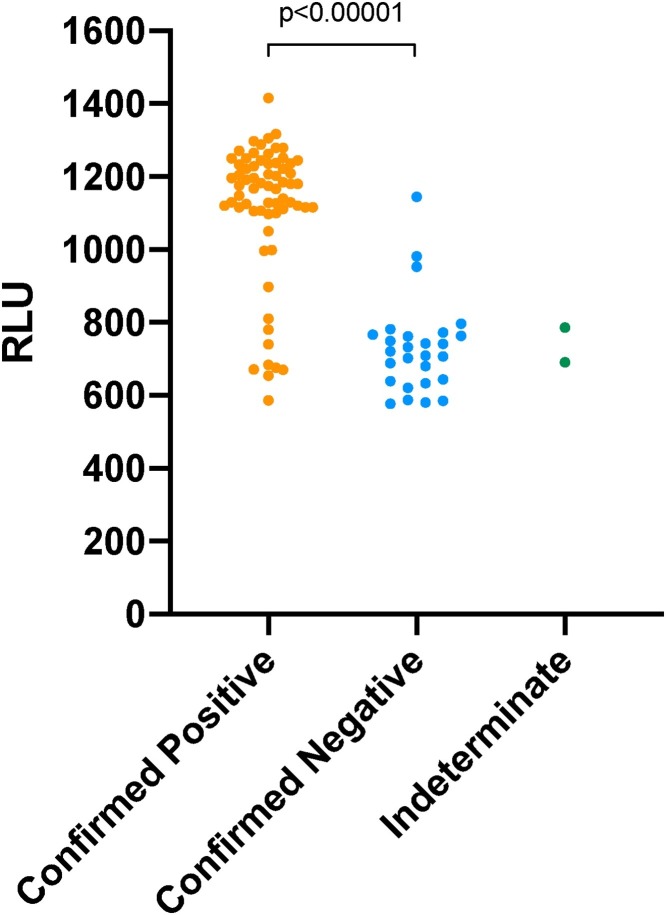
There was a highly significant difference between initial RLU and subsequent result of confirmatory testing, Fig. 1 . The median RLU for confirmed positives was 1176 (IQR 1109–1236) vs 721 (642–765) for those not confirming, P < 0.00001 (Mann-Whitney U Test).
Fig. 1.
Relative Light Unit output of Hologic Aptima SARS-CoV-2 assay results against confirmatory testing outcome: confirmed positive, confirmed negative or indeterminate (total = 96, confirmed = 67, not confirmed = 27, indeterminate = 2).
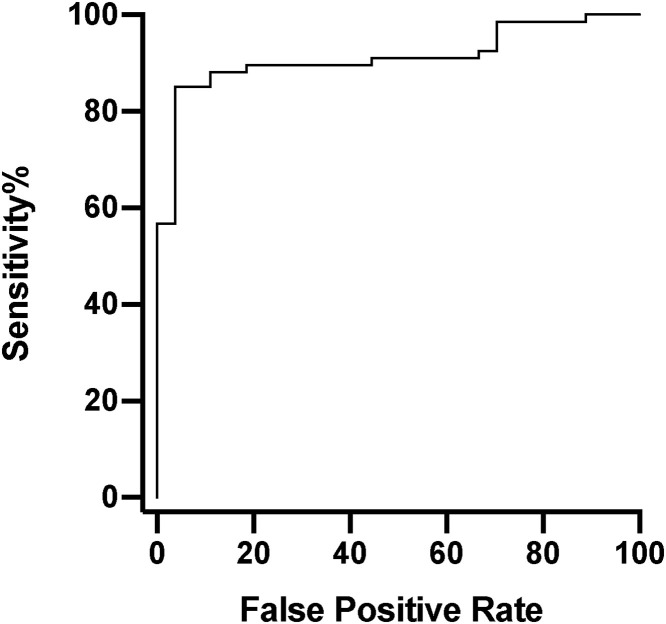
We plotted a receiver operating characteristic (ROC) curve of RLU for those tests that confirmed as positive against those that did not, Fig. 2 , area under curve (AUC) 0.92. This identified an optimal compromise between sensitivity and false positive rate at around RLU = 990. To err on the side of lower false positive rate and for ease of use, RLU = 1000 was selected as a potential threshold: with RLU ≥ 1000, 55/56 (98 %) confirmed, whereas with RLU < 1000 only 12/38 (32 %) confirmed.
Fig. 2.
Receiver operating characteristic curve of Relative Light Unit (RLU) output of initial Aptima SARS-CoV-2 assay results (n = 96, confirmed = 67, not confirmed = 27). Area under curve = 0.92. The optimised RLU for sensitivity and false positive rate is 989.
4. Discussion
Performing three individual tests on a sample increases TAT and adds a substantial work-load to laboratories. A balance must be sought between taking the time to ensure a valid result against the risk of increased transmission if patients are not isolated during this period.
We identified a highly significant difference between the initial RLU of samples that confirmed as positive and those that did not. This allowed us to ascertain a threshold RLU (1000) below which confirmatory testing was most likely to be required. Using this threshold would ensure that almost every false positive result was identified, yet drastically reduce the total number requiring confirmatory testing, easing laboratory workload and ensuring expedient results for the majority of positive patients. Our figures suggest confirmatory testing of samples run on the Aptima SARS-CoV-2 Assay could be reduced by 58 % (56/96) with this cut-off. From mid-September, as case numbers rose, this threshold was instigated in our laboratory.
Only a very small minority of our total results were false positives, 0.057 % (32/56,463), but when compared to newly positive results 30 % (32/108) were false. With increasing prevalence of COVID-19 the percentage of positives that are false will fall, but the absolute number will remain the same [6,7]. When performing such a high number of tests nationally, the potential for false positives, and therefore subsequent harm, becomes substantial.
Nosocomial infection has been a major feature of the COVID-19 pandemic, the true extent of which is unknown [8,9]. False positives can lead to nosocomial infection following placement with COVID-19 patients [3]. Timely and accurate release of SARS-CoV-2 test results is crucial for improving patient care, limiting infection control risks, and accurate reporting which directs national policy. Our improved and streamlined confirmatory testing pathway facilitates this process.
We acknowledge that in some cases a non-repeatable result may indicate a very low level of RNA which degrades over the time of confirmatory testing, potentially indicating very late or very early infection. Indeed, samples that did not confirm were often closer to the limit of detection, highlighting the difficulty of interpreting these results. To investigate this we reviewed the subsequent results of all 32 patients with false positive results: no patient went on to have a further positive test while 25 had a later negative test (of which 9 were tested within a period of 7 days, 8 between 7–10 days later and 6 at over 10 days). This provides evidence that our confirmatory testing identified true false positives.
Confirmatory testing can be a burden on a laboratory and becomes impractical as the number of positive tests rise. We have shown that it is possible to optimise this process, increasing accuracy while limiting impact on the laboratory. We suggest that every laboratory creates their own confirmatory testing pathways based on their own capacity, pressures and resources. A period of low prevalence provides an ideal opportunity to study these relatively new assays and streamline the confirmatory testing process as recommended by Public Health England [10]. It is imperative that confirmatory testing continues as far as possible, even as prevalence rises, to prevent harm from SARS-CoV-2 false positive results.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethical statement
This study was performed as part of a service evaluation and so does not require formal ethical review.
CRediT authorship contribution statement
Michael J. Wilson: Conceptualization, Methodology, Data curation, Formal analysis, Project administration, Writing - original draft, Writing - review & editing. Dominic Sparkes: Conceptualization, Methodology, Data curation, Formal analysis, Project administration, Writing - original draft, Writing - review & editing. Chloe Myers: Data curation, Project administration, Writing - review & editing. Anna A. Smielewska: Conceptualization, Data curation, Project administration, Writing - review & editing. Mir Mubariz Husain: Writing - review & editing. Christopher Smith: Writing - review & editing. Kathryn J. Rolfe: Writing - review & editing. Hongyi Zhang: Project administration, Writing - review & editing. Hamid Jalal: Conceptualization, Project administration, Writing - review & editing.
Declaration of Competing Interest
The authors have nothing to declare.
Acknowledgements
We are very grateful to the staff of the Cambridge Clinical Microbiology and Public Health Laboratory including the laboratory managers, biomedical scientists, and all staff involved in SARS-CoV-2 testing for their dedicated work. This work was undertaken in a regional laboratory and should not be taken as a recommendation from PHE.
References
- 1.Skittrall J.P., Wilson M., Smielewska A.A., Parmar S., Fortune M.D., Sparkes D., et al. Specificity and positive predictive value of SARS-CoV-2 nucleic acid amplification testing in a low prevalence setting. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GOV.UK, Public Health England, Coronavirus (COVID-19) in the UK [Internet]. Updated 14/12/2020 [cited 15/12/2020]. Available from: https://coronavirus.data.gov.uk/testing.
- 3.Surkova E., Nikolayevskyy V., Drobniewski F. False-positive COVID-19 results: hidden problems and costs. Lancet Respir. Med. 2020 doi: 10.1016/s2213-2600(20)30453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trémeaux P., Lhomme S., Abravanel F., Raymond S., Mengelle C., Mansuy J.M., et al. Evaluation of the Aptima™ transcription-mediated amplification assay (Hologic®) for detecting SARS-CoV-2 in clinical specimens. J. Clin. Virol. 2020:129. doi: 10.1016/j.jcv.2020.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorzalski A.J., Tian H., Laverdure C., Morzunov S., Verma S.C., VanHooser S., et al. High-Throughput Transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J. Clin. Virol. 2020:129. doi: 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudlow C., Diggle P., Warlow O., Seymour D., Gordon B., Walker R., et al. Testing for coronavirus (SARS-CoV-2) infection in populations with low infection prevalence: the largely ignored problem of false positives and the value of repeat testing. MedRxiv. 2020 [Google Scholar]
- 7.Watson J., Whiting P.F., Brush J.E. Interpreting a covid-19 test result. BMJ. 2020:369. doi: 10.1136/bmj.m1808. [DOI] [PubMed] [Google Scholar]
- 8.Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S., et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020:323. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GOV.UK, Public Health England Research and Analysis, SARS-CoV-2 RNA testing: assurance of positive results during periods of low prevalence [Internet]. Updated 16/10/2020 [cited 15/12/2020]. Available from: https://www.gov.uk/government/publications/sars-cov-2-rna-testing-assurance-of-positive-results-during-periods-of-low-prevalence.