Abstract
Biometric monitoring technologies (BioMeTs) are becoming increasingly common to aid data collection in clinical trials and practice. The state of BioMeTs, and associated digitally measured biomarkers, is highly reminiscent of the field of laboratory biomarkers 2 decades ago. In this review, we have summarized and leveraged historical perspectives, and lessons learned from laboratory biomarkers as they apply to BioMeTs. Both categories share common features, including goals and roles in biomedical research, definitions, and many elements of the biomarker qualification framework. They can also be classified based on the underlying technology, each with distinct features and performance characteristics, which require bench and human experimentation testing phases. In contrast to laboratory biomarkers, digitally measured biomarkers require prospective data collection for purposes of analytical validation in human subjects, lack well‐established and widely accepted performance characteristics, require human factor testing, and, for many applications, access to raw (sample‐level) data. Novel methods to handle large volumes of data, as well as security and data rights requirements add to the complexity of this emerging field. Our review highlights the need for a common framework with appropriate vocabulary and standardized approaches to evaluate digitally measured biomarkers, including defining performance characteristics and acceptance criteria. Additionally, the need for human factor testing drives early patient engagement during technology development. Finally, use of BioMeTs requires a relatively high degree of technology literacy among both study participants and healthcare professionals. Transparency of data generation and the need for novel analytical and statistical tools creates opportunities for precompetitive collaborations.
Measure what is measurable and make measurable what is not so. Galileo Galilei (1564–1642).
What is a biomarker? The word itself is an abbreviation of “biological marker,” which has been used in peer‐reviewed biomedical journals since the 1940 and 1950s in various contexts. 1 , 2 The abbreviated term “biomarker” was coined in 1980. 3 The Biomarkers, Endpoints, and other Tools (BEST) glossary defines a biomarker as a “defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions.” 4 This definition has been widely accepted since the National Institutes of Health Biomarkers Definitions Working Group defined the term in 1998. 5
Traditionally, there have been two major categories of biomarkers: laboratory and imaging‐based biomarkers. Recently, a new category has emerged: digitally measured biomarkers. 6 In this review, we examine the historical underpinnings of laboratory biomarkers and how they have influenced digitally measured biomarker development. To do so, we define laboratory biomarkers as measures produced by laboratory assays and digitally measured biomarkers as measures produced by biometric monitoring technologies (BioMeTs), 7 which are “connected digital medicine products that process data captured by mobile sensors using algorithms to generate measures of behavioral and/or physiological function” that may ultimately result in the identification and deployment of digitally measured biomarkers. Just like laboratory assays do not necessarily produce a biomarker, digital measures derived from BioMeTs are not digitally measured biomarkers by definition. There are multiple types of BioMeTs that can be used either alone or in concert to create behavioral or physiological end points (Figure 1 ). Currently, signal modalities that can be measured by BioMeTs include but are not limited to motion, pressure, biopotentials, skin impedance, light, and temperature. We limit the scope of our review to mobile sensors in direct contact with the body but contactless technologies that measure biometric signals, such as camera, WiFi, and Doppler radar‐based devices, or digitally captured data by imaging devices used in the hospitals, are out of scope because of fundamental differences in the means of data collection. BioMeTs come in a variety of form factors, such as watches, adhesive patches, headbands, rings, and clothing, which form the basis for the development of digital end points. As new technologies are developed, the abilities of BioMeTs will continue to expand.
Figure 1.
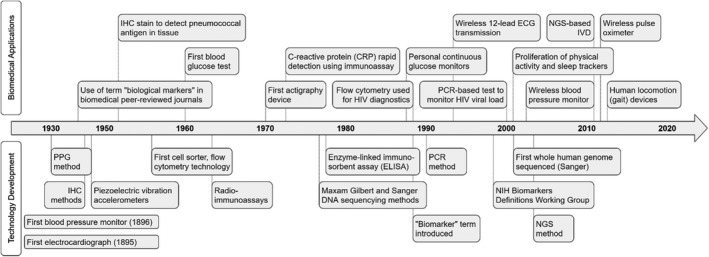
Timeline comparing a number of major technology developments (bottom) underlying laboratory biomarker assays and present‐day biometric monitoring technologies (BioMeTs) and examples of major biomedical applications based on those technology developments (top). ECG, echocardiogram; IHC, immunohistochemistry; IVD, in vitro diagnostics; NGS, next‐generation sequencing; NIH, National Institutes of Health; PCR, polymerase chain reaction; PPG, postprandial glucose.
Digitally measured biomarkers, being newcomers, have not yet gained the same degree of acceptance and name recognition in the biomedical field as laboratory or imaging‐based biomarkers for several reasons. First, the field is inundated with technology applications collecting various types of health‐related data of highly variable and often unknown quality. 8 Second, the field lacks commonly accepted language, frameworks, data standards, and methodologies to determine whether a certain application is ready and appropriate for use in human experimentation. Frameworks 9 , 10 and recommendations 11 have been proposed to answer this question and facilitate adoption of digitally measured biomarkers as tools for drug development and health care delivery; however, the field remains fragmented as it involves an interdisciplinary intersection of device engineering, software development, data science, ethical and regulatory bodies, clinicians, academic researchers, drug development companies, and others. Lack of consensus in the field leads to a high number of feasibility/pilot experiments, sometimes referred to as “pilotitis,” many of which yield inconclusive results and trigger further experiments. Some results are published 12 ; however, others are not disclosed, especially if the outcomes are negative. 13 A more concerted effort is needed from all stakeholders to establish BioMeT performance characteristics, human factor testing, and clinical interpretation for novel measurements. Moreover, adoption of new technologies depends on a large number of opinions and perspectives from patients, clinicians, regulators, and scientists. This necessitates that all groups be involved in the development and implementation of the approach in order to reach the user and the ultimate “goal” of the technology. 14
Laboratory biomarker foundations
The state of the digitally measured biomarkers field and its challenges are highly reminiscent of the field of laboratory biomarkers 15 2 decades ago. For example, the terms “biomarker” and “technology category” (e.g., enzyme‐linked immunosorbent assay, reverse transcription polymerase chain reaction, or immunohistochemistry), have been commonly used for laboratory methods (Figure 1 ), but widespread adoption of biomarkers as tools in drug development was not realized until unified definitions were created 5 and general principles of biomarker assay validation for the purposes of human experimentation were established. 16 Publication of the work by the National Institutes of Health (NIH) Biomarker Definitions Working Group was a turning point; it defined biomarker categories (diagnostic, disease staging, prognostic, or predictive) and also highlighted the role of biomarkers as surrogate end points, 5 creating a necessary framework for adoption of biomarkers in clinical trials. Importantly, that group defined the term “validation” as a “performance characteristics (i.e., sensitivity, specificity, and reproducibility) of a measurement or an assay technique.” The concept of validation was subsequently split into “analytical validation,” referring to biomarker performance characteristics and “clinical validation,” referring to association of a method readout to an outcome of interest. 5 This important work was complemented by the American Association of Pharmaceutical Sciences (AAPS) 2003 Biomarker Workshop, which developed the “fit for purpose” concept. The workshop used ligand binding assays as a use case to overcome the issue of “limited experience interpreting biomarker data and an unclear regulatory climate” 16 by defining assay performance characteristics and acceptance criteria to determine if a biomarker is fit for the purpose of supporting a certain end point in a clinical trial. In turn, those performance characteristics were based on those defined in the 2001 US Food and Drug Administration (FDA) Bioanalytical Method Validation pharmacokinetic assessments for conventional small molecule drugs.
The effort of the AAPS working group prompted expansion of the “fit for purpose” concept beyond ligand binding assays, leading to defined assay performance characteristics for other technologies, including immunohistochemistry, reverse transcription polymerase chain reaction, and mass spectrometry. There was also recognition of a need for a defined set of characteristics and stringency of performance verification to render a certain biomarker method valid for a specific purpose in clinical development. The burden of proof required for a particular biomarker to be considered as “valid” is based on the predefined purpose(s) and also tailored to its purpose distinguishing between pharmacodynamic, proof of mechanism, proof of concept, surrogate end points, and prognostic/predictive biomarkers. 17 The technologies were classified into bins (qualitative, quasi‐quantitative, and relative and definitive quantitative) and applicable assay characteristics were ascribed to a particular technology, revealing limitations of certain methods in terms of ability to quantify the amount of analyte in any given specimen. 17
Both concepts—analytical and clinical validation—were incorporated in the FDA guidance “Biomarker Qualification: evidentiary guidance for industry and FDA staff” 18 describing an evidentiary framework for all biomarker qualification submissions, regardless of the type of biomarker or context of use (COU). For the purpose of biomarker qualification, the framework defined analytical validation as “establishing that the analytical performance characteristics of a biomarker test, such as the accuracy and reproducibility, are acceptable for the proposed COU in drug development,” highlighting the difference with the biomarker’s usefulness. It defined three key elements for a biomarker method: (1) source or materials for measurement, (2) an assay for obtaining the measurement, and (3) methods and/or criteria for interpreting those measurements, 19 bringing together major advancements of the field in the last 2 decades.
With careful assay development supported by these guidance documents, the use of laboratory biomarkers in clinical development has become common across many therapeutic areas. Today, the procedure of developing and transferring biomarker assays for utilization in clinical trials in partnership with contract‐research laboratories, in addition to already existing validated assays, is routine. Most, if not all, clinical trial professionals would agree that laboratory biomarkers have made clinical development programs more efficient and more robust. 20
Extension to sensor technologies
Sensor‐based technologies share some historical trends with laboratory biomarker assays. BioMeTs have been available in the ecosystem for many years, but the appreciation of their potential and widespread interest started emerging only in the last decade. Sensor technologies, such as wireless echocardiogram (ECG), pulse oximetry, and accelerometers embedded in wearable devices, have existed for extended periods of time (Figure 1 ). However, a noticeable interest in and uptake of remote patient monitoring for healthcare management and clinical trials did not happen until smartphones and computer tablets became ubiquitously equipped with applications mediating BioMeT data synchronization and real‐time collection of patient responses about their disease conditions and quality of life incorporated into electronic patient reported outcome and e‐diaries. 21 Moreover, a number of digital wellness devices were directed toward consumers, offering a convenient combination of vital signs, physical activity, and estimated sleep data in a single device. These devices are user‐friendly, have an attractive form factor, and provide an easy way to digest data summaries in mobile applications, thus increasing the visibility and acceptance of BioMeT‐like technologies among users and study participants. The number of smartphone applications capturing different aspects of health grew exponentially, raising legitimate concerns about app quality, 22 security, user privacy, and data governance. 23 Unlike the laboratory biomarker field, the digitally measured biomarker field continues to be “characterized by irrational exuberance and excessive hype” 24 because of the lack of a commonly accepted framework for designing validation experiments and practical recommendations to interpret the validation data, similar to the work done by the NIH biomarker working group and AAPS biomarker working groups. Some elements outlined in the BEST framework and “Biomarker Qualification: evidentiary guidance for industry and FDA staff” may not apply because both were designed for laboratory and imaging 25 biomarkers, rather than digital. As of May 2020, the list of qualified biomarkers or pending submissions did not include any digitally measured biomarkers. 26 , 27
In 2017, the Clinical Trials Transformation Initiative mobile technology project 11 drafted the first relevant framework for clinical research using BioMeTs (termed digital technologies in the recommendation itself), which was finalized in 2018. This documentation adopted the verification and validation processes from the FDA guidance for software validation, 25 and provided recommendations on some experimental and operational aspects, such as device choice, data management, and statistical analysis that need to be considered for BioMeT use in clinical trials. Recently, the V3 framework was introduced to consolidate these efforts into a unified definition of verification, analytical validation, and clinical validation (V3) to determine if a BioMeT and/or digitally measured biomarker is fit‐for‐purpose. 7 Here, we summarize and leverage historical perspectives and lessons learned from laboratory biomarkers while considering recent advances in BioMeTs to inform the development and use of digitally measured biomarkers. Although differences between conventional laboratory biomarkers and digitally measured biomarkers are numerous, including the data transmission mode and associated security and data right concerns, we focus here on elements that need to be addressed as a part of the V3 process (Figure 2 ), to leverage experiences from laboratory biomarkers.
Figure 2.
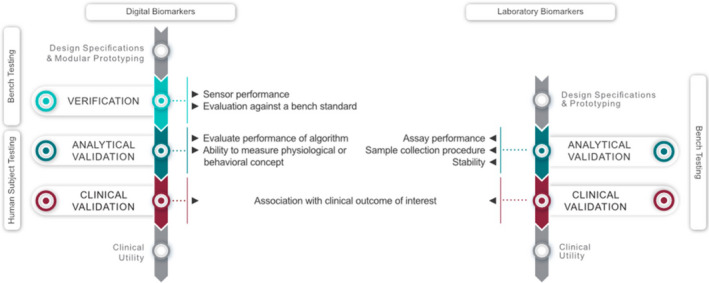
The V3 framework as applied to digital (left) and laboratory (right) biomarkers. Laboratory biomarkers go through an analytical and clinical validation step as defined in the Biomarkers, Endpoints, and other Tools framework. 95 Digitally measured biomarkers are derived from sensor technology (BioMeT) that needs to undergo verification, before the physiological or behavioral measures of interest can be analytically and clinically validated. 7 Whereas laboratory biomarkers can go through the process based on bench testing, digitally measured biomarkers are highly reliant on human subject testing. Figure adapted from ref. 7 with permission.
A COMPARISON OF DIGITALLY MEASURED BIOMARKERS WITH LABORATORY BIOMARKERS
Digitally measured and conventional laboratory biomarkers share a common foundation. First, most of the BEST definitions and the framework outlined in the FDA guidance on biomarker qualification, 26 such as COU, apply to digitally measured biomarkers (Table 1 ) as they share the same goals and roles in biomedical research. 28 The FDA has clarified that digitally measured biomarkers are not a separate class of biomarkers. 29 Second, both laboratory and digitally measured biomarkers can be classified into a number of technologies with distinct features and performance characteristics. Third, validation of a specific laboratory assay or BioMeT entails both bench and human experimentation testing phases, albeit these are not identical, and the terminology varies (Table 1 ). Fundamentally, both categories represent measurements capturing the features of biological processes, although the technological approaches for producing measurements are different.
Table 1.
Comparison of laboratory assay‐based biomarkers and BioMeT characteristics
| Comparison parameters | Laboratory biomarker assays | Digitally measured biomarkers derived from BioMeTs |
|---|---|---|
| Goals and role in biomedical research |
The BEST framework defines several biomarker types: 1) diagnostic, 2) monitoring of symptoms or disease progression, 3) pharmacodynamic/response, 4) predictive, 5) prognostic, 6) safety, and 7) susceptibility/risk biomarker. Examples of COU in drug development:
|
|
| Bench testing phase | Analytical validation ‐ establishing that the analytical performance characteristics of a biomarker test, such as the accuracy and reproducibility, are acceptable for the proposed COU in drug development 96 | Verification ‐ Evaluating and demonstrating the performance of a sensor technology within a BioMeT, and the sample‐level data it generates, against a prespecified set of criteria |
| Human subject testing phase |
Clinical validation ‐ establishes a biomarker’s relationship with the outcome of interest and confirms that it is acceptable for the proposed COU Clinical validation example: HbA1c could be used in drug development as a well‐validated surrogate for the short‐term clinical consequences of elevated glucose levels and long‐term vascular complications of diabetes mellitus |
Analytical validation ‐ Evaluating and demonstrating the performance of the algorithm, and the ability of this component of a BioMeT to measure, detect, or predict physiological or behavioral metrics Analytical Validation Example: Comparison of energy expenditure estimated by a fitness tracker against doubly labeled water. Clinical validation is the process that evaluates whether the digitally measured biomarker acceptably identifies, measures, or predicts a meaningful clinical, biological, physical, functional state, or experience Clinical validation example: 95th percentile of stride velocity can be used in DMD as a biomarker of disease response to treatment 78 |
| Stability over time | Example: Defining analyte stability in a defined type of specimen over a certain period of time under specified storage conditions | Example: Static and dynamic recalibration of an inertial measurement unit component inside a BioMeT to account for possible axis misalignment or inertial sensor alterations because of damage (e.g., device dropped) |
| Human factor testing | N/A ‐ other than defining the risk associated with sample collection procedure | Evaluation of human interaction with the BioMeT related to configuration, calibration, instructions, maintenance, user interface, and data synchronization |
| Data structure | Snapshot in time | Continuous or frequent (e.g., daily) data collection for extended periods of time |
| Product system, data, and network security | Vulnerabilities exist as in many cases a laboratory equipment involved in data generation is internet connected | BioMeTs transfer data over the internet, which introduces risks as actors could attack or assess products remotely and often in near‐real time. Security issues will need to be re‐assessed regularly as new technologies and vulnerabilities are identified |
| Data rights and governance | Data security and privacy protection requirements apply, but given the restricted scope of data sharing in the process of data generation, the vulnerabilities are limited | Often collect sensitive data. The ability of BioMeTs to collect and integrate multimodal data can lead to the unconsented identification or localization of an individual. Data rights and governance concerns should be pivotal to the BioMeT development and deployment. |
BEST, Biomarkers, Endpoints, and other Tools; BioMeT, biometric monitoring technology; COU, context of use; DMD, Duchenne Muscular Dystrophy; HbA1c, hemoglobin A1c; MOA, mechanism of action; N/A, not applicable.
The difference between these two categories are related to the fact that the methods of capturing the data have some essential differences. Unlike laboratory biomarker assays, analytical validation to ensure a BioMeT sensor correctly captures a physiological concept of interest typically requires prospective data collection from human subjects. Analytical validation of a biomarker assay can be done entirely in the laboratory with retrospectively collected human samples as long as the specimens were collected following informed consent and stored appropriately. Another key difference is that the laboratory biomarker assay characteristics that need to be established for a specific technology are relatively well‐defined (Table 2 ), whereas this concept is more variable for BioMeTs, not in the least because the sensor technologies used in BioMeTs are developing and proliferating rapidly. Here, we discuss BioMeTs and physiological or behavioral measures derived from them in the context of the sensor technology they rely on (Table 3 ). The examples in Table 2 and Table 3 highlight the differences in laboratory assay and BioMeT performance criteria. Although there are guidelines for establishing the accuracy for some categories of devices, such as blood pressure monitoring devices, 30 the acceptance of other sensor types is less well‐defined or not defined at all. For example, one problematic category of sensors is accelerometers. There is a lot of variability in technological solutions as well as physiological concepts measured, ranging from gait and balance assessments to pronation and supination and hand tremor. 31 , 32 , 33 Very often, analytical validation is performed by comparing to a “gold standard” in the field. For gait, this can include comparison to electronic walkways, which may capture the same end point and are also based on sensor recordings, yet walkways collect intermittent force/pressure sensor recordings, compared with continuous tracking of the body through space 34 with a BioMeT. Elsewhere, a comparison to a conventional standard, which is not based on an objective measurement by a sensor, but an established rating scale can be difficult, and yet essential for advancing the field of BioMeT‐based clinical research. Despite these potential limitations, rating scales are routinely used as the accepted de facto standard. An added complexity is that some of the scales, while asking specific questions, are actually not looking at a simple physiological end point in isolation. A simple example includes the use of a self‐reported physical activity diary compared with accelerometry over many days. 35 This study found poor convergent validity between the diary and a waist‐mounted commercial device, which was attributed to an overestimation of the intensity and duration of different types of activities recalled subjectively by the study cohort. However, study authors also noted that the accelerometers used in their study underestimated intensity and duration of certain types of activities because the commercial model was less sensitive to registering activities, such as walking up the stairs, cycling, and activities that mainly involve arm movements depending on wear location. For the given example, a more like‐for‐like result comparison between the accelerometer and indirect calorimetry (the reference standard in this scenario) or video may have shown greater utility of the accelerometer to quantify physical activity in the chosen cohort. Thus, scientific rigor pertaining to defining appropriate analytical validation parameters and appropriate reference/comparison tools (given a specific physiological construct of interest) is paramount, but unfortunately, is often lacking for BioMeTs.
Table 2.
Laboratory biomarker classification for assay validation characteristics purposes 17 (provides examples, not all‐inclusive list)
| Analytical technology category (example) | Assay characteristics | Assay controls and requirements |
|---|---|---|
| Definitive assay example (mass spectrometry) | Accuracy, trueness (bias), precision, sensitivity, LLOQ; ULOQ, specificity, dilution linearity, parallelism, assay range | Requires calibrators and uses a regression model to calculate absolute values for sample with an unknown amount of analyte |
| Relative quantitative assay example: LBA | Precision, trueness (bias) reproducibility, sensitivity, LLOQ; ULOQ, specificity, dilution linearity, parallelism, and assay range | Requires a standard curve and low/medium/high controls; quantitation is relative not absolute |
| Quasi‐quantitative assay (flow cytometry) | Precision, sensitivity, specificity, and assay range | No calibration standard, a continuous response as a characteristic of a test sample |
| Qualitative assay (IHC) | Reproducibility, sensitivity, and specificity | Discrete scoring scales or binary outcome (yes/no) |
IHC, immunohistochemistry; LBA, ligand‐binding assay; LLOQ, lower limit of quantification; ULOQ, upper limit of quantification.
Table 3.
Examples of organizing BioMeTs into categories based on signal modalities to establish common performance characteristics (provides examples, not all‐inclusive list)
| Physiological concept | Sensor | Verification | Analytical Validation |
|---|---|---|---|
| Step count | Accelerometer | Accuracy, precision, reliability of raw acceleration data by means of a shake table moving with known frequency and amplitude | Comparison of a step count data produced by an algorithm to a human rater counting steps |
| Blood pressure | Pressure sensor embedded in an inflatable air‐bladder cuff | Accuracy, precision, and reliability of pneumatic leakage, pressure transducer accuracy, and cuff durability | Comparison to an auscultatory standard or intra‐arterial blood pressure measurement with a predefined sample size with established validation criteria and reporting 97 |
| HR by ECG method | Electrode | ECG: inputting a sine wave with known frequency and amplitude and measuring how closely the device reproduces this known signal; HR: comparing the performance of a new HR algorithm on ECG databases with known and validated feature labels as specified in the relevant international standards | Comparison of HR to a previously analytically validated heart rate monitor |
| SpO2 (measuring oxygenated and deoxygenated hemoglobin) | Light source and detector | Inputting a known optical signal and measuring how closely the device reproduces this signal | Comparison of pulse oximeter values (SpO2) against arterial blood samples (SaO2). |
| Body temperature: | Thermistor | Comparison to a probe under defined range of temperature | Comparison of a number of sequential measurements under defined conditions to a temperature measured in a specific location 98 |
BioMeTs, biometric monitoring technologies; ECG, echocardiogram; HR, heart rate.
Human factor testing
BioMeTs require human factor and usability testing, 36 which needs to be incorporated judiciously into the evaluation process. This is one of the major distinctions with laboratory biomarkers—the subject interface is limited to the procedure or biological sample collection (Table 1 ). For BioMeTs, key components include completion of setup and configuration, calibration if required, using the device according to provided instructions, and/or performing maintenance, such as cleaning, charging, or replacing a battery. 36 Moreover, technical aspects of usability, such as the user interface, battery life, and data synchronization, should be tested and addressed appropriately, with patients and patient advocacy groups intimately involved as early as possible in the design process. BioMeTs used in clinical trials can be off‐the‐shelf (already existing and marketed solutions) or devices under development. For devices under development, usability testing can be completed at the end of the design phase, as required for marketed devices. 37 For already marketed BioMeTs, some components of usability testing should preferably be done in the first study involving human subjects to have an early understanding of usability issues, and to correct them if possible, before deployment in other human studies. Usability studies may need to be repeated if the COU changes; for example, an initial test in normal healthy volunteers with subsequent testing in a disease population. The following factors should be considered: disease characteristics, demographics, symptoms and known disease heterogeneity, and the scientific question(s) being addressed. This approach will ensure that the use of such BioMeTs will not impact the disease condition in a negative way (e.g., lead to skin irritation in a dermatological condition). A particular BioMeT of interest may not be “fit‐for‐purpose” if it fails usability testing, meaning that study participants are unable to generate valid data and/or adherence is suboptimal. Moreover, user literacy with digital sensors and corresponding mobile applications is critical for successful device deployment and data collection. The user facing materials, which may include manuals and hands‐on‐training for both clinical study participants and site staff, are a must have. 38
Data structure and the need for raw data
The other distinctive feature of digitally measured biomarkers is the multilayered data structure required for V3 evaluation, 7 interpretation, and reproducibility of results. 38 , 39 Data generated from BioMeTs can be classified into two categories: (i) raw data, also referred to as sample‐level data, which are recorded directly by a sensor and output with little if any further processing, and (ii) processed data, which have undergone analysis by an algorithm/software into different units, often summary statistics, such as the total number of steps or activity counts that can be aggregated at different time resolutions (e.g., minutes, hours, or days).
Access to sample‐level data is required for verification purposes, as the sensor output needs to be compared with that of a bench standard, such as a shake table for accelerometers 40 (Table 3 ). A review of raw data may also be required for particular applications (e.g., a review by a human reader to adjudicate if a certain cardiovascular event has taken place by examining an ECG waveform. 39 Availability of sample‐level data also offers the potential for re‐analysis as new or modified algorithms are developed.
There are challenges in obtaining sample‐level data from certain devices due to a number of reasons, including both technical and commercial. Technical reasons include operational difficulties due to the high volumes generated that often exceed the memory capacity on‐board. For example, tri‐axial accelerometry data sampled at 100 Hz creates 18,000 data points/minute and, therefore, firmware might compress those data into a less complex signal of 20 Hz while retaining the original units. Many actigraphy devices have raw acceleration data immediately processed into “activity counts,” resulting in a time series of much lower frequency, such as one for 30 seconds. In this scenario, sample‐level data are never accessible to the manufacturer or any user, as it is never written to memory. However, as memory capacity exponentially increases, it has become more common to collect and store raw high‐frequency data, 41 albeit with a trade‐off against battery life.
The advantage of processed data is that it allows a digital tool to collect information over very long time periods in the field—in some cases, this could be as long as several months. Depending on the mode of data transmission, another potential advantage is that a lower sampled signal can be sent via Bluetooth, whereas a high sampled signal might overwhelm this kind of transmission. The major drawback of this approach is that the units of processed data are usually specific to each manufacturer, such that the activity count of company A is not the same as the activity count of company B. As such, it is not possible to apply the same algorithm to data collected from different devices, and even if multiple algorithms are developed, their outputs may not be directly comparable despite purporting to measure the same construct, such as the number of steps in a given day. Thus, there is a need for harmonizing the units of the processed data across BioMeT platforms and standardizing derived summaries to facilitate more comprehensive cross‐study comparison and compatibility. Another potential drawback of processing data is that doing so may remove behavioral features of interest, such as gait parameters estimated from sub‐second raw data, 42 , 43 , 44 which could be clinically meaningful. Although complete harmonization and transparency of data processing and associated algorithms might be beneficial to the end user, manufacturers often prefer to enable protection and control over intellectual property thereby preventing competitor reverse‐engineered alternatives. Moreover, it lessens the technical know‐how required by more clinically oriented users who wish to avoid the burden of trying to store and interpret raw data.
Handling large amounts of data
BioMeTs are capable of generating an ever‐increasing amount of data, which requires retrospective or (near) real‐time processing. Unlike laboratory biomarkers that are usually sampled individually at predefined time points, BioMeT data are typically sampled at high rates, are often multimodal, and collected under ambulatory conditions leading to more complex and unpredictable environmental influences, thus raising new challenges for data analysis. A typical data processing workflow consists of 1) preprocessing steps (e.g., filtering of an ECG signal to remove unwanted artifacts), 2) analysis (e.g., calculation of heart rate (HR) from the preprocessed ECG), 3) presentation of the results (e.g., plotting of the minute‐by‐minute averaged HR over prolonged periods of time), and 4) transforming the continuous HR signal into a study end point for statistical analysis. The field of signal processing and assorted statistical techniques have built up a rich catalogue of tools to help “lift the signal from the noise” for this type of data. There is, however, a big gap between the complexity of data collected with BioMeTs and the available statistical methodology to leverage the richness of the data fully, and to extract the digitally measured biomarkers that are most sensitive to quantify safety, efficacy, or treatment effects. Analytic challenges are centered around the complexity of BioMeT data that are longitudinal, have different time scales (e.g., sub‐second level for accelerometers and minute‐level for continuous glucose monitoring (CGM)), exhibit huge intersubject and intrasubject heterogeneity across days and weeks of observation, 45 and which may follow daily, weekly, and/or seasonal cycles. 46 This is further complicated by the fact that many BioMeTs are capable of collecting multimodal data under uncontrolled real‐world conditions (e.g., simultaneously collected heart rate, accelerometry, and CGM in closed‐loop systems). Some of these analytical challenges have been actively addressed through the recent development of functional data approaches 47 , 48 providing deeper insights into the diurnal organization of composite biomarkers of physical activity, sleep, and circadian rhythms. Open‐source platforms for rapid dissemination of reproducible software, such as the GGIR package in R 49 and the Open Wearables Initiative, 50 help promote the development of standardized algorithms and digitally measured biomarkers.
Considering the heterogeneity and the granularity needed to investigate subtle signal changes and their correlation to disease onset and progression, traditional rule‐based approaches for data handling, manipulation, and synthesis can be slow to develop, complex to maintain, and sensitive to environmental influences. More recently, machine learning (ML) is gaining popularity as a complementary set of tools to build analytical models by letting them learn from large amounts of data, potentially alleviating some of these shortcomings. The general concept of ML is that if the programmer uses the right data to train the ML model for the problem under consideration, it will then be able to predict (“infer”) the outcome for that problem on new data. The initial challenge is to ensure the training data offers appropriate context and enough variability, ensuring a broad enough spectrum of normative data as well as incidences of interest (i.e., anomalies) that the ML methodology can adequately interpret. A subsequent challenge is that those ML‐derived insights are corroborated by suitable reference standards, offering an overall degree of trust to any ML‐based outcomes and decisions.
To date, approaches for the use of ML in healthcare remain cautious, primarily due to the fact that ML algorithms are often described as “black boxes” to signify that the user knows what goes in and what comes out, but is blinded to the specific ML functionality. In a way, this interpretation is understandable, as the trained ML model usually does not have readable source code in the typical sense. Although the ML model may generate verifiable outputs, an understanding of how it came to that conclusion can be difficult, sometimes even impossible, to come by. This can create uncertainty and mistrust, especially when considering something as sensitive as medical decision making. Additionally, there are a number of other potential pitfalls, especially when using deep learning: 1) the more complex the model topology, the more (labeled) training data is needed to create it, which can be difficult to acquire, 2) monolithic models are harder to verify and validate as intermediate steps are obscured, 3) the model is assumed to “learn” from scratch, ignoring relevant domain (tacit) knowledge, and 4) many medically relevant (and correctly identified) events are rare, creating so‐called unbalanced datasets. The first successful medical applications of ML are primarily situated in medical imaging 51 , 52 and histopathological screening, 53 , 54 especially for malignancies. But commonly used approaches, such as convolutional neural networks, have been successfully applied to BioMeT data. 55
Typically, ML approaches have been created by siloed teams of data scientists focusing on technological and technical development rather than ensuring transparency and translation (pragmatic implementation), which would support acceptance in the wider field. The latter can only be successful when considering multidisciplinary approaches— data scientists working with healthcare professionals to understand clinical limitations, as well as data scientists with human biology or medical backgrounds who intrinsically understand the problems they are trying to solve, formulating targeted approaches that are adequately validated, described, and open. Such approaches are being adopted by major European academic studies that are currently underway to utilize real‐world, free‐living data from individuals with conditions, such as neurodegenerative movement disorders and immune‐mediated inflammatory diseases. 56 , 57 Where ML has notable historical roots in the identification of abnormalities for cancer screening, the same approaches have been used to investigate useful markers from BioMeT data. 58 , 59
Once the challenges outlined above are successfully addressed, BioMeTs will facilitate establishing nuanced patient‐level normative values for physiological measures, such as HR and blood glucose level, and behavioral measures, such as the estimated daily number of steps and sleep duration. This will 1) help with more accurate estimation of patient‐level treatment interventions effects and allow for near real‐time tracking of recovery or functional decline, and 2) allow the development of individualized clinical risk prediction models to identify subjects with an increased risk for adverse health‐related events, such as falls in elderly, re‐admissions in different clinical subgroups, or relapse in multiple sclerosis.
Security concerns
Most BioMeTs transfer data over the internet, which introduces security‐related risks because an actor could attack and access the product remotely and often in near‐real time. 9 When using BioMeTs, responsible parties will need to protect the internet‐connected systems, data, and networks from unauthorized access and attacks, including human error, such as sending personal health information to an unapproved party. Additional concerns include potential unbinding in clinical trials, which should be considered carefully during the study design. Measures to ensure safe and secure systems include but are not limited to using principles of “safety by design,” collaborating with third parties when a vulnerability has been discovered, building products that capture evidence of tampering, and updating systems to decrease the risk of harm from code flaws or other issues. 60 Over the past few years, regulatory agencies, like the FDA, have issued a number of guidance documents for both premarket and postmarket security considerations for connected medical devices. 61 , 62 A BioMeT’s security risk will need to be continuously re‐assessed as new technologies and vulnerabilities are discovered. 9
Data rights and governance concerns
When considering the data rights from BioMeTs, the industry has started to coalesce around the term “governance” rather than “privacy,” as more individuals want to be empowered to choose how to share their data (e.g., for rare disease research), rather than defaulting to privacy and a lack of ability to choose to share. 9 As BioMeTs have one or more mobile components, are usually developed to be used outside the controlled environments typical for laboratory biomarkers, and often collect personal and/or sensitive data, data rights and governance should be pivotal to their development. Although exact definitions vary by country, personal data are generally considered to be directly or indirectly identifiable, whereas sensitive data covers many categories but includes health data, such as diagnoses, medical test results, and prescriptions. The ability of BioMeTs to collect and integrate multimodal data can lead to the unconsented identification or localization of an individual, 63 even though that may not be the intent of the investigator. For example, if an investigator is interested in using a smartphone application to generate a BioMeT of interest, they must be aware for the potential of an application to access geolocation data. In the European Union, the General Data Protection Regulation (GDPR) has set the stage for improved transparency, privacy protection, and portability of personal (health) data. In the United States, the more narrowly scoped Health Insurance Portability and Accountability Act (HIPAA) defines similar responsibilities when handling personal health information. Providers of BioMeTs have a responsibility to inform their users of the potential risks and the steps taken to mitigate those risks. We will not attempt to draw extensive parallels with the field of laboratory biomarkers as BioMeTs and digitally measured biomarkers have unique and much more extensive requirements that warrant a discussion of their own.
CASE STUDIES
Below, we consider examples of BioMeTs used or intended to be used in drug development across three areas: (1) next‐generation sequencing (NGS) based on in vitro diagnostics (IVD), (2) BioMeTs used to support clinical trials and labeling claims, and (3) hybrid solutions, combining laboratory and BioMeT features. This will highlight the challenges of qualifying these BioMeTs for clinical research while comparing and contrasting them with conventional laboratory biomarkers.
NGS‐based IVD
NGS‐based IVD tests provide an interesting case of how definitions, prior experience with genetic tests, and examples of test validation provided sufficient information to develop laboratory biomarker tests using a novel technology platform. This is because these tests use a novel, unprecedented technology platform while leveraging existing framework and definitions. 64 IVDs are laboratory tests performed on samples, such as blood or tissue that have been taken from the human body. To be marketed legally in the United States, they need to go through a clearance or approval process at the FDA. Many IVDs are used in clinical trials, often as companion diagnostics or their prototypes, to determine clinical trial eligibility criteria or to determine who should get a treatment. 65 The NGS‐based tests were radically different from other genomic‐based tests—they provided an opportunity to interrogate multiple genetic changes from very small amounts of DNA material derived from tumor samples in a timeframe compatible with clinical trial timelines. In 2017, after the approval of three different tumor profiling NGS‐based IVD devices, the FDA published a document highlighting the approach taken for the regulation of these devices and the analytical and clinical evidence used to support claims based on NGS‐based biomarkers. Yet, this work took several years, whereas prior to these approvals, a comprehensive validation study was performed to provide an example of assay performance characteristics and acceptance criteria. 66 Platform validation performance characteristics were established using DNA derived from a wide range of tissue types, including tissue types associated with CDx indications. One of the approved NGS tests is FoundationOneCDx (F1CDx), designed to detect genetic mutations in 324 genes and 2 genomic signatures in tumor tissue.
The challenges and risks included potential mismanagement of patients resulting from false results of the test. Patients with false‐positive results may undergo treatment with one of the therapies in the intended use statement without clinical benefit and may experience adverse reactions associated with the therapy; conversely, patients with false‐negative results may not be considered for treatment with the indicated therapy. The probable benefits of F1CDx were based on data collected in clinical trials to support the FDA premarket approval of the assay and the clinical benefit of the assay was demonstrated in a retrospective analysis of efficacy and safety data obtained from a randomized, double‐blind, placebo‐controlled study. The FDA has specific guidance on evaluating test performance and analytical test validation, including measuring the accuracy, precision, limit of detection, and specificity. 67 The prerequisites for this framework were: the information in the public domain about test analytical validation, which included validation parameters and assay performance, as well as demonstrated clinical validity based on the results from several clinical studies. Using a similar regulatory approach, in 2017, the FDA approved a cancer treatment based on such a common biomarker rather than the location in the body where the tumor originated. 68
BioMeTs used to support clinical trials and labeling claims
Remote assessment of sleep for drug development trials
Actigraphy has been used to estimate sleep parameters in the research setting for decades 69 , 70 establishing sleep parameters in both healthy populations and specific disease conditions 71 , 72 , 73 , 74 in natural settings for extended periods of time. This approach is less expensive and more accessible than in‐laboratory polysomnography, and is also better suited to monitoring night‐to‐night variability of sleep over long periods. Wrist‐worn actigraphy devices have been tested extensively for usability and have favorable operational characteristics, such as a long battery life, large memory, and extremely low maintenance, allowing noninvasive use over long time periods. Actigraphy device‐based end points have been used successfully to support successful regulatory submissions; for example, Bayer Healthcare used actigraphy methods for primary and secondary end points to expand the indications for Aleve PM. The sponsor performed a multicenter, double‐blind, randomized controlled trial in 712 participants with an advanced circadian phase with postoperative pain. Comparisons of different doses demonstrated statistically significant differences in sleep onset latency of up to 16 minutes, and in wake after sleep onset of up to 70 minutes. 75
Sleep measurements via actigraphy provides one of the best examples of BioMeT analytical validation. Multiple studies have been conducted in different sleep disorders comparing the output from actigraphy devices to polysomnography metrics, driving wide adoption of actigraphy‐based sleep parameters, characterizing them, and highlighting data limitations. One example 76 previously quantified a range of accelerometer‐based outcomes, such as non‐wear time and z‐angle (defined by authors as the dorsal‐ventral direction when the wrist is in the anatomic position) across 5‐second epochs to estimate the sleep period time window and sleep episodes. The work was subsequently used to examine genomewide associations from > 85,000 UK Biobank participants. 77 Ongoing pragmatic challenges include difficulties differentiating naps from daytime sedentary periods, and sleep from motionless wakefulness common in insomnia. 70
Stride velocity 95th centile in Duchenne Muscular Dystrophy
One of the recently published examples of successfully validated BioMeTs is the stride velocity 95th centile as a secondary end point in Duchenne Muscular Dystrophy (DMD), 78 providing a clear example of the roadmap to qualify a motion‐based measure derived from a BioMeT as a clinical end point. The COU is a validated wearable device for continuous monitoring of ambulation in patients with DMD 5 years of age and above. This is the first BioMeT, to our knowledge, approved for use in clinical trials and drug development with substantial information available in the public domain. Those proposing the BioMeT and stride velocity 95th centile (i.e., the applicant) asked for a qualification to support a primary end point as a digital clinical outcome assessment measure and not as a “patient reported outcome,” citing deficiencies currently used in clinical scales in DMD, such as the 6‐minute walk test, North Star Ambulatory Assessment, and four‐stair climb. The applicant provided data from a comprehensive set of experiments, which included biological plausibility, face validity, content validity, accuracy, reliability, concurrent validity, sensitivity to change, and known group discrimination. Nevertheless, only a secondary end point qualification was granted, as regulators cited the additional need for “data on quality of walking, fall, sway, real‐world stairs, time to stand, and correlation with patient well‐being” and longer follow‐up studies. The roadmap to this qualification was pioneering and highlighted a number of the potential pitfalls, namely the lack of a framework to design experiments for qualification for this specific application, clearly defined performance parameters, and acceptance criteria to interpret the data needed to obtain a qualification for use in drug development.
Hybrid solutions, combining laboratory and BioMeT features
Continuous glucose monitors
CGM provides an interesting example of a hybrid between conventional laboratory biomarkers and BioMeTs. Blood glucose (BG) concentration is a well‐established biomarker of disease activity for type 1 and type 2 diabetes mellitus. It is a useful pharmacodynamic biomarker to track glucose concentration over the course of treatment, and is also a safety biomarker for the purposes of drug development in early‐stage clinical trials. 79 For late‐stage drug development, including pivotal trials, glycated hemoglobin is an established surrogate end point that reflects mean glucose levels of the previous 2–3 months. 80 CGMs were developed as minimally invasive or implantable devices to enable longitudinal monitoring in the interstitial fluid to overcome the shortcomings of earlier generation blood glucose monitors that required up to four finger‐stick measurements throughout the day, an approach that often missed hyperglycemic and hypoglycemic events, 81 especially during sleep. CGMs have features of both laboratory biomarker assays and BioMeTs. The laboratory biomarker features include calibration vs. a well‐established gold standard, BG, reference ranges, and reference interval. A number of CGM instruments, designated as medical devices, are used in care management. 82 The most common principle is the measurement of interstitial fluid via the glucose‐oxidase electrochemical reaction 83 with interstitial fluid harvesting every 10 seconds and an average glucose value is recorded every 1–5 minutes 24 hours a day. 84
Although several BG sensing mechanisms have been utilized, analytic validation of the data obtained from the sensors includes quantification of accuracy using primarily the mean absolute relative difference between CGM sensor output and the reference standard highly accurate laboratory instrument, collected in a hospital setting. Notably, accuracy of commercial grade CGMs show a mean absolute relative difference of between 5% and 10% on BG levels (based on an ideal BG range of 70–140 mg/dL). 83 Analytical validation has been obtained on remote monitoring of BG levels, BG level variability, minimum and maximum glycemic values, and number of severe hypoglycemic episodes. 85 BioMeT features include the continuous nature of the data, highly complex usability testing, and the extensive user training required prior to using a CGM device. The challenges are the need of clearly defined end points to be used for drug development 86 and the need of analytical approaches to deal with continuous data, 87 including meta‐data for improved data interpretation (e.g., the influence of exercise or diet). 88
CONCLUSIONS AND RECOMMENDATIONS
User‐friendly, affordable, and scalable BioMeTs are improving our ability to directly impact disease management, especially for chronic conditions, and define a more personalized, inclusive, and preventive modern medicine. Never before have healthcare professionals had access to such high resolution, objective, and, in certain cases, habitual‐based data. Yet, proliferation of many BioMeTs is also driven by the ease in which they have entered the commercial market. This has caused mistrust and skepticism, as quite often data from BioMeTs have been reported as inaccurate or easily manipulated. 89 , 90 , 91 Thus, their obvious medical advantages need to outweigh the technical shortcomings by improving how they are used and regulated. In contrast to IVDs, there is no uniform requirement for analytical validation for all BioMeTs prior to the release to the market in the United States. Moreover, no device is cleared for the purpose of use in a clinical trial. 92 , 93 A more robust regulatory framework is needed and biomarker qualification evidentiary guidance from the FDA is a good first step. In the European Union, various initiatives are underway on national levels to certify BioMeTs so they can be safely used in clinical practice. 94 These quality labels will become increasingly important to generate trust. Here, we draw parallels to the development of laboratory biomarkers and how they could and should inform the current challenges facing BioMeTs and their digitally measured biomarkers.
The development of digitally measured biomarkers is a seemingly straightforward process once the hardware is created. Yet, steps required for complete and thorough verification, analytical validation, and clinical validation 7 (Figure 2 ) is long and complex and should be informed by an agreed set of standards for the type of BioMeT and its context of use, ensuring its fit‐for‐purpose. Although complete transparency is questionable in the commercial sector due to intellectual property concerns, some transparency should be evidenced to ensure minimal black‐box development, and, therefore, confidence and openness in how raw and processed data are acquired. Therefore, we make a number of recommendations for future considerations within the BioMeT and digitally measured biomarker field:
The existing biomarker qualification framework should be conceptually adopted and extended to cover digitally measured biomarkers, including appropriate descriptions as to what digitally measured biomarker tests entail, common vocabulary and standardized approaches to technology validation according to a specific sensor type, and the physiological or behavioral measure of interest.
Leverage experience from laboratory biomarkers to enable appropriate comparison with conventional standards to understand data behavior and limitations. We discussed that the performance criteria for laboratory biomarker assays cannot be translated directly to BioMeT‐derived physiological and behavioral measures. However, as more digitally measured biomarkers are validated according to the V3 framework, we will be able to draw lessons to harmonize their performance criteria across a wide variety of BioMeT signal modalities.
A major difference to laboratory biomarkers is that a BioMeT used to generate a specific digitally measured biomarker needs to interface either directly or indirectly with the patient. Therefore, engaging patients and patient advocacy groups as early as possible in the development process is of the utmost importance to ensure the technology will fit into the patient’s life with minimal disruption and will not negatively impact outcomes. The same holds true for regulatory authorities and health care providers who should be engaged as early as possible to provide input in the design process.
The literacy needs for intended users need to be considered carefully. Health care professionals should be equipped with basic tools to understand the language of digitally measured biomarker development, and to evaluate proposed digitally measured biomarkers for suitability in the clinic.
Transparently disclose, with sufficient level of detail, how raw data are acquired, processed, and digital results are calculated to determine if a BioMeT is fit‐for‐purpose.
Share insights derived from experiments that include a BioMeT in the public domain to move the field forward as a whole.
Develop novel but standardized statistical approaches and open ML tools to analyze and interpret large amounts of data generated by BioMeTs.
Funding
No funding was received for this work. The opinions expressed in this paper are those of the authors and should not be interpreted as the position of the US Food and Drug Administration. This publication is a result of a collaborative research performed under the auspices of the Digital Medicine Society (DiMe).
Conflict of Interest
A.G. is an editor with the publisher Elsevier; B.V. is an employee of Byteflies and owns company stock; J.P.B is a full‐time employee at Philips; N.G. is a full‐time employee at Samsung NeuroLogica. E.S.I. is an employee of Koneksa Health and owns company stock; C.A.N. is an employee of Pfizer, Inc. and owns company stock; V.P. is a full‐time employee at Takeda; W.A.W. is an Advisor of Koneksa and Elektra Labs, a consultant for Best Doctors/Teladoc and has research funding from Genentech and Pfizer; C.F.A. is a full‐time employee at Mitsubishi Tanabe Pharma America. All other authors declared no competing interests for this work. As Editor‐in‐Chief of Clinical & Translational Science, John A. Wagner was not involved in the review or decision process for this paper.
Acknowledgments
The authors would like to thank Jennifer Goldsack and Andrea Coravos for critically reading and commenting on the manuscript.
†Shared first authorship.
References
- 1. Porter, K.A. Effect of homologous bone marrow injections in x‐irradiated rabbits. Br. J. Exp. Pathol. 38, 401–412 (1957). [PMC free article] [PubMed] [Google Scholar]
- 2. Basu, P.K. , Miller, I. & Ormsby, H.L. Sex chromatin as a biologic cell marker in the study of the fate of corneal transplants. Am. J. Ophthalmol. 49, 513–515 (1960). [PubMed] [Google Scholar]
- 3. Aronson, J.K. Biomarkers and surrogate endpoints. Br. J. Clin. Pharmacol. 59, 491–494 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. US Food and Drug Administration . About biomarkers and qualification <https://www.fda.gov/drugs/cder‐biomarker‐qualification‐program/about‐biomarkers‐and‐qualification#what‐is>. Accessed May 11, 2020.
- 5. Biomarkers Definitions Working Group . Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69, 89–95 (2001). [DOI] [PubMed] [Google Scholar]
- 6. Coravos, A. , Khozin, S. & Mandl, K.D. Developing and adopting safe and effective digital biomarkers to improve patient outcomes. NPJ Digit. Med. 2, 14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldsack, J.C. et al Verification, analytical validation, and clinical validation (V3): the foundation of determining fit‐for‐purpose for Biometric Monitoring Technologies (BioMeTs). NPJ Digit. Med. 3, 55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roess, A. The promise, growth, and reality of mobile health — another data‐free zone. N. Engl. J. Med. 377, 2010–2011 (2017). [DOI] [PubMed] [Google Scholar]
- 9. Coravos, A. et al Modernizing and designing evaluation frameworks for connected sensor technologies in medicine. NPJ Dig. Med. 3, 37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Walton, M.K. et al Considerations for development of an evidence dossier to support the use of mobile sensor technology for clinical outcome assessments in clinical trials. Contemp. Clin. Trials 91, 105962 (2020). [DOI] [PubMed] [Google Scholar]
- 11. Clinical Trials Transformation Initiative (CTTI) . Advancing the use of digital health technologies for data capture & improved clinical trials <https://www.ctti‐clinicaltrials.org/projects/digital‐health‐technologies>. Accessed May 11, 2020.
- 12. Bakker, J.P. et al A systematic review of feasibility studies promoting the use of mobile technologies in clinical research. NPJ Digit. Med. 2, 47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matosin, N. , Frank, E. , Engel, M. , Lum, J.S. & Newell, K.A. Negativity towards negative results: a discussion of the disconnect between scientific worth and scientific culture. Dis. Model Mech. 7, 171–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Safi, S. , Thiessen, T. & Schmailzl, K.J. Acceptance and resistance of new digital technologies in medicine: qualitative study. JMIR Res. Protoc. 7, e11072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Izmailova, E.S. , Wagner, J.A. & Perakslis, E.D. Wearable devices in clinical trials: hype and hypothesis. Clin. Pharmacol. Ther. 104, 42–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee, J.W. et al Fit‐for‐purpose method development and validation for successful biomarker measurement. Pharm. Res. 23, 312–328 (2006). [DOI] [PubMed] [Google Scholar]
- 17. Cummings, J. , Raynaud, F. , Jones, L. , Sugar, R. & Dive, C. Fit‐for‐purpose biomarker method validation for application in clinical trials of anticancer drugs. Br. J. Cancer 103, 1313–1317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. US Food and Drug Administration . Biomarker qualification: evidentiary framework guidance for industry and FDA <https://www.fda.gov/media/119271/download>. Accessed May 11, 2020.
- 19. US Food and Drug Administration . Biomarker qualification: evidentiary framework <https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/biomarker‐qualification‐evidentiary‐framework>. Accessed May 16, 2020.
- 20. Menetski, J.P. et al The Foundation for the National Institutes of Health Biomarkers Consortium: past accomplishments and new strategic direction. Clin. Pharmacol. Ther. 105, 829–843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coons, S.J. , Eremenco, S. , Lundy, J.J. , O'Donohoe, P. , O'Gorman, H. & Malizia, W. Capturing patient‐reported outcome (pro) data electronically: the past, present, and promise of ePRO measurement in clinical trials. Patient 8, 301–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Larson, R.S. A path to better‐quality mhealth apps. JMIR Mhealth Uhealth 6, e10414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martínez‐Pérez, B. , de la Torre‐Díez, I. & López‐Coronado, M. Privacy and security in mobile health apps: a review and recommendations. J. Med. Syst. 39, 181 (2014). [DOI] [PubMed] [Google Scholar]
- 24. Steinhubl, S.R. & Topol, E.J. Digital medicine, on its way to being just plain medicine. NPJ Digit. Med. 1, 20175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. US Food and Drug Administration . General principles of software validation <https://www.fda.gov/media/73141/download>. Accessed May 11, 2020.
- 26. US Food and Drug Administration . Biomarker qualification submissions <https://www.fda.gov/drugs/cder‐biomarker‐qualification‐program/biomarker‐qualification‐submissions>. Accessed May 17, 2020.
- 27. US Food and Drug Administration . List of qualified biomarkers <https://www.fda.gov/drugs/cder‐biomarker‐qualification‐program/list‐qualified‐biomarkers>. Accessed May 17, 2020).
- 28. Babrak, L.M. et al Traditional and digital biomarkers: two worlds apart? Digit. Biomark. 3, 92–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. US Food and Drug Administration . Qualification process for drug development tools guidance for industry and FDA staff <https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/qualification‐process‐drug‐development‐tools‐guidance‐industry‐and‐fda‐staff>. Accessed May 11, 2020.
- 30. Stergiou, G.S. et al Recommendations and Practical Guidance for performing and reporting validation studies according to the Universal Standard for the validation of blood pressure measuring devices by the Association for the Advancement of Medical Instrumentation/European Society of Hypertension/International Organization for Standardization (AAMI/ESH/ISO). J. Hypertens. 37, 459–466 (2019). [DOI] [PubMed] [Google Scholar]
- 31. Lipsmeier, F. et al Evaluation of smartphone‐based testing to generate exploratory outcome measures in a phase 1 Parkinson's disease clinical trial. Mov. Disord. 33, 1287–1297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Godfrey, A. , Conway, R. , Meagher, D. & Olaighin, G. Direct measurement of human movement by accelerometry. Med. Eng. Phys. 30, 1364–1386 (2008). [DOI] [PubMed] [Google Scholar]
- 33. Rovini, E. , Maremmani, C. & Cavallo, F. How wearable sensors can support Parkinson's disease diagnosis and treatment: a systematic review. Front. Neurosci. 11, 555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Del Din, S. , Godfrey, A. & Rochester, L. Validation of an accelerometer to quantify a comprehensive battery of gait characteristics in healthy older adults and Parkinson's disease: toward clinical and at home use. IEEE J. Biomed. Health Inform. 20, 838–847 (2016). [DOI] [PubMed] [Google Scholar]
- 35. Armbrust, W. , Bos, G. , Geertzen, J.H.B. , Sauer, P.J.J. , Dijkstra, P.U. & Lelieveld, O. Measuring physical activity in juvenile idiopathic arthritis: activity diary versus accelerometer. J. Rheumatol. 44, 1249–1256 (2017). [DOI] [PubMed] [Google Scholar]
- 36. US Food and Drug Administration . Human factors and medical devices <https://www.fda.gov/medical‐devices/device‐advice‐comprehensive‐regulatory‐assistance/human‐factors‐and‐medical‐devices>. Accessed May 11, 2020.
- 37. US Food and Drug Administration . Applying human factors and usability engineering to medical devices <https://www.fda.gov/media/80481/download>. Accessed May 11, 2020.
- 38. Izmailova, E.S. et al Evaluation of wearable digital devices in a phase I clinical trial. Clin. Transl. Sci. 12, 247–256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izmailova, E.S. et al Continuous monitoring using a wearable device detects activity‐induced heart rate changes after administration of amphetamine. Clin. Transl. Sci. 12, 677–686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clarke, C.L. , Taylor, J. , Crighton, L.J. , Goodbrand, J.A. , McMurdo, M.E.T. & Witham, M.D. Validation of the AX3 triaxial accelerometer in older functionally impaired people. Aging Clin. Exp. Res. 29, 451–457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karas, M. et al Accelerometry data in health research: challenges and opportunities. Stat. Biosci. 11, 210–237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Espay, A.J. et al A roadmap for implementation of patient‐centered digital outcome measures in Parkinson's disease obtained using mobile health technologies. Mov. Disord. 34, 657–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Urbanek, J.K. et al Prediction of sustained harmonic walking in the free‐living environment using raw accelerometry data. Physiol. Meas. 39, 02NT02 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Urbanek, J.K. , Harezlak, J. , Glynn, N.W. , Harris, T. , Crainiceanu, C. & Zipunnikov, V. Stride variability measures derived from wrist‐ and hip‐worn accelerometers. Gait Posture 52, 217–223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goldsmith, J. , Zipunnikov, V. & Schrack, J. Generalized multilevel function‐on‐scalar regression and principal component analysis. Biometrics 71, 344–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Natarajan, A. , Pantelopoulos, A. , Emir‐Farinas, H. & Natarajan, P. Heart rate variability with photoplethysmography in 8 million individuals: results and scaling relations with age, gender, and time of day. bioRxiv, 772285 (2019). 10.1101/772285 [DOI] [Google Scholar]
- 47. Di, J. et al Joint and individual representation of domains of physical activity, sleep, and circadian rhythmicity. Stat. Biosci. 11, 371–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang, L. et al Multilevel matrix‐variate analysis and its application to accelerometry‐measured physical activity in clinical populations. J. Am. Stat. Assoc. 114, 553–564 (2019). [Google Scholar]
- 49. Migueles, J.H. , Rowlands, A.V. , Huber, F. , Sabia, S. & van Hees, V.T. GGIR: a research community–driven open source r package for generating physical activity and sleep outcomes from multi‐day raw accelerometer data. J. Measurement Phys. Behaviour 2, 188–196 (2019). [Google Scholar]
- 50. Open Wearables Initiative <https://www.owear.org/>. Accessed May 11, 2020.
- 51. Erickson, B.J. , Korfiatis, P. , Akkus, Z. & Kline, T.L. Machine learning for medical imaging. RadioGraphics 37, 505–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Currie, G. , Hawk, K.E. , Rohren, E. , Vial, A. & Klein, R. Machine learning and deep learning in medical imaging: intelligent imaging. J. Med. Imag. Rad. Sci. 50, 477–487 (2019). [DOI] [PubMed] [Google Scholar]
- 53. Blanc‐Durand, P. et al Abdominal musculature segmentation and surface prediction from CT using deep learning for sarcopenia assessment. Diagnost. Interven. Imaging (2020). 10.1016/j.diii.2020.04.011 [DOI] [PubMed] [Google Scholar]
- 54. Arcadu, F. , Benmansour, F. , Maunz, A. , Willis, J. , Haskova, Z. & Prunotto, M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. NPJ Digit. Med. 2, 92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hannun, A.Y. et al Cardiologist‐level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 25, 65–69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mobilise‐D <https://www.mobilise‐d.eu/>. Accessed May 11, 2020.
- 57. IDEA‐FAST <https://idea‐fast.eu/>. Accessed May 11, 2020.
- 58. Kubota, K.J. , Chen, J.A. & Little, M.A. Machine learning for large‐scale wearable sensor data in Parkinson's disease: concepts, promises, pitfalls, and futures. Mov. Disord. 31, 1314–1326 (2016). [DOI] [PubMed] [Google Scholar]
- 59. Randhawa, P. , Shanthagiri, V. & Kumar, A. A review on applied machine learning in wearable technology and its applications, in 2017 International Conference on Intelligent Sustainable Systems (ICISS), December 7–8, 2017. 2017, pp. 347–354.
- 60. Woods, B. , Coravos, A. & Corman, J.D. The case for a Hippocratic oath for connected medical devices: viewpoint. J. Med. Internet Res. 21, e12568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. US Food and Drug Administration . Content of premarket submissions for management of cybersecurity in medical devices <https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/content‐premarket‐submissions‐management‐cybersecurity‐medical‐devices>. Accessed May 11, 2020.
- 62. US Food and Drug Administration . Postmarket management of cybersecurity in medical devices <https://www.fda.gov/regulatory‐information/search‐fda‐guidance‐documents/postmarket‐management‐cybersecurity‐medical‐devices>. Accessed May 11, 2020.
- 63. Schukat, M. et al Unintended consequences of wearable sensor use in healthcare. Yearbook Med. Inform. 25, 73–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. US Food and Drug Administration . CDRH’S approach to tumor profiling next generation sequencing tests <https://www.fda.gov/media/109050/download>. Accessed May 11, 2020.
- 65. US Food and Drug Administration . In vitro diagnostics <https://www.fda.gov/medical‐devices/products‐and‐medical‐procedures/vitro‐diagnostics>. Accessed May 11, 2020.
- 66. Frampton, G.M. et al Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 31, 1023–1231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. US Food and Drug Administration . Considerations for Design, Development, and Analytical Validation of Next Generation Sequencing (NGS) ‐ Based In Vitro Diagnostics (IVDs) Intended to Aid in the Diagnosis of Suspected Germline Diseases <fda.gov/regulatory‐information/search‐fda‐guidance‐documents/considerations‐design‐development‐and‐analytical‐validation‐next‐generation‐sequencing‐ngs‐based>. Accessed May 11, 2020.
- 68. US Food and Drug Administration . FDA approves first cancer treatment for any solid tumor with a specific genetic feature <https://www.fda.gov/news‐events/press‐announcements/fda‐approves‐first‐cancer‐treatment‐any‐solid‐tumor‐specific‐genetic‐feature>. Accessed May 11, 2020.
- 69. American Sleep Disorders Association . Practice parameters for the use of actigraphy in the clinical assessment of sleep disorders. Sleep 18, 285–287 (1995). [DOI] [PubMed] [Google Scholar]
- 70. Sadeh, A. The role and validity of actigraphy in sleep medicine: an update. Sleep Med. Rev. 15, 259–267 (2011). [DOI] [PubMed] [Google Scholar]
- 71. Sadeh, A. & Acebo, C. The role of actigraphy in sleep medicine. Sleep Med. Rev. 6, 113–124 (2002). [DOI] [PubMed] [Google Scholar]
- 72. Smith, M.T. et al Use of actigraphy for the evaluation of sleep disorders and circadian rhythm sleep‐wake disorders: an American academy of sleep medicine systematic review, meta‐analysis, and GRADE assessment. J. Clin. Sleep Med. 14, 1209–1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cole, R.J. , Kripke, D.F. , Gruen, W. , Mullaney, D.J. & Gillin, J.C. Automatic sleep/wake identification from wrist activity. Sleep 15, 461–469 (1992). [DOI] [PubMed] [Google Scholar]
- 74. Sadeh, A. , Hauri, P.J. , Kripke, D.F. & Lavie, P. The role of actigraphy in the evaluation of sleep disorders. Sleep 18, 288–302 (1995). [DOI] [PubMed] [Google Scholar]
- 75. US Food and Drug Administration . DNP Clinical review <https://www.fda.gov/media/88284/download>. Accessed May 12, 2020.
- 76. van Hees, V.T. et al Estimating sleep parameters using an accelerometer without sleep diary. Sci. Rep. 8, 12975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jones, S.E. et al Genetic studies of accelerometer‐based sleep measures yield new insights into human sleep behaviour. Nat. Commun. 10, 1585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. European Medicines Agency . Qualification opinion on stride velocity 95th centile as a secondary endpoint in Duchenne Muscular Dystrophy measured by a valid and suitable wearable device <https://www.ema.europa.eu/en/documents/scientific‐guideline/qualification‐opinion‐stride‐velocity‐95th‐centile‐secondary‐endpoint‐duchenne‐muscular‐dystrophy_en.pdf>. Accessed May 12, 2020.
- 79. Foundation for the National Institutes of Health . Biomarkers consortium ‐ Workshop: Remote digital monitoring for medical product development <https://fnih.org/what‐we‐do/biomarkers‐consortium/programs/digitalmonitoring>. Accessed May 12, 2020.
- 80. Clinical Trials Transformation Initiative (CTTI) . Use case for developing novel endpoints generated using mobile technology: diabetes mellitus <https://www.ctti‐clinicaltrials.org/files/usecase‐diabetes.pdf>. Accessed May 12, 2020.
- 81. Klonoff, D.C. Continuous glucose monitoring: roadmap for 21st century diabetes therapy. Diabet. Care 28, 1231–1239 (2005). [DOI] [PubMed] [Google Scholar]
- 82. Scholten, K. & Meng, E. A review of implantable biosensors for closed‐loop glucose control and other drug delivery applications. Int. J. Pharm. 544, 319–334 (2018). [DOI] [PubMed] [Google Scholar]
- 83. Cappon, G. , Vettoretti, M. , Sparacino, G. & Facchinetti, A. Continuous glucose monitoring sensors for diabetes management: a review of technologies and applications. Diabet. Metab. J. 43, 383–397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kerr, D. & Fayers, K. Continuous real‐time glucose monitoring systems: time for a closer look. Pract. Diabet. Int. 25, 37–41 (2008). [Google Scholar]
- 85. Vigersky, R.A. Going beyond HbA1c to understand the benefits of advanced diabetes therapies. J. Diabet. 11, 23–31 (2019). [DOI] [PubMed] [Google Scholar]
- 86. Acciaroli, G. , Vettoretti, M. , Facchinetti, A. & Sparacino, G. Calibration of minimally invasive continuous glucose monitoring sensors: state‐of‐the‐art and current perspectives. Biosensors 8, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schnell, O. et al Role of continuous glucose monitoring in clinical trials: recommendations on reporting. Diabet. Technol. Ther. 19, 391–399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Castle, J.R. et al Randomized outpatient trial of single‐ and dual‐hormone closed‐loop systems that adapt to exercise using wearable sensor. Diabet. Care 41, 1471–1477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moran, S. , Nishida, T. & Nakata, K. Perceptions of a wearable ubiquitous monitoring device. IEEE Technol. Soc. Magazine 32, 56–64 (2013). [Google Scholar]
- 90. Fereidooni, H. et al"Breaking fitness records without moving: Reverse engineering and spoofing fitbit. International Symposium on Research in Attacks, Intrusions, and Defenses. (Springer, New York, NY, 2017) pp. 48‐69.
- 91. Rupp, M.A. , Michaelis, J.R. , McConnell, D.S. & Smither, J.A. The role of individual differences on perceptions of wearable fitness device trust, usability, and motivational impact. Appl. Ergon. 70, 77–87 (2018). [DOI] [PubMed] [Google Scholar]
- 92. Izmailova, E. S. , Ellis, R. & Benko, C. Remote monitoring in clinical trials during the covid‐19 pandemic. Clin. Transl. Sci.. 10.1111/cts.12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cohen, J.B. et al History and justification of a national blood pressure measurement validated device listing. Hypertension 73, 258–264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. German Federal Ministry of Health . Federal Institute for Drugs and Medical Devices (Bundesinstitut für Arzneimittel und Medizinprodukte ‐ BfArM). "Digital Health Applications (DiGA)." <www.bfarm.de/EN/MedicalDevices/DiGA/_node.html>. Accessed July 20, 2020.
- 95. US Food and Drug Administration . FDA‐NIH Biomarker Working Group . BEST (Biomarkers, EndpointS, and other Tools). Co‐published by National Institutes of Health (US). Accessed May 17, 2020.
- 96. US Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER), and Center for Biologics Evaluation and Research (CBER) . Biomarker Qualification: Evidentiary Framework," in "Drug Development Tools. [Online] <https://www.fda.gov/media/119271/download> (2018).
- 97. Stergiou, G.S. et al A universal standard for the validation of blood pressure measuring devices. Hypertension 71, 368–374 (2018). [DOI] [PubMed] [Google Scholar]
- 98. Ganio, M.S. et al Validity and reliability of devices that assess body temperature during indoor exercise in the heat. J. Athl. Train. 44, 124–135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]


