Abstract
Reduced expression of the uptake transporter, OCTN1 (SLC22A4), has been reported as a strong predictor of poor event‐free and overall survival in multiple cohorts of patients with acute myeloid leukemia (AML) receiving the cytidine nucleoside analog, cytarabine (Ara‐C). To further understand the mechanistic basis of interindividual variability in the functional expression of OCTN1 in AML, we hypothesized a mechanistic connection to DNA methylation‐based epigenetic repression of SLC22A4. We found increased basal SLC22A4 methylation was associated with decreased Ara‐C uptake in AML cell lines. Pre‐treatment with hypomethylating agents, 5‐azacytidine, or decitabine, restored SLC22A4 mRNA expression, increased cellular uptake of Ara‐C, and was associated with increased cellular sensitivity to Ara‐C compared with vehicle‐treated cells. Additionally, lower SLC22A4 methylation status was associated with distinct clinical advantages in both adult and pediatric patients with AML. These findings suggest a regulatory mechanism is involved in the interindividual variability in response to Ara‐C, and provides a basis for the integration of hypomethylating agents into Ara‐C‐based treatment regimens.
Acute myeloid leukemia (AML) is a blood disorder that is classified by abnormal proliferation and differentiation of myeloid cells within the bone marrow compartment. Despite advances in supportive care, the backbone of therapy has remained unchanged for the last 30 years, and consists of combination regimens of cytarabine (Ara‐C) and anthracyclines. 1 The efficacy and response to Ara‐C‐based treatment vary dramatically between individual patients with AML and are, in part, dependent on efficiency of Ara‐C uptake, 2 intracellular activation, 3 and deamination. 4 Because the transport of Ara‐C is the initial step to intracellular accumulation and subsequent cytotoxicity, defective uptake is a major contributor to clinical resistance observed with nucleoside analog‐based therapy in AML. 5
One hallmark of AML is dysregulation at the genetic and epigenetic level. 6 , 7 Previously completed clinical trials have demonstrated that an increase in overall methylation leads to worse patient outcomes in both adult 8 and pediatric patients with AML. 9 Currently, clinical trials are underway to modify this basal methylation profile by “epigenetically priming” AML cells in patients using US Food and Drug Administration (FDA)‐approved DNA methyltransferase inhibitors, such as 5‐azacytidine and decitabine, prior to starting first‐line therapy (NCT01177540 and NCT03164057). These clinical trials continue to progress although the mechanistic basis of the improved outcomes seen remains poorly understood.
Epigenetic regulation of uptake transporters has been previously demonstrated for OCT1 (SLC22A1), 10 OCT2 (SLC22A2), 11 OCT3 (SLC22A3), 12 MATE1 (SLC47A1), 13 and OCTN2 (SLC22A5). 14 Surprisingly, similar epigenetic regulatory mechanisms have not been reported for OCTN1 (SLC22A4), a transporter that is expressed on AML blasts that strongly predicts survival in patients with AML receiving first‐line therapy with Ara‐C. 15 Here, we report a connection between DNA methylation within the promoter region of SLC22A4 and its contribution to OCTN1 expression, Ara‐C accumulation in AML cells, and subsequent antileukemic effects in patients.
METHODS
Cell lines
HEK293T cells were maintained in DMEM, and all AML cells were maintained in RPMI 1640 medium, both supplemented with 10% fetal bovine serum. All cell lines were maintained at 37°C and 5% CO2, and regularly tested for mycoplasma contamination.
Reverse transcriptase polymerase chain reaction
RNA was extracted from cell lines using an EZNA Total RNA Extraction Kit (Omega Bio‐tek, Norcross, GA), and cDNA was generated from 2 µg of RNA using qScript XLT cDNA Supermix (Quantabio, Beverly, MA). Real‐time reverse transcriptase polymerase chain reaction was performed using TaqMan probes and Fast reagents (ThermoFisher Scientific, Waltham, MA). Reactions were carried out in triplicate, and normalized to GAPDH.
Cytotoxicity and cellular uptake assays
Ara‐C‐mediated cytotoxicity was determined as previously reported. 16 Further details are outlined in the Supplementary Material s and Methods. Uptake studies were performed using H3‐labeled substrates in serum‐free, phenol red‐free DMEM. Radioactivity was quantified using a Beckman LS 6500 Liquid Scintillation Counter (Indianapolis, IN). 17 Uptake was normalized to total protein content using a BCA kit (ThermoFisher Scientific).
OCTN1 dual reporter assay
Dual reporter assays were completed as previously described. 18 Further details are supplied in the Supplementary Material s and Methods. The reporter readout of each well was normalized by taking the ratio of Firefly luciferase to cypridina of each well. Individual replicate plates were combined by normalizing to 100% of the respective unmethylated readings within the same experiment.
Epigenetic priming and bisulfite‐sequencing
AML cells were plated at a density of 2.0 × 105 cells/mL in a 6‐well plate in media treated with varying concentrations of 5‐azacytidine or decitabine. The suspension media was changed daily to prevent aqueous instability of the demethylating agents. 19 On day 3, AML cells were washed, subjected to uptake, methylthiazol tetrazolium, reverse transcriptase polymerase chain reaction, and harvesting of genomic DNA. Bisulfite sequencing was carried utilizing the services from EpigenDx (Hopkinton, MA).
Clinical association studies
To evaluate biological concordance of clinical data with the cell line data, we tested the association of the M values of the probe sets overlapping with our in vitro analysis (cg05231888) with clinical outcomes in the AML02 clinical trial (NCT00136084), and the Cancer Genome Atlas LAML cohort (https://www.cancer.gov/tcga). For AML02, statistical analysis associated the M values of diagnostic leukemia samples with the presence of minimal residual disease following the first course of chemotherapy, event‐free and overall survival, and expression of SLC22A4 in diagnostic leukemia using the rank sum test, Cox regression, and Spearman’s correlation, respectively. The same methods were used to evaluate the association of diagnostic leukemia M values with SLC22A4 expression and overall survival in the Cancer Genome Atlas LAML cohort.
RESULTS
AML cells are differentially methylated within the SLC22A4 promoter region
In order to further understand the potential epigenetic regulation of the OCTN1 gene SLC22A4 through DNA methylation, we initially evaluated the predicted CpG islands in proximity to SLC22A4 using the UCSC Genome Browser. Of the three predicted CpG islands ( Figure 1a ), CpG122 was of the greatest interest given that it spanned the promoter area that was previously found to be a methylated region regulating the expression of other, phylogenetically linked organic cation transporters. 11 , 12 , 14 To build on our previously reported connection of OCTN1 function with the transport of nucleoside analogs, we investigated the methylation of SLC22A4 in AML, the disease‐specific context of interest. Using bisulfite‐sequencing, we compared the methylation status of the CpG122 in two phenotypically distinct AML cell lines, OCI‐AML3 and CHRF‐288‐11, which exhibited high Ara‐C uptake and low Ara‐C uptake, respectively ( Figure S1 a). High uptake (OCI‐AML3) was associated with an overall lower methylation profile within the promoter region, whereas in low uptake (CHRF‐288‐11) was associated with an overall higher methylation within this same region ( Figure 1b ).
Figure 1.
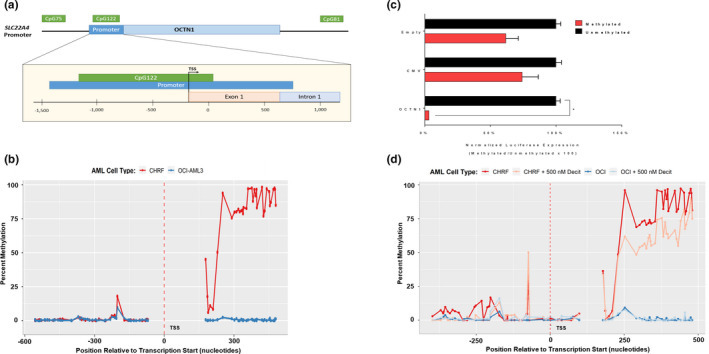
Methylation status of the SLC22A4 promoter region. (a) Cartoon depiction of the OCTN1 promoter region with overlapping CpG island. (b) Methylation profiles as determine by BS‐Seq of acute myeloid leukemia (AML) cell lines OCI‐AML3 and CHRF, with “high” and “low” cytarabine (Ara‐C) uptake respectively. (c) In vitro methylation luciferase assay of OCTN1, cytomegalovirus promoter regions. Relative luciferase readings shown as the ratio of Firefly luciferase (CpG‐free plasmids) to cypridina luciferase (pTK) expression. Data shown as the mean of two independent experiment plus SD. *P < 0.001. (d) Percent methylation as determined by BS‐Seq of OCTN1 promoter region following 3‐day treatment with 500 nM decitabine or aqueous control.
Methylation status affects the downstream expression of SLC22A4
To functionally characterize the SLC22A4 promoter activity, with and without methylation, we constructed a SLC22A4 promoter luciferase vector carrying the upstream section of CpG122. This specific vector backbone is free of any CpG islands, which could potentially be methylated and erroneously hinder luciferase expression. We confirmed that both the positive control (cytomegalovirus) and the OCTN1 promoter plasmid could drive the expression of Firefly luciferase. After in vitro methylation with an M.SssI methylase (Figure S2 ), OCTN1 showed a significant decrease in luciferase expression (39.1‐fold; P < 0.0001), whereas only modest changes were seen in cytomegalovirus (2.1‐fold) and the empty constructs (2.3‐fold) (Figure 1b ).
Pretreatment with DNA methyltransferase inhibitors decreases SLC22A4 methylation
Because differences in methylation were observed OCI‐AML3 and CHRF‐288‐11, as well as methylation repressed downstream transcription, we hypothesized that DNA methyltransferase inhibitors (DNMTi; i.e., hypomethylating agents) could modify the methylation status of the SLC22A4 promoter. In agreement with a previously completed clinical trial, 20 using physiologically relevant levels observed in patients, 21 we treated AML cells with DNMTi for 72 hours prior to downstream analysis and sequencing. These studies revealed that pretreatment with decitabine (500 nM) decreased methylation within the SLC22A4 promoter region in highly methylated CHRF‐288‐11 as determined by bisulfite‐sequencing ( Figure 1d ). These findings suggest that pretreatment with decitabine has the ability to decrease SLC22A4 methylation in AML cells.
DNA methyltransferase inhibitors affect OCTN1 expression and function
We next explored phenotypic changes that may impact cellular drug disposition following pretreatment with DNMTi. Treatment of CHRF‐288‐11 (high methylation cell line), with nanomolar concentrations (100 or 500 nM) of either 5‐azacytidine or decitabine was carried out. A significant concentration‐dependent increase in SLC22A4 mRNA transcripts compared with vehicle‐treated cells was associated with both concentrations of decitabine and 500 nM for 5‐azacytidine (Figure 2a ; P < 0.05), whereas similar treatment in OCI‐AML3 (low methylation cell line) had minimal effects on SLC22A4 mRNA expression. The observed changes in SLC22A4 expression following pretreatment with DNMTi correlated functionally with higher uptake of Ara‐C (Figure 2b ); CHRF‐288‐11 cells following pretreatment with 500 nM 5‐azacytidine or decitabine showed a 1.61 and 2.91 fold increase in uptake (P < 0.005 and P < 0.05), whereas OCI‐AML3 showed a 1.16 and 1.49 fold increase in uptake (P ≥ 0.05), respectively.
Figure 2.
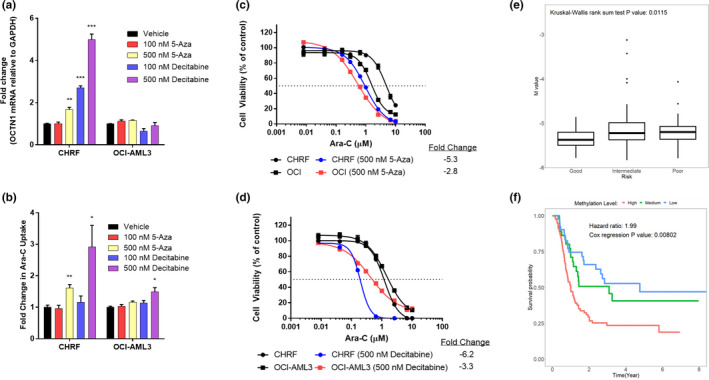
Epigenetic priming alters cytarabine (Ara‐C) activity in vivo, whereas SLC22A4 methylation status correlated with clinical outcome. (a) SLC22A4 mRNA expression (normalized to GAPDH), (b) fold change in [3H]‐Ara‐C uptake (1 μM; 15 minutes), (c) and (d) 72‐hour cell viability to Ara‐C measured using methylthiazol tetrazolium following 72 hour epigenetic priming with either 5‐azacytidine or decitabine treatment. Data is shown as the mean of two independent experiments (n = 3 each) ± SEM; *P ≤ 0.05, **P ≤ 0.005, ***P < 0.0005. Correlation of SLC22A4 methylation (cg05231888) from the Cancer Genome Atlas LAML dataset grouped by cytogenetic risk (e) and event‐free survival (f). Methylation probe cg05231888 overlap with the CpG island of SLC22A4 promoter region (CpG122).
Next, we evaluated the influence of increased intracellular accumulation of Ara‐C on changes in cellular sensitivity to Ara‐C. With 500 nM 5‐azacytidine pretreatment, sensitivity to Ara‐C was substantially increased in CHRF‐288‐11 cells (5.3‐fold), whereas more modest increases were noted in the low methylated OCI‐AML3 cells (2.8‐fold; Figure 2c ). Similar observations were made with decitabine (500 nM) in CHRF‐288‐11 cells (6.3‐fold) and OCI‐AML3 cells (3.3‐fold; Figure 2d ). Taken together, these findings are consistent with the notion that exposure of AML cells to hypomethylating agents increases SLC22A4 expression, leading to an increase in Ara‐C uptake, and subsequent increase in sensitivity to the antileukemic effects of Ara‐C.
Methylation within SLC22A4 correlates with clinical outcomes in AML
By accessing the results of our previously reported exploratory integrated analysis of the methylome, transcriptome, and outcomes of pediatric patients with AML, 22 we found that the canonical correlation of SLC22A4 expression and the methylation of 18 evaluated probes was 0.53 (P = 0.0002; Figure S3 a), indicating a strong empirical connection between OCTN1 expression and methylation. To more fully characterize this observation, we evaluated the association of each methylation probe in SLC22A4 with the expression of SLC22A4 and clinical outcomes. We found that increased methylation at the locus of the probe cg05231888, which lays within CpG122, consistently associated with decreased SLC22A4 expression, higher‐risk cytogenetics, and worse outcomes in both adult ( Figure 2e,f , Figure S3 b) and pediatric populations ( Figure S3 c–e).
Furthermore, because Ara‐C is also a substrate of SLC28A1, SLC28A3, and SLC29A1, we evaluated the overall survival association with transporter expression. We previously reported in the pediatric population that expression of SLC28A1 and SLC28A3 did not have a survival advantage but there was a significant survival advantage with SLC22A4 and SLC29A1 high expression. 15 When analyzing the adult population, we previously found the expression of SLC22A4 had a significant association with overall survival. 15 However, SLC29A1 expression did not have a significant association with overall survival as a single‐predictor (hazard ratio (HR) = 1.004, 95% confidence interval (CI) 0.90–1.91, P = 0.15) or after adjustment for risk group (HR = 1.33, 95% CI 0.92–1.94, P = 0.13). The expression of SLC28A3 was also not significantly associated with overall survival as a single predictor (HR = 0.98, 95% CI 0.92–1.04) or after adjustment for risk group (HR = 1.004, 95% CI 0.94–1.07, P = 0.91). The expression of SLC28A1 was marginally associated with better overall survival as a single predictor (HR = 0.95, 95% CI 0.90–1.007, P = 0.085) and was significantly associated with better overall survival after adjustment for risk group (HR = 0.94, 95% CI 0.889–0.998, P = 0.042). Taken together with our previous data, 15 advocates for the importance of SLC22A4 expression in overall survival.
DISCUSSION
The epigenetic regulation of xenobiotic uptake transporters contributes to a dynamic interplay between intracellular substrate concentrations and the extracellular environment of various cell types. Disruption of this sensitive balance has the potential to modify intracellular accumulation of substrates, and may contribute to variability in drug resistance or toxicity. This epigenetic regulation can be exploited in cancer cells by restoring the expression of drug uptake transporters in resistant cells, thereby impacting the accumulation of cytotoxic drugs. One such transporter, OCTN1, that has been profiled as an important transporter of Ara‐C uptake, 15 a mainstay in AML treatment, epigenetic regulation has not been previously evaluated. Recently, there has been some controversy as to whether OCTN1 influences the intracellular accumulation of Ara‐C. 23 Difference in experimental design and rapid metabolism of Ara‐C once in the cell has lead this controversy. 24 However, in our studies where total radioactivity is measured, rather than the parental compound, OCTN1 still impacts that total intracellular accumulation of Ara‐C.
Using the prior knowledge that expression of several transporters is regulated by the DNA methylation status of their corresponding upstream promoters, we hypothesized that OCTN1 might be regulated in a similar manner. In particular, methylation within the CpG island, CpG122, located in the SLC22A4 promoter region, could cause repression of the downstream transcription and subsequently leads to decreased intracellular accumulation of transported substrates. In addition, these methylation events can be prevented with the use of hypomethylating agents and transporter expression can be restored in AML cell lines.
To test this hypothesis, we examined the DNA methylation profile of CpG122 and found that cells exhibiting low intrinsic uptake of Ara‐C (CHRF‐288‐11) show increased SLC22A4 methylation as compared with cells exhibiting high intrinsic uptake of Ara‐C (OCI‐AML3). In addition, we found that the SLC22A4 promoter is highly sensitive to DNA methylation. We showed that exposure of CHRF‐288‐11 to hypomethylating agents decreases DNA methylation within this region, although complete restoration to basal levels of OCI‐AML3 was not achieved. This inability to completely restore methylation levels to unmethylated cell types points to the potential existence of other mechanisms of epigenetic regulation, such as histone methylation.
Reports have confirmed that DNA hypomethylating agents can sensitize various cancer cells to cytotoxic agents, 25 , 26 although the mechanistic basis for the observed increases in cytotoxicity, compared with single agent use, have remained largely elusive. An interesting observation was that decitabine appears to be more effective in our studies compared with 5‐azacytidine, despite both being categorized as hypomethylating agents. It has been reported that they act in two different manners, with 5‐azacytidine having an RNA involvement. 19 Thus, one could postulate that these difference and lack of RNA involvement for decitabine allows it to have a more hypomethylating effect rather than cytotoxic, allowing the cells to continue to survive in a weakened state making them more susceptible to subsequent treatment to Ara‐C. Our studies provide an additional reasonable mechanistic explanation for these prior findings and suggest that increases in cytotoxicity following epigenetic priming may stem, in part, from a restoration of the expression of transporters of relevance to the intracellular accumulation and subsequent cytotoxicity of chemotherapeutic agents. These findings are particularly relevant in light of our observations that increased methylation associates with decreased SLC22A4 and worse outcomes in the both the adult and pediatric populations. We also noted in a previous publication that high expression of SLC22A4 in AML blasts is associated with distinct survival advantages in patients. 17 The overall survival significance was greater in the adult population compared with the pediatric population, which could, in part, be due to the increased methylation observed as individuals age, leading them to be more susceptible to hypomethylating agents. 27
The methylation of CpG dinucleotides within SLC22A4 and subsequent inhibition of expression could potentially be occurring through two different mechanisms. First, the methylation of specific CpG dinucleotides may disrupt the binding of specific transcription factors that regulate SLC22A4, such as Sp1 and RUNX1. 28 Second, it is possible that methyl‐CpG binding proteins may be altered by the DNA methylation status of SLC22A4. 29 Further work is warranted to address the specific mechanisms contributing to the altered expression of SLC22A4 following DNA methylation. Changes in cell growth following epigenetic priming may also independently impact the cellular sensitivity to drugs. Indeed, treatment with hypomethylating agents has been shown to decrease doubling times in HL60 and T24 cells, 30 which would make the cells more resistant to the S‐phase dependent cytotoxicity of agents, such as Ara‐C. In contrast, we found that epigenetic priming is associated with increased sensitivity, suggesting that the observed phenotypes occur despite possible effects on changes in cell growth.
Collectively, our findings suggest that DNA methylation within the promoter region of SLC22A4 has a direct impact on the expression and function of an important organic cation transporter, and affects the sensitivity of AML cells to Ara‐C‐mediated cytotoxicity. These findings are in agreement with clinical data showing a distinct clinical advantage of low SLC22A4 methylation in both pediatric and adult patients with AML. The reported epigenetic regulation of OCTN1 may have immediate translational relevance, and provides a mechanistic explanation for previously observed potentiation of antileukemic properties associated with the combined use of hypomethylating agents and Ara‐C. In vivo studies beyond the scope of this paper could shed further light onto this mechanistic explanation. In addition, the described regulatory mechanism may help explain the variable pharmacodynamic responses seen in patients receiving treatment with xenobiotic substrates as well as the interindividual variation in the pharmacokinetic profile of dietary substrates of OCTN1, such as the antioxidant ergothioneine.
Funding
The work was supported in part by the National Institutes of Health (Grants R01CA215802 (to A.S.), R01CA238946 (to S.H.), R01CA138744 (to S.D.B.), and R01CA132946 (to J.K.L. and S.B.P.)) and by the OSU Comprehensive Cancer Center using Pelotonia funds.
Conflicts of Interest
The authors declared no competing interests for this work.
Authors Contributions
J.T.A., D.R.B., S.D.B., and A.S. wrote the manuscript. S.D.B., J.K.L., S.B.P., and A.S. designed the research. J.T.A., D.R.B., A.A.G., E.A.G., and S.H. performed the research. J.T.A., D.R.B., S.H., S.B.P., and L.S. analyzed the data.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
It is known that there is a large interindividual variability to cytarabine (Ara‐C) response within patients with acute myeloid leukemia (AML). It has been reported that the uptake transporter, OCTN1 (SLC22A4), plays a critical role in Ara‐C uptake. Reduced expression of OCTN1 has been reported as a strong predictor of poor event‐free survival and overall survival. The promoter regions of other uptake transporters in this class of transporters are known to be regulated by methylation.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study addresses the question as to what is the mechanism behind that variable expression seen in the uptake transporter, OCTN1 (SLC22A4) that has been reported as a strong predictor of survival in patients with AML receiving Ara‐C.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study demonstrates that the expression and function of the uptake transporter OCTN1 (SLC22A4) in AML is dependent on a DNA methylation‐based epigenetic mechanism that is sensitive to modulation by hypomethylating agents.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
This study highlights regulatory mechanisms that could explain the interindividual pharmacodynamics responses. Understanding these mechanisms will promote investigation into therapy strategies that could help decrease the interindividual variation in the pharmacokinetic profile.
Supporting information
Supplementary Material
Fig S1
Fig S2
Fig S3
Acknowledgments
The authors thank the Rehli Laboratory (University of Regensburg, Germany) for the CpG‐free luciferase plasmid, EpigenDx (Hopkinton, MA) for assistance in BS‐Seq analysis, and Dr. Moray Campbell for assistance with CpG island prediction.
References
- 1. Ferrara, F. & Schiffer, C.A. Acute myeloid leukaemia in adults. Lancet 381, 484–495 (2013). [DOI] [PubMed] [Google Scholar]
- 2. Gati, W.P. , Paterson, A.R. , Larratt, L.M. , Turner, A.R. & Belch, A.R. Sensitivity of acute leukemia cells to cytarabine is a correlate of cellular es nucleoside transporter site content measured by flow cytometry with SAENTA‐fluorescein. Blood 90, 346–353 (1997). [PubMed] [Google Scholar]
- 3. Bhalla, K. , Nayak, R. & Grant, S. Isolation and characterization of a deoxycytidine kinase‐deficient human promyelocytic leukemic cell line highly resistant to 1‐beta‐D‐ arabinofuranosylcytosine. Cancer Res. 44, 5029–5037 (1984). [PubMed] [Google Scholar]
- 4. Schroder, J.K. , Seidelmann, M. , Kirch, H.C. , Seeber, S. & Schutte, J. Assessment of resistance induction to cytosine arabinoside following transfer and overexpression of the deoxycytidylate deaminase gene in vitro. Leukemia Res. 22, 619–624 (1998). [DOI] [PubMed] [Google Scholar]
- 5. Obata, T. , Endo, Y. , Murata, D. , Sakamoto, K. & Sasaki, T. The molecular targets of antitumor 2'‐deoxycytidine analogues. Curr. Drug Targets 4, 305–313 (2003). [DOI] [PubMed] [Google Scholar]
- 6. Gutierrez, S.E. & Romero‐Oliva, F.A. Epigenetic changes: a common theme in acute myelogenous leukemogenesis. J. Hematol. Oncol. 6, 57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bullinger, L. et al Quantitative DNA methylation predicts survival in adult acute myeloid leukemia. Blood 115, 636–642 (2010). [DOI] [PubMed] [Google Scholar]
- 9. Ochs, M. et al Genome wide promoter methylation patterns predict AML subtype outcomes and identify novel pathways characterizing diagnostic and relapsed disease in children. Blood 120, 1287 (2012). [Google Scholar]
- 10. Schaeffeler, E. et al DNA methylation is associated with downregulation of the organic cation transporter OCT1 (SLC22A1) in human hepatocellular carcinoma. Genome Med. 3, 82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu, Y. et al Epigenetic activation of the drug transporter OCT2 sensitizes renal cell carcinoma to oxaliplatin. Sci. Transl. Med. 8, 348ra397 (2016). [DOI] [PubMed] [Google Scholar]
- 12. Chen, L. et al Genetic and epigenetic regulation of the organic cation transporter 3, SLC22A3. Pharmacogenom. J. 13, 110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka, T. , Hirota, T. & Ieiri, I. Relationship between DNA methylation in the 5' CpG Island of the SLC47A1 (multidrug and toxin extrusion protein MATE1) gene and interindividual variability in MATE1 expression in the human liver. Mol. Pharmacol. 93, 1–7 (2018). [DOI] [PubMed] [Google Scholar]
- 14. Qu, Q. et al Different involvement of promoter methylation in the expression of organic cation/carnitine transporter 2 (OCTN2) in cancer cell lines. PLoS One 8, e76474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drenberg, C.D. et al OCTN1 Is a high‐affinity carrier of nucleoside analogues. Cancer Res. 77, 2102–2111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drenberg, C.D. et al ABCC4 is a determinant of cytarabine‐induced cytotoxicity and myelosuppression. Clin. Transl. Sci. 9, 51–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drenberg, C.D. et al OCTN1 is a high‐affinity carrier of nucleoside analogues. Cancer Res. 77, 2102–2111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buelow, D.R. & Baker, S.D.D. Hypoxia reporter element assay. Bio. Protocol 8, e2951 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stresemann, C. & Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 123, 8–13 (2008). [DOI] [PubMed] [Google Scholar]
- 20. Scandura, J.M. et al Phase 1 study of epigenetic priming with decitabine prior to standard induction chemotherapy for patients with AML. Blood. 118, 1472–1480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gore, L. et al A multicenter, randomized study of decitabine as epigenetic priming with induction chemotherapy in children with AML. Clin. Epigenetics. 9, 108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamba, J.K. et al Integrated epigenetic and genetic analysis identifies markers of prognostic significance in pediatric acute myeloid leukemia. Oncotarget 9, 26711–26723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tschirka, J. , Kreisor, M. , Betz, J. & Grundemann, D. Substrate selectivity check of the ergothioneine transporter. Drug Metab. Disposit. 46, 779–785 (2018). [DOI] [PubMed] [Google Scholar]
- 24. Anderson, J.T. , Hu, S. , Fu, Q. , Baker, S.D. & Sparreboom, A. Role of equilibrative nucleoside transporter 1 (ENT1) in the disposition of cytarabine in mice. Pharmacol. Res. Perspect. 7, e00534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eramo, A. et al Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor‐related apoptosis‐inducing ligand‐mediated destruction. Cancer Res. 65, 11469–11477 (2005). [DOI] [PubMed] [Google Scholar]
- 26. Kanda, T. et al 5‐aza‐2'‐deoxycytidine sensitizes hepatoma and pancreatic cancer cell lines. Oncol. Rep. 14, 975–979 (2005). [PubMed] [Google Scholar]
- 27. Zjablovskaja, P. & Florian, M.C. Acute myeloid leukemia: aging and epigenetics. Cancers (Basel) 12, 103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maeda, T. , Hirayama, M. , Kobayashi, D. , Miyazawa, K. & Tamai, I. Mechanism of the regulation of organic cation/carnitine transporter 1 (SLC22A4) by rheumatoid arthritis‐associated transcriptional factor RUNX1 and inflammatory cytokines. Drug Metab. Dispos. 35, 394–401 (2007). [DOI] [PubMed] [Google Scholar]
- 29. Harikrishnan, K.N. et al Alleviating transcriptional inhibition of the norepinephrine SLC6A2 transporter gene in depolarized neurons. J. Neurosci. 30, 1494–1501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiu, X. et al Equitoxic doses of 5‐azacytidine and 5‐aza‐2'deoxycytidine induce diverse immediate and overlapping heritable changes in the transcriptome. PLoS One 5, e12994 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Fig S1
Fig S2
Fig S3


