Abstract
Aim
MicroRNA-21 is an oncogenic miRNA that modulates the expression of multiple cancer-related target genes. We conducted this meta-analysis to assess diagnostic role of circulating miR-21 in CRC, hoping to choose the best biomarker in CRC diagnosis.
Methods
We searched the PubMed, CNKI and WanFang database to identify records related to diagnostic role of circulating miR-21 in CRC. The search words were “microRNA-21”, “miRNA-21”, “colorectal cancer”, “colorectal carcinoma” and “diagnosis”. The searched articles were published before 14th July 2020.
Results
We got 18 studies to conduct this meta-analysis including 1129 blood specimens of CRC patients and 951 control specimens. The meta-analysis showed that the pooled sensitivity and specificity of circulating miR-21 for CRC diagnosis were 77% (95% CI, 70–82) and 83% (95% CI, 78–88). The combined positive likelihood ratio (PLR) was 4.20 (95% CI, 3.12–5.66) and the combined negative likelihood ratio(NLR) was 0.30 (95% CI, 0.23–0.38). The diagnostic odds ratio (DOR) was 16.48 (95% CI 10.09–26.91) and the area under the summary receiver operating characteristic curve (SROC) for the included studies was 0.87(95%CI, 0.84–0.90).
Conclusion
Our meta-analysis results suggest that circulating miR-21 has a potential diagnostic value with moderate sensitivity and good specificity for CRC.
Keywords: MicroRNA-21, colorectal cancer, diagnosis, meta-analysis
1. Introduction
Colorectal cancer (CRC) is a major cause of cancer-related death worldwide [1]. Despite prevention and screening, this disease has an increasing incidence and high mortality. It is the most frequently occurring malignancy in the word, accounting for more than one million cases and 500,000 deaths per year [2,3]. Several CRC screening tests, including faecal occult-blood test and colonoscopy, are frequently used in the detection of CRC. Nevertheless, none of these tests have been established as well-accepted screening tools due to their invasiveness, high cost, or low sensitivity [4]. Therefore, the search for a more sensitive, easy to be detected and representative biomarker is of great significance for CRC early diagnosis and monitoring.
MicroRNAs (miRNAs) are non-coding RNA molecules of about 20 nucleotides long that are involved in regulation of gene expression at the transcriptional and post-translational levels. MiRNAs are extensively implicated in many complex physiological processes such as cellular differentiation, proliferation, and apoptosis [5]. Numerous studies indicate that miRNAs can play an important role as reliable biomarkers for cancer detection and prognostic prediction, and even as novel targets for cancer therapy [6,7].
MiR-21 is an oncogenic miRNA that modulates the expression of multiple cancer-related target genes such as PTEN, TPM1, and PDCD and has been shown to be overexpressed in various human tumors [8–10]. Elevated expression of miR-21 in the tumour tissues of CRC has shown to be as an independent prognostic and predictive biomarker [11–14]. Therefore, there will be great significance to explore diagnostic role of circulating miR-21 in CRC. Although several studies have reported the diagnostic effect of miR-21 in blood samples of CRC patients, the conclusion is various and the number of included patients of each study is not enough [15,16]. Hence, we conducted this meta-analysis to assess diagnostic role of circulating miR-21 in CRC, hoping to choose the best biomarker in CRC diagnosis.
2. Materials and methods
This meta-analysis was performed following the guidelines of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement [17].
2.1. Search strategy
We searched the PubMed, China National Knowledge infrastructure (CNKI) and WanFang database to identify records related to diagnostic role of circulating miR-21 in CRC. The search words were “microRNA-21”, “miRNA-21”, “colorectal cancer”, “colorectal carcinoma” and “diagnosis”. The searched articles were published before 14th July 2020.
2.2. Inclusion criteria
(1) Patients were diagnosed with colorectal cancer. (2) MiRNA-21 expressions in blood specimens of CRC patients were detected by real time quantitative RT-PCR assay. (3) The included study at least reported sensitivity and specificity of miR-21 in diagnosis of CRC patients.
2.3. Exclusion criteria
(1) Letters, reviews, conference abstracts, animal experiments, fundamental research, and duplicated studies were excluded. (2) Studies that did not estimate the diagnostic role of miR-21 in CRC were excluded. (3) Studies whose data could not be used for meta-analysis were excluded.
2.4. Data extraction
According to inclusion and exclusion criteria, two reviewers independently screened all studies to extract data. Extracted data included the first author’s name, publication year, Country, sample size, source of miR-21, cancer type, true positive(TP), false positive(FP), false negative(FN), true negative(TN), sensitivity and specificity. Disagreements were resolved by the third-party adjudication.
2.5. Quality assessment
The quality of each included study was assessed by Quality Assessment of Diagnostic Accuracy Studies(QUADAS) tool.
2.6. Statistical analysis
STATA15.1 software was used to perform the statistical analysis for this meta-analysis. The sensitivity and specifificity of each study were used to construct a 2 × 2 contingency table. The bivariate meta-analysis model was employed to summarise the sensitivity, specifificity, positive likelihood ratio(PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) and generate the bivariate summary receiver operating characteristic curve. The pooled sensitivity, specificity and other related indexes across studies were calculated using a random-effects model. Heterogeneity between studies was evaluated using the chi-squared and I2 tests. A probability value of p < .05 was taken as being statistically significant and I2≥50% indicated the existence of significant heterogeneity [15]. The presence of publication bias was analysed by Deeks' funnel plot asymmetry test. Statistical significance was considered, if publication bias was present at p < .1 [16].
3. Results
3.1. Literature search and study characteristics
As shown in Figure 1, we identified 265 records from PubMed, CNKI and WanFang Databases. After reading titles and abstracts to exclude the duplicates and irrelevant studies, 34 studies remained to review full text to extract available data. As some data in studies could not be used, finally we got 18 studies to conduct meta-analysis to evaluate the diagnostic role of circulating miR-21 in CRC [18–35].
Figure 1.
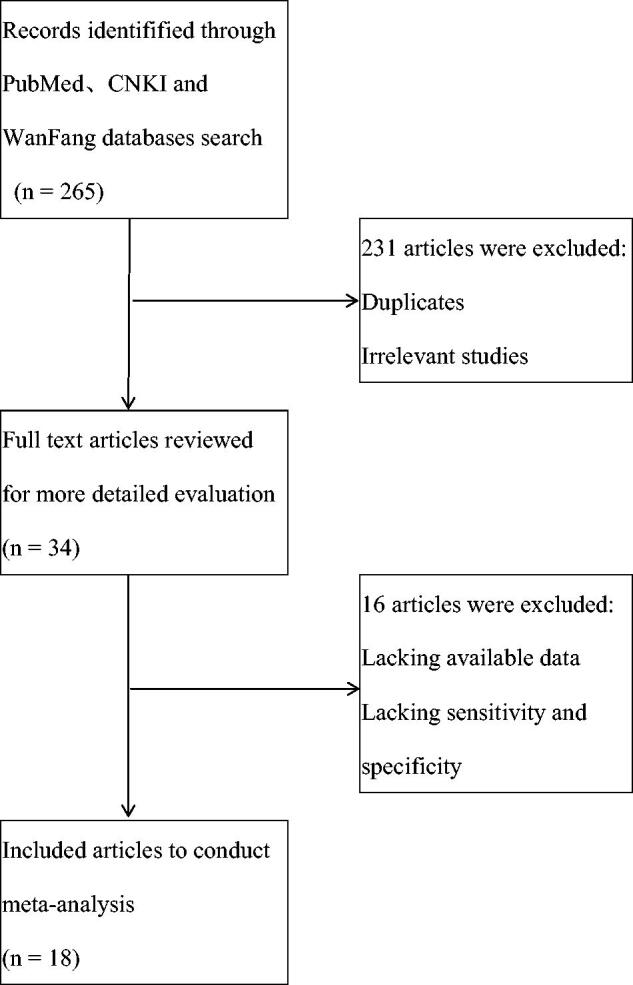
Study flow diagram.
The characteristics of included studies are shown in Table 1. We got 1129 blood specimens of CRC patients and 951 control specimens. The control specimens were from the age and sex matched healthy people. The publication year of included studies were from 2012 to 2020 and the 18 studies came from 8 different countries(China, Slovak, Russia, Japan, Egypt, America, Iran, Germany). Based on QUADAS scores, all included studies were evaluated as high quality.
Table 1.
Characteristics of included studies in this meta-analysis.
| Study (year) | Age (year) |
Gender (M/F) |
Country | Source of control | Source of miR-21 | Samples size |
Cancer type | Test method | Reference gene | Diagnosis parameters |
QUADAS scores | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | C | P | C | P | C | TP | FP | FN | TN | Sen | Spe | |||||||||
| Sazanov et al. [18] (2017) | NA | NA | Russia | Healthy volunteers | Plasma | 31 | 34 | CRC II-IV | qRT-PCR (SYBR Green) | U6 snRNA | 20 | 5 | 11 | 29 | 0.65 | 0.85 | 12 | |||
| Saliva | 34 | 34 | 33 | 3 | 1 | 31 | 0.97 | 0.91 | ||||||||||||
| Basati et al. [19] (2014) | 55.35 ± 10.13 | 55.00 ± 10.35 | 21/19 | 22/18 | Iran | Healthy volunteers | Serum | 40 | 40 | CRC | qRT-PCR (SYBR Green) | RNU6B | 31 | 9 | 9 | 31 | 0.77 | 0.78 | 13 | |
| Liu et al. [20] (2013) | NA | NA | China | Healthy volunteers | Serum | 200 | 80 | CRC | qRT-PCR (TaqMan) | miR-16 | 130 | 12 | 70 | 68 | 0.65 | 0.85 | 12 | |||
| Fouad et al. [21] (2017) | 49.54 ± 15.37 | 49.18 ± 16.17 | 26/24 | 25/25 | Egypt | Healthy volunteers | Serum | 50 | 50 | CRC | qRT-PCR(TaqMan) | RNU6B | 41 | 22 | 9 | 28 | 0.82 | 0.56 | 12 | |
| Sarlinova et al. [22] (2016) | 40-86 | 44-86 | 46/25 | 58/22 | Slovak | Healthy volunteers | Serum | 71 | 80 | CRC | qRT-PCR (TaqMan) | RNU48 | 51 | 26 | 20 | 54 | 0.72 | 0.68 | 12 | |
| Bastaminejad et al. [23] (2017) | NA | 21/19 | 21/19 | Iran | Healthy control | Serum | 40 | 40 | CRC | qRT-PCR (SYBR Green) | miR-16 | 34 | 11 | 6 | 29 | 0.86 | 0.73 | 12 | ||
| Stool | 40 | 40 | 34 | 8 | 6 | 32 | 0.86 | 0.81 | ||||||||||||
| Jin et al. [24] (2020) | 50.9 ± 9.5 | 42.32 ± 9.23 | 38/42 | 28/22 | China | Healthy control | Serum | 80 | 50 | CRC | qRT-PCR (ABI7500) | U6 snRNA | 73 | 7 | 7 | 43 | 0.91 | 0.86 | 13 | |
| Toiyama et al. [25] (2013) | NA | NA | Japan | Healthy volunteers | Serum | 186 | 53 | CRC | qRT-PCR (TaqMan) | cel-miR-39 | 154 | 5 | 32 | 48 | 0.83 | 0.91 | 12 | |||
| Sabry et al. [26] (2018) | 51.97 ± 12.18 | 48.47 ± 15.16 | 19/16 | 52/49 | Egypt | Healthy control | Serum | 35 | 101 | CRC | qRT-PCR (SYBR Green) | RNU6B | 32 | 5 | 3 | 96 | 0.91 | 0.95 | 13 | |
| Hua et al. [27] (2018) | 35-86 | 32-69 | 30/22 | 25/20 | China | Healthy volunteers | Serum | 52 | 45 | CRC | qRT-PCR (SYBR Green) | U6 snRNA | 44 | 9 | 8 | 36 | 0.85 | 0.80 | 13 | |
| Chen et al. [28] (2019) | 58.0 ± 16.5 | 56.5 ± 16.0 | 39/33 | 15/15 | China | Healthy control | Plasma | 72 | 30 | CRC | qRT-PCR | NA | 50 | 1 | 22 | 29 | 0.69 | 0.97 | 12 | |
| Song et al. [29] (2013) | 59.18 ± 11.1 | 55.44 ± 12.1 | 30/10 | 40/16 | China | Healthy control | Serum | 40 | 56 | CRC | qRT-PCR(TaqMan) | miR-16 | 30 | 17 | 10 | 39 | 0.75 | 0.69 | 13 | |
| Wang et al. [30] (2012) | 63 | 46 | 17/15 | 9/30 | China | Healthy individuals | Serum | 32 | 39 | CRC | qRT-PCR (SYBR Green) | miR-16 | 28 | 10 | 4 | 29 | 0.88 | 0.74 | 12 | |
| Ogata-Kawata et al. [31] (2014) | 35-65 | 35-65 | 55/33 | 3/8 | Japan | Physical screening individuals | Serum | 88 | 11 | CRC | qRT-PCR (TaqMan) | miR-451 | 54 | 1 | 34 | 10 | 0.61 | 0.91 | 13 | |
| Zhang et al. [32] (2014) | NA | NA | China | Normal control | Serum | 41 | 30 | CRC | qRT-PCR | U6 snRNA | 21 | 6 | 20 | 24 | 0.51 | 0.79 | 11 | |||
| Du et al. [33] (2014) | 61.1 ± 12.7 | 61.7 ± 12.6 | 30/19 | 30/19 | China | Healthy control | Plasma | 49 | 49 | CRC | NA | NA | 37 | 3 | 12 | 46 | 0.76 | 0.93 | 11 | |
| Luo et al. [34] (2013) | 67.1 ± 11.0 | 61.7 ± 6.4 | 25/25 | 25/25 | Germany | Healthy control | Plasma | 80 | 144 | CRC | qRT-PCR (TaqMan) | miR-16 | 42 | 27 | 38 | 117 | 0.52 | 0.81 | 13 | |
| Kanaan et al. [35] (2012) | 60 ± 11 | 61 ± 9 | 7/13 | 7/13 | America | Healthy control | Plasma | 30 | 30 | CRC | qRT-PCR (TaqMan) | U6 snRNA | 27 | 3 | 3 | 27 | 0.9 | 0.9 | 13 | |
M: Male; F: Female; P: Patient; C: Control; Sen: Sensitivity; Spe: Specificity; NA: Not Available.
3.2. Data analysis
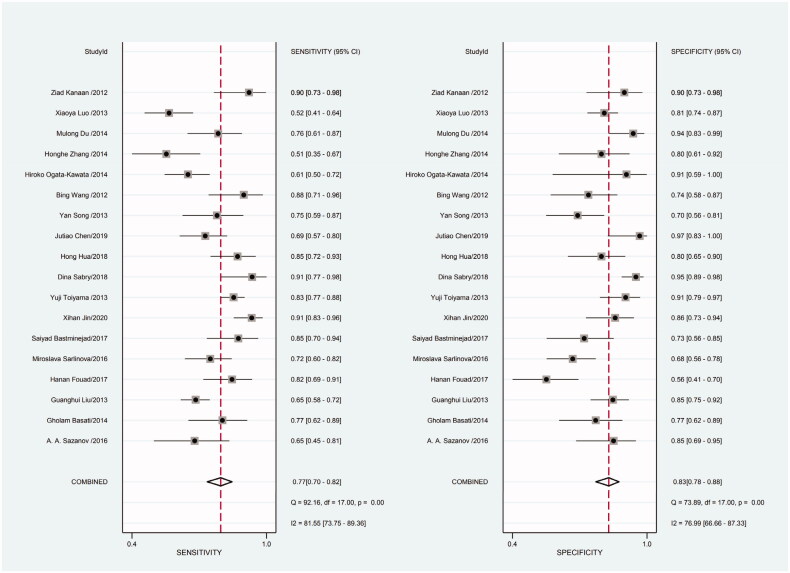
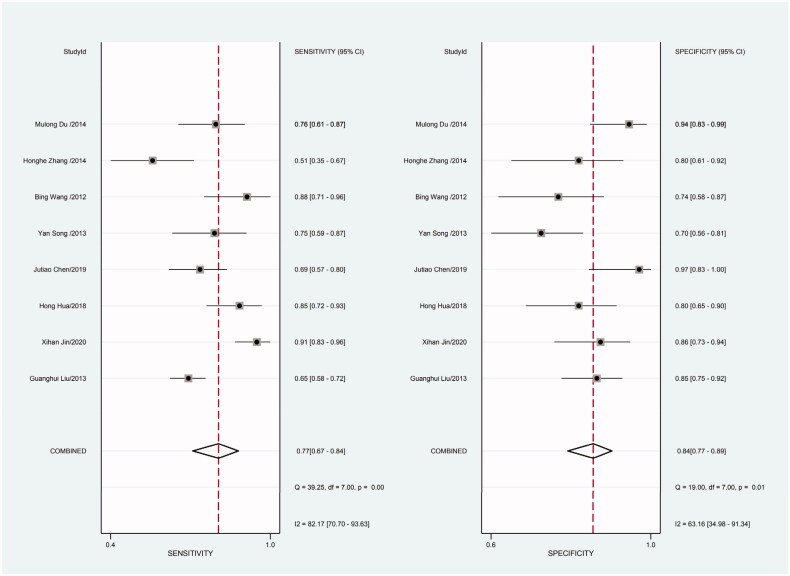
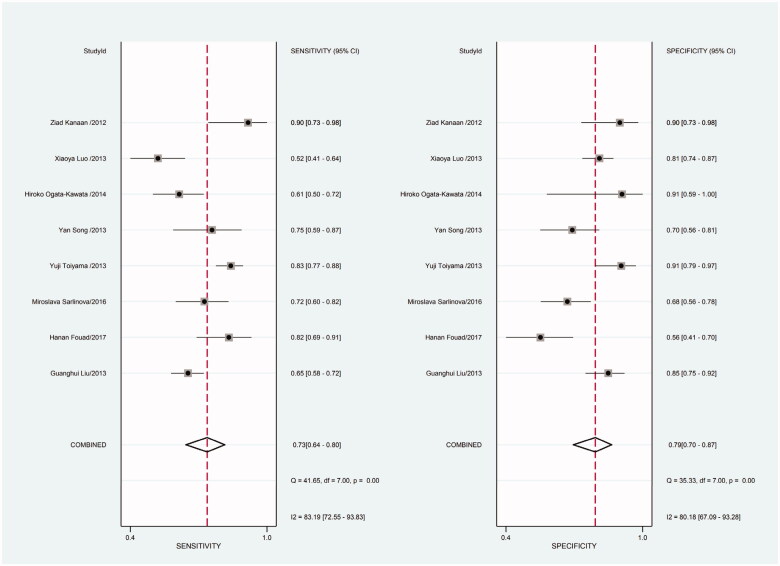
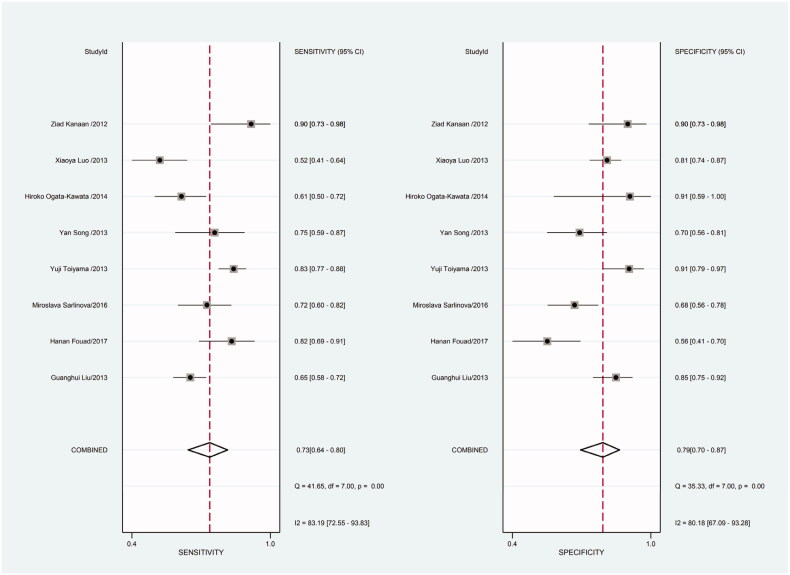
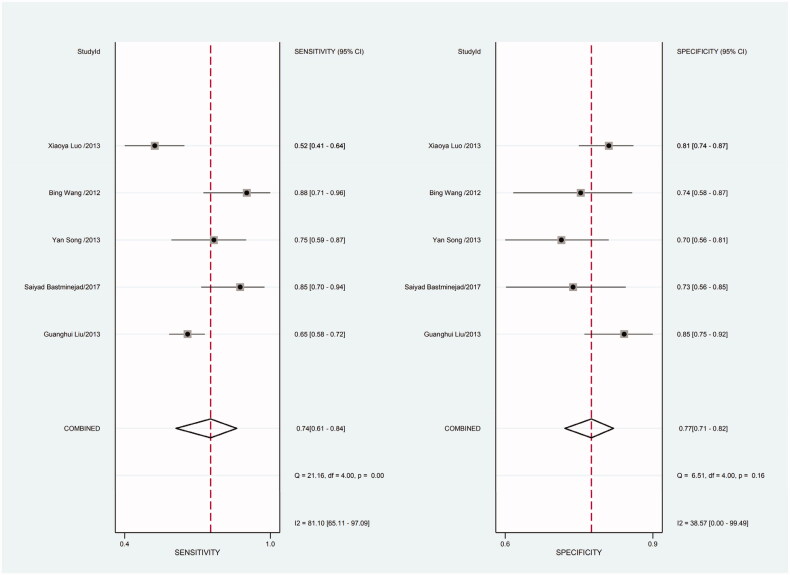
Heterogeneity in sensitivity and specificity was detected in the 18 studies (I2=81.55% and I2=76.99%, respectively), suggesting significant heterogeneity in sensitivity and specificity (Figure 2). For this reason the random-effects model was employed. The meta-analysis showed that the pooled sensitivity and specificity of circulating miR-21 for CRC diagnosis were 77% (95% CI, 70–82) and 83% (95% CI, 78–88), respectively.
Figure 2.
Forest plot of included studies assessing the sensitivity and specificity of circulating miR-21 in CRC.
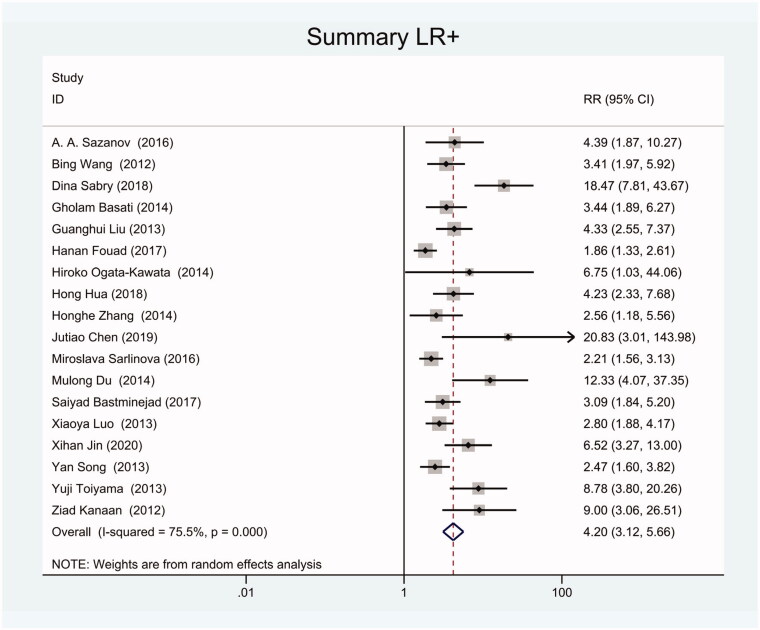
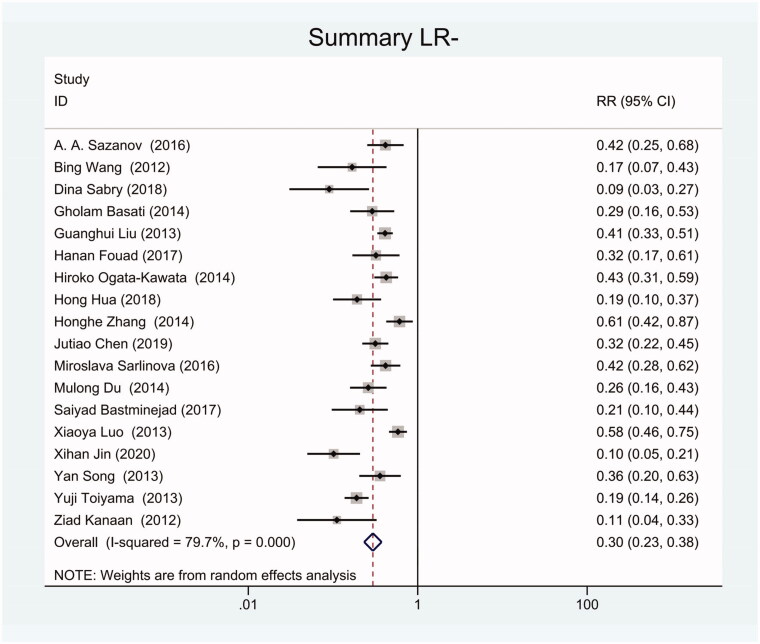
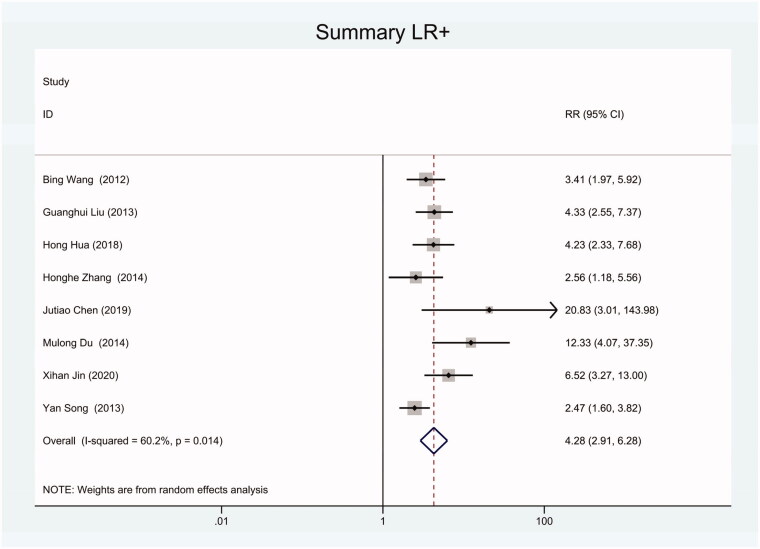
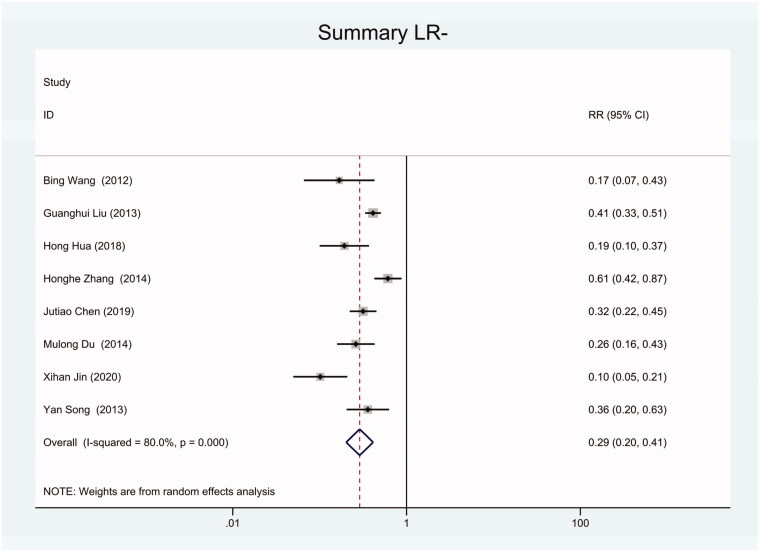
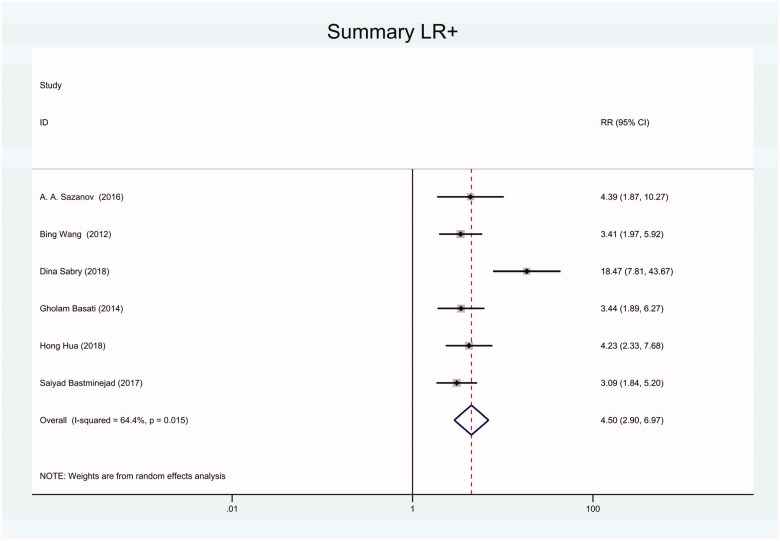
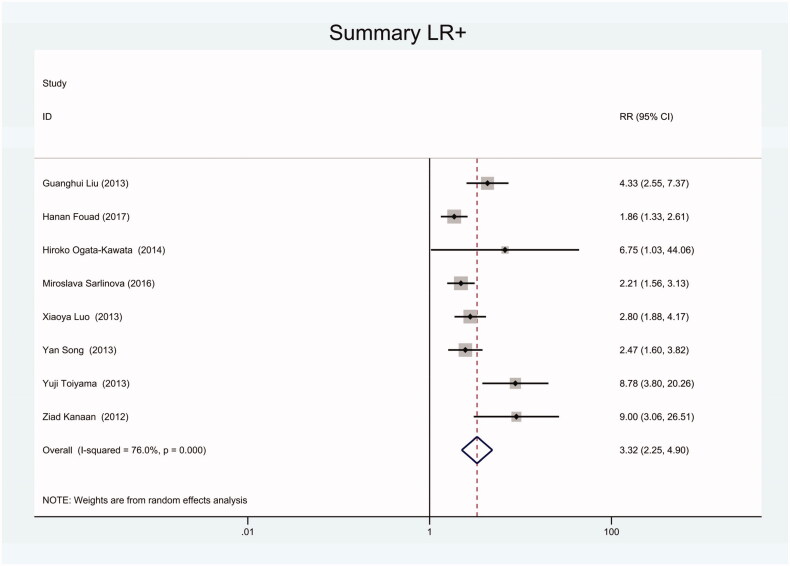
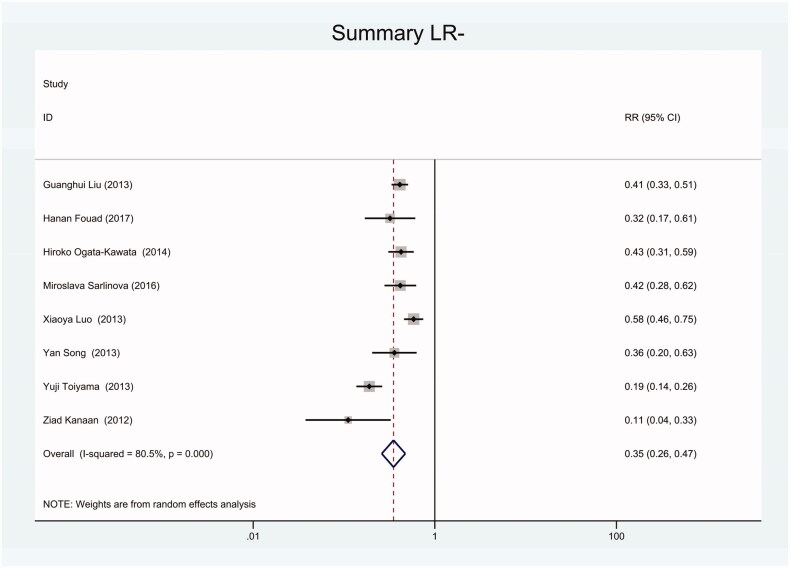
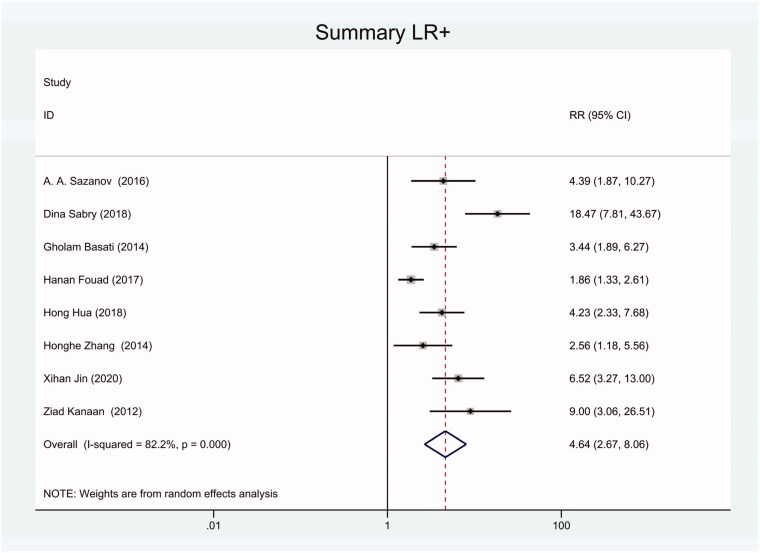
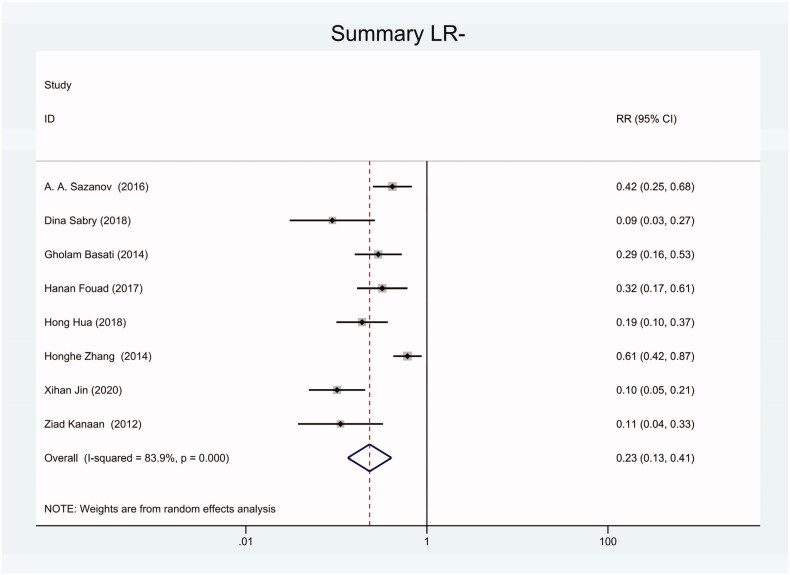
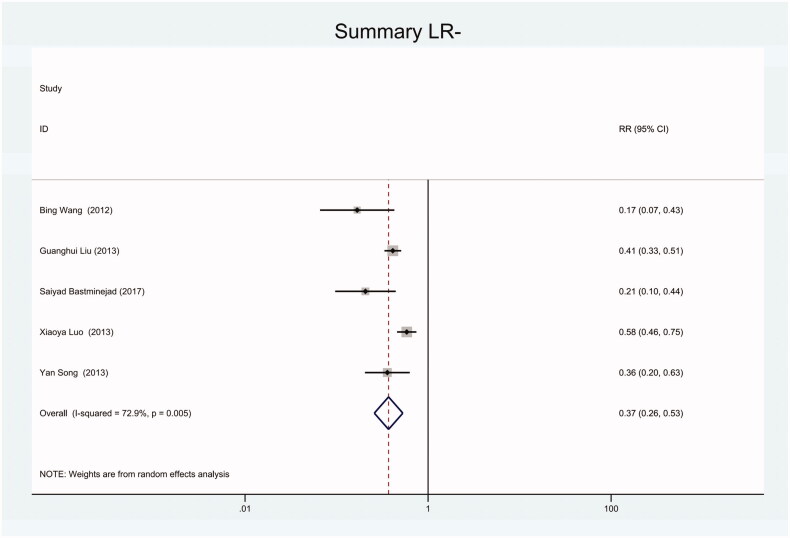
As the positive likelihood ratio(PLR) and negative likelihood ratio(NLR) have been considered more clinically valuable compared to the specificity and sensitivity. In this meta-analysis, the combined PLR was 4.20 (95% CI, 3.12–5.66) (Figure 3), which indicated that patients with CRC had a nearly four-fold greater chance of having an elevated miR-21 in blood specimens compared with healthy individuals. The combined NLR was 0.30 (95% CI, 0.23–0.38) (Figure 4).
Figure 3.
Forest plot of included studies assessing the PLR of circulating miR-21 in CRC.
Figure 4.
Forest plot of included studies assessing the NLR of circulating miR-21 in CRC.
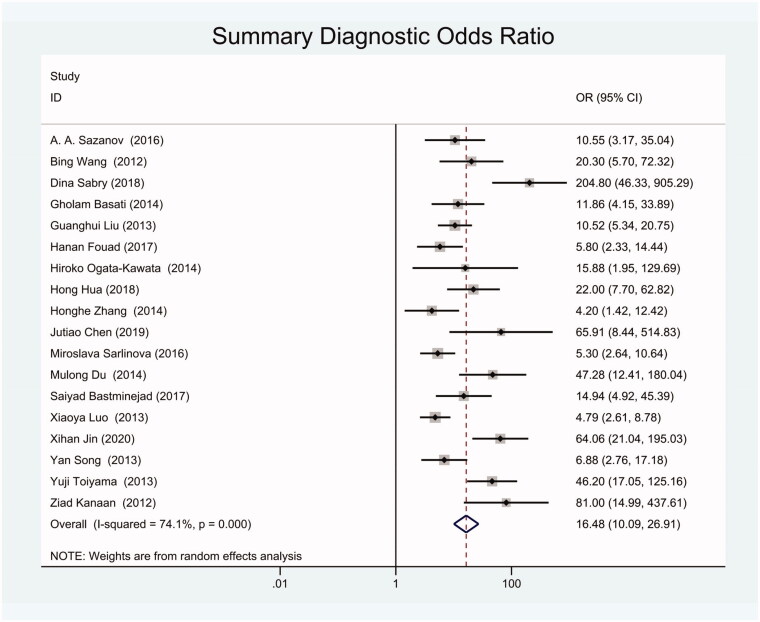
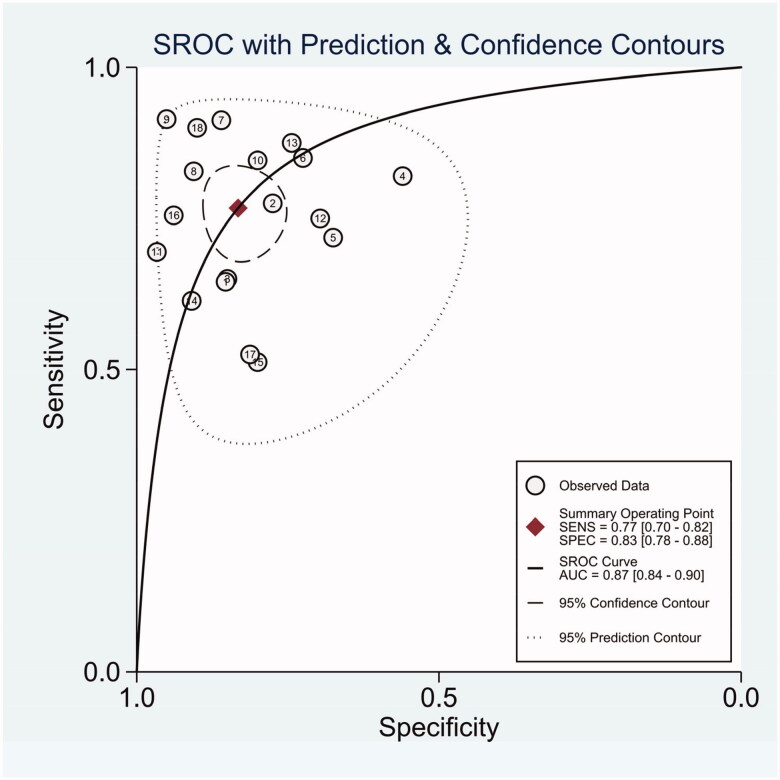
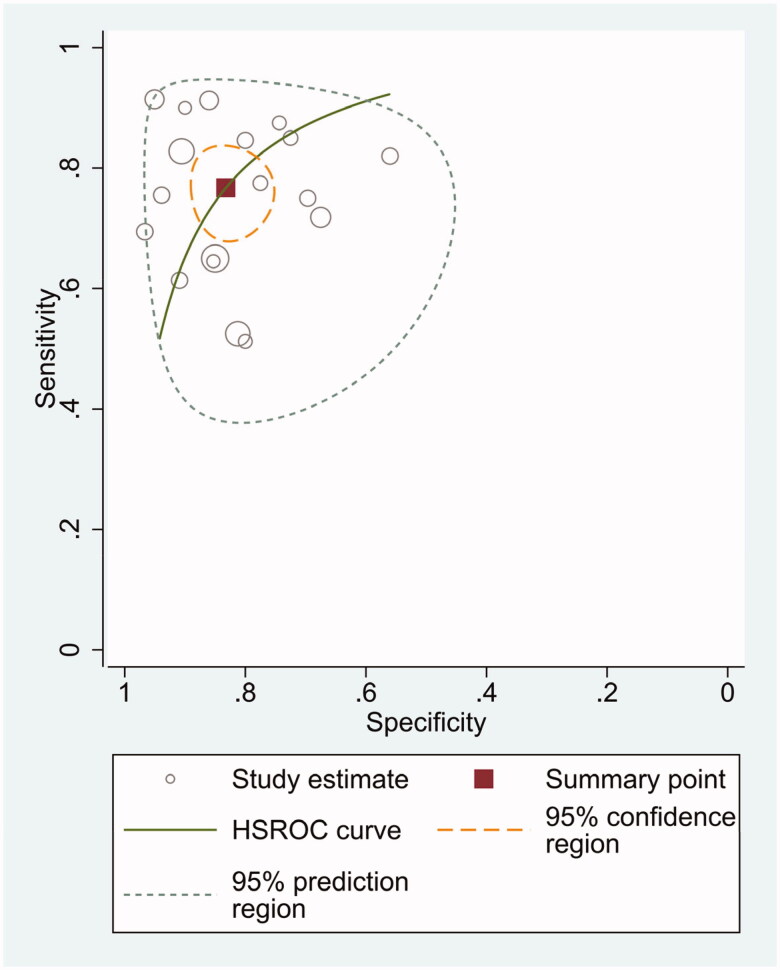
The diagnostic odds ratio (DOR) was 16.48 (95% CI 10.09–26.91) (Figure 5). The summary receiver operating characteristic curve (SROC) for the included studies showed the AUC was 0.87(95%CI, 0.84–0.90) (Figure 6), indicating a good diagnostic accuracy. Hierarchical summary receiver operating characteristics (HSROC) curve was shown in Figure 7, and the value of beta was 0.05(95%CI,−0.59–0.70), and the p-value was .872, indicating HSROC is symmetrical. The value of Lambda was 2.78 (95%CI, 2.27–3.29), indicating that circulating miR-21 is an accurate diagnostic marker for CRC.
Figure 5.
Forest plot of included studies assessing the DOR of circulating miR-21 in CRC.
Figure 6.
SROC curve for miR-21 in CRC diagnosis.
Figure 7.
HSROC curve for miR-21 in CRC diagnosis.
3.3. Publication bias
Deeks' funnel plots were employed to evaluate publication bias in this meta-analysis. As shown in Figure 8, the funnel plot presents symmetry and the p-value was .39, indicating there is no publication bias in this meta-analysis.
Figure 8.
Deeks’ tests for the assessment of publication bias in miR-21 assays.
4. Discussion
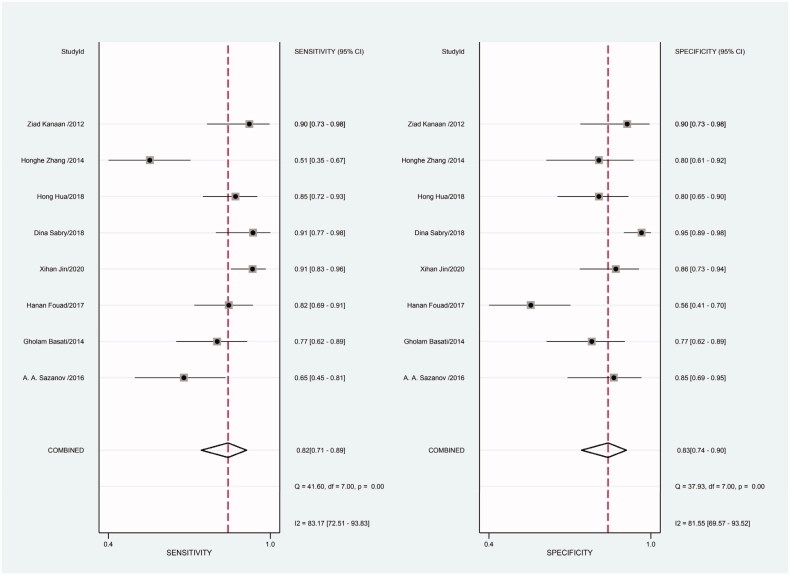
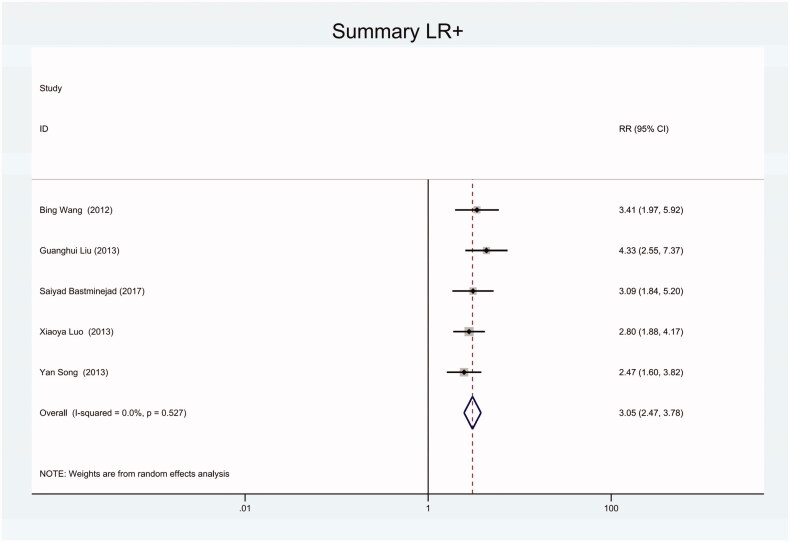
As there was heterogeneity in sensitivity and specificity in this meta-analysis, we employed random-effects model. Besides, we also conducted subgroup meta-analysis. There were 8 included studies coming from China, 6 studies using SYBR-Green qRT-PCR as test method, 8 studies using TaqMan qRT-PCR as test method, 8 studies applying U6 snRNA as internal reference gene and 5 studies applying miR-16 as internal reference gene. We conducted subgroup meta-analysis to observe the heterogeneity of each group respectively (Figures 9–23). The results shown that there was no heterogeneity in specificity in group using miR-16 as internal reference gene, and all the other groups had heterogeneity.
Figure 9.
Forest plot of included Chinese studies assessing the sensitivity and specificity of circulating miR-21 in CRC.
Figure 10.
Forest plot of included Chinese studies assessing the PLR of circulating miR-21 in CRC.
Figure 11.
Forest plot of included Chinese studies assessing the NLR of circulating miR-21 in CRC.
Figure 12.
Forest plot of included studies using SYBR-Green qRT-PCR as test method assessing the sensitivity and specificity of circulating miR-21 in CRC.
Figure 13.
Forest plot of included studies using SYBR-Green qRT-PCR as test method assessing the PLR of circulating miR-21 in CRC.
Figure 14.
Forest plot of included studies using SYBR-Green qRT-PCR as test method assessing the NLR of circulating miR-21 in CRC.
Figure 15.
Forest plot of included studies using TaqMan qRT-PCR as test method assessing the sensitivity and specificity of circulating miR-21 in CRC.
Figure 16.
Forest plot of included studies using TaqMan qRT-PCR as test method assessing the PLR of circulating miR-21 in CRC.
Figure 17.
Forest plot of included studies using TaqMan qRT-PCR as test method assessing the NLR of circulating miR-21 in CRC.
Figure 18.
Forest plot of included studies using U6 snRNA as internal reference gene assessing the sensitivity and specificity of circulating miR-21 in CRC.
Figure 19.
Forest plot of included studies using U6 snRNA as internal reference gene assessing the PLR of circulating miR-21 in CRC.
Figure 20.
Forest plot of included studies using U6 snRNA as internal reference gene assessing the NLR of circulating miR-21 in CRC.
Figure 21.
Forest plot of included studies using miR-16 as internal reference gene assessing the sensitivity and specificity of circulating miR-21 in CRC.
Figure 22.
Forest plot of included studies using miR-16 as internal reference gene assessing the PLR of circulating miR-21 in CRC.
Figure 23.
Forest plot of included studies using miR-16 as internal reference gene assessing the NLR of circulating miR-21 in CRC.
To explore the potential source of heterogeneity, we tried to find other factors in heterogeneity. We supposed whether gender and age might be factors in heterogeneity. To confirm our hypothesis, we performed meta regression to investigate the heterogeneity of covariates. Unfortunately, there was no statistically significant heterogeneity about gender and age in this meta-analysis (Table 2). We suspected the tumour stage of the patients might be the factor in heterogeneity, but most included studies in this meta-analysis did not provide details of the tumour stage. Therefore, we cannot analyse the heterogeneity from tumour stage data.
Table 2.
Possible sources of heterogeneity of covariants in the meta-regression analysis.
| Coef | p | 95%CI | |
|---|---|---|---|
| Age | −0.08 | .2 | (−0.2;0.05) |
| Gender | −0.81 | .17 | (−2.01;0.4) |
| Country | −0.1 | .48 | (−0.42;0.21) |
Coef: Coefficent.
Several studies have reported the meta-analysis result of circulating miR-21 as diagnostic marker in CRC [15,16], and we read these studies for reference. Compared with these meta-analysis published before, we included more studies to conduct this meta-analysis. Based on QUADAS scores, all included studies had high quality. The included studies were from 8 different countries with 1129 blood specimens of CRC patients and 951 control specimens, which enriched the meta-analysis data of miR-21 as diagnostic marker in CRC published before. Hence, this meta-analysis result was reliable and valuable.
5. Conclusion
Our meta-analysis results suggest that circulating miR-21 has a potential diagnostic value with moderate sensitivity and good specificity for CRC. However, more high quality studies about circulating miR-21 diagnostic role should be conducted in future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A.. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. [DOI] [PubMed] [Google Scholar]
- 3.Luo X, Burwinkel B, Tao S, et al. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20(7):1272–1286. [DOI] [PubMed] [Google Scholar]
- 4.Levin B, Lieberman DA, Andrews KS, American College of Radiology Colon Cancer Committee, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58(3):130–160. [DOI] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM.. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. [DOI] [PubMed] [Google Scholar]
- 6.Mishra PJ. MicroRNAs as promising biomarkers in cancer diagnostics. Biomark Res. 2014;2(19):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sempere LF. Recent advances in miRNA-based diagnostic applications. Expert Rev Mol Diagn. 2012;12(6):557–559. [DOI] [PubMed] [Google Scholar]
- 8.Meng F, Henson R, Wehbe-Janek H, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S, Si ML, Wu H, et al. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282(19):14328–14336. [DOI] [PubMed] [Google Scholar]
- 10.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. [DOI] [PubMed] [Google Scholar]
- 11.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profifiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200(2):154–160. [DOI] [PubMed] [Google Scholar]
- 13.Shibuya H, Iinuma H, Shimada R, et al. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79(3-4):313–320. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen BS, Jørgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28(1):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu F, Xu L, Wang M, et al. The accuracy of circulating microRNA-21 in the diagnosis of colorectal cancer: a systematic review and meta-analysis. Colorectal Dis. 2015;17(5):O100–107. [DOI] [PubMed] [Google Scholar]
- 16.Yu W, Wang Z, Shen L, et al. Circulating microRNA-21 as a potential diagnostic marker for colorectal cancer: a meta-analysis. Mol Clin Oncol. 2016;4(2):237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sazanov AA, Kiselyova EV, Zakharenko AA, et al. Plasma and saliva miR-21 expression in colorectal cancer patients. J Appl Genetics. 2017;58(2):231–237. [DOI] [PubMed] [Google Scholar]
- 19.Basati G, Emami Razavi A, Abdi S, et al. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Med Oncol. 2014;31(10):205. [DOI] [PubMed] [Google Scholar]
- 20.Liu G, Zhou Z, Chen R, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumor Biol. 2013;34(4):2175–2181. [DOI] [PubMed] [Google Scholar]
- 21.Fouad H, Sabry D, Morsi H, et al. XRCC1 gene polymorphisms and miR-21 expression in patients with colorectal carcinoma. Eurasian J Med. 2017;49(2):132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarlinova M, Halasa M, Mistuna D, et al. miR 21, miR 221 and miR 150 are deregulated in peripheral blood of patients with colorectal cancer. AR. 2016;36(10):5449–5454. [DOI] [PubMed] [Google Scholar]
- 23.Bastaminejad S, Taherikalani M, Ghanbari R, et al. Investigation of microRNA-21 expression levels in serum and stool as a potential non-invasive biomarker for diagnosis of colorectal cancer. IBJ. 2017;21(2):106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin X-H, Lu S, Wang A-F.. Expression and clinical significance of miR-4516 and miR-21-5p in serum of patients with colorectal cancer. BMC Cancer . 2020;20(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabry D, El-Deek SEM, Maher M, et al. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1α-VEGF signaling pathway. Mol Cell Biochem. 2019;454(1-2):177–189. [DOI] [PubMed] [Google Scholar]
- 27.Hua H, Hao H, Han C, et al. Value of combined detection of serum miR-21 and miR-92 in diagnosis and recurrence of colorectal cancer. Shandong Med. 2018; 58(8):81–83. [Google Scholar]
- 28.Chen J, Wang C, Yuan G, et al. Application of combined detection of miR-21, miR-210 and miR-92a in diagnosis of colorectal cancer. Chin J Health Lab Tec. 2019;2(10):1194–1198. [Google Scholar]
- 29.Song Y, He M, Shao D, et al. Study on the expression of serum miR-21 in patients with benign and malignant colorectal diseases. J Med Res. 2013;42(9):124–128. [Google Scholar]
- 30.Wang B, Zhang Q.. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138(10):1659–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PloS ONE. 2014;9(4):e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Li P, Ju H, et al. Diagnostic and prognostic value of microRNA-21 in colorectal cancer: an original study and individual participant data meta-analysis. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2783–2792. [DOI] [PubMed] [Google Scholar]
- 33.Du M, Liu S, Gu D, et al. Clinical potential role of circulating microRNAs in early diagnosis of colorectal cancer patients. Carcinogenesis. 2014;35(12):2723–2730. [DOI] [PubMed] [Google Scholar]
- 34.Luo X, Stock C, Burwinkel B, et al. Burwinkel Band Brenner H: identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PloS One. 2013;8(5):e62880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544–551. [DOI] [PubMed] [Google Scholar]