Abstract
Introduction
It has been shown that miR-192 is abnormally expressed in a variety of cancer types and participates in different kinds of signaling pathways. The role of miR-192 in the diagnosis and prognosis of cancer has not been verified. This article is aimed at exploring the diagnostic and prognostic value of miR-192 through a systematic review and meta-analysis.
Methods
A systematic search was performed through PubMed, Embase, Web of Science, and Cochrane Library databases up to June 16, 2020. A total of 16 studies were enrolled in the meta-analyses, of which 11 articles were used for diagnostic meta-analysis and 5 articles were used for prognostic meta-analysis. The values of sensitivity and specificity using miR-192 expression as a diagnostic tool were pooled in the diagnostic meta-analysis. The hazard ratios (HRs) of overall survival (OS) with 95 confidence intervals (CIs) were extracted from the studies, and pooled HRs were evaluated in the prognostic meta-analysis. Eleven studies including 667 cancer patients and 514 controls met the eligibility criteria for the diagnostic meta-analysis. Five studies including 166 patients with high miR-192 expression and 236 patients with low miR-192 expression met the eligibility criteria for the prognostic meta-analysis.
Results
The overall diagnostic accuracy was as follows: sensitivity 0.79 (95%CI = 0.75-0.82), specificity 0.74 (95%CI = 0.64-0.82), positive likelihood ratio 3.03 (95%CI = 2.11-4.34), negative likelihood ratio 0.29 (95%CI = 0.23-0.37), diagnostic odds ratio 10.50 (95%CI = 5.89-18.73), and area under the curve ratio (AUC) 0.82 (95%CI = 0.78-0.85). The overall prognostic analysis showed that high expression of miR-192 in patients was associated with positive survival (HR = 0.62, 95%CI : 0.41-0.93, p = 0.020).
Conclusion
Our results revealed that miR-192 was a potential biomarker with good sensitivity and specificity in cancers. Moreover, highly expressed miR-192 predicted a good prognosis for patients.
1. Introduction
Cancer is threatening human health and shorting human life. Around 1.8 million new cancer cases and 60 thousands of cancer deaths occurred in the United States based on the cancer statistics 2020 [1]. One reason for tumor death is that the tumor is already in its advanced stage as soon as it is discovered. In this case, it is important to find a marker that can detect tumors sensitively. However, a sensitive tumor biomarker is lacking in clinical practice. At present, more and more attention has been focused on microRNAs, which are highly conserved, short noncoding RNAs. MicroRNA binds to the 3′ untranslated region of target mRNA by base pairing matching, resulting in degradation of target mRNA or inhibition of protein translation, which is involved in biological progress such as cell growth, differentiation, proliferation, and apoptosis [2]. Besides, miRNAs have been reported to regulate the key characteristics of cancer, involving self-sufficiency in growth signal, antigrowth signal, evasion from apoptosis, limitless replicative potential, angiogenesis, invasion, and metastasis [3]. Therefore, miRNAs could be promising biomarkers for diagnosis and prognosis [4].
MicroRNA-192 was firstly confirmed by Lim et al. in 2003 [5]. It is reported to be overexpressed in gastric cancer [6], hepatocellular carcinoma [7], and neuroblastoma [8], but downregulated in colorectal cancer [9] and lymphoblastic leukemia [10]. MicroRNA-192 plays a critical role in cell proliferation, migration and invasion [11], apoptosis [12], and epithelial-to-mesenchymal transition [13]. More importantly, miR-192 has been consistently detected in sputum [14], cervical cancer tissue, serum [15], and urine [16], suggesting that miR-192 might be a valuable biomarker for cancer diagnosis and detection. However, no meta-analyses concerning association between miR-192 expression and cancer diagnosis and prognosis have been published. Here, we conducted the diagnostic and prognostic meta-analyses to assess the diagnostic and prognostic value of miR-192.
2. Materials and Methods
2.1. Literature Search Strategy
A systematic literature search was carried out in PubMed, Embase, Web of Science, and Cochrane Library databases up to June 2020. The first part was to screen articles that explored the diagnostic value of miR-192 in cancers. Both MeSH terms and free-text words were used in the search strategy. The following search keywords were used in combination: “neoplasms” or “tumor” or “cancer”, “diagnosis”, “ROC curve”, “sensitivity and specificity”, and “microRNA-192”. The second part was to screen articles that explored the prognostic value of miR-192 in cancers. The keywords were as follows: “neoplasms” or “tumor” or “cancer”, “survival”, “prognosis”, “recurrence”, and “microRNA-192”.
2.2. Inclusion and Exclusion Criteria
For diagnostic meta-analysis, studies were included for further evaluation if they meet the following criteria: (1) any types of cancers concerning miR-192, (2) inclusion of a diagnostic standard, (3) sufficient data (true positive, false positive, false negative, and true negative) for calculating the sensitivity and specificity, (4) studies based on humans, and (5) studies published in English. Exclusion criteria were as follows: (1) non-English articles; (2) other types of articles such as conference abstracts, reviews, meta-analysis, patents, case reports, comments, and letters; and (3) insufficient data for calculating the sensitivity and specificity.
For prognostic meta-analysis, studies were included for further evaluation if they meet the following criteria: (1) any types of cancers concerning miR-192, (2) inclusion of a diagnostic standard, (3) associations between the expression of miR-192 and prognosis of patients being determined, (4) hazard ratios (HRs) and their corresponding 95% confidence intervals (CIs) being evaluated, and (5) English publications. Exclusion criteria were as follows: (1) non-English articles; (2) other types of articles such as conference abstracts, reviews, meta-analysis, patents, case reports, comments, and letters; (3) insufficient data to calculate the HRs and 95% CIs; and (4) the prognostic data based on TCGA dataset.
2.3. Data Extraction and Quality Assessment
All studies were independently selected by two investigators (Lili Wang and Yuhan Liu), and uncertain data were reviewed by a third author (Chen Lyu). The following information was collected in diagnostic analysis: first author's name, publication year, nationality, ethnicity, cancer type, sample type, test method, cut-off, case number, sensitivity, and specificity. For the prognostic analysis, the following information was collected: first author's name, publication year, nationality, cancer type, cases of high expression of miR-192, cases of low expression of miR-192, the endpoint of follow-up, and HRs along with their corresponding 95% CIs.
The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS 2) tool [17] was used to assess the quality of articles included in the diagnostic meta-analysis. The Newcastle–Ottawa Scale (NOS) [18] was used to assess the articles in the prognostic meta-analysis.
2.4. Statistical Analysis
All data analyses were performed using Stata MP 16.0 software (StataCorp, College Station, TX). For the diagnostic meta-analysis, the pooled sensitivity, specificity, diagnostic odds ratio (DOR), positive likelihood ratio (PLR), and negative likelihood ratio (NLR) were generated through bivariate meta-analysis. The summary receiver operator characteristic (SROC) curve and the area under the curve (AUC) were calculated to evaluate the overall diagnostic value of miR-192 in cancers. The heterogeneity test was conducted using the chi-square-based Q test and Higgins I-squared statistic. That I2 > 50%, and p < 0.10 indicated heterogeneity among studies. Publication bias was evaluated by funnel plots and by Begg's and Deeks' tests. p < 0.05 suggests the existence of publication bias in studies. For the prognostic meta-analysis, the pooled HRs and 95% CIs were determined using the Z-test, with p < 0.05 defined as significant. HR > 1 indicated poor prognosis for patients with miR-192, while HR < 1 meant a protective effect for the prognosis of highly expressed miR-192. The methods for the assessment of heterogeneity and publication bias were the same as those for the diagnostic meta-analysis.
3. Results
3.1. Study Selection and Characteristics
A total of 387 articles were searched using the search strategy, of which 11 articles [14–16, 19–26] met the inclusion criteria for diagnostic meta-analysis (Figure 1) and included 667 cancer cases and 514 controls. The characteristic details of these articles are summarized in Table 1. The studies involved different types of cancer: non-small-cell lung cancer (n = 2, NSCLC), cholangiocarcinoma (n = 3, CCA), hepatocellular carcinoma (n = 1, HCC), pancreatic ductal adenocarcinoma (n = 1, PDAC), pancreatic cancer (n = 1, PC), cervical cancer (n = 1, CC), bladder cancer (n = 1, BC), and acute myeloid leukemia (n = 1, AML). The expression of miR-192 was evaluated through qRT-PCR in the tissue (n = 3), serum (n = 6), and urine (n = 2).
Figure 1.
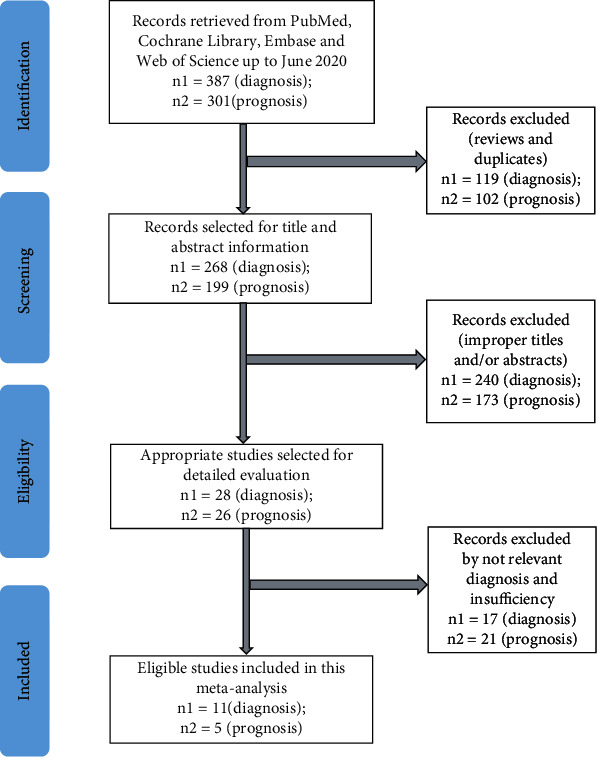
Study selection flowchart.
Table 1.
Main characteristics of the eligible studies for diagnostic meta-analysis.
| First author | Year | Country | Ethnicity | Cancer type | Sample type | Gender case/control male (female) | Up-/downregulation | Single/multiple miRNAs | Test method | Cut-off | Cases/controls | TP | FP | FN | TN | QUADAS-2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bagheri | 2017 | Iran | Asian | NSCLC | Tissue | 15 (2)/15 (2) | Down | Multiple | qRT-PCR | 0.53 | 17/17 | 12 | 9 | 5 | 8 | 5 |
| Wali | 2014 | USA | Caucasian | NSCLC | Tissue | 19 (18)/26 (13) | Up | Multiple | qRT-PCR | NA | 37/39 | 33 | 8 | 4 | 31 | 4 |
| Loosen | 2019 | Germany | Caucasian | CCA | Serum | 54 (40)/31 (9) | Up | Multiple | qRT-PCR | NA | 94/40 | 81 | 4 | 13 | 36 | 6 |
| Silakit | 2014 | Thailand | Asian | CCA | Serum | 32 (19) | Up | Single | qRT-PCR | 0.0054 | 51/30 | 38 | 8 | 13 | 22 | 4 |
| Silakit | 2015 | Thailand | Asian | CCA | Urine | 11 (11)/6 (15) | Up | Multiple | qRT-PCR | 0.936 | 22/21 | 14 | 7 | 8 | 14 | 4 |
| Zhu | 2017 | China | Asian | HCC | Serum | 39 (11)/27 (23) | Up | Multiple | qRT-PCR | 1.1 | 50/50 | 41 | 17 | 9 | 34 | 6 |
| Zhang | 2014 | China | Asian | PDAC | Serum | NA | Up | Multiple | qRT-PCR | 1.15 | 70/40 | 53 | 18 | 17 | 22 | 5 |
| Zou | 2019 | China | Asian | PC | Serum | NA | Up | Multiple | qRT-PCR | 2 | 93/71 | 72 | 31 | 21 | 40 | 5 |
| Farzanehpour | 2019 | Iran | Asian | CC | Tissue | NA | Up | Multiple | qRT-PCR | NA | 18/36 | 14 | 2 | 5 | 34 | 5 |
| Jiang | 2020 | China | Asian | BC | Urine | 82 (36)/79 (41) | Down | Single | qRT-PCR | NA | 118/120 | 91 | 26 | 27 | 94 | 5 |
| Tian | 2018 | China | Asian | AML | Serum | 57 (40) | Down | Single | qRT-PCR | NA | 97/50 | 77 | 9 | 20 | 41 | 5 |
NSCLC: non-small-cell lung cancer; CCA: cholangiocarcinoma; HCC: hepatocellular carcinoma; PDAC: pancreatic ductal adenocarcinoma; PC: pancreatic cancer; CC: cervical cancer; BC: bladder cancer; AML: acute myeloid leukemia; NA: not available; TP: true positive; FP: false positive; FN: false negative; TN: true negative; QUADAS-2: Quality Assessment of Diagnostic Accuracy Studies 2.
For the prognostic meta-analysis, 301 articles were obtained from four databases, and only 5 articles ([23, 27–30]) met the inclusion criteria (Figure 1) and included 166 high-miR-192 cases and 236 low-miR-192 cases. Details concerning the included articles are displayed in Table 2. The studies included several types of cancer: gastric cancer (n = 1, GC), colon cancer (n = 1, COC), small-cell lung cancer (n = 1, SCLC), hepatocellular carcinoma (n = 1, HCC), and acute myeloid leukemia (n = 1, AML). The expression of miR-192 was assessed through qRT-PCR using plasma (n = 1), tissue (n = 3), and serum (n = 1).
Table 2.
Main characteristics of the eligible studies for prognostic meta-analysis.
| Study | Year | Country | Cancer type | Sample type | Gender high/low male (female) | Up-/downregulation | Single/multiple miRNAs | Methods | Cases high/low | Cut-off | Survival | HR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chen 2014 | 2014 | China | GC | Plasma | NA | Up | Multiple | qRT-PCR | 29/33 | NA | OS | 0.894 (0.391-2.041) |
| Li 2018 | 2018 | China | CoC | Tissue | 20 (18)/20 (40) | Down | Single | qRT-PCR | 38/60 | 97.43 | OS | 0.500 (0.390-0.610) |
| Lian 2016 | 2016 | China | HCC | Tissue | NA | Down | Single | qRT-PCR | 24/71 | NA | OS | 0.267 (0.081-0.887) |
| Mancuso 2016 | 2016 | Italy | SCLC | Tissue | 32 (18) | Up | Multiple | qRT-PCR | 25/25 | 13.59 | OS | 1.330 (0.720-2.450) |
| Tian 2018 | 2018 | China | AML | Serum | 31 (19)/26 (21) | Down | Single | qRT-PCR | 50/47 | NA | OS | 0.490 (0.342-0.758) |
GC: gastric cancer; CoC: colon cancer; SCLC: small-cell lung cancer; HCC: hepatocellular carcinoma; AML: acute myeloid leukemia; high: high miR-192 expression; low: low miR-192 expression; NA: not available; HR: hazard ratio.
3.2. Quality Assessment of Studies
The quality of diagnostic meta-analysis was assessed using the QUADAS-2 tool. All the studies were scored between 4 and 6 points which represented moderate or high quality (Table 1). For the prognostic meta-analysis, the Newcastle–Ottawa Scale (NOS) tool was used to assess the quality of studies according to three elements: selection (0-4 points), comparability (0-2 points), and outcome (0-3 points). All the studies were assessed as moderate or high quality, with scores between 5 and 7 points (Table 3).
Table 3.
Newcastle–Ottawa Scale scores of studies.
| Study | Selection | Comparability | Outcome | Total |
|---|---|---|---|---|
| Chen 2014 | ★★★★ | ★★ | 6 | |
| Li 2018 | ★★★★ | ★ | ★★ | 7 |
| Lian 2016 | ★★★★ | ★ | 5 | |
| Mancuso 2016 | ★★★★ | ★★ | 6 | |
| Tian 2018 | ★★★★ | ★ | ★★ | 7 |
3.3. The Results of the Diagnostic Meta-Analysis
The sensitivity and specificity of 11 studies are presented in the forest plots as shown in Figure 2. There was no heterogeneity in the sensitivity (I2 = 9.63%, 95%CI = 0-61.53%), but significant heterogeneity in the specificity (I2 = 76.64%, 95%CI = 63.03%-90.24%). Overall, the sensitivity and specificity for the pooled data were 0.79 (95%CI = 0.75-0.82) and 0.74 (95%CI = 0.64-0.82). In addition, the pooled PLR was 3.03 (95%CI = 2.11-4.34), and the NLR was 0.29 (95%CI = 0.23-0.37) as shown in Figure 3. The DOR was 10.50 (95%CI = 5.89-18.73, Figure 4). The SROC curve is shown in Figure 5. The AUC for the miR-192 test method was 0.82 (95%CI = 0.78-0.85), suggesting that miR-192 has a relatively high diagnostic value. Fagan's nomogram was applied for assessing the clinical utility of the index test shown in Figure 6. When miR-192 was tested in patients with a pretest probability of cancer of 50%, the posttest probability of having cancer was improved to 75% by a positive result, while the posttest probability without cancer was dropped to 22% by a negative result. Taken together, miR-192 had a relatively moderate accuracy for identification of cancer patients.
Figure 2.
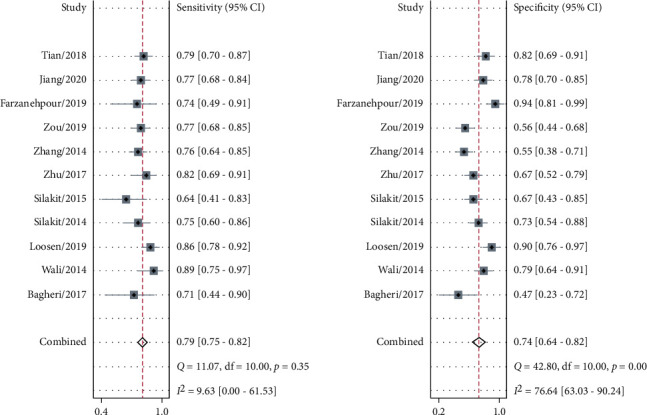
Forest plot of pooled sensitivity and specificity for 11 studies in the diagnostic meta-analysis.
Figure 3.
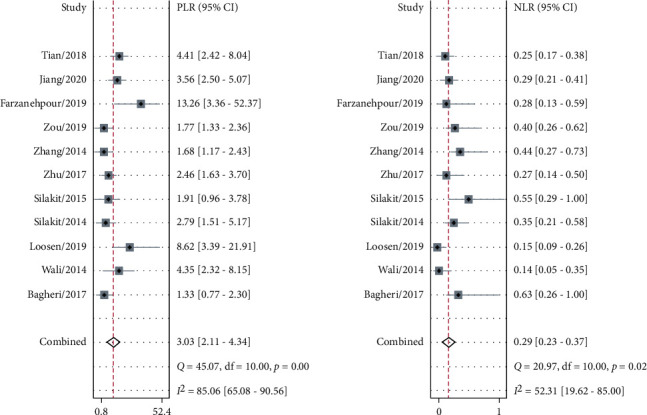
Forest plot of the positive likelihood ratio (PLR) and negative likelihood ratio (NLR) for miR-192 in the diagnostic meta-analysis.
Figure 4.
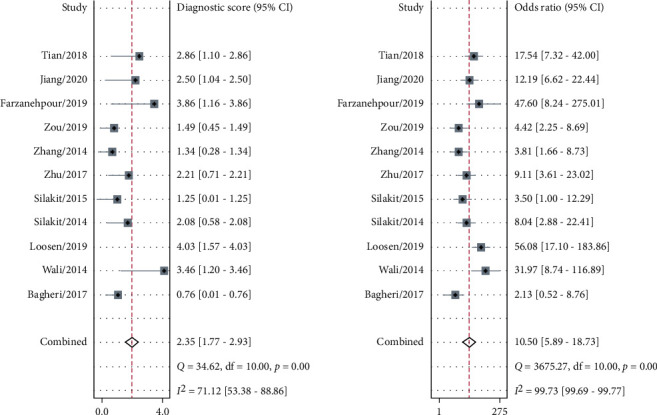
Forest plot of the diagnostic odds ratio for miR-192 in the diagnostic meta-analysis.
Figure 5.
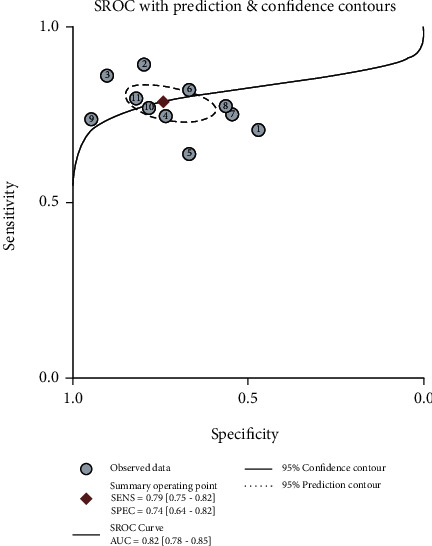
SROC curve for miR-192 in the diagnostic meta-analysis. SENS: pooled sensitivity; SPEC: pooled specificity; AUC: area under the curve.
Figure 6.
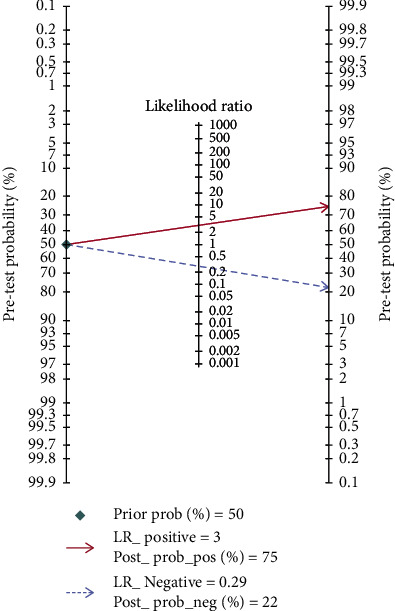
Fagan's nomogram was used to assess the post-test probabilities. LR: likelihood ratio.
3.4. Influence Analysis and Robustness Test
The goodness-of-fit and bivariate normality (Figures 7(a) and 7(b)) analyses suggested that the bivariate model was moderately robust. Influence analysis (Figure 7(c)) and outlier detection (Figure 7(d)) did not identify any outliers.
Figure 7.
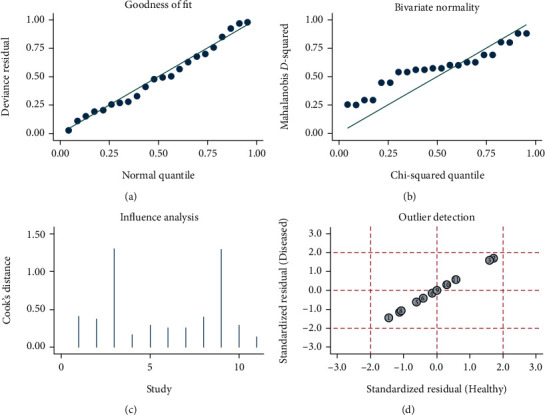
Sensitivity analyses: (a) goodness-of-fit; (b) bivariate normality; (c) influence analysis; (d) outlier detection.
3.5. Publication Bias
Deeks' funnel plot asymmetry test suggested that there was no significant publication bias (p = 0.82, Figure 8).
Figure 8.
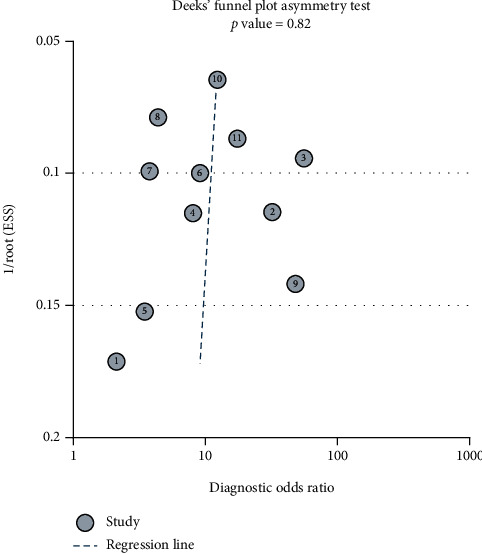
Deeks' funnel plot asymmetry test showed that the p value was 0.82 indicating that there was no publication bias.
3.6. Threshold Effect and Heterogeneity
The ROC plane showed the appearance of a nontypical shoulder arm suggesting no threshold effect existing (Figure 9). Spearman's correlation coefficient was –0.374 (p = 0.258), also indicating no threshold effect existing. The Galbraith radial plot showed that all the studies were in the 95% CI region suggesting no heterogeneity (Figure 10(a)). The bivariate boxplot showed that most studies were scattered in the middle region except three studies suggesting that there was heterogeneity between studies (Figure 10(b)). Due to only 11 studies included in the diagnostic meta-analysis, it was difficult to perform the subgroup and meta-regression analyses to investigate the sources of heterogeneity.
Figure 9.
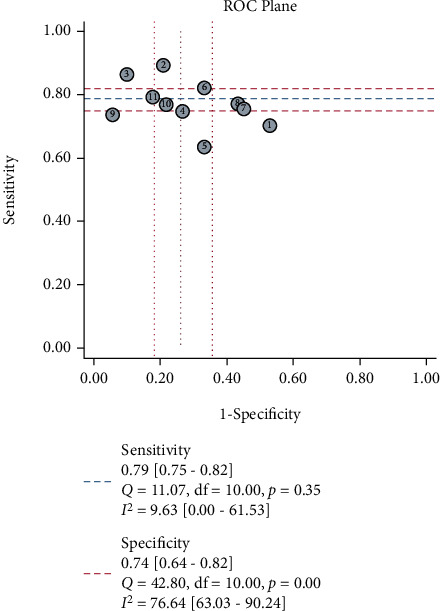
ROC plane showed the results of sensitivity, specificity, Q test, and I2 result.
Figure 10.
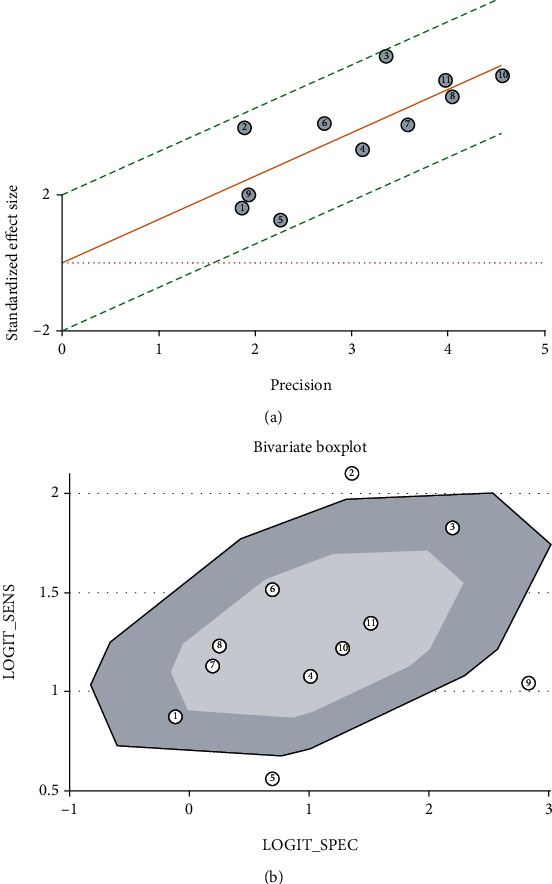
Heterogeneity test: (a) the Galbraith radial plot showed that all the studies were in the 95% CI region suggesting no heterogeneity; (b) there was heterogeneity for three studies beyond the middle region in the bivariate boxplot.
3.7. A Prognostic Meta-Analysis of the Relationship between miR-192 Expression and Prognosis in Cancers
Five studies were used to assess the OS shown in Figure 11. There was statistically significant heterogeneity (I2 = 65.9%, p = 0.019), so a random effects model was used. The pooled HR was 0.62 (95%CI = 0.41-0.93, p = 0.020), suggesting that a high level of miR-192 was associated with positive patients' survival. The funnel plot was symmetrical, and Begg's test (p = 0.806) also indicated that there was no publication bias (Figure 12).
Figure 11.
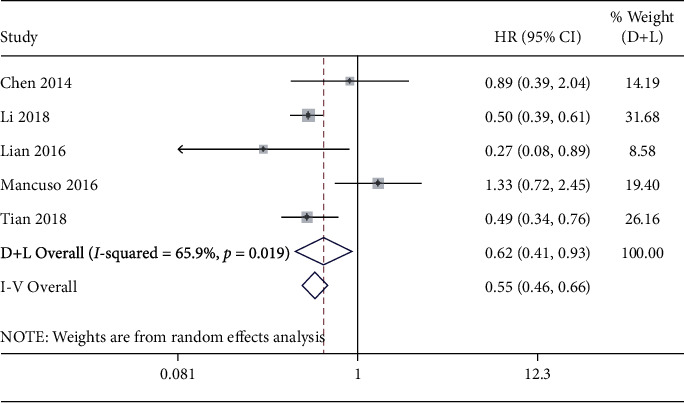
Forest plot of pooled HRs for miR-192 in the prognostic meta-analysis.
Figure 12.
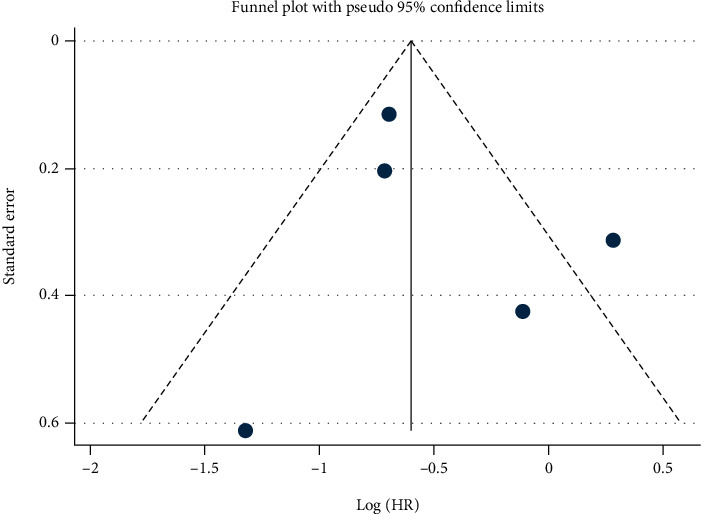
Begg's funnel plot for the prognostic meta-analysis.
4. Discussion
At present, imaging examination is used as a means of preliminary diagnosis and the final diagnosis still relies on the invasive biopsy. Once discovered, most cancers have entered the advanced stage, which causes great difficulties in treatment. Moreover, biopsy progress is invasive and may cause tumor dissemination [31]. Therefore, cancers might be detected at an early stage, if some biomarkers can be used for tumor screening clinically, providing the possibility of a cure. It is also possible to judge the prognosis by biomarkers, which is convenient, fast, and economical.
miRNA is a type of small noncoding RNA that plays important regulatory roles in gene expression and various biological processes [32]. Recently, miRNAs have been shown to have the potential to predict the diagnosis and prognosis of cancer patients [33], which could be used as diagnostic and prognostic biomarkers. miR-192 is one of those miRNAs reported to be a potential diagnostic and prognostic marker [23, 28].
Some studies have revealed that miR-192 was associated with the progression of cancers. miR-192 inhibits cell proliferation, induces apoptosis, and is a positive prognostic factor in human breast cancer [12], acute lymphoblastic leukemia [34], osteosarcoma [11, 35], and colon cancer [9]. However, miR-192 also induces the proliferation and is a poor prognostic factor in neuroblastoma [8], gastric cancer [6], NSCLC [14], cholangiocarcinoma [19], hepatocellular carcinoma [25], pancreatic cancer [26], cervical cancer [15], bladder cancer [16], and AML [23].
The roles of miR-192 in different types of cancers are controversial. In breast cancer, BMP-6 inhibits cell proliferation through upregulating miR-192 [36], and miR-192 inhibits the proliferation and induces the apoptosis in breast cancer through targeting caveolin 1 [12]. In acute lymphoblastic leukemia, overexpression of miR-192 results in cell proliferation arrest and apoptosis increasing in ALL cells through upregulating P53, BAX, and CASP3 [34]. Upregulation of miR-192 suppresses the progress of osteosarcoma through targeting USP1 [35], TCF7 [37], and XIAP [38]. miR-192 also benefits the prognosis of colon cancer patients by regulating SRPX2 [39], Rab-2A [40], RhoA-ROCK-LIMK2 [41], farnesoid X receptor [42], RB1/E2F1 pathway [43], and NOD2 [44]. miR-192 also suppresses the growth of bladder cancer cells via targeting Yin Yang 1 [45]. However, miR-192 serves as a poor prognosis marker in other cancer types including neuroblastoma targeting Dicer1 [8], gastric cancer targeting RAB11-FIP2 [46] and SMG-1 [47], NSCLC targeting the FGFR3/RB1 pathway [48], hepatocellular carcinoma targeting SEMA3A [49], and pancreatic cancer targeting SIP1 [50]. The summary of different interactions of miR-192 with proteins or genes in different cancer types is shown in Table 4. miR-192 has been reported to be induced or inhibited by different agents. Some agents such as nicotine [48], baicalin [51], sinomenine [52], astragaloside IV [13], captopril and spironolactone [53], paclitaxel [54], and doxorubicin [55] can upregulate miR-192, while other agents including metabolites of intestinal microflora [41], curcumin [56, 57], and lactobacillusin [58] can downregulate the expression of miR-192 (Table 5).
Table 4.
Proteins interacting with miR-192 in different cancer types.
| Prognosis | Cancer type | miR-192-related regulatory proteins | References |
|---|---|---|---|
| Positive | Breast cancer | BMP-6/miR-192/caveolin 1 | [12, 36] |
| ALL | miR-192/P53, BAX, CASP3 | [34] | |
| OS | miR-192/USP1, TCF7, XIAP | [35, 37, 38] | |
| Colon cancer | miR-192/SRPX2, Rab-2A, RhoA-ROCK-LIMK2, farnesoid X receptor, RB1-E2F1, NOD2 | [39–44] | |
| HCC | miR-192/SLC39A6-SNAIL | [29] | |
| BC | miR-192/transcription factor Yin Yang 1 | [45] | |
|
| |||
| Negative | NB | miR-192/Dicer1 | [8] |
| GC | miR-192/RAB11-FIP2, SMG-1 | [46, 47] | |
| NSCLC | miR-192/FGFR3-RB1 | [48] | |
| PDAC | miR-192/SIP1 | [50] | |
ALL: acute lymphoblastic leukemia; OS: osteosarcoma; HCC: hepatocellular carcinoma; NB: neuroblastoma; BC: bladder cancer; GC: gastric cancer; NSCLC: non-small-cell lung carcinoma; PDAC: pancreatic ductal adenocarcinoma.
Table 5.
Agents that induce or inhibit miR-192.
This study is the first meta-analysis regarding the diagnostic and prognostic value of miR-192 in various cancers. The results are as follows: AUC 0.82 (95%CI = 0.78-0.85), sensitivity 0.79 (95%CI = 0.75-0.82), and specificity 0.74 (95%CI = 0.64-0.82), demonstrating that miR-192 might be used as a novel biomarker for the detection of cancers. The pooled DOR was 10.50 (95%CI = 5.89-18.73) suggesting that miR-192 is reliably used in cancer diagnosis.
The pooled PLR was 3.03 (95%CI = 2.11-4.34), and NLR was 0.29 (95%CI = 0.23-0.37), meaning that cancer patients had a higher 3.03-fold probability of being miR-192 positive compared to control patients, and the probability of a negative result in patients was 0.29 times that in nonpatients. Fagan's nomogram revealed that when a pretest probability of 50% was specified, the positive posttest probability would increase to 75%, and the negative posttest probability would decrease to 22%. The results suggest that miR-192 is reliable for the detection and diagnosis in NSCLC, CCA, HCC, PDAC, PC, CC, BC, and AML.
The pooled HR was 0.62 (95%CI = 0.41-0.93, p = 0.020), suggesting that a high level of miR-192 was associated with positive patients' survival in GC, CoC, HCC, SCLC, and AML.
In the diagnostic meta-analysis, the sensitivity had no heterogeneity (I2 = 9.63%, 95%CI = 0-61.53%), but there was significant heterogeneity in the specificity (I2 = 76.64%, 95%CI = 63.03%-90.24%). One reason for the heterogeneity perhaps was that the ethnicity of patients in most studies was Asian, which may result in the bias. Secondly, the cut-off values in studies were different and some of them were not mentioned. Thirdly, different sample types might contribute to the heterogeneity. However, the ROC plane represented a nontypical shoulder arm-like appearance, and Spearman's correlation coefficient was -0.374 (p = 0.258), suggesting that there was no threshold effect. Therefore, the threshold effect was not a major cause of heterogeneity.
For the prognostic meta-analysis, there was also heterogeneity (I2 = 65.9%, p = 0.019). However, meta-regression analyses and subgroup analysis cannot be performed because of insufficient study numbers.
The present study has several limitations: firstly, the number of studies available for meta-analysis was small; secondly, the kind of ethnicity was monotonous; thirdly, the values of cut-off in the studies were partial and different; finally, it is difficult to conduct subgroup analysis, such as the influence of gender and tumor stage on the results due to the limited data of original articles.
5. Conclusions
In conclusion, this study demonstrates that miR-192 has a moderate diagnostic value to distinguish cancer patients from controls and also can be a promising positive prognostic biomarker in some types of cancer.
Acknowledgments
The study was supported by the LMU Clinic. The author Lili Wang is funded by the China Scholarship Council (CSC No. 201706270196).
Data Availability
All data generated or analyzed during this study are included in this article. More information concerning the data can be obtained from the corresponding author.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA: a Cancer Journal for Clinicians. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Sato F., Tsuchiya S., Meltzer S. J., Shimizu K. MicroRNAs and epigenetics. The FEBS Journal. 2011;278(10):1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 3.Negrini M., Nicoloso M. S., Calin G. A. MicroRNAs and cancer--new paradigms in molecular oncology. Current Opinion in Cell Biology. 2009;21(3):470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Ghafouri-Fard S., Shoorei H., Taheri M. Role of microRNAs in the development, prognosis and therapeutic response of patients with prostate cancer. Gene. 2020;759:p. 144995. doi: 10.1016/j.gene.2020.144995. [DOI] [PubMed] [Google Scholar]
- 5.Lim L. P., Glasner M. E., Yekta S., Burge C. B., Bartel D. P. Vertebrate microRNA genes. Science. 2003;299(5612):p. 1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 6.Jin Z., Selaru F. M., Cheng Y., et al. MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 2011;30(13):1577–1585. doi: 10.1038/onc.2010.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan Y., Ge G., Pan T., et al. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9(9, article e107986) doi: 10.1371/journal.pone.0107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg-Gorenshtein G., Guedj A., Shichrur K., et al. miR-192 directly binds and regulates Dicer1 expression in neuroblastoma. PLoS One. 2013;8(11, article e78713) doi: 10.1371/journal.pone.0078713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song B., Wang Y., Kudo K., Gavin E. J., Xi Y. miR-192 regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clinical Cancer Research. 2008;14(24):8080–8086. doi: 10.1158/1078-0432.CCR-08-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schotte D., De Menezes R. X., Akbari Moqadam F., et al. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96(5):703–711. doi: 10.3324/haematol.2010.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang G., Mi Y., Mei Y., et al. MicroRNA-192 inhibits the proliferation, migration and invasion of osteosarcoma cells and promotes apoptosis by targeting matrix metalloproteinase-11. Oncology Letters. 2018;15(5):7265–7272. doi: 10.3892/ol.2018.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P., Feng Y., Zhang H., et al. MicroRNA‑192 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting caveolin 1. Oncology Reports. 2019;42(5):1667–1676. doi: 10.3892/or.2019.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y., Zhang L., Wang Y., Fan Q., Cong Y. Astragaloside IV attenuates renal fibrosis through repressing epithelial-to-mesenchymal transition by inhibiting microRNA-192 expression: <i>in vivo</i> and <i>in vitro</i> studies. American Journal of Translational Research. 2019;11(8):5029–5038. [PMC free article] [PubMed] [Google Scholar]
- 14.Bagheri A., Khorshid H. R. K., Mowla S. J., et al. Altered miR-223 expression in sputum for diagnosis of non-small cell lung cancer. Avicenna Journal of Medical Biotechnology. 2017;9(4):189–195. [PMC free article] [PubMed] [Google Scholar]
- 15.Farzanehpour M., Mozhgani S. H., Jalilvand S., et al. Serum and tissue miRNAs: potential biomarkers for the diagnosis of cervical cancer. Virology Journal. 2019;16(1):p. 116. doi: 10.1186/s12985-019-1220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang F., Li C., Han J., Wang L. Diagnostic value of combination of microRNA-192 in urinary sediment and B-ultrasound for bladder cancer. Technology in Cancer Research & Treatment. 2020;19, article 153303381989457 doi: 10.1177/1533033819894573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiting P. F., Rutjes A. W., Westwood M. E., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 18.Wells G. A. S. B., O'Connell D., Peterson J., Welch V., Losos M., Tugwell P. The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. 2012, http://wwwohrica/programs/clinical_epidemiology/oxfordasp.
- 19.Loosen S. H., Lurje G., Wiltberger G., et al. Serum levels of miR-29, miR-122, miR-155 and miR-192 are elevated in patients with cholangiocarcinoma. PLoS One. 2019;14(1, article e0210944) doi: 10.1371/journal.pone.0210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wali R. K., Hensing T. A., Ray D. W., et al. Buccal microRNA dysregulation in lung field carcinogenesis: gender-specific implications. International Journal of Oncology. 2014;45(3):1209–1215. doi: 10.3892/ijo.2014.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silakit R., Loilome W., Yongvanit P., et al. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: a prospective prognostic indicator. Journal of Hepato-Biliary-Pancreatic Sciences. 2014;21(12):864–872. doi: 10.1002/jhbp.145. [DOI] [PubMed] [Google Scholar]
- 22.Silakit R., Loilome W., Yongvanit P., et al. Urinary microRNA-192 and microRNA-21 as potential indicators for liver fluke-associated cholangiocarcinoma risk group. Parasitology International. 2017;66(4):479–485. doi: 10.1016/j.parint.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Tian C., Zhang L., Li X., Zhang Y., Li J., Chen L. Low miR-192 expression predicts poor prognosis in pediatric acute myeloid leukemia. Cancer Biomarkers. 2018;22(2):209–215. doi: 10.3233/CBM-170657. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Zhao C. Y., Zhang S. H., et al. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncology Reports. 2014;31(3):1157–1164. doi: 10.3892/or.2013.2960. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H. T., Liu R. B., Liang Y. Y., et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver International. 2017;37(6):888–896. doi: 10.1111/liv.13356. [DOI] [PubMed] [Google Scholar]
- 26.Zou X., Wei J., Huang Z., et al. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Medicine. 2019;8(6):2810–2822. doi: 10.1002/cam4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q., Ge X., Zhang Y., et al. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncology Reports. 2014;31(4):1863–1870. doi: 10.3892/or.2014.3004. [DOI] [PubMed] [Google Scholar]
- 28.Li P., Ou Q., Braciak T. A., Chen G., Oduncu F. S. MicroRNA-192-5p is a predictive biomarker of survival for stage IIIB colon cancer patients. Japanese Journal of Clinical Oncology. 2018;48(7):619–624. doi: 10.1093/jjco/hyy019. [DOI] [PubMed] [Google Scholar]
- 29.Lian J., Jing Y., Dong Q., et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7(3):2672–2683. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancuso G., Bovio E., Rena O., et al. Prognostic impact of a 3-microRNA signature in cytological samples of small cell lung cancer. Cancer Cytopathology. 2016;124(9):621–629. doi: 10.1002/cncy.21729. [DOI] [PubMed] [Google Scholar]
- 31.Paranjape T., Slack F. J., Weidhaas J. B. MicroRNAs: tools for cancer diagnostics. Gut. 2009;58(11):1546–1554. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Wu Q., Yang Z., Shi Y., Fan D. miRNAs in human cancers: the diagnostic and therapeutic implications. Current Pharmaceutical Design. 2014;20(33):5336–5347. doi: 10.2174/1381612820666140128204914. [DOI] [PubMed] [Google Scholar]
- 34.Sayadi M., Ajdary S., Nadali F., Rostami S., Edalati Fahtabad M. Tumor suppressive function of microRNA-192 in acute lymphoblastic leukemia. Bosnian Journal of Basic Medical Sciences. 2017;17(3):248–254. doi: 10.17305/bjbms.2017.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S., Xiong M., Dai G., et al. MicroRNA-192-5p suppresses the initiation and progression of osteosarcoma by targeting USP1. Oncology Letters. 2018;15(5):6947–6956. doi: 10.3892/ol.2018.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu F., Meng X., Tong Q., et al. BMP-6 inhibits cell proliferation by targeting microRNA-192 in breast cancer. Biochimica et Biophysica Acta. 2013;1832(12):2379–2390. doi: 10.1016/j.bbadis.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Zhang S., Xu Y., et al. Upregulation of miR-192 inhibits cell growth and invasion and induces cell apoptosis by targeting TCF7 in human osteosarcoma. Tumour Biology. 2016;37(11):15211–15220. doi: 10.1007/s13277-016-5417-z. [DOI] [PubMed] [Google Scholar]
- 38.Li H., He L., Tuo Y., Huang Y., Qian B. Circular RNA hsa_circ_0000282 contributes to osteosarcoma cell proliferation by regulating miR-192/XIAP axis. BMC Cancer. 2020;20(1):p. 1026. doi: 10.1186/s12885-020-07515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J., Xu J., Zhang R. SRPX2 regulates colon cancer cell metabolism by miR-192/215 via PI3K-Akt. American Journal of Translational Research. 2018;10:483–490. [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng X. F., Liu K. X., Wang X. M., Zhang R., Li X. MicroRNA‑192 acts as a tumor suppressor in colon cancer and simvastatin activates miR‑192 to inhibit cancer cell growth. Molecular Medicine Reports. 2019;19(3):1753–1760. doi: 10.3892/mmr.2019.9808. [DOI] [PubMed] [Google Scholar]
- 41.Huang Y. L., Li X. H., Ma H., Yue H. Y., Hu X. Y. Metabolites of intestinal microflora upregulate miR-192-5p to suppress proliferation of colon cancer cells via RhoA-ROCK-LIMK2 pathway. European Review for Medical and Pharmacological Sciences. 2020;24(4):1794–1806. doi: 10.26355/eurrev_202002_20357. [DOI] [PubMed] [Google Scholar]
- 42.Krattinger R., Bostrom A., Schioth H. B., Thasler W. E., Mwinyi J., Kullak-Ublick G. A. MicroRNA-192 suppresses the expression of the farnesoid X receptor. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2016;310(11):G1044–G1051. doi: 10.1152/ajpgi.00297.2015. [DOI] [PubMed] [Google Scholar]
- 43.Kang D. W., Lee S. W., Hwang W. C., et al. Phospholipase D1 acts through Akt/TopBP1 and RB1 to regulate the E2F1-dependent apoptotic program in cancer cells. Cancer Research. 2017;77(1):142–152. doi: 10.1158/0008-5472.CAN-15-3032. [DOI] [PubMed] [Google Scholar]
- 44.Chuang A. Y., Chuang J. C., Zhai Z., Wu F., Kwon J. H. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflammatory Bowel Diseases. 2014;20(1):126–135. doi: 10.1097/01.MIB.0000436954.70596.9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji D., Jiang L., Li Y. miR-192-5p suppresses the growth of bladder cancer cells via targeting Yin Yang 1. Human Cell. 2018;31(3):210–219. doi: 10.1007/s13577-018-0201-6. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Peng Y., Huang Y., et al. Inhibition of the miR-192/215-Rab11-FIP2 axis suppresses human gastric cancer progression. Cell Death & Disease. 2018;9(7):p. 778. doi: 10.1038/s41419-018-0785-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Peng Y., Huang Y., et al. SMG-1 inhibition by miR-192/-215 causes epithelial-mesenchymal transition in gastric carcinogenesis via activation of Wnt signaling. Cancer Medicine. 2018;7(1):146–156. doi: 10.1002/cam4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du X., Qi F., Lu S., Li Y., Han W. Nicotine upregulates FGFR3 and RB1 expression and promotes non-small cell lung cancer cell proliferation and epithelial-to-mesenchymal transition via downregulation of miR-99b and miR-192. Biomedicine & Pharmacotherapy. 2018;101:656–662. doi: 10.1016/j.biopha.2018.02.113. [DOI] [PubMed] [Google Scholar]
- 49.Yan-Chun L., Hong-Mei Y., Zhi-Hong C., Qing H., Yan-Hong Z., Ji-Fang W. MicroRNA-192-5p promote the proliferation and metastasis of hepatocellular carcinoma cell by targeting SEMA3A. Applied Immunohistochemistry & Molecular Morphology. 2017;25(4):251–260. doi: 10.1097/PAI.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 50.Zhao C., Zhang J., Zhang S., et al. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncology Reports. 2013;30(1):276–284. doi: 10.3892/or.2013.2420. [DOI] [PubMed] [Google Scholar]
- 51.Kang C., Wang L., Kang M., Liu X., Fu Y., Gao J. Baicalin alleviates 6-hydroxydopamine-induced neurotoxicity in PC12 cells by down-regulation of microRNA-192-5p. Brain Research. 2019;1708:84–92. doi: 10.1016/j.brainres.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y., Yu C., Zhang H. Lipopolysaccharides-mediated injury to chondrogenic ATDC5 cells can be relieved by sinomenine via downregulating microRNA-192. Phytotherapy Research. 2019;33(7):1827–1836. doi: 10.1002/ptr.6372. [DOI] [PubMed] [Google Scholar]
- 53.Ebadi Z., Moradi N., Kazemi Fard T., et al. Captopril and spironolactone can attenuate diabetic nephropathy in Wistar rats by targeting microRNA-192 and microRNA-29a/b/c. DNA and Cell Biology. 2019;38(10):1134–1142. doi: 10.1089/dna.2019.4732. [DOI] [PubMed] [Google Scholar]
- 54.Sun L., Zhang D., Liu F., et al. Low-dose paclitaxel ameliorates fibrosis in the remnant kidney model by down-regulating miR-192. The Journal of Pathology. 2011;225(3):364–377. doi: 10.1002/path.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rizzo S., Cangemi A., Galvano A., et al. Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget. 2017;8(42):71924–71932. doi: 10.18632/oncotarget.18028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin H., Qiao F., Wang Y., Xu Y., Shang Y. Curcumin inhibits cell proliferation and induces apoptosis of human non-small cell lung cancer cells through the upregulation of miR-192-5p and suppression of PI3K/Akt signaling pathway. Oncology Reports. 2015;34(5):2782–2789. doi: 10.3892/or.2015.4258. [DOI] [PubMed] [Google Scholar]
- 57.Ye M., Zhang J., Zhang J., Miao Q., Yao L., Zhang J. Curcumin promotes apoptosis by activating the p53-miR-192-5p/215-XIAP pathway in non-small cell lung cancer. Cancer Letters. 2015;357(1):196–205. doi: 10.1016/j.canlet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 58.Archambaud C., Nahori M. A., Soubigou G., et al. Impact of lactobacilli on orally acquired listeriosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(41):16684–16689. doi: 10.1073/pnas.1212809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article. More information concerning the data can be obtained from the corresponding author.


