Abstract
Background
Natural killer (NK) cells provide important immune protection from cancer and are a key requirement for particular immunotherapies. There is accumulating evidence that NK cells become dysfunctional during cancer. Overcoming NK cell exhaustion would be an important step to allow them to function optimally in a range of NK cell therapies, including those that depend on autologos circulating NK cells. We have previously demonstrated that NK cells undergo a normal metabolic reprogramming in response to cytokine activation and that this is required for optimal function. The objective of this work was to investigate if cellular metabolism of circulating NK cells is dysregulated in patients with metastatic breast cancer and if so, to gain insights into potential mechanisms underpinning this. Such discoveries would provide important insights into how to unleash the full activity of NK cells for maximum immunotherapy output.
Methods
Single-cell analysis, metabolic flux and confocal analysis of NK cells from patients with metastatic breast cancer and healthy controls
Results
In addition to reduced interferon-γ production and cytotoxicity, peripheral blood NK cells from patients had clear metabolic deficits including reduced glycolysis and oxidative phosphorylation. There were also distinct morphologically alterations in the mitochondria with increased mitochondrial fragmentation observed. Transforminggrowth factor-β (TGFβ) was identified as a key driver of this phenotype as blocking its activity reversed many metabolic and functional readouts. Expression of glycoprotein-A repetitions predominant (GARP) and latency associated peptide (LAP), which are involved with a novel TGFβ processing pathway, was increased on NK cells from some patients. Blocking the GARP–TGFβ axis recapitulated the effects of TGFβ neutralization, highlighting GARP as a novel NK cell immunotherapy target for the first time.
Conclusions
TGFβ contributes to metabolic dysfunction of circulating NK cells in patients with metastatic breast cancer. Blocking TGFβ and/or GARP can restore NK cell metabolism and function and is an important target for improving NK cell-based immunotherapies.
Keywords: killer cells, natural, immunity, innate, immune evation, immunologic surveillance, breast Neoplasms
Introduction
Natural killer (NK) cells are cytotoxic lymphocytes with important roles in the immune responses to cancer.1 They provide a key primary immune defense against cancer and have shown great potential for immunotherapy.2 3 NK cells are currently used for both autologos and allogeneic immunotherapy, and offer advantages over T cells for chimeric antigen receptor (CAR)-based cell therapy.4 However, one limiting factor is that during cancer, NK cells themselves may become dysfunctional,5 6 reducing the effectiveness of NK cell mediated therapies. The impact of the cancer environment on NK cells is a profound and systemic one, as circulating NK cells, the source of cells for adoptive immunotherapy, also have impaired functions.7–9 Given that systemic and not intratumoral, immune activation has recently been shown to predict successful antibody mediated immunotherapy outcome,10 understanding how and why peripheral blood NK cells are impaired during cancer is an important step towards restoring their functions for improved immunotherapy.
Significant progress has been made in understanding how cellular metabolism regulates immune cell function. We have begun to define the normal metabolic changes that NK cells undergo in response to stimulation.11–15 These changes are important for growth and proliferation but also impact on NK cell effector functions. Here, we hypothesized that impaired metabolism underpins metabolic dysfunction of circulating human NK cells during cancer. Support for this comes from observations that intertumoral CD8 T cells from murine cancer models and from human tumors have distinct metabolic changes including fragmented mitochondria16 17 and this has also recently been described for tumor infiltrating NK cells.18
Herein, we show that peripheral NK cells from patients with metastatic breast cancer had impaired production of interferon-γ (IFNγ), reduced expression of TNF-related apotosis-inducing ligand (TRAIL) and reduced cytotoxicity against K562 tumor cells. Importantly, this observed NK cell dysfunction was associated with distinct metabolic defects including an altered mitochondrial phenotype and impaired oxidative phosphorylation (OXPHOS) response on cytokine stimulation. In terms of identifying a mechanism that contributes to metabolic dysfunction, we found that transforming growth factor-β (TGFβ), which we have previously demonstrated to be a homeostatic regulator of normal NK cell metabolism,19 significantly contributed to the pathological dysfunction of NK cell metabolism and function in circulating NK cells from patients with metastatic breast cancer. Crucially, both NK cell metabolic and functional parameters were significantly improved when TGFβ, including NK cell derived, was neutralized. Furthermore, we identified that glycoprotein-A repetitions predominant (GARP),20 a receptor which anchors endogenously produced latent TGFβ, is constitutively overexpressed, along with latency associated peptide (LAP), on NK cells of some patients. Targeting GARP/TGFβ complexes on purified patient NK cells recapitulated the effects of TGFβ neutralization. These data reveal a potential new pathway of endogenous TGFβ-dependent inhibition of NK cells as an important mechanism leading to NK cell dysfunction in cancer. Understanding mechanisms of dysregulated NK cell metabolism during cancer will allow specific targeting to improve NK cell function and their usefulness as a cancer immunotherapy.
Materials and methods
Cell culture
Peripheral blood mononuclear cells (PBMC) were isolated by Lymphoprep (Axis-Shield) gradient. For seahorse analysis, NK cells were purified using the EasySep negative selection purification kit (STEMCELL) as per the manufacturer’s instructions. Unless stated otherwise, 5×106 cells/mL PBMC or purified NK cells were incubated at 37°C for 18 hours in RPMI V.1640 GlutaMAX medium (Life Technologies/Invitrogen) supplemented with 10% fetal calf serum (FCS), 1% penicillin/streptomycin (Invitrogen). Cells were stimulated with interleukin (IL)2 (500 IU/mL; National Cancer Institute) or IL12 (30 ng/mL; Miltenyi Biotec) and IL15 (100 ng/mL; National Cancer Institute). Where indicated, cells were cultured with or without IgG1 isotype control (5 or 10 µg/mL), anti-TGFβ MAb (5 µg/mL, 1D11, BioTechne/R&D Systems), anti-GARP MAb (10 µg/mL, Sophie Lucas lab, Belgium) or recombinant human TGFβ (10 µg/mL, Bio-Techne).
Patients and healthy controls
Blood samples were obtained from normal healthy donors (all female, mean age 45 years) or from patients with breast cancer (all female, mean age 58 years) from whom informed written consent had been obtained. Patients had metastatic breast cancer with mixed hormone status (table 1). They had not received conventional chemotherapy within the last year. Some patients were on one or more of the following therapies: fulvestrant, zometa, letrozole, trastuzumab, palbociclib, pertuzumab, xgeva, tamoxifen, decapeptyl and goserelin.
Table 1.
| Hormone status | No. patients |
| PR+ | 0 |
| PR+ HER2+ | 1 |
| ER− HER2− PR− | 2 |
| ER+ HER2+ | 6 |
| ER+ | 7 |
| HER2+ | 8 |
| ER+HER+ PR+ | 15 |
| ER+ PR+ | 37 |
Flow cytometry analysis
Cells were stained for 30 min at 4 °C with saturating concentrations of titered Abs CD56 (HCD56/NCAM16.2), CD3 (SK7/UCHT1), granzyme B (GB11), IFNγ (B27), CD71 (M-A172), CD69 (L78), CD98 (UM7F8), NKp44 (p44-8.1), TRAIL (RiK-2), pSMAD2/3 (O72-670), CD25 (M-A251) (eBioscience or BD Pharmingen); S6 ribosomal protein phosphorylated on serine 235/6 (pS6), and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) phosphorylated on Thr37/46 (236B4, Cell Signaling Technology), LAP (TW4-2F8) and GARP (7B11). A viability dye was included in every panel (LIVE/DEAD Near-IR, Bio Sciences). Cells were prepared, stained and analyzed as previously described.21
Mitochondrial membrane potential (MMP) was measured via staining of cells for 30 min with tetramethylrhodamine methyl ester (TMRM, 100 nM—Thermo Fisher), oligomycin (2 µM) and carbonyl cyanide p-trifluoro-methoxyphenyl hydrazone (FCCP, 2 µM) were used as positive and negative controls, respectively. Mitochondrial mass was measured via staining of cells for 30 min with MitoTracker Red (100nM—Thermo Fisher) or MitoTracker Green (100nM—Thermo Fisher). Adenosine triphosphate (ATP) synthase analysis was performed by detecting the expression of the ATP5B subunit of the (3D5, Abcam) via intracellular flow cytometry staining. Mitochondrial superoxide levels were measured via staining of cells for 15 min with MitoSOX (Thermo Fisher). Rotenone (20 µM) and MitoTEMPO (2.5 µM) were used as positive and negative controls, respectively.
Metabolism analysis
Determination of oxygen consumption rate (OCR) representing OXPHOS or extracellular acidification rate (ECAR) indicating glycolysis was detected by XFp extracellular flux analyzer (Agilent Technologies). NK cells were purified from PBMC using the EasySep negative selection purification kit (STEMCELL) as per the manufacturer’s instructions; purity was routinely >90% CD56+CD3− NK cells. Cells were stimulated for 18 hours in the presence or absence of IL2. In order to adhere NK cells to the bottom of the seahorse plate, cell plates were coated with Cell-Tak (6 µg/mL). The Cell-Tak was diluted in sodium bicarbonate (0.1M) with 0.15% (v/v) NaOH (1M) and added to the bottom of each well (25 µl). It was left at room temperature (RT) for a minimum of 20 min, removed from the plate and each well was washed two times with sterile ddH2O. NK cells were washed two times in GlutaMAX seahorse media supplemented with glucose (1M), adjusted to pH 7.4. NK cells were added to each well (2.5×105 cells, 180 µl), while seahorse media were used in the blank wells. The cell plate was centrifuged at 200g for 3 min with no brake, and then placed in a non-CO2 incubator for 30 min prior to metabolic analysis. During the assay, the following inhibitors were added in order—oligomycin (2 µM), FCCP (0.5 µM), rotenone (100 nM)+antimycin A (4 µM) and 2-deoxyglucose (30 mM).
Confocal imaging and analysis of mitochondrial morphology
Purified NK cells (8×105 cells and >90% purity) were stained using Mitospy CMX Ros (250 nM, Biolegend) for 30 min at 37°C and fixed in 2% paraformaldehyde (PFA, Sigma) for 15 min at RT, prior to nuclear staining with 4',6-diamidino-2-phenylindole, dihydrochloride (300 nM, Thermo Fischer Scientific) for 5 min at room temperature. NK cells were mounted using Mowiol (Sigma) and imaged on a Leica SP8 inverted motorized microscope equipped with a ×63/1.4 N.A. oil objective and 405 nm diode and Leica white laser lines. Z-stacks at 0.2 µm increments were captured using an HyD detector in conjunction with Leica LAS X acquisition software.
Analysis of mitochondrial morphology
Image analysis was performed on the maximum intensity projections of the z-stack. NK cell mitochondrial morphology was assessed by blind scoring of a mixed healthy and patient dataset (total number of cells=84). Six volunteers blindly scored whether morphology was predominantly fragmented or tubular, and if mitochondria were predominantly single or networked, as per an ordinal scale provided in online supplemental figure 1. The mode of each cell score was collated and the per cent morphology score was calculated for each individual donor as follows: (number of cells with each score divided by the total number of donor cells)×100. The per cent morphology scores were then averaged in healthy and patient groups and compared using a two-way analysis of variance (ANOVA).
jitc-2020-002044supp001.pdf (2.5MB, pdf)
Mitochondrial fluorescence and morphology were also quantified in Fiji (http://fiji.sc/Fiji) by delineating the mitochondria as regions of interest (ROI) and measuring their integrated density or circularity, respectively, using the following formulae: Integrated density: (raw integrated density of ROI)×(area of ROI). Circularity=4π (area of ROI÷〖perimeter〗2 of ROI). A circularity value of 1.0 indicates a perfect circle, while values approaching 0.0 indicates an elongated polygon.22
Enzyme-linked immunosorbent assay (ELISA)
ELISA kit was purchased from Invivogen. Plates were coated with capture antibody at 4°C overnight. Wells were blocked with ELISA coating buffer at RT for 1 hour. To activate latent TGFb, all samples were acid activated with 1M HCl for 10 min at RT. 1M NaOH was then used to neutralize the reaction. Samples/standards were added to wells in the appropriate dilutions and incubated at RT for 2 hour. Biotinylated detection antibody was added and the plate was incubated for 1 hour. Horseradish peroxidase (HRP)-conjugated streptavidin was added and incubated for 30 min at RT. Tetramethylbenzidine was added to each well. The reaction was stopped using 1M H3PO4. Optical density (OD) values were determined by measuring absorbance values at 450 nm using a SpectraMax Microplate Reader (Molecular Devices).
Cytotoxicity assay
K562 cells were counted and washed thoroughly out of media two times using phosphate buffered saline (PBS). The cells were resuspended at a concentration of 2×106 cells/mL in PBS+20 µM calcein acetoxymethyl (AM) dye. Following 30 min incubation, labeled K562 cells were thoroughly washed in wash buffer (20% FCS in PBS) three times after which they were resuspended at a concentration of 0.3×106 cells/mL and allowed to rest for 1 hour at 37°C. PBMC were counted, washed and plated in 96 well plates according to the Effector to Target (E:T) ratios of 10:1, 20:1 and 40:1, in triplicate. K562 cells were washed again following the 1 hour rest and resuspended at 0.3×106 cells/mL in RPMI and plated in with the NK cells. K562s were plated on their own without NK cells, to measure spontaneous release. Additionally, K562 cells alone were plated in the presence of 8 µl of 10% Triton X to measure maximum release of calcein AM.
Statistics
GraphPad Prism V.8.00 (GraphPad Software) was used for statistical analysis. Normality was determined using the D’Agostino-Pearson omnibus test. Data were then analyzed using a Student’s t-test when two data sets were being compared, or a one-way/two-way ANOVA test when more than two data sets were being compared.
Results
Functional defects in NK cells from patients with breast cancer
We first defined NK cell functional responses and subset distribution in our cohort of patients with metastatic breast cancer compared with healthy controls (see table 1 for details of the patient cohorts). The overall frequency of NK cells in patient versus control PBMC was similar, though there was a decrease in the frequency of CD56bright NK cells in patient NK cells (figure 1A). Cytokines are commonly used to activate NK cells. While both IL2 and IL12/IL15 upregulated the expression of the activation marker CD69 equivalently (figure 1B), significantly fewer patient NK cells expressed CD25 in response to either IL2 or IL12/15 compared with healthy control NK cells (figure 1C). While TNFα production was similar, patient NK cells had a clear defect in the production of IFNγ in both CD56dim and CD56bright NK cell subsets in response to IL12/IL15 stimulation when compared with healthy control cells (figure 1D–F). In terms of cytotoxicity, granzyme B levels of expression were similar but cytokine-induced TRAIL expression was significantly lower on CD56dim NK cells from patients compared with controls (figure 1G, H). In terms of direct killing, NK cells from healthy donors were able kill standard K562 target cells efficiently as expected (representative cytokine-induced killing over a range of effector:target ratios are shown in figure 1I) demonstrating that their killing machinery is working well. However, in stark contrast, NK cells from patients failed to induce even minimal killing (figure 1I, J), Overall, profound defects in both cytotoxicity and cytokine production were found in circulating NK cells from patients with metastatic breast cancer.
Figure 1.
Natural killer (NK) cells from patients with metastatic breast cancer have reduced activation and function. Freshly isolated peripheral blood mononuclear cells (PBMC) from healthy donors and patients were stimulated with interleukin (IL)2 (500 IU/mL), or with IL12 (30ng/mL and IL15 (100 ng/mL) for 18 hours as indicated. (A) PBMC were stained for CD56 and CD3 directly ex vivo and analyzed by flow cytometry to identify the frequency of NK cells (CD56+ CD3−) and the CD56bright subset. (B–H) PBMC were stimulated as indicated and stained for CD69, CD25, TNFα, TRAIL, IFNγ or granzyme B and analyzed by flow cytometry. (I, J) K562 tumor cells were stained with calcein AM and co-cultured with NK cells for 4 hours at varying ratios as indicated. Dots represent individual donors and bars show the mean (n=4–40). Samples were compared using an unpaired Student’s t-test or a one-way analysis of variance, *p<0.05. ns, not significant.
Metabolic defects in NK cells from patients with breast cancer
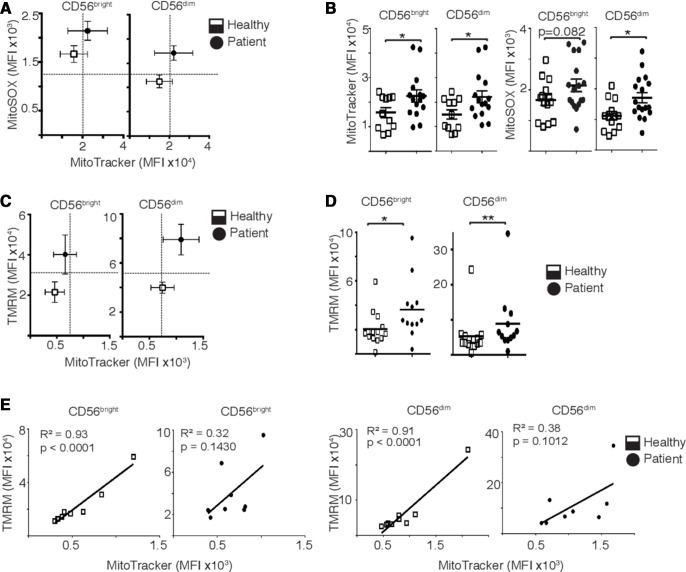
As we have previously demonstrated that metabolic reprogramming is required for NK cell effector functions, we investigated whether metabolism was dysregulated in circulating NK cells from patients with metastatic breast cancer. Direct ex vivo analysis, without any experimental manipulation, showed evidence of mitochondrial alterations in patient NK cells with increased mitochondrial mass alongside elevated levels of mitochondrial reactive oxygen species production and increased MMP compared with healthy controls (figure 2A–D). It was striking that while an increase in mitochondrial mass correlated with increased MMP in healthy controls, as might be predicted, this was not the case for mitochondria for patient NK cells where pathology disrupted this relationship, as evidenced by R2 correlation values (figure 2E).
Figure 2.
Natural killer (NK) cells from patients with metastatic breast cancer have dysfunctional mitochondria. (A, B) NK cells were stained directly ex vivo with MitoSOX (1.5 µM) or MitoTracker Red (100 nM) and mean flourescence intensity (MFI) measured by flow cytometry. (C, D) NK cells were stained directly ex vivo with TMRM (100 nM) or MitoTracker Green (100 nM) and MFI measured. (E) Linear correlation between mitochondrial polarization and mitochondrial mass. Bars show the mean±SEM (n=9–17), and individual donors are shown by dots. Samples were compared using an unpaired Student’s t-test, *p<0.05, **p<0.01.
We also performed parallel confocal microscopy analysis that revealed differences in mitochondrial structure; while mitochondria from healthy controls were generally more fused with elongated morphologies, those from patients had disrupted structures with more punctate mitochondria, a morphology indicative of decreased mitochondrial fusion (figure 3A, B). This was supported by quantitative analysis where circularity scores (n=1 for perfect circle associated with extreme fragmented phenotype) and length of individual mitochondria were significantly higher from patient compared with healthy control NK cell mitochondria (figure 3C). Furthermore, patient NK cells had increased numbers of mitochondria per cell, and integrated density values (akin to mean flourescence intensity intensity, MFI) were also significantly increased in patient NK cells (figure 3D) suggesting increased MMP, mitochondrial mass or increases in both parameters, further supporting the data generated in figure 2.
Figure 3.
Natural killer (NK) cells from patients with metastatic breast cancer have altered mitochondrial structure. (A) Representative confocal images of ex vivo healthy and patient NK cells stained with MitoSpy CMX Ros (250 nM) for 30 min at 37°C and 4',6-diamidino-2-phenylindole, dihydrochloride (DAPI) (300 nM). Images shown are the maximum intensity projection of z-stacks taking at 0.2 µm increments. Red=Mitospy CMX Ros, Blue=DAPI. Scale bar=5 µm. (B) Ordinal scoring of NK cell mitochondrial morphology. Per cent morphology score was determined by blinded ordinal scoring by six volunteers (n=84 cells, n=3 donors). Bars show the mean±SEM. Samples were compared by two-way ANOVA. (C) Pooled analysis of circularity and mitochondrial length measurements. (D) Number of mitochondria per cell and integrated density measurement quantified per NK cell from healthy control or patients with breast cancer (n=5 donors). Samples were compared using a two-way analysis of variance (B) or an unpaired Student’s t-test (C, D). *p<0.05, **p<0.01, ***p<0.001.
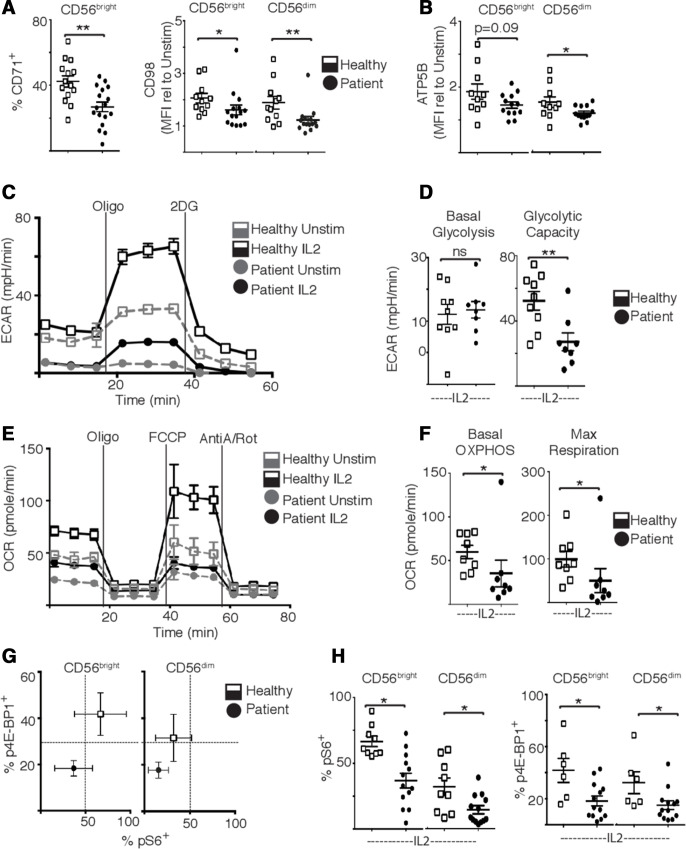
These metabolic defects observed directly ex vivo argue that patient NK cells would have impaired metabolic responses to subsequent cytokine stimulation. Indeed, patient NK cells had impaired upregulation of metabolic machinery in response to cytokine stimulation including the nutrient receptors CD71 and CD98, and the ATP synthase subunit ATP5b (figure 4A, B). Additionally, while cytokine-induced basal glycolysis levels were similar, patient NK cells had significantly reduced glycolytic capacity and reduced rates of flux through the OXPHOS pathway compared with NK cells from healthy controls (figure 4C–F). Concurrent with these overall decreased metabolic rates, IL2 stimulated patient NK cells also had reduced levels of mammalian target of rapamycin complex 1 (mTORC1) activity, an important metabolic regulator in NK cells, measured as levels of phosphorylated 4E-BP1 and S6 ribosomal protein (figure 4G, H).
Figure 4.
Natural killer (NK) cells from patients with metastatic breast cancer have impaired metabolism and mammalian target of rapamycin complex 1 activity. (A) Peripheral blood mononuclear cells (PBMC) were stimulated with IL2 (500 IU/mL) for 18 hours and NK cells (CD56+ CD3−) stained for CD71 and CD98 expression. (B) PBMC were stimulated with interleukin (IL)2 for 18 hours and NK cell stained for ATP5B expression. (C) Representative extracellular acidification rate (ECAR) trace of 18 hours unstimulated and IL2 stimulated healthy donor and patient purified NK cells. (D) Pooled data of basal glycolysis and glycolytic capacity. (E) Representative oxygen consumption rate (OCR) trace of 18 hours unstimulated and IL2 stimulated healthy donor and patient purified NK cells. Inhibitors added at times indicated were oligomycin (Oligo), carbonyl cyanide p-trifluoro-methoxyphenyl hydrazone (FCCP) and antimycin-A/rotenone (AntiA/Rot) (F) Pooled data of basal oxidative phosphorylation (OXPHOS) and maximal respiration. (G, H) NK cells were stimulated with IL2 and stained for pS6 and p4E-binding protein 1. Bars show the mean±SEM (n=6–13). Samples were compared using an unpaired Student’s t-test, *p<0.05, **p<0.01, ns, not significant.
NK cells from some patients with metastatic breast cancer overexpress the cell surface, TGFβ sequestering receptor, GARP
Ourselves and others have previously published that TGFβ inhibits cytokine-induced metabolic reprogramming in human NK cells.19 23 We therefore hypothesized that TGFβ may contribute to the dysfunction observed in circulating NK cells from patients, particularly as it contributes to the immunosuppressive environment found in many cancer types.24 It is often found at higher circulating levels in cancer although in human breast cancer, the literature is conflicting.25–27 To investigate if TGFβ might be a mechanism leading to dysfunction in metastatic breast cancer, we first measured TGFβ levels in the serum of patients and healthy controls and found no significant difference (figure 5A). Given that there was no apparent systemic increase in TGFβ, it was considered important to investigate for any evidence of intrinsic TGFβ signaling in NK cells in patients. Using a well-described anti-phospho-SMAD2/3 to demonstrate canonical TGFβ signaling, we found that patient NK cells trended towards higher constitutive levels of pSMAD2/3 in both CD56dim and CD56bright NK cells (figure 5B, C). A similar pattern of expression was found for LAP, a protein which binds TGFβ homodimers intracellularly to form the TGFβ small latency complex. As LAP is cotranscribed at the N-terminal region of the TGFβ gene product, expression of LAP indicates active TGFβ transcription. Thus, while not reaching statistical significance, there was a trend towards increased expression of LAP on NK cells from patients compared with controls, for both CD56dim and CD56bright subsets, suggesting that these NK cells intrinsically produce both TGFβ and LAP proteins (figure 5D). We next considered whether there might be locally produced TGFβ that could impact on NK cells metabolism and function. It is described that T-regs produce TGFβ that acts in an autocrine manner through a mechanism involving the sequestering of latent TGFβ (as small latency complex) at the cell surface by the transmembrane protein, GARP.20 28 Although there were a few exceptions, most healthy donors expressed low levels of GARP on the surface of their NK cells (with no difference between levels on CD56dim or CD56bright subsets) (figure 5H). However, it was striking that patients with metastatic breast cancer expressed significantly higher levels of GARP on both CD56dim and CD56bright NK cell subsets compared with controls (figure 5E). Cytokines could increase expression of GARP, particularly on CD56bright cells (online supplemental figure 2). In general, LAP and GARP expression mirrored each other as can be seen in representative plots in figure 5F. Expression patterns ranged from a subset of cells expressing both LAP and GARP, a general overall shift in the population, to a dramatic upregulation with almost all NK cells expressing both. In order to see if higher GARP expression was associated with intrinsic TGFβ signaling, we stratified patients according to ‘high’ or ‘low’ GARP expression and looked at pSMAD2/3 phosphorylation in the two groups. As can be seen in figure 5G, patients with higher GARP expression on their NK cells had statistically significantly higher TGFβ signaling, in both CD56dim and CD56bright subsets. In combination, the overexpression of TGFβ machinery and higher intrinsic TGFβ signaling in NK cells from patients compared with controls suggest that GARP may serve as a novel mechanism for autocrine inhibition of NK cell metabolism and function during cancer.
Figure 5.
Glycoprotein-A repetitions predominant (GARP) expression is associate with intracellular transforming growth factor-β (TGFβ) signaling in natural killer (NK) cells from patients with metastatic breast cancer. (A) The concentration of TGFβ in the plasma of healthy donors and patients with metastatic breast cancer was measured by ELISA as per the manufacturer’s instructions. (B–G) PBMC were isolated from healthy donors and patients directly ex vivo and NK cells were stained for pSMAD2/3, latency associated peptide (LAP) and GARP expression. (H) Patients were divided into those with high GARP expression (MFI >500) or low GARP expression (MFI <500) and pSMAD2/3 levels were compared. Bars show the mean±SEM (n=10–14). Samples were compared using an unpaired Student’s t-test, *p<0.05. MFI, mean flourescence intensity.
TGFβ restricts the metabolism and function of patient NK cells
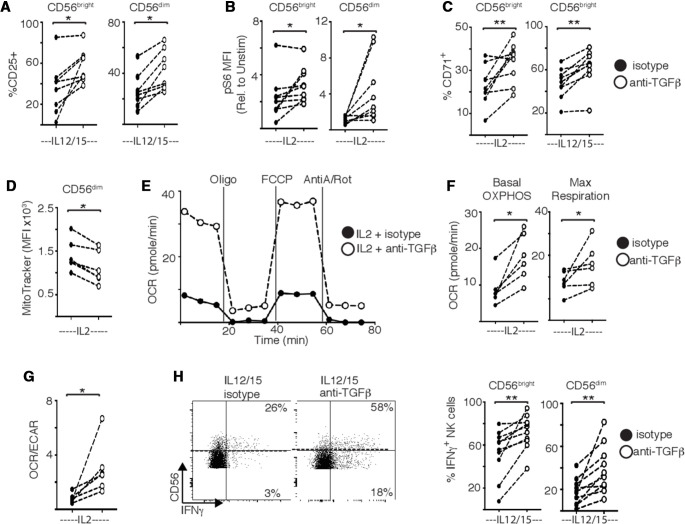
Our data support that NK cell metabolism and function are severely impacted during metastatic breast cancer and that locally produced TGFβ could potentially drive these defects in patient NK cells. Indeed, neutralizing TGFβ with anti-TGFβ antibodies had a clear restorative impact on patient NK cells in vitro. First, neutralizing TGFβ rescued both CD25 expression and mTORC1 activity in cytokine stimulated patient NK cells (figure 6A, B). This finding was supported by increased expression of the transferrin receptor CD71, a downstream target of mTORC1 signaling (figure 6C). Neutralizing TGFβ also significantly restored features of dysregulated mitochondrial metabolism in NK cells from patients, including the reduction of mitochondrial mass in patient NK cells (figure 6D). Blocking TGFβ also increased OXPHOS and maximal respiration in patient NK cells stimulated with IL2 (figure 6E, F). Overall, there was a shift in the OCR/ECAR ratio indicating an increased reliance on oxidative glucose metabolism compared with glycolysis (figure 1G). It is important to note that this latter experiment was performed using purified NK cells, further supporting endogenous TGFβ from NK cells as a contributing factor. Finally, while blocking TGFβ had minimal impact on IFNγ production by healthy NK cells (see online supplemental figure 3), it significantly increased its production in patient NK cells (figure 6H). To our knowledge, this is the first demonstration that neutralizing TGFβ can significantly contribute to restoring some of the profound metabolic defects found in peripheral blood NK cells of cancer patients.
Figure 6.
Targeting transforming growth factor-β (TGFβ) restores the function and metabolism of natural killer (NK) cells from patients with breast cancer. Peripheral blood mononuclear cells from patients with breast cancer were stimulated with interleukin (IL)2 (500 IU/mL) (A–D), or with IL12 (30 ng/mL) and IL15 (100 ng/mL) (B, E, F) for 18 hours in the presence of isotype control or anti-TGFβ antibody (5 µg/mL). (A–C) Patient NK cells were stained for CD25, pS6 and CD71 and analyzed by flow cytometry. (D) Patient NK cells were stained with MitoTracker Green (100 nM) and analyzed by flow cytometry. (E) Representative oxygen consumption rate (OCR) trace. Inhibitors added at times indicated were oligomycin (Oligo), carbonyl cyanide p-trifluoro-methoxyphenyl hydrazone (FCCP) and antimycin-A/rotenone (AntiA/Rot). (F) Pooled data for basal oxidative phosphorylation (OXPHOS) and maximal respiration. (G) Ratio of OCR:extracellular acidification rate (ECAR) in patient NK cells. (H) Representative dot plot for interferon-γ (IFNγ) production and pooled data for IFNγ production. Individual donors are shown by a dot (n=8–14). Samples were compared using a paired Student’s t-test, *p<0.05, **p<0.01.
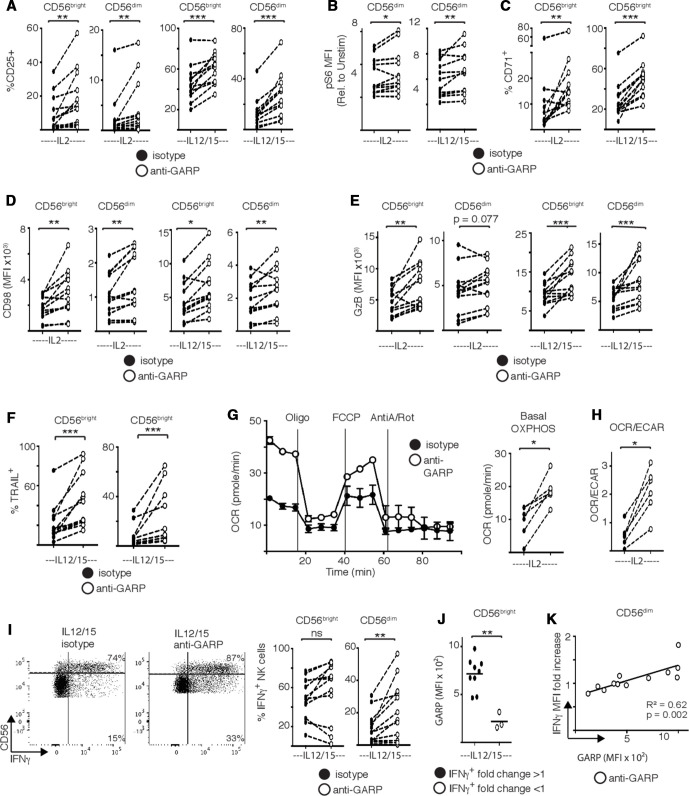
In order to support data from purified NK cells which demonstrate that NK cell-derived TGFβ contributes to the defects observed and to further investigate GARP expression by NK cells as a potential mechanism explaining this, we obtained a well-characterized monoclonal antibody which specifically blocks the activities of GARP when bound to LAP.29 Experiments were performed on purified NK cells that were stimulated by cytokine in the presence of either an anti-GARP or an isotype control antibody. For a range of activation (CD25), signaling (mTORC1), nutrient receptor (CD71, CD98) and cytotoxicity markers (granzyme B, TRAIL), inclusion of anti-GARP restored these activities in dysregulated patient NK cells. Direct measurement of metabolic flux clearly shows that anti-GARP allowed dysfunctional NK cells from patients with cancer to reengage in OXPHOS. As anti-GARP did not restore glycolysis (if anything, it decreased already low levels of glycolysis, data not shown), its inclusion effectively switched these NK cells to OXPHOS as the primary pathway of glucose metabolism as indicated by the OCR/ECAR ratio (figure 7E). In terms of IFNγ production, results were more variable. The frequency of CD56dim but not CD56bright cells producing IFNγ increased significantly in the presence of anti-GARP (figure 7F). Analysis identified that increased IFNγ expression correlated with GARP expression (figure 7G, H), supporting that GARP contributes to NK cell dysfunction and that blocking its activity restores function in patient NK cells. This research demonstrates the potential in targeting metabolic pathways as a route towards restoring functional responses in ‘exhausted’ NK cells from patients with cancer.
Figure 7.
Targeting the glycoprotein-A repetitions predominant (GARP)–transforming growth factor-β axis restores the function and metabolism of natural killer (NK) cells from patients with breast cancer. NK cells were purified from freshly isolated peripheral blood mononuclear cells from patients with breast cancer and stimulated with interleukin (IL)2 (500 IU/mL), or with IL12 (30 ng/mL) and IL15 (100 ng/mL) as indicated in the presence of isotype control or anti-GARP antibody (10 µg/mL). (A–F) Patient NK cells were stained for CD25, pS6, CD71, CD98, granzyme B and TRAIL and analyzed by flow cytometry. (D) Patient NK cells were stained with MitoTracker Green (100 nM) and analyzed by flow cytometry. (G) Representative oxygen consumption rate (OCR) trace and pooled data for basal oxidative phosphorylation (OXPHOS). (H) Ratio of OCR:extracellular acidification rate (ECAR) in patient NK cells. (I) Representative dot plot for interferon-γ (IFNγ) production and pooled data for IFNγ production. (J) Patients were stratified in those for which anti-GARP treatment increased CD56bright NK cell IFNγ production (fold >1) or decreased IFNγ production (fold <1) and GARP expression was compared. Linear correlation between the fold increase in IFNγ induced by anti-GARP treatment and GARP expression in CD56dim patient NK cells. Individual donors are shown by a dot (n=8–14). Samples were compared using a paired Student’s t-test, *p<0.05, **p<0.01, ***p<0.001.
Discussion
This study explored the concept that altered metabolism underpins metabolic dysfunction of circulating NK cells during human cancer. Indeed, when peripheral NK cells were analyzed from patients with metastatic breast cancer, the ability of these NK cells to produce IFNγ was significantly impaired and importantly, this IFNγ deficit was associated with distinct metabolic defects. Patient NK cells had numerous mitochondrial abnormalities including altered mitochondrial structure and they failed to increase metabolic rates of glycolysis and OXPHOS in response to cytokine stimulation. Our metabolic data in patient NK cells parallels some of the metabolic features of exhausted T cells studied using murine models of chronic viral infection or cancer, and identified in human tumors.16 17 30 31 In particular, the striking fragmentation of mitochondria in NK cells from peripheral patient NK cells is similar to that observed in tumor infiltrating CD8 T cells from patients with renal cell carcinoma,17 and more recently in tumour infiltrating NK (TINK) cells from hepatic cellular carcinoma tumors.18 Importantly, in this study the observed mitochondrial defects are in peripheral NK cells demonstrating that tumors can affect NK cells that are distal from the tumor. This study is the first to describe metabolic defects in peripheral lymphocytes in patients with cancer.
A key finding of this study is that the neutralization of TGFβ cytokine with a blocking antibody was sufficient to restore some of the metabolic defects found in patient NK cells and to increase IFNγ production, suggesting that TGFβ is a key mechanism driving NK cell dysfunction in breast cancer. TGFβ cytokine can directly inhibit both murine and human NK cell metabolism and in particular has a strong inhibitory effect on NK cell mitochondrial respiration.19 23 32 These TGFβ-mediated effects involve multiple mechanisms including the inhibition of mTORC1 signaling as well as direct effects on mitochondrial metabolism.19 32 We originally focused on TGFβ as it has been well documented to inhibit NK cells and to promote an immunosuppressive environment facilitating cancer progression through a variety of mechanisms. It can be produced by tumor cells, tumor associated cells, and also found within tumour-derived extracellular vesicles that act to inhibit NK cell anti-tumor functions.33–37 Circulating TGFβ levels have been shown to be elevated in various cancers including in patients with breast cancer38 39 but equally, there are reports of no change.26 We found no change in circulating TGFβ levels in our cohort. Therefore, the source of TGFβ in our ‘reversal of exhaustion’ experiments was both distal to the tumor and present within PBMC. It has been previously been shown that NK cells can produce TGFβ.40 Additionally, there is evidence that there is increased frequency of TGFβ-producing NK cells in patients with breast cancer, suggesting a role for autocrine TGFβ signaling.41 Our experiments also support this but go further to provide a new mechanism which is likely to function during pathological situations such as cancer. GARP has previously been described to be expressed on activated T-regs, tumor cells and platelets where it can capture and activate latent TGFβ released.28 42 While there is heterogeneity in patients, some had clear overexpression of both LAP and GARP. Confidence is provided by the mirroring patterns of expression and the co-expression of the molecules. Indeed, blocking TGFβ in a culture of purified NK cells resulted in a significant rescue of NK cell OXPHOS, which further supports a inhibitory role for local and endogenous production of TGFβ by peripheral NK cells during cancer. Specific blocking of GARP on purified NK cells also strongly rescued NK cell metabolism and function providing direct evidence of a role for GARP in this process. Our data provide evidence that circulating NK cells in patients with cancer can actively contribute to their own metabolic and functional decline and identifies LAP/GARP as a new mechanism contributing to this.
Understanding the mechanisms by which metabolism regulates NK cell function and how these are dysregulated during cancer will allow their strategic targeting to improve cancer therapies. This could include in vivo use of pharmaceuticals or biologics that inhibit TGFβ signaling. Indeed, some of these have already shown promise in clinical trials.43–45 Furthermore, overexpression of GARP and/or LAP-1 on immune cells may help to personalize therapy and stratify patients likely to benefit from this approach. As we move into an era where cellular therapies for example, CAR-NK cells enter mainstream use, genetic manipulations, such as downregulation of TGFβ receptor expression or GARP, are likely to result in products that are more metabolically robust and clinically effective.
Acknowledgments
We would like to thank all the participants who provided blood for this study and the phlebotomists that facilitated them. We would also like to thank Barry Moran and Gavin McManus, from the flow cytometry and confocal microscopy units, respectively, for their on-going support.
Footnotes
Correction notice: This article has been corrected since it first published. The provenance and peer review statement has been included.
Contributors: KS designed, performed and analyzed the patient experiments and writing of the manuscript. EW designed and performed the confocal experiments, preparation of figure and contribution to the paper. VZ-B designed, performed and analyzed experiments and writing of the manuscript. SM, SC, MC and CG identified, recruited and consented patients; took blood samples, anonymized samples and provided patient information. JK contributed to design of patient recruitment strategy, ethics approval, identification of patients and responsibility for clinical information. SL provided anti-GARP antibody and helped with data analysis. DF contributed to conceptual design of work, design of experiments, analysis of data, writing of manuscript. CMG contributed to conceptual design of work, design of experiments, analysis of data, writing of manuscript.
Funding: VZ-B is supported by the Brazilian Government under the Science without Borders Program (Grant BEX 13446134). DF is supported by Science Foundation Ireland Grant 13/CDA/2161. KS and CMG are support by the National Children’s Research Center (grant A/18/5).
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Ethics for this study was provided by the Research Ethics Committee (REC) of School of Biochemistry and Immunology in Trinity College Dublin and by the REC of St. James Hospital, Dublin, Ireland.
References
- 1.Miller JS, Lanier LL. Natural killer cells in cancer immunotherapy. Annu Rev Cancer Biol 2019;3:77–103. 10.1146/annurev-cancerbio-030518-055653 [DOI] [Google Scholar]
- 2.Tanaka J, Miller JS. Recent progress in and challenges in cellular therapy using NK cells for hematological malignancies. Blood Rev 2020;44:100678. 10.1016/j.blre.2020.100678 [DOI] [PubMed] [Google Scholar]
- 3.Suen WC-W, Lee WY-W, Leung K-T, et al. Natural killer cell-based cancer immunotherapy: a review on 10 years completed clinical trials. Cancer Invest 2018;36:431–57. 10.1080/07357907.2018.1515315 [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Hermanson DL, Moriarity BS, et al. Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell 2018;23:181–92. 10.1016/j.stem.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlsten M, Järås M. Natural killer cells in myeloid malignancies: immune surveillance, NK cell dysfunction, and pharmacological opportunities to bolster the endogenous NK cells. Front Immunol 2019;10:2357. 10.3389/fimmu.2019.02357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung PS, Jang JW. Natural killer cell dysfunction in hepatocellular carcinoma: pathogenesis and clinical implications. Int J Mol Sci 2018;19. 10.3390/ijms19113648. [Epub ahead of print: 19 Nov 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mamessier E, Sylvain A, Thibult M-L, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest 2011;121:3609–22. 10.1172/JCI45816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espí A, Arenas J, García-Granero E, et al. Relationship of curative surgery on natural killer cell activity in colorectal cancer. Dis Colon Rectum 1996;39:429–34. 10.1007/BF02054059 [DOI] [PubMed] [Google Scholar]
- 9.Kastelan M, Kovacić K, Tarle R, et al. Analysis of NK cell activity, lymphocyte reactivity to mitogens and serotest PSA and TPS values in patients with primary and disseminated prostate cancer, PIN and BPH. Anticancer Res 1997;17:1671–5. [PubMed] [Google Scholar]
- 10.Spitzer MH, Carmi Y, Reticker-Flynn NE, et al. Systemic immunity is required for effective cancer immunotherapy. Cell 2017;168:487–502. 10.1016/j.cell.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiner CM. Nk cell metabolism. J Leukoc Biol 2019;105:1235–42. 10.1002/JLB.MR0718-260R [DOI] [PubMed] [Google Scholar]
- 12.Keppel MP, Saucier N, Mah AY, et al. Activation-Specific metabolic requirements for NK cell IFN-γ production. J Immunol 2015;194:1954–62. 10.4049/jimmunol.1402099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mah AY, Rashidi A, Keppel MP, et al. Glycolytic requirement for NK cell cytotoxicity and cytomegalovirus control. JCI Insight 2017;2. 10.1172/jci.insight.95128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer JR, Salzillo TC, Chakravarti N, et al. Education-dependent activation of glycolysis promotes the cytolytic potency of licensed human natural killer cells. J Allergy Clin Immunol 2019;143:346–58. 10.1016/j.jaci.2018.06.047 [DOI] [PubMed] [Google Scholar]
- 15.Loftus RM, Assmann N, Kedia-Mehta N, et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat Commun 2018;9:2341. 10.1038/s41467-018-04719-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharping NE, Menk AV, Moreci RS, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 2016;45:701–3. 10.1016/j.immuni.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 17.Siska PJ, Beckermann KE, Mason FM, et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2017;2. 10.1172/jci.insight.93411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X, Qian Y, Fu B, et al. Mitochondrial fragmentation limits NK cell-based tumor immunosurveillance. Nat Immunol 2019;20:1656–67. 10.1038/s41590-019-0511-1 [DOI] [PubMed] [Google Scholar]
- 19.Zaiatz-Bittencourt V, Finlay DK, Gardiner CM. Canonical TGF-β signaling pathway represses human NK cell metabolism. J Immunol 2018;200:3934–41. 10.4049/jimmunol.1701461 [DOI] [PubMed] [Google Scholar]
- 20.Liénart S, Merceron R, Vanderaa C, et al. Structural basis of latent TGF-β1 presentation and activation by GARP on human regulatory T cells. Science 2018;362:952–6. 10.1126/science.aau2909 [DOI] [PubMed] [Google Scholar]
- 21.Keating SE, Zaiatz-Bittencourt V, Loftus RM, et al. Metabolic reprogramming supports IFN-γ production by CD56bright NK cells. J Immunol 2016;196:2552–60. 10.4049/jimmunol.1501783 [DOI] [PubMed] [Google Scholar]
- 22.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viel S, Marçais A, Guimaraes FS-F, et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal 2016;9:ra19. 10.1126/scisignal.aad1884 [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Pang Y, Moses HL. Tgf-Beta and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol 2010;31:220–7. 10.1016/j.it.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong FM, Anscher MS, Murase T, et al. Elevated plasma transforming growth factor-beta 1 levels in breast cancer patients decrease after surgical removal of the tumor. Ann Surg 1995;222:155–62. 10.1097/00000658-199508000-00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakefield LM, Letterio JJ, Chen T, et al. Transforming growth factor-beta1 circulates in normal human plasma and is unchanged in advanced metastatic breast cancer. Clin Cancer Res 1995;1:129–36. [PubMed] [Google Scholar]
- 27.Grau AM, Wen W, Ramroopsingh DS, et al. Circulating transforming growth factor-beta-1 and breast cancer prognosis: results from the Shanghai breast cancer study. Breast Cancer Res Treat 2008;112:335–41. 10.1007/s10549-007-9845-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran DQ, Andersson J, Wang R, et al. Garp (LRRC32) is essential for the surface expression of latent TGF-beta on platelets and activated Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A 2009;106:13445–50. 10.1073/pnas.0901944106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuende J, Liénart S, Dedobbeleer O, et al. Monoclonal antibodies against GARP/TGF-β1 complexes inhibit the immunosuppressive activity of human regulatory T cells in vivo. Sci Transl Med 2015;7:284ra56. 10.1126/scitranslmed.aaa1983 [DOI] [PubMed] [Google Scholar]
- 30.Bengsch B, Johnson AL, Kurachi M, et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8(+) T Cell Exhaustion. Immunity 2016;45:358–73. 10.1016/j.immuni.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schurich A, Pallett LJ, Jajbhay D, et al. Distinct Metabolic Requirements of Exhausted and Functional Virus-Specific CD8 T Cells in the Same Host. Cell Rep 2016;16:1243–52. 10.1016/j.celrep.2016.06.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimeloe S, Gubser P, Loeliger J, et al. Tumor-derived TGF-β inhibits mitochondrial respiration to suppress IFN-γ production by human CD4+ T cells. Sci Signal 2019;12. 10.1126/scisignal.aav3334 [DOI] [PubMed] [Google Scholar]
- 33.Dalal BI, Keown PA, Greenberg AH. Immunocytochemical localization of secreted transforming growth factor-beta 1 to the advancing edges of primary tumors and to lymph node metastases of human mammary carcinoma. Am J Pathol 1993;143:381–9. [PMC free article] [PubMed] [Google Scholar]
- 34.Cai J, Xia L, Li J, et al. Tumor-Associated macrophages derived TGF-β‒Induced epithelial to mesenchymal transition in colorectal cancer cells through Smad2,3-4/Snail signaling pathway. Cancer Res Treat 2019;51:252–66. 10.4143/crt.2017.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erdogan B, Webb DJ. Cancer-Associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem Soc Trans 2017;45:229–36. 10.1042/BST20160387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J, Schlößer HA, Wang Z, et al. Tumor-Derived extracellular vesicles inhibit natural killer cell function in pancreatic cancer. Cancers 2019;11. 10.3390/cancers11060874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczepanski MJ, Szajnik M, Welsh A, et al. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011;96:1302–9. 10.3324/haematol.2010.039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanović V, Todorović-Raković N, Demajo M, et al. Elevated plasma levels of transforming growth factor-beta 1 (TGF-beta 1) in patients with advanced breast cancer: association with disease progression. Eur J Cancer 2003;39:454–61. 10.1016/S0959-8049(02)00502-6 [DOI] [PubMed] [Google Scholar]
- 39.Tas F, Karabulut S, Yasasever CT, et al. Serum transforming growth factor-beta 1 (TGF-β1) levels have diagnostic, predictive, and possible prognostic roles in patients with melanoma. Tumour Biol 2014;35:7233–7. 10.1007/s13277-014-1984-z [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y, Yang M, Sun X, et al. IL-10+ NK and TGF-β+ NK cells play negative regulatory roles in HIV infection. BMC Infect Dis 2018;18:80. 10.1186/s12879-018-2991-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostapchuk YO, Cetin EA, Perfilyeva YV, et al. Peripheral blood NK cells expressing HLA-G, IL-10 and TGF-β in healthy donors and breast cancer patients. Cell Immunol 2015;298:37–46. 10.1016/j.cellimm.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 42.Metelli A, Wu BX, Fugle CW, et al. Surface expression of TGFβ docking receptor GARP promotes oncogenesis and immune tolerance in breast cancer. Cancer Res 2016;76:7106–17. 10.1158/0008-5472.CAN-16-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodon J, Carducci MA, Sepulveda-Sánchez JM, et al. First-In-Human dose study of the novel transforming growth factor-β receptor I kinase inhibitor LY2157299 monohydrate in patients with advanced cancer and glioma. Clin Cancer Res 2015;21:553–60. 10.1158/1078-0432.CCR-14-1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melisi D, Garcia-Carbonero R, Macarulla T, et al. Galunisertib plus gemcitabine vs. gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br J Cancer 2018;119:1208–14. 10.1038/s41416-018-0246-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faivre S, Santoro A, Kelley RK, et al. Novel transforming growth factor beta receptor I kinase inhibitor galunisertib (LY2157299) in advanced hepatocellular carcinoma. Liver Int 2019;39:1468–77. 10.1111/liv.14113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002044supp001.pdf (2.5MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.