ABSTRACT
Bacteria–fungi interactions (BFIs) are essential in ecosystem functioning. These interactions are modulated not only by local nutritional conditions but also by the physicochemical constraints and 3D structure of the environmental niche. In soils, the unsaturated and complex nature of the substrate restricts the dispersal and activity of bacteria. Under unsaturated conditions, some bacteria engage with filamentous fungi in an interaction (fungal highways) in which they use fungal hyphae to disperse. Based on a previous experimental device to enrich pairs of organisms engaging in this interaction in soils, we present here the design and validation of a modified version of this sampling system constructed using additive printing. The 3D printed devices were tested using a novel application in which a target fungus, the common coprophilous fungus Coprinopsis cinerea, was used as bait to recruit and identify bacterial partners using its mycelium for dispersal. Bacteria of the genera Pseudomonas, Sphingobacterium and Stenotrophomonas were highly enriched in association with C. cinerea. Developing and producing these new easy-to-use tools to investigate how bacteria overcome dispersal limitations in cooperation with fungi is important to unravel the mechanisms by which BFIs affect processes at an ecosystem scale in soils and other unsaturated environments.
Keywords: bacteria–fungi interactions, additive printing, soil, horse dung, Coprinopsis
Fungal highway columns produced by additive printing.
INTRODUCTION
The spatial structure of a natural soil environment plays a central role in modulating microbial activity (Nunan et al. 2003; Wolf et al. 2013). Soil is a complex matrix in which solid, liquid and gaseous phases are intermixed and organized stochastically (Or et al. 2007; Tecon and Or 2017). In this unsaturated matrix, substrate bioavailability is not only determined by the nature of the substrate but also by its accessibility (Semple et al. 2007). This is not only relevant in soils but also in other complex and heterogeneous matrices in which microbial activity is an important driver of function, for instance during the process of fermentation of food products (Zhang et al. 2018a) or for the functioning of specific animal tissues (Mikaelyan, Meuser and Brune 2017).
Many bacteria are able to move using different mechanisms, often requiring the production of specific appendages such as flagella or pili (Harshey 2003). However, in the case of soils, an essential point that tightly controls the rate of bacterial dispersal is the level of hydration (Or et al. 2007). In the case of flagellar motility, experiments in a porous matrix with different hydration levels have demonstrated that both flagellar velocity and bacterial dispersal are impaired in conditions of disconnected liquid films (Tecon and Or 2016). In contrast, the filamentous lifestyle of fungi results in an ideal structure for colonizing heterogeneous soil environments (Soufan et al. 2018). This is aided by the two specialized protein families, hydrophobins and adhesins. The first allows fungal hyphae to dynamically adapt to the water–air interface, while the second helps in surface anchoring (Linder et al. 2005). Experiments performed with the fungus-like oomycete Pythiumultimum have shown the existence of a liquid layer along hyphae (Furuno et al. 2010), and several studies have shown that some bacteria can use this water film for their dispersal in an interaction known as fungal highways (Kohlmeier et al. 2005). This bacterial–fungal interaction (BFI) has been shown to be relevant in the context of pollutant degradation (Kohlmeier et al. 2005; Wick et al. 2007; Banitz et al. 2013), biologically induced carbonate formation (Martin et al. 2012), endospore germination (Worrich et al. 2017), rhizobia-nodule formation (Zhang et al. 2020) and in the structuring of simplified microbial communities (Zhang et al. 2018a; Christofides et al. 2020).
In order to investigate this dispersal mechanism at a laboratory scale, a sampling device referred to as a fungal highway column was designed in a previous study, to select and isolate both bacteria and their fungal highway partners directly from soils (Simon et al. 2015). The device consists of a 4-cm hourglass-shaped column, experimentally determined to have the optimal shape to avoid the formation of a continuous liquid film inside the column during transport or when placed horizontally in soils. Each extremity is then filled with two 5-mm thick media slices—the attracting medium and the target medium. At the neck of the hourglass, a physical barrier mimicking soil structure (soil solid particles and pores) was created by the addition of 1-mm glass beads. The device also included a 25-µm filtering mesh placed at the extremity where the attracting medium is in contact with the environmental sample, to prevent the dispersal of bacteria by soil microfauna entering the column (Simon et al. 2015). The target medium, which can be colonized only by bacteria using fungal highways, can be retrieved for analyses focusing on this specific BFI. However, one major limitation for the routine and standardized use of such a device as a scientific instrument has been the variability and time-consuming manual production of these devices, performed only by experienced scientists.
Additive manufacturing, also known as 3D printing, is a manufacturing method in which material is added, usually layer by layer, to create a final product, as opposed to subtractive manufacturing that starts with a block of material and removes pieces, usually through machining, until the final product remains (Horn and Harrysson 2012). In many cases, the additive approach allows for more intricate designs, less material waste, greater reproducibility and faster production times, all of which contribute to its growing use in many areas of biology (He et al. 2016). Another advantage of additive manufacturing is the ability to move directly from a digital design to a finished part (Neches et al. 2016). Although the use of 3D printing in microbial ecology is not yet widespread, the increasing affordability and flexibility of this technology will help promote its use to generate novel scientific tools. Moreover, together with the development of microfluidics and single-cell metabolomics, 3D printing is one of the technologies promising to bridge the gap between axenic cultivation and co-cultivation of mixed cultures (Nai and Meyer 2018).
Additive manufacturing allows the production of devices with detailed geometrical control of the physical environment in which microbial communities can engage on different levels of spatially dependent interactions (Connell et al. 2013). This is highly relevant in many scientific areas and, more specifically, to investigate the effect of physical habitat structure on cell-to-cell interactions and microbial activity. The spatial control of the physical arena within which interactions take place is essential to represent the complex microenvironments in which the wealth of mechanisms by which microorganisms engage in cooperative or competitive relationships is established (Connell et al. 2013).
In this study, we present the design and testing of a fungal highway column generated by additive manufacturing. These 3D printed fungal highway columns (hereafter referred to as 3D columns) were tested in experiments designed to assess whether pre-inoculation with a particular fungal species can be used to identify bacteria dispersing on its hyphae. For this, either the target medium or the substrate (environmental sample) was pre-inoculated with a model fungus that was then used as bait for the enrichment and colonization of the target medium by bacteria present in an environmental sample. The experiments were conducted with the common coprophilous fungus Coprinopsis cinerea (Basidiomycota), a model organism widely used to investigate fundamental aspects of fungal biology (Kamada et al. 2010; Stajich et al. 2010). The experimental and molecular knowledge base available for this fungus makes it an ideal candidate for exploring interkingdom interactions. Previous studies suggest that C. cinerea produces active compounds that allow it to antagonize bacteria in the laboratory (Künzler 2018; Kombrink et al. 2019). However, positive interactions with other microbes in its habitat are also possible. As a saprophytic fungus, C. cinerea thrives in organic-rich degrading materials such as dung or compost (Kues 2000; de Mattos-Shipley et al. 2016), and co-exists with other organisms, including a number of bacteria (Slade et al. 2016). Therefore, we hypothesized that this fungus would be able to establish positive interactions with these bacteria. To explore this, we used 3D columns to assess the diversity of the potential bacterial partners that are present in dung and that are able to disperse on the hyphae of this fungus.
MATERIALS AND METHODS
3D column design and production
The 3D columns were developed on SolidWorks® computer-aided design (CAD) software (Dassault Systèmes SolidWorks Corporation, Waltham, MA, USA). The dimensions of the model were based on the hand-assembled fungal highway columns used in prior experiments (Simon et al. 2015). In lieu of manually twisting the column to produce an hourglass shape, the 3D design incorporated the desired hourglass shape to limit variability between the printed columns. Additionally, to replace the glass beads (1 mm diameter) from the previous design (Simon et al. 2015), an internal body-centered cubic lattice structure with a 2 mm unit cell and 0.5 mm strut diameter was incorporated at the hourglass portion of the column, extending 20 mm along the height of the column. The addition of the internal lattice provides a uniform, regular internal structure that was not easily achieved with glass beads. The lattice structure is also easily tunable in CAD software. Negative space provided by the internal lattice structure may be increased by reducing the strut size, or by increasing the size of the unit cell. Finally, the 3D column model included two threaded caps, one for each end of the column, as well as a threaded ring on one end of the otherwise symmetrical column. The purpose of the ring was to fasten a 25-µm mesh filter on the end of the column in contact with the sample environmental matrix to secure the attracting medium. CAD and 3D printing were well suited to the development of the 3D columns, due to the ease of prototyping, manufacture, and the commercial availability of a suitable material.
The column and caps were printed on an Asiga Freeform Pico Plus 39 digital light processing (DLP) 3D printer (Asiga Germany, Erfurt, Germany). DLP is a 3D printing process that polymerizes photosensitive resins into polymers by exposing a thin layer of resin to a 2D projected light image; in this case, UV light was used. The procedure then continues by lifting the polymerized layer on the build platform out of the resin bath and then lowering to create a thin film of resin between the previous printed layer and the build platform. Subsequently, UV light projects a 2D image onto the next layer, repeating until the build is complete. DLP was chosen over alternative 3D printing methods because it was suitable for producing the desired part dimensions with minimal porosity or surface texture. In order for the printed 3D columns to be feasible for the desired application, the criteria used to select the photosensitive resin were: biocompatibility, ability to undergo autoclave sterilization (20 min at 121°C and 1 atm), translucent appearance and the ability to cure with the desired part accuracy. The selected material, Dental SG Resin (Formlabs, Somerville, MA, USA), fulfilled each of the criteria. Dental SG Resin is a biocompatible resin, evaluated in accordance with ISO 10993-1:2018 (Formlabs 2019). This material was initially designed for dental professionals to utilize DLP technology for rapidly manufacturing surgical guides.
The Asiga Freeform Pico Plus 39 print parameters were adjusted to produce parts with the desired precision within a reasonable amount of time. This was achieved with a 0.05 mm slice thickness, and 0.8 s exposure time. The main production constraint with the Pico Plus 39 printer was the size of the build platform, 48.47 mm × 30.45 mm, with a maximum part height of 76.00 mm. This limited production to three columns at a time, followed by three additional builds to print the caps to complete each column. After printing, the caps and columns were submerged in a bath of isopropyl alcohol for 15 min at room temperature to rinse off excess resin. The parts were then air dried and post-cured by exposing them to light (405 nm) for 30 min at 60°C. The post-curing process ensured that there was no residual monomer. During post-cure, the parts also underwent further crosslinking to achieve maximum mechanical strength and thermal stability. Prior to use, the columns were sterilized by autoclaving 21 min at 121°C. We also evaluated whether these devices could be re-used after autoclaving by testing their sterility after use in the experiments with horse dung. For this, agar plugs were placed as attracting and target medium and tested for regrowth as indicated in the section concerning subculturing.
Fungal and bacterial strains and cell culture conditions
The homothallic strain AmutBmut (A43mut B43mut pab1.2) of the fungus C. cinerea (Basidiomycota) was kindly provided by Dr Markus Künzler from the Swiss Institute of Technology, Zurich (ETHZ). For the experiments performed in this study, carboxymethyl cellulose medium [CMC; composed of (NH4)2SO4 1.0 g.L−1, MgSO4 × 7H2O 1.0 g.L−1, CaCl2 × 2H2O 1.0 g.L−1, FeCl3 0.2 g.L−1, K2HPO4 1.0 g.L−1 (autoclaved separately), Casitone (Difco Laboratories, Detroit, MI, USA) 2.0 g.L−1, carboxymethyl cellulose 15.0 g.L−1, agar 6.0 g.L−1] was used for the routine maintenance of the fungus. Three bacterial strains were used to test the ability of the fungus to engage on fungal highways. Those corresponded to the flagellated bacteria P. putida KT2440, Cupriavidus necator JMP289, and Cupriavidus oxalaticus. All these bacteria produce fluorescent proteins constitutively (P. putida and C. necator are GFP tagged; C. oxalaticus is mCherry tagged). Pseudomonas putida KT2440 was kindly provided by Dr Arnaud Dechesne (Technical University of Denmark). Cupriavidus necator JMP289 was kindly provided by Prof. Jan van der Meer (University of Lausanne). Cupriavidus oxalaticus was tagged in-house using insertion with a MiniTn7 system. All three bacterial strain have been shown to disperse on the mycelium of other fungal species in previous experiments (data not shown). For regular maintenance, bacteria were grown on nutrient agar (NA) medium, composed of 8 g.L−1 nutrient broth (Biolife Italiana S.r.l., Milano, Italy) and 15 g.L−1 technical agar (Biolife). All media were sterilized by autoclaving for 21 min at 121°C.
Bacterial dispersal tests
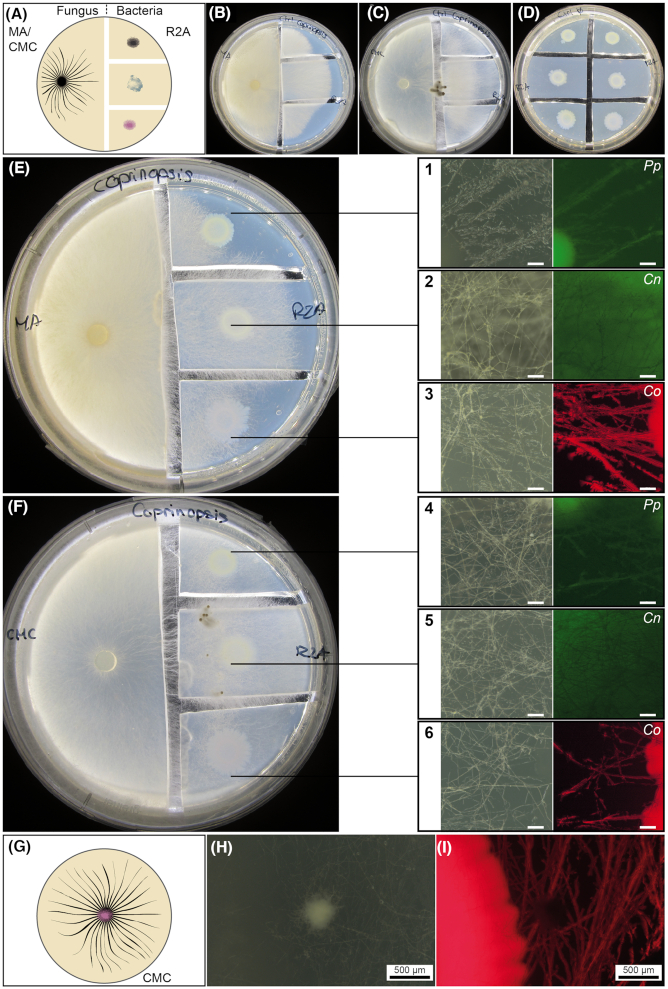
Tests for bacterial dispersal in the presence and absence of a fungal network were conducted on compartmentalized culture media in Petri dishes (Bravo et al. 2013) (Fig. 2A) using CMC and malt agar (MA; malt extract 12 g.L−1; agar 15 g.L−1) media for fungal growth, and R2A medium (Reasoner and Geldreich 1985) for bacterial growth. R2A medium was composed of 0.5 g.L−1 yeast extract, 0.5 g.L−1 Bacto Peptone, 0.5 g.L−1 casamino acids, 0.5 g.L−1 glucose, 0.5 g.L−1 soluble starch, 0.3 g.L−1 Na pyruvate, 0.3 g.L−1 K2HPO4, 0.05 g.L−1 MgSO4.7H2O and 15 g.L−1 purified agar. After fungal colonization of the bacterial compartment, fungal highway bacterial dispersal was assessed by observing their fluorescent signal on the fungal mycelium with a Nikon C-BD230 stereomicroscope with different filters for bright field (Photonic PL3000 cold light as source; Nikon C-SHG1; Nikon Instruments Inc. Melville, NY, USA), GFP and mCherry (mercury light source for fluorescence images; Nikon C-SHG1). A second test was conducted by placing C. cinerea and C. oxalaticus in direct contact on CMC in order to confirm that the fungus is capable of engaging on fungal highways in this medium and upon co-inoculation with the bacterium .
Figure 2.
Validation of the ability of bacteria to colonize the mycelial network of the fungus C. cinerea. Dispersal of fluorescently labeled Cupriavidus oxalaticus (Co), Cupriavidus necator (Cn) and Pseudomonas putida (Pp) was tested in two-compartment Petri dishes with malt agar (MA) and carboxymethyl cellulose (CMC) (fungal growth), and R2A (bacterial growth) media or by co-inoculation in CMC. (A) Schematic representation of the experimental design in which the fungus and bacteria were inoculated in physically separated compartments. To test bacterial dispersal, agar pieces containing the bacterial inoculum were physically separated by cutting out agar slices forming a gap that must be connected by fungal hyphae. Macroscopic images demonstrating the colonization by C. cinerea grown on MA (B) or CMC (C) of the R2A media without bacteria. (D) Control with bacteria only grown on R2A. Macroscopic images demonstrating the colonization by C. cinerea grown on MA (E) or CMC (F) of R2A media pre-inoculated with bacteria. Bacterial dispersal was visually assessed by stereoscopic observations (right-hand panels). The colocalization of the fluorescence with the hyphae (bright field) indicates the colonization of the fungal mycelial network by the fluorescently labeled bacteria. The pictures correspond to close-up images taken from the R2A medium. The scale bar in the close-up images (in white) corresponds to 50 µm. (G) Schematic representation of the experimental design in which the fungus and bacterium were co-inoculated directly in the same medium. (H and I) Images showing the colonization of the fungal mycelium when C. cinerea was co-inoculated in CMC together with C. oxalaticus. The white spot observed in the bright-field image corresponds to a fungal primordium, which was not colonized by the bacteria as seen in the fluorescent image.
Preparation of the columns and sampling
The columns and the 25-µm mesh filter used to avoid dispersal associated with microfauna were sterilized separately by autoclaving for 21 min at 121°C prior to assembly. CMC medium was poured into 90-mm Petri dishes. A slice of the exact inner diameter of the column (cut directly with the column edge) was then placed horizontally inside the two extremities of the column to serve as attracting and target medium. On the side of the column in contact with the environmental matrix to sample (bottom side) a piece of nylon mesh tissue (25 µm pore size) was placed in contact to the attracting medium and fixed using the second screw in the column. Three treatments were applied to the 3D columns: Treatment 1 corresponded to the target medium already colonized with C. cinerea and the 3D columns inserted in dung. Treatment 2 consisted of the target medium left uninoculated and the 3D columns inserted in dung previously colonized with C. cinerea. Finally, treatment 3 (uninoculated control treatment) consisted of an uninoculated target medium and dung . All treatments were performed in triplicate, leading to nine 3D columns. Pre-colonized CMC target media consisted of CMC media in which C. cinerea has been growing for 14 days. Fresh horse dung was provided by a horse owner from the Jura region in Switzerland and was stored at 4°C prior to use. Pre-inoculation of the dung with C. cinerea was done by placing a piece of CMC pre-colonized with the fungus and letting it colonize the surface dung for 10 days before applying the columns. The dung was placed in Magenta boxes and the 3D columns were then inserted into the dung substrate. These boxes were incubated for 21 days at room temperature in the dark.
Reculturing
After incubation, the target culture medium of each of the nine columns was cut into two equal pieces. The first half piece was transferred onto a fresh 90-mm diameter CMC Petri dish, while the other half piece was frozen for direct DNA extraction. CMC plates were incubated for 72 h at room temperature and in the dark and observed as indicated previously but using the bright-field mode of the Nikon C-BD230 stereoscope. The colonized agar from the recultured samples was cut into small pieces and all biomass was collected randomly in equal amounts into three 1.5-mL Eppendorf tubes. One of these tubes was used for DNA extraction.
DNA extraction, sequencing and analysis
Just prior to DNA extraction, the agar was ground in liquid nitrogen. Then, DNA was extracted using the FastDNA spin kit for soil (MP Biomedicals LLC, Solon, OH, USA), following the standard protocol provided with the kit for the target agar samples. In the case of the horse dung samples, a more forceful approach was used. Three successive bead-beating cycles were applied, but after each bead-beating step the supernatant was recovered and DNA was purified separately to avoid over shredding. After that, the three individual extractions were pooled by ethanol precipitation (Wunderlin et al. 2013). DNA quantification was performed using the Qubit® dsDNA HS Assay Kit on a Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Purified DNA extracts were sent to Fasteris (Geneva, Switzerland) for 16S rDNA and ITS amplicon sequencing using an Illumina MiSeq platform (Illumina, San Diego, CA, USA), generating 250 bp paired-end reads. For the 16S rDNA, the V3–V4 region was amplified using the universal primers Bakt_341F (5′-CCT ACG GGN GGC WGC AG-3′) and Bakt_805R (5′-GAC TAC HVG GGT ATC TAA TCC-3′) (Herlemann et al. 2011). The primers ITS3_KYO2 (5′-GAT GAA GAA CGY AGY RAA-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) (Toju et al. 2012) were used for the amplification of the ITS2 region.
Sequence analysis
Bacterial and fungal taxonomic profiling analysis was performed using QIIME2 microbiome analysis package (Bolyen et al. 2019). Data were provided as pre-demultiplexed and pre-trimmed sequences. Based on QIIME2’s quality plots, the 16S rDNA and ITS sequences were truncated at 230 bp during the denoising step performed using the DADA2 pipeline. Bacterial taxonomy attribution was computed using a classifier trained using QIIME2 tools (Naive Bayesian classifier) on the last release (GG13_5) of Greengenes database (http://greengenes.lbl.gov) for the V3–V4 regions (341F-805R) of the 16S rDNA. Fungal taxonomy attribution was computed using a classifier trained using QIIME2 on the last release of UNITE database for ITS amplicons (https://files.plutof.ut.ee/public/orig/98/AE/98AE96C6593FC9C52D1C46B96C2D9064291F4DBA625EF189FEC1CCAFCF4A1691.gz; https://unite.ut.ee/repository.php).
Exported QIIME2 outputs were merged within R software V3.5.0 (https://www.r-project.org/) using the Phyloseq package (McMurdie and Holmes 2013). ‘Bottom-up’ stream plots and combined bar plots for diversity were prepared for all available samples. Unassigned OTUs were not taken into account for the stream plots. For the latter, minor genera percentage reflects the sum of the percentages of under-represented genera.
RESULTS
Design of the 3D printed fungal highway columns
The design of the 3D columns focused on conserving all the key features of the manually built device: the possibility to apply any type of attracting and target media at each of the extremities of the column; an hourglass shape to avoid the formation of a continuous water film; and a built-in modular lattice that simulates the presence of soil particles and air-filled gaps. The final model consisted of a 4-cm-long column with a connecting path of 2 mm × 3 mm. A regular lattice covered half of the length of the column (Fig. 1A; Video S1, Supporting Information). These two features, the narrow connecting path and the soil simulating cross, were generated in the manually built columns by first twisting a plastic cylinder and then adding glass beads at each end. As such, those were the two most challenging elements of the production and were entirely integrated in the built-in design of the printed version. The top of the 3D column can be sealed with a removable lid; the bottom includes a ring to affix an appropriate mesh to prevent the transport of bacteria by soil microfauna along with a removable lid to open and close the end of the column in contact with the environmental sample. After 3D printing and sterilization, the assemblage and preparation of the 3D column is similar to the procedure used in the production of the manually built version (Simon et al. 2015). In order to assemble the columns, the researcher must insert two pieces of agar-based medium (or a similar gelified medium) at each extreme of the column (Fig. 1B). In addition, at the extreme of the column that will be in contact with the environmental sample, a mesh with a suitable pore size (for instance 25 µm) should be placed with the help of the additional ring. Thereafter, the column can be placed on the environmental sample and incubated for a period of 2–3 weeks. After the incubation, the target medium (on top of the column) can be used for reculturing of the bacterial–fungal species attained in this compartment or for direct DNA/RNA-based analysis (Fig. 1B). Moreover, an additional advantage of the 3D columns was the possibility to construct the columns from autoclavable materials (see the 'Materials and Methods' section), allowing for their re-use after autoclaving. We tested this and were able to use the same 3D columns in three independent experiments without cross-contamination (data not shown).
Figure 1.
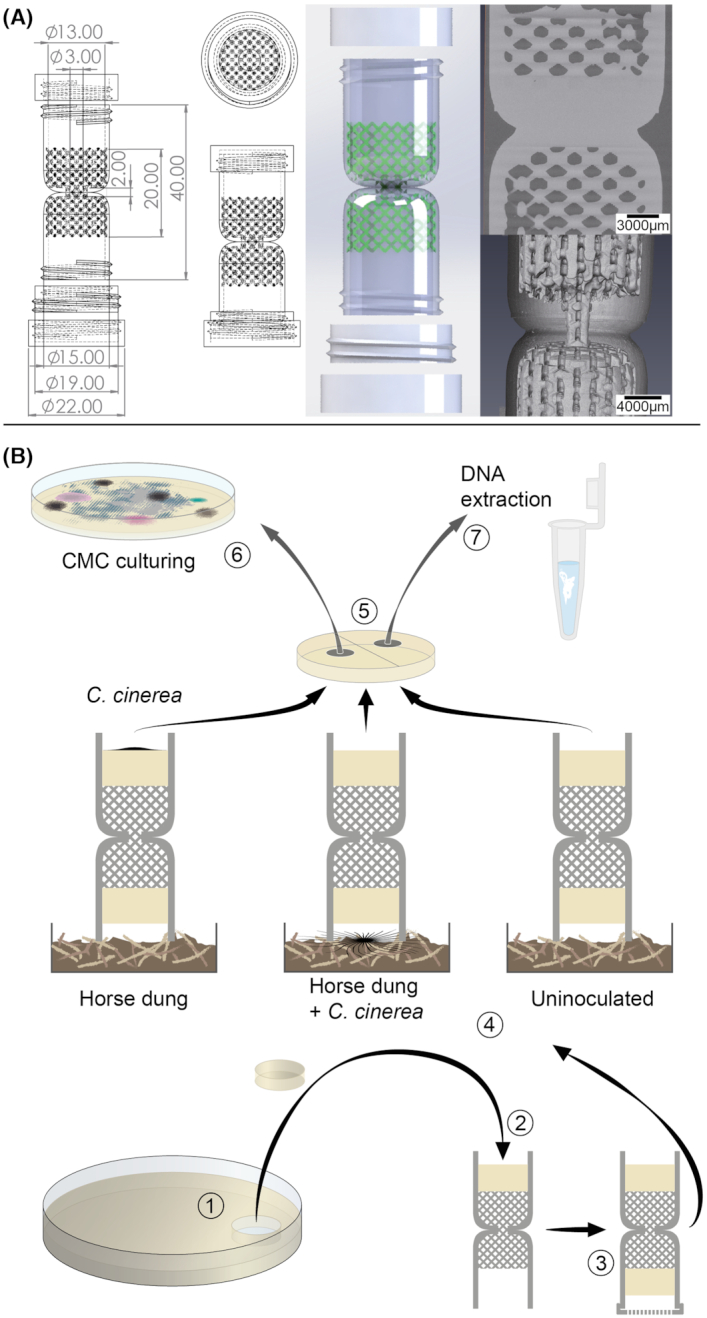
Design and use of the 3D printed columns. (A) Schematic representation of the design for additive printing including the dimensions of the different components. In the 3D model, the internal regular lattice structure consists of a 2 mm unit cell and 0.5 mm strut diameter replacing the glass beads from the previous design (Simon et al. 2015). The structure of the lattice can be appreciated in the video included in the Supporting Information. (B) Once printed, the columns can be autoclaved and any kind of agar-based medium (pieces usually cut from a Petri dish; step 1) can be placed on the top (2) and on the bottom of the lattice (3) as target and attracting media, respectively. Once completed with a filtering mesh in direct contact with the attracting medium, the columns can be placed on the environmental sample (4). After incubation, the target medium (5) can be collected from the columns and used to isolate bacteria and fungi by reculturing [in this case carboxymethyl cellulose (CMC) medium as indicated in the text; step 6] or for direct DNA extraction (7).
Testing of the dispersal of bacteria on the hyphae of C. cinerea
The model organism selected to validate the use of the 3D columns was the common coprophilous fungus C. cinerea. Although many aspects of the biology of C. cinerea are known, one element that has never been established is its ability to engage in fungal highway interactions with bacteria. Therefore, in order to verify this ability, an initial series of bacterial dispersal tests were conducted. In the first experiment, C. cinerea and model soil bacteria (Cupriavidus oxalaticus, Cupriavidus necator and Pseudomonas putida) were inoculated onto physically separated media to avoid direct competition for the same carbon source, but still triggering fungal highway dispersal (Fig. 2A). These bacteria were selected because they are known to disperse on hyphae of other fungi (Pion et al. 2013). The fungus was grown on two types of carbon-rich media as substrate: malt agar (MA; Fig. 2B) and carboxymethyl cellulose (CMC; Fig. 2C), and bacteria were grown on the nutrient poor Reasoner's 2 agar (R2A) medium (Fig. 2D). After the fungus had colonized the bacteria-containing R2A medium and reached the bacterial inoculum, bacterial colonization of the surface of the fungal mycelium that was distant from the bacterial inoculum was verified with a stereoscope by visualizing the fluorescently labeled bacteria. This colonization is the result of the active dispersal of the bacteria on the fungal hyphae. In this test, all three bacterial species colonized areas of the mycelium that were physically distant from the bacterial inoculum, indicating that they can disperse on the mycelial network (Fig. 2E and F). Based on these results, bacterial dispersal was also tested when the two organisms were co-inoculated on the same medium and without a physical gap between each organism. For this, C. cinerea was co-inoculated with C. oxalaticus in CMC medium (Fig. 2G). This bacterium was selected because the red fluorescent tag afforded easy visualization of the BFIs in CMC. In the co-inoculation system, bacterial dispersal from the point of the inoculation and along the fungal mycelium was also observed (Fig. 2H and I). These preliminary results demonstrate that C. cinerea can engage in fungal highways with model soil bacteria.
Enrichment of BFI with the 3D columns using C. cinerea as bait
To enrich potential dung-inhabiting bacterial partners able to disperse using the mycelial network of C. cinerea, 3D columns were prepared with three different treatments. Two treatments consisted of pre-inoculating the fungus either on the CMC target medium or in non-sterile horse dung. This was done prior to the placement of the columns in the dung as the environmental sample. These treatments were compared with a third control treatment consisting of uninoculated 3D columns/dung substrate, in which the original microbial communities were expected to be unaltered (Fig. 3A). After 21 days of incubation, the target medium and the initial dung material were collected to assess the presence of bacteria and fungi, both by reculturing on CMC medium or by direct DNA extraction followed by amplicon sequencing of fungal or bacterial genetic markers as shown in Fig. 1B (recultivation as shown in step 6).
Figure 3.
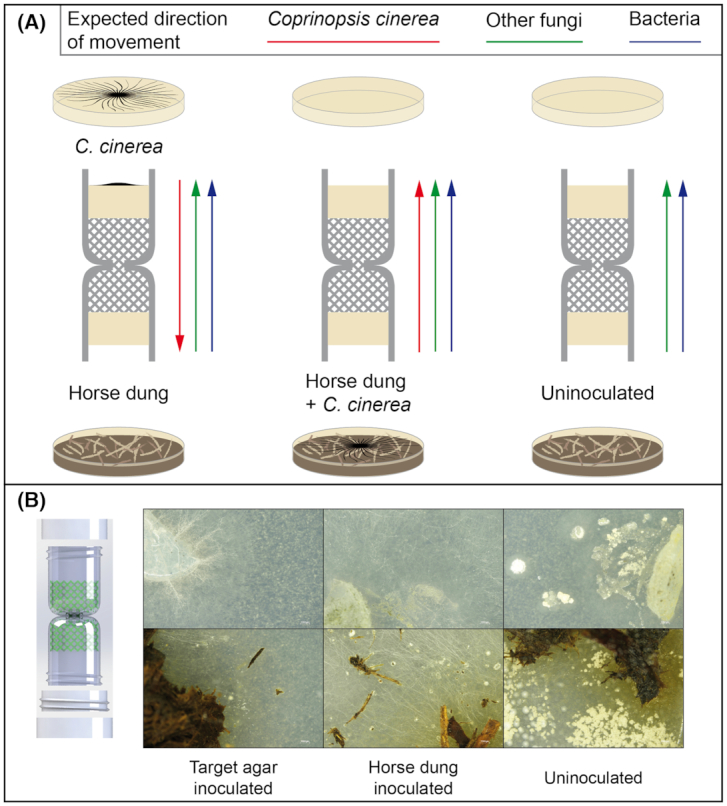
Schematic representation of the experimental design and macroscopic images illustrating the results after reculturing of the target medium and dung samples. (A) In the experimental design, the target medium consisted of either CMC medium pre-colonized by C. cinerea or CMC medium only. The attracting medium was always CMC. The columns were placed on horse dung or on horse dung pre-inoculated with C. cinerea. The combinations of these resulted in three treatments in which the expected direction of dispersal from C. cinerea, other fungi and bacteria dispersing using fungal highways is expected to vary. (B) Examples of stereoscopy images of CMC medium inoculated with samples from the target medium (top) or horse dung (bottom), for the respective treatments.
After reculturing on CMC, further subculturing for isolation or morphological identification of pure strains was not attempted. Nonetheless, the presence of bacterial colonies and filamentous fungi growing on CMC was recorded for each condition based on their macroscopic morphology (Table 1; Fig. S1, Supporting Information). Despite the fact that fungi and bacteria were always recovered from the dung samples (Fig. 3B; Table 1), consistent bacterial and fungal regrowth from the target medium was not observed for all the columns. After the reculturing of the target agar from the 3D columns pre-inoculated with C. cinerea in the target medium, strong fungal growth was observed in only one of the replicates, while a very weak fungal growth was only observed next to the piece of agar recovered from the columns in the other two. In contrast, bacterial growth was observed in all the replicates, but it was weaker in the replicate with strong fungal regrowth. In the columns with the treatment consisting of the pre-inoculation with C. cinerea in dung, growth of both fungi and bacteria was observed after subculturing in two of the columns, but in one of these two replicates (R2), the bacteria that grew in CMC had the morphology of actinobacteria. In the third replicate, fungal regrowth was negative but actinobacteria-like bacteria grew in CMC. Finally, in the samples from the uninoculated treatment, only one of the three replicates was clearly colonized by both fungi and bacteria, while in the other two, only actinobacteria-like colonies were observed. It is not possible to establish whether mycelial growth by actinobacteria allows them to colonize the target agar without interacting with fungi, or whether the fungi associated with those did not survive during subculturing.
Table 1.
Colonization and recovery of fungi and bacteria in the 3D columns. The colonization and recovery were assessed by reculturing (growth) or by DNA extraction based on the cultures (culture) or agar/dung samples (direct). The success in the amplification and sequencing of ITS (fungi) and 16S rRNA (bacteria) genes is also indicated for DNA extracts obtained from the cultures or agar/dung samples.R = replicate number; w = weak growth; + = positive; − = negative; a = actinobacterial-like bacterial colonies.
| Growth | DNA extraction (ng/µL) | ITS | 16S rDNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | Sample | R | Fungi | Bacteria | Culture | Direct | Culture | Direct | Culture | Direct |
| TM agar | Horse dung | 1 | + | + | 2.11 | 44.2 | + | + | + | + |
| 2 | + | + | 32.2 | 76.1 | + | + | + | + | ||
| 3 | + | + | 5.14 | 74.1 | + | + | + | + | ||
| Target agar | 1 | w | + | 0.167 | <0.5 ng/mL | − | + | + | + | |
| 2 | w | + | 0.374 | <0.5 ng/mL | − | + | + | + | ||
| 3 | + | w | 1.94 | 1.35 | + | + | + | − | ||
| HD | Horse dung | 1 | + | + | 29.8 | 110 | + | + | + | + |
| 2 | + | + | 14.3 | 188 | + | + | + | + | ||
| 3 | + | + | 37.8 | 84.7 | + | + | + | + | ||
| Target agar | 1 | − | a | 9.43 | 3.18 | − | - | + | − | |
| 2 | + | a | 11.8 | 0.294 | + | − | + | − | ||
| 3 | + | + | 2.74 | 0.1 | + | + | + | + | ||
| Uninoculated | Horse dung | 1 | + | + | 15.8 | 40.2 | + | + | + | + |
| 2 | + | + | 9.07 | 44.6 | + | + | + | + | ||
| 3 | + | + | 23.5 | 40.4 | + | + | + | + | ||
| Target agar | 1 | − | a | 0.264 | <0.5 ng/mL | − | − | + | − | |
| 2 | + | + | 17.4 | 2.74 | + | + | + | + | ||
| 3 | − | a | 0.383 | <0.5 ng/mL | − | − | + | − | ||
Diversity analysis of fungal and bacterial communities enriched in the 3D columns
In order to gain insight into the diversity of the microbial communities in dung and those colonizing the columns, amplicon sequencing of fungi Internal transcribed spacer (ITS) or bacteria (16S ribosomal RNA (rRNA) gene) was performed. This analysis was made for both the communities that were recultured in CMC, as well as the total community present in the target medium and the dung. In the case of the communities recovered from CMC, material from the entire plate was collected and used as a pooled sample for DNA extraction of what was then defined as the culturable fraction of the community. This community was contrasted with the total uncultured community that was assessed by extracting DNA directly from the target medium or the dung sample without prior recultivation.
Enough DNA for amplicon sequencing was obtained from DNA extractions from all dung samples (both for the culturable and total uncultured community in the three treatments; Table 1). Likewise, in the target medium samples in which microbial regrowth was observed, enough DNA for amplicon sequencing was also obtained. In contrast, amplification and sequencing of DNA extracts of the target medium without subculturing were only partially successful. In the 3D columns pre-inoculated with C. cinerea in the target agar, the ITS was amplifiable in all the replicates, while 16S rRNA gene amplification failed in one replicate (replicate R3; weak bacterial regrowth). In the treatment consisting of the pre-inoculation with C. cinerea in dung and in the treatment without pre-inoculation, amplification for the ITS and 16S rDNA was only possible in one replicate each (R3 and R2, respectively).
The uneven rate of colonization and/or survival in the target agar together with the failure in the amplification of fungal and/or bacterial markers in some of the replicates, limited our ability to perform a general comparison of the fungal and bacterial diversity between the different treatments. Nonetheless, the evaluation of potential bacterial associates to C. cinerea, as well as of the fungal and bacterial diversity in horse dung and the BFI associated with fungal highways in this environment could be made.
In total, 12 operational taxonomic units (OTUs) affiliated to Coprinopsis were detected. The percentage of identity between the OTUs and reference ITS sequences for C. cinerea varied from 96–100%. Six OTUs were over 98% identical to a common reference sequence and corresponded to the most abundant OTUs in the samples in which C. cinerea was pre-inoculated or in which the fungus presumably migrated (Table S1, Supporting Information). In addition and as expected, based on the ITS sequences that could be classified, Coprinopsis was detected as the most abundant fungal genus in the target medium pre-inoculated with C. cinerea (mean relative abundance 39%; Table 2). This was the case for the total community analysis (uncultured) and the one replicate in which fungal growth was observed after reculturing (44.4% ITS relative abundance; Fig 4; Fig. S2, Supporting Information). In the replicate in which Coprinopsis was detected both after reculturing and by direct DNA extraction, Coprinopsis was also detected in the horse dung sample (relative abundance of 2.3%) and the OTUs corresponded to those detected in high abundance in the target medium in which the fungus was pre-inoculated (Table S1, Supporting Information). Thus, the baiting strategy based on the pre-inoculation of the target medium appears to be successful allowing the fungus to migrate through the column and to reach the dung.
Table 2.
Relative abundance of ITS OTUs classified as Coprinopsis spp. The relative abundance was estimated based on sequencing results from DNA from the culturable fraction of the community (culture) or agar/dung samples (direct). NA = not applicable (no sequence data available).
| Relative abundance | ||||
|---|---|---|---|---|
| Treatment | Sample | R | Culture | Direct |
| TM agar | Horse dung | 1 | 0 | 0 |
| 2 | 0 | 0.08 | ||
| 3 | 0 | 2.92 | ||
| Target agar | 1 | NA | 43.97 | |
| 2 | NA | 35.48 | ||
| 3 | 44.46 | 39.57 | ||
| HD | Horse dung | 1 | 0.03 | 0.09 |
| 2 | 0 | 0.47 | ||
| 3 | 0 | 2.3 | ||
| Target agar | 1 | NA | NA | |
| 2 | 0.03 | NA | ||
| 3 | 0 | 0 | ||
| Uninoculated | Horse dung | 1 | 0 | 0.31 |
| 2 | 0 | 0.1 | ||
| 3 | 0 | 0 | ||
| Target agar | 1 | NA | NA | |
| 2 | 0 | 0 | ||
| 3 | NA | NA | ||
Figure 4.
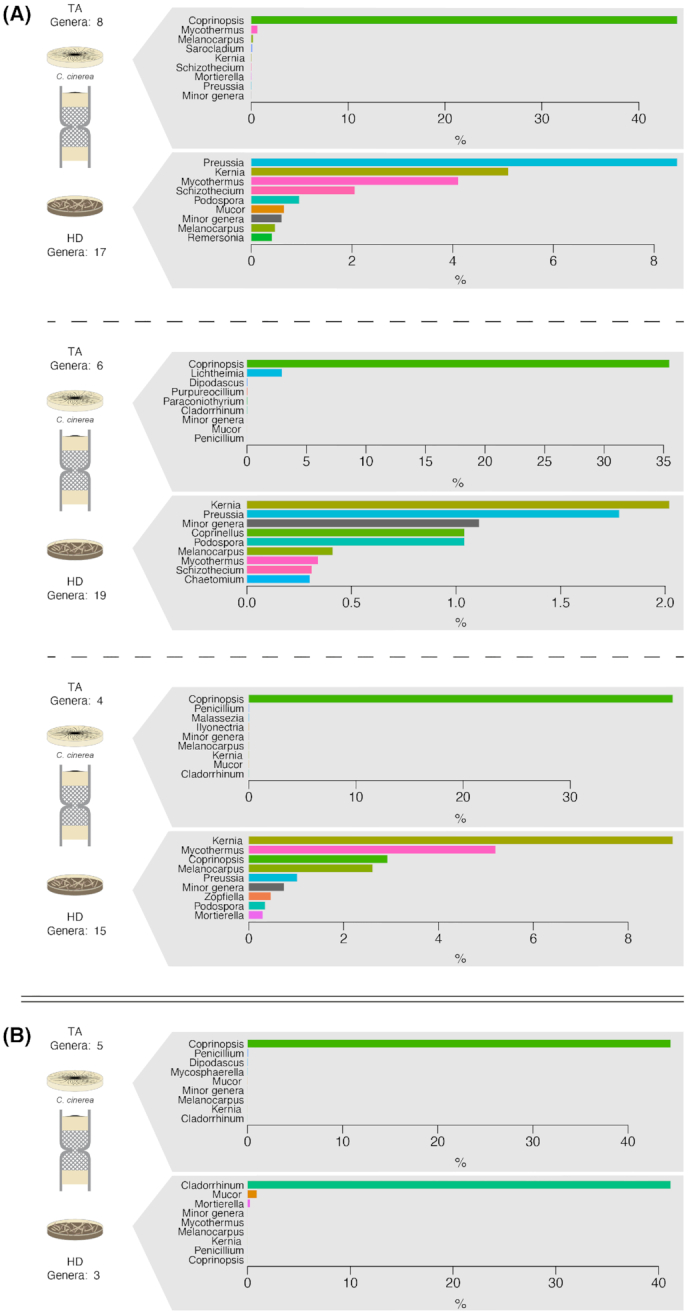
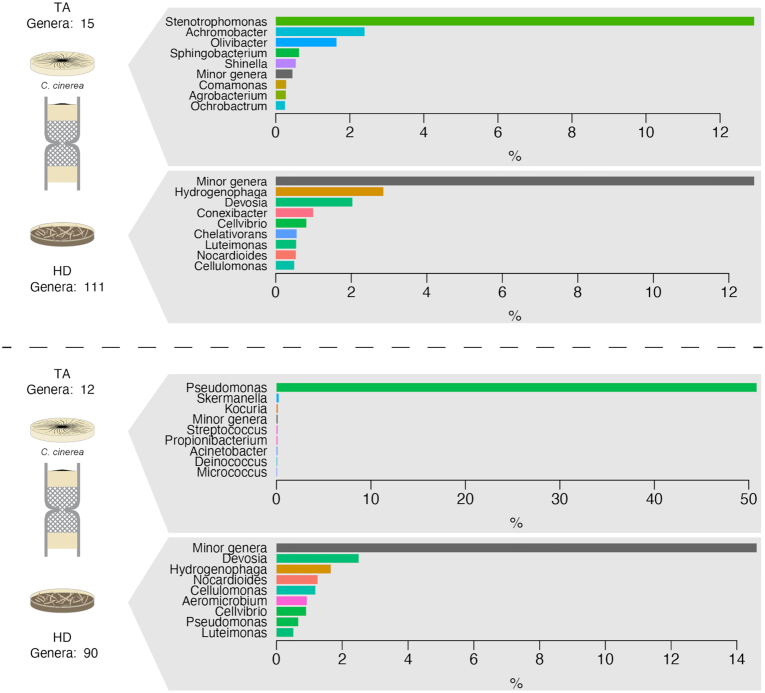
Fungal community analysis based on the sequencing of ITS comparing the communities in horse dung and the target medium. The treatment corresponds to columns in which C. cinerea was pre-inoculated in the target medium. Each plot represents the comparison in the relative abundance of the most dominant fungal genera in horse dung (bottom) and target medium (up) per column replicate. The analysis corresponded to DNA extractions of the total community (A; three replicates) or of the culturable community (B; one replicate). To increase readability, the relative abundance of the eight most abundant genera is shown, and other genera were grouped in a ‘Minor genera’ category. A complete representation of the community composition is shown in Fig. S2 (Supporting Information).
When the fungus was pre-inoculated in the horse dung, OTUs related to C. cinerea only represented a small fraction of the fungal community in dung (<0.5% of the mean relative abundance; Table 2). The OTUs detected were the same as those found in the pre-inoculated agar (Table S1, Supporting Information). Kernia, Melanocarpus, Mycothermus, Preussia and Podospora were the most abundant representatives of the total fungal community in dung (Fig. S3, Supporting Information), while Cladorrhinum dominated the fungal community in the dung samples after reculturing (Fig. S4, Supporting Information). However, none of these fungal genera were detected in the target medium of the same columns and under this configuration, either Coprinopsis did not colonize the columns or was outcompeted by other fungi that instead attained the target medium (Penicillium spp.).
Bacteria affiliated with the genera Pseudomonas and Stenotrophomonas were highly enriched in the target medium samples in which Coprinopsis was also detected (Fig. 5; Fig. S5, Supporting Information). Moreover, the results for the bacterial community composition illustrate the highly selective nature of fungal highway interactions, in association with a specific fungus. A strong reduction in richness was observed in the bacterial genera detected in the target media relative to the richness of the dung communities (reduction from 115 to 15 and from 90 to 12 in the number of detected genera in the two columns in which data for the bacterial communities were recovered; Fig. 5).
Figure 5.
Bacterial community analysis based on the sequencing of 16S rDNA comparing the communities in horse dung and the target medium. The treatment corresponds to columns in which C. cinerea was pre-inoculated in the target medium. Each plot represents the comparison in the relative abundance of the most dominant bacterial genera in horse dung (bottom) and target medium (up) per column replicate. The analysis corresponded to DNA extractions of the total community (A) or of the culturable community (B), represented each by one replicate. To increase readability, the relative abundance of the eight most abundant genera is shown, and other genera were grouped in a ‘Minor genera’ category. A complete representation of the community composition is shown in Fig. S5 (Supporting Information).
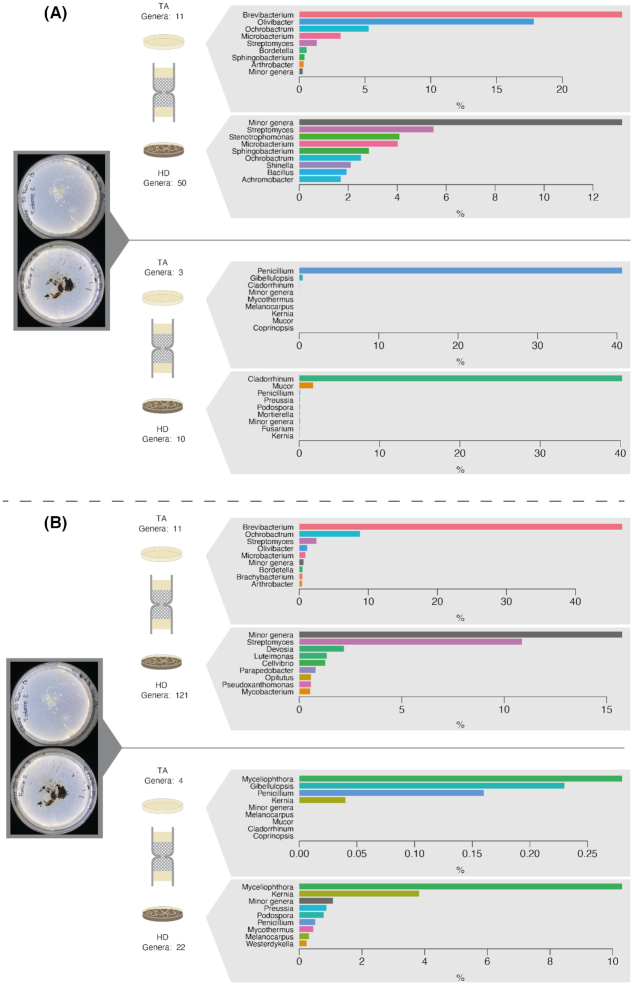
Finally, regarding the uninoculated control, the replicate showing the simultaneous colonization of the target medium (and thus the 3D columns) by fungi and bacteria provides a first glimpse of the potential fungal highway-like BFI naturally occurring in horse dung. Only a small fraction of the fungi that reached the target medium were also identified in the direct DNA extraction (Lasiosphaeriaceae, Ascomycota) or culturing (Penicillium) of the horse dung. In contrast, regarding the bacterial partners, those were similar for the total and the cultured communities and were dominated by bacteria belonging to the genera Brevibacterium,Ochrobactrum,Olivibacter,Microbacterium and Streptomyces (Fig. 6; Fig. S6, Supporting Information).
Figure 6.
Bacterial and fungal community analysis based on the sequencing of the 16S rDNA and ITS genes comparing the communities in horse dung and the target medium in the uninoculated treatment. Each plot represents the comparison in the relative abundance of the most dominant genera of bacteria (upper top-half of the figure) and fungi (bottom half of the figure) in horse dung (lower panel) and target medium (upper panel). The analysis corresponded to DNA extractions of the total community (A) or of the culturable community (B). To increase readability, the relative abundance of the eight most abundant genera is shown, and other genera were grouped in a ‘Minor genera’ category. A complete representation of the community composition is shown in Fig. S6 (Supporting Information).
DISCUSSION
The principal objective of this study was to present and validate a version of fungal highway columns produced by additive printing (3D columns). The success rate of the 3D columns to enrich BFI from horse dung corresponded to about two-thirds of colonized columns, which was also the case with the original manually built columns (Simon et al. 2015). In addition, the high throughput offered by additive printing offers the possibility to test additional applications with higher replication numbers. In the past, the fungal highway columns have been used to enrich cultivable bacterial–fungal pairs of organisms potentially interacting in soils (Simon et al. 2015) or to create an inventory of the diversity of bacteria and fungi engaging in fungal highways in soils with different physicochemical properties (Simon et al. 2017). In this study, we did not simply replicate the use of the columns to compare the diversity of bacteria and fungi interacting via fungal highways in different soils (Simon et al. 2017), but rather, we used them to selectively enrich for bacterial partners with the ability to interact with a specific fungus. Although in this study, we did not continue the isolation of specific organisms and the verification of their ability to interact (for instance, to disperse on the fungus tested), this can be easily incorporated in the experimental design. The motivation for this type of application is the enrichment of specific consortia based on a selective pressure applied either at the level of the attracting medium, the target medium or both. This is possible thanks to the fact that the 3D model retains the flexibility provided in the hand-made model and, more specifically, the possibility to include any type of medium.
For this novel application, we selected the common saprophytic and coprophilous fungus C. cinerea. Unlike many mushroom forming fungi, C. cinerea is easy to culture in the laboratory in defined media and it can complete its entire lifecycle in a very short period of time (Kamada et al. 2010; Stajich et al. 2010). The AmutBmut mutant strain is a self-compatible homothallic (self-fertile) mutant with a simplified life cycle (Swamy, Uno and Ishikawa 1984; Liu et al. 2006) and is thus widely used in laboratory experiments around the world. While it might have lost competitiveness compared to wild strains freshly isolated from horse dung, this strain offers the possibility to investigate different developmental stages (including the formation of fruiting bodies) in the laboratory. Coprinopsis cinerea has been widely recognized by its ability to produce compounds to antagonize bacteria (Künzler 2018; Kombrink et al. 2019), including important antimicrobial peptides (Essig et al. 2014) or enzymes that disrupt bacterial activity (Stockli et al. 2017). In some cases, it has been demonstrated that these defense mechanisms are inducible by competition with bacteria (Stockli et al. 2019) or in response to fungivorous attack of predatory nematodes (Schmieder et al. 2019).
Positive interactions with other microbial partners could be as important in the ecology of C. cinerea as the negative interactions described above. Positive interactions can also be relevant in biotechnology (Bouws, Wattenberg and Zorn 2008), in which co-cultures of cooperative organisms may constitute a source of novel bioactive compounds (Egan, Thomas and Kjelleberg 2008; Netzker et al. 2018). For instance, the pre-treatment of rice straw with C. cinerea has been proposed as a cost-effective and environmentally friendly approach to improve biofuel production or animal feeding (Zhang et al. 2018a). Further advancement for this application could come from identifying bacterial species naturally associated with this fungus and that improve its cellulolytic activity. In our proof-of-concept test, the primary aim was not to isolate new strains of C. cinerea, but to test if the 3D columns could provide information on natural partners to this coprophilous species in horse dung. Our results show that the best way to bait for partners of this fungus is by pre-inoculating the target medium in which Coprinopsis (presumably the strain inoculated) dominated the fungal communities. The results also indicated that in this condition, Pseudomonas and Stenotrophomonas were consistently associated with Coprinopsis. Previous studies have suggested that Stenotrophomonas are among the nematode-associated bacteria that induce a defense in response to the attack of Coprinopsis by fungivorous nematodes (Tayyrov et al. 2019), while Pseudomonas induce a negative impact on growth in direct competition (Essig et al. 2014). The results obtained here, suggested that bacteria of these genera can also interact with Coprinopsis to colonize new environments, and in the case of Pseudomonas spp. could be a way to improve cellulose degradation, as Pseudomonas are among the bacterial genera effective in the degradation of cellulose (Behera et al. 2017) or other complex polymers (Pathak and Navneet 2017).
More generally, the comparison of the diversity of fungi and bacteria in the target media and dung samples illustrate how the 3D printed columns induce selection on microbiota's propagation, highlighting colonization processes through fungal highways interactions. Using horse dung as an environmental matrix, we were able to detect and enrich an entirely novel set of BFI as compared to soils (Simon et al. 2015; Simon et al. 2017), which were largely dominated by Mucoromycota (direct DNA extraction) (Simon et al. 2017) or Fusarium (reculturing) (Simon et al. 2015). Overall, this study shows that 3D columns are a flexible and useful scientific tool that can be used to investigate fungal highways in a multitude of systems. The ecology of many of the fungi detected in this study is largely unknown, but some of the microorganisms identified can become significant model organisms. For instance, Cladorrhinum, a fungal genus that was enriched in dung samples, is also of interest given its potential use as a biocontrol agent (Barrera et al. 2019). In the case of other fungi whose ecology is still poorly described, such as Kernia spp. for instance, their enrichment using the 3D columns in combination with methods such as shotgun metagenomic sequencing, could be a first approach for understanding their ecology and relevance in ecosystem functioning.
Finally, one aspect that was surprising in our study and that is in strong contrast with previous studies on fungal highways is the fact that we detected almost exclusively the presence of actinobacteria after reculturing of the target medium in several treatment and replicates (Table 1). Although the dispersal of actinobacteria using fungal highways has not been extensively investigated, a previous study suggested that actinobacteria could not use hyphae of fungi found on cheese rinds for dispersal (Zhang et al. 2018b). However, our results contradict this and show that diverse genera of actinobacteria (both filamentous and not) can disperse through the device. Unfortunately in those columns with actinobacterial growth, fungi could not be recovered and thus it is not possible to assess whether this dispersal is associated with fungal highways. Nevertheless, this is a significant finding that should be explored further.
CONCLUSION
Despite the interest generated by the fungal highway columns after the publication of the initial study describing their design and testing (Simon et al. 2015), the development of a routine and standardized manner to produce them would certainly promote their use. Thanks to the progress made here, by simplifying their production using additive manufacturing, the 3D fungal highway columns can become a regular tool for investigating fungal highways. Both, through the direct dispersal of bacteria on hyphae, and the impact of hyphae on the transfer of nutrients and water (Worrich et al. 2017), fungal highways are an essential component of soil functioning and microbial activity. The device presented here will certainly contribute to expanding our understanding and knowledge of the organisms involved in this BFI in soils and other unsaturated environments with a similar 3D nature.
ACKNOWLEDGMENTS
The authors want to thank Dr Markus Kuenzler for his comments on earlier versions of this manuscript.
FUNDING
This study was supported by the U.S. Department of Energy, Office of Science, Biological and Environmental Research Division, under award number LANLF59T.
Supplementary Material
Contributor Information
Pilar Junier, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Guillaume Cailleau, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Ilona Palmieri, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Celine Vallotton, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Olivia C Trautschold, Materials Science and Technology, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Thomas Junier, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland; Vital-IT Group, Swiss Institute of Bioinformatics, CH, 1015, Lausanne, Switzerland.
Christophe Paul, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Danae Bregnard, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Fabio Palmieri, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Aislinn Estoppey, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Matteo Buffi, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Andrea Lohberger, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Aaron Robinson, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Julia M Kelliher, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Karen Davenport, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Geoffrey L House, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Demosthenes Morales, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
La Verne Gallegos-Graves, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Armand E K Dichosa, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Simone Lupini, Civil and Environmental Engineering, University of Houston, Houston, TX 77004, USA.
Hang N Nguyen, Civil and Environmental Engineering, University of Houston, Houston, TX 77004, USA.
Jamey D Young, Department of Chemical and Biomolecular Engineering, and Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 37212, USA.
Debora F Rodrigues, Civil and Environmental Engineering, University of Houston, Houston, TX 77004, USA.
A Nicholas G Parra-Vasquez, Materials Science and Technology, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
Saskia Bindschedler, Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, CH, 2000, Neuchâtel, Switzerland.
Patrick S G Chain, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM 87544, USA.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
Conflict of Interest
None declared.
REFERENCES
- Banitz T, Johst K, Wick LYet al. Highways versus pipelines: contributions of two fungal transport mechanisms to efficient bioremediation. Environ Microbiol Rep. 2013;5:211–8. [DOI] [PubMed] [Google Scholar]
- Barrera VA, Martin ME, Aulicino Met al. Carbon-substrate utilization profiles by Cladorrhinum (Ascomycota). Rev Argent Microbiol. 2019;51:302–6. [DOI] [PubMed] [Google Scholar]
- Behera BC, Sethi BK, Mishra RRet al. Microbial cellulases: diversity & biotechnology with reference to mangrove environment: a review. J Genet Eng Biotechnol. 2017;15:197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MRet al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouws H, Wattenberg A, Zorn H. Fungal secretomes: nature's toolbox for white biotechnology. Appl Microbiol Biotechnol. 2008;80:381–8. [DOI] [PubMed] [Google Scholar]
- Bravo D, Cailleau G, Bindschedler Set al. Isolation of oxalotrophic bacteria able to disperse on fungal mycelium. FEMS Microbiol Lett. 2013;348:157–66. [DOI] [PubMed] [Google Scholar]
- Christofides SR, Bettridge A, Farewell Det al. The influence of migratory Paraburkholderia on growth and competition of wood-decay fungi. Fungal Ecology. 2020;45:100937. [Google Scholar]
- Connell JL, Ritschdorff ET, Whiteley Met al. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A. 2013;110:18380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mattos-Shipley KM, Ford KL, Alberti Fet al. The good, the bad and the tasty: the many roles of mushrooms. Stud Mycol. 2016;85:125–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S, Thomas T, Kjelleberg S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr Opin Microbiol. 2008;11:219–25. [DOI] [PubMed] [Google Scholar]
- Essig A, Hofmann D, Munch Det al. Copsin, a novel peptide-based fungal antibiotic interfering with the peptidoglycan synthesis. J Biol Chem. 2014;289:34953–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formlabs . Surgical guide material data sheet. FLSGAM01-V1, 2019. [Google Scholar]
- Furuno S, Pazolt K, Rabe Cet al. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ Microbiol. 2010;12:1391–8. [DOI] [PubMed] [Google Scholar]
- Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–73. [DOI] [PubMed] [Google Scholar]
- Herlemann DP, Labrenz M, Jurgens Ket al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011;5:1571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu Y, J-Z Fuet al. Developments of 3D printing microfluidics and applications in chemistry and biology: a review. Electroanalysis. 2016;28:1658–78. [Google Scholar]
- Horn TJ, Harrysson OL. Overview of current additive manufacturing technologies and selected applications. Sci Prog. 2012;95:255–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada T, Sano H, Nakazawa Tet al. , Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea. Fungal Genet Biol. 2010;47:917–21. [DOI] [PubMed] [Google Scholar]
- Kohlmeier S, Smits TH, Ford RMet al. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–6. [DOI] [PubMed] [Google Scholar]
- Kombrink A, Tayyrov A, Essig Aet al. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019;13:588–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kues U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64:316–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler M. How fungi defend themselves against microbial competitors and animal predators. PLoS Pathog. 2018;14:e1007184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder MB, Szilvay GR, Nakari-Setala Tet al. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29:877–96. [DOI] [PubMed] [Google Scholar]
- Liu Y, Srivilai P, Loos Set al. An essential gene for fruiting body initiation in the basidiomycete Coprinopsis cinerea is homologous to bacterial cyclopropane fatty acid synthase genes. Genetics. 2006;172:873–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Guggiari M, Bravo Det al. Fungi, bacteria and soil pH: the oxalate-carbonate pathway as a model for metabolic interaction. Environ Microbiol. 2012;14:2960–70. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelyan A, Meuser K, Brune A. Microenvironmental heterogeneity of gut compartments drives bacterial community structure in wood- and humus-feeding higher termites. FEMS Microbiol Ecol. 2017;93:fiw210. [DOI] [PubMed] [Google Scholar]
- Nai C, Meyer V. From axenic to mixed cultures: technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 2018;26:538–54. [DOI] [PubMed] [Google Scholar]
- Neches RY, Flynn KJ, Zaman Let al. On the intrinsic sterility of 3D printing. PeerJ. 2016;4:e2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzker T, Flak M, Krespach MKet al. Microbial interactions trigger the production of antibiotics. Curr Opin Microbiol. 2018;45:117–23. [DOI] [PubMed] [Google Scholar]
- Nunan N, Wu K, Young IMet al. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol Ecol. 2003;44:203–15. [DOI] [PubMed] [Google Scholar]
- Or D, Smets BF, Wraith JMet al. Physical constraints affecting bacterial habitats and activity in unsaturated porous media: a review. Adv Water Res. 2007;30:1505–27. [Google Scholar]
- Pathak VM, Navneet. Review on the current status of polymer degradation: a microbial approach. Bioresource Bioprocess. 2017;4:15. [Google Scholar]
- Pion M, Bshary R, Bindschedler Set al. Gains of bacterial flagellar motility in a fungal world. Appl Environ Microbiol. 2013;79:6862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder SS, Stanley CE, Rzepiela Aet al. Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Curr Biol. 2019;29:217–28.e4. [DOI] [PubMed] [Google Scholar]
- Semple KT, Doick KJ, Wick LYet al. Microbial interactions with organic contaminants in soil: definitions, processes and measurement. Environ Pollut. 2007;150:166–76. [DOI] [PubMed] [Google Scholar]
- Simon A, Bindschedler S, Job Det al. Exploiting the fungal highway: development of a novel tool for the in situ isolation of bacteria migrating along fungal mycelium. FEMS Microbiol Ecol. 2015;91:fiv116. [DOI] [PubMed] [Google Scholar]
- Simon A, Herve V, Al-Dourobi Aet al. An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol Ecol. 2017;93:fiw217. [DOI] [PubMed] [Google Scholar]
- Slade EM, Roslin T, Santalahti Met al. Disentangling the ‘brown world’ faecal–detritus interaction web: dung beetle effects on soil microbial properties. Oikos. 2016;125:629–35. [Google Scholar]
- Soufan R, Delaunay Y, Gonod LVet al. Pore-scale monitoring of the effect of microarchitecture on fungal growth in a two-dimensional soil-like micromodel. Front Environ Sci. 2018;6. 10.3389/fenvs.2018.00068. [DOI] [Google Scholar]
- Stajich JE, Wilke SK, Ahren Det al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci U S A. 2010;107:11889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockli M, Lin CW, Sieber Ret al. Coprinopsis cinerea intracellular lactonases hydrolyze quorum sensing molecules of Gram-negative bacteria. Fungal Genet Biol. 2017;102:49–62. [DOI] [PubMed] [Google Scholar]
- Stockli M, Morinaka BI, Lackner Get al. Bacteria-induced production of the antibacterial sesquiterpene lagopodin B in Coprinopsis cinerea. Mol Microbiol. 2019;112:605–19. [DOI] [PubMed] [Google Scholar]
- Swamy S, Uno I, Ishikawa T. Morphogenetic effects of mutations at the A and B incompatibility factors in Coprinus cinereus. Microbiology. 1984;130:3219–24. [Google Scholar]
- Tayyrov A, Stanley CE, Azevedo Set al. Combining microfluidics and RNA-sequencing to assess the inducible defensome of a mushroom against nematodes. BMC Genomics. 2019;20:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R, Or D. Bacterial flagellar motility on hydrated rough surfaces controlled by aqueous film thickness and connectedness. Sci Rep. 2016;6:19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R, Or D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol Rev. 2017;41:599–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H, Tanabe AS, Yamamoto Set al. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One. 2012;7:e40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick LY, Remer R, Wurz Bet al. Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ Sci Technol. 2007;41:500–5. [DOI] [PubMed] [Google Scholar]
- Wolf AB, Vos M, de Boer Wet al. Impact of matric potential and pore size distribution on growth dynamics of filamentous and non-filamentous soil bacteria. PLoS One. 2013;8:e83661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrich A, Stryhanyuk H, Musat Net al. Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments. Nat Commun. 2017;8:15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlin T, Junier T, Roussel-Delif Let al. Stage 0 sporulation gene A as a molecular marker to study diversity of endospore-forming Firmicutes. Environ Microbiol Rep. 2013;5:911–24. [DOI] [PubMed] [Google Scholar]
- Zhang W, Li XG, Sun Ket al. Mycelial network-mediated rhizobial dispersal enhances legume nodulation. ISME J. 2020;14:1015–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu S, Cai Let al. Improved treatment and utilization of rice straw by Coprinopsis cinerea. Appl Biochem Biotechnol. 2018a;184:616–29. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kastman EK, Guasto JSet al. Fungal networks shape dynamics of bacterial dispersal and community assembly in cheese rind microbiomes. Nat Commun. 2018b;9:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.