Abstract
Cellular metabolism regulates immune cell activation, differentiation and effector functions but current metabolic approaches lack single-cell resolution and simultaneous characterization of cellular phenotype. Here, we developed an approach to characterize the metabolic regulome of single cells together with their phenotypic identity. The method, single-cell metabolic regulome profiling (scMEP), quantifies proteins that regulate metabolic pathway activity using a high-dimensional antibody-based approach. We employed mass cytometry (CyTOF) to benchmark scMEP against bulk metabolic assays by reconstructing the metabolic remodeling of in vitro-activated naïve and memory CD8+ T cells. We applied the approach to clinical samples and identified tissue-restricted, metabolically repressed cytotoxic T cells in human colorectal carcinoma. Combining our method with imaging mass spectrometry (MIBI-TOF), we uncovered the spatial organization of metabolic programs, which indicated exclusion of metabolically repressed immune cells from the tumor-immune boundary. Overall, our approach enables robust approximation of metabolic and functional states in individual cells.
Immune cells dynamically execute highly context-dependent functions, including migration into affected tissues, exponential expansion and secretion of effector molecules. All of these diverse capacities are enabled and coordinated by dynamic changes in cellular metabolism1–3. Pharmacological targeting of selected metabolic pathways can thus be used to influence specific aspects of immune cell behavior, e.g. direct the balance between effector and regulatory functionality4,5. Such therapeutic modulation has been shown to improve antitumor responses6–8, ameliorate autoimmune diseases9,10 and is a promising option for many other diseases11.
So far, approximation of the cellular metabolic state has been mostly based on quantification of metabolites and intermediates of selected metabolic pathways. Typically in bulk assays, mass spectrometry12 is used to quantify metabolite abundances and to trace isotopically enriched metabolites through metabolic pathways13. Alternatively, an approach termed extracellular flux analysis measures oxygen consumption and acidification of the extracellular milieu as proxies for OXPHOS and glycolytic activity, respectively. Together, these technologies have yielded invaluable insight into cellular metabolism and they continue to provide the basis for many studies in the field of immunometabolism.
Still, significant challenges and open questions related to metabolic heterogeneity and its relationship with cell identity remain. Firstly, while several metabolic features have been shown to direct T cell differentiation14, a more comprehensive understanding of the coordination within and between metabolic pathways as well as the interplay with other cellular processes would allow to better direct T cell differentiation for various therapeutic uses. Furthermore, given the recently highlighted metabolic differences between physiologically activated cells and in vitro models15, there is a need to analyze metabolic states directly ex vivo. Especially analysis of limited samples from human clinical material, which has been challenging using traditional technologies, could determine tissue-specific metabolic states as well as their potential modulation in human diseases, particularly cancer16,17. Moreover, multimodal analysis of metabolic immune cell states within their physiological microenvironment18 or in metabolically challenging contexts could reveal novel therapeutic targets and guide clinical decisions19. An ideal technological solution to approach these questions would bridge the gap between highly multiplexed, single-cell phenotyping platforms and the bulk determination of metabolic state, thus enabling the study of cellular metabolism directly from ex vivo human clinical samples with sparse material while determining important metabolic and functional relationships.
To address this need, we have developed an approach, termed single-cell metabolic regulome profiling (scMEP), that enables quantification of metabolic features of individual cells by capturing the composition of the metabolic regulome using antibody-based proteomic platforms. We assessed over 110 antibodies against metabolite transporters, metabolic enzymes, regulatory modifications (e.g. protein phosphorylation), signaling molecules and transcription factors across eight metabolic axes and on a variety of sample formats and tissue types. Utilizing these antibodies in multiplexed mass cytometry20 assays demonstrated that heterogeneous populations such as human peripheral blood can be metabolically analyzed in a highly robust manner and that cell identity is reflected in lineage-specific metabolic regulome profiles. Furthermore, we benchmarked scMEP against conventional extracellular flux analysis, demonstrating close agreement of metabolic regulome expression with glycolytic and respiratory activity. We investigated the tissue-specificity of metabolic characteristics of human cytotoxic T cell subsets isolated from clinical samples, including colorectal carcinoma and healthy adjacent colon. This analysis revealed the metabolic heterogeneity of physiologically activated CD8+ T cell subsets, including subsets expressing the T cell exhaustion-associated molecules CD39 and PD1. Finally, we adopted scMEP to multiplexed imaging of human tissue samples by MIBI-TOF21,22 which revealed the spatial organization of metabolic T cell states as well as exclusion of clinically relevant CD8+ T cell subsets from the tumor-immune boundary.
Overall, our scMEP approach enables the study of cellular metabolic states in combination with phenotypic identity. We expect this to deepen our understanding of cellular metabolism in homeostatic and dysfunctional settings, across heterogeneous cell populations and in situ, together providing a new lens through which to understand and impact human disease.
Results
Targeted quantification of the metabolic regulome discriminates human immune populations
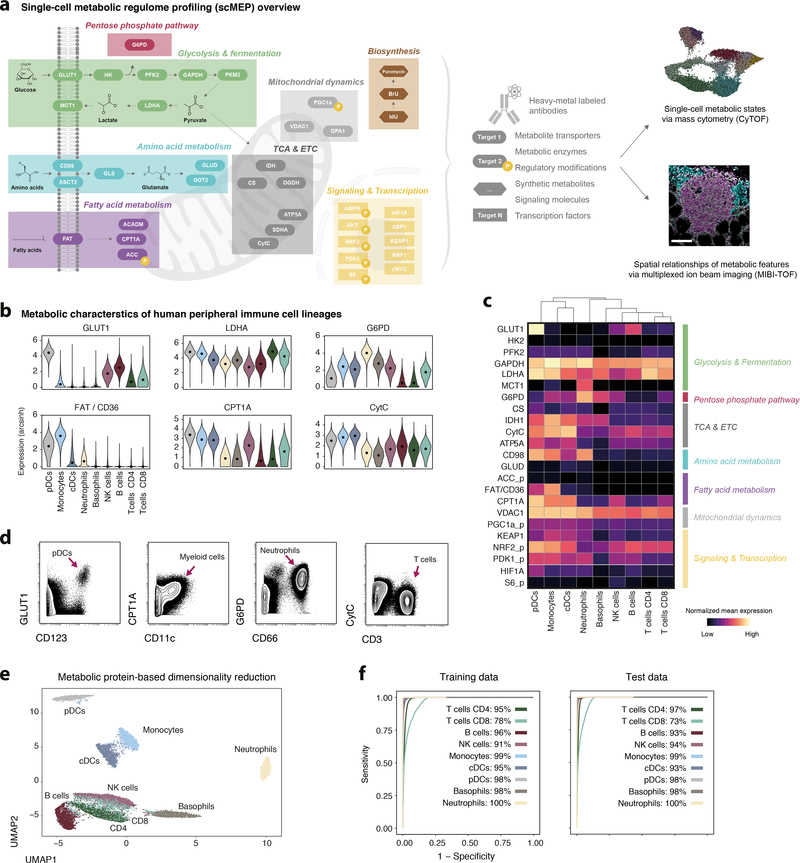
Much like the epigenetic regulome governs transcription by controlling gene accessibility, the balance of cellular metabolites, and thus the metabolic state, is influenced by the molecular machinery that regulates these pathways. As such, we sought to quantify the abundance of metabolite transporters, rate-limiting metabolic enzymes and their regulatory modifications (e.g. phosphorylation), modifiers of mitochondrial dynamics, as well as transcription factors and signaling molecules that drive specific metabolic programs, here collectively referred to as the cellular metabolic regulome. To accomplish this, we assessed the performance of over 110 commercially available antibodies by mass cytometry, immunohistochemistry (IHC) and MIBI-TOF (Supplementary Figure 1a,b). Following this screening, we selected a subset of metabolic antibodies (N = 41) that were used in varying combinations and in conjunction with phenotypic antibodies in this study (Fig. 1a).
Fig. 1: Single-cell metabolic regulomes organize the human immune system.
a, Conceptual overview of the scMEP approach. Important regulators of metabolic activity were identified and respective antibodies were conjugated to heavy-metal isotopes for their use in mass cytometry (CyTOF) and MIBI-TOF. For a full account of all probes tested in this study see Supplementary Table 1. Scale bar = 100 μm. b, Whole blood of healthy individuals (N = 5, for donor characteristics see Supplementary Table 2) was fixed and stained with an antibody panel of 23 metabolic and 22 immunological antibodies. Cell populations were identified through FlowSOM clustering (Supplementary Figure 2a,b) and annotated into the major immune cell lineages. Shown are examples of (asinh transformed) expression values across identified peripheral immune cell lineages. Black dots represent population medians. c, Normalized (99.9th percentile) mean expression of all assessed metabolic regulators across immune cell lineages. d, Examples of metabolic regulator expression across immune cell lineages. Shown are live, single, CD45+ cells of one representative individual. e, Cells from all five donors were subsampled for equal representation of all immune cell lineages and all donors. Only metabolic regulators (23 features) were used as input data to the UMAP-based dimensionality reduction. Cells are colored by their lineage identity determined as in b. f, L1 regularized linear regression (using only metabolic features) was trained on a subset of donors (N = 3) and tested on a separate set of donors (N = 2). Stated numbers report balanced accuracy for the indicated population.
Given the importance of metabolic networks to immune cell function, we first asked whether functional specialization within the human immune system might be reflected in lineage-specific metabolic regulomes. We obtained whole blood and identified the major immune cell lineages through phenotypic markers (Supplementary Figure 2a,b), thus enabling in silico subset selection and comparison of metabolic regulomes of cell lineages without the need for prior isolation or enrichment (Fig. 1b). We observed lineage-specific metabolic states that were in agreement with previously established functional roles (Fig. 1c,d). For example, plasmacytoid dendritic cells (pDCs) expressed high levels of several regulators of glycolysis (e.g. glucose transporter GLUT1/SLC2A1), and fatty acid metabolism (e.g. fatty acid translocase FAT/CD36) which both have been shown to impact pDC functionality, including hallmark interferon production23. In line with their metabolic quiescence in the absence of antigen, lymphocytes (T and B cells) expressed lower levels of many metabolic proteins and intermediate levels of proteins within the tricarboxylic acid cycle (TCA) and the electron transfer chain (ETC), crucial for basal respiration. In general, lineage-specific expression of metabolic enzymes was found to be reproducible across different donors as well as independent experiments and was stable during standard blood collection and storage (Supplementary Figure 2c–h).
Given these robust and lineage-specific profiles, we hypothesized that scMEP could be used to infer cell identity independently of cell lineage markers. We found that metabolic features were largely able to separate immune cell lineages, with expected overlap between CD4+ and CD8+ T cells and to a lesser extent NK cells (Fig. 1e). In addition, metabolic features could predict immune cell identity for a large fraction of cells (94% average in training and test data, Fig. 1f). Together, this demonstrates the suitability and robustness of scMEP to study metabolic regulation and its relationship to single-cell specialization and it revealed remarkable cell type-specific metabolic diversification within the human immune system.
Benchmarking single-cell metabolic regulomes with metabolic flux
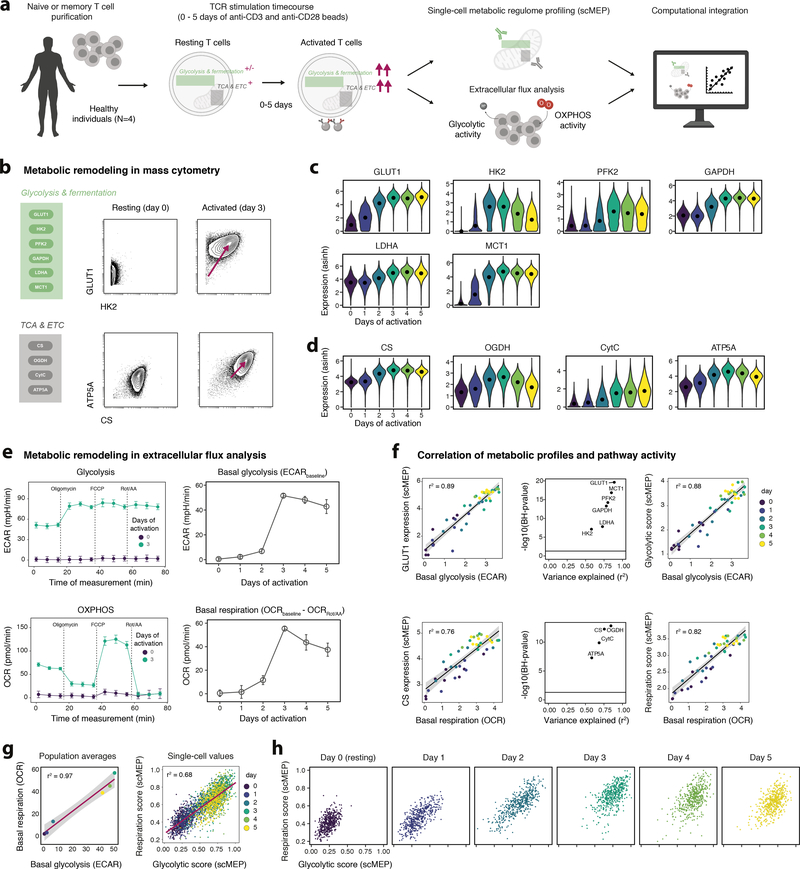
Assessing metabolic enzyme abundances has provided insight into the regulation of cellular metabolism in many scenarios24–26 and multiple studies have identified enzymes that determine metabolic pathway activity27. To establish the relationship between antibody-based, single-cell metabolic features and pathway activity, we activated human naïve and memory T cells (including CD4+ and CD8+) and analyzed them by mass cytometry and extracellular flux analysis (Fig. 2a). Focusing on glycolytic and TCA/ETC components, mass cytometry-based scMEP analysis recapitulated several established hallmarks of metabolic remodeling upon TCR engagement28–30, including increased surface expression of GLUT1, as well as elevated intracellular levels of several key glycolytic enzymes including hexokinase 2 (HK2), phosphofructokinase 2 (PFK2) and critical downstream enzymes such as lactate dehydrogenase A (LDHA) and the monocarboxylate transporter 1 (MCT1, Fig. 2b,c). Likewise, TCA/ETC proteins such as citrate synthase (CS), oxoglutarate dehydrogenase (OGDH), the cytochrome complex (CytC) and ATP synthase (ATP5A) were upregulated upon TCR engagement (Fig. 2b,d). Of note, upregulation of metabolic enzymes exceeded activation-induced increases in cell size and could, to a large extent, not be explained by mitochondrial content (Supplementary Figure 3a–e).
Fig. 2: Single-cell metabolic regulome profiles of T cell activation dynamics.
a, Experimental setup to benchmark scMEP mass cytometry analysis with bulk metabolic analysis by extracellular flux analysis (Seahorse). b, Examples of mass cytometry-quantified expression levels of glycolytic (top) and TCA/ETC (bottom) enzymes following no (resting, left) and 3 days (right) of activation of naïve T cells. Examples show data of one representative experiment (out of N = 4 independent experiments). c, Expression levels of important determinants of glycolysis on naïve CD8+ T cells. Black dots indicate population medians. d, Expression levels of TCA/ETC components on naïve CD8+ T cells as in c. e, Extracellular flux analysis of bulk cell populations from the same donor as in a-c. Extracellular acidification rate (ECAR; top) and oxygen consumption rate (OCR; bottom) for each measurement following injections of mitochondrial modifiers. FCCP = fluoro-carbonyl cynade phenylhydrazon, Rot = Rotenone, AA = antimycin A. Shown is data from one individual (out of N = 4 independent experiments). Circles and error bars represent mean±s.d. for three technical replicates (wells). f, Correlation between glycolytic and oxidative enzymes (left panel) and basal glycolysis (top row) or basal respiration (bottom row). Mass cytometry and extracellular flux analysis values were asinh transformed. Circles represent mean population values for each donor and are based on technical replicates (single cells in mass cytometry and three replicate wells in flux analysis). Black lines and r2 values represent results of a linear regression model, with black shading representing the 95% confidence interval (CI). Log10 of (Benjamini-Hochberg; BH) false discovery rate (FDR)-adjusted P-values and r2 values from linear regression models (middle panel). Black line indicates a BH-corrected P-value of 0.05. Protein-based scMEP scores (right) represent the mean expression of all metabolic enzymes within a pathway. Each circle represents the mean scMEP score of a T cell population (naïve or memory). g, Linear correlation of mean (based on three technical replicates) flux analysis values (left) and single-cell scMEP scores (right) of naïve CD8+ T cell populations, calculated as in f. Shown is data from one individual (out of N = 4 independent experiments). Red lines and r2 values represent results of a linear regression model, with black shading representing the 95% CI. h, scMEP scores as in g, visualized for each day.
Performing extracellular flux analysis on the same samples, we found that as expected, levels of basal glycolysis and OXPHOS increased upon TCR engagement (Fig. 2e and Supplementary Figure 4a). Comparison to scMEP showed that the analyzed glycolytic enzymes were robustly correlated with glycolytic flux across several donors and independent experiments (linear regression mean r2 = 0.77, Fig. 2f). A similarly strong correlation was observed between expression of TCA/ETC components and OXPHOS activity (linear regression mean r2 = 0.72, Fig. 2f). Given this correlation, we hypothesized that it would be possible to utilize scMEP-based high-dimensional co-expression patterns to derive in silico scores representing glycolytic and OXPHOS activity. Of note, these scMEP-based glycolytic and OXHPOS scores strongly and robustly predicted respective metabolic activity across multiple donors, activation time points, and independent experiments (Supplementary Figure 4b–f). In addition to T cells, we validated mass cytometry-based scMEP values against extracellular flux analysis in differentially activated human monocyte-derived macrophages which again demonstrated agreement between these assays and highlights that scMEP analysis faithfully recapitulates metabolic pathway activity across various stimulation conditions and multiple human immune cell types (Supplementary Figure 5a–h).
Extracellular flux analysis suggested that upon TCR engagement, naïve T cell populations simultaneously increase their glycolytic and respiratory activity (Fig. 2g). However, given the nature of bulk measurements, it remained unclear whether there is metabolic heterogeneity and specialization of a subset of cells towards a glycolytic or oxidative phenotype. scMEP scores calculated for each cell independently revealed simultaneous upregulation of the glycolytic and oxidative machinery on the single-cell level (Fig. 2g), together indicating that these cells simultaneously engage multiple metabolic pathways to support their wide-ranging bioenergetic demands31. Furthermore, our single-cell analysis revealed previously obscured metabolic heterogeneity within each timepoint, with T cells activated for two days spanning almost the entire range of possible glycolytic and respiratory scMEP scores (Fig. 2h). Cells activated for three days displayed a more homogeneous upregulation of glycolytic and TCA/ETC enzymes, suggesting eventual convergence of metabolic remodeling and potentially indicating the presence of a series of metabolic and cellular checkpoints32. Together, this data validates the close relation of scMEP-based quantification of the cellular metabolic regulome with pathway activity assessed through a well-established orthogonal method and demonstrates the ability of scMEP to enable the discovery of metabolic heterogeneity at the single-cell level.
Integrative modeling of T cell activation identifies checkpoints of metabolic switching
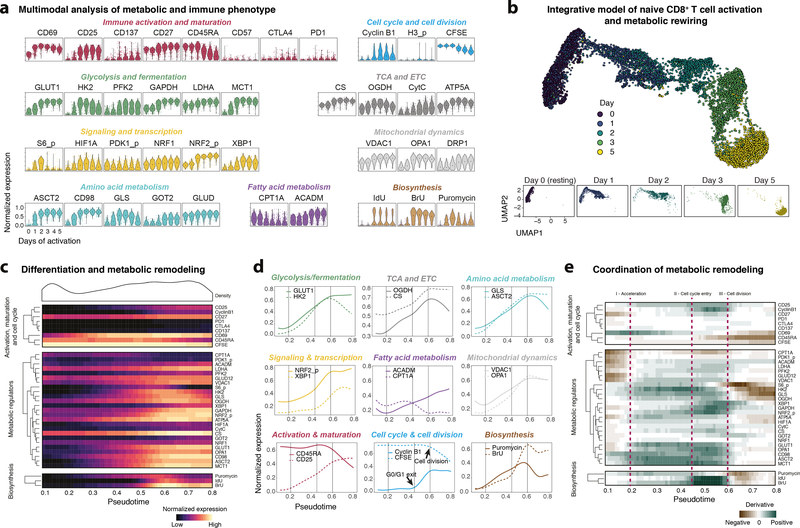
In order to study the metabolic remodeling of human T cells and its relation to cell activation, differentiation, and proliferation, we expanded our analysis to incorporate a broader set of metabolic pathways and other cellular features. To do so, we simultaneously analyzed 48 non-redundant, biological (non-technical) parameters across millions of single cells (Fig. 3a). Indicating the initiation of metabolic remodeling in activated naïve T cells, we observed early phosphorylation of signaling molecules (e.g. ribosomal protein S6) and induction of transcription factors (e.g. Hypoxia-inducible factor 1-alpha HIF1A) which was closely followed by upregulation of metabolic enzymes (GLUT1, HK2 and LDHA, Fig. 3a), together reminiscent of previously described temporal optimization patterns in which expression hierarchy matches enzyme order in metabolic pathways33.
Fig. 3: Integrative modeling of metabolic rewiring reveals determinants of human T cell activation.
a, Normalized (99.9th percentile) expression of metabolic and phenotypic proteins by naïve CD8+ T cells across different days of activation. Shown are cells from one representative donor (N = 3 independent donors). Black dots indicate population medians. b, Cells were subsampled for equal representation of the indicated days of activation. Metabolic and other features (with the exception of IdU and H3 phosphorylation) were used as input to UMAP dimensionality reduction and visualization. Cells are colored by their day of activation (top) and shown separately for each day of activation (bottom). c, Cells as in b were used as input to SCORPIUS trajectory inference using the same features as in b. Data was grouped into 100 bins based on pseudotime. Heatmap depicts mean (scaled) expression levels of the indicated feature in the according pseudotime bin. Density (top) shows cell distribution along the pseudotime axis. d, Examples of continuous (smoothed) protein expression along pseudotime, calculated as in c and grouped by metabolic pathway. Vertical lines indicate important inflection points in the trajectory. Inflection points were chosen based on coordinated changes in the slope (see e) across the indicated metabolic regulators and divide T cell metabolic remodeling into distinct stages. e, Slope (first derivative) of protein expression across pseudotime as in c.
We next integrated the high-dimensional information of all features to visualize the metabolic and phenotypic progression of naïve CD8+ T cells upon TCR stimulation. This indicated a continuous progression of immune activation and metabolic rewiring across multiple days with recently activated cells (day 1 and 2) spanning larger areas of the phenotypic and metabolic space compared to cells from later days (day 3 and 5, Fig. 3b and Supplementary Figure 6a). Consistent with previous reports34, these observations indicate that, like cell phenotype and cellular transcriptional profile, naïve CD8+ T cell metabolism is most plastic in the earliest phases of antigen experience.
To further explore the temporal coordination of metabolic remodeling in conjunction with cell phenotype, we inferred a pseudotime axis representing progression of cellular differentiation and metabolic remodeling (Fig. 3c). Pseudotime correlated well with time of activation and was robust across different trajectory algorithms as well as independent donors (Supplementary Figure 6b–e). We found metabolic protein expression within pathways to be highly coordinated, especially during early remodeling (Supplementary Figure 6f,g). In addition to intra-pathway coordination, we found early synchronization of metabolic protein expression trajectories across various metabolic and other cellular pathways (Fig. 3d,e). Their divergence at later stages potentially indicates redirection of metabolic intermediates into different metabolic pathways. For example, cells maintained high expression of the glutamine transporter ASCT2 but downregulated the glutamine-to-glutamate converting enzyme GLS, possibly increasing glutamine availability for nucleotide biosynthesis35.
Utilizing the change of metabolic feature expression over pseudotime (first derivative), we defined three inflection points during metabolic remodeling of naïve human CD8+ T cells (Fig. 3e). The first inflection point (pseudotime 0.2) was marked by a coordinated and accelerated upregulation of metabolic proteins in various pathways (e.g. concerted induction of GLUT1, ASCT2, OGDH, VDAC1; Fig. 3e) leading up to the second inflection point (pseudotime 0.45), which was characterized by initiation of RNA synthesis (BrU incorporation) and activation of cellular stress responses (NRF2_p and XBP1). Further, we observed reduced expression of carnitine palmitoyltransferase (CPT1A), a rate-limiting enzyme of fatty acid oxidation. Interestingly, this second inflection point coincides with cells exiting the G0/G1 cell cycle phase (Fig. 3d,e). The third metabolic inflection point (pseudotime 0.6) was defined by stabilized or decreasing expression levels of metabolic proteins (e.g. GLUT1 and ASCT2) as well as peak translational activity (puromycin incorporation). Of note, this inflection point coincided with the first cell division, determined through reduction in CFSE signal. Interestingly, VDAC1 (also porin, located in the outer mitochondrial membrane) peaked at this point, suggesting an increase in mitochondrial mass prior to the first cell division, followed by cell division-dependent dilution36. Further underlining this crucial interplay between cell cycle progression, cell division and metabolic activity37, activation of T cells in the presence of low-dose oligomycin revealed activation time-dependent influences on metabolic rewiring that led to suppressed T cell division (Supplementary Figure 7a–d).
Naïve and memory T cells are known to differentially engage metabolic pathways. Comparing their metabolic regulator expression using scMEP confirmed that memory CD8+ T cells displayed increased GLUT1, elevated TCA/ETC proteins (e.g. CS), higher fatty acid oxidation components (CPT1A), and greater mitochondrial mass (VDAC1, Supplementary Figure 8a–d), all of which are well established features of memory T cells. Taken together, scMEP facilitated in-depth and multimodal analysis of early T cell activation which revealed regulatory pathway coordination and identified distinct phases of T cell metabolic remodeling as well as their relationship to transitions between cellular states.
Tissue-specific metabolic regulome profiles define human T cell subsets
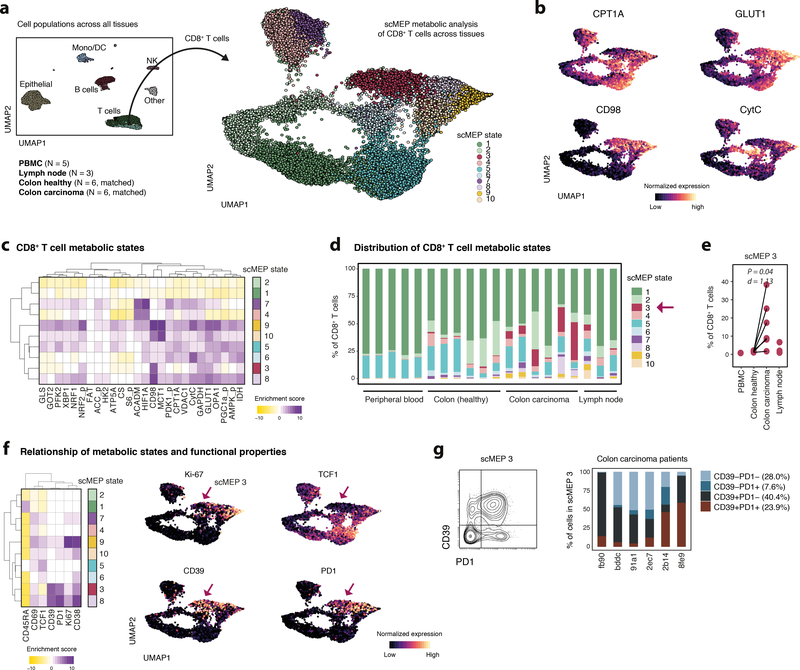
In addition to cell-intrinsic factors, metabolic states are influenced by the tissue in which they reside15, underlining the importance of determining metabolic states directly ex vivo and in clinical samples. To investigate potential tissue-dependent metabolic influences in the context of human cancer, we prepared single-cell suspensions from tissue resections of colorectal carcinoma, including tumor (N = 6) and healthy adjacent tissue from the same patients (N = 6, see Supplementary Table 2). In addition, we included unrelated healthy donor PBMCs (N = 5) and lymph node biopsies (N = 3). Samples were stained with 18 phenotypic and 27 metabolic antibodies (see Supplementary Table 1) and analyzed by mass cytometry. We identified all major cell lineages (Supplementary Figure 9a–f) but focused our downstream analysis on CD8+ T cells (for CD4+ T cells see Supplementary Figure 10a–f). Employing automated clustering via FlowSOM, we grouped CD8+ T cells into ten distinct metabolic states, based exclusively on their metabolic features (Fig. 4a–c). Identified metabolic CD8+ T cell scMEP states included subsets characterized by low metabolic protein expression (scMEP1&2) suggesting reduced metabolic activity, subsets with elevated expression of a broad range of regulators (including CD98, GLUT1, PFK2, MCT1, CytC; scMEP9&10), indicating increased metabolic demands, and subsets with characteristic expression patterns, e.g. increased ACADM/HIF1A (scMEP4&7) or CD98 (scMEP3&8) which indicate more specific, context-dependent metabolic adaptions.
Fig. 4: Cytotoxic T cell metabolic states reflect tissue of residence.
Healthy donor PBMC (N = 5), lymph node biopsies (N = 3) as well as single-cell suspensions from colorectal carcinoma (N = 6) and matched adjacent healthy sections (N = 6, see Supplementary Table 2) were barcoded, stained and acquired on a mass cytometer. a, Major cell lineages from all samples and tissues were identified through FlowSOM clustering. UMAP-dimensionality reduction was calculated using subsampled data from all lineages and all available features. Cells are colored by their FlowSOM-based lineage definition (left). Total CD8+ T cells from all samples were selected and metabolic regulators were used to define 10 scMEP states using FlowSOM. UMAP-dimensionality reduction was calculated using subsampled data and only metabolic features. Cells are colored by their scMEP state (right). b, UMAP visualization of CD8+ T cell scMEP states as in a, colored by normalized expression of the indicated metabolic proteins. c, Marker enrichment modeling (MEM) was used to visualize enrichment (purple) or depletion (yellow) of metabolic protein expression across CD8+ T cell scMEP states. d, Frequencies of scMEP states across individual samples. e, Statistical comparison of scMEP state frequencies (see also Supplementary Figure 9f). P-values were calculated using a two-sided, paired t-test between healthy and malignant colon sections from the same patient. Welch correction was applied to account for potentially differing variances. Effect size is represented as Cohen’s d. Estimate = −13.8, t-statistic = −2.76, CI −26.7 to −0.97, 5 degrees of freedom, BH FDR = 0.198. f, MEM of immunological markers (not used for metabolic clustering) across scMEP states (left). UMAP visualization as in a,b with cells colored by their normalized expression value of the indicated marker. g, Biaxial representation of cells from scMEP3 pooled from all colorectal carcinoma samples (left). Frequencies of cells within scMEP3, gated as PD1+ and CD39+ across all colorectal carcinoma samples (right).
Determining the tissue distribution of these phenotypes, we found that peripheral T cells primarily consisted of the metabolically low scMEP1 (mean 76.6% of peripheral CD8+ T cells) with smaller numbers of scMEP5 (19.7%) and negligible frequencies of all other metabolic states (all < 3%; Fig. 4d). Compared to peripheral blood, tissue infiltrating CD8+ T cells displayed greater metabolic heterogeneity with a more diverse range of phenotypes. Interestingly, we found a specific metabolic phenotype (scMEP3) to be enriched in cells isolated from colorectal carcinoma (mean 14.6% of total CD8+ T cells) versus cells from healthy adjacent sections (mean 0.8%, P = 0.0396, FDR = 0.198) and all other tissues (PBMCs 0.1%, lymph nodes 2.4%; Fig. 4e and Supplementary Figure 9f). This subset was characterized by enriched expression of CD98 and lower levels of several metabolic enzymes across various pathways (GLS, GOT2, PFK2, ATP5A, CS, S6_p, CPT1A, PGC1a_p).
Since CD8+ T cell scMEP states were defined using exclusively metabolic features, we next investigated their immunological characteristics (Fig. 4f). scMEP metabolic states associated clearly with distinct immunological phenotypes. For example, metabolically low states (scMEP1&5) displayed features of resting cells (CD69-, CD38- and Ki-67low) with naïve (CD45RA+ and TCF1high, scMEP1) or memory phenotypes (CD45RA- and TCF1low, scMEP5). scMEP9, which displayed high overall expression of metabolic proteins, showed signs of recent activation (CD69+, CD38+ and Ki-67high), again indicating the increased metabolic demands of cycling cells.
Of note, the tumor-associated metabolic T cell state (scMEP3) was significantly enriched in exhaustion-associated molecules programmed death 1 (PD1) and CD39 (both not used for the initial definition of scMEP states). Besides scMEP3, we also observed enrichment of CD39+ and/or PD1+ cells (termed CD39/PD1 cells) in scMEP8. With the exception of increased CD98 and minor enrichments in GLUT1 and GAPDH, cells from scMEP3 but not scMEP8 showed decreased expression of metabolic regulators (i.e. reduced ATP5A, CS, CytC and PGC1a_p) and were thus termed metalow CD39/PD1 cells. Interestingly, lower mitochondrial capacity as indicated here is a characteristic of T cell exhaustion6,38. Furthermore, metalow CD39/PD1 cells (scMEP3) were TCF1low which has been proposed to specifically identify terminally exhausted T cells39. In comparison, cells from scMEP8 retained TCF1 expression and were characterized by higher levels of many metabolic regulators (thus termed metahigh CD39/PD1 cells), which might be indicative of different functional capacities of these subsets. Lastly, while the metalow subset (scMEP3) was strongly enriched in CD39 and PD1, it also included a fraction of cells negative for both markers (mean 28%, Fig. 4g), suggesting that integration of the metabolic state could be employed as an additional dimension to functionally define T cell capacities. In summary, these analyses demonstrate the unique capability of scMEP to identify metabolic states of low abundance cell populations directly from sparse clinical samples, revealing important relationships between cellular phenotype and metabolism in human disease.
Cellular metabolism is related to spatial organization in human tissue compartments
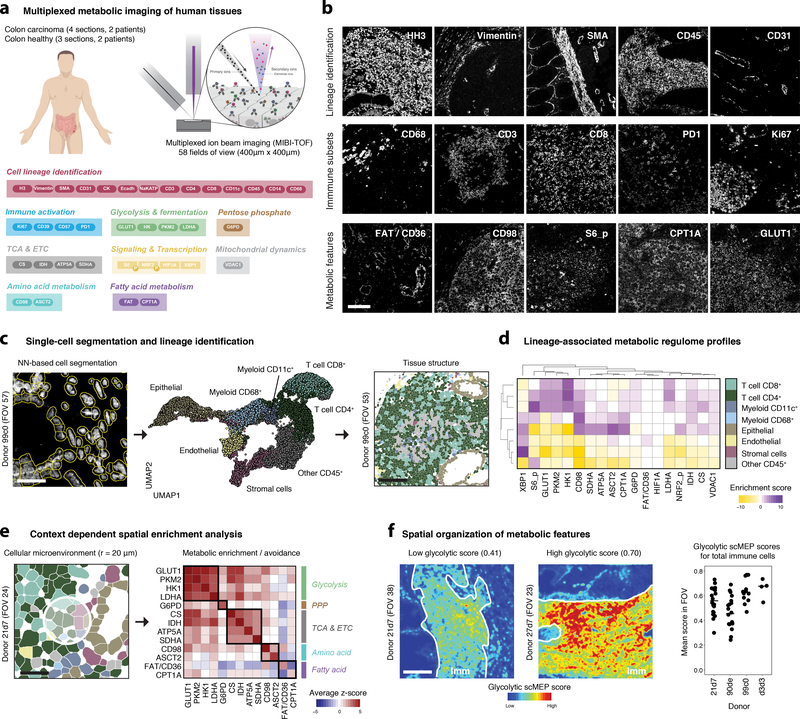
In addition to lineage intrinsic factors, activation status and tissue of residence, a cell’s metabolism is influenced by local nutrient availability and other microenvironmental conditions16,17. To analyze metabolic regulation directly in human tissues, we adapted the scMEP approach to a high-dimensional imaging platform, specifically MIBI-TOF. Having assessed the performance of all metabolic antibodies in traditional IHC (Supplementary Figure 11) and MIBI-TOF (Fig. 5a,b and Supplementary Figure 12a), we obtained human tissue samples (standard, archival formalin-fixed, paraffin-embedded (FFPE) blocks), including sections from colorectal carcinoma patients (N = 4) and non-malignant control sections from independent donors (N = 3, different to patients analyzed in Fig. 4, see Supplementary Table 2). Sections were stained with a combination of lineage and metabolic antibodies (Fig. 5a and Supplementary Table 1). In total, we acquired 58 fields of view (FOV; 400 μm by 400 μm, resolution ~400 nm), each comprised of 36 antibody-dimensions, thus allowing us to determine cell lineage, subset, and activation status, together with metabolic characteristics (Fig. 5b).
Fig. 5: Imaging-based scMEP analysis reveals spatial influences on the organization of metabolic features.
a, FFPE colon-sections from colorectal carcinoma patients (N = 2) and healthy controls (N = 2) were stained with a panel of 36 antibodies. A total of 58 fields of view (FOV), each 400 μm by 400 μm, were acquired by MIBI-TOF. b, Exemplary grayscale images of features used for lineage identification (top), immune activation and subsets (middle) and their metabolic characterization (bottom). See also Supplementary Figure 12. Scale bar = 100 μm. c, A pixel-based classifier was applied to automatically identify single-cells within these images (left, scale bar = 25 μm). FlowSOM was used to identify the main cell lineages based on their lineage marker expression values. Single-cell data was projected onto two-dimensions using UMAP and colored by their cell lineage identity (middle). Clustered single-cell data can be mapped back onto the original segmented images to investigate spatial influences (right, scale bar = 100 μm). d, Metabolic regulome profiles of cell lineages as identified in c are represented as MEM scores. e, Cellular microenvironments were defined as cells present within a 20 μm radius (based on cell centroids) of any given index cell. Colors indicate cell lineage as in d (left, scale bar = 25 μm). Within all such groups, spatial enrichments were calculated by comparing the distributions of metabolic protein expression with a random subsampling of the same cell lineage composition. Enrichments (red) and avoidances (blue) are visualized as average z-scores across all FOVs. Black outlines indicate proteins within the same metabolic pathway (right). f, Spatial scMEP scores for a given metabolic pathway were calculated by averaging (and blurring) pixel-based expression values of all metabolic markers within a pathway. Areas of immune cell infiltration were outlined manually based on CD45 staining (left and middle, scale bar = 100 μm). Circles represent mean glycolytic scMEP scores for all CD45+ cells within a FOV (right). Black lines indicate donor means.
From these images, we identified single cells and clustered them into the main cell lineages (Fig. 5c and Supplementary Figure 12b–d). In agreement with our mass cytometry analysis (see Fig. 4), distinct cell lineages displayed lineage-specific metabolic protein expression patterns with potentially activation-induced glycolytic expression in T cells and high metabolic protein levels in epithelial cells from colorectal carcinoma tissue (Fig. 5d and Supplementary Figure 12e). To interrogate the spatial organization of metabolic features, we used context-dependent spatial enrichment (CDSE) analysis40 which indicated the presence of spatially enriched metabolic features (Fig. 5e). For example, GLUT1high cells were highly enriched around other GLUT1high cells, independent of cell lineage. This analysis also revealed spatial enrichments of enzymes within the same metabolic pathway (e.g. GLUT1high cells enriched around PKM2high cells), suggesting the existence of environmental niches that enable or drive certain cellular metabolic behavior. We found such spatial enrichment for glycolysis, respiratory and amino acid pathways in contrast to fatty acid metabolism where FAT/CD36 but not CPT1A was enriched on endothelial cells, potentially indicating their role in tissue uptake of fatty acids but not their oxidation41.
In analogy to single-cell scMEP scores (see Fig. 2), we calculated spatial, pixel-based scMEP scores to visualize the spatial distribution of metabolic programs directly in images (Fig. 5f and Supplementary Figure 12f). Imaging several regions within a tissue section, we found that scMEP scores of total immune cells varied across but also within donors, together reinforcing the concept of local, microenvironment-driven influences on metabolic polarization that can be revealed by image-based scMEP analysis.
The tumor-immune boundary represents a unique metabolic niche
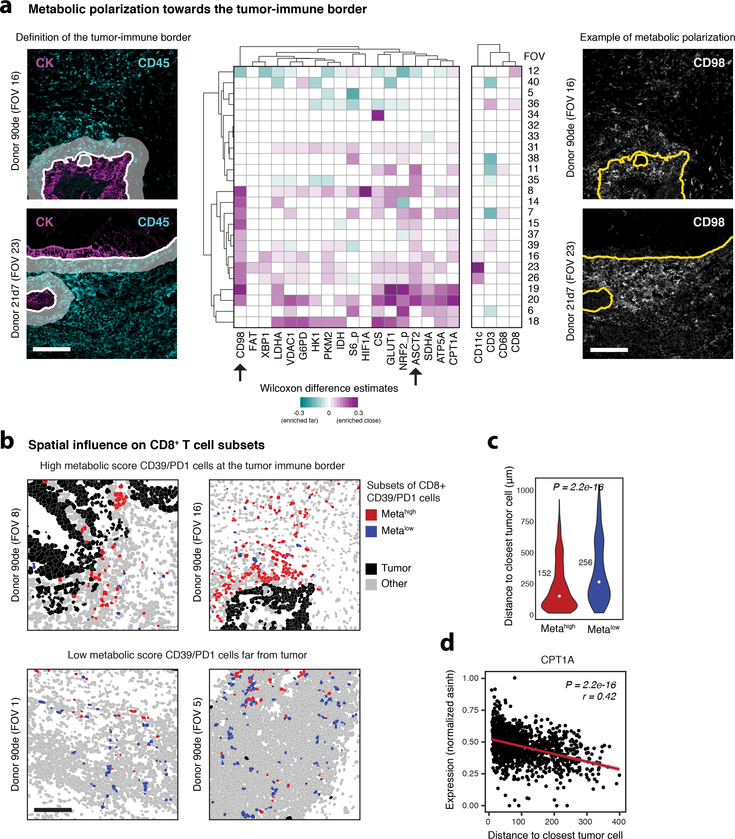
Metabolic competition within the tumor microenvironment is known to influence immune cell metabolism42. To investigate whether malignant epithelial cells directly modulate neighboring immune cells, we computationally identified a tumor-immune border40 and compared immune cells close (within 20 μm) to cells located further away from this boundary (Fig. 6a). A large fraction of FOVs (17 out of 24) displayed metabolic polarization of immune cells towards the tumor, which was dominated by increased expression of CD98 and ASCT2 and which could not be not explained by variations in immune cell lineage (Fig. 6a). CD98 and ASCT2 have shown prognostic value in human cancer43,44 and it will be of great interest to see whether integrating their expression with tissue features or multidimensional co-expression of other metabolic proteins will improve diagnostic power.
Fig. 6: Metabolic polarization at the tumor-immune boundary in human colorectal carcinoma.
a, Immune cells within a 20 μm radius of malignant epithelial cells were classified as located within the tumor-immune border (left, scale bar = 100 μm). Shown is data from all 24 FOVs that contain a tumor-immune boundary. Two-sided Wilcoxon-rank sum test (FDR-corrected using the BH approach to adjust for multiple hypothesis testing) were used to compare cells close to the border with cells further from the border in each FOV that contained cells of both categories. Heatmap shows Wilcoxon rank sum test-based estimates (representing the median of the difference between samples of the two groups) of enrichment for enriched (magenta) and decreased (cyan) expression on immune cells within the border. Non-significant (BH-adjusted P-value > 0.05) estimates were colored white (middle). Exemplary grayscale images of two FOVs showing polarization of CD98 towards the tumor immune border, indicated in yellow. Scale bar = 100 μm (right). b, CD8+ T cells expressing high levels of CD39 and/or PD1 were clustered into two subsets based on their metabolic features (see Supplementary Figure 13). The two subsets (metahigh and metalow) were visualized in the original images (left top and bottom, scale bar = 100 μm). c, Two-sided Wilcoxon rank sum test of distance to closest malignant epithelial cell for CD39/PD1 cells stratified by metabolic phenotype (P = 2.2e-16). Numbers indicate median distance. d, Linear regression between normalized asinh expression of metabolic enzymes (e.g. CPT1A) and distance to closest tumor cell (bottom right). Red line and values indicate linear regression model (P = 2.2e-16).
Again focusing on CD8+ T cells, in=depth analysis of metabolic features revealed a variety of diverse subsets (Supplementary Figure 13a,b), akin to our suspension-based analysis. As before, we identified two metabolically divergent subsets of CD39/PD1 cells (metahigh and metalow, Supplementary Figure 13c,d). Integrating spatial information unique to imaging data revealed that metalow CD39/PD1 cells were located further away from the tumor-border than their metahigh counterparts (Fig. 6b,c) and that the expression of metabolic feature on these CD39/PD1 cells was correlated to distance to closest tumor cell (most pronounced for CPT1A, Fig. 6d). Together, this analysis suggests that only CD39/PD1 cells that were distal and unengaged with the tumor appeared metabolically suppressed as opposed to more metabolically active cells at the tumor-immune interface. These observations potentially reconcile the ambivalent nature of CD39 and PD1 expression, which is associated with exhaustion and dysfunction but at the same time T cell activation45–47.
In summary, these spatial analyses revealed specific exclusion of metabolic immune cell subsets from the tumor-immune boundary, demonstrating the influence of tissue architecture on metabolic regulation that goes beyond what can be observed using conventional deep phenotyping of cell identity alone. Incorporating this new lens of single-cell metabolism into translational research promises better control of cellular alterations and dysfunction in human disease.
Discussion
Biological tissues possess great heterogeneity, thus necessitating the use of single-cell platforms for their in-depth study48. Technological advances are pushing the frontier of single-cell analyses on multiple levels, including the genome49, transcriptome50 as well as aspects of the epigenome51 and proteome20. Here, we presented scMEP, an approach that utilizes antibody-based assays to analyze metabolic regulation in combination with cellular identity on the single-cell level. Focusing on the metabolic regulome allowed us to define metabolic states directly from limited, ex vivo, human material. For example, we performed metabolic analyses of fewer than 1000 (median 842) human tumor-infiltrating CD8+ T cells per sample (Fig. 4). Recent studies have highlighted significant metabolic differences between such physiologically activated cells and in vitro models15. Our scMEP approach is applicable to fixed cells and FFPE tissues, offering the opportunity to analyze metabolic states from existing clinical cohorts and thus enabling the identification of features associated with clinical outcome or therapeutic success.
Instead of individual metabolites, scMEP quantifies the abundance of metabolic regulators. Many of these factors are known to directly drive or correlate with metabolic flux25,27,28 and we demonstrated their correlation with metabolic pathway activity (Fig. 2). Nevertheless, , as indicated in our analysis of oligomycin inhibition, scenarios might exist in which external factors (e.g. synthetic inhibitors) drive a divergence between regulator expression and momentary pathway activity. Such inherently interesting exceptions would however be easily identified by subsequent validation and orthogonal technologies.
We validated a large number of antibodies (Supplementary Table 1) to provide a resource for the implementation and potential adjustments of scMEP. Of interest for pre-clinical researchers, a large fraction (~70%) of the tested metabolic antibodies are reactive with murine epitopes. All antibodies were validated post heavy-metal conjugation which potentially affects antibody-binding affinity. Presented antibody performances are therefore assay-specific and might reflect the impact of the employed conjugation chemistry.
Mass cytometry and MIBI-TOF both allow simultaneous quantification of >40 features, permitting analysis of multiple metabolic pathways. However, given its antibody-based nature, the scMEP approach can be transferred to other high-dimensional probe-based platforms, e.g. flow cytometry. Analysis of metabolic aspects by single-cell RNA sequencing using with novel analytical approaches could offer additional insights52. Especially once challenges related to RNA stability following fixation and permeabilization are resolved, antibody-sequencing hybrid technologies53,54 would present an exciting platform to implement scMEP, combining the unbiased nature of RNA sequencing with the large dynamic range of protein expression and the ability to assess post-transcriptional and post-translational regulation offered by antibody-based technologies55. In addition, metabolic profiling could be further extended by combining single-cell analysis of metabolic regulation with determination of epigenetic features51.
We demonstrated that scMEP can drive the discovery of biologically and clinically relevant findings. Reconstruction of metabolic remodeling of T cells (Fig. 3) could serve as a framework to design metabolic interventions in a phase-specific manner to direct in vitro differentiation of chimeric antigen receptor (CAR) T cells56 or cells used in adoptive cell transfer (ACT) therapy57. Applying scMEP to human clinical material revealed the presence of tissue-specific metabolic T cell subsets (Fig. 4), including two metabolically diverging subsets expressing CD39 and PD1 which were expanded in human colorectal carcinoma. Follow-up image-based scMEP via MIBI-TOF (Fig. 5) revealed that the metabolically-repressed CD39/PD1 cells were excluded from the tumor-immune boundary, a complex multicellular structure known to regulate immune function40. Although surface expression of CD39 and PD1 indicate T cell exhaustion/dysfunction, they can be expressed more broadly. The here identified association of metabolic phenotype with CD39/PD1 expression and TCF1 downregulation, as well as the tumor-specific expansion of this metabolic subset and its exclusion from the tumor-immune boundary suggest that incorporation of metabolic profiling to identify functionally diverse T cell states could further improve clinical stratification, e.g. to better predict response to immunotherapy.
In summary, we here presented a robust approach to study the metabolism of single cells using antibody-based multiplex technologies. The application of scMEP should enable a better understanding of human immune cell biology and benefit the identification of disease-associated metabolic alterations that could serve as potential biomarkers and therapeutic targets for a variety of human diseases.
Online methods
Human samples
De-identified peripheral blood samples from healthy human subjects (see Supplementary Table 2) were obtained and experimental procedures were carried out in accordance with the guidelines of the Stanford Institutional Review Board (IRB). Written informed consent was obtained from all subjects. Fresh whole human blood in heparin collection tubes or leukoreduction system chamber contents (Terumo BCT) were obtained via the Stanford Blood Center. PBMCs were isolated via Ficoll (GE Healthcare) density gradient centrifugation.
FFPE tissue samples were obtained from the tissue repository of the Stanford Department of Pathology. Colorectal carcinoma and healthy adjacent tissue samples for mass cytometry (see Supplementary Table 2) were collected fresh after resection and transported for processing on ice in cell culture medium (CCM: RPMI-1640 (life technologies), 10% FBS, 1x L-glutamine, 1x penicillin/streptomycin (Thermo Fisher)). Samples were minced and processed using the MACS tumor dissociation kit (Miltenyi Biotec) as recommended. All viable single-cell suspensions were frozen in FBS supplemented with 10% DMSO and stored in liquid nitrogen.
In vitro cell activation
Cryopreserved PBMC samples where thawed into 10 ml of cold CCM supplemented with 0.025 U/ml benzonase (Sigma) and washed once (250 g, 4 ºC). Pan T cells were enriched through negative selection using magnetic beads (Pan T cell Isolation Kit, Miltenyi Biotec). Isolated T cells (including CD4+ and CD8+) were CFSE labeled by incubating them with 80 μm CSFE (Thermo Fisher) in CCM for 5 min at RT as described previously34. Labeled cells were quenched with warm CCM and washed three times by centrifuging for 5 min at 250 g. After washing, samples were divided for naïve and memory T cell isolation. Naïve T cells were enriched by depleting CD45RO expressing T cells using magnetic beads and memory T cells were isolated by depleting CD45RA expressing T cells. Using this approach, all cells used in subsequent assays were negatively isolated. Cells were counted using an automated cell counting system and distributed into a 24-well plate in CCM at 1×106 cells/well. For naïve T cells, 5 ng/ml of IL-2 was added to the culture and memory T cells were supplemented with 5 ng/ml IL-7 and IL-15 (all Peprotech). Cells were activated in a reverse time-course so that their total time in culture was identical and to ensure that all cells finish their indicated activation period at the same day to enable extracellular flux analysis. To do so, anti-CD3/anti-CD28 beads (Dynabeads, Thermo Fisher) were added in a 1:1 cell-to-bead ratio on the respective day and cells were incubated at 37 ºC, 5% CO2 for up to 5 days. At the end of the activation period, cells from the same condition (same wells) were divided up and entered into the mass cytometry and extracellular flux analysis workflows.
Activation of human macrophages
Human macrophages were generated from isolated monocytes obtained from buffy coats of healthy donors (Sanquin, Amsterdam) in the presence of 25 ng/mL human macrophage-colony stimulating factor (M-CSF, Miltenyi) in Iscove’s modified Dulbecco’s medium (IMDM, Life Technologies) with 10% FCS, penicillin (100 U/mL), streptomycin (100 μg/mL) as described before59. On day 6, cells were harvested, seeded at 106 cells/mL, and stimulated 24 hr with 10 ng/ml LPS (Sigma-Aldrich) or remained untreated before metabolic analysis.
Live/dead discrimination and cell fixation
Whole blood was fixed, and erythrocytes were lysed using Lyse/Fix Buffer (BD Biosciences) as suggested by the supplier. Cryopreserved single-cell suspensions (viable PBMC, lymph node and tumor biopsy samples) where thawed into 10 ml of cold CCM supplemented with 0.025 U/ml benzonase (Sigma) and washed once (250 g, 4 ºC). In vitro activated T cell and macrophage populations (not cryopreserved) were washed once in CCM and directly processed further. For live/dead cell discrimination, monoisotopic cisplatin-194 (Fluidigm) was pre-conditioned for 48 h at 37 ºC, aliquoted and stored at −20 ºC. Viability staining was performed by resuspending cells in 1 ml of low-barium PBS and adding cisplatin-194 to a final concentration of 500 nM, followed by incubation for 5 min at RT and washing with cell staining medium (CSM: low-barium PBS with 0.5 % BSA and 0.02 % sodium azide (all Sigma)). Cells were fixed with 1.6% paraformaldehyde (PFA) in PBS for 10 min at RT and washed twice with CSM. Fixed cells were either entered directly into the staining workflow or cryopreserved by resuspending them in CSM supplemented with 10% DMSO and storing them at −80 ºC.
Heavy-metal conjugation of antibodies
Antibodies were conjugated to heavy-metal ions with commercially available MaxPar (Fluidigm) or MIBItag (IonPath) reagents using an optimized conjugation protocol60. In short, antibodies were reduced with 4 mM TCEP (Thermo Fisher) for 30 min at 37 ºC and washed two times. For conjugations using MaxPar reagents, metal chelation was performed by adding metal solutions (final 0.05 M) to chelating polymers and incubating for 40 min at RT. Metal-loaded polymers were washed twice using a 3 kDa MWCO microfilter (Millipore) by centrifuging for 30 min, 12,000 g at RT. For conjugations using MIBItag reagents, pre-loaded polymers were obtained, and no loading reactions needed to be performed. For both approaches, antibody buffer exchange was performed by washing purified antibody through a 50 kDa MWCO microfilter (Millipore) and centrifuging for 10 min, 12,000 g at RT. Partially reduced antibodies and metal-loaded polymers were incubated together for 90 min at 37 ºC. Conjugated antibodies were washed four times and collected by two centrifugations (2 min, 1,000 g, RT) into an inverted column in a fresh 1.6 ml collection tube. Protein content was assessed by NanoDrop (Thermo Fisher) measurement, antibody stabilization buffer (Candor Bioscience) was added to a final volume of at least 50 v/v % and antibodies were stored at 4 ºC. Conjugations to 139La, 209Bi (Sigma Aldrich), and 140Ce (Trace Sciences International) were performed following the above protocol, using either Fluidigm or IonPath polymers. 139La-conjugated antibodies were lyophilized immediately after conjugation and reconstituted prior to staining. 209Bi conjugations are following a slightly modified protocol60. Conjugations of antibodies to platinum-isotopes were performed following polymer-independent protocols61,62.
Palladium barcoding and staining with heavy-metal conjugated antibodies
To eliminate technical variability during staining or acquisition, individual samples within one experiment were palladium-barcoded as described previously63 and combined into a composite sample before further processing and staining. Cell-surface antibody master-mix in CSM was filtered through a pre-wetted 0.1 μm spin-column (Millipore) to remove antibody aggregates and added to the samples. After incubation for 30 min at RT, cells were washed once with CSM. To enable intracellular staining, cells were permeabilized by incubating with ice-cold MeOH for 10 min on ice and washed to times with CSM to remove any residual MeOH. Intracellular antibody master-mix in CSM was added to the samples and incubated for 1 h at RT. Cells were washed once with CSM and resuspended in intercalation solution (1.6% PFA in PBS and 0.5 μM rhodium-intercalator (Fluidigm)) for 20 min at RT or overnight at 4 °C. Before acquisition, samples were washed once in CSM and twice in ddH2O and filtered through a cell strainer (Falcon). Cells were then resuspended at 1 × 106 cells/mL in ddH2O supplemented with 1x EQ four element calibration beads (Fluidigm) and acquired on a CyTOF2 mass cytometer (Fluidigm).
Antibody validation workflow
Mass cytometry-based antibody validation was performed on a range of cell lines, immune populations found in whole blood and T cells with or without TCR activation. First, various leukemic, embryonic and carcinoma cell lines62 were cultured in standard conditions, fixed, palladium-barcoded and subsequently stained with heavy-metal conjugated antibodies as described above. Whole blood was processed as described above and stained with a combination of metabolic antibodies and cell lineage markers (CD45, CD3, CD4, CD8, CD45RA, CD66, CD14, CD19, CD20, HLA-DR, CD56, CD57, CD11c, CD123, FceRI, CD235ab as established previously64) to identify the major immune cell types through manual gating. Human T cells were either rested or activated with anti-CD3/anti-CD38-beads for 72 h (see above), fixed and palladium-barcoded before staining with metabolic antibodies. Metabolic antibodies were initially used at a concentration of 2 μg/ml which was subsequently adjusted if necessary. For all populations, median arsinh values were calculated and positive staining was defined as a median of at least 10 ion counts (asinh transformed value >1.5) of any subpopulation. Where available, cell-lineage specific expression and induction upon activation were compared to previously determined values for the given cell population25,26,65.
To validate antibodies on tissues, control tonsil and liver FFPE tissues were stained with the indicated metal-conjugated antibodies as described below and their performance was validated through traditional IHC and MIBI-TOF. Detectable staining was determined through visual inspection of both IHC and grayscale MIBI-TOF images. For intra-assay quality control, IHC and MIBI-TOF images were visually compared and in addition, related to previously determined staining patterns65.
Extracellular flux analysis
Extracellular flux analysis of T cell activation was performed by adopting previously outlined protocols66. In short, in vitro activated T cells were spun onto a Cell-Tak (Thermo Fisher) coated XF96 cell culture microplate (Agilent) with a density of 100,000 or 150,000 cells/well and rested in Seahorse XF RMPI 1640 medium supplemented with 2 mM L-glutamine, 2 mM sodium pyruvate and 25 mM glucose (all Agilent) for 1 h in a non-CO2 incubator at 37 ºC. ECAR and OCR were measured using a Seahorse XF96 extracellular flux analyzer (Agilent). Oligomycin (1 μM), fluoro-carbonyl cyanide phenylhydrazone (FCCP; 1.5 μM) and rotenone (0.5 μM) together with antimycin A (0.5 μM) were sequentially injected to establish baseline parameters. Extracellular flux analysis of macrophages was performed as outlined previously67, including an additional injection of 25 mM glucose. Raw data was imported into the R environment in order to calculate basal glycolysis respiration rates as described68. Data was normalized by cell number. For linear regression between extracellular flux analysis values and mass cytometry values, both were asinh transformed with a cofactor of 5.
Mass cytometry data preprocessing
Raw mass cytometry data was first bead-normalized to remove acquisition-related influences on marker expression using the premessa R package. Next, barcoded cells were assigned back to their initial samples using their unique palladium barcode combination. Normalized data was uploaded onto cytobank.org69 or cellengine.com to identify single, live cells by excluding remaining beads (140Ce and 153Eu) and manually gating on DNA (103Rh) and viability (194Pt) channels. Data was subsequently imported into the R environment, asinh transformed (cofactor 5) and normalized to the 99.9th percentile of each respective channel before downstream analysis.
Clustering and data visualization
Pre-processed single-cell (mass cytometry and MIBI-TOF) data was clustered using the FlowSOM R package70 and the indicated input channels. Resulting clusters were either manually annotated with the main cell lineages based on their lineage marker profiles or, if the underlying number and identity of clusters was unknown (e.g. for clustering on metabolic features) the metaclustering function of the FlowSOM package was used. Given their widespread expression across cell lineages, differences in metabolic protein expression between different populations and clusters were visualized by marker enrichment modeling58. UMAP embeddings were calculated using the R uwot implementation with the following parameters: n_neighbors = 15, min_dist = 0.02.
Calculation of metabolic scMEP scores
To calculate single-cell scMEP scores, expression values (debarcoded, bead-normalized, asinh transformed and percentile-normalized as described above) from all metabolic enzymes within a given pathway (glycolysis, respiration, amino acid metabolism and fatty acid metabolism) were summed and divided by the number of channels within the pathway.
To calculate image-based scMEP scores, pixel-based expression values from pre-processed data were blurred with a gaussian filter (sigma = 6) and asinh transformed. Next, pixel values from images within a given pathway were summed and finally percentile normalized to the 99th percentile.
Trajectory analysis of metabolic remodeling
Pre-processed data was randomly subsampled to represent all indicated days of activation equally. Pseudotime was calculated using the SCORPIUS71 and Slingshot72 algorithms, given their documented robustness across different datasets73, making use of the dynverse R implementation. All indicated channels were used as input dimensions to both algorithms and we did not define priors. Mitotic (M phase) cells and were excluded from this analysis given their drastically different metabolic characteristics. Strongly cell phase-dependent markers (IdU incorporation and H3 phosphorylation74) were not used as input dimensions for the trajectory calculation. Resulting pseudotime was scaled from 0–1. While resting cells (day 0) and cells from day 5 were included in the trajectory calculation to allow identification of starting and end points, we were mostly interested in early T cell activation (pseudotime 0.1–0.8), thus focusing downstream analysis on this period which additionally constitutes the most robust part of the trajectory as determined by comparison between SCORPIUS and Slingshot trajectories.
Analysis of scMEP repeatability and robustness
To determine the robustness of the scMEP approach, metabolic regulator expression values of samples from the same healthy donors were stained and analyzed in two separate experiments and compared by linear regression using the lm() function. Hierarchical clustering using the R function hclust() was performed using the same input data. For the training phase of immune cell lineage prediction from metabolic features, cells were first compensated using the CATALYST package75 and an in-house determined isotopic purity matrix to rule out slight signal overlap contributing to this prediction. Next, 20,000 cells were randomly subsampled from three healthy donors to create an L1 regularized linear regression model using the glmnet R package76. For the test phase, data was derived from two independent healthy donors not included in the training data. Prediction of T cell maturation status from scMEP profiles was performed similarly by randomly subsampling cells from both conditions into separate training and test data. Metabolic heterogeneity was defined as the cellular Euclidean distance to the average expression levels of the given population34. Only (pre-processed) metabolic feature expression values were used for this calculation.
Staining for multiplexed ion beam imaging
Tissue sections (4 μm) were cut from colorectal carcinoma and control colon FFPE tissue blocks using a microtome and mounted on silanized gold-coated slides (IONpath). Mounted tissue sections were incubated at 70°C for 20 min and deparaffinized with three washes of fresh xylene followed by rehydration with successive washes of ethanol 100% (2x), 95% (2x), 80% (1x), 70% (1x), and distilled water. Washes were performed using a Leica ST4020 Linear Stainer (Leica Biosystems) programmed to three dips per wash for 30 s each. Rehydrated sections were immersed in epitope retrieval buffer (Target Retrieval Solution, pH 9, DAKO Agilent), incubated at 97 °C for 40 min and cooled down to 65 °C using Lab vision PT module (Thermofisher Scientific). Slides were washed with MIBI wash buffer (low-barium PBS IHC Tween buffer (Cell Marque) containing 0.1% (w/v) BSA (Thermofisher Scientific)). Slides were then placed into a Sequenza staining rack (Thermofisher Scientific) and sections were blocked for 1 h with blocking buffer (1X TBS IHC Wash Buffer with Tween 20 (Cell Marque) + 2% donkey serum, 0.1% cold fish skin gelatin (Sigma), 0.1% Triton X-100, and 0.05% sodium azide). Metal-conjugated antibody mix was prepared in 3% (v/v) donkey serum TBS IHC wash buffer and filtered using a centrifugal filter with a 0.1 mm PVDF membrane (Merck Millipore). Sections were stained with the antibody mix, incubating overnight at 4°C in the Sequenza staining rack. After incubation, slides were washed twice with MIBI wash buffer and fixed for 5 min in 2% glutaraldehyde solution (Electron Microscopy Sciences) in low-barium PBS. Slides were then rinsed briefly in low-barium PBS and then dehydrated with successive washes of Tris 0.1 M (pH 8.5) (3x), distilled water (2x), and ethanol 70% (1x), 80% (1x), 95% (2x), 100% (2x). Slides were immediately dried in a vacuum chamber for at least 1 h prior to imaging.
Immunohistochemistry
All MIBI-TOF antibodies were validated by DAB chromogenic IHC. The protocol for IHC closely followed the MIBI-TOF staining protocol, with minor changes. Before assembly of slides into the Sequenza staining rack and blocking, endogenous peroxidase activity was quenched by incubation in 3% H2O2 for 30 min and sections were washed with H2O on an orbital shaker for 5 min. Sections were stained with MIBI antibodies individually, and detected with ImmPRESS universal (Anti-Mouse/Anti-Rabbit) secondary antibody kit (Vector labs) and ImmPACT DAB Substrate kit (Vector Labs), according to the manufacturer’s guidelines.
Multiplexed ion beam imaging acquisition
Quantitative imaging was performed using a custom designed MIBI-TOF mass spectrometer (IONpath), as previously described21,40, with an image size of 400 μm2 and 1024 × 1024 pixels. The entire cohort of 58 FOVs was acquired over a 24 h period of continuous imaging, yielding a total of 2088 single channel images.
Imaging data pre-processing and single-cell segmentation
Multiplexed imaging data was preprocessed as described before40. In short, for each pixel, MS spectra were converted into pixel counts by extracting a mass range from atomic mass unit amu-0.25 to amu±0. Background (due to absence of tissue or high gold signal) was removed, and noise was filtered out using a k-nearest-neighbor approach. To segment single cells from images, we trained a convolutional neural network77 using annotated training data from a variety of different cancer types. The network output was fed into the watershed algorithm to produce individual cells. This mask was used to extract per-cell counts for each marker in each image. Counts were normalized by cell size to account for different sampling of cells in the given plane. Normalized data was imported into the R environment and transformed using an inverse hyperbolic sine (asinh) cofactor of 0.05 (adjusted due to cell size normalization).
Context-dependent spatial enrichment analysis
The context-dependent spatial enrichment (CDSE) approach40 was used to identify structured patterns of metabolic protein expression in the tissue. For each pair of metabolic features, X and Y, the number of times cells positive for protein X was within a 50-pixel (~20 um) radius of cells positive for protein Y was counted. Thresholds for positivity were customized to each feature individually. A null distribution was produced by performing 1000 bootstrap permutations where the locations of cells positive for protein Y were randomized. Randomizations retained the distribution of cells positive for protein Y across major lineage categories: immune, endothelial, epithelial, and fibroblast. A z-score was calculated comparing the number of true cooccurrences of cells positive for protein X and Y relative to the null distribution. For each pair of metabolic proteins X and Y the average z-score was calculated across malignant and control tissues separately.
Visualization
Plots were created using the ggplot2 R package. Schematic representations were created with biorender (https://biorender.io/). Figures were prepared in Illustrator (Adobe).
Statistical analysis
All statistical tests used in this study are described in detail in the according figure legends. In general, we selected two-tailed tests over their one-tailed alternatives. Where indicated, we applied Welch’s correction to t-tests to account for unequal variances. For comparisons of cell populations derived from the same individual, we performed matched t-tests. To account for multiple hypotheses testing, we applied the BH procedure to report the associated FDR. Additional information can be found in the Life Science Reporting Summary.
Data availability
Single-cell mass cytometry (CyTOF) datasets for metabolic analysis of human whole blood populations, in vitro T cell activation and analysis of metabolic states in human tissues as well as MIBI-TOF imaging data of colorectal carcinoma and healthy colon are publicly available at https://doi.org/10.5281/zenodo.3951613.
Code availability
All custom R scripts associated with this manuscript will be made available upon request.
Supplementary Material
Acknowledgements
We thank L. Keren for insightful discussions and invaluable feedback as well as the Nakamura lab at the Gladstone Institutes for access to their Seahorse XF analyzer. Further, we thank A. Tsai for advice and help with clinical samples. This study was supported by an EMBO Long-Term Fellowship ALTF 1141-2017 (F.J.H.), the Novartis Foundation for medical-biological Research 16C148 (F.J.H.) and the Swiss National Science Foundation SNF Early Postdoc Mobility P2ZHP3-171741 (F.J.H.). In addition, we received support from the NIH 1DP2OD022550-01 (S.C.B.), 1R01AG056287-01 (S.C.B.), 1R01AG057915-01 (S.C.B.) and 1U24CA224309-01 (S.C.B.).
Footnotes
Declaration of Interests
The authors declare no competing interest.
References
- 1.Klein Geltink RI, Kyle RL & Pearce EL Unraveling the Complex Interplay Between T Cell Metabolism and Function. Annu. Rev. Immunol 36, 461–488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olenchock BA, Rathmell JC & Vander Heiden MG Biochemical Underpinnings of Immune Cell Metabolic Phenotypes. Immunity 46, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck MD, Sowell RT, Kaech SM & Pearce EL Metabolic Instruction of Immunity. Cell 169, 570–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu T et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature 548, 228–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel CH, Leone RD, Horton MR & Powell JD Targeting metabolism to regulate immune responses in autoimmunity and cancer. Nat. Rev. Drug Discov 18, 669–688 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Scharping NE et al. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 45, 374–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishton RJ, Sukumar M & Restifo NP Metabolic Regulation of T Cell Longevity and Function in Tumor Immunotherapy. Cell Metab. 26, 94–109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leone RD et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angiari S et al. Pharmacological Activation of Pyruvate Kinase M2 Inhibits CD4+ T Cell Pathogenicity and Suppresses Autoimmunity. Cell Metab. 31, 391–405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornberg MD et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee C-F et al. Preventing Allograft Rejection by Targeting Immune Metabolism. Cell Rep. 13, 760–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dettmer K, Aronov PA & Hammock BD Mass spectrometry-based metabolomics. Mass Spectrom. Rev 26, 51–78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buescher JM et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol 34, 189–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verbist KC et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma EH et al. Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8+ T Cells. Immunity 51, 856–870 (2019). [DOI] [PubMed] [Google Scholar]
- 16.O’Sullivan D, Sanin DE, Pearce EJ & Pearce EL Metabolic interventions in the immune response to cancer. Nat. Rev. Immunol 19, 324–335 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Li X et al. Navigating metabolic pathways to enhance antitumour immunity and immunotherapy. Nat. Rev. Clin. Oncol 16, 425–441 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Muir A & Vander Heiden MG The nutrient environment affects therapy. Science 360, 962–963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bourgeois T et al. Targeting T Cell Metabolism for Improvement of Cancer Immunotherapy. Front. Oncol 8, 237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–96 (2011).21551058 [Google Scholar]
- 21.Keren L et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv 5, eaax5851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmann FJ & Bendall SC Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat. Rev. Rheumatol 16, 87–99 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saas P, Varin A, Perruche S & Ceroi A Recent insights into the implications of metabolism in plasmacytoid dendritic cell innate functions: Potential ways to control these functions. F1000Research 6, 456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakus D, Gizak A, Deshmukh A & Wiśniewski JR Absolute Quantitative Profiling of the Key Metabolic Pathways in Slow and Fast Skeletal Muscle. J. Proteome Res 14, 1400–1411 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Geiger R et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howden AJM et al. Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol 20, 1542–1554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanner LB et al. Four Key Steps Control Glycolytic Flux in Mammalian Cells. Cell Syst. 7, 49–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R et al. The Transcription Factor Myc Controls Metabolic Reprogramming upon T Lymphocyte Activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finlay DK et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8 + T cells. J. Exp. Med 209, 2441–2453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frauwirth KA et al. The CD28 Signaling Pathway Regulates Glucose Metabolism. Immunity 16, 769–777 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Slack M, Wang T & Wang R T cell metabolic reprogramming and plasticity. Mol. Immunol 68, 507–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R & Green DR Metabolic checkpoints in activated T cells. Nat. Immunol 13, 907–915 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Zaslaver A et al. Just-in-time transcription program in metabolic pathways. Nat. Genet 36, 486–491 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Good Z et al. Proliferation tracing with single-cell mass cytometry optimizes generation of stem cell memory-like T cells. Nat. Biotechnol 37, 259–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altman BJ, Stine ZE & Dang CV From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer M et al. Early effector maturation of naïve human CD8 + T cells requires mitochondrial biogenesis. Eur. J. Immunol 48, 1632–1643 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Icard P, Fournel L, Wu Z, Alifano M & Lincet H Interconnection between Metabolism and Cell Cycle in Cancer. Trends Biochem. Sci 44, 490–501 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Bengsch B et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8 + T Cell Exhaustion T Cell Exhaustion. Immunity 45, 358–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blank CU et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol 19, 665–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keren L et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son N-H et al. Endothelial cell CD36 optimizes tissue fatty acid uptake. J. Clin. Invest 128, 4329–4342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang C-H et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaira K et al. Prognostic significance of l-type amino acid transporter 1 (LAT1) and 4F2 heavy chain (CD98) expression in stage I pulmonary adenocarcinoma. Lung Cancer 66, 120–126 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Shimizu K et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br. J. Cancer 110, 2030–2039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canale FP et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8+ T Cells. Cancer Res. 78, 115–128 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Raczkowski F et al. CD39 is upregulated during activation of mouse and human T cells and attenuates the immune response to Listeria monocytogenes. PLoS One 13, e0197151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon S & Labarriere N PD-1 expression on tumor-specific T cells: Friend or foe for immunotherapy? Oncoimmunology 7, e1364828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heath JR, Ribas A & Mischel PS Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov 15, 204–216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linnarsson S & Teichmann SA Single-cell genomics: coming of age. Genome Biol. 17, 97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papalexi E & Satija R Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol 18, 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Schwartzman O & Tanay A Single-cell epigenomics: techniques and emerging applications. Nat. Rev. Genet 16, 716–726 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Xiao Z, Dai Z & Locasale JW Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun 10, 3763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahi P, Kim SC, Haliburton JR, Gartner ZJ & Abate AR Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci. Rep 7, 44447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mair F et al. A Targeted Multi-omic Analysis Approach Measures Protein Expression and Low-Abundance Transcripts on the Single-Cell Level. Cell Rep. 31, 107499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadelain M Chimeric Antigen Receptors: A Paradigm Shift in Immunotherapy. Annu. Rev. Cancer Biol 1, 447–466 (2017). [Google Scholar]
- 57.Rosenberg SA & Restifo NP Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 58.Diggins KE, Greenplate AR, Leelatian N, Wogsland CE & Irish JM Characterizing cell subsets using marker enrichment modeling. Nat. Methods 14, 275–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van den Bossche J et al. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Rep. 17, 684–696 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Hartmann FJ et al. Scalable Conjugation and Characterization of Immunoglobulins with Stable Mass Isotope Reporters for Single-Cell Mass Cytometry Analysis. Methods Mol. Biol 1989, 55–81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mei HE, Leipold MD & Maecker HT Platinum-conjugated antibodies for application in mass cytometry. Cytom. Part A 89, 292–300 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Hartmann FJ, Simonds EF & Bendall SC A Universal Live Cell Barcoding-Platform for Multiplexed Human Single Cell Analysis. Sci. Rep 8, 10770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zunder ER et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc 10, 316–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartmann FJ et al. Comprehensive Immune Monitoring of Clinical Trials to Advance Human Immunotherapy. Cell Rep. 28, 819–831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uhlen M et al. Tissue-based map of the human proteome. Science 347, 1260419–1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 66.van der Windt GJW, Chang C-H & Pearce EL Measuring Bioenergetics in T Cells Using a Seahorse Extracellular Flux Analyzer. Curr. Protoc. Immunol 113, 3.16B.1–3.16B.14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van den Bossche J, Baardman J & de Winther MPJ Metabolic Characterization of Polarized M1 and M2 Bone Marrow-derived Macrophages Using Real-time Extracellular Flux Analysis. J. Vis. Exp 105, e53424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mookerjee SA, Gerencser AA, Nicholls DG & Brand MD Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem 292, 7189–7207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kotecha N, Krutzik PO & Irish JM Web-Based Analysis and Publication of Flow Cytometry Experiments. Curr. Protoc. Cytom 53, 10.17.1–10.17.24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Gassen S et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry. A 87, 636–45 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Cannoodt R et al. SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. bioRxiv (2016) doi: 10.1101/079509. [DOI] [Google Scholar]
- 72.Street K et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saelens W, Cannoodt R, Todorov H & Saeys Y A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ & Nolan GP Single-cell mass cytometry adapted to measurements of the cell cycle. Cytom. Part A 81, 552–566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chevrier S et al. Compensation of Signal Spillover in Suspension and Imaging Mass Cytometry. Cell Syst. 6, 612–620 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Friedman J, Hastie T & Tibshirani R Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 77.Bannon D et al. Dynamic allocation of computational resources for deep learning-enabled cellular image analysis with Kubernetes. bioRxiv (2019) doi: 10.1101/505032. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell mass cytometry (CyTOF) datasets for metabolic analysis of human whole blood populations, in vitro T cell activation and analysis of metabolic states in human tissues as well as MIBI-TOF imaging data of colorectal carcinoma and healthy colon are publicly available at https://doi.org/10.5281/zenodo.3951613.