Abstract
BACKGROUND & AIMS:
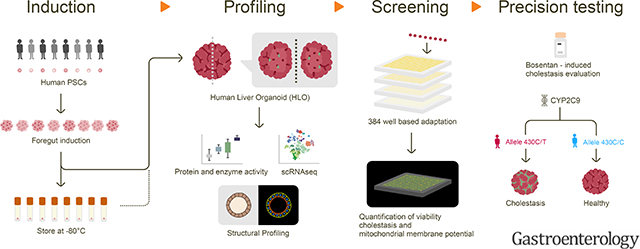
Preclinical identification of compounds at risk of causing drug induced liver injury (DILI) remains a significant challenge in drug development, highlighting a need for a predictive human system to study complicated DILI mechanism and susceptibility to individual drug. Here, we established a human liver organoid (HLO) based screening model for analyzing DILI pathology at organoid resolution.
METHODS:
We first developed a reproducible method to generate HLO from storable foregut progenitors from pluripotent stem cell (PSC) lines with reproducible bile transport function. The qRT-PCR and single cell RNA-seq determined hepatocyte transcriptomic state in cells of HLO relative to primary hepatocytes. Histological and ultrastructural analyses were performed to evaluate micro-anatomical architecture. HLO based drug-induced liver injury assays were transformed into a 384 well based high-speed live imaging platform.
RESULTS:
HLO, generated from 10-different pluripotent stem cell lines, contain polarized immature hepatocytes with bile canaliculi-like architecture, establishing the unidirectional bile acid transport pathway. Single cell RNA-seq profiling identified diverse and zonal hepatocytic populations that in part emulate primary adult hepatocytes. The accumulation of fluorescent bile acid into organoid was impaired by CRISPR-Cas9 based gene editing and transporter inhibitor treatment of BSEP. Furthermore, we successfully developed an organoid based assay with multiplexed readouts measuring viability, cholestatic and/or mitochondrial toxicity with high predictive values for 238 marketed drugs at 4 different concentrations (Sensitivity: 88.7%, Specificity: 88.9%). LoT positively predicts genomic predisposition (CYP2C9*2) for Bosentan-induced cholestasis.
CONCLUSIONS:
Liver organoid-based Toxicity screen (LoT) is a potential assay system for liver toxicology studies, facilitating compound optimization, mechanistic study, and precision medicine as well as drug screening applications.
Keywords: Liver organoid, pluripotent stem cell, cholestasis, DILI
Lay Summary
Human liver organoid (HLO) based screening system, termed LoT assay, will offer an opportunity to analyze cholestatic pathology specific to developing drugs and genetic susceptibility otherwise inaccessible at preclinical stage.
Graphical Abstract
Introduction
Billions of dollars are lost annually from drug development in the pharmaceutical industry due to the failures of drug candidates identified in initial screens, and nearly a third of drugs are withdrawn from the market 1. Further, despite the promising efficacy, a failure of drug candidates results in tremendous loss of a patient’s treatment opportunity. Preclinical studies generally consist of an in vitro evaluation as a primary efficacy screen to identify a “hit” compound, followed by safety studies in vitro and in vivo to assess the mechanisms of metabolism and toxicology. This inefficiency can be explained by the substantial lack of physiologically relevant preclinical models in evaluating drug induced liver injury (DILI) in humans and thus, an urgent need to develop an in vitro screening model for the evaluation of the vast amounts of continuously growing compound libraries.
Primary hepatocytes are a highly polarized metabolic cell type, and form a bile canaliculi structure with microvilli-lined channels, separating the peripheral circulation from the bile acid secretion pathway. The most upstream aspects of DILI include detoxification of drugs or their reactive metabolites by hepatocytes and excretion into bile canaliculi through transporters such as multi-drug resistance-associated protein (MRP) transporters, suggesting the need to reconstruct these uniquely organized structures as a crucial property of hepatocytes in vivo for predicting DILI pathology. However, there are considerable differences in drug toxicity profiles between the current simplified culture model with the use of isolated primary human hepatocytes or hepatic cell lines, and in vivo physiology, resulting in failed translation of drugs or drug discontinuation, for example, Troglitazone, Nefazodone and Tolcapone (https://livertox.nlm.nih.gov/index.html). The determination of toxicological properties thus mainly relies on animals as an essential step towards drug development, however, due to the pronounced differences in physiology between humans and animals, there is a significant lack of fidelity to human outcomes 2, 3.
We recently reported a method to generate human liver organoids (HLO) from pluripotent stem cells (PSCs) with a potential to model inflammatory diseases 4. However, it remains elusive whether the functional bile canaliculi-like structure, an essential component for modeling defective bile excretion, is formed and can be used towards a drug toxicology analysis. Additionally, while minimizing batch-differences, the enhancement of the assay throughput will be critical before being translated into preclinical studies. Herein we developed a reproducible liver organoid protocol using stably expandable foregut cells from human stem cells, i.e. iPSC and ESC. We established a live imaging based dynamic detection assay for bile acid uptake and excretion in modified HLO, that can be prevented by bile acid transporter gene knockout. This assay platform is amenable for large scale compound screening and annotated 238 drugs with multiplexed readouts. Furthermore, HLO were shown to model genotype-specific susceptibility5 to Bosentan induced cholestasis with multiple iPSC. This robust assay, named Liver organoid-based Toxicity screen (LoT), provides functional readout developed in human liver organoids, and will facilitate diagnosis, functional studies, drug development and personalized medicine.
Materials and methods
Human PSCs.
Maintenance of PSCs T (TkDA3) human iPSC clone used in this study was kindly provided by K. Eto and H. Nakauchi. 12 (1231A3) and 13 (1383D6) were gifted by Kyoto University (Japan). CW10150, CW10027, CW10077 and WD90, 91, 92 were purchased from Coriell (NJ, USA). iPSC_285.0, iPC_18.4 and iPSC_54.1 were obtain from patient skin fibroblast/PBMC and reprogrammed into iPSC by CCHMC pluripotent stem cell core. Human H1 embryonic stem cell clone used in this study was kindly provided by WiCell institute. Human iPSC lines were maintained as described previously6, 7. All human iPSC lines were maintained as described previously (Takebe et al., 2017). Briefly, undifferentiated hiPSCs were cultured on Laminin 511E8-flagment (Nippi, Japan) coated dishes in Stem Fit medium (Ajinomoto Co, Japan) with 100ng/ml bFGF (R&D Systems, MN, USA) at 37 °C in 5% CO2 with 95% air.
Foregut induction.
hiPSCs were differentiated into foregut using previously described method with modifications. 8. In brief, hiPSCs were detached by Accutase (Thermo Fisher Scientific Inc., MA, USA) and were seeded on Laminin coated tissue culture plate with 100,000 cells/cm2. Medium was changed to RPMI 1640 medium (Life Technologies) containing 100 ng/mL Activin A (R&D Systems) and 50 ng/mL bone morphogenetic protein 4 (BMP4; R&D Systems) at day 1, 100 ng/mL Activin A and 0.2% fetal calf serum (FCS; Thermo Fisher Scientific Inc.) at day 2, and 100 ng/mL Activin A and 2% FCS at day 3. On Day4–6, cells were cultured in Advanced DMEM/F12 (Thermo Fisher Scientific Inc.) with B27 (Life Technologies) and N2 (Gibco, CA, USA) containing 500 ng/ml fibroblast growth factor (FGF4; R&D Systems) and 3 μM CHIR99021 (Stemgent, MA, USA). Cells were maintained at 37 °C in 5% CO2 with 95% air and the medium was replaced every day. The foregut cells were detached by Accutase and then frozen in Cell Banker 1(Nippon Zenyaku Kogyo Co., Ltd., Japan). The foregut cells can be stored in −80°C or LN2 for long term storage. The differentiation protocol is illustrated in Figure. 1A.
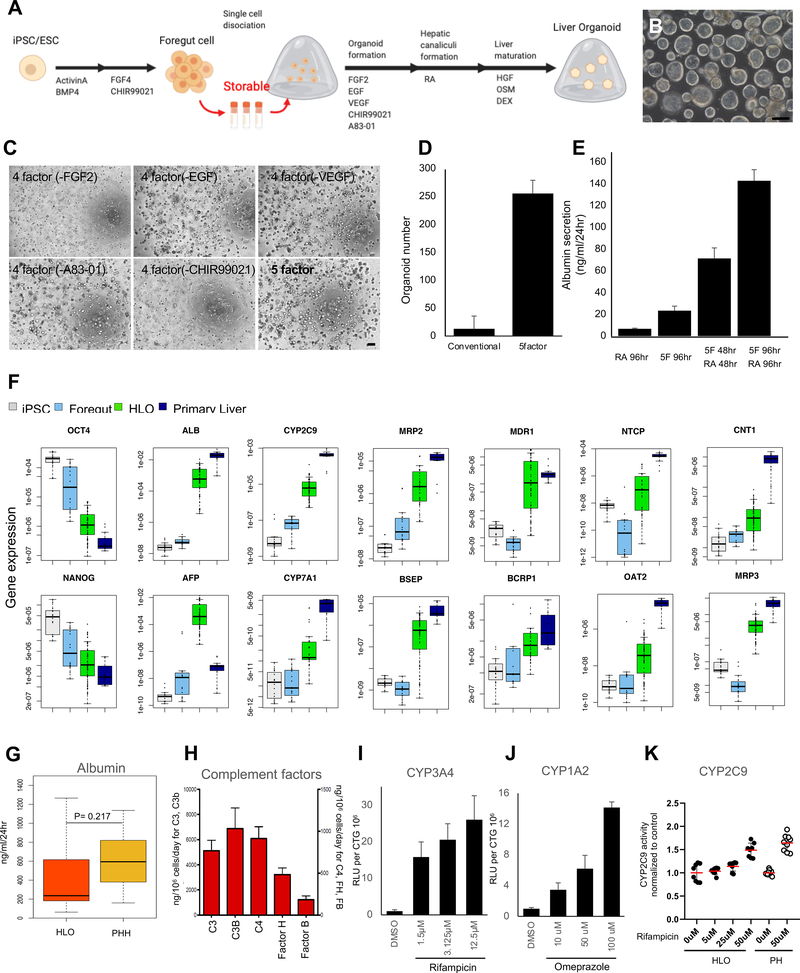
Figure 1. Generation of human liver organoid (HLO) from storable PSC-derived foregut cells.
A. Overview of our differentiation method for liver organoids.
B. Representative Image of HLO at Day 20. Bar = 200 μm.
C. Phase-contrast image of HLO in different culture conditions with 5 factors, which were FGF2, VEGF, EGF, CHIR-99021 (GSK-3 inhibitor) and A83–01 (TGF-β inhibitor). Bar = 200 μm
D. The number of HLO cultured with 5 factors. Conventional: Treated with only RA.
E. The secretion of albumin from HLO in culture condition with treatment of different factors.
F. Quantitative RT-PCR analysis of representative gene related to hepatic function and pluripotency marker. Undifferentiated iPSCs (iPSC, n=8), posterior foregut organoid (Foregut, n=8), human liver organoid (HLO, n=16) and Primary Liver (Human primary hepatocytes, n=8).
G. The secretion of albumin in HLO (n=92) and primary hepatocytes (n=14).
H. The secretion of complement factors in HLO. n=4.
I. CYP3A4 activation by treatment of Rifampicin. n = 5.
J. CYP1A2 activation by treatment of Omeprazole. n = 5.
K. The CYP2C9 induction capacity after Rifampicin treatment in HLO and Primary Hepatocytes (PHH). Bars represent the mean ± SD, n=7 for HLO, n=9 for primary hepatocytes.
Generation of HLO.
The frozen foregut cells were thawed quickly and then centrifuged at 1200 rpm for 3 minutes. Cells was resuspended in Matrigel (Corning, In., NY, USA). A total of 100,000 cells were embedded in 50μl Matrigel drop on the dishes in organoid formation media with 5 factors for 4days. After organoid formation, the media was switched to liver specification media for 4days. After the liver specification step, organoids were harvested from Matrigel by scratching and pipetting. Then organoids were re-embedded in Matrigel on the Ultra-low attached plate (Corning) in liver maturation media for 10 days. Cultures for HLO induction were maintained at 37 °C in 5% CO2 with 95% air and the medium was added every 2 days. (see Supplementary Methods)
Large scale screening with 238 test compounds
Hepatotoxicity library (Enzo SCREEN-WELL®); Each drug was diluted into 4 doses (100μM, 10μM, 1μM and 0.1μM) and dispersed in 384-well plates using Eppendorf epMotion® 5075 liquid handler. Each plate included on-board controls for toxicity (e.g. Penicillin V, Lactic acid) and delivery vehicle (DMSO). Human iPSCs (TkDA3) were differentiated into liver organoids as described previously. At Day 15, embedded liver organoids were re-plated in floating culture by disrupting Matrigel using gentle pipetting. At Day 21, floating cultured liver organoids were seeded at ~15 organoids per well into 384-well plates (Corning high content imaging plate, Cat.4681) which contained test compounds. CLF (5nM) was added before imaging at 24 hrs. Multi-channel fluorescence and bright field images were acquired by Nikon Ti-E SpectraX Widefield Microscope equipped with high-speed, triggerable Lumencor SpectraX LED light engine under 40x magnification. Imaging was processed in ~7 min per plate. Measurement of image intensity were obtained from each individual organoid (n = approximately 10). Cell viability was tested using the CellTiter-Glo® luminescent cell viability assay (Promega) and quantified by a BioTek® Synergy H1 plate reader after 72 hrs. Each experiment was repeated in triplicate. The data were given from five independent experiments.
Image data analysis.
Nikon Elements analysis software was used to identify fluorescent pixels expressed within organoids based on intensity gaiting, size, and circularity filtering. Fluorescent data is taken as an intensity average so to normalize for any changes in organoid size. Moreover, as each organoid imaged within a well contains its own analyzed fluorescent intensity data, an average and standard deviation of the intensity data across all organoids in a single well is then taken so to account for variation in organoid number. Although each HLO size were relatively uniform (Supplementary Figure. 5), to account for variation in HLO size and number during cell titer glow, a custom image analysis script was developed in Matlab using Canny edge detection, watershed segmentation, and filtering based on size and circularity to identify total cellular area per well as the normalizing factor for the luminescent intensity (CellTiter-Glo) data characterizing organoid viability. Pixels were regarded as comprising part of an organoid if a pixel appeared as positive (value of 1 in binary images) in greater than or equal to 2 sets of processed images. Each well’s CTG value was calculated as luminescent signal / organoid area. All data points reflected on the 2D or 3D plots were normalized to the control (DMSO) wells.
Statistical analysis.
Statistical significance was determined using unpaired Student’s t-test, Dunnett’s test or one-way ANOVA with Dunnett’s multiple comparison post-hoc test. P < 0.05 was considered significant.
Results
Generation and characterization of polarized liver organoids from multiple human iPSC
We first established a new method to effectively obtain uniform organoids in a large quantity using human iPSC-derived foregut (FG) spheroids 4 (Figure. 1A). Briefly, foregut cells are dissociated into single cells on day7. In this stage, dissociated foregut cells can be cryopreserved at −80 degrees. Fresh or thawed cells were embedded into Matrigel, followed by four days of organoid formation medium composed of 5 factors, which were FGF2, VEGF, EGF, a GSK-3 inhibitor (CHIR99021) and a TGF-β inhibitor (A83–01), with ascorbic acid (AA) 9 (Supplementary Figure. 1A–B). On the following day, we exposed the cultures to four days of retinoic acid (RA), which enhances the formation of the bile canaliculi and the pericanalicular sheaths 10. Next, we switched to hepatocyte maturation medium (See Methods), and organoids with intraluminal structure were efficiently produced (Figure. 1B) and 5 factors provided the larger number of HLOs than conventional factors, which was only treated with RA (Figure. 1C–D). We confirmed that HLO were formed from multiple iPSC lines (Supplementary Figure. 1C) under 5 factors. Furthermore, we demonstrate that HLO cultured in the organoid formation medium with 5 factors, then RA for 96 hours has the highest albumin secretion (Figure. 1E).
Gene expressions and functional profiling in HLOs
To profile hepatic lineage in HLO, the expressions of some representative genes related to hepatic functional liability were analyzed by the quantitative polymerase chain reaction (qPCR). The expression of hepatic marker genes was upregulated in HLO, such as albumin (ALB), alpha-fetoprotein (AFP), CYP2C9 and CYP7A1 (Figure. 1F). Genes related to polarity and transporter activities such as multidrug resistance-associated protein 2 (MRP2), bile salt export pump (BSEP), Multiple drug resistance 1 (MDR1), BCRP1, Na+-taurocholate co-transporting polypeptide (NTCP), organic anion transporter 2 (OAT2), concentrative nucleoside transporter-1 (CNT1) and MRP3 were also increased at their expressions. Moreover, this was accompanied by progressive decreases in genes related to the undifferentiated state, such as NANOG and OCT4. The gene expression of NANOG, OCT4, ALB, CYP2C9, MRP3 and TDO2 in HLOs generated from different 6 donors were comparable (Supplementary Figure. 2A). The albumin secretion capacity of HLO was shown in Figure. 1G. HLO and primary hepatocytes albumin secretion levels were 488.2±517.5 and 630.49±292.5, respectively, which were not statistically different. Although the ALB secretion levels in HLOs and primary liver were comparable, gene expression of ALB in both were different presumably due to different cell compositions in HLO and primary liver. In addition, to investigate the difference of HLOs generated from foregut with or without frozen process, the albumin secretion level in both HLOs were tested and comparable (Supplementary Figure. 2B), suggested that the influence of frozen process on foregut stage was not large for hepatic functions in HLO. Hepatocyte specific proteins such as complement factors were also confirmed in HLO culture supernatant by ELISA (Figure. 1H). Finally, at a functional level, we demonstrate the major CYP induction responses in HLO, i.e. CYP3A4 and CYP1A2 after treatment with Rifampicin and Omeprazol, respectively (Figure. 1/–J). Additionally, we tested CYP2C9 inducibility in HLO. After the treatment with Rifampicin CYP2C9 reactivity in HLO was comparable to that of primary hepatocytes (Figure. 1K).
Single cell RNA sequencing profiling in HLO.
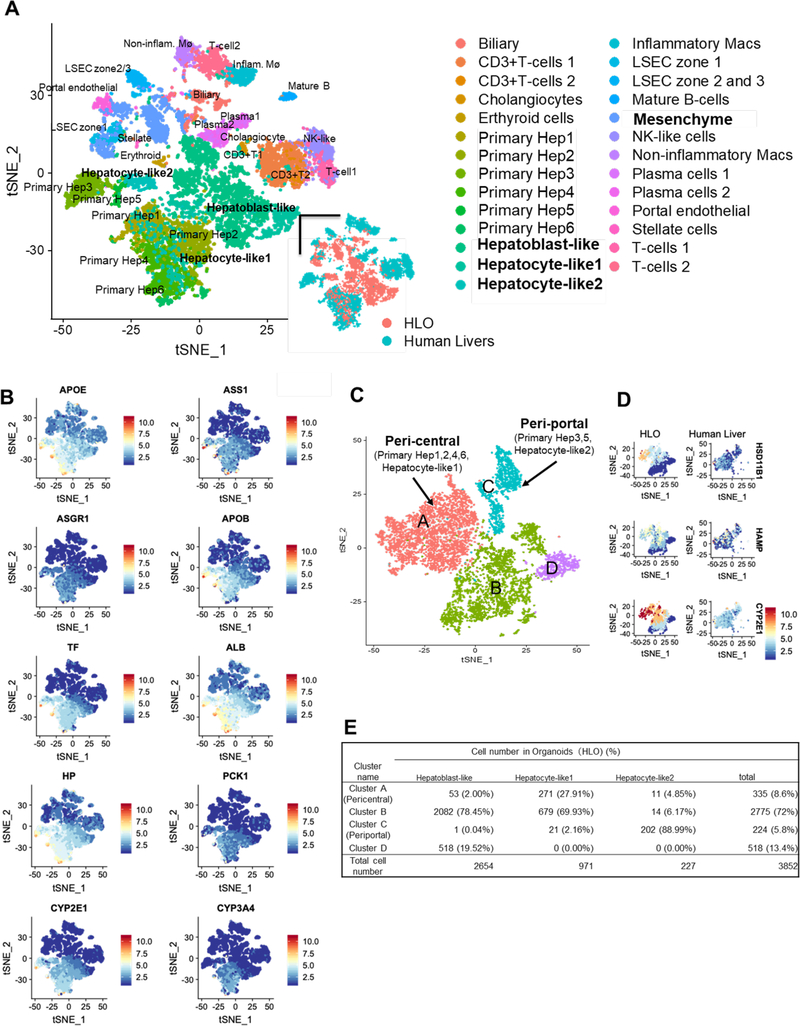
To compare the differentiation status with those of primary hepatocytes, we used single-cell RNA-seq (scRNA-seq) to measure the transcriptome of HLO derived cells (5177 cells) in HLO at day 20 and conducted integrated analysis with primary human liver-derived cells derived from independent 5 donors 11 (Figure. 2A and Supplementary Figure 3). t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis revealed distinct populations consisting of a parenchymal (74.41%), and a non-parenchymal population (25.59%) in HLO (Figure. 2B and 2C), which express characteristic markers of hepatic stellate cells, portal endothelial cells and cholangiocytes in primary samples as seen in our previous method 4. Next, we investigated gene expressions related to peri-central, peri-portal, mesenchyme and endothelial, and compared to those of human primary livers (Supplementary Figure 3A). Of them, although HLO derived cells include non-parenchymal population, subsets of hepatic cells, termed as hepatocyte-like1, 2 cells, are nearly identical to primary hepatocytes with remaining half of cells representing hepatoblast-like state. In the gene set of mesenchyme and endothelial, the gene expression in HLOs shared similarity to primary liver tissues. To further probe the hepatocytic identity, we isolated the hepatocytic population from both HLO and primary liver samples and conducted tSNE analysis. 88.99% of hepatocyte-like2 cells were in the peri-portal primary hepatocyte population, whereas 27.91% of hepatocyte-like1 cells were in the peri-central primary hepatocyte population (Figure. 2D–E). Zone 3 (centrilobular region in the liver) play a role in drug metabolism and detoxification. Four zonal peri-central markers, recently reported in human scRNA-seq 11, are notably expressed at similar levels to primary hepatocytes which include a key drug metabolism enzyme, CYP2C9 (Figure. 1F). Pathway analysis indicated highly enriched gene sets in peri-central and peri-portal population in HLO correlate with lipid/drug metabolism, cytochrome P450 and cholesterol biosynthetic process, respectively. Gene expression levels in these pathways in HLOs were similar to those in primary liver (Supplementary Figure. 3B). Collectively, our human liver organoid model harbors diverse and zonal hepatocytic populations that in part emulate primary adult hepatocyte profiles.
Figure 2. Single cell RNA sequencing profiling of hepatic cells in HLO.
A. Integrated tSNE map of 5177 HLO cells and 8439 primary human liver cells. Each point indicates a single cell which are colored by cell type (upper and right bottom) or original source.
B. In integrated tSNE map of HLO and primary hepatocytes, the feature plots showed expression of hepatic marker genes. Color bar indicates scaled gene expression level.
C. Integrated tSNE map and clustering of 3852 hepatoblast-like and hepatocyte-like cells in HLO and 3507 hepatocytes in primary human liver identify zonal characters in A and C. The inset indicates cell cycle phase of each cell.
D. The feature plots showed expression of four zonal peri-central markers of hepatocytic clusters.
E. The table shown percentage of hepatoblast-like and hepatocyte-like cell of HLO in each cluster.
Structural profiling with micro-anatomical characterization in HLOs
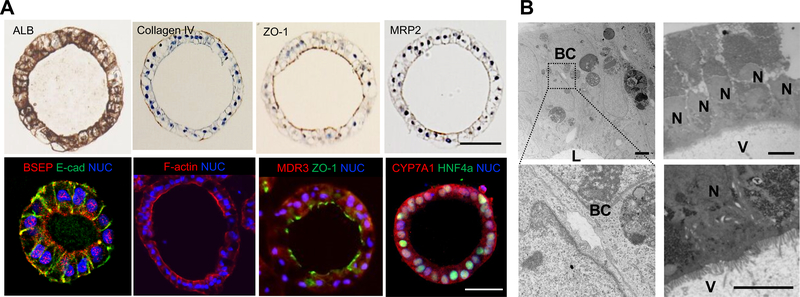
Immunohistochemistry analysis revealed albumin and type IV collagen staining in the epithelial cells of HLO (Figure. 3A). In addition, immunostaining of ZO-1(zonula occludens), MRP2, BSEP, F-actin and MDR3 demonstrated that these proteins preferentially localized in the intraluminal region with staining hepatocyte nuclear factor-4a (HNF4a). CYP7A1 was also positively stained in HLO. The bile canaliculus is the smallest intrahepatic secretory channel and the canalicular lumen consists of a space formed by a modified apical region of the opposing plasma membranes of contiguous hepatocytes 12. In addition, it is delimited by tight junction complexes and the microvilli are located on the inside of the canalicular lumen 12. Similarly, transmission electron microscopy (TEM) analysis of HLO confirmed the bile canaliculus-like structure in between the hepatocyte-like cells (Figure. 3B. Left). TEM analysis also revealed HLO contained microvilli directed towards the lumen (Figure. 3B. Right). Given with immunohistochemistry analysis resulting that MDR3, MRP2, BSEP and ZO-1 stained the intraluminal lining, it was suggested that these HLO have polarized characteristics. Consistent with these anatomical features, qRT-PCR analysis revealed that HLO had gene expression of BSEP and NTCP (Figure. 1F). Therefore, the HLO contained polarized human hepatocytes separated from the internal lumen surrounded by canaliculi-like structures, which reflects a unique micro-anatomical architecture resembling in vivo hepatic tissues.
Figure 3. Structural profiling of HLO.
A. Immunostaining for Albumin (ALB), Collagen IV, ZO-1, MRP2, E-cadherin (E-cad), BSEP, F-actin, MDR3, CYP7A1 and HNF4a in HLO. Nuclei were stained with DAPI (blue). Bars, 50 μm.
B. Transmission electron micrograph of an HLO showing microvilli (V) intra-luminal surface; V: Microvilli, BC: Bile canaliculi-like structure, L: Lumen, N: nuclei. Bars, 10 μm.
Bile acid producing and transport properties in HLOs
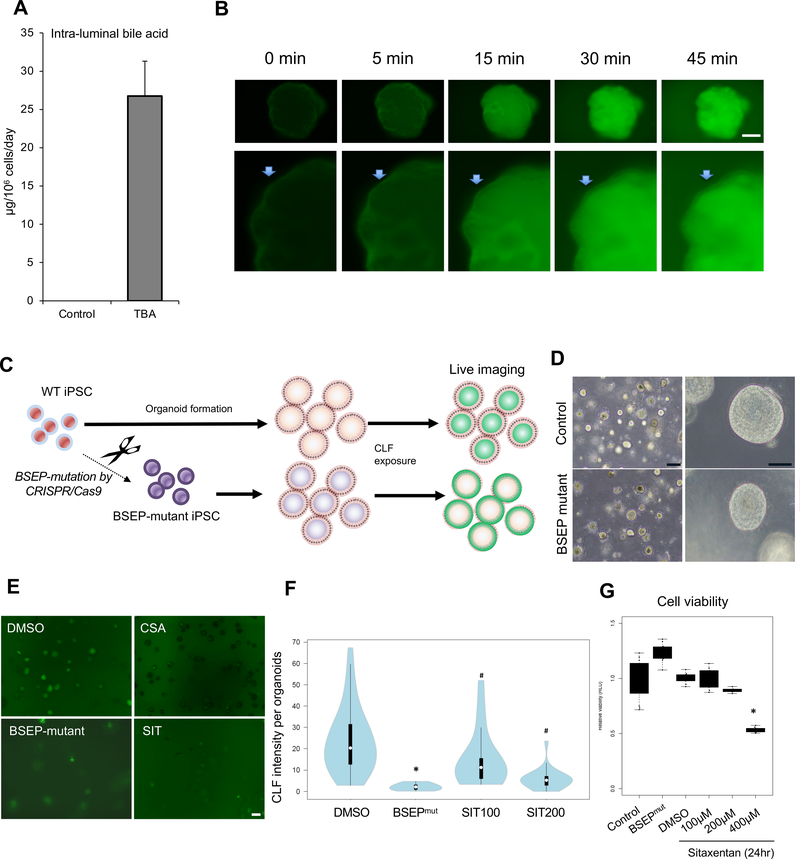
Next, in order to determine bile acid (BA) production capacity, we conducted a BA ELISA on intra-luminal fluid collected from organoid cultures. As Figure. 1F and Figure. 3A, CYP7A1 which catalyzes the initial step in cholesterol catabolism and bile acid synthesis, was expressed and stained positively in HLO, respectively, suggesting the activation of BA synthesis pathway in HLO. The level of total BA pool of intra-luminal fluid was 26.7 μg/day/106 cells (approximately 125 μmol/L in single HLO with a 200 μm diameter) (Figure. 4A) and, surprisingly, the BA concentration was comparable to that in primary hepatocytes derived from sandwich culture (approximately 40 μg/day/106 cells, 10 μmol/L in culture supernatant) in previous reports 13. Bile acid excretion is the major determinant of bile flow, therefore, defects in this system may result in impaired bile secretion (cholestasis) associated with various liver disease pathologies. Efflux transport proteins located in the apical (canaliculi) membranes of hepatocytes play an important role in the hepatic elimination of many endogenous and exogenous compounds, including drugs and metabolites 14. BSEP and MRP2 mediate canaliculi bile salt transport in humans. As the positive expression of key proteins for bile transport in Figure. 3A, we next wondered if the HLO can actively transport bile acid into its lumen. To monitor the dynamics on bile transport activity in HLO, fluorescein diacetate (FD), which was used to investigate an active transport mechanism from the sinusoidal membrane to the bile canalicular membrane in vitro15, was applied. HLO incubated with FD demonstrated that FD was transported into inside HLO with changing the intensity sequentially in 45 min (Figure. 4B). In addition, the fluorescent bile acid cholyl-lysyl-fluorescein (CLF) and cholylglycylamido-fluorescein (CGamF), an indicator for bile acid transport activity 16 and a bile salt analog 17, respectively, were found to be reproducibly excreted from outside and accumulated into the intra-lumen of HLO (Supplementary Figure. 4A–B). To determine the specificity deeper, we have developed an iPSC line carrying a defunctionalized BSEP allele constructed by the CRISPR-Cas9 based gene editing approach (Figure. 4C). Although the morphology of BSEP-mutated iPSC-HLO was similar to control HLO (Figure. 4D), BSEP-mutated HLO failed to accumulate fluorescent bile acid compared with parental control HLO (Figure. 4E). Similarly, chemical BSEP inhibitor sitaxentan at viable dosing inhibited CLF transport in normal HLO (Figure. 4F–G). These data suggest that HLO have the ability to uptake bile acid from the outside and efflux them inside the HLO. Thus, HLO do not merely have the canaliculi-like morphology but also possess bile acid production and secretion activity, suggesting that the bile acids transport pathway is functionally constructed.
Figure 4. Bile transport property in HLO.
A. Total bile acid (TBA) secretion level inside HLO at day 27. Bars represent the mean ± SEM, n=4.
B. Sequential images for efflux of FD toward the inside of the organoid from outside. Arrow: Surface of an organoid. Bar = 50 μm.
C. Overview of the method to investigate transporter BSEP activity in organoids.
D. Representative phase-contrast image of BSEP-mutated HLO. Bar = 200 μm (Left image), 50 μm (Right image).
E. Representative image of bile acid uptake in BSEP-mutated HLO, CSA and sitaxentan (SIT) which were BSEP inhibitors after 30 min of culture in the presence of CLF. Bar = 200 μm
F. Graph indicated CLF intensity levels in each HLO. White circles show the medians: box limits indicate the 25th and 75th percentiles as determined by R software: whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles: polygons represent density estimates of data and extend to extreme values. BSEPmut: BSEP mutant. SIT100: sitaxentan 100 μM, SIT200: 200 μM. *: p <0.01 on t-test. #: p <0.05 on Dunnett’s test.
G. The viability of HLO. *: <0.01 on Dunnett’s test.
High-throughput drug induced cholestasis evaluation in HLO
During drug discovery, in the early stages of DILI investigation, functional and quantifiable assays are critical for lead compound generation. However, current models suffer from oversimplified readouts such as only viability despite multiple factors being involved such as bile transport defects. To address these limitations, we developed a 384 well based high-speed live imaging platform that we refer to as a human liver organoid (HLO)-based high throughput toxicity screening (LoT) model (Figure. 5A). Our LoT system can evaluate 15–20 organoids per well with a unique imaging algorithm (Figure. 5B) and has functionally validated 238 marketed drugs including 32 negative control and 206 reported DILI compounds with 4 different concentrations based on dual readouts: viability and cholestatic function. In the system, CLF transport inhibition induced by representative cholestatic drugs was observed. First, we screened the viability in response to the treatment at multiple doses comparing with previous reports (See Supplementary Table 1). To evaluate the predictability of LoT system, we extracted clinical maximum drug concentration (Cmax) value of each compound listed in Supplementary Table 118–20 and analyzed the data at a concentration closest over Cmax. As a result, the sensitivity and specificity were 88.7% and 88.9% (or 69.0% and 100%), respectively, and provided the comparable or higher values to previous reports on primary hepatocyte-based models 18, 20–22 (Supplementary Table 2 and Figure. 5C). Interestingly, Indomethacin (Cmax: 8.38 μM) and Zileuton (Cmax: 13.12 μM) induced toxicity were successfully detected at 10 μM and 100 μM in cytotoxicity assay, respectively, while these drugs were previously difficult to be detected in other platforms (27, 28). Of them, drugs known to induce cholestasis tended to reduce CLF intensity, suggesting that significant bile transport inhibition effects can be modeled in organoid imaging analysis (Supplementary Table 3 and Figure. 5D). Together, our LoT system developed here is potentially available for the early drug discovery process due to its throughput and relevance to human data.
Figure 5. Large scale-organoid based toxicity (LoT) screen identifies compounds with toxic potential.
A. Schematic of the DILI screen workflow with image of quantification strategy for CLF and cell viability.
B. Representative images of 384-well drug screen plate with quantification of CLF at 24 hours. CLF mono-fluorescence converted to intensity plot by imageJ. Right upper image: negative compound (sucrose), right bottom image: positive compounds (CSA). See Supplementary Table 1.
C. Violin plot of CLF intensity in HLO treated with compounds relative to control.
D. Scatter plot of the viability and CLF intensity. Sen: sensitivity (true positive / true positive + false negative), Spe: specificity (true negative / true negative + false positive). Circle dots: the data of DILI positive compounds. Triangle dot: the data of DILI negative compounds. n = 5.
E. CLF intensity of representative drugs categorized into Negative and Cholestatic groups.
Revisiting mechanistic classification of DILI compounds by LoT system
We next set out to establish in-depth investigation model with the use of organoids. To monitor MMP mitochondrial health alongside with bile transport, we multiplexed the readouts in the intact organoids. After defining an optimal dose for mechanistic study of 10 FDA approved drugs that does not affect viability (Supplementary Results and Supplementary Figure 6), we quantified the mitochondrial health assessment by MMP. After treatment of TCs for 24h, dose-dependent increases in MMP were observed with treatment of Tolcapone (2–8 fold change, p < 0.01), Diclofenac (7–13 Fold change, p < 0.05 or 0.01), CSA (3–7 fold change, p < 0.01) and Nefazodone (4–42 fold change, p < 0.01) (Figure. 6A–C). In addition, Troglitazone also increased MMP in HLO (3–5 fold change, p < 0.05), although dose-dependence was not observed. On the other hand, after treatment of Bosentan, Entacapone and Pioglitazone, increases in MMP were not clearly observed even in multiple doses. Severe manifestations of human DILI are multifactorial, and are highly associated with combinations of known mechanisms of DILI such as mitochondrial and BSEP inhibition 23. We further analyzed the relationship among survival, cholestasis and mitochondrial stress. To benchmark our toxicity assay against the conventional in vitro assay systems, we chose the TCs and their concentrations based on published reports focused on cholestatic toxicity and mitochondrial stress using primary hepatocyte systems 24, 25. Of note, drugs with dual potency at 24 hours (cholestasis and mitochondrial stress) such as CSA, TRO and NEFA significantly lowered cell viability at 72 hours relative to TOL, DICLO and BOS (Figure. 6D and Supplementary Figure 6A). These data are comparable to clinical data demonstrating that dual potencies were highly associated with the severity of DILI consistent in previous reports23. Additionally, we also note that Entacapone treatment at 130 μM decreased organoid viability (from 85% at 24h to 64% at 72h). Entacapone requires extensive binding to plasma proteins, mainly to albumin, to induce DILI26. It would be interesting to further investigate the mechanisms of Entacapone toxicity beyond cholestasis and mitochondrial health using our LoT system, as toxicity mechanisms are ill-defined. Taken together, the LoT system is a useful human model system for the major mechanistic classifications of DILI, and a testing platform for further delineating unknown complex mechanisms.
Figure 6. Mechanistic toxicological approach using DILI HLO model.
A. Upper: Image of FD transport inhibition after treatment of 9 training compounds for 5 min. Bottom: Image of mitochondria membrane potential (MMP) on TMRM after treatment of 9 training compounds (TC).
B. Quantification of transport inhibition after treatment of TC, Bars represent the mean ± SD, *: p<0.05, **: p<0.01, n= 4–6. C. Quantification of MMP change after treatment of TC, Bars represent the mean ± SD, *: p<0.05, **: p<0.01, n= 4–5. CON: Control sample, STP: Streptomycin, TOL: Tolcapone, DICLO: Diclofenac, BOS: Bosentan, CSA: Cyclosporin A, TRO: Troglitazone, NEFA: Nefazodone, ENTA: Entacapone, PIO: Pioglitazone.
C. Quantification of transport inhibition after treatment of training compounds, Bars represent the mean ± SD, *: p<0.05, **: p<0.01, n= 4–6. C. Quantification of MMP change after treatment of training compounds, Bars represent the mean ± SD, *: p<0.05, **: p<0.01, n= 4–5. CON: Control sample, STP: Streptomycin, TOL: Tolcapone, DICLO: Diclofenac, BOS: Bosentan, CSA: Cyclosporin A, TRO: Troglitazone, NEFA: Nefazodone, ENTA: Entacapone, PIO: Pioglitazone.
D. Analysis between viability for 72h after treatment of drugs and dual risk parameters, drug-induced cholestasis potential and mitochondria toxicity potential. Cholestasis and Mitochondria toxicity (Mito-tox) indexes were derived from data in Figure. 6 B–C. The size of circles indicated the magnitude of viability decreases.
E. Overview of evaluation of drug-induced cytotoxicity on vulnerable organoid model. Profiling of vulnerable model on lipid accumulation (Blue: nuclei, Green: Lipid, Red: F-actin)
F. ROS production (Blue: nuclei, Green: ROS) and Mitochondria detection (Blue: nuclei, Red: Mitochondria). Image of organoids at 24h after drugs treatment.
G. Viability assessment on lipid accumulation-induced vulnerable organoid model. Bars represent the mean ± SD, *: p<0.05, n= 5–6. CON: control, STP: Streptomycin, TRO: Troglitazone, PIO: Pioglitazone, NAC: N-acetylcysteine.
DILI incidence is known to be often confounded by a number of host factors. For instance, growing evidence suggested that obesity and nonalcoholic fatty liver disease greatly increase the risk of hepatotoxicity both in rodents and humans when combined with certain drugs such as acetaminophen (APAP) 27, 28. Therefore it seems important to foresee DILI potential in such a “vulnerable” condition with a patient even in the subclinical phase. Here, we established a lipotoxic organoid model by exposure to an unsaturated fatty acid, oleic acid (Figure. 6E). At 3 days after oleic acid treatment lipid accumulation in HLO was intense (Figure. 6E). The oxidation of fatty acids is an important source of reactive oxygen species (ROS), which leads to depletion of ATP and nicotinamide dinucleotide, and induces DNA damage in fatty livers 29. Consistent with this, ROS production was enhanced in lipid treated HLO (Figure. 6F and Supplementary figure. 7A). Additionally, fatty acids induced massive swelling of liver mitochondria (Figure. 6F and Supplementary figure 7B) similar to a published phenotype 30. Since this lipotoxic organoid model is a vulnerable condition with increased ROS, HLO were treated with two thiazolidinediones, Troglitazone (0–50 μM) and Pioglitazone (200 μM) for 24h and cell viability was assessed. We observed massive fragmentation of organoids due to cell death after treatment of Troglitazone during lipotoxic conditions (Supplementary figure 8), confirmed by subsequent cell viability analysis (−40 % cell viability compared to control, p < 0.05) (Figure. 6G). As treatment of Pioglitazone alone did not affect viability in HLO with/without lipotoxic condition, lipotoxic HLO possibly highlights DILI vulnerability with positive and negative predictive power (Figure. 6G).
Next, we investigated whether HLO can recover from DILI-like conditions using a compound with therapeutic potential. As a proof-of-concept, we used N-acetylcysteine (NAC), an antioxidant, to inhibit ROS production based on literature reports that intravenous NAC improves survival in patients with non-acetaminophen-related acute liver failure 31 and diminished the Troglitazone-induced cytotoxicity 32. As expected, cell viability was significantly improved by NAC, suggesting that NAC rescued cell death in HLO even in vulnerable conditions (Figure. 6G and Supplementary figure 8). This LoT system potentially serves as a preclinical tool in identifying an effective compound in alleviating DILI-like condition under multi-drug regimens.
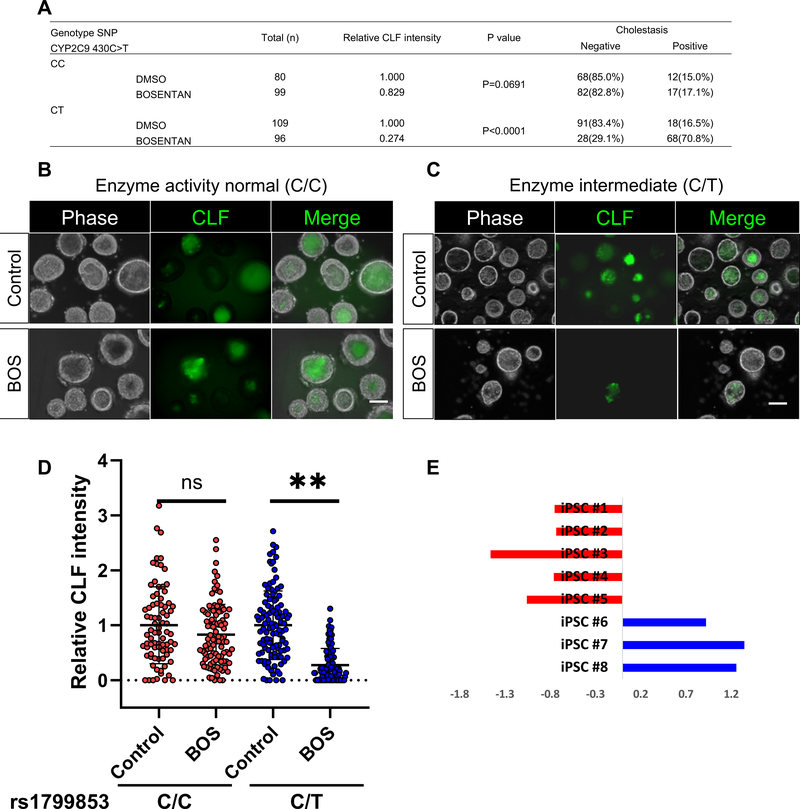
Bosentan induced cholestasis specific to CYP2C9*2 iPSC-liver organoids
As the drug induced events are highly variable, an iPSC based organoid approach is promising for the potential assessment of individual susceptibility5. To determine the clinical relevance of our system, we employed pharmacogenomic insights that affect drug-induced bile transport inhibition potential. We genotyped 8-different iPSC lines with/without the well-known susceptibility gene variant (i.e. CYP2C9*2 activity intermediate) to the Bosentan induced DILI, and compared the HLO lines’ cholestatic potential. Interestingly, CLF excretion into HLO was severely impaired (Positive rate: CC, 17.1% of positive, CT, 70.8% of positive) by Bosentan in CY2C9*2 carrier HLO but not in 3-different iPSC derived HLO without CYP2C9*2 (Figure. 7A–E). These results indicate that the organoid based cholestasis assay both negatively and positively predicts CYP2C9 mediated variation for drug-induced cholestasis as seen in humans33. These results indicate that the organoid based cholestasis assay predicts some aspects of human genetic variation for drug-induced cholestasis.
Figure 7. Bosentan induced cholestasis is specific to CYP2C9*2 HLO.
A. The table indicates the possession of Enzyme activity normal(C/C) / intermediate (C/T) alleles, CYP2C9*2 to Bosentan-induced liver injury in iPSC lines and the ratio of CLF efflux difference in each group.
B. Images of CLF transport activity and inhibition by Bosentan in HLO derived from donor with enzyme intermediate allele. Bar = 100 μm.
C. Images of CLF transport activity and inhibition by Bosentan in HLO derived from donor with enzyme active allele. Bar = 100 μm.
D. CLF intensity levels in HLOs derived from iPSC lines. **: p <0.0001, t-Test. NS: not significant. In the box plots, the top and bottom of the box represent the 75th and 25th percentiles, the center line represents the median. Dot indicates the data from each organoid.
E. CLF intensity levels in organoids from 8 iPSC lines. The data were shown as Z-score in each sample.
Discussion
Human adult stem cell derived liver organoids 34, 35 restores mature hepatocyte profiles which can repopulate in vivo injured liver. The morphology of HLO with lumen and bile canaliculi-like structure within cells share dissimilarity and similarity to primary organoids. For example, iPSC-HLO exhibits hollow-like structure with large lumen, whereas primary tissue contains cell chords with chicken wire appearance of canaliculi. Nevertheless, both systems contain the canalicular lumen consisting a space formed by a modified apical region of the opposing plasma membranes of contiguous hepatocytes11, 12, and distribution of tight junction complexes and the microvilli located on the inside of the lumen. Gene editing and pharmco-inhibitor assays confirm internalization of the bile acid analogues into HLO utilizes BSEP, which is a known transporter for bile acid excretion. Given that half of our hepatocyte like cells in HLO display an immature status by scRNA-seq, PSC-derived HLO exhibited relatively immature transcriptome signatures at bulk compared to adult liver tissue derived organoid based approaches similar to other published literatures36, 37. Interestingly, it is known that the human fetal liver has a functional drug metabolizing enzyme system even in the early gestational period, this unique characteristic is remarkably specific to human but not animal species 38. Additionally, our scRNA-seq identified subsets of the populations in HLO represents Zone1 (peri-portal region in the liver) and Zone3 (centrilobular region in the liver) hepatocyte-like cells. Differential signatures between immature versus mature zonal hepatocytes in HLO warrant future investigations so as to pinpoint signaling logics to continue the maturation of hepatocyte-like cells in HLO.
The major advantages of the LoT assay include: the use of patient’s iPSC, the storable feature at foregut stage, assay throughput, and the multiplexed readouts for analyzing interplay between other factors such as mitochondrial stress. As mentioned above, retrospective studies revealed that multiple cell stress potentials were associated with the incidence of DILI 23 and the LoT assay showed comparable results with that study, since cell viability was decreased dependent on dual readouts; mitochondrial and cholestatic stress. Oxidative stress plays an important role in cell death and has been linked to the development of cholestatic liver injury 39. Hydrophobic bile acids accumulate intracellularly during cholestasis and interfere with normal mitochondrial electron transport, inhibiting the activity of respiratory complexes I and III and consequently reducing adenosine triphosphate synthesis 40, resulting mitochondrial dysfunction-induced apoptosis 41. In line with these findings, our correlational analysis of these dual readouts indicated cholestatic stress was the more dominating factor for liver injury compared with mitochondria stress. Notably, different susceptibilities to Bosentan that were associated with gene variants were recapitulated by our LoT system using multi-donor iPSC-derived HLO. Due to difficulty and inefficiency in primary liver tissue sampling34 particularly from healthy individuals without diseases, iPSC-derived HLO generated from healthy donors will be a valuable tool for precision studies as was preliminarily demonstrated here with CYP2C9*2 carrier identification. More importantly, recently evolving large scale iPSC bank with gene-sequencing data will enable an imaginable cohort coupled with genomic stratification 42. LoT might serve as an exciting strategy for the pharmaceutical industry by providing essential insights to minimize the potential for DILI.
Limitations for this study
LoT system is currently oversimplified model that lacks adaptive immune components. For this reason, to fully predict other types of DILIs including idiosyncratic-DILI and immune mediated-DILI observed in patients, additional modification is crucial. In addition, the present study was conducted at a single institution. To facilitate the application into preclinical toxicology analysis, 384 well based LoT screening assay needs to be validated in multiple independent institutions with blinded compound testing to further enhance reproducibility and credibility. Although CYP2C9 genetic variation dependent cholestasis exacerbation was shown within over 8 donors LoT assay, this interpretation cannot exclude the possibility of donor-dependent differences rather than genotype-dependence. To formerly prove the causative relationship, isogenic base editing approach in iPSC will be highly informative.
Supplementary Material
What You Need to Know.
Background and Context
Preclinical identification for drug induced liver injury (DILI) represents a major challenge due to complex pathogenesis and highly variable individual susceptibility to drug.
New Findings
We established a human liver organoid (HLO) based screening model for analyzing DILI pathology and provided the possibility to assess different susceptibility based on polymorphism at organoid resolution.
Limitations
This study was performed in at a single institution with limited compound numbers.
Impact
Self-organized HLO with human hepatocyte-like properties including bile transport function predicts known cholestatic DILI drug activity, and have potential to predicts genomic predisposition for DILI.
Acknowledgments
The authors would like to express their sincere gratitude to Drs Jim Wells, Jorge Bezerra, Aaron Zorn and their lab members for their support and excellent technical assistance, and Asuka Kodaka for graphical abstract illustration and Mary Koch for kind administrative/technical assistance. This work was supported by Cincinnati Children’s Research Foundation grant, Ohio TVSF grant, CCHMC Innovation Acceleration fund and PRESTO grant from Japan Science and Technology Agency (JST) to TT. This work was also supported by an NIH grant UG3 DK119982, Cincinnati Center for Autoimmune Liver Disease Fellowship Award, PHS Grant P30 DK078392 (Integrative Morphology Core and Pluripotent Stem Cell and Organoid Core) of the Digestive Disease Research Core Center in Cincinnati, Takeda Science Foundation award, Mitsubishi Foundation award and AMED JP19fk0210037, JP19bm0704025, JP19fk0210060, JP19bm0404045, and JSPS JP18H02800, 19K22416. TT is a New York Stem Cell Foundation – Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Takebe T, Taniguchi H. Human iPSC-derived miniature organs: a tool for drug studies. Clin Pharmacol Ther 2014;96:310–3. [DOI] [PubMed] [Google Scholar]
- 2.Yang K, Woodhead JL, Watkins PB, et al. Systems pharmacology modeling predicts delayed presentation and species differences in bile acid-mediated troglitazone hepatotoxicity. Clin Pharmacol Ther 2014;96:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leslie EM, Watkins PB, Kim RB, et al. Differential inhibition of rat and human Na+-dependent taurocholate cotransporting polypeptide (NTCP/SLC10A1)by bosentan: a mechanism for species differences in hepatotoxicity. J Pharmacol Exp Ther 2007;321:1170–8. [DOI] [PubMed] [Google Scholar]
- 4.Ouchi R, Togo S, Kimura M, et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inoue H, Nagata N, Kurokawa H, et al. iPS cells: a game changer for future medicine. EMBO J 2014;33:409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takebe T, Enomura M, Yoshizawa E, et al. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell Stem Cell 2015;16:556–65. [DOI] [PubMed] [Google Scholar]
- 7.Takebe T, Zhang RR, Koike H, et al. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat Protoc 2014;9:396–409. [DOI] [PubMed] [Google Scholar]
- 8.Spence JR, Mayhew CN, Rankin SA, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang RR, Koido M, Tadokoro T, et al. Human iPSC-Derived Posterior Gut Progenitors Are Expandable and Capable of Forming Gut and Liver Organoids. Stem Cell Reports 2018;10:780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falasca L, Favale A, Serafino A, et al. The effect of retinoic acid on the re-establishment of differentiated hepatocyte phenotype in primary culture. Cell Tissue Res 1998;293:337–47. [DOI] [PubMed] [Google Scholar]
- 11.MacParland SA, Liu JC, Ma XZ, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukada N, Ackerley CA, Phillips MJ. The structure and organization of the bile canalicular cytoskeleton with special reference to actin and actin-binding proteins. Hepatology 1995;21:1106–13. [PubMed] [Google Scholar]
- 13.Ni X, Gao Y, Wu Z, et al. Functional human induced hepatocytes (hiHeps) with bile acid synthesis and transport capacities: A novel in vitro cholestatic model. Sci Rep 2016;6:38694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kock K, Brouwer KL. A perspective on efflux transport proteins in the liver. Clin Pharmacol Ther 2012;92:599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yumoto AU, Watanabe S, Hirose M, et al. Structural and functional features of bile canaliculi in adult rat hepatocyte spheroids. Liver 1996;16:61–6. [DOI] [PubMed] [Google Scholar]
- 16.Keitel V, Burdelski M, Vojnisek Z, et al. De novo bile salt transporter antibodies as a possible cause of recurrent graft failure after liver transplantation: a novel mechanism of cholestasis. Hepatology 2009;50:510–7. [DOI] [PubMed] [Google Scholar]
- 17.Mork LM, Isaksson B, Boran N, et al. Comparison of culture media for bile Acid transport studies in primary human hepatocytes. J Clin Exp Hepatol 2012;2:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khetani SR, Kanchagar C, Ukairo O, et al. Use of micropatterned cocultures to detect compounds that cause drug-induced liver injury in humans. Toxicol Sci 2013;132:107–17. [DOI] [PubMed] [Google Scholar]
- 19.O’Brien PJ, Irwin W, Diaz D, et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch Toxicol 2006;80:580–604. [DOI] [PubMed] [Google Scholar]
- 20.Vorrink SU, Zhou Y, Ingelman-Sundberg M, et al. Prediction of Drug-Induced Hepatotoxicity Using Long-Term Stable Primary Hepatic 3D Spheroid Cultures in Chemically Defined Conditions. Toxicol Sci 2018;163:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proctor WR, Foster AJ, Vogt J, et al. Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch Toxicol 2017;91:2849–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu JJ, Henstock PV, Dunn MC, et al. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol Sci 2008;105:97–105. [DOI] [PubMed] [Google Scholar]
- 23.Aleo MD, Luo Y, Swiss R, et al. Human drug-induced liver injury severity is highly associated with dual inhibition of liver mitochondrial function and bile salt export pump. Hepatology 2014;60:1015–22. [DOI] [PubMed] [Google Scholar]
- 24.Oorts M, Baze A, Bachellier P, et al. Drug-induced cholestasis risk assessment in sandwich-cultured human hepatocytes. Toxicol In Vitro 2016;34:179–186. [DOI] [PubMed] [Google Scholar]
- 25.Bort R, Ponsoda X, Jover R, et al. Diclofenac toxicity to hepatocytes: a role for drug metabolism in cell toxicity. J Pharmacol Exp Ther 1999;288:65–72. [PubMed] [Google Scholar]
- 26.Fisher A, Croft-Baker J, Davis M, et al. Entacapone-induced hepatotoxicity and hepatic dysfunction. Mov Disord 2002;17:1362–5; discussion 1397–1400. [DOI] [PubMed] [Google Scholar]
- 27.Fromenty B Drug-induced liver injury in obesity. J Hepatol 2013;58:824–6. [DOI] [PubMed] [Google Scholar]
- 28.Michaut A, Le Guillou D, Moreau C, et al. A cellular model to study drug-induced liver injury in nonalcoholic fatty liver disease: Application to acetaminophen. Toxicol Appl Pharmacol 2016;292:40–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest 2004;114:147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zborowski J, Wojtczak L. Induction of Swelling of Liver Mitochondria by Fatty Acids of Various Chain Length. Biochim Biophys Acta 1963;70:596–8. [DOI] [PubMed] [Google Scholar]
- 31.Lee WM, Hynan LS, Rossaro L, et al. Intravenous N-acetylcysteine improves transplant-free survival in early stage non-acetaminophen acute liver failure. Gastroenterology 2009;137:856–64, 864 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rachek LI, Yuzefovych LV, Ledoux SP, et al. Troglitazone, but not rosiglitazone, damages mitochondrial DNA and induces mitochondrial dysfunction and cell death in human hepatocytes. Toxicol Appl Pharmacol 2009;240:348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markova SM, De Marco T, Bendjilali N, et al. Association of CYP2C9*2 with bosentan-induced liver injury. Clin Pharmacol Ther 2013;94:678–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015;160:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu H, Gehart H, Artegiani B, et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018;175:1591–1606 e19. [DOI] [PubMed] [Google Scholar]
- 36.Zachos NC, Kovbasnjuk O, Foulke-Abel J, et al. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J Biol Chem 2016;291:3759–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dedhia PH, Bertaux-Skeirik N, Zavros Y, et al. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology 2016;150:1098–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rane A, Sjoqvist F. Drug metabolism in the human fetus and newborn infant. Pediatr Clin North Am 1972;19:37–49. [DOI] [PubMed] [Google Scholar]
- 39.Serviddio G, Pereda J, Pallardo FV, et al. Ursodeoxycholic acid protects against secondary biliary cirrhosis in rats by preventing mitochondrial oxidative stress. Hepatology 2004;39:711–20. [DOI] [PubMed] [Google Scholar]
- 40.Krahenbuhl S, Talos C, Fischer S, et al. Toxicity of bile acids on the electron transport chain of isolated rat liver mitochondria. Hepatology 1994;19:471–9. [DOI] [PubMed] [Google Scholar]
- 41.Bernardi P The permeability transition pore. Control points of a cyclosporin A-sensitive mitochondrial channel involved in cell death. Biochim Biophys Acta 1996;1275:5–9. [DOI] [PubMed] [Google Scholar]
- 42.Koido M, Kawakami E, Fukumura J, et al. Polygenic architecture informs potential vulnerability to drug-induced liver injury. Nature Medicine 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.