Abstract
Background & Aims
Liver CRIg+ (complement receptor of the immunoglobulin superfamily) macrophages play a critical role in filtering bacteria and their products from circulation. Translocation of microbiota-derived products from an impaired gut barrier contributes to the development of obesity-associated tissue inflammation and insulin resistance. However, the critical role of CRIg+ macrophages in clearing microbiota-derived products from bloodstream in the context of obesity is largely unknown.
Methods
We performed studies with CRIg−/−, C3−/−, cGAS−/−, and their wildtype littermate mice. CRIg+ macrophage population and bacterial DNA abundance were examined in both mouse and human liver by either flow cytometric or immunohistochemistry analysis. Gut microbial DNA containing extracellular vesicles (mEVs) were adoptively transferred into CRIg−/−, C3−/−, or WT mice, and tissue inflammation and insulin sensitivity were measured in these mice. After cocultured with gut mEVs, cellular insulin responses and cGAS/STING-mediated inflammatory responses were evaluated.
Results
Gut mEVs can reach metabolic tissues in obesity. Liver CRIg+ macrophages efficiently clear mEVs from the bloodstream through a C3-dependent opsonization mechanism, while obesity elicits a marked reduction in CRIg+ macrophage population. Depletion of CRIg+ cells results in the spread of mEVs into distant metabolic tissues and subsequently exacerbating tissue inflammation and metabolic disorders. Additionally, in vitro treatment of obese mEVs directly triggers inflammation and insulin resistance of insulin target cells. Depletion of microbial DNAs blunts the pathogenic effects of intestinal EVs. Furthermore, cGAS/STING pathway is crucial for microbial DNA-mediated inflammatory responses.
Conclusions
Deficiency of CRIg+ macrophages and leakage of intestinal EVs containing microbial DNA contribute to the development of obesity-associated tissue inflammation and metabolic diseases.
Keywords: microbial DNA, extracellular vesicle, obesity, tissue inflammation
Introduction
Insulin resistance is an antecedent, pathophysiological defect in the great majority of patients with Type 2 diabetes mellitus (T2DM) 1, 2. Obesity is the main driver of insulin resistance in humans worldwide 3. The ongoing obesity epidemic is driving a parallel rise in the prevalence of T2DM. It has become clear that obesity-induced chronic, subacute tissue inflammation, particularly when it occurs in adipose tissue and liver, can cause insulin resistance 4,5.
Obesity is characterized by an impaired and defective gut barrier which results in the translocation of microbiota-derived products into the circulation and distant organs of host 6-10. The microbiota component and function change dramatically with diet and the development of obesity 11. Recent studies have highlighted the pathogenic effects of obesity-associated microbial metabolites on the development of tissue inflammation and metabolic disorders 12-17. In addition to microbial metabolites, emerging evidence indicates that microbial DNAs are enriched in the circulation and various metabolic tissues of both obese humans and mice 18-22. Recent studies suggest that these circulating microbial DNAs can be biomarkers predicting the development of metabolic diseases 18, 21. However, whether these circulating microbial DNAs are parts of the influx of microbiota-derived products into the portal system or due to the translocation of microbiota through obese disrupted gut barrier remains unclear. Various types of extracellular vesicle (EV) serve as vehicles to transport a variety of cargoes, including RNAs, DNAs, lipids, and proteins between the neighbor or distant cells 23. Budding events of microbial EVs and their release into the circulation of host have been reported for a wide range of microbiota species 24, 25. Thus, this led us to hypothesize that microbiota-derived bacterial DNA-containing EVs can pass through the disrupted intestinal barrier, subsequently exacerbating obesity-associated tissue inflammation and insulin resistance.
The liver plays a critical role in filtering bacteria and their byproducts in the blood flow from the portal vein draining the intestine. However, obesity elicits increased levels of endotoxins in circulation, supporting the notion that obesity reduces the clearance of bacterial products in liver 7,26. The ability of liver to remove bacterial components from bloodstream is mainly attributed to the phagocytosis of Kupffer cells, majority of which express a complement receptor of the immunoglobulin superfamily (CRIg) 27, 28. Previous studies have demonstrated that CRIg expression is critical for Kupffer cells to efficiently bind and phagocytize complement C3-opsonized bacterial products 28-31. However, the critical role of CRIg+ Kupffer cells in clearing microbiota-derived products from bloodstream in the context of obesity is largely unknown.
Here, we show that the leakage of gut microbial DNA-containing EVs (mEVs) results in a marked accumulation of bacterial DNAs in metabolic tissues in obesity, subsequently fueling obesity-associated tissue inflammation and insulin resistance. High fat diet feeding remarkably decreases the population of CRIg+ Kupffer cells which exert pronounced effect on blocking the spread of mEVs into distant metabolic tissues. In addition, complement component C3 is required for the interaction between CRIg+ macrophages and mEVs, whereas CRIg+ Kupffer cells fail to clear mEVs from circulation in C3 knockout mice. mEV engulfment can trigger inflammatory responses and insulin resistance of insulin target cells, both in vivo and in vitro. In contrast, depletion of microbial DNAs blunts the ability of obese intestinal EVs to induce inflammation and insulin responses, indicating that microbial DNAs are the key functional cargoes within these EVs. Finally, the cGAS/STING pathway, one of the key sensors for bacterial DNAs, is crucial for mEV-mediated cellular inflammation and insulin resistance.
Materials and Methods
Glucose tolerance and insulin tolerance tests
For glucose tolerance tests, mice received one dose of dextrose (1 g/kg body weight) via i.p. injection after 12 hr of fasting. For insulin tolerance tests, mice were fasted for 6 hr and then i.p. injected with insulin (0.35 units/kg body weight for HFD mice; 0.175 units/kg body weight for NCD mice).
In vivo EV trafficking assays
PKH26-labeled EVs (5x109 EVs per mouse) were delivered to either NCD or HFD recipient mice through either injection into tail vein or jejunum section. After 2 hours or 16 hours EV injection, parts of Liver, skeletal muscle, and eWAT were collected for detecting the appearance of PKH26 red fluorescence.
Depletion DNA of intestinal EVs
The intestinal EV pellet was dissolved in 100 μL PBS. As previously described 34-36, these EVs were loaded into a Gene Pulser/micropulser Cuvettes (Bio-Rad) for electroporation (GenePulser Xcell electroporator, Bio-Rad) and then treated with DNase I (300U) for 30 mins, 37°C.
Quantification of bacterial DNA using real-time PCR
Levels of bacterial DNAs were assessed by qPCR using the Femto Bacterial DNA Quantification Kit by following the manufacturer’s instructions (Cat. No. D4301, Zymo Research).
In vivo and in vitro EV treatment
For in vitro treatment, 0.5x106 3T3-L1 cells or hepatocytes were treated with 5x108 EVs for 24 hours and then used for either glucose uptake or output assays. For in vivo treatment, recipient mice were tail vein injected with 5x109 EVs twice per week. Intestinal EVs derived from obese C3KO mice were intravenously injected into NCD C3KO recipient mice (5x109 EVs/mouse).
Statistical Analysis
Tests used for statistical analyses are described in the figure legends. To assess whether the means of two groups are statistically different from each other, unpaired two-tailed Student’s t test was used for statistical analyses using Prism8 software (GraphPad software v8.0; Prism, La Jolla, CA). P values of 0.05 or less were considered to be statistically significant. Degrees of significance were indicated in the figure legends. For the results of glucose and insulin tolerance tests, statistical comparisons between every two groups at each time point were performed with unpaired two-tailed Student’s t test.
Results
Gut mEVs are translocated into the circulation and metabolic tissues of obese host.
To examine whether obesity results in accumulation of bacterial DNAs in host, we probed 16s rRNA abundance in the key metabolic tissues. As shown in Figure 1A and Figure S1A, bacterial DNAs were rarely detected in the liver, skeletal muscle, and adipose tissue of healthy lean mice, whereas 16s rRNAs were highly enriched in the key metabolic tissues of mice after a 16-week high fat diet (16wks HFD) feeding regimen. Consistently, obese human liver also contained a greater level of bacterial DNAs than in healthy normal human liver, as evidenced by a robust fluorescent signal in obese human liver stained with 16s rRNA probes (Figure 1B). qPCR analysis also indicates that obese human hepatocytes contained a greater abundance of bacterial DNAs than healthy human hepatocytes (Figure S1B). In addition, the circulating EVs of 16wks HFD-fed mice or obese human contained greater abundance of bacterial DNAs, compared to lean/healthy plasma EVs (Figure 1C). More importantly, in plasma samples we found that the majority of bacterial DNAs were transported with EVs, as indicated by a marked reduction in 16s rRNA abundance in plasma after EV depletion (Figures 1D and S1C). Taken together, these results indicate that there is an enrichment of bacterial DNAs in metabolic tissues in obesity.
Figure 1. Obesity induces the translocation of microbial DNA containing EVs from gut into circulation.
The abundance of 16s rRNA and CRIg in the liver of NCD or 16wks HFD WT mice (A) and healthy lean or obese human liver (B). (C) The abundance of 16s rRNA within plasma EVs of both NCD and 16wks HFD-fed WT mice, and healthy and obese humans. (D) 16s rRNA abundance in plasma, EV fraction, and EV-free fraction of 16wks HFD WT mice. (E) 16s rRNA abundance within the intestinal EVs of NCD or 16wks HFD WT mice. (F) The levels of PKH26 red fluorescence, 16s rRNA, and CRIg in liver after injection of PKH26-labeled intestinal EVs into the jejunum sections of both NCD and 16wks HFD WT mice. (G) Top 20 microbial DNAs with the most significant abundance difference between NCD and 16wks HFD WT gut lumen EVs. Data are presented as mean ± SEM. n=6-8 per group (C); n=5 per group (D), n=4 per group (E), n=6 per group (G). (A, B, and F) scale bar=50 μm. *P<0.05, Student’s t test. mEV, microbial DNA-containing extracellular vesicle.
Obesity is characterized by an impaired and defective gut integrity, subsequently resulting in the translocation of microbiota-derived products into the circulation of host. We next addressed whether intestinal EVs harboring microbial DNAs can be penetrated into the circulation and then transported into the key metabolic tissues of host. We purified EVs from the small intestinal lumen contents of 16wks HFD fed WT mice, and the characteristics of these EVs were evaluated by measuring particle size, morphology, and EV-related protein markers (Figures S1D and S1E). Furthermore, qPCR analysis with 16s rRNA primers suggests that microbial DNAs are one of the cargoes within these intestinal EVs (Figure 1E). Interestingly, obese intestinal EVs harbored more bacterial DNAs than the intestinal EVs of lean mice (Figure 1E). Bacterial DNA abundance were similar within EVs derived from different lumen sections of small intestine (Figure S1F). We further validated whether intestinal mEVs pass through gut barrier. After labeled with the PKFI26 red fluorescent dye, mEVs isolated from 16wks HFD WT mice were injected (5x109 EVs per mouse) into the jejunum section of either lean or obese WT recipient mice. As a result of obesity-induced gut barrier breach, PKH26 mEVs were readily leaked into the key metabolic tissues of obese recipients, as demonstrated by the appearance of strong red fluorescence in liver, skeletal muscle, and epididymal fat after 16 hours injection of PKH26 mEVs (Figure 1F, Figure S1G). By contrast, the intact gut barrier of lean recipient mice prevented the penetration of PKH26-labeled mEVs into metabolic tissues of host (Figure 1F, Figure S1G). In addition, in the liver of obese recipients, most of PKH26 red fluorescent signals were co-localized with the green fluorescence-conjugated 16s rRNAs, thus indicating microbial DNAs are one of the key cargoes within intestinal EVs (Figure 1F).
To comprehensively characterize the microbial DNA composition within intestinal EVs, we conducted 16s rRNA gene sequencing of bacterial DNAs from lean and obese intestinal EVs. Sequences were clustered based on similarity into operational taxonomic units and compared against a reference 16s rRNA database to identify their bacterial classifications. As shown in Figures 1G and S1H and Table S1, comparison between the microbial DNA abundance within lean and obese mEVs revealed that DNAs derived from obesity-associated phyla such as Firmicutes were remarkably enriched in obese mEVs, while lean mEVs carried higher abundance of Bacteroidetes DNAs. Thus, these results have shown that parts of microbial DNAs within mEVs mirror the components of small intestine microbiota 12.
Liver CRIg+ macrophages are the critical effectors filtering mEVs from circulation.
CRIg+ macrophages residing in liver serve as the key player clearing bacteria and its byproducts from bloodstream. To assess the role of liver CRIg+ macrophages in blocking the penetration of mEVs into the metabolic tissues of host, PKH26-labeled obese mEVs (5x109 EVs per mouse) were delivered into lean recipient mice through an intravenous injection. As expected, PKH26 mEVs quickly accumulated in the liver of lean mice after 2 hours injection (Figure 2A). More importantly, most of injected mEVs co-localized with CRIg+ cells in liver, as shown by overlapping PKH26 red fluorescence with the CRIg signals, suggesting the ability of CRIg+ macrophages to capture mEVs (Figure 2A). In line with findings from previous studies, CRIg was mainly expressed on Kupffer cells, whereas, we found that a prolonged HFD feeding regimen resulted in a remarkable decrease in the population of CRIg+ macrophages in liver (Figures 1A and 2B). We also found that obese human liver contained notably less amount of CRIg+ cells than in healthy lean human liver (Figures 1B and S2A). However, there was a comparable population of CRIg+ Kupffer cells between lean and 4wks HFD WT mice (Figure 2B). To further validate the importance of CRIg+ Kupffer cells on preventing the spread of mEVs into distant metabolic tissues, PKH26-labeled obese mEVs (5x109 EVs per mouse) were intravenously injected into lean CRIgKO mice. As shown in Figures 2A and 2C, knockout of CRIg resulted in more mEVs passing through liver and subsequently accumulating in the skeletal muscle and epididymal fat of lean recipient mice, whereas, a minimal level of PKH26 red fluorescence was observed in the distant metabolic tissues of lean WT mice. In addition, 4wks HFD fed CRIgKO mice exhibited greater levels of 16s rRNA abundance in the key metabolic tissues than in the 4wks HFD WT mice (Figure 2D). Thus, these data suggest that CRIg+ macrophages play a crucial role in filtering circulating mEVs.
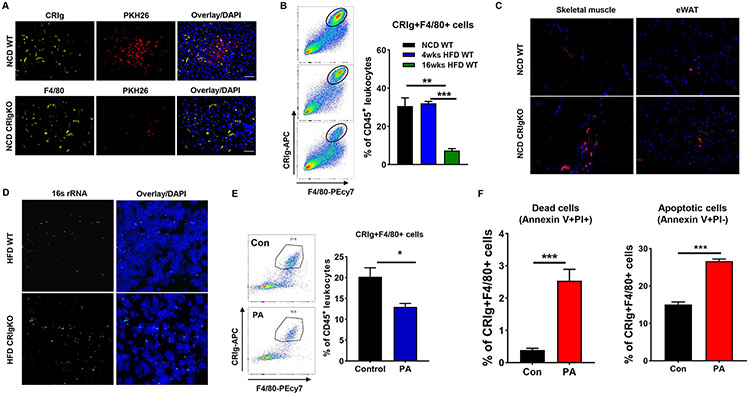
Figure 2. Liver CRIg+ macrophages exert profound function clearing intestinal mEVs from circulation.
(A) The levels of PKH26 red fluorescence, CRIg, and F4/80 in the liver of either NCD WT or CRIgKO mice intravenously injected with PKH26-labeled mEVs. (B) Effect of obesity on liver CRIg+ macrophage population. (C) The abundance of PKH26 red fluorescence in the skeletal muscle and epididymal fat of both NCD WT and CRIgKO mice after tail vein injection of PKH26 mEVs. (D) The abundance of 16s rRNA in the liver of both 4wks HFD WT and CRIgKO mice. (E) Effect of palmitate acid (PA, daily 0.8g PA/mouse, 2 weeks) treatment on the liver CRIg+ macrophage population of NCD WT mice. (F) Effect of PA (500 μM) treatment on the apoptosis of liver CRIg+F4/80+ cells. Data are presented as mean ± SEM. n=4 per group (B, E, and F). (A, C, and D) scale bar=50 μm. *P<0.05, **P<0.01, ***P<0.001, Student’s t test.
We further addressed how obesity causes a marked reduction in CRIg abundance. We first ruled out the impact of microbiota and its byproducts on CRIg expression, as evidenced by significantly lower CRIg abundance in the liver of 16wks HFD-fed germ-free mice than in normal chow diet (NCD)-fed germ-free mice (Figures S2B and S2C). In addition, treatment of obese mEVs had negligible effect on the population of CRIg+ Kupffer cells in NCD WT mice (Figure S2D). We next tested the effect of lipids, which are highly enriched in HFD, on CRIg expression. After 2 weeks oral gavage of palmitate acid/corn oil mixture into NCD WT mice, we found that the proportion of CRIg+ macrophages was significantly decreased, compared to the control mice without lipid treatment (Figure 2E). In addition, in vitro treatment of palmitate acid induced the apoptosis of CRIg+F4/80+ macrophages (Figure 2F).
The leakage of gut mEVs causes tissue inflammation and insulin resistance.
Emerging evidence indicates that gut microbiota-derived products can exacerbate tissue inflammation and insulin resistance in obesity. Given that the absence of CRIg+ macrophages can result in accumulation of microbial DNAs in the key metabolic tissues of host, we next evaluated the roles of CRIg+ macrophages in regulating obesity-associated tissue inflammation and insulin resistance. After 4 weeks HFD feeding, CRIgKO mice had similar body weight with WT littermates (Figure S3A). However, compared to HFD WT mice, the phenotypes of HFD CRIgKO mice were most pronounced in worse glucose intolerance and insulin resistance, as measured by glucose and insulin tolerance tests (Figures 3A and 3B). Consistent with these results, ablation of CRIg led to decreased extents of insulin responses, as indicated by less insulin-stimulated AKT phosphorylation in the liver, skeletal muscle, and epididymal adipose tissue of HFD CRIgKO mice (Figure S3B). As expected, HFD feeding can result in an increase in proinflammatory adipose tissue macrophage (ATM) population, while more M1-like (CD11b+F4/80+CD11c+CD206-) ATMs were recruited into visceral adipose tissue of obese CRIgKO mice (Figure S3C). In addition, the liver of HFD CRIgKO mice accumulated more recruited liver macrophages (CD11b+Ly6c+F4/80-), compared to HFD WT mice (Figure S3D). Consistently, qPCR analysis also suggests that HFD CRIgKO mice had greater levels of proinflammatory cytokines in liver and epididymal adipose tissue (Figure S3E). Overall, these results indicate that depletion of CRIg can exacerbate obesity-induced tissue inflammation and insulin resistance.
Figure 3. Obese intestinal mEVs cause tissue inflammation and insulin resistance in obesity.
(A and B) Glucose and insulin tolerance tests (GTTs and ITTs) were performed in 4 weeks HFD WT and CRIgKO mice. (C and D) After 4 weeks of adoptive transfer of obese mEVs, GTTs and ITTs were measured in NCD CRIgKO recipient mice. (E) Insulin stimulated AKT phosphorylation levels at S473 in the liver, skeletal muscle (SKL), epididymal white fat (eWAT) of NCD CRIgKO mice after 4 weeks obese mEV treatment. (F) Glucose infusion rate (GIR), insulin-stimulated glucose disposal rate (IS-GDR), hepatic glucose production (HGP), and the percentage of suppression of free fatty-acid levels (FFA suppression) during hyperinsulinemic-euglycemic clamp studies. (G and H) Glucose tolerance and insulin sensitivity of NCD WT recipient mice treated with obese mEVs. Data are presented as mean ± SEM. n=8 per group (A, B, and F); n=5 per group (C, D, G, and H). *P<0.05, **P<0.01, Student’s t test.
To assess the pathogenic effects of obese mEVs on the incidence of tissue inflammation and insulin resistance, obese mEVs were intravenously injected (5x109 EVs per mouse, twice per week) into either NCD CRIgKO or WT recipient mice. After 4 weeks treatment, all mice had comparable body weight (Figure S3F). Importantly, treatment of obese mEVs led to impaired glucose tolerance and insulin sensitivity of NCD CRIgKO mice, compared to NCD CRIgKO control mice, as measured by glucose and insulin tolerance tests (Figures 3C and 3D). Assessment of AKT phosphorylation in response to insulin stimulation suggested that tissue insulin sensitivities were significantly reduced after 4 weeks obese mEV treatment (Figures 3E). The effects of obese mEVs on insulin sensitivity were further confirmed by hyperinsulinemic-euglycemic clamp studies. NCD CRIgKO treated with obese mEVs exhibited lower glucose infusion rates, decreased insulin-stimulated glucose disposal rates, and a lower degree to insulin-mediated suppression of hepatic glucose production and circulating free fatty acid levels (Figure 3F). In addition, flow cytometric analysis suggested that treatment of obese mEVs increased the population of proinflammatory macrophages in both visceral adipose tissue and liver, accompanied by greater levels of proinflammatory cytokines (Figures S3G-S3J). In NCD WT recipient mice, obese mEV treatment had minimal effects on the population of CRIg+ Kupffer cells (Figure S2D). Concomitantly, NCD WT mice treated with obese mEVs exhibited comparable metabolic phenotypes and tissue inflammation with NCD WT controls (Figures 3G and 3H, Figures S3K-S3N). Thus, in the absence of CRIg+ macrophages, obese mEVs can induce tissue inflammation and insulin resistance.
The ability of CRIg+ macrophage to capture microbial products requires the complement component C3-mediated opsonization. Thus, we further addressed the crucial role of C3 protein in mediating the interaction between CRIg+ macrophages and gut mEVs. While circulating C3 levels were comparable between obese and lean WT mice, the abundance of C3 bound with plasma EVs was greater in the context of obesity (Figure 4A, Figure S4A). In line with previous findings, we observed that knockout of C3 blunted the capacity of CRIg+ Kupffer cells to recognize obese mEVs, as evidenced by less PKH26 red fluorescence co-localizing with CRIg+ cells in the liver of NCD C3KO mice injected with PKH26-labeled C3KO mEVs (Figure 4B). Consequently, more intestinal C3KO mEVs were leaked into skeletal muscle and adipose tissue of NCD C3KO recipient mice (Figure S4B). Concomitant with the penetration of mEVs into these metabolic tissues, treatment of obese C3KO mEVs impaired glucose tolerance and insulin sensitivity of NCD C3KO mice, although high abundance of CRIg+ cells resided in the liver of these mice (Figures 4B-4D). Consistently, compared to control NCD C3KO mice, the levels of insulin-stimulated AKT phosphorylation were decreased in the key metabolic tissues of mEVs-treated NCD C3KO mice (Figure 4E). Obese C3KO mEV treatment also activated tissue inflammation of NCD C3KO mice, as evidenced by switching macrophage activation towards a proinflammatory M1-like state and greater abundance of proinflammatory cytokines in both adipose tissue and liver (Figures 4F and 4G). Thus, these results suggest the important role of complement component C3 in the ability of CRIg+ macrophages to capture mEVs.
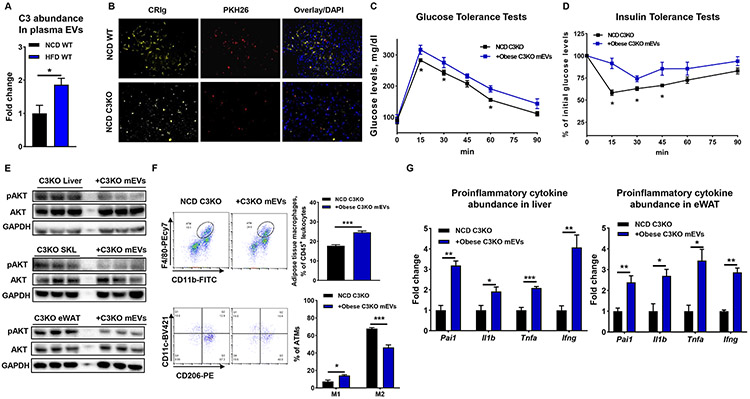
Figure 4. Complement component C3 is critical for the ability of CRIg+ macrophages to capture mEVs.
(A) C3 abundance in plasma EVs of both NCD WT and 16wks HFD WT mice. (B) Effect of C3 knockout on the ability of CRIg+ macrophage to capture mEVs. (C and D) After 4 weeks treatment with obese mEVs, glucose tolerance and insulin sensitivity of NCD C3KO recipient mice were measured. (E) The levels of insulin-stimulated AKT phosphorylation in liver, SKL, and eWAT of NCD C3KO recipient mice treated with obese mEVs. After 4 weeks injection with obese mEVs, the population of ATMs and their activation (F) and proinflammatory cytokine abundance in the liver and eWAT (G) of NCD C3KO mice. Data are presented as mean ± SEM. n=6 per group (A); n=4 per group (C, D, F, and G). (B) scale bar=50 μm. *P<0.05, **P<0.01, ***P<0.001, Student’s t test.
Gut mEVs can trigger cellular inflammation and insulin resistance.
Given the notable in vivo effects of obese mEVs on insulin sensitivity, we next evaluated the direct impact of obese mEVs on insulin target cells by conducting in vitro studies of insulin action in adipocytes and hepatocytes. Consistent with the effective transportation of mEVs into metabolic tissues, the appearance of a strong red fluorescence in 3T3-L1 adipocytes or hepatocytes indicated efficient uptake of PKH26-labeled obese mEVs (Figures S5A and S5B). As shown in Figure 5A, co-culturing obese mEVs with 3T3-L1 adipocytes resulted in a significant decrease in insulin-stimulated glucose uptake. In primary hepatocytes isolated from lean WT mice, glucagon treatment enhanced hepatic glucose production, whereas insulin can effectively repress this glucagon-induced glucose output (Figure 5B). However, treatment of obese mEVs blunted the ability of insulin to suppress glucagon stimulation on hepatic glucose production (Figure 5B). In line with these findings, we observed that less amount of insulin-stimulated phosphorylation of AKT in these insulin target cells treated with obese mEVs (Figures 5C and 5D). Consistently, we also observed that obese mEV treatment impaired insulin sensitivity of healthy human hepatocytes (Figures S5C and S5D). We also found that obese mEV treatment led to increased levels of proinflammatory cytokines in these cells (Figures 5E and 5F). Therefore, obese intestinal mEVs exert pathogenic effects on cellular inflammatory activation and insulin sensitivity.
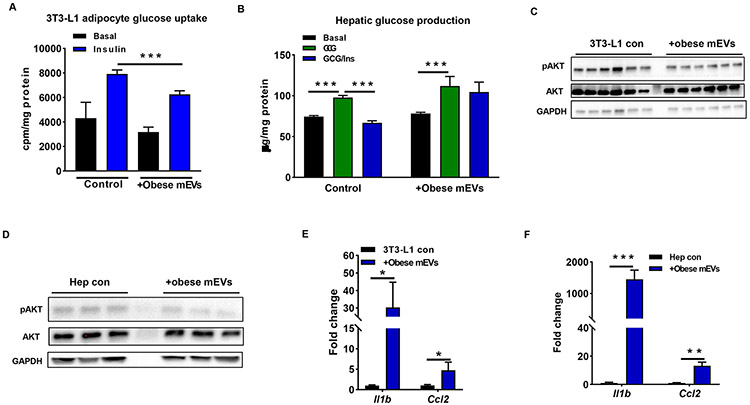
Figure 5. Obese intestinal mEVs induce cellular inflammation and insulin resistance.
Effects of obese mEVs on 3T3-L1 adipocyte glucose uptake (A) and hepatic glucose output (B). The levels of insulin-stimulated AKT phosphorylation of 3T3-L1 adipocytes (C) and primary mouse hepatocytes (D) after obese mEV treatment. The induction of obese mEVs on proinflammatory cytokine expression in 3T3-L1 adipocytes (E) and primary mouse hepatocytes (F). Data are presented as mean ± SEM. n=6 per group (A and B); n=4 per group (E and F). *P<0.05, ***P<0.001, Student’s t test. GCG, glucagon; Ins, insulin.
Microbial DNAs are the key pathogenic cargoes within intestinal EVs that induce tissue inflammation and insulin resistance.
The findings that intestinal EVs delivered microbial DNAs into metabolic tissues raise the possibility that microbiota-derived EVs within these obese intestinal EVs exert profound regulation on the tissue inflammation and insulin resistance of host. We also observed that gut mEVs derived from lean WT mice can cause metabolic disorders (Figures S6A and S6B). To address the role of microbial DNAs within gut EVs, we collected intestinal EVs from 16wks HFD fed germ-free mice (GF EVs) and delivered these EVs into NCD CRIgKO mice through tail vein injection (5x109 EVs per mouse, twice per week). In addition, to make DNA-free gut EVs for in vivo experiment, we depleted microbial DNA cargoes in intestinal EVs by electroporating obese mEVs and then treating these EVs with DNase (Figure S6C). All mice had similar body weight after 4 weeks treatment of EVs (Figure S6D). Consistently, obese mEV treatment caused decreased glucose tolerance and insulin sensitivity, whereas NCD CRIgKO recipients treated with either GF EVs or DNA-free EVs had comparable metabolic responses with the control mice without EV treatment (Figures 6A and 6B). In addition, GF EVs or DNA-free EVs did not trigger tissue inflammation in the adipose tissue of NCD CRIgKO recipients, as shown by similar proinflammatory adipose tissue macrophage population among these mice (Figure S6E).
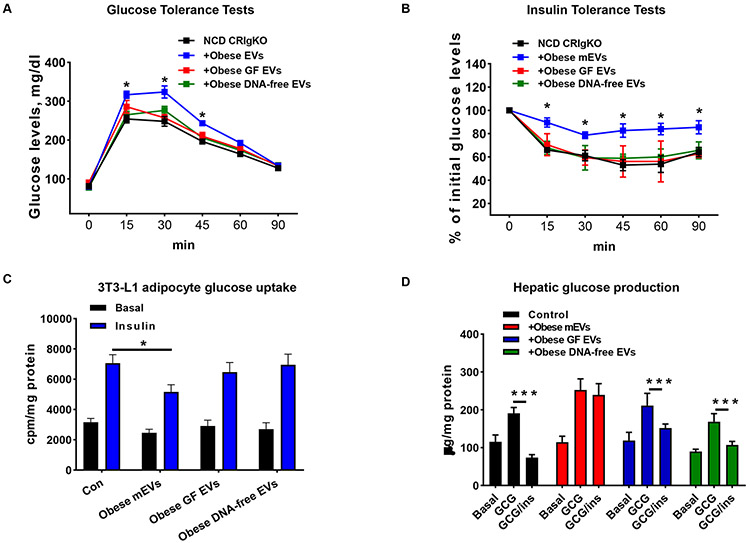
Figure 6. Microbial DNAs are important cargoes for the intestinal EV effects.
(A and B) the glucose tolerance and insulin sensitivity of NCD CRIgKO recipient mice treated with either obese mEVs, obese germ-free (GF) EVs, or obese DNA-free EVs. (C and D) Effects of intestinal EVs on 3T3-L1 glucose uptake (C) and hepatic glucose output (D). Data are presented as mean ± SEM. n=6 per group (A-D). *P<0.05, ***P<0.001, Student’s t test.
Addition of obese mEVs into 3T3-L1 adipocytes led to a reduction in insulin-stimulated glucose uptake, compared to the control cells without EV treatment (Figure 6C). By contrast, co-culturing GF EVs with 3T3-L1 adipocytes had a negligible effect on cellular insulin sensitivity, as evidenced by similar levels of glucose uptake between GF EVs or DNA-free EVs-treated cells and control cells (Figure 6C). In contrast to the suppressive effect of obese mEVs on hepatic insulin sensitivity, the glucagon-stimulated glucose production of primary mouse hepatocytes treated with either GF EVs or DNA-free EVs was effectively blocked by insulin, thus indicating that GF EVs did not affect hepatic insulin responses (Figure 6D). Overall, there results indicate that microbial DNAs are the key pathogenic cargoes within obese intestinal EVs that induce tissue inflammation and insulin sensitivity.
Microbial DNAs within intestinal EVs trigger the activation of cGAS/STING pathway that enhances inflammatory responses.
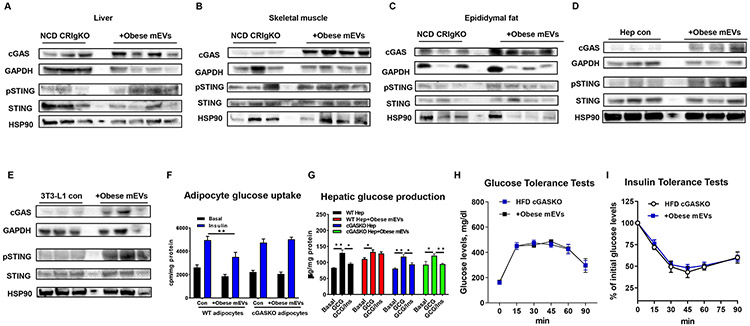
Bacterial DNAs can activate the cGAS/STING pathway that subsequently initiates inflammatory action. In addition, obese mEV treatment can activate inflammatory status in both in vivo and in intro experiments, whereas depletion of microbial DNAs prevented these effects of obese mEVs (Figure 6). Thus, we further assessed whether obese mEVs mediated the inflammatory responses of target cells through a cGAS/STING-mediated mechanism. As a result of a marked accumulation of microbial DNAs after obese mEV treatment, all metabolic tissues of NCD CRIgKO mice contained greater abundance of cGAS and phosphorylated STING than in the control CRIgKO mice without mEV treatment, thus demonstrating that obese mEVs trigger the cGAS/STING-mediated inflammatory responses (Figures 7A-7C). We also observed that obese C3KO mEV treatment enhanced the activation of cGAS/STING signaling in NCD C3KO mice (Figure S7A). In line with these in vivo phenotypes, obese mEV treatment elevated the levels of cGAS expression and STING phosphorylation in insulin target cells (Figures 7D and 7E). We observed that obese human liver contained greater levels of cGAS and phosphorylated STING than that in healthy human hepatocytes (Figure S7B). In addition, obese mEV treatment caused elevated activation of cGAS/STING pathway in healthy human hepatocytes (Figures S5D and S7C). There were greater levels of cGAS and phosphorylated STING in the liver, skeletal muscle, and epididymal fat of HFD CRIgKO mice, compared to HFD WT mice (Figure S7C). Consistent with the minimal effects of GF EVs or DNA-free EVs on the tissue inflammation of recipients, depletion of microbial DNAs completely blocked the ability of obese mEVs to initiate activation of cGAS/STING pathway in the key metabolic tissues (Figures S7D and S7E). To further confirm the critical role of cGAS in the pathogenic effects of obese mEVs, we prepared cGASKO primary adipocytes and hepatocytes from lean cGASKO mice. As shown in Figures 7F and 7G, knockout of cGAS had negligible effects on cellular insulin responses, while deletion of cGAS prevented the suppressive effect of obese mEVs on insulin action (Figures 7F and 7G). Consistently, obese mEV treatment had minimal effects on the metabolic responses of HFD/obese cGASKO mice (Figures 7H and 7I, S7F-S7H). Thus, these results demonstrate the important role of cGAS/STING-mediated pathways in the ability of obese mEVs to regulate inflammation and insulin sensitivity of target cells.
Figure 7. The cGAS/STING pathway plays a critical role in the ability of mEVs to trigger inflammatory responses.
(A-C) The levels of cGAS and STING phosphorylation in the liver, skeletal muscle, and epididymal fat of NCD CRIgKO recipient mice after 4 weeks treatment of obese mEVs. (D and E) Effects of obese mEVs on the activation of cGAS/STING pathway in hepatocytes and adipocytes. (F and G) The influence of cGAS ablation on the responses of adipocytes and hepatocytes to obese mEV treatment. (H and I) Glucose tolerance and insulin sensitivity of 8wks HFD cGASKO mice after 4wks treatment with obese mEVs. Data are presented as mean ± SEM. n=6 per group (F and G); n=5 per group (H and I) *P<0.05, **P<0.01, Student’s t test.
Discussion
In this study, we have shown that the obesity-induced leakage of gut microbial DNA-containing EVs can trigger cellular inflammation and impair glucose tolerance and insulin sensitivity. CRIg+ macrophages exert profound function on preventing the infiltration of mEVs into the key metabolic tissues, whereas obesity significantly decreases the population of CRIg+ macrophages. Thus, when lean insulin sensitive CRIgKO mice are treated with obese mEVs, they exhibit elevated tissue inflammation, concomitant with systemic insulin resistance and glucose intolerance. In contrast, obese mEV treatment has minimal effects on the metabolic phenotypes of lean WT recipients. C3 is crucial for CRIg+ macrophages to recognize intestinal mEVs, as evidenced by the penetration of injected mEVs into the key metabolic tissues and a subsequent decrease in insulin sensitivity and glucose tolerance of lean C3KO mice. In vitro experiments show that treatment with obese mEVs directly triggers cellular inflammatory responses and decreases insulin signaling in adipocytes and hepatocytes. However, depletion of microbial DNAs blocks the ability of obese mEVs to regulate the tissue inflammation and metabolic responses of host. Finally, we demonstrate that accumulation of microbial DNAs, as a result of intestinal mEV delivery, activates cGAS/STING-mediated inflammatory signaling, whereas ablation of cGAS blunts the cellular responses to the enrichment of microbial DNAs. Taken together, these studies suggest that microbial DNAs encapsulated with intestinal EVs are pathogenic for the development of obesity-associated tissue inflammation and metabolic disorders.
As a result of obesity-induced gut barrier disruption, various microbiota-derived products can be translocated into the circulation of host 11. In current study, we found that a greater abundance of microbial DNAs was accumulated in various metabolic tissues, including liver, skeletal muscle, and adipose tissue of obese individuals, thus suggesting a leakage of microbial DNAs from gut into host. Indeed, in line with findings from previous reports, HFD-fed mice contained greater levels of microbial DNAs in blood and tissues than lean mice 18, 19, 21. However, how these microbial DNAs are transported beyond the gut lumen remains debated. In current study, in both human and mouse plasma samples we found that microbial DNAs were mainly enriched within circulating EVs, as a result of the leakage of microbial DNA-containing EVs from gut. Bradley et al. suggest that bacteria can be bound by platelets and rapidly removed from bloodstream 48. In line with this observation, previous studies have also shown that bacterial DNAs are mainly found in the blood buffy coat layer which contains platelets 19.
In addition to intact gut barrier, liver serves as an important blood-filtering organ that can prevent the spread of microbial products into distant organs. Our study highlights the critical role of CRIg+ macrophages, majority of which are Kupffer cells 28, in blocking the invasion of gut microbial DNA-containing EVs into metabolic tissues. Notably, prolonged obesity can reduce the population of CRIg+ macrophages residing in the liver of both humans and mice. We further confirmed that treatment with palmitate acid can decrease the population of liver CRIg+ macrophages in both in vivo and in vitro experiments. Thus, obesity-induced loss of CRIg+ cells allows for the translocation of microbial DNAs/gut EVs into host metabolic tissues. In addition to CRIg, previous studies have suggested that the scavenger receptor A plays an important role in the interaction between Kupffer cells and invaded bacteria 49. However, we observed that liver failed to clean gut mEVs in the experiment with NCD CRIgKO mice intravenously injected with PKH26-labeled gut mEVs, suggesting that scavenger receptor A may not interact with gut mEVs.
Complement component C3-mediated opsonization plays a central role in the ability of CRIg+ macrophages to recognize foreign particles from the bloodstream 28, 37-40. In agreement with the requirement of C3 opsonization for the clearing function of CRIg+ macrophages, ablation of C3 resulted in much less interaction between mEVs and CRIg+ macrophages in the liver of lean C3KO mice, accompanied by more mEVs infiltrating into distant organs. A previous study by Broadley et al. suggests that, before C3-mediated opsonization, Kupffer cells rapidly remove Listeria from bloodstream through a scavenger receptor-dependent manner 48. In addition, Zeng et al. suggest that hepatic CRIg+ macrophages interact with Gram-positive bacteria through directly binding with bacterial lipoteichoic acid (LTA) component 27. While we did not evaluate the function of scavenger receptor and LTA, the liver of NCD C3KO mice failed to block gut mEVs after injection with PKH26-labeled mEVs. This support the critical role of C3 in the ability of CRIg+ macrophages to capture mEVs.
Emerging evidence indicates that EVs encapsulating various cargoes exert profound regulation on the development of obesity-associated tissue inflammation and insulin resistance. Previous studies, including ours, have shown that EVs derived from immune cells or insulin-targeting cells play a critical role in regulating the functions of neighbor cells or the cells residing at distal site through transferring various cargoes including non-coding RNAs, proteins, and lipids 23, 33. While several studies have reported the functions of microbiota-derived EVs in various disease models 25,42,43, our novel findings from current study reveal the pathogenic effects of microbial DNAs-encapsulating gut EVs on host tissue inflammation and metabolic disorders.
Bacterial DNAs are pathogenic molecules that can trigger the inflammatory responses of host 41. As previously reported, microbiota can secrete various types of EVs which are main components of intestinal EVs 25, 42, 43. In our study, 16s rRNA sequencing analysis suggests that parts of microbial DNA contents, such as Firmicutes-associated DNAs, within intestinal EVs mirror the composition of microbiota in small intestine. Previous studies have reported that Proteobacteria-related DNAs are enriched in non-alcoholic fatty human liver 21,50, while our data showed no significant difference in this bacterial DNA abundance within both NCD and HFD gut EVs derived from mouse small intestine. It is possible that mEVs leaked from other intestinal sections may harbor high abundance of Proteobacteria DNAs and contribute to the accumulation of Proteobacteria-related DNAs within obese liver. In addition to EVs, gut microbial DNAs may be translocated into host tissues through other manners20. More importantly, our data reveal a new mechanism by which microbial DNAs induce tissue inflammation and metabolic disorders after leakage of gut mEVs. Ortiz et al. also report that translocation of microbial DNAs may contribute to the inflammatory responses and insulin resistance of morbid obese patients 22. However, these results can not exclude the potential functions of other cargoes of intestinal EVs such as protein and lipids which have been reported their profound functions in previous studies 23. In addition, Jing et al. suggest that EVs derived from intestinal epithelial cells exert profound regulation on intestinal tract immune balance that is important for the development of inflammatory bowel disease 51.
In line with the critical roles of cGAS/STING pathway in sensing bacterial DNAs and triggering cellular inflammatory responses 44,45, microbial DNA-containing intestinal EVs elevated the activation of cGAS/STING signaling in both in vivo and in vitro experiments. Ablation of cGAS blunts the cellular responses to the treatment of obese intestinal mEVs, suggesting that cGAS is required for intestinal mEV-induced cellular abnormalities. Bai et al. have reported that mitochondrial DNA release can induce the activation of cGAS/STING pathway, contributing to adipose tissue inflammation in obesity 46. In addition, depletion of STING can attenuate liver inflammation and fibrosis 47. Emerging evidence indicates that the nucleotide-binding oligomerization domain (NOD) proteins are important mediators detecting bacterial peptides and initiating cellular inflammatory responses 52. However, whether microbial EVs contain sufficient amount of NOD ligands for NOD-associated signaling activation is still unknown.
In summary, we find that microbial DNA-containing intestinal EVs can penetrate through obese disrupted gut barrier and exacerbate tissue inflammation and metabolic disorders. CRIg+ macrophages contribute to the ability of liver to filter gut microbial DNAs encapsulating EVs, whereas obesity elicits decreased CRIg+ macrophage population and subsequently facilitates the spread of microbial DNA-containing EVs into various metabolic tissues. It is recognized that various microbiota-derived products are pathogenic for obesity-associated tissue inflammation and insulin resistance. Based on these studies, we suggest a new mechanisms whereby leaky microbial DNAs trigger tissue inflammation in obesity.
Supplementary Material
What You Need to Know.
Background and Context
As a hallmark of obesity, microbiota-derived products can be translocated through disrupted gut barrier into host circulation, subsequently exacerbating obesity-associated tissue inflammation and insulin resistance.
New Findings
Obesity reduces liver CRIg+ macrophages that clear circulating microbiota EVs. Infiltration of microbiota EVs exacerbates tissue inflammation and insulin resistance. Microbial DNAs are responsible for the microbiota EV effects.
Limitations
This study was performed in mice and human liver samples. Further studies are needed in humans.
Impact
Strategies to increase liver CRIg+ macrophages might be developed for treatment of obesity-associated tissue inflammation and insulin resistance.
Lay Summary.
Gut microbiota extracellular vesicles can translocate through obese disrupted gut barrier. Liver CRIg+ macrophages can clear circulating microbiota-derived extracellular vesicles that exacerbate tissue inflammation and insulin resistance in obesity.
Acknowledgements
We thank Dr. Wenxian Fu for providing CRIg KO mice; the integrative genomics core of City of Hope for the 16s rRNA sequencing; Vanessa Taupin for electron microscope; Jennifer Santini for confocal microscope.
Grant support: This study was funded by the UCSD School of Medicine Microscopy Core grant (P30 NS047101), the National Natural Science Foundation of China (No. 81500436 to Z. L.), and the U.S. National Institute of Diabetes and Digestive and Kidney Diseases R00 award (R00DK115998 to W. Y.) and the UCSD/UCLA Diabetes Research Center Pilot and Feasibility grant (P30 DK063491 to W. Y.).
Transcript profiling: Bacterial 16s rRNA sequencing raw data supporting these studies can be found at the Sequence Read Archive database under accession number PRJNA625759.
Abbreviations:
- CRIg
complement receptor of the immunoglobulin superfamily
- mEVs
microbial DNA containing intestinal extracellular vesicles
- T2DM
Type 2 diabetes mellitus
- WT
wild type
- HFD
high-fat diet
- NCD
normal chow diet
- GF
germ-free
- PA
palmitate acid
- eWAT
epididymal white adipose
- ATMs
adipose tissue macrophages
- SVC
stromal vascular cell
- ISH
in situ hybridization
- RT-PCR
reverse transcriptase-polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have declared that no conflict of interest exists.
References
- 1.Lee YS, Wollam J, Olefsky JM. An Integrated View of Immunometabolism. Cell 2018;172:22–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roden M, Shulman GI. The integrative biology of type 2 diabetes. Nature 2019;576:51–60. [DOI] [PubMed] [Google Scholar]
- 3.Johnson AM, Olefsky JM. The origins and drivers of insulin resistance. Cell 2013;152:673–84. [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature 2017;542:177–185. [DOI] [PubMed] [Google Scholar]
- 5.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest 2017;127:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy M, Kolodziejczyk AA, Thaiss CA, et al. Dysbiosis and the immune system. Nat Rev Immunol 2017;17:219–232. [DOI] [PubMed] [Google Scholar]
- 7.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761–72. [DOI] [PubMed] [Google Scholar]
- 8.Jin X, Yu CH, Lv GC, et al. Increased intestinal permeability in pathogenesis and progress of nonalcoholic steatohepatitis in rats. World J Gastroenterol 2007;13:1732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson AM, Costanzo A, Gareau MG, et al. High fat diet causes depletion of intestinal eosinophils associated with intestinal permeability. PLoS One 2015;10:e0122195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thaiss CA, Levy M, Grosheva I, et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science 2018;359:1376–1383. [DOI] [PubMed] [Google Scholar]
- 11.Tilg H, Zmora N, Adolph TE, et al. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol 2020;20:40–54. [DOI] [PubMed] [Google Scholar]
- 12.Wollam J, Riopel M, Xu YJ, et al. Microbiota-Produced N-Formyl Peptide fMLF Promotes Obesity-Induced Glucose Intolerance. Diabetes 2019;68:1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018;359:1151–1156. [DOI] [PubMed] [Google Scholar]
- 14.Koh A, Molinaro A, Stahlman M, et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018;175:947–961 el7. [DOI] [PubMed] [Google Scholar]
- 15.Rothhammer V, Borucki DM, Tjon EC, et al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018;557:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puri P, Liangpunsakul S, Christensen JE, et al. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 2018;67:1284–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lelouvier B, Servant F, Paisse S, et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: A pilot analysis. Hepatology 2016;64:2015–2027. [DOI] [PubMed] [Google Scholar]
- 20.Amar J, Chabo C, Waget A, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med 2011;3:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anhê FF, Jensen BAFI, Varin TV et al. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat Metab 2020;2:233–242. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz S, Zapater P, Estrada JL, et al. Bacterial DNA translocation holds increased insulin resistance and systemic inflammatory levels in morbid obese patients. J Clin Endocrinol Metab 2014;99:2575–83. [DOI] [PubMed] [Google Scholar]
- 23.Mathieu M, Martin-Jaular L, Lavieu G, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 2019;21:9–17. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Defourny KAY, Smid EJ, et al. Gram-Positive Bacterial Extracellular Vesicles and Their Impact on Health and Disease. Front Microbiol 2018;9:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chelakkot C, Choi Y, Kim DK, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med 2018;50:e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pendyala S, Walker JM, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology 2012;142:1100–1101 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Z, Surewaard BG, Wong CH, et al. CRIg Functions as a Macrophage Pattern Recognition Receptor to Directly Bind and Capture Blood-Borne Gram-Positive Bacteria. Cell Host Microbe 2016;20:99–106. [DOI] [PubMed] [Google Scholar]
- 28.Helmy KY, Katschke KJ Jr., Gorgani NN, et al. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell 2006;124:915–27. [DOI] [PubMed] [Google Scholar]
- 29.He JQ, Katschke KJ Jr., Gribling P, et al. CRIg mediates early Kupffer cell responses to adenovirus. J Leukoc Biol 2013;93:301–6. [DOI] [PubMed] [Google Scholar]
- 30.Gorgani NN, He JQ, Katschke KJ Jr., et al. Complement receptor of the Ig superfamily enhances complement-mediated phagocytosis in a subpopulation of tissue resident macrophages. J Immunol 2008;181:7902–8. [DOI] [PubMed] [Google Scholar]
- 31.Gorgani NN, Thathaisong U, Mukaro VR, et al. Regulation of CRIg expression and phagocytosis in human macrophages by arachidonate, dexamethasone, and cytokines. Am J Pathol 2011;179:1310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo JB, Riopel M, Cabrales P, et al. Knockdown of Ant2 Reduces Adipocyte Hypoxia And Improves Insulin Resistance in Obesity. Nat Metab 2019;1:86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying W, Riopel M, Bandyopadhyay G, et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017;171:372–384 e12. [DOI] [PubMed] [Google Scholar]
- 34.Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol Pharm 2015;12:3650–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamichhane TN, Jay SM. Production of Extracellular Vesicles Loaded with Therapeutic Cargo. Methods Mol Biol 2018;1831:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Usman WM, Pham TC, Kwok YY, et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun 2018;9:2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pangburn MK, Muller-Eberhard HJ. Relation of putative thioester bond in C3 to activation of the alternative pathway and the binding of C3b to biological targets of complement. J Exp Med 1980;152:1102–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walport MJ. Complement. Second of two parts. N Engl J Med 2001;344:1140–4. [DOI] [PubMed] [Google Scholar]
- 39.Walport MJ. Complement. First of two parts. N Engl J Med 2001;344:1058–66. [DOI] [PubMed] [Google Scholar]
- 40.Reis ES, Mastellos DC, Hajishengallis G, et al. New insights into the immune functions of complement. Nat Rev Immunol 2019;19:503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hornef MW, Wick MJ, Rhen M, et al. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat Immunol 2002;3:1033–40. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Giardino Torchia ML, Lawson GW, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012;12:509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bitto NJ, Chapman R, Pidot S, et al. Bacterial membrane vesicles transport their DNA cargo into host cells. Sci Rep 2017;7:7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn J, Barber GN. STING signaling and host defense against microbial infection. Exp Mol Med 2019;51:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science 2019;363. [DOI] [PubMed] [Google Scholar]
- 46.Bai J, Cervantes C, Liu J, et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc Natl Acad Sci U S A 2017;114:12196–12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo X, Li H, Ma L, et al. Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Macrophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology 2018;155:1971–1984 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broadley SP, Plaumann A, Coletti R, et al. Dual-track clearance of circulating bacteria balances rapid restoration of blood sterility with induction of adaptive immunity. Cell Host Microbe 2016;20:36–48. [DOI] [PubMed] [Google Scholar]
- 49.Ono K, Nishitani C, Mitsuzawa H, et al. Mannose-binding lectin augments the uptake of lipid A, Staphylococcus aureus, and Escherichia coli by Kupffer cells through increased cell surface expression of scavenger receptor A. J Immunol 2006;177:5517–23. [DOI] [PubMed] [Google Scholar]
- 50.Sookoian S, Salatino A, Castano GO et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut 2020;69:1483–1491. [DOI] [PubMed] [Google Scholar]
- 51.Jiang L, Shen Y, Guo D, et al. EpCAM-dependent extracellular vesicles from intestinal epithelial cells maintain intestinal tract immune balance. Nat Commun 2016;7:13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caruso R, Warner N, Inohara N, et al. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity 2014;41:898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.