Abstract
IgG4 autoimmune diseases (IgG4-AID) are an emerging group of autoimmune diseases that are caused by pathogenic autoantibodies of the IgG4 subclass. It has only recently been appreciated, that members of this group share relevant immunobiological and therapeutic aspects even though different antigens, tissues and organs are affected: glomerulonephritis (kidney), pemphigus vulgaris (skin), thrombotic thrombocytopenic purpura (hematologic system) muscle-specific kinase (MuSK) in myasthenia gravis (peripheral nervous system) and autoimmune encephalitis (central nervous system) to give some examples. In all these diseases, patients’ IgG4 subclass autoantibodies block protein-protein interactions instead of causing complement mediated tissue injury, patients respond favorably to rituximab and share a genetic predisposition: at least five HLA class II genes have been reported in individual studies to be associated with several different IgG4-AID. This suggests a role for the HLA class II region and specifically the DRβ1 chain for aberrant priming of autoreactive T-cells toward a chronic immune response skewed toward the production of IgG4 subclass autoantibodies. The aim of this review is to provide an update on findings arguing for a common pathogenic mechanism in IgG4-AID in general and to provide hypotheses about the role of distinct HLA haplotypes, T-cells and cytokines in IgG4-AID.
Keywords: IgG4 autoimmune disease, MHC, autoimmunity, HLA-DRB1, HLA-DQB1, etiology, HLA class II
Introduction to IgG4-AID
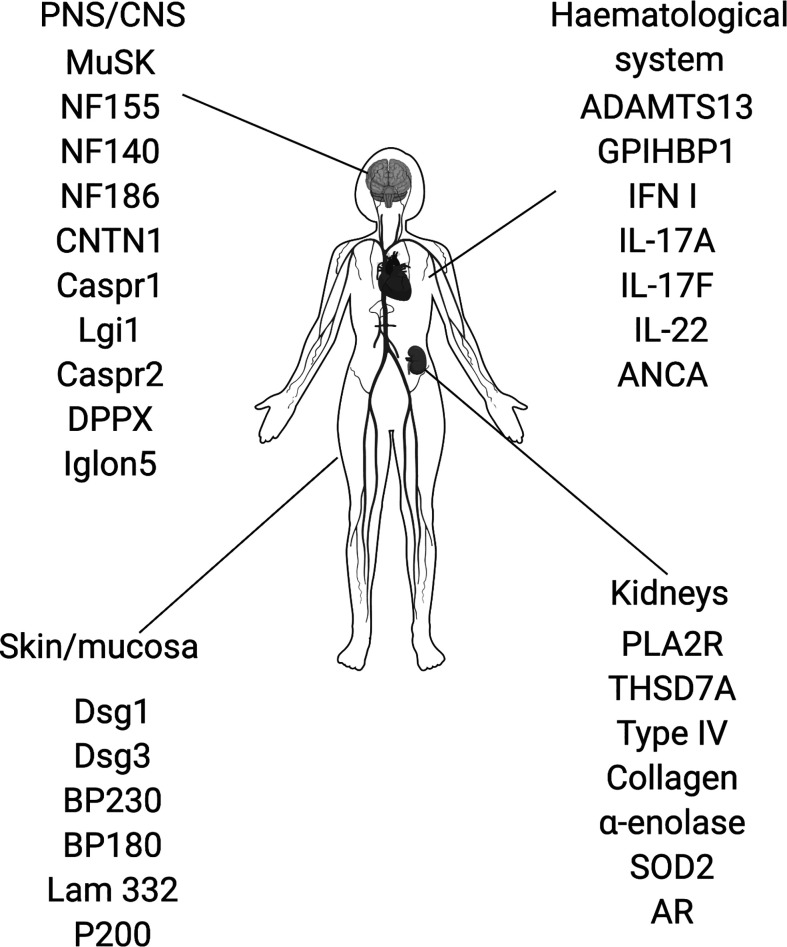
IgG4 autoimmune diseases (IgG4-AID) are a group of autoimmune diseases that are mediated by antigen-specific autoantibodies of the IgG4 subclass (1, 2). The best known IgG4 autoimmune diseases include pemphigus (3), MuSK myasthenia gravis (4), thrombotic-thrombocytopenic purpura (5), Goodpasture syndrome (6) and membranous nephropathy (7, 8). IgG4 is the least common subclass of IgG, it is immunologically inert as it does not activate complement or engage activating Fcγ receptors, which is also why therapeutic monoclonal antibodies are frequently modeled on the IgG4 subclass (9, 10). Since IgG4 is also produced in response to strong or chronic stimulation with antigen to down-regulate an overshooting immune response, it is often considered a protective antibody subclass, competing with other antibody classes and subclasses, e.g. IgG1 or IgE, for binding of antigen [reviewed here (9, 11, 12):]. Therefore, IgG4 is not considered as a pathogenic player in autoimmunity, and is an unlikely antibody to cause harm. Yet in recent years, more and more diseases were found to be primarily associated with IgG4 subclass autoantibodies targeting different autoantigens (2, 13). IgG4 autoimmune diseases were first appreciated as a group in 2015 (13). The number of suspected IgG4-AID is constantly growing and to date, 29 candidate antigens are known [ Figure 1 , (14)]. Most antigens are located in the central and peripheral nervous system, but there are also antigens in the skin and mucosa, kidneys and in the hematological system ( Figure 1 ). The group of IgG4-AID is defined by several characteristics beyond mere antibody isotype predominance: 1) the IgG4 autoantibodies target transmembrane/extracellular or soluble antigens, 2) are directly pathogenic, meaning that they exert their effect without complement- or cell-dependent cytotoxicity by blocking of binding sites for direct interaction partners, ligands or substrates, 3) all IgG4-AID have a low prevalence affecting less than 5 per 10,000, and 4) mostly respond favorable to treatment with the B-cell depleting agent rituximab, an observation that has been puzzling because long-lived antibody producing plasma cells do not carry CD20 and are therefore not depleted. Another more recent observation points into the same direction of IgG4-AID sharing common immunobiological mechanisms: Most of them are associated with similar variants of the HLA class II region ( Table 1 ). In this review, we summarize the information available for the group of IgG4-AID and hypothesize on common immunopathogenesis.
Figure 1.
Proposed IgG4 autoantibody targets. ADAMTS13, disintegrin and metalloproteinase with thrombospondin motifs 13; ANCA, antineutrophil cytoplasmic antibodies; AR, Aldose reductase; BP180; BP230, bullous pemphigoid antigen 180/230; CNTN1, contactin 1; Caspr1/2, contactin associated protein1/2; Dsg1/3, desmoglein 1/3; DPPX, dipeptidyl-peptidase-like protein 6; GPIHBP1, glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1; IFN I, type I interferon, IgLON5, IgLON family member 5; IL-17A/17F/22, interleukin-17A/17F/22; Lam332, Laminin 332; LGI1, leucine-rich, glioma inactivated 1; MuSK, muscle-specific kinase; NF140/155/186; Neurofascin 140/155/186; PLA2R, phospholipase A2 receptor; SOD2, manganese superoxide dismutase; THSD7A, thrombospondin type-1 domain- containing 7A.
Table 1.
Reported HLA-associations of IgG4 autoimmune diseases.
| Disease/antigen | Affected organ | Reported HLA association | DQB1*05 | DRB1*04 | DRB1*11 | DRB1*14 | DRB1*15 | Protective alleles/No association | References |
|---|---|---|---|---|---|---|---|---|---|
| Class I IgG4 autoimmune diseases | |||||||||
| MuSK-MG/MuSK | PNS | DQB1*05, DRB1*14, DRB1*16, DQB*03, DQB1*0502, DQB1*0301/0313, DQA1*0101 (Iran) | ✓ | ✓ | DQA1* alleles (except Iran) HLA-DRB1∗03 (Turkey) DRB1*13 (Serbia) |
(15–23) | |||
| Pemphigus foliaceus, fogo selvagem (and p. vulgaris of mucocutaneous type)/Desmoglein 1 | Skin/mucosa | PF: HLA-DRB1*04, HLA-DRB1*14, DRB1*1404 DQB1*0503, HLA-A10 FS: DRB1*01 DRB1*0404, *1402, *1406 or *0102 |
✓ | ✓ | ✓ | N/A | (24–30) | ||
| Pemphigus vulgaris/Desmoglein 3 | Skin/mucosa | HLA-DRB1* 0402, HLA-DRB1*1401/*1402, HLA-DQB1*05:03, HLA-DQB1*03 HLA-DRB1*03 DQB1*03:02, DQB1*0503 |
✓ | ✓ | ✓ | DQB1*05:01, DQB1*02, DQB1*06:01, and DQB1*03:03 | (27–29, 31, 32) | ||
| Thrombic thrombocytopenic purpura/ADAMTS13 | Blood/hematological system | HLA-DRB1*11, HLA-DRB1*04, HLA-DRB1*14, HLA-DRB4, HLA-DQB1*03, and HLA-DQB1*05/*06/*02, HLA-A*03 DRB1∗15, HLA-DQB1*02:02 |
✓ | ✓ | ✓ | ✓ | ✓ | HLA-DRB1*03DRB1*07-DQB1*02 and DRB1 *13-DQB1 *06 HLA-DRB1*04 (John study) |
(33–37) |
| CIDP/Neurofascin 155 | PNS | HLA-DRB15*01 and *02 | ✓ | N/A | (38) | ||||
| CIDP/CNTN1 (Contactin 1)/ | CNS | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Class II IgG4 autoimmune diseases | |||||||||
| Encephalitis Morvan’s syndrome/LGI1 | CNS | HLA-DRB1*07:01 and linked alleles (HLA-DRB1*07:01- DQB1*02:02) HLA-DR7 and HLA-DRB4 HLA-B*44:03, C*07:06 |
✓ | N/A | (39–42) | ||||
| Membranous nephropathy/PLA2R | Kidneys | DRB1*15:01, DRB3*0202, | ✓ | N/A | (7, 43, 44) | ||||
| CNS and PNS disorders/CASPR2 | CNS/PNS | HLA-DRB1*11:01 (Linkage: DRB1*11:01-DQA1*05:01-DQB1*03:01) | ✓ | N/A | (39) | ||||
| Membranous nephropathy/THSD7A | Kidneys | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| GPIHBP1 autoantibody syndrome/GPIHBP1 | Blood/hematological system | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| CIDP/CASPR1 | PNS | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Mucous membrane pemphigoid/Laminin 332 | Skin/mucosa | Unclear/N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Class III IgG4 autoimmune diseases | |||||||||
| Goodpasture syndrome/Type IV collagen | Kidneys | HLA-DRB1*15:01 (DRB1*0404) DRB1∗15:02 | ✓ | ✓ | N/A | (6, 45–48) | |||
| IgLON5 Parasomnia/IgLON5 | CNS | HLA-DRB1*10:01, HLA-DQB1*05:01 | ✓ | N/A | (49) | ||||
| Bullous pemphigoid/Bulloid pemphigoid antigen 180/230 | Skin/mucosa | DQB1*0301, DRB1*04, DRB1*11, DQB1*0501 and DRB1*1001, DQA1*05, |
✓ | ✓ | ✓ | HLA-DQA1*01:02/03, HLA-DQB1*02:02, and HLA-DRB1*07:01 | (50–53) | ||
| Granulomatosis with Polyangiitis/ANCA | Blood/hematological system | HLA-DPB1*0401, HLA-DR1, HLA-DR4, HLA-DRB1*0901-HLA DQB1*0303, HLA-DR9, HLA-DR13, HLA-DRB1*1202, HLA-DRB1*15, HLA-DRB1*0405 | ✓ | ✓ | (54–56) | ||||
| Autoimmune polyendocrine syndrome type 1 (APECED)/IFN I, IL-17A, IL-17F and IL-22 | Blood/hematological system | DRB1*03:01, DRB1*04:01, DQA1*03:01, DQA1*05:01, DQB1*02:01, DQB1*03:02 Subtypes: Addison’s disease: HLA-DRB1*03, Alopecia: HLA-DRB1*04- DQB1*0302 |
✓ | Subtype with type 1 diabetes: HLA-DRB1*15-DQB1*0602 | (54–56) | ||||
| CIDP/Neurofascin 140/186 | PNS | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Anti-laminin γ1/Anti-P200 pemphigoid/P200 (laminin γ1) | Skin/mucosa | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| DPPX encephalitis/DPPX | CNS | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Membranous nephropathy/αenolase, SOD2, AR/ | Kidneys | N/A | N/A | N/A | N/A | NA/ | N/A | N/A | N/A |
Notably the study populations are from various different geographic locations and contain participants of diverse ethnicities, which may influence HLA associations. CIDP, chronic inflammatory demyelinating polyneuropathy; N/A, data not available; CNS, central nervous system; PNS, peripheral nervous system.
Known IgG4 Autoantibody-Associated Disease: A Colorful Bouquet
There are many distinct forms of IgG4-AID with a wide variety of symptoms, caused by IgG4 autoantibodies that may target one of four main organs: 1) the skin and mucosa, 2) the peripheral and central nervous system, 3) the kidneys and 4) the hematological system ( Figure 1 ). The pathogenic mechanism of these antibodies is based on an IgG4-mediated blocking of protein-protein and/or cell-cell interaction. The detailed description of all IgG4 -AID and their disease mechanisms would by far exceed the scope of this review and was therefore published separately (14), but we would like to briefly summarize the key mechanisms of a few example diseases.
Probably the most commonly known IgG4-AID targets skin and mucous membranes: pemphigus vulgaris. This disease is hallmarked by acantholysis and blister formation caused by the loss of keratinocyte adherence. The targets of IgG4 autoantibodies are the desmogleins (Dsg1 or 3) located at the desmosomes. Desmogleins interact closely with each other to maintain the adherence of the cells, and thereby the tissue architecture of skin and mucosa. The binding of IgG4 to desmoglein 1 or 3 leads to a loss of interaction between desmogleins, which causes the disruption of the cell adhesion and in consequence to the formation of blisters (57–59).
Some IgG4-AID cause central and/or peripheral nervous system dysfunction, depending on the target antigen localization. Antibodies targeting the CNS synaptic scaffolding protein LGI1 disrupt synaptic signaling by blocking the trans-synaptic clustering of receptors and channels mediated by this antigen. This leads to amnesia, epileptic seizures and psychiatric symptoms (60–62).
In the peripheral nervous system, IgG4 autoantibodies may, for example, recognize the protein neurofascin 155 (NF155). NF155 is important for attaching the myelin sheath lamellae to the axon at the paranodes adjacent to the nodes of Ranvier. This leads to disruption of action potentials (conduction slowing and block) and consecutively to chronic neuropathies [chronic inflammatory demyelinating polyneuropathy; CIDP (63–67)]. In IgG4-AID, which target synaptic antigens localized in the peripheral and central nervous system, e.g. CASPR2, combinations of central nervous system dysfunction (limbic encephalitis) and peripheral nervous system dysfunction (neuromyotonia, pain) may occur (68–70).
The signal for muscle contraction is transmitted by the release of the neurotransmitter acetylcholine from the motoneuron to the muscle, where it is bound by acetylcholine receptors (AChR). Muscle-specific kinase (MuSK) is a key regulator of the neuromuscular junction that is part of a signal transduction pathway required for dense clustering of acetylcholine receptors (AChR) at the neuromuscular junction. IgG4 autoantibodies against MuSK block its interaction with another protein in the signal transduction pathway, leading to reduced densities of AChR (71, 72). As a consequence, there is less efficient neuromuscular transmission. This means that not every signal from the motoneuron to move the muscle leads to a muscle contraction, which translates into a severe, fatigable weakness of skeletal muscles.
Autoantibodies against kidney antigens PLA2R andTHSD7A are found in patients with membranous nephropathy, where the pathogenic mechanisms are still unclear, PLA2R and THSD7A are expressed in cells of the glomerulus (podocytes) inside of renal corpuscles. The glomerulus filters the blood, so the adhesion between the cells is important for the function of the glomerulus, but we know very little about the role of PLA2R and THSD7A and what the autoantibodies exactly do. THSD7A-IgG4 was hypothesized to block protein-protein interactions of THSD7 and cell-cell interactions, therefore affecting the glomerular filtration and causing proteinuria (73–75). Potentially affected binding partners of PLA2R are collagen IV (76), which is important for cell adhesion, or the At2-complex (14, 77), which is important to maintain cell architecture and affect podocyte function. Perhaps a blocking of PLA2R interaction with these binding partners affects cell adhesion and/or cell architecture. Interestingly it is also thought that a specific subtype of complement activation, via mannose-binding lectins may occur (78–80).
Thrombotic thrombocytopenic purpura (TTP) is another well-known disease that is mediated by IgG4. Patients with TTP have IgG4 autoantibodies against a protease located in the blood, ADAMTS13. The normal function of ADAMTS13 is to cleave and inactivate the multimeric forms of the von Willebrand factor (vWF), which is a protein that is (in its multimeric form) required for blood clotting, e.g. following injury. Blocking of ADAMTS13 function by IgG4 leads to accumulation of multimeric vWF, which then binds to platelets. This leads to the formation of microthrombi that may block capillaries and therefore cause vascular occlusion. This is especially severe if the thrombi occur in brain, kidneys or lungs (17, 81, 82). A different antigen in the hematological system is GPIHBP1, which is a protein expressed in capillary endothelial cells and which have the main function to transport lipoprotein lipase (LPL) into the blood. LPL breaks down lipids, so when it is not transported into the blood because IgG4 binding to its transporter GPIHBP1, the patients will suffer from hypertriglyceridemia (83, 84).
Role of HLA Class II Polymorphism in IgG4-AID: Emerging Genetic Predisposition
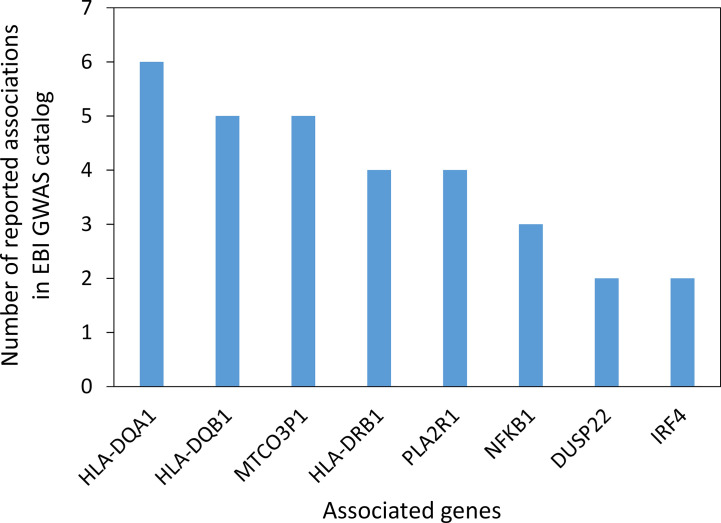
In the search for common immunopathogenesis, we wanted to investigate mutations or variants of genes that may contribute to a genetic predisposition for the development of IgG4-AID. We therefore investigated whether there are candidate genes that have been identified in genome wide association studies (GWAS) of individual IgG4-AID. Data on pemphigus vulgaris, pemphigus foliaceus, membranous glomerulonephritis and eosinophilic granulomatosis with polyangiitis were identified in the NHGRI-EBI catalog of human genome-wide association studies (85). The most frequently reported associations were with variants in the HLA class II region: HLA-DQA1, HLA-DQB1, HLA-DRB1 ( Figure 2 , Supplementary Table 1 ).
Figure 2.
Genes with potential association to IgG4-AID reported in the EBI GWAS catalog (85). Data represents 17 studies investigating four Ig4G-AIDs (Pemphigus vulgaris, Pemphigus foliaceus, Membranous glomerulonephritis, Eosinophilic granulomatosis with polyangiitis; median of 4 studies per disease). The GWAS catalog reports multiple associations per disease and multiple genes mapped to the association signal, so reported genes do not necessarily represent independent associations. The gene MTCO3P1 is a pseudogene located next to HLA-DQB1 in the HLA class II region. Only genes reported more than once are shown. [GWAS catalog accessed on 03.11.2020).
Most autoimmune diseases are known to have a genetic association with the HLA class II region, and it is thought that the presence of specific HLA alleles may cause a genetic predisposition to develop specific autoimmune diseases or groups of autoimmune diseases (86–88). Therefore, we also looked at reports of associations with HLA class II genes, as it may be possible that there are one or more HLA alleles that may be specifically associated with one or more IgG4-AIDs. Interestingly, we observed that many studies reported a genetic association of individual IgG4-AID with HLA-class II variation: HLA*DRB1*14, 04,15 or 11, and/or HLA-DQB1*05 ( Table 1 ).
For example, the HLA-DRB1*14 supertype was found associated with MuSK MG, pemphigus and thrombotic thrombocytopenic purpura, HLA-DQB1*05 in Iglon5 parasomnia, thrombotic thrombocytopenic purpura and MuSK MG (15, 16, 18, 19, 89), HLA-DRB1*15 is associated with NF155 antibody positive CIDP, pemphigus and PLA2R antibody positive MN [in a Chinese population, up to 99% of patients with PLA2R antibodies carried either DRB1*15:01 or DRB3*02:02, and individuals with these alleles have a 99-fold increased risk of PLA2R-associated MN (43)]. Alleles of the DRB1 and DQB1 genes were associated with most IgG4 autoimmune diseases, suggesting that the HLA class II genes play a role not only in typical autoimmune diseases, but also in the induction of an IgG4 specific autoimmune response. The next question is, whether there is a specific allele associated with a class switch toward IgG4. Looking at IgG4-related diseases in comparison, also the HLA-DRB1 locus was found to be associated in two out of six studies (90), in one of these studies HLA-DRB1*15 was associated with increased IgG4 levels (91). Whether specific alleles are truly associated with an increased predisposition to induce a class switch toward IgG4 and to develop IgG4-AID cannot yet be concluded, but a systematic review and meta-analysis are currently being conducted in our research groups to address this possibility.
A Potential Explanation for the Gradient: Association of Prevalence and Genetic Background in Europe
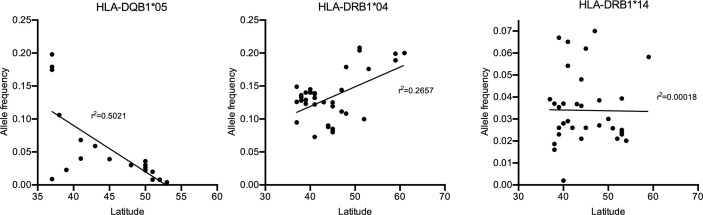
If specific HLA class II alleles would contribute to a genetic predisposition for IgG4-AID, it may be possible that geographical differences in HLA class II genotype may correlate with an increased disease prevalence. Interestingly, both in MuSK-MG (92) and in pemphigus vulgaris (93), a north-south gradient can be observed in Europe, with higher prevalence in southern countries such as Italy and fewer cases in northern countries such as Finland or Norway. Potential reasons may be environmental or genetic factors. If there were also geographical differences in HLA class II genotype frequencies, it would underline the relevance of the HLA class II alleles on disease susceptibility. To test this hypothesis, we have plotted the frequency of the HLA-DQB1*05:02 allele as well as of HLA-DRB1*04 and *014 {data obtained from the Allele Frequency Net Database [AFND, (94)], Table 2 } against latitude in European countries ( Figure 3 ). There is a trend toward increased frequency of HLA-DQB1*05:02 in countries with lower latitude, such as Italy, which also show increased frequency of MuSK MG (92) and pemphigus (93), while there is a reduced frequency for HLA-DRB1*04 and no apparent change in HLA-DRB1*14 in countries with low latitudes.
Table 2.
Frequency of the HLA-DQB1*05:02 allele in European countries sorted by latitude. Source: The Allele Frequency Net Database [AFND (94)].
| Country/Region (defined by AFND) | % of population carrying allele | Allele frequency | Cohort size | Geographical location |
|---|---|---|---|---|
| Ireland Northern pop 2 | 0.8 | 0.004 | 122 | 54_32_N_5_55_W |
| Ireland South | 1.6 | 0.008 | 250 | 53_20_N_6_15_W |
| Netherlands UMCU | 1.6 | 0.0078 | 64 | 52_5_N_5_10_E |
| Netherlands | 5.5 | 447 | 52_0_N_7_0_E | |
| England pop 3 | 4 | 0.02 | 61 | 51_30_N_0_7_W |
| England pop 6 | 1.7 | 0.008 | 177 | 51_30_N_0_7_W |
| Germany Essen | 5.2 | 0.026 | 174 | 51_27_N_7_0_E |
| Czech Republic pop 3 | 7.2 | 0.036 | 180 | 50_7_N_14_24_E |
| Belgium | 6.1 | 0.03 | 99 | 50_50_N_4_21_E |
| Belgium pop 2 | 4.5 | 0.022 | 715 | 50_50_N_4_21_E |
| Czech Republic pop 2 | 5.1 | 99 | 50_5_N_14_28_E | |
| Czech Republic Romani | 17.6 | 34 | 50_5_N_14_28_E | |
| Germany pop 3 | 3.6 | 111 | 50_48_N_8_46_E | |
| Austria | 6 | 0.03 | 200 | 48_13_N_16_21_E |
| France West | 3 | 100 | 48_0_N_2_0_W | |
| Slovenia pop 2 | 8.6 | 140 | 46_3_N_14_30_E | |
| France Southeast | 7.7 | 0.039 | 130 | 46_0_N_5_0_E |
| Italy Bergamo | 11.9 | 0.059 | 101 | 45_42_N_9_40_E |
| Italy North pop 3 | 13.4 | 0.068 | 97 | 43_0_N_12_0_E |
| Italy Rome | 7 | 0.04 | 100 | 41_54_N_12_27_E |
| Spain (Catalunya, Navarra, Extremadura, Aaragón, Cantabria, | 4.5 | 0.0228 | 4,335 | 41_22_N_2_11_E |
| Italy Sardinia pop2 | 20.6 | 0.106 | 1,129 | 39_14_N_9_3_E |
| Portugal Azores Terceira Island | 1.8 | 0.0088 | 130 | 38_43_N_27_13_W |
| Greece | 31 | 0.198 | 48 | 37_58_N_23_43_E |
| Greece pop 8 | 31.3 | 0.1747 | 83 | 37_58_N_23_43_E |
| Greece pop2 | 32.6 | 0.179 | 120 | 37_58_N_23_43_E |
| Greece pop3 | 30.1 | 246 | 37_58_N_23_43_E | |
| Spain Malaga | 3.8 | 160 | 36_43_N_4_25_W | |
| Spain Malaga Romani | 10 | 80 | 36_43_N_4_25_W |
Figure 3.
Allele frequency of the HLA-DQB1*05:02 allele in Europe [Source: The Allele Frequency Net Database (AFND)].
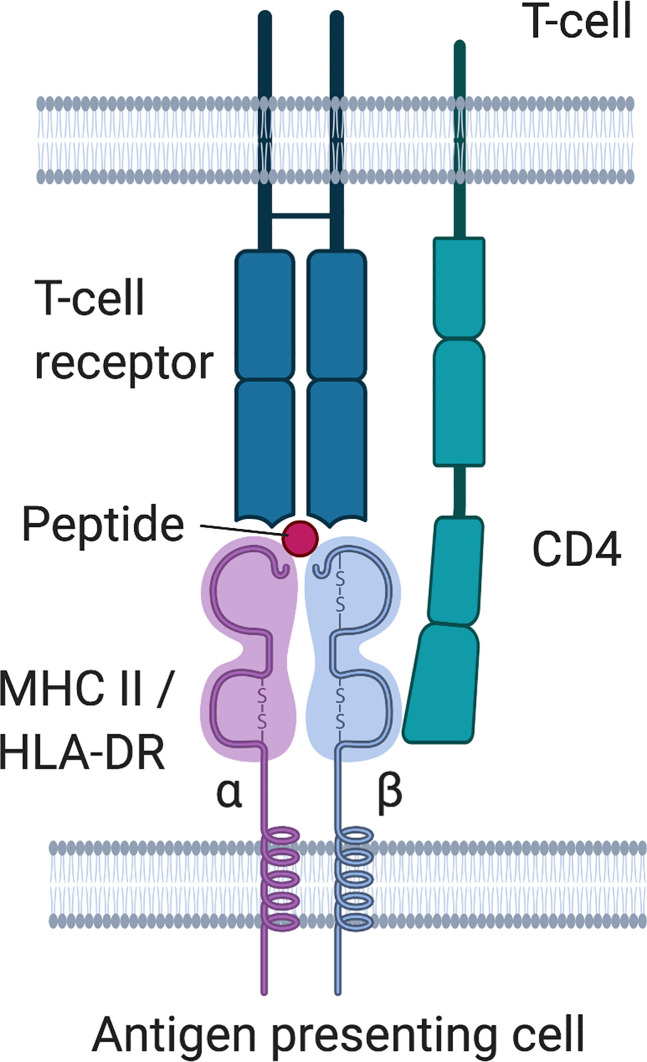
Figure 4.
HLA class II molecule structure and interaction with T-cell receptor.
Immunogenic Peptides and how to Present Them: Are There Distinct Shared Sequences in Antigen Peptides and HLA Class II Peptide Binding Grooves?
Next we wanted to investigate whether there is any evidence that may support a functional role of HLA class II molecules for disease susceptibility. One possibility is that distinct HLA class II alleles may play a role in inducing a distinct IgG4 specific T-cell response by the presentation of specific immunogenic peptides (that perhaps are best presented by peptide-binding grooves that have a distinct sequence). This would mean that the HLA haplotype may play a role in the susceptibility to develop IgG4-AIDs via specific sequences in the peptide binding groove and/or specific sequences in the presented peptide or in the T-cell receptor (TCR). There is some evidence for this hypothesis.
HLA Class II Alleles in IgG4-AID
We investigated if there is evidence for any interesting structural characteristics of the peptide-binding groove of IgG4-AID associated HLA class II molecules. Two risk alleles for membranous nephropathy with PLA2R antibodies were identified in a Han Chinese population in DRB1*15:01 and DRB1*03:01 (43), either of which were found in 73.6% of the patient cohort (compared to 33% in the healthy control population), and that contained specific amino acid variants in the peptide-binding groove of the HLA class II molecules which is responsible for antigen presentation to the T-cell receptor (43, 44). Specifically important were arginine at position 13 (arginine13) in HLA-DRB1*15:01 and alanine at position 71 (alanine71) in DRB1*03:01 for interactions with T cell epitopes of PLA2R. The role of arginine13 in HLA-DRB1*15:01 is also mentioned as relevant in another IgG4-AID. IgG4 autoantibodies against the α3(IV)NC1 in Goodpasture disease also have a strong association with HLA-DRB1*15:01, and that arginine13 and proline11 of the DRB1*15:01 encoded DRβ1 chain were associated with disease susceptibility. Furthermore isoleucine137, tryptophan140, glycine142, phenylalanine143 and phenylalanine145 were presented in peptide-binding pockets 1, 4, 6, 7, and 9, and specifically arginine13 in pocket 4 interacts with tryptophan140 of the peptide and forms a hydrogen bond (45). Different alleles, HLA-DRB1*04:04,14:02 and 14:06 all share a sequence in the hypervariable region in residues 67 to 74 (LLEQRRAA) and these alleles were reported to be linked to fogo selvagem, an endemic form of pemphigus vulgaris (24). HLA-DRB1*04 is also associated with other IgG4-AID ( Table 1 ). In pemphigus, the carrier frequency of the HLA-DRB1*04 allele is very high, for example it is found in 60% of patients from Brazil and in 71% of Dutch patients, with similar and lower allele frequencies in the healthy population in both countries [Brazil: 39%, Netherlands: 38%, (96)]. Analysis of polymorphic residues that form the binding pockets of the DRβ1 chain of HLA-DRB1*04 and *14 alleles in pemphigus vulgaris show that they contain similar sequences, as well as in HLA-DRB1*04, *14 and *01 in pemphigus foliaceus. Of note are structural similarities between DRB1*04:04 and 01:02 that lead to similar binding pockets and may bind similar peptides [ Table 3 , (25)]. These may present the same peptide from Dsg1, while in pemphigus vulgaris, two distinct types of peptide may be presented by DRB1*04:02 or DRB1*14/04:06. Furthermore, DRB1*14/04:06 may be able to present both Dsg1 and Dsg3 peptides, which may be the reason for antibodies against both antigens in pemphigus vulgaris with muco-cutaneous clinical phenotype (25). Overall it is likely that HLA-DR is important in antigen presentation in pemphigus. HLA- DRB1*04:02 is relevant for the selection of specific self-peptides in pemphigus vulgaris (97), autoreactive T-cells proliferated in response to antigen presented by HLA-DR in Fogo selvagem and this response could be blocked by anti–HLA-DR antibodies, but not by anti–HLA-DQ or anti–HLA-DP antibodies (98).
Table 3.
Similarities between the two IgG4-AID-associated HLA variants DRB1*0404 and DRB1*0102 in the protein sequence of the two HLA variants (a) and in the amino acid motifs of the peptides bound by these two HLA variants (b).
| a) Residues characterizing the binding pockets of DRB1*0404 and DRB1*0102 | |||||
| DRB1* | P1 | P4 | P6 | P7 | P9 |
| 0404 | V | H Q R A Y | V | D Y Y L R | E Y D Y |
| 0102 | V | F Q R A Y | L | E C Y L R | W S D Y |
| b) Amino acid motifs of peptides bound to pockets of DRB1*0404 and DRB1*0102 | |||||
| DRB1* | P1 | P4 | P6 | P7 | P9 |
| 0404 | VLMIF | ALIV/WDEFY/no KR | STNQR | ADHIL/MNPST | ALV/QGSTK |
| 0102 | VLMIF | ALIV/MNQ/no HKR | AGSTP | ALV/INFY | |
Table adapted from (25). Negative charged amino acid (D, E) are in bold, positively charged amino acids (R) are underlined. P1-P9: binding pockets.
The Role of Peptides Bound to the HLA Class II
If indeed individual HLA class II associations predispose to disease development, could this be mediated by certain immunogenic peptides of the target antigens? Therefore we were interested in the sequences of the IgG4 antigen peptides that are presented by HLA class II molecules and recognized by the T-cell receptor. Several studies investigated T-cell peptides and their (potential) interaction with the binding pockets of the HLA class II, other studies provide at least a list of predicted peptides ( Table 4 ). In Goodpasture disease, two overlapping peptides of α3(IV)NC2 that may bind HLA-DRB1*15:01 were investigated: DIPPCPHGWISLWKG and HGWISLWKGFSFIMF, with glycine142 identified as relevant for binding present in both peptides (45, 99). Several Dsg3 peptides binding to HLA-DRB1*04:02 (one of the alleles associated with pemphigus vulgaris) were identified (101–103) and several were validated by in vitro tests with patient derived peripheral blood mononuclear cells [PBMCs, (100)]. T-cell responses were elicited by several peptides ( Table 4 ), with one peptide, Dsg3190-204 inducing reactivity in all samples and also led to secretion of high levels of IL- 4 and IL-10 (100), which are cytokines for IgG4 class switch.
Table 4.
An (incomplete) list of predicted and validated peptides of IgG4 autoantigens presented in the MHC II in the context of IgG4-AID identified from published literature.
| HLA allele | Peptide | Comment | Antigen | Reference |
|---|---|---|---|---|
| DRB1*15:01 | HGWISLWKGFSFIMF | Tryptophan140 forms hydrogen bond with arginine13 in pocket 4, glycine142 relevant for binding | α3(IV)NC2 | (45) |
| DRB1*15:01 | DIPPCPHGWISLWKG | Cysteine132, tryptophan136, lysine 141, and glycine 142 are relevant for binding | α3(IV)NC2 | (99) |
| DRB1*04:02 | LNSKIAFKIVS QEPA | Dsg3190-204,Elicited immune response in PBMC of all patients in study, IL-4 and IL-10 secretion | Dsg3 | (100) |
| DRB1*04:02 | TPMFLLSRNTGEVRT | Elicited immune response in PBMCs of a subset of patients | Dsg3 | (100) |
| DRB1*04:02 | CECNIKVKDVNDNFP | Dsg3 | (100) | |
| DRB1*04:02 | QSGTMRTRHSTGGTN | Dsg3 | (100) | |
| DRB1*04:02 | VKILDI NDNPPVFSQQ IFMGEIEENS ASNSLVMILN ATDADEPNHL NS | Induced proliferation of T cells from PV patients | Dsg3 | (101) |
| DRB1*04:02 | A DKDGEGLSTQ CECNIKVKDV NDNFPMFRDS QYSARIEENI LSSELLRFQV TDLDEEYTDNWLA | Dsg3 | (101) | |
| DRB1*04:02 | DSQNNRCEMPR SLTLEVCQCD NRGICGTSYPTTSPGTRYGR PHSG | Dsg3 | (101) | |
| DRB1*04:02 | IFMGEIEENSASNSLVM | Predicted peptide | Dsg3 | (102) |
| DRB1*04:02 | GIAFRPASKTFTVQKGI | Predicted peptide | Dsg3 | (103) |
| DRB1*11:01 | GDMLLLWGRLTWRKM | Immunodominant T-cell epitope | ADAMTS13 | (104) |
| DRB1*11/DRB1*15 | FINVAPHAR | Core motif presented by DRB1*11 and DRB1*15, recognized by patient CD4+ T cells | ADAMTS13 | (105, 106) |
| HLA-DRB1*11/DRB1*03 | ASYILIRD | core motif recognized by CD4+ T-cells | ADAMTS13 | (105, 107) |
| HLA-DQ/DR | IHALATNMG | Predicted peptides presented by HLA-DR | ADAMTS13 | (105) |
| HLA-DR | LIRDTHSLR | ADAMTS13 | (105) | |
| HLA-DR | LKTLP-PARC | ADAMTS13 | (105) | |
| HLA-DQ/DR | RGPGQADCAVAIGRPLG | Predicted peptide presented by both HLA-DR and -DQ | ADAMTS13 | (105) |
| HLA-DR | SRRQLLSLLSAGRAR | Predicted peptide presented by multiple HLA-DR alleles | ADAMTS13 | (105) |
| HLA-DR | FSEGFLKA-QASLRGQYW | ADAMTS13 | (105) | |
| DRB1*11:01 | YRFRNKKMK | Predicted core motifs for DRB1*11:01 | CASPR2 | (39) |
| YITLELKKAKLV | ||||
| YFCKMSRLLNT | ||||
| FKTLTPWGV | ||||
| FLGCIRSLRMNGV | ||||
| FLKLDHYPS | ||||
| FNQIAPLKAALR | ||||
| LIRYMFRHKG | ||||
| DRB1*07:01 | FLFTPSLQLLL | Predicted core motifs for DRB1*07:01 | LGI1 | (39) |
| FRGLKSLIHLSL | ||||
| FVVADSSKA | ||||
| FYSHQSLHA | ||||
| YQWNKATQL | ||||
| FTHVSINKR | ||||
| FLFASSFKGN |
In studies on thrombotic thrombocytopenic purpura (TTP), several T-cell peptides were identified in ADAMTS13 ( Table 4 ), with the ADAMTS13 1239-1253 peptide (GDMLLLWGRLTWRKM) as the single immunodominant CD4+ T-cell epitope that was bound both by HLA-DRB1*15:01 and DRB1*11:01 in ELISA and also by T-cells from a strain of mice expressing human HLA-DRB1*01:01 (104). A recent study looking into peptides presented on HLA-DR and HLA-DQ (105) identified 12 peptides presented by DR and 8 by DQ. The study identified an overlap and also differences in the repertoire of ADAMTS13 peptides that are presented on HLA-DQ and HLA-DR. Most DR peptides derived from the CUB-domain of ADAMTS13. DQ-presented peptides mostly derived from the cysteine-rich, TSP2, and TSP2-linker 1 of ADAMTS13, which demonstrates that haplotypes have distinct peptide repertoires. Nevertheless there are also peptides which could be presented by both DR and DQ ( Table 4 ). Furthermore, polymorphisms in the antigen may play a role in disease susceptibility. In pemphigus foliaceus a coding synonymous T/C single nucleotide polymorphism (SNP) at position 809 in the DSG1 gene was found in French patients (108). This mutation is silent, but the SNP was hypothesized to have a role in alternative splicing (109). In a different study, alternative splicing of DSG1 led to a 101-bp insertion at the 3′ end of DSG1-intron 6 and introducing a stop codon in the nucleotide sequence, thus coding for a truncated isoform of Dsg1 containing a peptide that was presented by DRβ1*01:02 and activated autoreactive T-cells (110).
Is There Evidence for Selective T-Cell Receptor VDJ Usage?
Peptides presented in the HLA class II molecules are recognized by the T-cell receptor (TCR). Therefore we also wanted to study whether the structure of the TCR may play a role for the recognition of IgG4 antigens, and therefore investigated the literature for evidence of specific TCR VDJ usage (which directly would influence structure of the TCR) in MuSK MG and pemphigus foliaceus. There was no evidence for a preference for a specific Vβ gene in pemphigus foliaceus (111), but in pemphigus vulgaris, different Vβ genes were used, with cells reacting to peptide Dsg3(AA145-192/VKILDI NDNPPVFSQQ IFMGEIEENS ASNSLVMILN ATDADEPNHL NS), (AA240-303, A DKDGEGLSTQ CECNIKVKDV NDNFPMFRDS QYSARIEENI LSSELLRFQV TDLDEEYTDN WLA) or (AA570-614, D SQNNRCEMPR SLTLEVCQCD NRGICGTSYP TTSPGTRYGR PHSG) being linked to Vβ13, 7 or 17 genes, respectively, and Dsg3(AA145-192) and (AA240-303) were also linked to Vα22 or 10, respectively (112).In a study with 13 MuSK MG patients, MuSK responsive T-cells from HLA-DQB1*05+ MuSK-MG patients had a restricted set of TCR VJ rearrangements. Two common motifs in TRBV29 were identified as GXGQET and TEHQET, these were shared in 4 patients (113). There is not enough data available to make any assumptions yet, and overall there is a need for more studies investigating the TCR-peptide-HLA class II interaction in the different IgG4-AID.
Hypothetical Etiology and Mechanisms in IgG4-Autoantibody Associated Diseases: A Unifying Theory?
The HLA class II gene region is linked to immunopathogenesis of autoimmune diseases (88, 114, 115) and we are specifically interested whether a subset of HLA alleles may be specifically linked with IgG4-autoimmunity. The presentation of autoantigens to developing T-cells relies strongly on the highly polymorphic β chain of HLA-DR ( Box 1 ). Perhaps the structure of the peptides presented by HLA-DR β1 variants contribute to the generation of IgG4 autoantibodies later on by aberrantly priming T-cells, by the type of peptide that is presented, by the conformation of the peptide in the peptide-binding groove or by altered interaction with the T-cell receptor or CD4. In line with this theory is the observation of one study that risk alleles HLA-DRB1*15:01 and HLA-DRB3*02:02 for PLA2R-IgG4 associated MN (DRB1*15:01 also being associated with further IgG4 AID such as Goodpasture disease or TTP, Table 1 ) contain specific amino acid variants in the peptide-binding groove of the DR β chain responsible for antigen presentation to the TCR (43).
Box 1 . HLA- class II molecules.
HLA class II molecules are located at the cell surface of professional antigen-presenting cells and epithelial cells in the thymus, and present peptides from extracellular proteins to CD4+ T helper cells or developing T- cells, which then in turn may activate B-cells and stimulate the production of (auto)antibodies. There are three main HLA II molecules, HLA-DR, HLA-DQ and HLA-DP. HLA-DR consists of an α- and a β -chain. The chains consist of two domains, the α1 and α2 as well as the β1 and β2 domain, with the α1 and β1 domain forming a groove that presents the peptide. The α chain is encoded by a single gene (DRA1), but the β -chain can be coded by four different genes (DRB1, 3, 4, or 5). HLA molecules are highly variable, and HLA-DRB1 is one of the most polymorphic genes with at least 1883 different alleles reported in 2015 (88). Each individual inherits two alleles of DRB1, and additionally variants from DRB3, 4, 5, depending on the parental haplotypes (i.e. whether these genes are present on the parental haplotypes or not). DRB1 is present on all HLA haplotypes, but DRB3, 4, 5 genes are not. HLA-DRB1 is a gene that codes for the β1 -chain of the DR isotype of the HLA class II molecule ( Figure 4 ), and the first field of digits (marked by the semicolon) after the gene name indicate the allele group (alleles with similar sequence and properties), while the second field of digits indicates specific alleles encoding distinct molecule variants. Due to the high variability in the sequence, especially in the peptide binding groove, HLA molecules derived from different alleles generally display distinct peptide repertoires. Therefore, two individuals with different HLA alleles would display different peptides, even from the same protein. HLA-DR also plays a role in central tolerance mechanisms as it has been linked to presentation of autoantigens to developing T-cells in the thymus and is associated with a range of autoimmune diseases, e.g. rheumatoid arthritis, diabetes mellitus type I, multiple sclerosis or systemic lupus erythematosus, suggesting an aberrant presentation of self-peptide to autoreactive T cells in the thymus (95). This may lead to inefficient negative selection of autoreactive T-cells or aberrant education of regulatory T-cells, therefore disrupting central and peripheral tolerance mechanisms and predisposing to autoimmunity. HLA-DRB1 is also associated with HLA-DQB1 due to a strong linkage disequilibrium. An excellent review on HLA-DRB1 and autoimmunity can be found here (88).
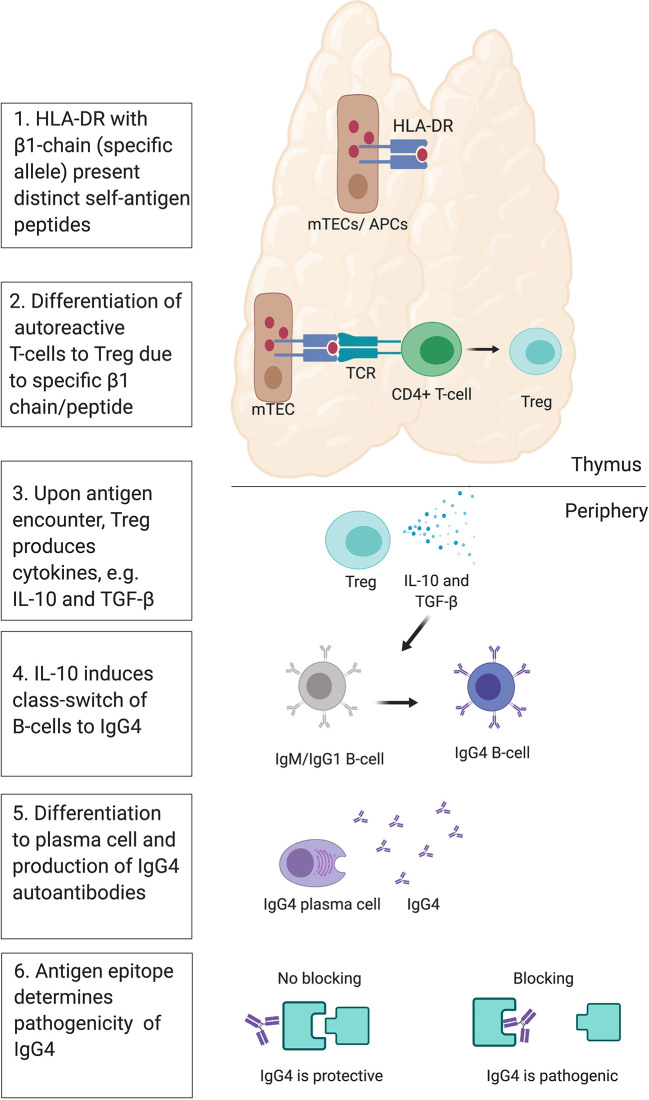
The HLA is responsible for the fate of T-cells, e.g. conventional pro-inflammatory T cells (Tconvs) or regulatory T cells (Tregs). A recent study with humanized mice investigated the T-cell response to self-antigen presented by HLA-DR molecules with different β chain alleles, HLA-DR1 or HLA-DR15 in mice (116). The two HLA class II molecules presented distinct peptide repertoires and the presentation via HLA-DR15 was associated with the generation of Tconv, while presentation via HLA-DR1 instead led to the development of Tregs that expressed tolerogenic cytokines such as IL-10 and TGF- β (116). Similar conclusions were drawn from a different study where mouse MHC II alleles (H2-Aj and H2-Ab) influenced the T-cell repertoire, including Treg and Tconv (117). This links the polymorphism in the HLA-DR molecules to the fate of T-cells and differentiation to Tregs and production of IgG4 via IL-10 (118–121). Moreover, blocking of DR15 prevented onset and attenuated severity of disease in animals in a follow-up study (122), suggesting that the HLA class II antigen presentation pathway has potential as a novel therapeutic target. Under normal circumstances, the generation of Tregs and a class switch of autoantibodies to IgG4 would protect from autoimmunity. IL-10 mediated class switch to protective IgG4 has been demonstrated in the context of allergen immunotherapy (123). In non-IgG4 autoimmune diseases, there may be a failure in this central tolerance mechanisms, leading to the survival of autoreactive CD4+ T-cells and in the production of non-IgG4 autoantibodies. This suggests that in IgG4-AID, a normally protective tolerance mechanism is at work, and the pathogenic effects of blocking IgG4 may just be an unlucky side effect ( Figure 5 ).
Figure 5.
Theory of IgG4 induction. Presentation of distinct self- peptides via HLA class II in medullary thymic epithelial cells (mTECs) or antigen-presenting cells (APCs) to developing autoreactive CD4+ T-cells via the T-cell receptor (TCR) may influence their fate toward differentiation into regulatory T-cells (Tregs). This may be influenced by polymorphisms in the beta-chain of the HLA-DR molecule, and/or the type of peptide presented to the TCR. In the periphery, Tregs may respond to their autoantigen by producing anti-inflammatory cytokines IL-10 and TGF-beta. IL-10 may then induce a class switch of autoreactive B-cells toward IgG4. The B-cell may then differentiate into plasma cells, producing autoreactive IgG4. IgG4 then bind their self-antigen, and depending on the location of the epitope, may cause a blocking effect and induce pathogenicity or not.
Potential Factors Influencing IgG4 Antibody Production
IgG4 subclass autoantibodies are the main cause of IgG4-AID and directly pathogenic, Therefore we were interested whether there is evidence for a mechanism linking specific HLA class II alleles to Tregs to IL-10 and finally to IgG4 autoantibodies. There are contradictory observations. Supporting evidence: 1) MuSK MG patients with the HLA-DRB1*14 type had elevated antibody titers and higher levels of IL-17A and IL-10 than MuSK MG patients with different haplotype (124). The higher IgG4 titers of MuSK autoantibodies in patients carrying HLA-DRB1*14 than those in the other patients suggest a role for HLA in the production of the antibodies. The differences in IL-10 and IL-17A support the role of DRB1 in the etiopathogenesis of this autoimmune response (124).
2) In addition, active immunization of mice with MuSK antigen leads to production of MuSK antibodies predominantly of the IgG1 subclass (which is the mouse equivalent to human IgG4), IL-4 (which induces IgG1- production in mice) and IL-10 (125). 3) Also IL-4 and IL-10 levels were increased in an adoptive transfer model of pemphigus vulgaris (126), and 4) IL-10 levels are elevated in serum of pemphigus patients (127, 128), serum of MuSK-MG patients (129) and serum of patients with thrombotic thrombocytopenic purpura (130), but in the latter study the IgG subclass levels were not entirely clear, with some patients also demonstrating high levels of complement fixing antibodies and complement C3a and C5a, suggesting that IgG4 was not the predominant subclass. More evidence for a role of IL-10 comes from an association between genetic variants/haplotypes of IL-10 and the occurrence of pemphigus vulgaris in patients from Argentina and Slovakia (131, 132).
What is not in line with the hypothesis that the DR haplotype directly leads to increased IgG4 antibody levels is that one study of TTP showed increased ADAMTS13 antibody levels in patients with the protective DR7-DQ2 and DR13-DQ6 haplotypes, though the relevance is not clear (133). Furthermore, there is evidence against the hypothesis that increased Treg and IL-10 levels (measured in the blood) may lead to IgG4 class switch and production of pathogenic IgG4 autoantibodies, as 1) total Treg numbers were not increased in MuSK MG (134), even decreased in pemphigus vulgaris [while Th17 T-cells were increased (135)] and 2) IL-10+ antigen-specific Tregs are only found in 17% of patients with pemphigus vulgaris, but in 80% of healthy controls, which suggests (at least in pemphigus) a protective role of Tregs and IL-10 (136). 2) B-cells from MuSK patients did not produce increased levels of IL-10 in culture (129, 137). 3) Further studies suggest a protective role of IL-10 in pemphigus, as IL-10−/− mice were more susceptible to disease and IL-10 protected mice from blister formation (138), and decreased IL-10 production was associated with pemphigus vulgaris in a subgroup of patients (131). 4) Also numbers of IL-10 producing Bregs (B10 cells) were reduced in MuSK MG patients (139), as well as pemphigus patients in one study (140), but increased in pemphigus patients in another study (141). 5) Interestingly, increased levels of IL-10 producing Bregs was associated with favorable response after B-cell depletion therapy with Rituximab in pemphigus (142), which may indicate a protective role for IL-10 and Bregs, but this could also have been an unrelated side-effect of Rituximab-induced increase in cytokines that stimulate IL-10 production (143, 144).
Taken together, there may not be a simple correlation between Tregs, Bregs, IL-10 and IgG4 autoantibodies, since IL-10 does have multiple function in immune regulation, and may have both anti- and pro-inflammatory effects (145), also in the context of IgG4-autoimmunity (146). Perhaps not increased systemic levels of IL-10, but local expression of IL-10 is important, as seen in studies on rheumatoid arthritis, where local administration of IL-10 to sites of inflammation was protective (147), but systemic application was not (148). More studies are required, and ideally also a systematic review and meta-analysis of cytokine levels in IgG4 autoimmune diseases and/or disease models.
Conclusion
In recent years, we could observe a growing number of rare, severe autoimmune diseases mediated by pathogenic autoantibodies of the IgG4 subclass. While these diseases were previously considered to be unrelated, as they may target different organs, there is (at least circumstantial) evidence that ties them together and suggests that similar immune mechanisms may be at work. These potential mechanisms need to be addressed in large comparative studies, and, once identified, may form the basis for new treatment approaches. These may provide a rationale for repurposing already existing drugs and treatment strategies used in well-known diseases such as MuSK MG or pemphigus for treatment of the very rare and the less-well studied forms of IgG4 autoimmune diseases. Newly identified shared immune mechanisms may also provide new druggable targets and ideas for novel treatments.
Author Contributions
IK conceived the idea, wrote the first draft of the manuscript, and provided the figures. VY, JT, KL, ST, ET, and TL contributed text. IK, FL, TL, and ET revised the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
IK was funded by a Hertha Firnberg Fellowship by the Austrian Science Fund, Austria, project number: T996-B30. TL was supported by the German Research Foundation (DFG, grant LE 2593/3-1). FL is supported by German Ministry of Education and Research (01GM1908A) and E-Rare Joint Transnational research support (ERA-Net, LE3064/2-1).
Conflict of Interest
JT and ST have shares in the research and diagnostic laboratory Tzartos NeuroDiagnostics, Athens. FL discloses having received speaker honoraria from Grifols, Teva, Biogen, Bayer, Roche, Novartis, Fresenius, travel funding from Merck, Grifols, and Bayer and serving on advisory boards for Roche, Biogen, and Alexion.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The figures were created by Biorender software.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.605214/full#supplementary-material
References
- 1. Huijbers MG, Plomp JJ, van der Maarel SM, Verschuuren JJ. IgG4-mediated autoimmune diseases: a niche of antibody-mediated disorders. Ann N Y Acad Sci (2018) 1413(1):92–103. 10.1111/nyas.13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koneczny I. A New Classification System for IgG4 Autoantibodies. Front Immunol (2018) 9:97:97. 10.3389/fimmu.2018.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kasperkiewicz M, Ellebrecht CT, Takahashi H, Yamagami J, Zillikens D, Payne AS, et al. Pemphigus. Nat Rev Dis Primers (2017) 3:17026. 10.1038/nrdp.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evoli A, Alboini PE, Damato V, Iorio R, Provenzano C, Bartoccioni E, et al. Myasthenia gravis with antibodies to MuSK: an update. Ann N Y Acad Sci (2018) 1412(1):82–9. 10.1111/nyas.13518 [DOI] [PubMed] [Google Scholar]
- 5. Kremer Hovinga JA, Coppo P, Lammle B, Moake JL, Miyata T, Vanhoorelbeke K. Thrombotic thrombocytopenic purpura. Nat Rev Dis Primers (2017) 3:17020. 10.1038/nrdp.2017.20 [DOI] [PubMed] [Google Scholar]
- 6. Hellmark T, Segelmark M. Diagnosis and classification of Goodpasture’s disease (anti-GBM). J Autoimmun (2014) 48-49:108–12. 10.1016/j.jaut.2014.01.024 [DOI] [PubMed] [Google Scholar]
- 7. Beck LH., Jr. PLA2R and THSD7A: Disparate Paths to the Same Disease? J Am Soc Nephrol (2017) 28(9):2579–89. 10.1681/ASN.2017020178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salant DJ. Unmet challenges in membranous nephropathy. Curr Opin Nephrol Hypertension (2019) 28(1):70–6. 10.1097/MNH.0000000000000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aalberse RC, Stapel SO, Schuurman J, Rispens T. Immunoglobulin G4: an odd antibody. Clin Exp Allergy (2009) 39(4):469–77. 10.1111/j.1365-2222.2009.03207.x [DOI] [PubMed] [Google Scholar]
- 10. van der Neut Kolfschoten M, Schuurman J, Losen M, Bleeker WK, Martinez-Martinez P, Vermeulen E, et al. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science (2007) 317(5844):1554–7. 10.1126/science.1144603 [DOI] [PubMed] [Google Scholar]
- 11. Lighaam LC, Rispens T. The Immunobiology of Immunoglobulin G4. Semin l=Liver Dis (2016) 36(3):200–15. 10.1055/s-0036-1584322 [DOI] [PubMed] [Google Scholar]
- 12. Koneczny I. A New Classification System for IgG4 Autoantibodies. Front Immunol (2018) 9:97. 10.3389/Fimmu.2018.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huijbers MG, Querol LA, Niks EH, Plomp JJ, van der Maarel SM, Graus F, et al. The expanding field of IgG4-mediated neurological autoimmune disorders. Eur J Neurol (2015) 22(8):1151–61. 10.1111/ene.12758 [DOI] [PubMed] [Google Scholar]
- 14. Koneczny I. Update on IgG4-mediated autoimmune diseases: New insights and new family members. Autoimmun Rev (2020) 19(10):102646. 10.1016/j.autrev.2020.102646 [DOI] [PubMed] [Google Scholar]
- 15. Ehsan S, Amirzargar A, Yekaninejad MS, Mahmoudi M, Mehravar S, Moradi B, et al. (DRB1, DQA1, DQB1) alleles and haplotypes with myasthenia gravis and its subgroups in the Iranian population. J Neurol Sci (2015) 359(1-2):335–42. 10.1016/j.jns.2015.11.021 [DOI] [PubMed] [Google Scholar]
- 16. Alahgholi-Hajibehzad M, Yilmaz V, Gulsen-Parman Y, Aysal F, Oflazer P, Deymeer F, et al. Association of HLA-DRB1 *14, -DRB1 *16 and -DQB1 *05 with MuSK-myasthenia gravis in patients from Turkey. Hum Immunol (2013) 74(12):1633–5. 10.1016/j.humimm.2013.08.271 [DOI] [PubMed] [Google Scholar]
- 17. Thomas MR, de Groot R, Scully MA, Crawley JT. Pathogenicity of Anti-ADAMTS13 Autoantibodies in Acquired Thrombotic Thrombocytopenic Purpura. Ebiomedicine (2015) 2(8):942–52. 10.1016/j.ebiom.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong Y, Li HF, Romi F, Skeie GO, Gilhus NE. HLA and MuSK-positive myasthenia gravis: A systemic review and meta-analysis. Acta Neurol Scand (2018) 138(3):219–26. 10.1111/ane.12951 [DOI] [PubMed] [Google Scholar]
- 19. Bartoccioni E, Scuderi F, Augugliaro A, Chiatamone Ranieri S, Sauchelli D, Alboino P, et al. HLA class II allele analysis in MuSK-positive myasthenia gravis suggests a role for DQ5. Neurology (2009) 72(2):195–7. 10.1212/01.wnl.0000339103.08830.86 [DOI] [PubMed] [Google Scholar]
- 20. Saruhan-Direskeneli G, Hughes T, Yilmaz V, Durmus H, Adler A, Alahgholi-Hajibehzad M, et al. Genetic heterogeneity within the HLA region in three distinct clinical subgroups of myasthenia gravis. Clin Immunol (2016) 166-167:81–8. 10.1016/j.clim.2016.05.003 [DOI] [PubMed] [Google Scholar]
- 21. Kanai T, Uzawa A, Kawaguchi N, Sakamaki T, Yoshiyama Y, Himuro K, et al. HLA-DRB1*14 and DQB1*05 are associated with Japanese anti-MuSK antibody-positive myasthenia gravis patients. J Neurol Sci (2016) 363:116–8. 10.1016/j.jns.2016.02.031 [DOI] [PubMed] [Google Scholar]
- 22. Cebi M, Durmus H, Yilmaz V, Yentur SP, Aysal F, Oflazer P, et al. Relation of HLA-DRB1 to IgG4 autoantibody and cytokine production in muscle-specific tyrosine kinase myasthenia gravis (MuSK-MG). Clin Exp Immunol (2019) 197(2):214–21. 10.1111/cei.13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niks EH, Kuks JB, Roep BO, Haasnoot GW, Verduijn W, Ballieux BE, et al. Strong association of MuSK antibody-positive myasthenia gravis and HLA-DR14-DQ5. Neurology (2006) 66(11):1772–4. 10.1212/01.wnl.0000218159.79769.5c [DOI] [PubMed] [Google Scholar]
- 24. Moraes ME, Fernandez-Vina M, Lazaro A, Diaz LA, Filho GH, Friedman H, et al. An epitope in the third hypervariable region of the DRB1 gene is involved in the susceptibility to endemic pemphigus foliaceus (fogo selvagem) in three different Brazilian populations. Tissue Antigens (1997) 49(1):35–40. 10.1111/j.1399-0039.1997.tb02707.x [DOI] [PubMed] [Google Scholar]
- 25. Loiseau P, Lecleach L, Prost C, Lepage V, Busson M, Bastuji-Garin S, et al. HLA class II polymorphism contributes to specify desmoglein derived peptides in pemphigus vulgaris and pemphigus foliaceus. J Autoimmun (2000) 15(1):67–73. 10.1006/jaut.2000.0388 [DOI] [PubMed] [Google Scholar]
- 26. Saha M, Harman K, Mortimer NJ, Binda V, Black MM, Kondeatis E, et al. Sporadic pemphigus foliaceus and class II human leucocyte antigen allele associations in the white British and Indo-Asian populations in the UK. Clin Exp Dermatol (2019) 44(3):290–4. 10.1111/ced.13774 [DOI] [PubMed] [Google Scholar]
- 27. Zhang SY, Zhou XY, Zhou XL, Zhang Y, Deng Y, Liao F, et al. Subtype-specific inherited predisposition to pemphigus in the Chinese population. Br J Dermatol (2019) 180(4):828–35. 10.1111/bjd.17191 [DOI] [PubMed] [Google Scholar]
- 28. Hashimoto K, Miki Y, Nakata S, Matsuyama M. HLA-A10 in pemphigus among Japanese. Arch Dermatol (1977) 113(11):1518–9. 10.1001/archderm.113.11.1518 [DOI] [PubMed] [Google Scholar]
- 29. Lombardi ML, Mercuro O, Ruocco V, Lo Schiavo A, Lombari V, Guerrera V, et al. Common human leukocyte antigen alleles in pemphigus vulgaris and pemphigus foliaceus Italian patients. J Invest Dermatol (1999) 113(1):107–10. 10.1046/j.1523-1747.1999.00626.x [DOI] [PubMed] [Google Scholar]
- 30. Brochado MJ, Nascimento DF, Campos W, Deghaide NH, Donadi EA, Roselino AM, et al. and class II associations in pemphigus foliaceus and pemphigus vulgaris patients from a prevalent Southeastern Brazilian region. J Autoimmun (2016) 72:19–24. 10.1016/j.jaut.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 31. Pollmann R, Schmidt T, Eming R, Hertl M. Pemphigus: a Comprehensive Review on Pathogenesis, Clinical Presentation and Novel Therapeutic Approaches. Clin Rev Allergy Immunol (2018) 54(1):1–25. 10.1007/s12016-017-8662-z [DOI] [PubMed] [Google Scholar]
- 32. Li S, Zhang Q, Wang P, Li J, Ni J, Wu J, et al. Association between HLA-DQB1 polymorphisms and pemphigus vulgaris: A meta-analysis. Immunol Invest (2018) 47(1):101–12. 10.1080/08820139.2017.1385622 [DOI] [PubMed] [Google Scholar]
- 33. Dere G, Yavuz IH, Ozaydin Yavuz G, Bayram Y, Gunes Bilgili S, Ozturk M. Assessment of HLA-A, HLA-DR, and HLA-DQ alleles in patients with pemphigus vulgaris from eastern of Turkey. J Cosmet Dermatol (2020) 19(9):2432–37. 10.1111/jocd.13298 [DOI] [PubMed] [Google Scholar]
- 34. Lombardi AM, Pulini S, Passeri C, Iuliani O, Salutari P, Businaro A, et al. Familial acquired thrombotic thrombocytopenic purpura: immunogenetic link with HLA-DRB1*11 and DQB1*03 antigens. Br J Haematol (2018) 183(2):317–9. 10.1111/bjh.14975 [DOI] [PubMed] [Google Scholar]
- 35. Coppo P, Busson M, Veyradier A, Wynckel A, Poullin P, Azoulay E, et al. HLA-DRB1*11: a strong risk factor for acquired severe ADAMTS13 deficiency-related idiopathic thrombotic thrombocytopenic purpura in Caucasians. J Thromb Haemost (2010) 8(4):856–9. 10.1111/j.1538-7836.2010.03772.x [DOI] [PubMed] [Google Scholar]
- 36. Sinkovits G, Szilagyi A, Farkas P, Inotai D, Szilvasi A, Tordai A, et al. The role of human leukocyte antigen DRB1-DQB1 haplotypes in the susceptibility to acquired idiopathic thrombotic thrombocytopenic purpura. Hum Immunol (2017) 78(2):80–7. 10.1016/j.humimm.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 37. John ML, Hitzler W, Scharrer I. The role of human leukocyte antigens as predisposing and/or protective factors in patients with idiopathic thrombotic thrombocytopenic purpura. Ann Hematol (2012) 91(4):507–10. 10.1007/s00277-011-1384-z [DOI] [PubMed] [Google Scholar]
- 38. Martinez-Martinez L, Lleixa MC, Boera-Carnicero G, Cortese A, Devaux J, Siles A, et al. Anti-NF155 chronic inflammatory demyelinating polyradiculoneuropathy strongly associates to HLA-DRB15. J Neuroinflamm (2017) 14(1):224. 10.1186/s12974-017-0996-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Binks S, Varley J, Lee W, Makuch M, Elliott K, Gelfand JM, et al. Distinct HLA associations of LGI1 and CASPR2-antibody diseases. Brain (2018) 141(8):2263–71. 10.1093/brain/awy109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mueller SH, Farber A, Pruss H, Melzer N, Golombeck KS, Kumpfel T, et al. Genetic predisposition in anti-LGI1 and anti-NMDA receptor encephalitis. Ann Neurol (2018) 83(4):863–9. 10.1002/ana.25216 [DOI] [PubMed] [Google Scholar]
- 41. van Sonderen A, Roelen DL, Stoop JA, Verdijk RM, Haasnoot GW, Thijs RD, et al. Anti-LGI1 encephalitis is strongly associated with HLA-DR7 and HLA-DRB4. Ann Neurol (2017) 81(2):193–8. 10.1002/ana.24858 [DOI] [PubMed] [Google Scholar]
- 42. Kim TJ, Lee ST, Moon J, Sunwoo JS, Byun JI, Lim JA, et al. Anti-LGI1 encephalitis is associated with unique HLA subtypes. Ann Neurol (2017) 81(2):183–92. 10.1002/ana.24860 [DOI] [PubMed] [Google Scholar]
- 43. Le WB, Shi JS, Zhang T, Liu L, Qin HZ, Liang S, et al. HLA-DRB1*15:01 and HLA-DRB3*02:02 in PLA2R-Related Membranous Nephropathy. J Am Soc Nephrol (2017) 28(5):1642–50. 10.1681/ASN.2016060644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cui Z, Xie LJ, Chen FJ, Pei ZY, Zhang LJ, Qu Z, et al. MHC Class II Risk Alleles and Amino Acid Residues in Idiopathic Membranous Nephropathy. J Am Soc Nephrol (2017) 28(5):1651–64. 10.1681/ASN.2016020114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie LJ, Cui Z, Chen FJ, Pei ZY, Hu SY, Gu QH, et al. The susceptible HLA class II alleles and their presenting epitope(s) in Goodpasture’s disease. Immunology (2017) 151(4):395–404. 10.1111/imm.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phelps RG, Rees AJ. The HLA complex in Goodpasture’s disease: a model for analyzing susceptibility to autoimmunity. Kidney Int (1999) 56(5):1638–53. 10.1046/j.1523-1755.1999.00720.x [DOI] [PubMed] [Google Scholar]
- 47. Yang R, Cui Z, Zhao J, Zhao MH. The role of HLA-DRB1 alleles on susceptibility of Chinese patients with anti-GBM disease. Clin Immunol (2009) 133(2):245–50. 10.1016/j.clim.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 48. Kitagawa W, Imai H, Komatsuda A, Maki N, Wakui H, Hiki Y, et al. The HLA-DRB1*1501 allele is prevalent among Japanese patients with anti-glomerular basement membrane antibody-mediated disease. Nephrol Dial Transplant (2008) 23(10):3126–9. 10.1093/ndt/gfn179 [DOI] [PubMed] [Google Scholar]
- 49. Gaig C, Graus F, Compta Y, Hogl B, Bataller L, Bruggemann N, et al. Clinical manifestations of the anti-IgLON5 disease. Neurology (2017) 88(18):1736–43. 10.1212/WNL.0000000000003887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fang H, Shen S, Zheng X, Dang E, Zhang J, Shao S, et al. Association of HLA class I and class II alleles with bullous pemphigoid in Chinese Hans. J Dermatol Sci (2018) 89(3):258–62. 10.1016/j.jdermsci.2017.11.014 [DOI] [PubMed] [Google Scholar]
- 51. Setterfield J, Theron J, Vaughan RW, Welsh KI, Mallon E, Wojnarowska F, et al. Mucous membrane pemphigoid: HLA-DQB1*0301 is associated with all clinical sites of involvement and may be linked to antibasement membrane IgG production. Br J Dermatol (2001) 145(3):406–14. 10.1046/j.1365-2133.2001.04380.x [DOI] [PubMed] [Google Scholar]
- 52. Zakka LR, Reche P, Ahmed AR. Role of MHC Class II genes in the pathogenesis of pemphigoid. Autoimmun Rev (2011) 11(1):40–7. 10.1016/j.autrev.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 53. Oyama N, Setterfield JF, Powell AM, Sakuma-Oyama Y, Albert S, Bhogal BS, et al. Bullous pemphigoid antigen II (BP180) and its soluble extracellular domains are major autoantigens in mucous membrane pemphigoid: the pathogenic relevance to HLA class II alleles and disease severity. Br J Dermatol (2006) 154(1):90–8. 10.1111/j.1365-2133.2005.06998.x [DOI] [PubMed] [Google Scholar]
- 54. Watts RA, MacGregor AJ, Mackie SL. HLA allele variation as a potential explanation for the geographical distribution of granulomatosis with polyangiitis. Rheumatol (Oxford) (2015) 54(2):359–62. 10.1093/rheumatology/keu321 [DOI] [PubMed] [Google Scholar]
- 55. Wu Z, Wu Q, Xu J, Chen S, Sun F, Li P, et al. HLA-DPB1 variant rs3117242 is associated with anti-neutrophil cytoplasmic antibody-associated vasculitides in a Han Chinese population. Int J Rheum Dis (2017) 20(8):1009–15. 10.1111/1756-185X.12561 [DOI] [PubMed] [Google Scholar]
- 56. Alberici F, Martorana D, Bonatti F, Gioffredi A, Lyons PA, Vaglio A. Genetics of ANCA-associated vasculitides: HLA and beyond. Clin Exp Rheumatol (2014) 32(3 Suppl 82):S90–7. [PubMed] [Google Scholar]
- 57. Futei Y, Amagai M, Ishii K, Kuroda-Kinoshita K, Ohya K, Nishikawa T. Predominant IgG4 subclass in autoantibodies of pemphigus vulgaris and foliaceus. J Dermatol Sci (2001) 26(1):55–61. 10.1016/S0923-1811(00)00158-4 [DOI] [PubMed] [Google Scholar]
- 58. Rock B, Martins CR, Theofilopoulos AN, Balderas RS, Anhalt GJ, Labib RS, et al. The pathogenic effect of IgG4 autoantibodies in endemic pemphigus foliaceus (fogo selvagem). N Engl J Med (1989) 320(22):1463–9. 10.1056/NEJM198906013202206 [DOI] [PubMed] [Google Scholar]
- 59. Evangelista F, Roth AJ, Prisayanh P, Temple BR, Li N, Qian Y, et al. Pathogenic IgG4 autoantibodies from endemic pemphigus foliaceus recognize a desmoglein-1 conformational epitope. J Autoimmun (2018) 89:171–85. 10.1016/j.jaut.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ohkawa T, Fukata Y, Yamasaki M, Miyazaki T, Yokoi N, Takashima H, et al. Autoantibodies to epilepsy-related LGI1 in limbic encephalitis neutralize LGI1-ADAM22 interaction and reduce synaptic AMPA receptors. J Neurosci (2013) 33(46):18161–74. 10.1523/JNEUROSCI.3506-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Petit-Pedrol M, Sell J, Planaguma J, Mannara F, Radosevic M, Haselmann H, et al. LGI1 antibodies alter Kv1.1 and AMPA receptors changing synaptic excitability, plasticity and memory. Brain (2018) 141(11):3144–59. 10.1093/brain/awy253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bastiaansen AEM, van Sonderen A, Titulaer MJ. Autoimmune encephalitis with anti-leucine-rich glioma-inactivated 1 or anti-contactin-associated protein-like 2 antibodies (formerly called voltage-gated potassium channel-complex antibodies). Curr Opin Neurol (2017) 30(3):302–9. 10.1097/WCO.0000000000000444 [DOI] [PubMed] [Google Scholar]
- 63. Kouton L, Boucraut J, Devaux J, Rajabally YA, Adams D, Antoine JC, et al. Electrophysiological features of chronic inflammatory demyelinating polyradiculoneuropathy associated with IgG4 antibodies targeting neurofascin 155 or contactin 1 glycoproteins. Clin Neurophysiol (2020) 131(4):921–7. 10.1016/j.clinph.2020.01.013 [DOI] [PubMed] [Google Scholar]
- 64. Ng JK, Malotka J, Kawakami N, Derfuss T, Khademi M, Olsson T, et al. Neurofascin as a target for autoantibodies in peripheral neuropathies. Neurology (2012) 79(23):2241–8. 10.1212/WNL.0b013e31827689ad [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Devaux JJ, Miura Y, Fukami Y, Inoue T, Manso C, Belghazi M, et al. Neurofascin-155 IgG4 in chronic inflammatory demyelinating polyneuropathy. Neurology (2016) 86(9):800–7. 10.1212/WNL.0000000000002418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kadoya M, Kaida K, Koike H, Takazaki H, Ogata H, Moriguchi K, et al. IgG4 anti-neurofascin155 antibodies in chronic inflammatory demyelinating polyradiculoneuropathy: Clinical significance and diagnostic utility of a conventional assay. J Neuroimmunol (2016) 301:16–22. 10.1016/j.jneuroim.2016.10.013 [DOI] [PubMed] [Google Scholar]
- 67. Labasque M, Hivert B, Nogales-Gadea G, Querol L, Illa I, Faivre-Sarrailh C. Specific contactin N-glycans are implicated in neurofascin binding and autoimmune targeting in peripheral neuropathies. J Biol Chem (2014) 289(11):7907–18. 10.1074/jbc.M113.528489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bien CG, Mirzadjanova Z, Baumgartner C, Onugoren MD, Grunwald T, Holtkamp M, et al. Anti-contactin-associated protein-2 encephalitis: relevance of antibody titres, presentation and outcome. Eur J Neurol (2017) 24(1):175–86. 10.1111/ene.13180 [DOI] [PubMed] [Google Scholar]
- 69. Lai M, Huijbers MG, Lancaster E, Graus F, Bataller L, Balice-Gordon R, et al. Investigation of LGI1 as the antigen in limbic encephalitis previously attributed to potassium channels: a case series. Lancet Neurol (2010) 9(8):776–85. 10.1016/S1474-4422(10)70137-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Patterson KR, Dalmau J, Lancaster E. Mechanisms of Caspr2 antibodies in autoimmune encephalitis and neuromyotonia. Ann Neurol (2018) 83(1):40–51. 10.1002/ana.25120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Koneczny I, Cossins J, Waters P, Beeson D, Vincent A. MuSK myasthenia gravis IgG4 disrupts the interaction of LRP4 with MuSK but both IgG4 and IgG1-3 can disperse preformed agrin-independent AChR clusters. PloS One (2013) 8(11):e80695. 10.1371/journal.pone.0080695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Huijbers MG, Zhang W, Klooster R, Niks EH, Friese MB, Straasheijm KR, et al. MuSK IgG4 autoantibodies cause myasthenia gravis by inhibiting binding between MuSK and Lrp4. Proc Natl Acad Sci USA (2013) 110(51):20783–8. 10.1073/pnas.1313944110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Herwig J, Skuza S, Sachs W, Sachs M, Failla AV, Rune G, et al. Thrombospondin Type 1 Domain-Containing 7A Localizes to the Slit Diaphragm and Stabilizes Membrane Dynamics of Fully Differentiated Podocytes. J Am Soc Nephrol (2019) 30(5):824–39. 10.1681/ASN.2018090941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tomas NM, Hoxha E, Reinicke AT, Fester L, Helmchen U, Gerth J, et al. Autoantibodies against thrombospondin type 1 domain-containing 7A induce membranous nephropathy. J Clin Invest (2016) 126(7):2519–32. 10.1172/JCI85265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tomas NM, Meyer-Schwesinger C, von Spiegel H, Kotb AM, Zahner G, Hoxha E, et al. A Heterologous Model of Thrombospondin Type 1 Domain-Containing 7A-Associated Membranous Nephropathy. J Am Soc Nephrol: JASN (2017) 28(11):3262–77. 10.1681/ASN.2017010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Skoberne A, Behnert A, Teng B, Fritzler MJ, Schiffer L, Pajek J, et al. Serum with phospholipase A2 receptor autoantibodies interferes with podocyte adhesion to collagen. Eur J Clin Invest (2014) 44(8):753–65. 10.1111/eci.12292 [DOI] [PubMed] [Google Scholar]
- 77. Fresquet M, Jowitt TA, McKenzie EA, Ball MD, Randles MJ, Lennon R, et al. PLA2R binds to the annexin A2-S100A10 complex in human podocytes. Sci Rep (2017) 7(1):6876. 10.1038/s41598-017-07028-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hayashi N, Okada K, Matsui Y, Fujimoto K, Adachi H, Yamaya H, et al. Glomerular mannose-binding lectin deposition in intrinsic antigen-related membranous nephropathy. Nephrol Dial Transplant (2018) 33(5):832–40. 10.1093/ndt/gfx235 [DOI] [PubMed] [Google Scholar]
- 79. Bally S, Debiec H, Ponard D, Dijoud F, Rendu J, Faure J, et al. Phospholipase A2 Receptor-Related Membranous Nephropathy and Mannan-Binding Lectin Deficiency. J Am Soc Nephrol (2016) 27(12):3539–44. 10.1681/ASN.2015101155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ma H, Sandor DG, Beck LH., Jr. The role of complement in membranous nephropathy. Semin Nephrol (2013) 33(6):531–42. 10.1016/j.semnephrol.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kremer Hovinga JA, Lammle B. Role of ADAMTS13 in the pathogenesis, diagnosis, and treatment of thrombotic thrombocytopenic purpura. Hematol Am Soc Hematol Educ Program (2012) 2012:610–6. 10.1182/asheducation-2012.1.610 [DOI] [PubMed] [Google Scholar]
- 82. Tsai HM, Lian EC. Antibodies to von Willebrand factor-cleaving protease in acute thrombotic thrombocytopenic purpura. N Engl J Med (1998) 339(22):1585–94. 10.1056/NEJM199811263392203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Beigneux AP, Miyashita K, Ploug M, Blom DJ, Ai M, Linton MF, et al. Autoantibodies against GPIHBP1 as a Cause of Hypertriglyceridemia. New Engl J Med (2017) 376(17):1647–58. 10.1056/NEJMoa1611930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hu X, Dallinga-Thie GM, Hovingh GK, Chang SY, Sandoval NP, Dang TLP, et al. GPIHBP1 autoantibodies in a patient with unexplained chylomicronemia. J Clin Lipidol (2017) 11(4):964–71. 10.1016/j.jacl.2017.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res (2019) 47(D1):D1005–D12. 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet (2016) 17(3):160–74. 10.1038/nrg.2015.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhong H, Zhao C, Luo S. HLA in myasthenia gravis: From superficial correlation to underlying mechanism. Autoimmun Rev (2019) 18(9):102349. 10.1016/j.autrev.2019.102349 [DOI] [PubMed] [Google Scholar]
- 88. Arango MT, Perricone C, Kivity S, Cipriano E, Ceccarelli F, Valesini G, et al. HLA-DRB1 the notorious gene in the mosaic of autoimmunity. Immunol Res (2017) 65(1):82–98. 10.1007/s12026-016-8817-7 [DOI] [PubMed] [Google Scholar]
- 89. Nikolic AV, Andric ZP, Simonovic RB, Rakocevic Stojanovic VM, Basta IZ, Bojic SD, et al. High frequency of DQB1*05 and absolute absence of DRB1*13 in muscle-specific tyrosine kinase positive myasthenia gravis. Eur J Neurol (2015) 22(1):59–63. 10.1111/ene.12525 [DOI] [PubMed] [Google Scholar]
- 90. Ishikawa Y, Terao C. Genetic analysis of IgG4-related disease. Mod Rheumatol (2019) 30(2):1–7. 10.1080/14397595.2019.1621000 [DOI] [PubMed] [Google Scholar]
- 91. Berntsen NL, Klingenberg O, Juran BD, Benito de Valle M, Lindkvist B, Lazaridis KN, et al. Association Between HLA Haplotypes and Increased Serum Levels of IgG4 in Patients With Primary Sclerosing Cholangitis. Gastroenterology (2015) 148(5):924–7 e2. 10.1053/j.gastro.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Vincent A, Leite MI, Farrugia ME, Jacob S, Viegas S, Shiraishi H, et al. Myasthenia gravis seronegative for acetylcholine receptor antibodies. Ann N Y Acad Sci (2008) 1132:84–92. 10.1196/annals.1405.020 [DOI] [PubMed] [Google Scholar]
- 93. Vodo D, Sarig O, Sprecher E. The Genetics of Pemphigus Vulgaris. Front Med (2018) 5:226:226. 10.3389/fmed.2018.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res (2020) 48(D1):D783–D8. 10.1093/nar/gkz1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Alvarez I, Collado JA, Colobran R, Carrascal M, Ciudad MT, Canals F, et al. Central T cell tolerance: Identification of tissue-restricted autoantigens in the thymus HLA-DR peptidome. J Autoimmun (2015) 60:12–9. 10.1016/j.jaut.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 96. de Sena Nogueira Maehara L, De-Souza-Santana FC, Porro AM, Marcos EVC, Ura S, Nolte IM, et al. HLA class II alleles of susceptibility and protection in Brazilian and Dutch pemphigus foliaceus. Br J Dermatol (2018) 178(3):e212–e4. 10.1111/bjd.16022 [DOI] [PubMed] [Google Scholar]
- 97. Tong JC, Bramson J, Kanduc D, Chow S, Sinha AA, Ranganathan S. Modeling the bound conformation of Pemphigus vulgaris-associated peptides to MHC Class II DR and DQ alleles. Immunome Res (2006) 2:1. 10.1186/1745-7580-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lin MS, Fu CL, Aoki V, Hans-Filho G, Rivitti EA, Moraes JR, et al. Desmoglein-1-specific T lymphocytes from patients with endemic pemphigus foliaceus (fogo selvagem). J Clin Invest (2000) 105(2):207–13. 10.1172/JCI8075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gu QH, Jia XY, Li JN, Chen FJ, Cui Z, Zhao MH. The critical amino acids of a nephritogenic epitope on human Goodpasture autoantigen for binding to HLA-DRB1*1501. Mol Immunol (2017) 88:1–9. 10.1016/j.molimm.2017.05.011 [DOI] [PubMed] [Google Scholar]
- 100. Wucherpfennig KW, Yu B, Bhol K, Monos DS, Argyris E, Karr RW, et al. Structural basis for major histocompatibility complex (MHC)-linked susceptibility to autoimmunity: charged residues of a single MHC binding pocket confer selective presentation of self-peptides in pemphigus vulgaris. Proc Natl Acad Sci USA (1995) 92(25):11935–9. 10.1073/pnas.92.25.11935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lin MS, Swartz SJ, Lopez A, Ding X, Fernandez-Vina MA, Stastny P, et al. Development and characterization of desmoglein-3 specific T cells from patients with pemphigus vulgaris. J Clin Invest (1997) 99(1):31–40. 10.1172/JCI119130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hertl M, Amagai M, Sundaram H, Stanley J, Ishii K, Katz SI. Recognition of desmoglein 3 by autoreactive T cells in pemphigus vulgaris patients and normals. J Invest Dermatol (1998) 110(1):62–6. 10.1046/j.1523-1747.1998.00086.x [DOI] [PubMed] [Google Scholar]
- 103. Riechers R, Grotzinger J, Hertl M. HLA class II restriction of autoreactive T cell responses in pemphigus vulgaris: review of the literature and potential applications for the development of a specific immunotherapy. Autoimmunity (1999) 30(3):183–96. 10.3109/08916939908993852 [DOI] [PubMed] [Google Scholar]
- 104. Gilardin L, Delignat S, Peyron I, Ing M, Lone YC, Gangadharan B, et al. The ADAMTS13(1239-1253) peptide is a dominant HLA-DR1-restricted CD4(+) T-cell epitope. Haematologica (2017) 102(11):1833–41. 10.3324/haematol.2015.136671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hrdinova J, Verbij FC, Kaijen PHP, Hartholt RB, van Alphen F, Lardy N, et al. Mass spectrometry-assisted identification of ADAMTS13-derived peptides presented on HLA-DR and HLA-DQ. Haematologica (2018) 103(6):1083–92. 10.3324/haematol.2017.179119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sorvillo N, van Haren SD, Kaijen PH, ten Brinke A, Fijnheer R, Meijer AB, et al. Preferential HLA-DRB1*11-dependent presentation of CUB2-derived peptides by ADAMTS13-pulsed dendritic cells. Blood (2013) 121(17):3502–10. 10.1182/blood-2012-09-456780 [DOI] [PubMed] [Google Scholar]
- 107. Verbij FC, Turksma AW, de Heij F, Kaijen P, Lardy N, Fijnheer R, et al. CD4+ T cells from patients with acquired thrombotic thrombocytopenic purpura recognize CUB2 domain-derived peptides. Blood (2016) 127(12):1606–9. 10.1182/blood-2015-10-668053 [DOI] [PubMed] [Google Scholar]
- 108. Martel P, Gilbert D, Drouot L, Prost C, Raux G, Delaporte E, et al. A polymorphic variant of the gene coding desmoglein 1, the target autoantigen of pemphigus foliaceus, is associated with the disease. Genes Immun (2001) 2(1):41–3. 10.1038/sj.gene.6363718 [DOI] [PubMed] [Google Scholar]
- 109. Martel P, Gilbert D, Busson M, Loiseau P, Lepage V, Drouot L, et al. Epistasis between DSG1 and HLA class II genes in pemphigus foliaceus. Genes Immun (2002) 3(4):205–10. 10.1038/sj.gene.6363839 [DOI] [PubMed] [Google Scholar]
- 110. Mouquet H, Farci S, Joly P, Maillere B, Leblond J, Drouot L, et al. A truncated alternative spliced isoform of human desmoglein 1 contains a specific T cell epitope binding to the pemphigus foliaceus-associated HLA class II DRbeta1*0102 molecule. J Immunol (2006) 177(9):6517–26. 10.4049/jimmunol.177.9.6517 [DOI] [PubMed] [Google Scholar]
- 111. Gebhard KL, Veldman CM, Wassmuth R, Schultz E, Schuler G, Hertl M. Ex vivo analysis of desmoglein 1-responsive T-helper (Th) 1 and Th2 cells in patients with pemphigus foliaceus and healthy individuals. Exp Dermatol (2005) 14(8):586–92. 10.1111/j.0906-6705.2005.00329.x [DOI] [PubMed] [Google Scholar]
- 112. Hacker-Foegen MK, Fairley JA, Lin MS. T cell receptor gene usage in desmoglein-3-specific T lymphocytes from patients with pemphigus vulgaris. J Invest Dermatol (2003) 121(6):1365–72. 10.1111/j.1523-1747.2003.12601.x [DOI] [PubMed] [Google Scholar]
- 113. Marino M, Maiuri MT, Di Sante G, Scuderi F, La Carpia F, Trakas N, et al. T cell repertoire in DQ5-positive MuSK-positive myasthenia gravis patients. J Autoimmun (2014) 52:113–21. 10.1016/j.jaut.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 114. Miyadera H, Tokunaga K. Associations of human leukocyte antigens with autoimmune diseases: challenges in identifying the mechanism. J Hum Genet (2015) 60(11):697–702. 10.1038/jhg.2015.100 [DOI] [PubMed] [Google Scholar]
- 115. Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA variation and disease. Nat Rev Immunol (2018) 18(5):325–39. 10.1038/nri.2017.143 [DOI] [PubMed] [Google Scholar]
- 116. Ooi JD, Petersen J, Tan YH, Huynh M, Willett ZJ, Ramarathinam SH, et al. Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature (2017) 545(7653):243–7. 10.1038/nature22329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Logunova NN, Kriukova VV, Shelyakin PV, Egorov ES, Pereverzeva A, Bozhanova NG, et al. MHC-II alleles shape the CDR3 repertoires of conventional and regulatory naive CD4(+) T cells. Proc Natl Acad Sci USA (2020) 117(24):13659–69. 10.1073/pnas.2003170117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jeannin P, Lecoanet S, Delneste Y, Gauchat JF, Bonnefoy JY. IgE versus IgG4 production can be differentially regulated by IL-10. J Immunol (1998) 160(7):3555–61. [PubMed] [Google Scholar]
- 119. Meiler F, Zumkehr J, Klunker S, Ruckert B, Akdis CA, Akdis M. In vivo switch to IL-10-secreting T regulatory cells in high dose allergen exposure. J Exp Med (2008) 205(12):2887–98. 10.1084/jem.20080193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Meiler F, Klunker S, Zimmermann M, Akdis CA, Akdis M. Distinct regulation of IgE, IgG4 and IgA by T regulatory cells and toll-like receptors. Allergy (2008) 63(11):1455–63. 10.1111/j.1398-9995.2008.01774.x [DOI] [PubMed] [Google Scholar]
- 121. Aversa G, Punnonen J, Cocks BG, de Waal Malefyt R, Vega F, Jr., Zurawski SM, et al. An interleukin 4 (IL-4) mutant protein inhibits both IL-4 or IL-13-induced human immunoglobulin G4 (IgG4) and IgE synthesis and B cell proliferation: support for a common component shared by IL-4 and IL-13 receptors. J Exp Med (1993) 178(6):2213–8. 10.1084/jem.178.6.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Huynh M, Eggenhuizen PJ, Olson GL, Rao NB, Self CR, Sun Y, et al. HLA-DR15-specific inhibition attenuates autoreactivity to the Goodpasture antigen. J Autoimmun (2019) 103:102276. 10.1016/j.jaut.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 123. Nouri-Aria KT, Wachholz PA, Francis JN, Jacobson MR, Walker SM, Wilcock LK, et al. Grass pollen immunotherapy induces mucosal and peripheral IL-10 responses and blocking IgG activity. J Immunol (2004) 172(5):3252–9. 10.4049/jimmunol.172.5.3252 [DOI] [PubMed] [Google Scholar]
- 124. Cebi M, Durmus H, Yilmaz V, Yentur SP, Aysal F, Oflazer P, et al. Relation of HLA-DRB1 to IgG4 autoantibody and cytokine production in muscle-specific tyrosine kinase myasthenia gravis (MuSK-MG). Clin Exp Immunol (2019) 197(2):214–21. 10.1111/cei.13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ulusoy C, Kim E, Tuzun E, Huda R, Yilmaz V, Poulas K, et al. Preferential production of IgG1, IL-4 and IL-10 in MuSK-immunized mice. Clin Immunol (2014) 151(2):155–63. 10.1016/j.clim.2014.02.012 [DOI] [PubMed] [Google Scholar]
- 126. Takahashi H, Amagai M, Nishikawa T, Fujii Y, Kawakami Y, Kuwana M. Novel system evaluating in vivo pathogenicity of desmoglein 3-reactive T cell clones using murine pemphigus vulgaris. J Immunol (2008) 181(2):1526–35. 10.4049/jimmunol.181.2.1526 [DOI] [PubMed] [Google Scholar]
- 127. Bhol KC, Rojas AI, Khan IU, Ahmed AR. Presence of interleukin 10 in the serum and blister fluid of patients with pemphigus vulgaris and pemphigoid. Cytokine (2000) 12(7):1076–83. 10.1006/cyto.1999.0642 [DOI] [PubMed] [Google Scholar]
- 128. Satyam A, Khandpur S, Sharma VK, Sharma A. Involvement of T(H)1/T(H)2 cytokines in the pathogenesis of autoimmune skin disease-Pemphigus vulgaris. Immunol Invest (2009) 38(6):498–509. 10.1080/08820130902943097 [DOI] [PubMed] [Google Scholar]
- 129. Yilmaz V, Oflazer P, Aysal F, Durmus H, Poulas K, Yentur SP, et al. Differential Cytokine Changes in Patients with Myasthenia Gravis with Antibodies against AChR and MuSK. PloS One (2015) 10(4):e0123546. 10.1371/journal.pone.0123546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Westwood JP, Langley K, Heelas E, Machin SJ, Scully M. Complement and cytokine response in acute Thrombotic Thrombocytopenic Purpura. Br J Haematol (2014) 164(6):858–66. 10.1111/bjh.12707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Eberhard Y, Burgos E, Gagliardi J, Vullo CM, Borosky A, Pesoa S, et al. Cytokine polymorphisms in patients with pemphigus. Arch Dermatol Res (2005) 296(7):309–13. 10.1007/s00403-004-0528-6 [DOI] [PubMed] [Google Scholar]
- 132. Javor J, Chmurova N, Parnicka Z, Ferencik S, Grosse-Wilde H, Buc M, et al. TNF-alpha and IL-10 gene polymorphisms show a weak association with pemphigus vulgaris in the Slovak population. J Eur Acad Dermatol Venereol: JEADV (2010) 24(1):65–8. 10.1111/j.1468-3083.2009.03260.x [DOI] [PubMed] [Google Scholar]
- 133. Sinkovits G, Szilagyi A, Farkas P, Inotai D, Szilvasi A, Tordai A, et al. Concentration and Subclass Distribution of Anti-ADAMTS13 IgG Autoantibodies in Different Stages of Acquired Idiopathic Thrombotic Thrombocytopenic Purpura. Front Immunol (2018) 9:1646:1646. 10.3389/fimmu.2018.01646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Yi JS, Guidon A, Sparks S, Osborne R, Juel VC, Massey JM, et al. Characterization of CD4 and CD8 T cell responses in MuSK myasthenia gravis. J Autoimmun (2014) 52:130–8. 10.1016/j.jaut.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Asothai R, Anand V, Das D, Antil PS, Khandpur S, Sharma VK, et al. Distinctive Treg associated CCR4-CCL22 expression profile with altered frequency of Th17/Treg cell in the immunopathogenesis of Pemphigus Vulgaris. Immunobiology (2015) 220(10):1129–35. 10.1016/j.imbio.2015.06.008 [DOI] [PubMed] [Google Scholar]
- 136. Veldman C, Hohne A, Dieckmann D, Schuler G, Hertl M. Type I regulatory T cells specific for desmoglein 3 are more frequently detected in healthy individuals than in patients with pemphigus vulgaris. J Immunol (2004) 172(10):6468–75. 10.4049/jimmunol.172.10.6468 [DOI] [PubMed] [Google Scholar]
- 137. Yilmaz V, Oflazer P, Aysal F, Parman YG, Direskeneli H, Deymeer F, et al. B cells produce less IL-10, IL-6 and TNF-alpha in myasthenia gravis. Autoimmunity (2015) 48(4):201–7. 10.3109/08916934.2014.992517 [DOI] [PubMed] [Google Scholar]
- 138. Toto P, Feliciani C, Amerio P, Suzuki H, Wang B, Shivji GM, et al. Immune modulation in pemphigus vulgaris: role of CD28 and IL-10. J Immunol (2000) 164(1):522–9. 10.4049/jimmunol.164.1.522 [DOI] [PubMed] [Google Scholar]
- 139. Guptill JT, Yi JS, Sanders DB, Guidon AC, Juel VC, Massey JM, et al. Characterization of B cells in muscle-specific kinase antibody myasthenia gravis. Neurol(R) Neuroimmunol Neuroinflamm (2015) 2(2):e77. 10.1212/NXI.0000000000000077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kabuto M, Fujimoto N, Takahashi T, Tanaka T. Decreased level of interleukin-10-producing B cells in patients with pemphigus but not in patients with pemphigoid. Br J Dermatol (2017) 176(5):1204–12. 10.1111/bjd.15113 [DOI] [PubMed] [Google Scholar]
- 141. Zhu HQ, Xu RC, Chen YY, Yuan HJ, Cao H, Zhao XQ, et al. Impaired function of CD19(+) CD24(hi) CD38(hi) regulatory B cells in patients with pemphigus. Br J Dermatol (2015) 172(1):101–10. 10.1111/bjd.13192 [DOI] [PubMed] [Google Scholar]
- 142. Colliou N, Picard D, Caillot F, Calbo S, Le Corre S, Lim A, et al. Long-term remissions of severe pemphigus after rituximab therapy are associated with prolonged failure of desmoglein B cell response. Sci Trans Med (2013) 5(175):175ra30. 10.1126/scitranslmed.3005166 [DOI] [PubMed] [Google Scholar]
- 143. Vallerskog T, Heimburger M, Gunnarsson I, Zhou W, Wahren-Herlenius M, Trollmo C, et al. Differential effects on BAFF and APRIL levels in rituximab-treated patients with systemic lupus erythematosus and rheumatoid arthritis. Arthritis Res Ther (2006) 8(6):R167. 10.1186/ar2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yang M, Sun L, Wang S, Ko KH, Xu H, Zheng BJ, et al. Novel function of B cell-activating factor in the induction of IL-10-producing regulatory B cells. J Immunol (2010) 184(7):3321–5. 10.4049/jimmunol.0902551 [DOI] [PubMed] [Google Scholar]
- 145. Saxena A, Khosraviani S, Noel S, Mohan D, Donner T, Hamad AR. Interleukin-10 paradox: A potent immunoregulatory cytokine that has been difficult to harness for immunotherapy. Cytokine (2015) 74(1):27–34. 10.1016/j.cyto.2014.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Cho MJ, Ellebrecht CT, Payne AS. The dual nature of interleukin-10 in pemphigus vulgaris. Cytokine (2015) 73(2):335–41. 10.1016/j.cyto.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Schwager K, Kaspar M, Bootz F, Marcolongo R, Paresce E, Neri D, et al. Preclinical characterization of DEKAVIL (F8-IL10), a novel clinical-stage immunocytokine which inhibits the progression of collagen-induced arthritis. Arthritis Res Ther (2009) 11(5):R142. 10.1186/ar2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. van Roon J, Wijngaarden S, Lafeber FP, Damen C, van de Winkel J, Bijlsma JW. Interleukin 10 treatment of patients with rheumatoid arthritis enhances Fc gamma receptor expression on monocytes and responsiveness to immune complex stimulation. J Rheumatol (2003) 30(4):648–51. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.