Abstract
Cytological features such as cell size and intracellular morphology provide fundamental information on cell status and hence may provide specific information on changes that arise within biological tissues. Such information is usually obtained by invasive biopsy in current clinical practice, which suffers several well-known disadvantages. Recently, novel MRI methods such as IMPULSED (imaging microstructural parameters using limited spectrally edited diffusion) have been developed for direct measurements of mean cell size non-invasively. The IMPULSED protocol is based on using temporal diffusion spectroscopy (TDS) to combine measurements of water diffusion over a wide range of diffusion times to probe cellular microstructure over varying length scales. IMPULSED has been shown to provide rapid, robust, and reliable mapping of mean cell size and is suitable for clinical imaging. More recently, cell size distributions have also been derived by appropriate analyses of data acquired with IMPULSED or similar sequences, which thus provides MRI-cytometry. This review summarizes the basic principles, practical implementations, validations, and example applications of MR cell size imaging based on TDS and demonstrates how cytometric information can be used in various applications. In addition, the limitations and potential future directions of MR cytometry are identified including the diagnosis of nonalcoholic steatohepatitis of the liver and the assessment of treatment response of cancers.
Keywords: diffusion, MRI, cell size, IMPULSED, dMRI, cytometry, oscillating gradient
1. Introduction
Cell size is a fundamental feature of cells and plays important roles in basic cellular functions. For example, cells monitor cell size and geometry and use this information to control cell division (1,2); Cell shrinkage is a hallmark of programmed cell death (i.e., apoptosis) (3); hepatocytes exhibit ballooning associated with cell swelling in nonalcoholic steatohepatitis; and cancer immunotherapy induces shifts in the mean cell size of a population of cells as small immune cells infiltrate targeted tumors (4). Measurements of cell size and their changes over time can thus have high clinical significance to provide key information at the cellular level, which can lead to more sensitive and specific information on biological tissues. Except for blood analyses of anemia (5,6) or cancer (7), cell size measurements in solid organs are not widely used in routine clinical practice. One major reason is that biopsy is an invasive procedure that has many well-known disadvantages including side effects and an inability to reflect the spatial heterogeneity of whole organs. It is desirable to develop an imaging method that can map cell size non-invasively.
Diffusion MRI (dMRI) is a widely used imaging method that relies on detecting the hindrance and/or restriction to the free diffusion of water molecules in tissues, and which thereby probes tissue microstructure indirectly. Because the degree of restriction depends on the density and spacing of obstacles such as cell membranes, dMRI provides a unique means to probe cell size and density. However, metrics derived from conventional dMRI, particularly the apparent diffusion coefficient (ADC), represent an averaged diffusion property that is influenced by several microstructural features simultaneously. Hence, ADC conveys non-specific information on a variety of tissue properties, including but not limited to sizes of nuclei (5), organelles (6), cells (2), cell membrane permeability (3), intra- and extracellular diffusion coefficients (4), and intracellular volume fraction (5). This sensitivity allows ADC to reflect a variety of pathological variations in biological tissues, but it reduces the specificity to individual tissue properties such as cell size. Moreover, different tissue properties at different length scales may have concurrent, opposing influences on ADC that may cancel out each other, which may result in a reduced sensitivity to pathological variations (7).
Quantitative diffusion modeling is a promising way to disentangle the complex information in dMRI and enhance the specificity to e.g., cell size. Tissues are complex, heterogeneous media containing multiple compartments of water in different microenvironments. These compartments may have different sizes, shapes, connectedness and intrinsic diffusivities, each of which behave differently in their signal changes in response to different diffusion times and diffusion weighting factors (b values). Different compartments may be classified into e.g., intra- and extracellular spaces, based on their particular diffusion and geometrical properties. By using a carefully selected set of data acquisitions and fitting quantitative models to dMRI data, microstructural parameters such as cell size can be obtained. A considerable number of recent studies (8–16) have attempted to derive quantitative indices of tissue microstructure using this strategy. The VERDICT (vascular, extracellular and restricted diffusion for cytometry in tumors) (8–10), IMPULSED (imaging microstructural parameters using limited spectrally edited diffusion) (12,13,17), and POMACE (pulsed and oscillating gradient MRI for assessment of cell size and extracellular space) methods (18) have been developed for mapping mean cell sizes in tumors. Among these methods, VERDICT and IMPULSED have been successfully implemented in patients with prostate (9) and breast (12) cancer, respectively, within clinically feasible scan times. Studies have reported the agreement between VERDICT MRI and 3D histology in fresh and fixed prostate specimens (19), but VERDICT derived cell sizes have not been validated, possibly due to the lack of clear cell boundaries in the haematoxylin and eosin (H&E) stained sections. By contrast, the IMPULSED derived cell size has been comprehensively validated using computer simulations in silico, cells in vitro, and animals in vivo using Na+/K+-ATPase (ab76020, Abcam) staining for pronounced visualization of cell boundaries (13,17).
However, the above imaging methods report only mean cell sizes of a population, and mean sizes are often inadequate to characterize tissue microstructure. It is well-known that there is significant heterogeneity in cell size particularly in tumors (2). Cell size distributions may provide important additional information on tissue status for diagnosis or monitoring responses to interventions (20–22). Several attempts have been reported to estimate non-parametric compartment size distributions without a priori knowledge of the distribution function in phantoms of glass capillaries (23), fixed tissues ex vivo (24–26), and animal allografts in vivo (27). However, these approaches either used special preclinical hardware or focused on animal applications only. Therefore, there is a need to develop an imaging method for mapping cell size distributions in humans. Recently, an extension of IMPULSED termed MRI-cytometry has been reported to fulfill this requirement (28). With comprehensive validations in preclinical models and demonstrations of applications in breast cancer patients, MRI-cytometry has been shown to be a relatively fast (~ 7 mins) and reliable imaging method suitable for reporting cell size distributions in clinical applications.
Both the IMPULSED and MRI-cytometry approaches are based on Temporal Diffusion Spectroscopy (TDS), which connotes the acquisition of dMRI data with a range of diffusion times expressed in terms of components of a diffusion spectrum. TDS is a general approach which uses different gradient waveforms that have different spectral contents. TDS has been comprehensively reviewed previously (29) so this review will focus on how MR cell size imaging is possible with IMPULSED and MRI-cytometry using the theory of TDS. Briefly, conventional dMRI uses pulsed gradient spin echo (PGSE) sequences that acquire signals after relatively long diffusion times (> 30 ms in practice), corresponding to low frequency components of the diffusion spectrum. TDS also uses oscillating gradient spin echo (OGSE) sequences that can probe much shorter diffusion times, such as 5 ms on regular clinical 3 Tesla MRI scanners (12) and ~1.6 ms on preclinical MRI scanners (13,17) which correspond to higher frequency components of the diffusion spectrum. Because the sensitivity of diffusion to different length scales is dependent on diffusion time (29), using a broader range of diffusion times provides a more comprehensive information on microstructure at different length scales. In particular, the shorter diffusion times achieved with the OGSE sequence increase the sensitivity of acquired signals to intracellular diffusivities, which is usually challenging with the conventional PGSE sequence only. Quantitative measurements of tissue microstructure (e.g., cell sizes and cell densities) are then obtained by fitting a combination of OGSE and PGSE signals to a simple but realistic model of restricted and hindered diffusion in solid tissues. For organs that have dense vasculature (e.g., livers and tumors), the signal decay from Intravoxel Incoherent Motion (IVIM), such as blood in the microcirculation, cannot be ignored. Although measuring perfusion and its related parameters using quantitative IVIM models have been actively investigated (30,31), its clinical values have been questioned (e.g., in need of impractical high SNRs for reliable fittings, and lack of careful assessment and validation of connections between IVIM-theory-based parameters and physiological factors) as to its suitability for tissue characterization (32). Therefore, MR cell size imaging with IMPULSED and MRI-cytometry deliberately remove the influences of IVIM signals prior to the extraction of microstructural parameters.
In this article, we provide a comprehensive review of the theory, development, implementation, validation, and reported applications of MR cell size imaging with IMPULSED and MRI-cytometry. The pulse sequences, recommended acquisition parameters, technical challenges, data analysis strategies, and possible future directions are provided. We aim to provide a detailed handbook from basic theory to advanced applications for implementing MR cell size imaging with IMPULSED and/or MRI-cytometry in research or clinical applications.
2. Theory
2.1. One-compartment model
Diffusion time dependent ADC(tdiff) has long been used to probe the microstructure of porous media and biological tissues (29,33). With the inclusion of OGSE acquisitions, the tdiff range can be significantly extended, so that measurements of the temporal diffusion spectrum ADC(f) provide more comprehensive information, where f is the oscillating gradient frequency. ADC(f) has been fit to either a power law (i.e., ~ e−θ) (34,35) or a linear function (i.e., ADC = DDR · f + ADC0) (7,36–39) to obtain the dispersion rate DDR, which has been shown to be correlated with average compartment size in selected media (36,37). To attempt quantification of microstructure over a broader range of tdiff, a simplified one-compartment model of tumors has been considered as (4,40)
| [1] |
where ADCsphere is the analytical form of ADC of the fluid inside an impermeable sphere at the frequency of f, is the apparent restriction size, and D0 and D∞ are ADC values when f → 0 and f → ∞, respectively. Note that is associated with mean cell size and D0 and D∞ are mainly determined by the extracellular tortuosity, and intracellular diffusion coefficient, respectively.
Because only a few ADC measurements at different f’s are required, Eq.[1] provides a rapid means to obtain averaged dimension sizes (41), and has been implemented in a few anti-cancer treatment studies (40,42). However, Eq.[1] is an oversimplified one-compartment model so that the measured dimension size reflects but does not equal mean cell size due to e.g., the influence of extracellular water. To better describe complex biological tissues, more realistic and specific multi-compartment models are required.
2.2. General multi-compartment model
The dMRI signal attenuations in biological tissues can be considered consisting of three main components: restricted diffusion in cells, hindered diffusion in the extravascular extracellular space, and dephasing from blood (i.e., the intra-voxel incoherent motion (IVIM) (43)), namely,
| [2] |
where TE is echo time, b is the diffusion weighting factor, tdiff is the effective diffusion time, SIVIM is the signal affected by IVIM and fIVIM is its fraction, T2,in, T2,ex, and T2,blood are T2 relaxation times in intra-, extra-cellular spaces and the IVIM component (blood in capillaries), respectively. is the total signal affected by diffusion in tissues, Sin and Sex are dMRI signals arising from the intra- and extra-cellular spaces, respectively, and vin is the proton volume fraction of intracellular space. It is usually challenging to directly fit Eq.[2] to dMRI data and hence a simplification is needed. Note that the transcytolemmal water exchange is ignored here for simplicity but its influence should be evaluated (see below).
2.2.1. Influence of IVIM
If the blood volume fraction is small such as in normal brains, fIVIM ≪ 1 and hence Eq.[2] degenerates to a simple two-compartment model. However, fIVIM cannot be ignored in tissues with high blood volume fractions such as in some tumors and livers (44). If signals are acquired with only long tdiff values using PGSE sequences only, fIVIM is much less sensitive to tdiff and hence a three-compartment model can be fit directly including a constant IVIM component (8). However, a broadened tdiff range achieved by using additional OGSE sequences no longer allows the approximation of tdiff-independence because fIVIM varies significantly with tdiff especially when short tdiff’s are involved (45). For example, a liver study found out that, if the IVIM effect is not removed, a longer tdiff causes a larger signal decay than that of a shorter tdiff, resulting in a patently erroneous conclusion that ADC at long tdiff is larger than the ADC at short tdiff (46).
Based on theory (43), SIVIM = exp(−b · D*) where D* is the pseudo-diffusion coefficient originated from blood flow and self-diffusion of blood in a randomly orientated blood capillary network. For non-flow-compensated PGSE acquisitions with a relatively long diffusion time, D* is dominated by signal dephasing resulting from blood flow, which is usually 10 – 50 times larger than an intrinsic diffusion coefficient. SIVIM decays much faster than Stissue and becomes negligible when b ≥ 0.2 ms/μm2. In addition, the Gaussian diffusion approximation is usually valid up to b ⩽ 1 ms/μm2. For flow-compensated OGSE acquisitions, D* is dominated by the self-diffusion of blood, which is close to the tissue water diffusion coefficient (47). Therefore, this provides a window of b values in which dMRI signal decays should be mono-exponential. By fitting a mono-exponential model to the data with b values in this window, the y-intercepts obtained by extrapolating the signal curves represent IVIM-weighted Stissue(b = 0). By normalizing signals with the y-intercepts, the IVIM effect can be removed. We need to emphasize that, because the IVIM effect is tdiff dependent, this procedure should be applied to each diffusion decay curve with the same tdiff. Figure 1 shows a representative case how the IVIM effect was removed in a healthy human liver.
Figure 1.
Typical Diffusion-weighted signals for voxels from a single slice of healthy human liver. (A) raw DW signals; (B) Corrected DW signals with the removal of the IVIM effect.
2.2.2. Influence of T2 relaxation
Most multi-compartment diffusion models ignore the possible differences in relaxation properties between intra- and extracellular spaces (8,17,18). This assumption is particularly relevant in tumors. For example, a quantitative magnetization transfer (qMT) study suggests that the relatively higher transcytolemmal water exchange rate in tumors enables enough proton mixing to yield homogenous T1 and MT values in the intra- and extracellular spaces (48). Moreover, a multi-echo T2 spectrum measurement showed that the majority (93.2 ± 6.2%) of total MRI signals arise from a single peak of T2 (76.4 ± 9.3 ms) in rodent brain tumors at 7 Tesla (49), suggesting small differences between intra- and extracellular spaces in tumors. Therefore, all relaxation terms are considered to be ignored in Eq.[2] for tumors.
2.2.3. Influence of transcytolemmal water exchange
It is well-known that the cell membranes are semipermeable and allow water molecules pass across, producing transcytolemmal water exchange. Early diffusion NMR studies have shown that such water exchange can significantly affect ADC measurements (50) and a recent study suggests transcytolemmal water exchange can be negligible only when the tdiff is at least one order of magnitude smaller than the resident lifetime of intracellular water, which is not the case for most PGSE measurements (51). Moreover, transcytolemmal water exchange is significantly higher in various neural diseases (52,53) and cancer (54), especially in apoptotic regions (55), and hence it should not be ignored in multi-compartment diffusion models.
However, a recent study with both simulations and experiments reported that, although enhanced transcytolemmal water change leads to a remarkable underestimation of intracellular water fraction, it has negligible influences on the estimation of mean cell size at least for tdiff ⩽ 50 ms (56). This is encouraging since our focus is MR cell size imaging and hence the transcytolemmal water exchange can be ignored in our model.
2.3. Simplified two-compartment model
After the removal of the IVIM effect and ignoring relaxation and transcytolemmal water exchange as shown above, the dMRI signal becomes
| [3] |
Eq.[3] is a simplified two-compartment model and can be fit regularly. With analytical forms of Sin and Sex linking to specific microstructural parameters, data can be fit to Eq.[3] to extract microstructural information including cell size. However, it is not a trivial problem to find appropriate analytical forms of Sin and Sex, particularly due to their strong dependence on tdiff.
2.3.1. Intracellular signal – Sin
The intracellular diffusion-weighted signal Sin reflects the intracellular microenvironment. There are numerous intracellular organelles of varying sizes, so precise descriptions of intracellular diffusion over all time scales are complicated. However, it usually requires very short tdiff to probe variations in diffusion caused by small intracellular structures or specific organelles. For example, it has been shown that tdiff < 0.5 ms (OGSE frequency > 500 Hz) is required detect intracellular diffusion differences in cells at different stages of mitosis (57) and tdiff < 0.14 ms (OGSE frequency > 1800 Hz) is needed to probe specific organelle changes in e.g., cytoskeleton (6). Because usually much longer tdiff values are used in practical dMRI measurements, such as tdiff ⩾ 1.6 ms in animal studies (13,17) and tdiff ⩾ 5 ms in human studies (12,28), the overall intracellular diffusion does not resolve individual contributions (58) arising from within single cells, and can be considered as a membrane-confined “homogenous” fluid with a single intracellular diffusion coefficient Din.
2.3.2. Extracellular signal – Sex
Extracellular diffusion has previously been considered as hindered diffusion and described using a constant diffusivity, such as in tumors (8,9). However, other studies have suggested that there is a significant tdiff-dependence in extracellular diffusion. The dependence of extracellular diffusion coefficient Dex on OGSE gradient frequency f can be described using a power-law as Dex = Dex0 + const · fθ when the effective tdiff is long (34), which has been observed recently in OGSE imaging in healthy human subjects with fitted θ = 0.5 and f up to 60 Hz (59). However, more generally the dependence on tdiff or f may vary. The range of f achievable in OGSE acquisitions is usually limited to a narrow range of f in practice. In such a narrow range, previous studies have suggested the extracellular diffusion coefficient shows an approximately linear dependence on the gradient frequency f, i.e., Dex = Dex0 + βex · f, where βex describes the rate of change of Dex with f, which is proportional to the inverse of diffusion time (7). This linear approximation has been used extensively in previous pre-clinical cell size imaging studies in tumors (4,13,60).
2.3.3. Analytical forms
The above sections have demonstrated that it is appropriate to assume a simplified dMRI model with practically feasible tdiff ranges that incorporates intracellular dMRI signals Sex arising from membrane-confined fluid with a single Din, and extracellular dMRI signals Sex arising from tdiff-dependent hindered diffusion. However, biological tissues are heterogeneous and contain a variety of cells with different sizes and other microstructures. Two practical approaches, IMPULSED and MRI-cytometry, address the inhomogeneity of cell sizes, Din’s, and Dex’s, in different ways and may be used to map mean cell size and cell size distribution, respectively.
2.4. IMPULSED
The variations in size and shape of cells complicate modeling. Although most epithelial tissues show a striking regularity in cell size in normal tissues, cell size heterogeneity occurs during neoplastic growth (2). To simplify the problem while keeping the key microstructural information, numerous studies have assumed cells to be hollow spheres with a cell-volume-weighted mean cell size (diameter) (8,12,13,18). For a few regular geometries (i.e., planes, cylinders, and spheres), the analytical expressions that link dMRI signals to the sequence parameters and underlying biophysical properties have been reported for PGSE acquisitions (61) and OGSE acquisitions with sine and cosine waveforms (62), square and general trapezoid-sine waveforms (63), and the trapezoid-cosine waveforms that are widely used in OGSE imaging studies in humans (12). The analytical equations for cosine waveforms (widely used in preclinical imaging) and cosine-modulated trapezoidal waveform with one oscillation n = 1 (widely used in clinical imaging) are provided in the Appendix. Readers are encouraged to refer to the above references for other details. Note that previous studies have suggested the assumption of spherical cells works effectively for non-spherical cells as well to obtain cell sizes of cell-volume-matched spheres (17).
Eq.[3] becomes
| [4] |
where Sin is given by the appropriate analytical intracellular equation of water diffusion in an impermeable sphere, as reported previously (61) (12,62,63). Eq.[4] can be fit to dMRI data to extract microstructural parameters that are summarized in Table 1. Note that βex can be fit reliably in preclinical imaging studies with f up to 150 Hz but not in human imaging studies with f only up to 50 Hz. Therefore, a constant mean extracellular diffusivity is usually assumed in human imaging studies on regular clinical MRI scanners to enhance the fitting precision of other metrics (12). Moreover, there are a few other quantities that can be calculated from IMPULSED data, such as the apparent three-dimensional cell density ρ that can be estimated as based on a face-centered-cubic packing approximation (13).
Table 1.
Summary of IMPULSED derived metrics and corresponding biophysical features. Note that cell density can be calculated from d and vin.
| IMPULSED metrics | Biophysical features |
|---|---|
| Mean cell size (cell volume weighted) | |
| vin | Apparent intracellular volume fraction |
| Mean intracellular diffusivity | |
| Dex0 | Mean extracellular diffusivity at long tdiff |
| βex | Mean extracellular diffusivity change rate with tdiff |
2.5. MRI-cytometry
Although the IMPULSED method reports the mean cell size, it is well-known that cell sizes are heterogeneous in biological tissues, so it is desirable to map cell size distribution non-invasively. MRI-cytometry has been recently described to fulfill this need (28). Instead of assuming mean values for all fitting metrics, MRI-cytometry considers there are distributions of all microstructural parameters in every image voxel. Each cell has a cell size d and an intracellular diffusion coefficient Din, both of which can differ from cell to cell. Thus, each image voxel contains cells characterized by Pin (d, Din), the normalized distribution function of the number of cells with a diameter d and an intracellular diffusivity Din. Similarly, extracellular space is considered to contain a large number of spin packets that are characterized by Pex(Dex0, βex), the normalized distribution function of the number of spin packets with Dex0 and βex. Therefore, the dMRI signals can be written as
| [5] |
and , v(d) = πd3/6 is cell volume, ρin and ρex are the T2-weighted intra- and extracellular dMRI signals per unit volume, respectively, and Sin is intracellular signal attenuation of an impermeable spherical cell. The analytical equations for Sin linking geometric features (d and Din) to dMRI signals (12,62). The marginal distribution P(d) obtained from P(d, Din) is the cell size distribution. The metrics achievable using IMPULSED, such as , , vin, and , can all be obtained by MRI-cytometry. For example, the non-cell-volume-weighted mean cell size where N is the total number cell sizes discretized in the fittings.
3. Data acquisition
Although IMPULSED and MRI-cytometry make different assumptions and perform different data analyses, both are based on the simplified two-compartment diffusion model (i.e., Eq.[3]) that can be addressed with the same data acquisitions with multiple b values and tdiff values using different pulse sequences.
3.1. Pulse sequences
IMPULSED combines both PGSE and OGSE acquisitions to cover a broader range of tdiff, providing more comprehensive information on tissue microstructure than single measurements of ADC (13,17). Figure 2 shows the typical pulse sequences containing both PGSE and OGSE sequences as implemented on regular clinical MRI scanners (12). G is the gradient strength, δ is the duration of each diffusion gradient, Δ is the separation of two gradients, tr is the gradient rise time, tp is the duration time of the first gradient plateau, and for OGSE sequences, t3 = tp + tr/2 and N is the number of cycles in each diffusion gradient. The trapezoid-cosine waveform is the preferred OGSE waveform on clinical MRI scanners because it maximizes the diffusion weighting (64).
Figure 2.
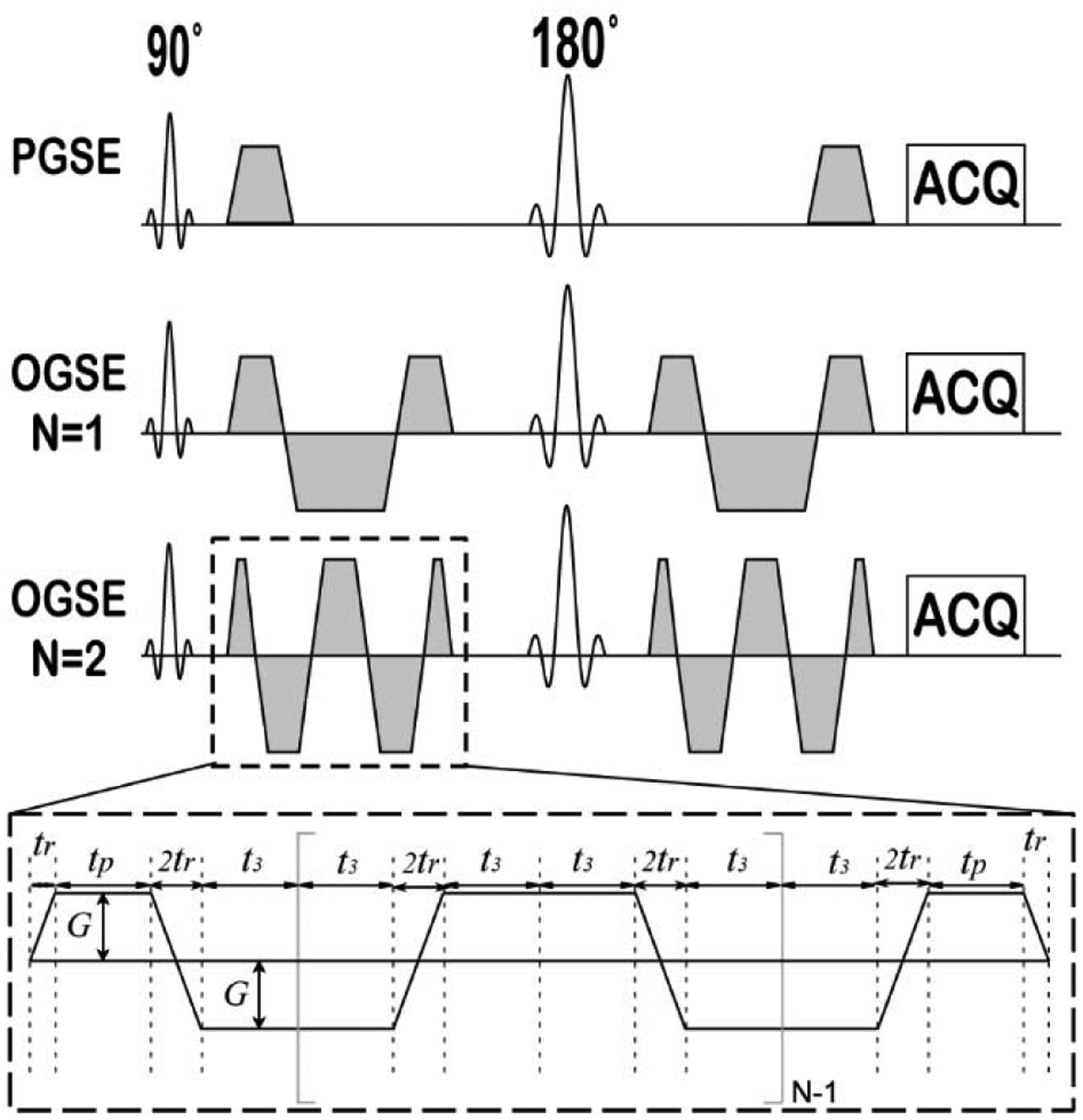
Pulse sequences used in the IMPULSED data acquisitions.
3.2. Imaging parameters
As for typical dMRI, single-shot spin-echo echo-planar imaging (EPI) is usually used for IMPULSED acquisitions but any other dMRI acquisition approaches can be incorporated in the IMPULSED. For example, the recently developed diffusion-prepared 3D gradient spin-echo sequence can reduce the scan time and enhance the SNR of OGSE acquisitions (65).
The maximum gradient strength is a key factor that determines the maximum b values for a specific diffusion time, which thus limits the shortest effective diffusion time, and the highest oscillating frequency, that can be achieved. For example, a gradient strength < 80 mT/m and slew rate ≈ 100 mT/m/sec is commonly achievable for state-of-the-art clinical scanners. With these limitations, the highest oscillating frequency is ~ 50 Hz with a maximum b value of ~300 s/mm2. Table 2 lists the typical diffusion parameters used in our previous clinical and preclinical studies, which have proven successful for assessing cell sizes in different types of cancers (4,13,60). Computer simulations suggest the clinical protocol can reliably fit mean cell sizes from 6 to 20 μm (12). Other imaging parameters (e.g., TE, TR, number of b values, number of acquisitions, and acquisition data matrix) should be determined with considerations of the total scan time and SNR obtainable. The SNR requirement for reliable IMPULSED fittings will be discussed later.
Table 2.
Typical diffusion parameters used in the temporal diffusion spectroscopy acquisitions.
| δ /Δ [ms] | N | f [Hz] | b [s/mm2] | Gmax [mT/m] | ||
|---|---|---|---|---|---|---|
| Clinical (e.g., breast cancer (4)) | PGSE | 12/74 | N/A | N/A | 0,250,500,750,1000,1400,1800 | 80 |
| OGSE | 40/50.5 | 1 | 25 | 0,250,500,750,1000 | ||
| 2 | 50 | 0,100,200,300 | ||||
| Pre-clinical (e.g., xenografts (13)) | PGSE | 12/48 | N/A | N/A | 0,375,750,1125,1500 | 360 |
| OGSE | 20/25 | 1 | 50 | 0,375,750,1125,1500 | ||
| 2 | 100 | 0,375,750,1125,1500 | ||||
| 3 | 150 | 0,330,660,990,1321 | ||||
4. Data analysis
The data analysis procedures of IMPULSED and MRI-cytometry are similar to other quantitative diffusion models. Here we summarize the procedure as below.
4.1. Pre-processing
The following are the pre-processing procedures that have been found useful.
Denoising (optional): Diffusion-weighted images can be denoised e.g. using the nonlocal means (NLM) method (66) or the random matrix theory (67), which can significantly enhance SNR for more reliable fittings. For example, the dwidenoise tool in the MRtrix3 software (https://www.mrtrix.org/) performs denoising on DW images (68).
Gibbs ringing removal (optional): Gibbs ringing artifacts can be removed to reduce the interactions of e.g., interfaces of CSF (69), such as using the mrdegibbs tool in the MRtrix3 software.
Preprocessing: All the DW images can be co-registered with the non-DW (b = 0) image using e.g. the FSL software (70) for correcting any displacement caused by body motions during imaging, eddy current and susceptibility induced image distortions (71).
Model selection: Each voxel signal may go through a model selection process using the Bayesian Data-Analysis toolbox (72) to determine which signal model (either constant ADCs across different tdiff, or tdiff dependent TDS model, or just noise) is statistically more justified. Imaging voxels that do not contain apparent cellular structures at length scales that are inside the range to which TDS is sensitive (e.g., edema and necrosis) will be excluded from further TDS parametric mapping.
Constraints: All fitting parameters can be constrained with physiologically relevant values. For example, 0 ≤ vin ≤ 1, , , and 0 ≤ βex ≤ 10 μm2, where Dfree is the diffusivity of free water (=3.07 μm2/ms at 37°C), d ≤ 25 μm because the root-mean-square displacement of free diffusion at 37°C is 21 μm, and βex ≤ 10 μm2 is determined from numerous studies in vivo (42).
4.2. Data fittings of IMPULSED
Multiple fitting methods have been used in our previous publications, such as traditional nonlinear regression algorithms (13), and Bayesian probability theory-based grid search method (73). The latter has been suggested as the optimal method for making inferences about data (74,75) but at a cost of increased processing time (proportional to mn, where m is the grid number and n is the number of fitting parameters).
4.3. Data fittings of MRI-cytometry
After discretizing d, Din, Dex0, and βex with N, M, P, and Q possible values, respectively, a regularized non-negative least-squares (NNLS) approach can be used to fit Eq. [5] to dMRI data for MRI-cytometry. Due to the complication caused by v(d), MRI-cytometry used a two-step approach: first fit the cell-volume-weighted Pvw(d, Din) and then fit the non-cell-volume-weighted P(d, Din). Specifically, a dictionary containing all possible intra- and extracellular signal forms is constructed and Eq. [5] can be rewritten as
| [6] |
where , , M is the basis signal matrix (dictionary) containing both intra- and extracellular signal forms: min (n,m∣k) = sin (d(n), Din(m)∣bk, fk), and mex (p,q∣k) = exp[−bk(Dex0(p) + βex(q) · f)]. A regularized non-negative least-square problem formulation is given as
| [7] |
where I is a unity matrix with a size of N × M + P × Q and ξ is a regularization penalty parameter that is empirically determined as 0.01 in human studies (28). Pvw(d, Din) and Pex(Dex0, βex) can be obtained by fitting Eq.[7] using the NNLS approach.
In the second step, the regularized non-negative least-square problem focuses on intracellular signals Sin and the non-cell-volume-weighted P(d, Din) can be obtained. The details of the data fittings can be found in (28).
5. Validation
Comprehensive validations, including computer simulations in silico, cells in vitro, and animals in vivo, have been performed to validate both IMPULSED and MRI-cytometry in preclinical studies. These validations suggest that IMPULSED and MRI-cytometry can provide reliable mapping of mean cell size and cell size distribution, respectively, in human imaging with short (~ 7 mins) scan time. The following section will focus on the validations of the IMPULSED method. The details of the validations of MRI-cytometry can be found in (28).
5.1. Computer simulations in silico
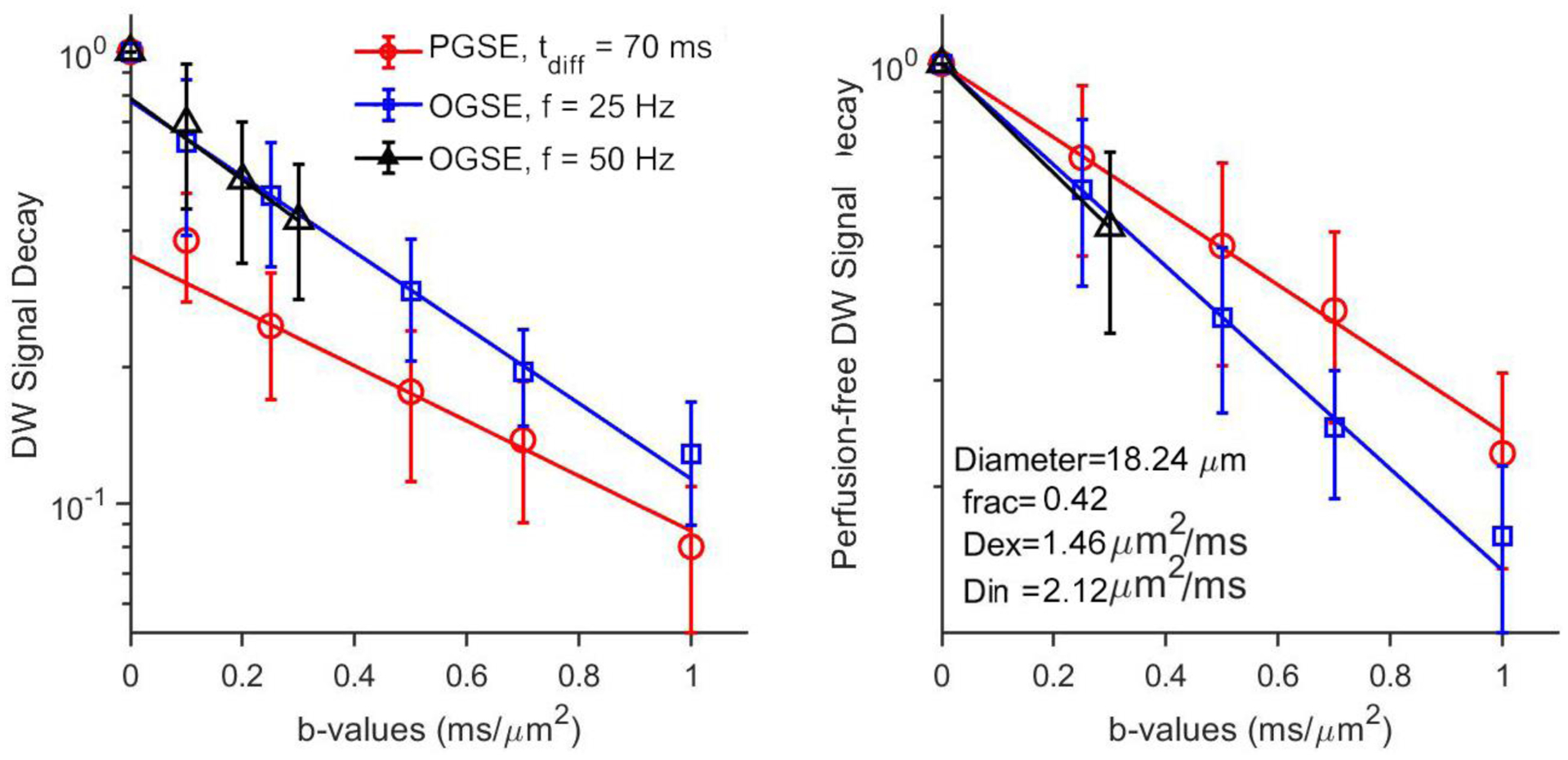
Computer simulations were performed to investigate the accuracy of IMPULSED measurements in a two-compartment model system mimicking tightly packed cells of diameters ranging from 2.5 to 25 μm with various intracellular volume fractions (43%, 51%, and 62%) (13). As shown in Figure 3, a combination of PGSE (tdiff = 48 ms) and 40, and 80 Hz OGSE acquisitions provided the best accuracy of estimating cell size and intracellular volume fraction when the cell size increased from 2.5 to 25 μm, and such accuracy is not affected by cell densities. Imaging protocols with OGSE acquisitions alone underestimated the sizes of large cells.
Figure 3.
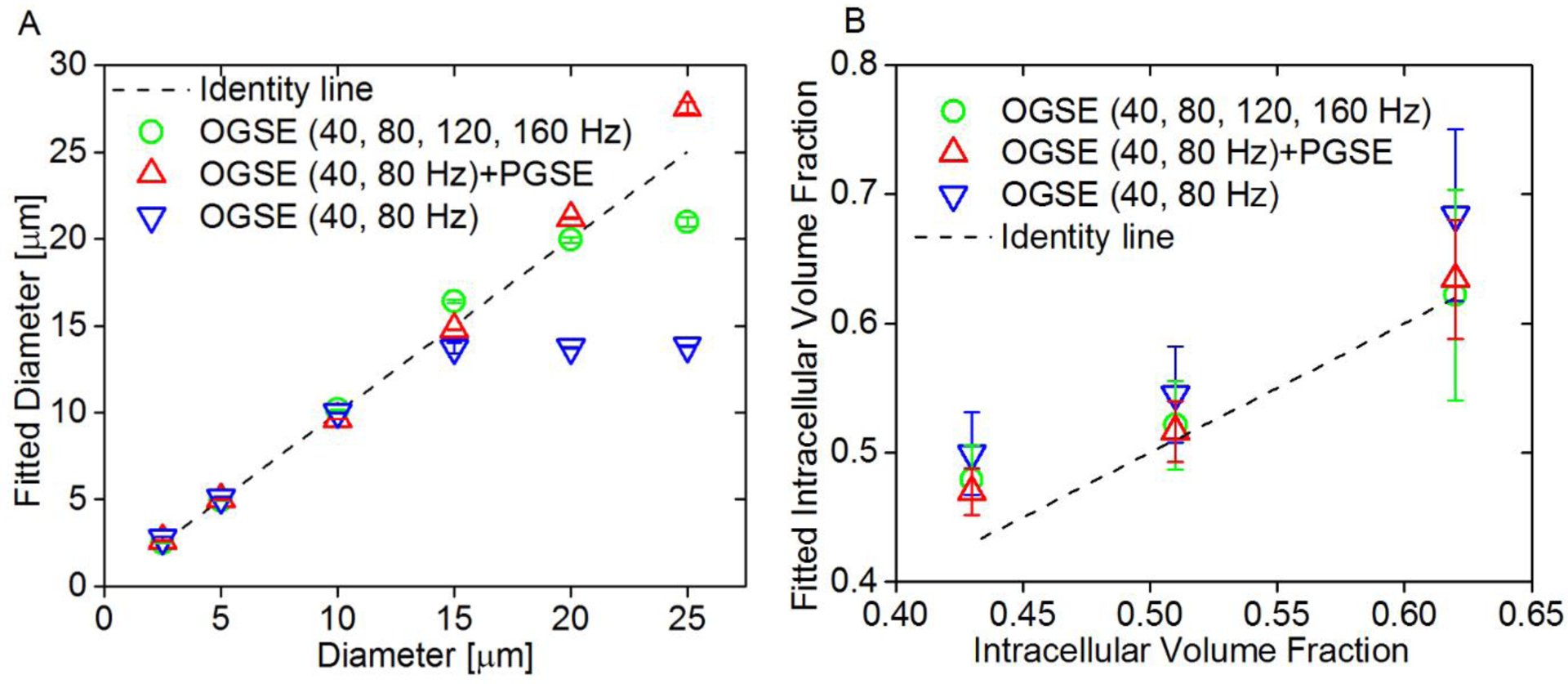
(A) Correlation between fitted diameters and preset diameters in simulations. Fitted diameters generated from three different combinations of OGSE and PGSE signals, mean ± std (n=3, with three different intracellular volume fractions: 43%, 51%, and 62%) vs. preset diameters. The dot line represents the identity line. (B) Fitted intracellular volume fraction generated from three different combinations of OGSE and PGSE signals, mean ± std (n=6, with cell size ranging from 2.5 to 25 μm) vs. preset intracellular volume fractions. The dot line represents the identity line.
5.2. Cultured cells in vitro
Cultured cells (murine erythroleukemia (MEL) and human promyelocytic leukemia K562 cells) of different sizes and at different densities, as well as MDA-MBA-231 cells before and after anti-cancer treatment with Abraxane, were used to validate the IMPULSED method (13). Abraxane is an FDA-approved mitotic inhibitor drug that interrupts cell division during the mitotic (M) phase of the cell cycle when two sets of fully formed chromosomes are supposed to separate into daughter cells. Cells are thereby trapped in the M phase and then undergo apoptosis. Note that cells significantly increase their sizes during the M phase, and hence the cell size can be used as a biomarker to monitor the efficacy of Abraxane treatment. Light microscopy was used to examine the cells. Figure 4 shows the excellent agreement between the sizes derived from experimental IMPULSED and microscopic measurements represented by green bands. Note the derived cell diameters are independent of the cell density, and the estimated intracellular diffusion did not change, while the value of the extracellular diffusion accurately tracked the cell density as expected.
Figure 4.
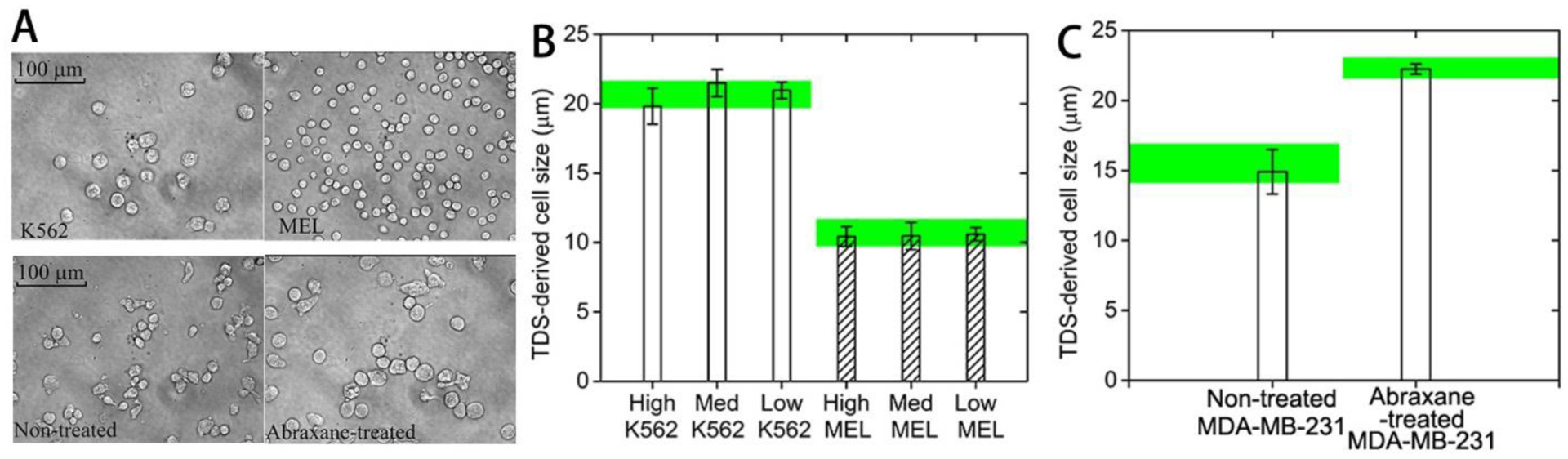
Microscopic pictures of K562, MEL, and MDA-MB-231 before and after anti-mitotic treatment. (B) IMPULSED-derived cell sizes for K562 and MEL at three different cell densities and for MDA-MB-231 before and after anti-mitotic treatment. Noted that the green bands represent the microscopy-derived cell sizes.
5.3. Animal xenografts in vivo
The IMPULSED method was also validated in vivo for mapping mean cell size and cellularities in vivo using three types of human colon cancers (DiFi, HCT116, and SW620). The parametric maps show similar patterns to the H&E stained pictures (Figure 5A). The measured cell sizes were very close to histological results. The IMPULSED-derived cellularity was strongly correlated (Spearman’s correlation coefficient Corr = 0.81, p < 0.001) with histology-derived cellularity and showed heterogeneity within the tumors (Figure 5B).
Figure 5.
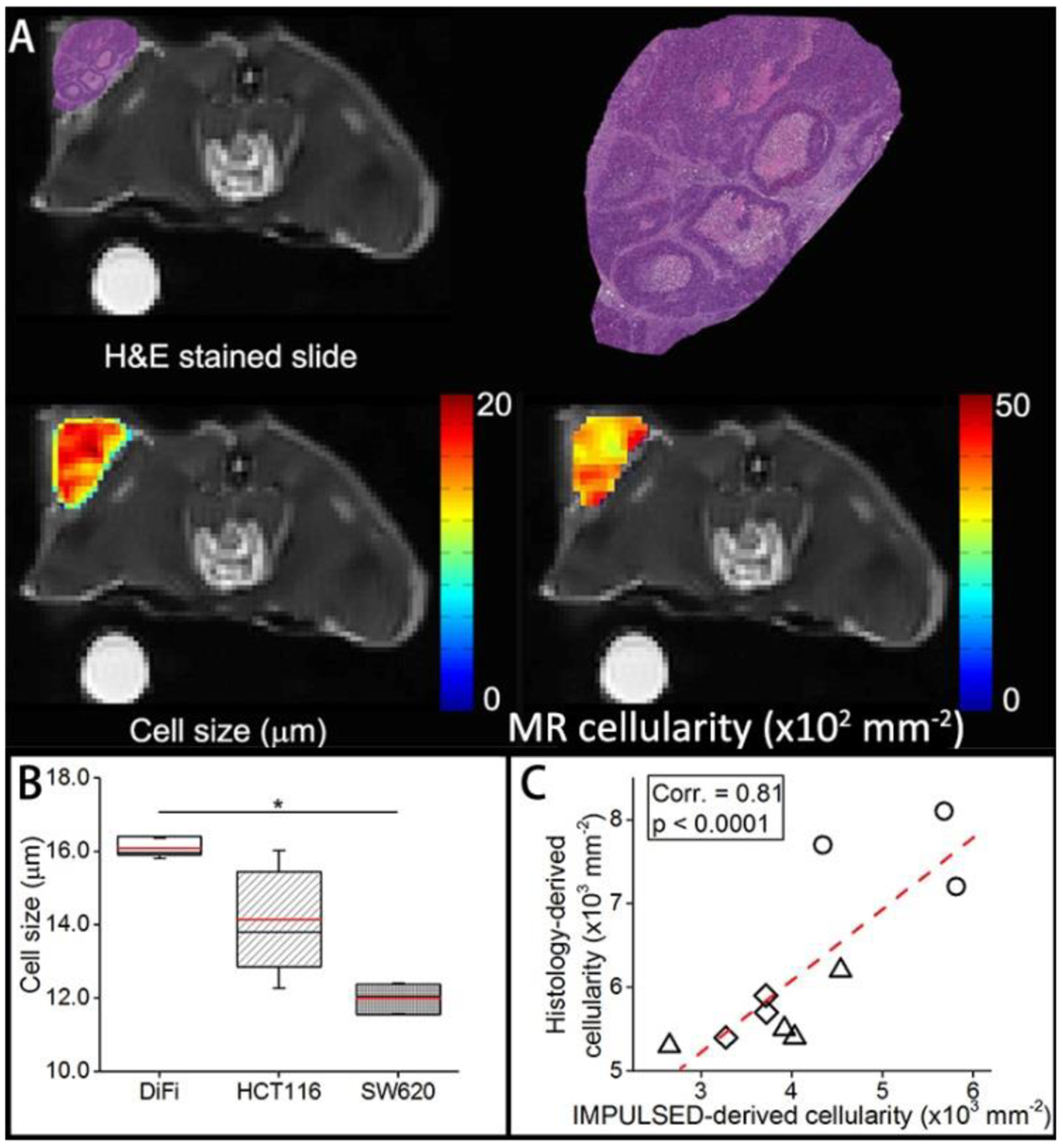
H&E stained histological image, IMPULSED-derived cell size and cell density maps of a representative slice through tumor, overlaid on T2-weighted MR images. (B). Box-and-whisker plot of the fitted cell size for three types of tumors. The 25th-75th percentiles are blocked by the box, the black and red bands inside the box are the median and mean, respectively, and the whiskers mark the SD. C. Correlation between histological-derived cellularities and IMPULSED-derived apparent cellularities for all the tumors.
5.4. Enhanced transcytolemmal water exchange
All the above studies used Eq.[3] that assumes no water exchange. The good agreement between the mean cell size obtained from the IMPULSED method and histology in those studies suggests the neglect of exchange is reasonable. However, the above studies were based on intact cells and tumors. Because cell membrane permeability increases after many anti-cancer treatments, it is unclear if such an assumption is still valid with a higher transcytolemmal water exchange rate. This was investigated by Li et al. (56) using both computer simulations and cell experiments in vitro. The computer simulations showed that, over a very broad range of intracellular water lifetimes τin (50 – ∞ ms) and mean cell sizes d (5 – 20 μm), transcytolemmal water exchange shows negligible influences on the IMPULSED derived d and Din, while vin may be significantly underestimated. This conclusion was confirmed in a cultured cell experiment in vitro by selectively changing cell membrane permeability using different concentrations of saponins while keeping all other cellular microstructure the same. Figure 6 summarizes the dependence of IMPULSED derived d, vin, and Din on intracellular water lifetime τin and cell membrane permeability Pm. The results suggest mean cell size can be reliably fit for a broad range of transcytolemmal water exchange rates, while vin is significantly underestimated due to this exchange. These findings are consistent with histology results obtained in animals in vivo.
Figure 6.
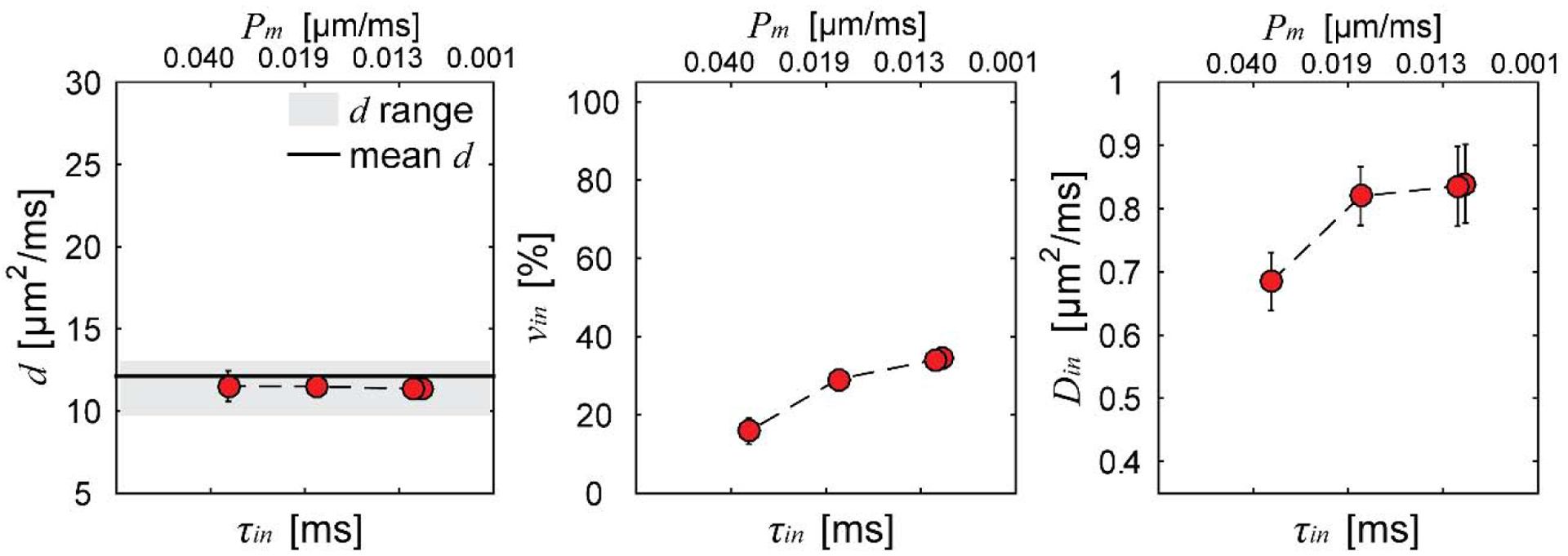
Summary of IMPULSED fitted microstructural parameters versus intracellular water lifetime τin and cell membrane permeability Pm. Error bars in each subfigure denote across-sample STD. The range indicates histology-derived mean cell diameter ± STD of all cells, and mean d is the volume-weighted cell diameter.
6. Applications in cancer – preclinical proof of concept
One of the potential applications for MR cell size imaging is that cell size could provide earlier and more specific assessments of tumor therapeutic responses to various treatments. Because changes at the cellular level (e.g., cell size) and sub-cellular level (e.g., cytoplasm condensation in apoptosis or organelle duplication during mitosis) are usually early responses of cancer cells to treatment, characterization of cell size and intracellular diffusivity may provide a unique means to assess tumor status. Figure 7 summarizes the recently published studies that use IMPULSED to characterize changes in cell morphology when cells undergo different pathological processes. These cell size changes usually occur earlier than changes in tumor volume, the current gold standard of effective cancer treatments. Three studies that treat tumors by inducing apoptosis (42), mitotic arrest (40), and T cell infiltration (4), respectively, will be highlighted below to show how the IMPULSED method can be used to monitor cell size variations following different treatments.
Figure 7.
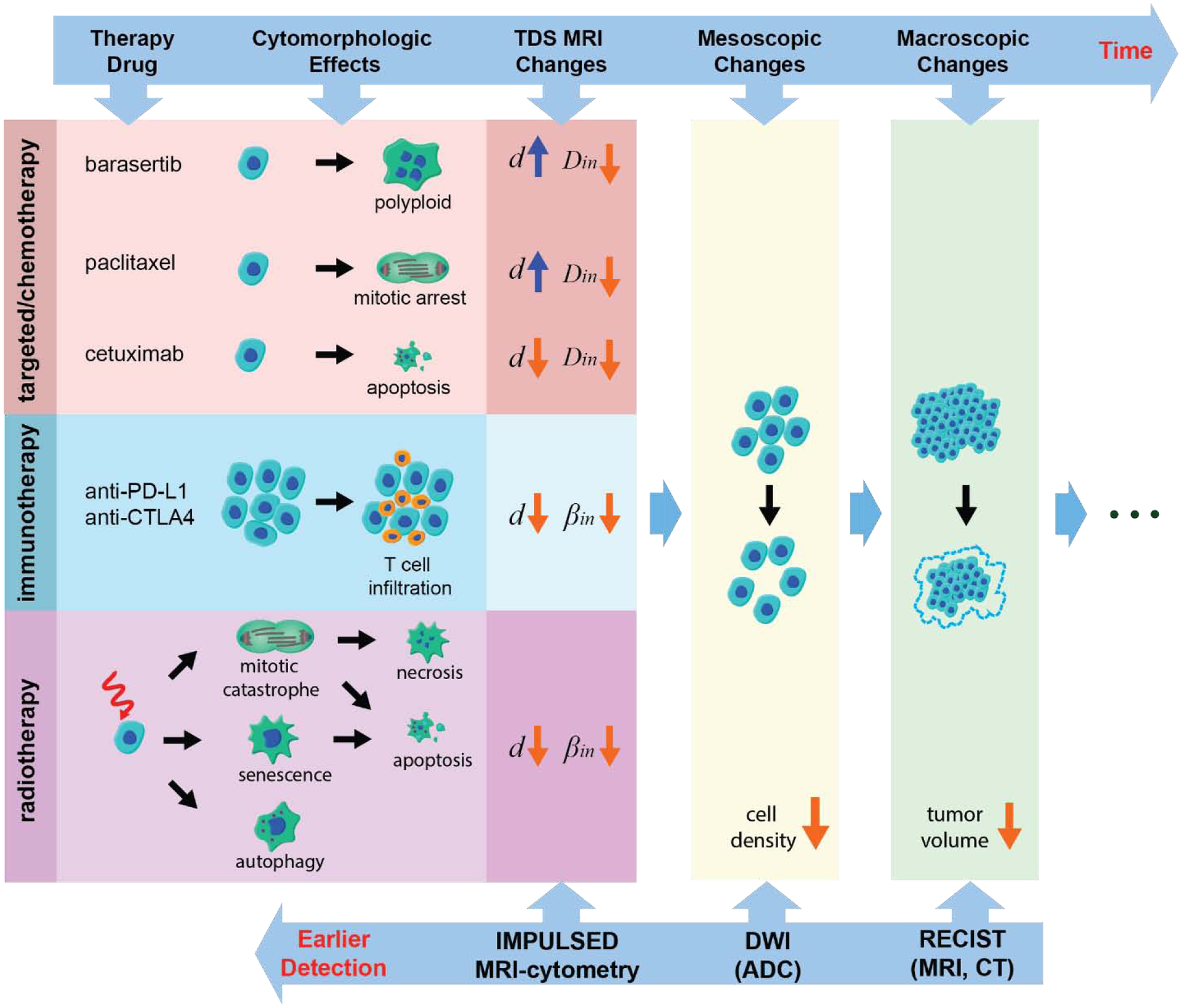
Diagram of various cell morphology changes responding to anti-cancer treatment. All listed therapy approaches and corresponding TDS MRI results are reported previously: barasertib (7), paclitaxel (40), cetuximab (42), anit-PD-L1/anti-CTLA4 (4), and radiation (60)
6.1. Apoptosis
Cell shrinkage, cytoplasm condensation, and DNA fragmentation are hallmarks of cell apoptosis. Their corresponding changes in biophysical features, such as the decrease in mean cell size and the decrease in mean intracellular diffusivity , can be detected by IMPULSED. This in turns provides a specific means to detect treatment-induced apoptosis in tumors. Two types of human colorectal cell lines, DiFi (responder (76)) and HCT116 (non-responder (77)), were treated with cetuximab, an epidermal growth factor receptor (EGFR) inhibitor used for treatment of colon cancers with wild-type KRAS gene. A one-compartment TDS model was used in this study. The temporal diffusion spectrum was measured using a combination of PGSE (δ/Δ = 4/12 ms) and OGSE (50 to 350 Hz with an interval of 50 Hz) acquisitions. Three parameters were fitted: (averaged dimension size), D0 (ADC when f → 0), and D∞ (ADC when f → ∞).
The mean cell size for cetuximab-treated DiFi tumors decreased significantly after the first two treatments and decreased even more after further treatments, indicating apoptotic cell shrinkage. Nuclear condensation and fragmentation slow the water diffusion within the cells, and therefore lowered D∞ for DiFi tumors after four treatments. D0 is sensitive to cell density changes as a result of cell death. The combination of these parameters (, D∞, and D0) may provide a specific in vivo assessment of treatment-induced apoptosis in solid tumors. In addition, changes reported by these parameters occurred earlier than changes in conventional tumor size measurements.
6.2. Mitotic arrest
Paclitaxel is a widely used cytotoxic chemotherapy drug for various cancers. As an inhibitor of microtubule depolymerization, it causes cells to arrest in a mitotic phase and eventually leads to cell death (78). During the mitotic phase, cell size d usually increases significantly and the duplicated intracellular organelles cause more hindrances to water mobility and hence produces significantly decreased Din. Therefore, measurements of the changes in d and Din provides a specific means to probe treatment-induced mitotic arrest.
Two types of human ovarian cell lines (OVCAR-8 as a responder to Nab-paclitaxel, and NCI/ADR-RES as a resistant type) were treated with either vehicle (PBS) or Nab-paclitaxel, and treatment responses of both in vitro and in vivo cases were investigated using the one-compartment TDS model. We found that Nab-paclitaxel induced acute cell size increases in responding tumors (confirmed by flow cytometry and light microscopy) in cell culture. Nab-paclitaxel-induced mitotic arrest was quantified histologically by measuring the mitotic index in vivo using a mitosis-specific marker (antiphosphohistone H3). The fitted was able to detect and quantify the increases in tumor cell sizes, which were not detected by ADC using conventional diffusion-MRI. All the MR results had a high degree of consistency with other flow, microscopy, and histological data. Moreover, with an appropriate analysis, the Nab-paclitaxel-responsive tumors in vivo could be easily distinguished from all the other drug-vehicle-treated tumors and Nab-paclitaxel-resistant tumors. This study confirms that TDS detects antimitotic-therapy-induced microstructural variations (notably, increases in cell sizes) in solid tumors in vivo before changes in tissue cellularity or conventional diffusion MRI metrics.
6.3. T-cell infiltration
There is increasing interest of assessing tumor responses to immunotherapy as it has been rapidly adopted as a treatment option for a wide variety of cancers (79–82). However, it takes significantly longer for conventional MRI to characterize successful response to immunotherapy (83). For example, checkpoint inhibitor treatments that block PD1 and CTLA4 receptors lead to cytotoxic T cell infiltration into tumors, which may lead to transient tumor enlargement due to the increased number of T cells, followed by shrinkage or long-term stability of tumor size (84). Because conventional standard-of-care RECIST (version 1.1) (85) criteria are based on tumor volume, this may misdiagnose effective response as disease progression i.e., pseudo-progression (83,86,87). There is a need to develop a specific imaging method to assess tumor response to immunotherapy. T cells are typically 5–10 μm in diameter (88–91), which is significantly smaller than regular cancer cells (10–20 μm) (92). The increased fraction of smaller T cells in tumors during T cell infiltration results in a decrease in the mean cell size, which can be measured by MR cell size imaging. This provides a unique means to detect T cell infiltration without any radiation-sensitive labeling (93).
We began preclinical assessment of immune-checkpoint blockade (ICB) of anti-PD-1 and anti-CTLA4 in a mouse model of colon adenocarcinoma (MC38). This cell line was chosen because MC38 is sensitive to anti-PD-1 alone, while most murine models (of melanoma, lung, and head & neck) are resistant to monotherapy. 34 C57/BL6 mice were subcutaneously injected with 1×106 MC38 cells. After tumors were MRI visible (day 7), mice were imaged at 7, 10, 13, and 16 days. Either dual therapy (200 ug/dose IP, n=19) or mIgG (200 ug/dose IP, n=15) alone were administered after the first 3 imaging sessions on 7, 10, and 13 days. The IMPULSED method was used to measure the mean cell size and other microstructural metrics (see Table 1).
The average cell sizes of ICB responders as measured by IMPULSED are significantly smaller than those of control IgG-treated tumors or ICB non-responders, at 16 days post injection (dpi) (Figure 8B), associated with increasing T cells in ICB responders (Figure 8A). We subsequently validated the MR findings by performing immunohistochemical analyses on tumor tissues collected at 16 dpi. Strong membranous Na+/K+-ATPase and CD3+ staining in the tumor allows us to segment tumor cells and T cells easily and calculate mean cell sizes and T cell fractions for all the slides (Figure 8C). As shown in Figure 8D, histology-derived mean cell sizes including both tumor cells and T cells have a strong negative correlation (r = −0.9, p < 0.001) with T cell fractions. The mean cell size decreases about 1 μm with a 10% increase of CD3+ T cell fraction. IMPULSED-derived cell sizes show a negative correlation (r = −0.64, p < 0.001) with T cell fractions (Figure 8E). IMPULSED-derived cell sizes show a moderate correlation (r = 0.52, p = 0.008) with histology-derived cell sizes (Figure 8F). All these results suggest that mean cell size is an indicator of immunotherapy-induced T cell infiltration and the IMPULSED method has the potential to assess immunotherapy response via mapping mean cell size.
Figure 8.
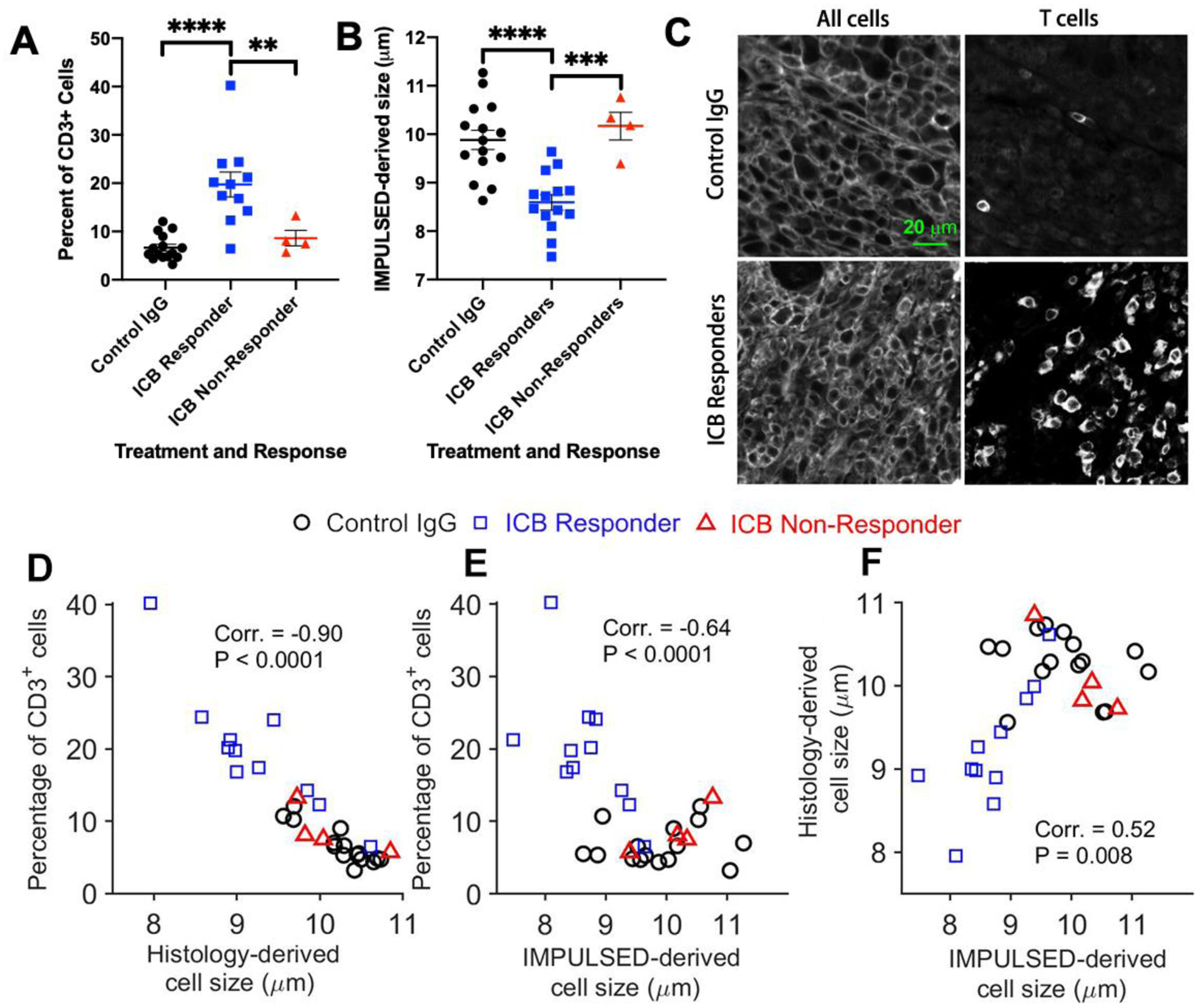
(A) Tumors from ICB responders had a significantly higher percent of CD3+ T cells as measured by immunohistochemistry compared to ICB non-responders and control IgG-treated mice; (B) ICB responding tumors had a significantly smaller mean cell size as measured by IMPULSE than tumors from control or ICB non-responders; (C). Examples of Immunohistochemical analyses of MC38 tumors treated with either control IgG (top) or checkpoint inhibitors (bottom); (D). Validation of IMPULSED-derived cell sizes using histology.
7. Translation to human imaging
The above validation and application studies used animal scanners with fewer hardware restrictions than clinical devices, such as maximum gradient strength Gmax up to 360 mT/m. However, Gmax is limited ⩽ 80 mT/m on most regular clinical MRI scanners. A few strategies have been used to implement IMPULSED acquisitions on human systems, including
use of the cosine-modulated trapezoidal gradient waveform to maximize b values (64);
extending gradient durations δ to 40 ms or more to enhance b values;
fixing Din as a constant because of the limited range of tdiff resulting in a decreased sensitivity to intracellular diffusion (12);
keeping Dex as tdiff independent to enhance the fitting accuracy and precision (12).
Another major limitation is the overall scan time, which should be as short as possible for clinical applications. To date, IMPULSED has been implemented for mapping mean cell size in human livers (73) and breast tumors (12). MRI-cytometry has been implemented in breast cancer patients as well (28).
7.1. Breast cancer
IMPULSED was implemented to study breast cancer patients on a Philips 3T scanner. Acquisition sequence parameters were TR/TE = 4500/103ms; FOV = 192×192mm; reconstructed in-plane resolution = 1.3×1.3 mm; 10 or 20 slices; slice thickness = 5 mm; NEX = 2; single-shot EPI; SENSE factor = 3; fat suppression with SPAIR; and dynamic stabilization were used to minimize dMRI signal drifts during scanning. Images were acquired with two opposite diffusion gradient directions for each axis and the geometric means were used as final images to mitigate the cross-terms between diffusion and background gradients. All diffusion sequence parameters were the same as in Table 2. The total scan time ≈ 7 mins.
Figure 9 shows representative IMPULSED-derived parametric maps of d, vin, and Dex of a human breast tumor overlaid on a high-resolution anatomical image. As the average SNR of b0 images is ~ 20, the intracellular diffusion coefficient Din was fixed to 1.58 μm2/ms for improved accuracies of other three parameters (detailed explanation will be given in the discussion). Significantly lower intracellular volume fraction and increased extracellular diffusion coefficients were observed at the center of the tumor, suggesting the presence of a necrotic core. All the fitted values of cell sizes, densities, and diffusion coefficients are within reasonable ranges.
Figure 9.
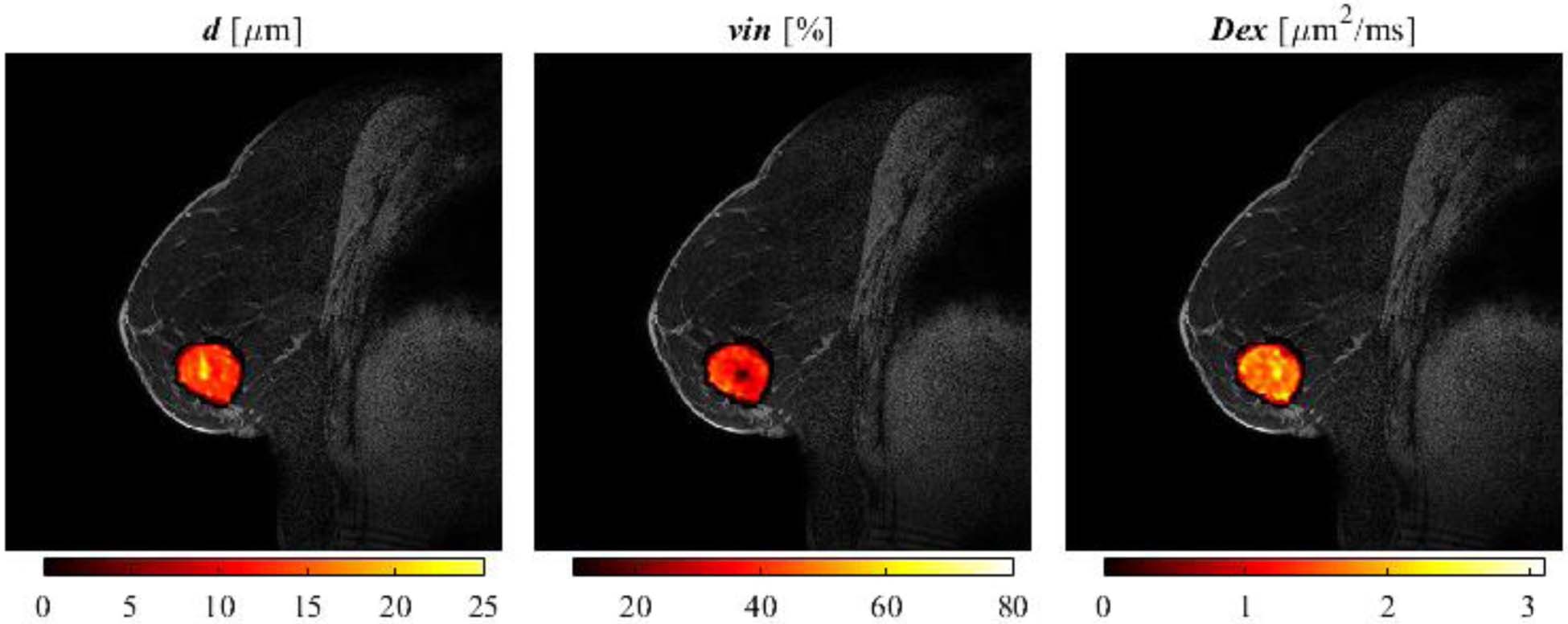
Representative IMPULSED-derived maps of mean cell size d (left), intracellular volume fraction vin (middle), and extracellular diffusion coefficient Dex (rigth) overlaid on a high-resolution fat-suppressed anatomical image of a breast cancer patient.
Figure 10 shows representative cell size distributions P(d) and MRI-Cytometry derived vin and parametric maps of a human breast tumor. SNR was ~45 on T2w b = 0 images. Four examples of cell size distributions from four representative image voxels are provided. For the voxels in the viable tumors (#1, #2, and #4 shown in Figure 10a, e, and g), the shapes of P(d) are similar to each other. However, voxel #2 shows hyperintensity on the T2-weighted b = 0 image and reduced vin. This might be due to increased extracellular water. By contrast, P(d) appears very differently as a small peak at large cell sizes (> 20 μm) for voxel #3 in the necrotic region. This is because the necrotic core consists of mainly fluid and cell debris with few restrictions. This leads to a small fitted intracellular volume fraction vin (< 10%) and large fitted cell sizes ≥ 20.5 μm, the root-mean-square-displacement of free water at body temperature 37°. Figure 10d and h demonstrate the MRI-Cytometry derived vin and parametric maps, which show low vin and high in the necrotic core compared with viable tumors. Note that transcytolemmal water exchange may be responsible for the underestimation of vin, but it may have minor influence on , as we reported previously (56).
Figure 10.
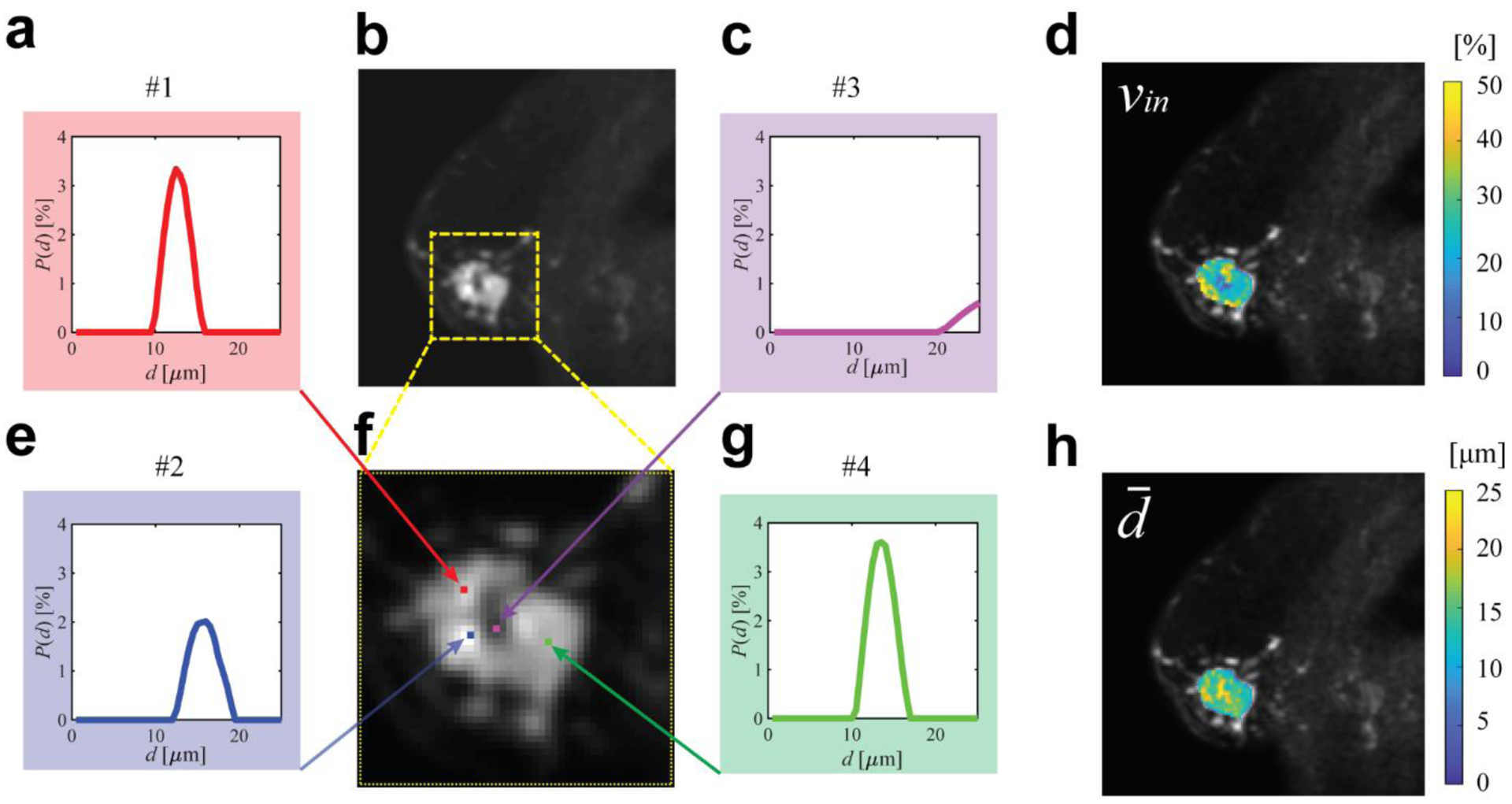
Representative cell size distributions and MRI-Cytometry derived parametric maps of a breast tumor. (b) is T2-weighted b = 0 image of the tumor and (f) is an enlarged view. (a,c,e,g) are four examples of cell size distributions. (d) and (h) are MRI-Cytometry derived vin and maps of the same tumor.
7.2. Liver
Another reported application of IMPULSED is liver imaging in human subjects. The size and density of hepatocytes, their variations and their changes over time, are fundamental characteristics of liver tissues, and diagnostic biomarkers of several normal and pathological processes. For example, hepatocytes shrink during early apoptosis and they swell during hepatocyte polyploidization; hepatocytes exhibit ballooning associated with cell swelling in nonalcoholic steatohepatitis; inflammation induces shifts in the mean cell size as small immune cells infiltrate the liver. Measurements of hepatocyte sizes and densities can thus have high clinical significance but currently can be obtained only by liver biopsy.
In a recent study, the IMPULSED method was performed for mapping mean cell size d of healthy subject livers shown in Figure 11. MR imaging was performed using a Philips 3T scanner with a dStream TorsoCardiac coil. In addition to the diffusion parameters shown in Table 2, other imaging parameters were: acquired in-plane resolution = 4×4 mm2, slice thickness=10 mm; NEX=2; respiratory-gated, single shot EPI; SENSE factor=2; fat suppression with Spectral Attenuated Inversion Recovery (SPAIR). The total scan time ≈ 12 mins. The fitted average cell size for a healthy subject is 17±4.6 μm, consistent with reported human hepatocyte sizes (94,95). The intracellular volume fraction is 37±18.4%, which is smaller than literature values (~ 70%) (96). Underestimation of intracellular volume fraction has been also observed in our previous IMPULSED studies in tumors (13,17). It is due to ignoring water exchange between intra- and extra-cellular spaces as discussed in section 2.2.3.
Figure 11.
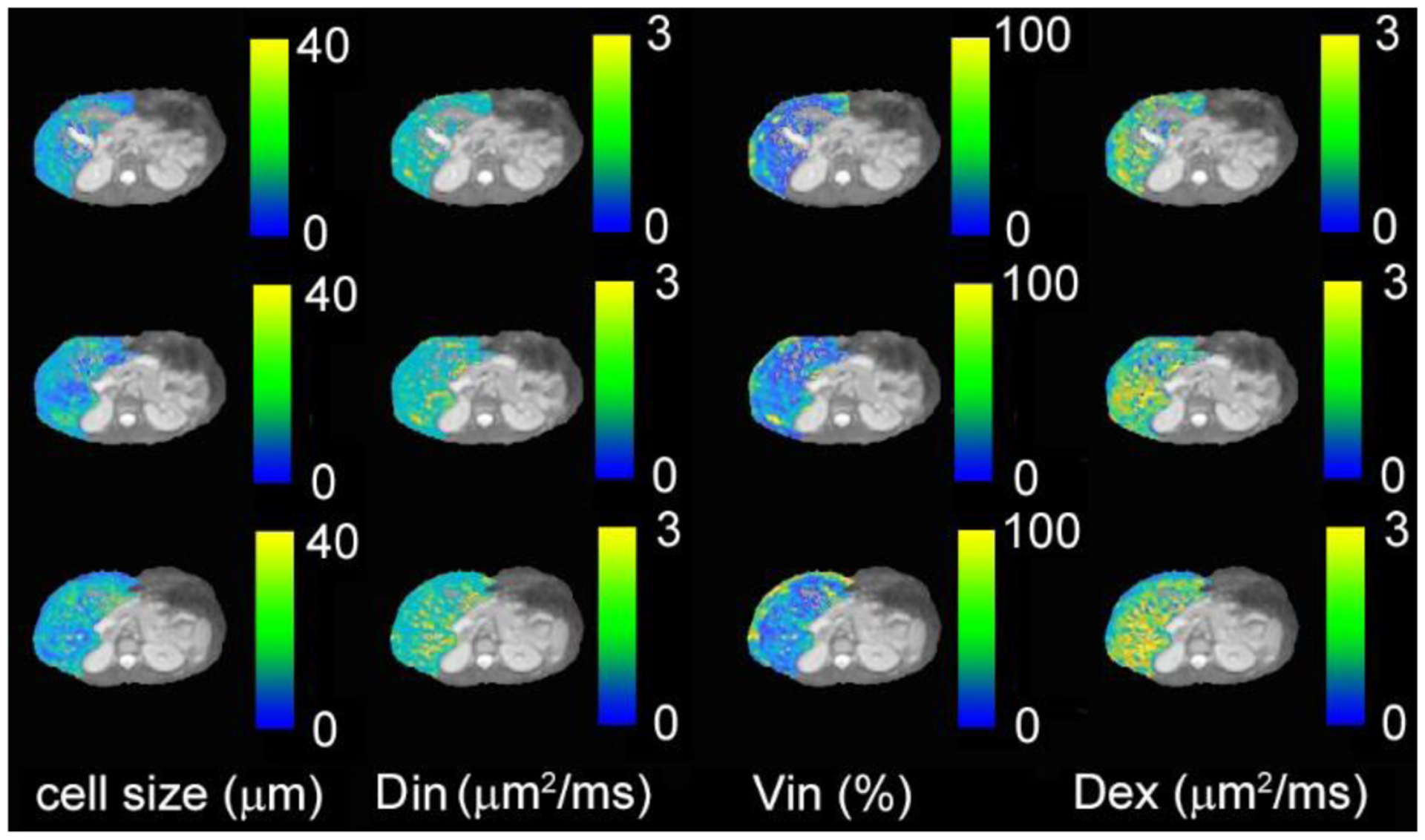
IMPULSED-derived maps of mean cell size d, intracellular diffusion coefficient Din, apparent intracellular volume fraction vin, and extracellular diffusion coefficient Dex overlaid on a high-resolution fat-suppressed anatomical image for three continuous slices of liver from a healthy human subject.
8. Discussion
8.1. Cell density
Cell density is calculated as the quotient of intracellular volume fraction and IMPULSED-derived cell volume. Our previous studies suggest that intracellular volume fraction vin is intrinsically underestimated by IMPULSED and fitted values decrease rapidly with shorter intracellular water lifetime τin, whereas accurate estimates of mean cell diameter may be obtained accurately independent of transcytolemmal water exchange with sufficient SNR (eg, 50). Accordingly, the cell density is likely to be underestimated. The clinical value of such an apparent cell density (including effects from the cell membrane permeability) needs to be further investigated. Future studies should aim to establish a quantitative relationship between the magnitude of membrane permeability and bias of vin estimates. For example, we can modify the analysis to include water exchange between intra and extracellular spaces, such as using the Karger model (97), or apply another independent measurement of water exchange effects, such as filter exchange imaging (54).
8.2. Intracellular diffusion
Intracellular diffusion is complex due to the existence of cell membranes and a variety of organelles and cytoplasmic contents. Cell membranes are usually believed to dominate the restriction to water diffusion, but organelles have also been reported to affect diffusion measurements significantly. For example, the nucleus, the largest organelle containing chromosomes is bounded by membranes (the nuclear envelope) which restrict water diffusion and allows dMRI to be sensitive to nuclear size and nuclear-to-cytoplasm ratio (5). Changes in individual organelles (6) or overall intracellular microenvironment during cell division (57) have also been shown to contribute to the variations of ADC measurements. All these studies suggest the need for complex modeling of intracellular diffusion. However, based on the theory of IMPULSED, we can adjust tdiff in experiments to tune down the detection sensitivity to intracellular organelles while increasing sensitivity to cell size, which in turn makes it possible to measure cell sizes accurately.
Previous studies assessing intracellular diffusion were achieved with short tdiff ranges such as < 0.5 ms (i.e., f > 500 Hz using OGSE). This is far beyond the shortest tdiff that is achievable on human MRI using current gradient coils (39). For tdiff > 10 ms on current regular clinical MRI scanners, the characteristic length of diffusion measurements is if diffusivity D = 2 μm2/ms. For typical cells with size < 10 μm (e.g., red blood cells and T cells) and many cancer cells such as breast cancer cells < 16 μm, this results in a significantly reduced sensitivity to small organelles. Instead, a single diffusion coefficient, Din, can be defined to represent an averaged diffusion property caused by all organelles and cytoplasm. However, if the cell size is large (e.g., hepatocyte size ~ 20 μm) with a large nuclear size (e.g., 5–7 μm in normal hepatocyte), the approximation of a single Din is questionable. However, about 7 – 10 % surface area of a nucleus is occupied by nuclear pore complexes (98,99), which allow small proteins and molecules with molecular weight < 40 kDa to translocate between cytoplasm and cell nucleus by passive diffusion (100,101). Therefore, the nuclear membrane has been considered to permit nearly free passage of water molecules between the nucleus and cytosol (101–104). With this assumption, it has been demonstrated in a simulation study that MR diffusion measurements at diffusion times ranging from 1 – 70 ms are not sensitive to variations in the ratio of nuclear volume to cell volume from 0 to 30%, covering the range of nuclear-to-cell-volume ratios reported previously and found in our histology analysis of nuclear size (5). Therefore, for tdiff from 5–70 ms typically used on clinical MRI scanners, it is reasonable to assume a single Din to describe the overall diffusion inside cells in most applications.
8.3. Extracellular diffusion
To date, the IMPULSED method assumes extracellular diffusion to be linearly dependent on gradient frequency, i.e., Dex~βex · f when f > 50 Hz or Dex is a constant independent on f when f < 50 Hz. Such assumptions have been used in various studies and gave results consistent with histology. However, the exact tdiff or f dependence of Dex in tumors or livers is still unclear. In theory, in the short-time range limit tdiff → 0, it is well know that or f−1/2 (105). In the long-time range, Dex ~ f3/2 for short-range disordered media (106). Therefore, there are transitions in the tdiff dependence in the intermediate time range. Because IMPULSED covers a broad range of tdiff (e.g., 5 – 70 ms on regular clinical MRI scanners), the tdiff dependence may vary across different tdiff values. This is particularly important for liver imaging since liver cell size is typically large (~ 20 μm) so that the tdiff range used in the acquisitions may cover both short and intermediate time range. This could complicate the modeling of extracellular diffusion and further investigation is needed.
However, if an even narrower range of f e.g., 25 – 50 Hz is implemented, the dependence of Dex on f reduces significantly and the assumption of a constant Dex may work better. In fact, our previous preclinical studies (SNR ~ 50 and oscillating frequency up to 150 Hz) (13) found much larger variations (relative standard deviation ~ 60%) in fitted β maps compared with much smaller variations (relative standard deviation ~ 8%) in the fitted cell size maps. The variation of parametric maps is a combination of tissue inhomogeneities and fitting uncertainties due to multiple factors (e.g., insufficient SNR levels, inefficient fitting algorithms, and insufficient sensitivity to the fitted parameters). The dramatic difference between variations of βex and cell size maps from the same tissue strongly suggested that diffusion measurements even with oscillating frequencies up to 150 Hz do not provide enough sensitivity for reliable fitting of the linear dependence of Dex on f. A direct comparison using simulations showed that Dex can be treated as a constant for f up to 50 Hz to result in more reliable and accurate fittings of cell size on clinical scanners (4).
9. Conclusion
MR cell size imaging using temporal diffusion spectroscopy provides a reliable and practical means for mapping cell size non-invasively. Because the variation in cell size is usually a part of the first cellular response to diseases or interventions, MR cell size imaging provides a unique and specific means to assess biological tissue status and response to treatments. Towards clinical applications, two approaches have been developed based on TDS, i.e., IMPULSED and MRI-cytometry, for non-invasive mapping of mean cell size and cell size distribution, respectively. Both methods have been comprehensively validated and show promise as useful ways to characterize tissues in a variety of disease models.
11. Acknowledgments
This work was funded by NIH Grants/Award Numbers: K25CA168936, R01CA109106, R01CA173593, UL1TR002243, S10OD021771, U01CA142565, F32CA216942, UL1TR000445, and P30CA068485; American Cancer Society, Grant/Award Number: IRG#58-009-56.
12. Appendix
The analytical signal decay of an impermeable structure using the cosine-modulated gradient waveform is derived previously (62) as
| [A1] |
where S0 is non-diffusion-weighted signal, γ is the gyromagnetic ratio, G is gradient strength, f gradient frequency, D is intra-structural, intrinsic diffusion coefficient, δ gradient duration of a single waveform, and Δ the separation of two gradients. ak and Bk are structure dependent parameters and have been derived analytically for simple geometries such as planes, cylinders, spheres (107) and spherical shells (62). For diffusion inside an impermeable sphere, and where R is radius and μk is the kth root of and J is a Bessel function of the first kind (μk = 2.08, 5.94, 9.21, …).
For the cosine-modulated trapezoidal OGSE sequence with one oscillation n = 1 shown in Figure 2, the analytical signal decay has been derived as (12)
| [A2] |
where tr is the gradient rise time, tp is the duration of the first gradient plateau in the gradient waveform, and all other parameters are the same as in Eq.[A1]. Equations for n = 2 and 3 can be found in (12).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.
13 References
- 1.Moseley JB, Nurse P. Cell division intersects with cell geometry. Cell 2010;142(2):184–188. [DOI] [PubMed] [Google Scholar]
- 2.Ginzberg MB, Kafri R, Kirschner M. Cell biology. On being the right (cell) size. Science 2015;348(6236):1245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bortner CD, Cidlowski JA. A Necessary Role for Cell Shrinkage in Apoptosis. Biochemical Pharmacology 1998;56:1549–1559. [DOI] [PubMed] [Google Scholar]
- 4.Jiang X, Dudzinski S, Beckermann KE, Young K, McKinley E, J McIntyre O, Rathmell JC, Xu J, Gore JC. MRI of tumor T cell infiltration in response to checkpoint inhibitor therapy. Journal for ImmunoTherapy of Cancer 2020;8(1):e000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Does MD, Gore JC. Sensitivity of MR diffusion measurements to variations in intracellular structure: effects of nuclear size. Magn Reson Med 2009;61(4):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colvin DC, Jourquin J, Xu J, Does MD, Estrada L, Gore JC. Effects of intracellular organelles on the apparent diffusion coefficient of water molecules in cultured human embryonic kidney cells. Magn Reson Med 2011;65(3):796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Li K, Smith RA, Waterton JC, Zhao P, Chen H, Does MD, Manning HC, Gore JC. Characterizing tumor response to chemotherapy at various length scales using temporal diffusion spectroscopy. PloS one 2012;7(7):e41714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, Lythgoe MF, Alexander DC. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res 2014;74(7):1902–1912. [DOI] [PubMed] [Google Scholar]
- 9.Panagiotaki E, Chan RW, Dikaios N, Ahmed HU, O’Callaghan J, Freeman A, Atkinson D, Punwani S, Hawkes DJ, Alexander DC. Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Investigative radiology 2015;50(4):218–227. [DOI] [PubMed] [Google Scholar]
- 10.Bonet-Carne E, Johnston E, Daducci A, Jacobs JG, Freeman A, Atkinson D, Hawkes DJ, Punwani S, Alexander DC, Panagiotaki E. VERDICT-AMICO: Ultrafast fitting algorithm for non-invasive prostate microstructure characterization. NMR in biomedicine 2019;32(1):e4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McHugh DJ, Hubbard Cristinacce PL, Naish JH, Parker GJM. Towards a ‘resolution limit’ for DW-MRI tumor microstructural models: A simulation study investigating the feasibility of distinguishing between microstructural changes. Magnetic resonance in medicine 2019;81(4):2288–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Jiang X, Li H, Arlinghaus LR, McKinley ET, Devan SP, Hardy BM, Xie J, Kang H, Chakravarthy AB, Gore JC. Magnetic resonance imaging of mean cell size in human breast tumors. Magnetic resonance in medicine 2020;83(6):2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X, Li H, Xie J, McKinley ET, Zhao P, Gore JC, Xu J. In vivo imaging of cancer cell size and cellularity using temporal diffusion spectroscopy. Magnetic resonance in medicine 2017;78(1):156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shemesh N, Westin CF, Cohen Y. Magnetic resonance imaging by synergistic diffusion-diffraction patterns. Physical review letters 2012;108(5):058103. [DOI] [PubMed] [Google Scholar]
- 15.Laun FB, Kuder TA, Semmler W, Stieltjes B. Determination of the defining boundary in nuclear magnetic resonance diffusion experiments. Physical review letters 2011;107(4):048102. [DOI] [PubMed] [Google Scholar]
- 16.Kuder TA, Bachert P, Windschuh J, Laun FB. Diffusion pore imaging by hyperpolarized xenon-129 nuclear magnetic resonance. Physical review letters 2013;111(2):028101. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magnetic resonance in medicine 2016;75(3):1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynaud O, Winters KV, Hoang DM, Wadghiri YZ, Novikov DS, Kim SG. Pulsed and oscillating gradient MRI for assessment of cell size and extracellular space (POMACE) in mouse gliomas. NMR in biomedicine 2016;29(10):1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey C, Bourne RM, Siow B, Johnston EW, Brizmohun Appayya M, Pye H, Heavey S, Mertzanidou T, Whitaker H, Freeman A, Patel D, Shaw GL, Sridhar A, Hawkes DJ, Punwani S, Alexander DC, Panagiotaki E. VERDICT MRI validation in fresh and fixed prostate specimens using patient-specific moulds for histological and MR alignment. NMR in biomedicine 2019;32(5):e4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price - Jones C The diameters of red cells in pernicious anaemia and in anaemia following haemorrhage. The Journal of Pathology and Bacteriology 1922;25(4):487–504. [Google Scholar]
- 21.Evans TC, Jehle D. The red blood cell distribution width. J Emerg Med 1991;9 Suppl 1:71–74. [DOI] [PubMed] [Google Scholar]
- 22.Montagnana M, Danese E. Red cell distribution width and cancer. Annals of translational medicine 2016;4(20):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini D, Komlosh ME, Basser PJ, Nevo U. Nonparametric pore size distribution using d-PFG: comparison to s-PFG and migration to MRI. J Magn Reson 2014;246:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini D, Komlosh ME, Holtzclaw LA, Nevo U, Basser PJ. White matter microstructure from nonparametric axon diameter distribution mapping. Neuroimage 2016;135:333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shemesh N, Alvarez GA, Frydman L. Size Distribution Imaging by Non-Uniform Oscillating-Gradient Spin Echo (NOGSE) MRI. PLoS One 2015;10(7):e0133201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anaby D, Morozov D, Seroussi I, Hametner S, Sochen N, Cohen Y. Single- and double-Diffusion encoding MRI for studying ex vivo apparent axon diameter distribution in spinal cord white matter. NMR Biomed 2019:e4170. [DOI] [PubMed] [Google Scholar]
- 27.Duchene G, Abarca-Quinones J, Leclercq I, Duprez T, Peeters F. Insights into tissue microstructure using a double diffusion encoding sequence on a clinical scanner: Validation and application to experimental tumor models. Magn Reson Med 2020;83(4):1263–1276. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Jiang X, Devan SP, Arlinghaus LR, McKinley ET, Xie J, Zu Z, Wang Q, Chakravarthy AB, Wang Y, Gore JC. MRI-cytometry: Mapping non-parametric cell size distributions using diffusion MRI. Magn Reson Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gore JC, Xu JZ, Colvin DC, Yankeelov TE, Parsons EC, Does MD. Characterization of tissue structure at varying length scales using temporal diffusion spectroscopy. Nmr in Biomedicine 2010;23(7):745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Bihan D What can we see with IVIM MRI? NeuroImage 2019;187:56–67. [DOI] [PubMed] [Google Scholar]
- 31.Koh DM, Collins DJ, Orton MR. Intravoxel incoherent motion in body diffusion-weighted MRI: reality and challenges. AJR American journal of roentgenology 2011;196(6):1351–1361. [DOI] [PubMed] [Google Scholar]
- 32.van Zijl P, Knutsson L. In vivo magnetic resonance imaging and spectroscopy. Technological advances and opportunities for applications continue to abound. J Magn Reson 2019;306:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanner JE. Self diffusion of water in frog muscle. Biophysical journal 1979;28(1):107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novikov DS, Jensen JH, Helpern JA, Fieremans E. Revealing mesoscopic structural universality with diffusion. Proc Natl Acad Sci U S A 2014;111(14):5088–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tetreault P, Harkins KD, Baron CA, Stobbe R, Does MD, Beaulieu C. Diffusion time dependency along the human corpus callosum and exploration of age and sex differences as assessed by oscillating gradient spin-echo diffusion tensor imaging. Neuroimage 2020;210:116533. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Jiang X, Wang F, Xu J, Gore JC. Structural information revealed by the dispersion of ADC with frequency. Magn Reson Imaging 2015;33(9):1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Li H, Li K, Harkins KD, Jiang X, Xie J, Kang H, Dortch RD, Anderson AW, Does MD, Gore JC. Fast and simplified mapping of mean axon diameter using temporal diffusion spectroscopy. NMR in biomedicine 2016;29(4):400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aggarwal M, Jones MV, Calabresi PA, Mori S, Zhang J. Probing mouse brain microstructure using oscillating gradient diffusion MRI. Magnetic resonance in medicine 2012;67(1):98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan ET, Shih RY, Mitra J, Sprenger T, Hua Y, Bhushan C, Bernstein MA, McNab JA, DeMarco JK, Ho VB, Foo TKF. Oscillating diffusion-encoding with a high gradient-amplitude and high slew-rate head-only gradient for human brain imaging. Magn Reson Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Li H, Zhao P, Xie J, Khabele D, Xu J, Gore JC. Early Detection of Treatment-Induced Mitotic Arrest Using Temporal Diffusion Magnetic Resonance Spectroscopy. Neoplasia 2016;18(6):387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li JR, Calhoun D, Poupon C, Le Bihan D. Numerical simulation of diffusion MRI signals using an adaptive time-stepping method. Physics in medicine and biology 2014;59(2):441–454. [DOI] [PubMed] [Google Scholar]
- 42.Jiang X, McKinley ET, Xie J, Li H, Xu J, Gore JC. In vivo magnetic resonance imaging of treatment-induced apoptosis. Scientific reports 2019;9(1):9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Bihan D, Turner R. The capillary network: a link between IVIM and classical perfusion. Magn Reson Med 1992;27(1):171–178. [DOI] [PubMed] [Google Scholar]
- 44.Taouli B, Koh DM. Diffusion-weighted MR imaging of the liver. Radiology 2010;254(1):47–66. [DOI] [PubMed] [Google Scholar]
- 45.Wu D, Zhang J. Evidence of the diffusion time dependence of intravoxel incoherent motion in the brain. Magn Reson Med 2019;82(6):2225–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Sun A, Li H, Saiviroonporn P, Wu EX, Guo H. Stimulated echo diffusion weighted imaging of the liver at 3 Tesla. Magnetic resonance in medicine 2017;77(1):300–309. [DOI] [PubMed] [Google Scholar]
- 47.Funck C, Laun FB, Wetscherek A. Characterization of the diffusion coefficient of blood. Magnetic resonance in medicine 2018;79(5):2752–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li K, Li H, Zhang XY, Stokes AM, Jiang X, Kang H, Quarles CC, Zu Z, Gochberg DF, Gore JC, Xu J. Influence of water compartmentation and heterogeneous relaxation on quantitative magnetization transfer imaging in rodent brain tumors. Magnetic resonance in medicine 2016;76(2):635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dortch RD, Yankeelov TE, Yue Z, Quarles CC, Gore JC, Does MD. Evidence of multiexponential T2 in rat glioblastoma. NMR in biomedicine 2009;22:609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanner JE. Transient diffusion in a system partitioned by permeable barriers - application to NMR measurements with a pulsed field gradient. J Chem Phys 1978;69(4):1748–1754. [Google Scholar]
- 51.Li H, Jiang X, Xie J, McIntyre JO, Gore JC, Xu J. Time-Dependent Influence of Cell Membrane Permeability on MR Diffusion Measurements. Magnetic resonance in medicine 2016;75(5):1927–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volles MJ, Lansbury PT Jr. Vesicle permeabilization by protofibrillar alpha-synuclein is sensitive to Parkinson’s disease-linked mutations and occurs by a pore-like mechanism. Biochemistry 2002;41(14):4595–4602. [DOI] [PubMed] [Google Scholar]
- 53.Moftakhar P, Lynch MD, Pomakian JL, Vinters HV. Aquaporin expression in the brains of patients with or without cerebral amyloid angiopathy. Journal of neuropathology and experimental neurology 2010;69(12):1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilsson M, Latt J, van Westen D, Brockstedt S, Lasic S, Stahlberg F, Topgaard D. Noninvasive mapping of water diffusional exchange in the human brain using filter-exchange imaging. Magn Reson Med 2013;69(6):1573–1581. [DOI] [PubMed] [Google Scholar]
- 55.Bailey C, Moosvi F, Stanisz GJ. Mapping water exchange rates in rat tumor xenografts using the late-stage uptake following bolus injections of contrast agent. Magn Reson Med 2014;71(5):1874–1887. [DOI] [PubMed] [Google Scholar]
- 56.Li H, Jiang X, Xie J, Gore JC, Xu J. Impact of transcytolemmal water exchange on estimates of tissue microstructural properties derived from diffusion MRI. Magnetic resonance in medicine 2017;77(6):2239–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu J, Xie J, Jourquin J, Colvin DC, Does MD, Quaranta V, Gore JC. Influence of cell cycle phase on apparent diffusion coefficient in synchronized cells detected using temporal diffusion spectroscopy. Magnetic resonance in medicine 2011;65(4):920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novikov DS, Fieremans E, Jespersen SN, Kiselev VG. Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation. NMR in biomedicine 2019;32(4):e3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arbabi A, Kai J, Khan AR, Baron CA. Diffusion dispersion imaging: Mapping oscillating gradient spin-echo frequency dependence in the human brain. Magnetic resonance in medicine 2020;83(6):2197–2208. [DOI] [PubMed] [Google Scholar]
- 60.Jiang X, Xu J, Gore JC. Quantitative temporal diffusion spectroscopy as an early imaging biomarker of radiation therapeutic response in gliomas: A preclinical proof of concept. Adv Radiat Oncol 2019;4(2):367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neuman CH. Spin-echo of spins diffusing in a bounded medium. J Chem Phys 1974;60(11):4508–4511. [Google Scholar]
- 62.Xu J, Does MD, Gore JC. Quantitative characterization of tissue microstructure with temporal diffusion spectroscopy. Journal of magnetic resonance 2009;200(2):189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ianus A, Siow B, Drobnjak I, Zhang H, Alexander DC. Gaussian phase distribution approximations for oscillating gradient spin echo diffusion MRI. J Magn Reson 2013;227:25–34. [DOI] [PubMed] [Google Scholar]
- 64.Van AT, Holdsworth SJ, Bammer R. In vivo investigation of restricted diffusion in the human brain with optimized oscillating diffusion gradient encoding. Magn Reson Med 2014;71(1):83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu D, Liu D, Hsu YC, Li H, Sun Y, Qin Q, Zhang Y. Diffusion-prepared 3D gradient spin-echo sequence for improved oscillating gradient diffusion MRI. Magnetic resonance in medicine 2020. [DOI] [PubMed] [Google Scholar]
- 66.Wiest-Daessle N, Prima S, Coupe P, Morrissey SP, Barillot C. Non-local means variants for denoising of diffusion-weighted and diffusion tensor MRI. Medical image computing and computer-assisted intervention: MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention 2007;10(Pt 2):344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veraart J, Novikov DS, Christiaens D, Ades-Aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. NeuroImage 2016;142:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh CH, Connelly A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 2019;202:116137. [DOI] [PubMed] [Google Scholar]
- 69.Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn Reson Med 2016;76(5):1574–1581. [DOI] [PubMed] [Google Scholar]
- 70.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 71.Andersson JLR, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 2016;125:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bretthorst GL. Bayesian-Analysis .1. Parameter-Estimation Using Quadrature Nmr Models. J Magn Reson 1990;88(3):533–551. [Google Scholar]
- 73.Jiang X, Xu J, Gore JC. Mapping hepatocyte size in vivo using temporal diffusion spectroscopy MRI. Magn Reson Med 2020;84(5):2671–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baierlein R, Jaynes ET - Papers on Probability, Statistics and Statistical Physics - Rosenkrantz,Rd. Am J Phys 1984;52(2):190–191. [Google Scholar]
- 75.Jaffe AH. Probability theory - The logic of science. Science 2003;301(5638):1329–1330. [Google Scholar]
- 76.Manning HC, Merchant NB, Foutch AC, Virostko JM, Wyatt SK, Shah C, McKinley ET, Xie J, Mutic NJ, Washington MK, LaFleur B, Tantawy MN, Peterson TE, Ansari MS, Baldwin RM, Rothenberg ML, Bornhop DJ, Gore JC, Coffey RJ. Molecular imaging of therapeutic response to epidermal growth factor receptor blockade in colorectal cancer. Clin Cancer Res 2008;14(22):7413–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jhawer M, Goel S, Wilson AJ, Montagna C, Ling YH, Byun DS, Nasser S, Arango D, Shin J, Klampfer L, Augenlicht LH, Perez-Soler R, Mariadason JM. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res 2008;68(6):1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clemons M, Leahy M, Valle J, Jayson G, Ranson M, Howell A. Review of recent trials of chemotherapy for advanced breast cancer: the taxanes. European journal of cancer (Oxford, England: 1990) 1997;33(13):2183–2193. [DOI] [PubMed] [Google Scholar]
- 79.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer 2012;12(4):252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hodi FS, Ballinger M, Lyons B, Soria JC, Nishino M, Tabernero J, Powles T, Smith D, Hoos A, McKenna C, Beyer U, Rhee I, Fine G, Winslow N, Chen DS, Wolchok JD. Immune-Modified Response Evaluation Criteria In Solid Tumors (imRECIST): Refining Guidelines to Assess the Clinical Benefit of Cancer Immunotherapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2018;36(9):850–858. [DOI] [PubMed] [Google Scholar]
- 81.Seymour L, Bogaerts J, Perrone A. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics (vol 18, pg e143, 2017). Lancet Oncol 2019;20(5):E242–E242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haslam A, Prasad V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for and Respond to Checkpoint Inhibitor Immunotherapy Drugs. JAMA network open 2019;2(5):e192535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiou VL, Burotto M. Pseudoprogression and Immune-Related Response in Solid Tumors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2015;33(31):3541–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okazaki T, Honjo T. PD-1 and PD-1 ligands: from discovery to clinical application. International immunology 2007;19(7):813–824. [DOI] [PubMed] [Google Scholar]
- 85.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 86.Kwak JJ, Tirumani SH, Van den Abbeele AD, Koo PJ, Jacene HA. Cancer immunotherapy: imaging assessment of novel treatment response patterns and immune-related adverse events. Radiographics: a review publication of the Radiological Society of North America, Inc; 2015;35(2):424–437. [DOI] [PubMed] [Google Scholar]
- 87.Borcoman E, Nandikolla A, Long G, Goel S, Le Tourneau C. Patterns of Response and Progression to Immunotherapy. American Society of Clinical Oncology educational book American Society of Clinical Oncology Annual Meeting 2018;38:169–178. [DOI] [PubMed] [Google Scholar]
- 88.Teague TK, Munn L, Zygourakis K, McIntyre BW. Analysis of lymphocyte activation and proliferation by video microscopy and digital imaging. Cytometry 1993;14(7):772–782. [DOI] [PubMed] [Google Scholar]
- 89.Pollizzi KN, Waickman AT, Patel CH, Sun IH, Powell JD. Cellular size as a means of tracking mTOR activity and cell fate of CD4+ T cells upon antigen recognition. PloS one 2015;10(4):e0121710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. Journal of immunology 2001;167(12):6869–6876. [DOI] [PubMed] [Google Scholar]
- 91.Tasnim H, Fricke GM, Byrum JR, Sotiris JO, Cannon JL, Moses ME. Quantitative Measurement of Naive T Cell Association With Dendritic Cells, FRCs, and Blood Vessels in Lymph Nodes. Frontiers in immunology 2018;9:1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shashni B, Ariyasu S, Takeda R, Suzuki T, Shiina S, Akimoto K, Maeda T, Aikawa N, Abe R, Osaki T, Itoh N, Aoki S. Size-Based Differentiation of Cancer and Normal Cells by a Particle Size Analyzer Assisted by a Cell-Recognition PC Software. Biological & pharmaceutical bulletin 2018;41(4):487–503. [DOI] [PubMed] [Google Scholar]
- 93.McCracken MN, Tavare R, Witte ON, Wu AM. Advances in PET Detection of the Antitumor T Cell Response. Advances in immunology 2016;131:187–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de la Iglesia FA, Sturgess JM, McGuire EJ, Feuer G. Quantitative microscopic evaluation of the endoplasmic reticulum in developing human liver. The American journal of pathology 1976;82(1):61–70. [PMC free article] [PubMed] [Google Scholar]
- 95.Duarte MI, Andrade HF, Jr., Mariano ON, Corbett CE, Sesso A. Baseline volume data of human liver parenchymal cell. Journal of submicroscopic cytology and pathology 1989;21(2):275–279. [PubMed] [Google Scholar]
- 96.Vekemans K, Braet F. Structural and functional aspects of the liver and liver sinusoidal cells in relation to colon carcinoma metastasis. World journal of gastroenterology 2005;11(33):5095–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karger J, Pfeifer H, Heink W Principles and application of self-diffusion measurements by nuclear magnetic resonance. Advanced Magnetic Resonance 1988;12:1–89. [Google Scholar]
- 98.Pflanzer RRR. Human Physiology (3rd Ed.): Saunders College Publishing; 1996. [Google Scholar]
- 99.Kirschner RH, Rusli M, Martin TE. Characterization of the nuclear envelope, pore complexes, and dense lamina of mouse liver nuclei by high resolution scanning electron microscopy. The Journal of cell biology 1977;72(1):118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia-Gonzalez A, Jacchetti E, Marotta R, Tunesi M, Rodriguez Matas JF, Raimondi MT. The Effect of Cell Morphology on the Permeability of the Nuclear Envelope to Diffusive Factors. Frontiers in physiology 2018;9:925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alberts B BD, Lewis J, Raff M, Roberts K, Watson JD Molecular biology of the cell, 3rd edn: Garland; 1994. [Google Scholar]
- 102.Zelenina M, Brismar H. Osmotic water permeability measurements using confocal laser scanning microscopy. European biophysics journal: EBJ 2000;29(3):165–171. [DOI] [PubMed] [Google Scholar]
- 103.Maul GG, Deaven L. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. The Journal of cell biology 1977;73(3):748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Allen TD CJ, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci 2000;13:1651–1659. [DOI] [PubMed] [Google Scholar]
- 105.Latour L, Mitra P. Time-dependent diffusion coefficient of fluids in porous media as a probe of surface-to-volume ratio. Journal of Magnetic Resonance, Series A 1993;101:342–346. [Google Scholar]
- 106.Burcaw LM, Fieremans E, Novikov DS. Mesoscopic structure of neuronal tracts from time-dependent diffusion. Neuroimage 2015;114:18–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stepišnik J Time-dependent self-diffusion by NMR spin-echo. Physica B: Condensed Matter 1993;183(4):343–350. [Google Scholar]


