Abstract
Natural mitochondrial DNA (mtDNA) mutations enable the inference of clonal relationships among cells. mtDNA can be profiled along with measures of cell state, but has not yet been combined with the massively parallel approaches needed to tackle the complexity of human tissue. Here, we introduce a high-throughput, droplet-based mitochondrial single-cell Assay for Transposase Accessible Chromatin with sequencing (mtscATAC-seq), a method that combines high-confidence mtDNA mutation calling in thousands of single cells with their concomitant high-quality accessible chromatin profile. This enables the inference of mtDNA heteroplasmy, clonal relationships, cell state, and accessible chromatin variation in individual cells. We reveal single-cell variation in heteroplasmy of a pathologic mtDNA variant, which we associate with intra-individual chromatin variability and clonal evolution. We clonally trace thousands of cells from cancers, linking epigenomic variability to subclonal evolution and infer cellular dynamics of differentiating hematopoietic cells in vitro and in vivo. Taken together, our approach enables the study of cellular population dynamics and clonal properties in vivo.
Introduction
Mitochondria play a central role in metabolism and are unique organelles that carry their own genome, often in high copy number, encoding a subset of proteins, tRNAs, and rRNAs essential to their function. Mutations in the mitochondrial genome are associated with a multitude of clinical phenotypes that are estimated to affect ~1 in 4,300 individuals, making them among the most common inherited metabolic disorders1. Critically, the fraction of mitochondrial genomes carrying a specific variant, heteroplasmy, may dictate the degree of disease severity in affected patients1,2,3. Furthermore, the high mutation rate (~2–10x that of nuclear DNA), leads to accumulation of somatic mtDNA mutations that may contribute to aging phenotypes1. While genomic approaches are emerging to quantify heteroplasmy, the majority of sequencing assessments have been based on bulk cell populations, limiting detection of somatic mutations in individual cells4,5.
Recently, we and others have shown that single-cell sequencing approaches can detect heteroplasmic or homoplasmic mutations, which we further leveraged as natural genetic markers in clone and lineage tracing of human cells, while also measuring cell state6,7. Due to the small size of the mitochondrial genome (16.6 kb) and its higher copy number per cell, retrospective inference of cellular relationships by somatic mtDNA mutations is significantly more cost-effective and robust compared to mutation detection in the nuclear genome by single-cell whole-genome sequencing8. Moreover, single-cell RNA- and ATAC-seq (scRNA/ATAC-seq) allow concomitant mtDNA mutation detection along with the transcriptional or accessible chromatin cell state. While this presents a powerful system for clonal/ lineage tracing in humans in vivo, only modest-throughput single-cell genomic assays had sufficient coverage of mitochondrial sequences for reliable mutation detection, whereas the massively parallel methods needed to draw meaningful conclusions on many biological systems had insufficient mitochondrial coverage6.
As recently reported droplet-based scATAC-seq techniques enable the profiling of accessible chromatin in thousands of cells per experiment9,10, we hypothesized that with appropriate modification, they may facilitate the enrichment of transposase-accessible mtDNA6. However, these protocols rely on processing of nuclei, thereby depleting mitochondria and resulting in only ~1% of reads mapping to mtDNA, compared to 20–50% in the original ATAC-seq protocol11,12, a level that is inadequate for single-cell mutation calling and clonal inference.
Here, we establish mtscATAC-seq, a massively parallel protocol for high and uniform single-cell mitochondrial genome coverage that retains high-quality chromatin accessibility data, and combine it with computational methods to identify rare, clonal mtDNA mutations in healthy and diseased cells. We demonstrate the wide applicability of mtscATAC-seq to quantify single-cell mitochondrial genotypes in the context of mitochondrial disease and clonally trace thousands of human cells in vitro and in vivo. Given the multi-omic nature, we envision the broad utility and applicability of mtscATAC-seq to enhance our understanding of mtDNA genotype-phenotype correlations and reconstruct clonal dynamics across diverse areas of human health and disease.
Results
Development and validation of mtscATAC-seq
To develop mtscATAC-seq, we modified the droplet-based scATAC-seq workflow of the widely used 10x Genomics platform to improve mtDNA yield and genome coverage. As most scATAC-seq protocols use nuclei, depleting cytoplasmic mitochondria, we turned to processing whole cells to retain mtDNA. We reasoned that mild lysis or permeabilization of cells would be required for the Tn5 enzyme to integrate adapters into accessible nuclear chromatin and mtDNA. Moreover, as cells contain multiple mitochondria, which may be more readily released upon lysis or permeabilization, we reasoned that fixation should minimize mixing of mtDNA between cells. Finally, we aimed to identify conditions retaining high-quality chromatin accessibility data.
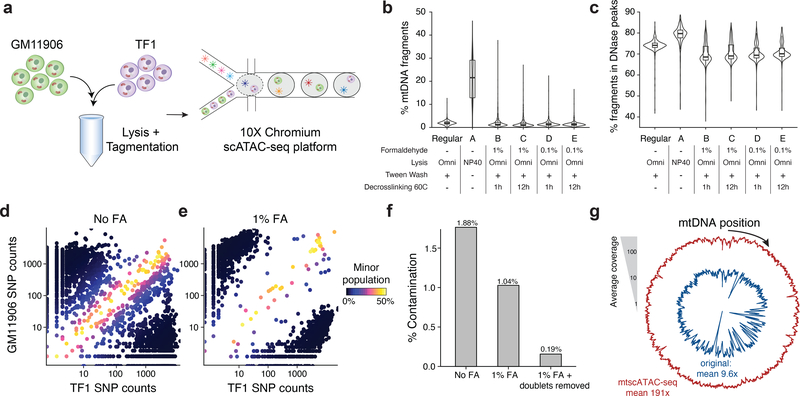
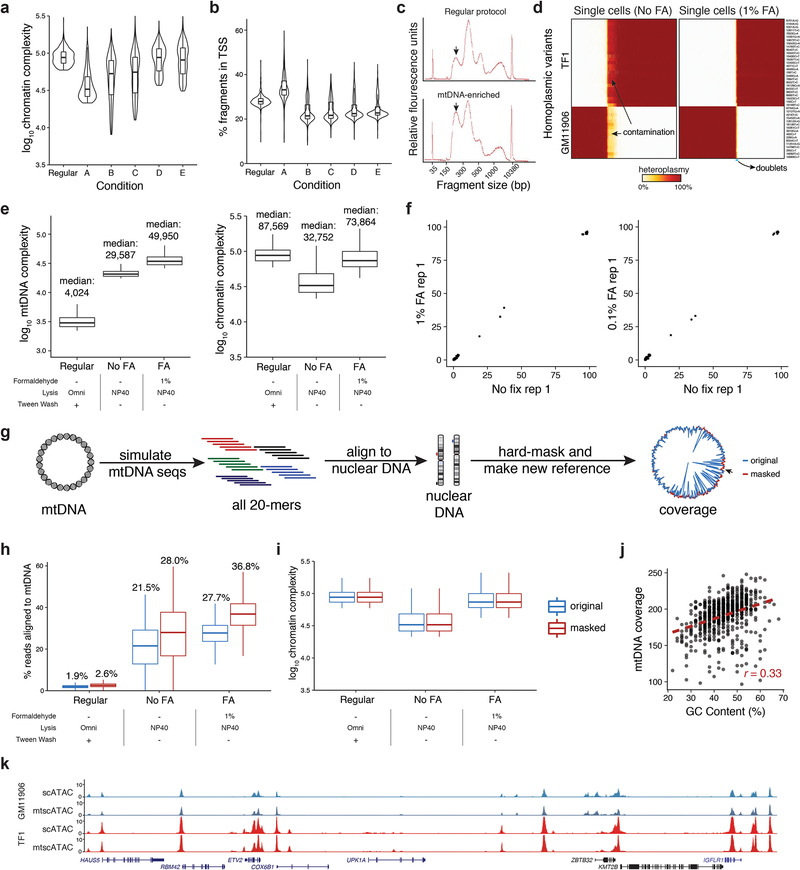
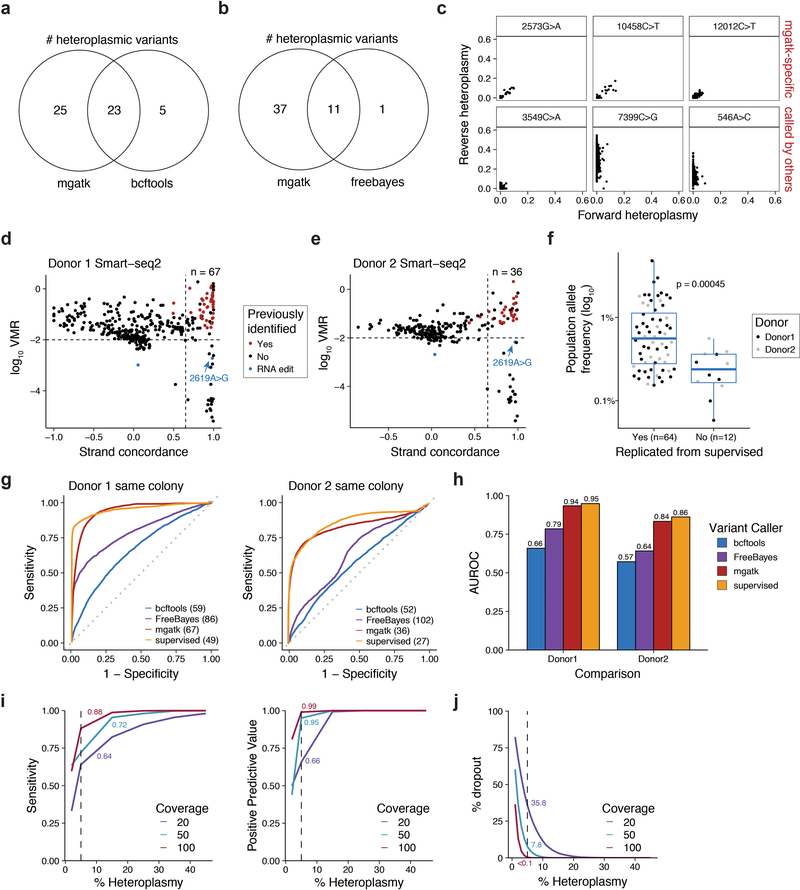
We systematically tested for conditions that satisfy these features in a mixture of two cell lines (GM11906 and TF1; Fig. 1a) by evaluating mtDNA abundance, cross-contamination, and mtDNA and chromatin fragment complexity. Because each cell line harbored private homoplasmic mutations, we sensitively detected mtDNA abundance, cell doublets, and possible mtDNA crosstalk due to cell lysis/ permeabilization and tagmentation that occurs in a pool. Omitting digitonin and Tween-20 in the lysis and wash buffers (“Condition A”) yielded substantially more mtDNA fragments per single-cell (median 21.5%) than the recommended protocol (1.9%; Fig. 1b; Supplementary Table 1; Methods), consistent with earlier observations11,12. These conditions retain high-quality chromatin accessibility data: while per-cell complexity of nuclear fragments slightly decreased (Extended Data Fig. 1a), other metrics associated with scATAC-seq data quality improved (Fig. 1c; Extended Data Fig. 1b). BioAnalyzer traces confirmed an increased ratio of nucleosome free to mononucleosome fragments, consistent with the increased recovery of mtDNA (Extended Data Fig. 1c). Based on 43 high-confidence homoplasmic mtDNA variants private to each cell line, ~8.7% of barcodes carried otherwise cell type-specific homoplasmic variants at intermediate (60%−90%) heteroplasmy, indicating contamination of mtDNA fragments between cells (Fig. 1d; Extended Data Fig. 1d; Methods). Because this contamination may occur due to the release of mitochondria during processing, we added a formaldehyde (FA) fixation step. Indeed, fixation with 0.1 or 1% FA led to a ~3x reduction in mtDNA fragment cross-contamination (Fig. 1e,f; Extended Data Fig. 1d), a 69% increase in mtDNA fragment complexity, and restoration of chromatin library complexity (Extended Data Fig. 1e). After removing cell doublets, the empiric rate of contamination was 0.19% (Fig. 1f; Methods), which is consistent with the order of magnitude for short-read sequencing error13. Importantly, FA treatment did not introduce additional mtDNA mutations (Extended Data Fig. 1f).
Figure 1 -. Optimization of a high-throughput single-cell mitochondrial DNA genotyping platform with concomitant accessible chromatin measurements.
(a) Schematic of cell line mixing experiment between indicated two human hematopoietic cell lines. (b) Distribution of percentage of mtDNA reads per single cell for screened conditions. (c) Distribution of percentage of reads mapping to annotated DNase hypersensitivity peaks (nuclear reads only) per single cell. Each condition in panels (b,c) represents the top 1,000 cells (based on chromatin complexity) from one experiment. (d) Mitochondrial SNP mixing depiction of variants for the TF1 or GM11906 cell line for “Condition A” as in (b). Both axes are log10 transformed. (e) Same as (d) but for “Condition A” with 1% FA treatment. (f) Summary of contamination (percent of reads from minor cell population) for FA treated and untreated comparison. Each bar represents the mean over one experiment. (g) Depiction of overall mitochondrial genome coverage improvements from three biotechnological and computational optimizations (mtscATAC-seq) compared to the original protocol. Boxplots for (b,c): center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range.
Furthermore, we observed regions of lower coverage across the mitochondrial genome, which we determined were due to high homology (and thus low mappability) to nuclear mitochondrial DNA segments (NUMT). We reasoned that due to the high mtDNA copy number and the high Tn5 accessibility of mtDNA, ambiguous fragments could be confidently assigned to the mitochondrial genome with a low false positive rate. Utilizing a compendium of DNase hypersensitivity data14,15 and additional public scATAC-seq data, we estimated that only ~1 accessible fragment from NUMTs would be detected per cell (Methods), such that these are unlikely to be a confounding element in heteroplasmy estimation. We therefore developed a computational approach that effectively assigns reads that map to both the mitochondrial and nuclear genome strictly to mtDNA, facilitating near-uniform coverage without altering chromatin complexity (Fig. 1g; Extended Data Fig. 1g–i). Some residual variation in coverage remained after reference genome masking and was correlated with GC content of the mtDNA genome (r=0.33; Extended Data Fig. 1j), likely reflecting PCR amplification and Tn5 insertion bias16.
Overall, mtscATAC-seq combines fixation, modified lysis, and computational analysis of multi-mapping reads, leading to a ~20-fold increase in mean mtDNA coverage per cell (from 9.6x to 191.0x; Fig. 1g) and in fraction of mtDNA reads (median per cell from 1.9% to 36.8%; Extended Data Fig. 1h) with only modest reduction in chromatin complexity (median per cell from 87,569 to 73,864; Extended Data Fig. 1e) and in reads mapping to pre-annotated DNase hypersensitivity peaks (from 74.1% to 72.3%), retaining cell type-specific accessible chromatin peaks (93.8% of 77,704 peaks; Extended Data Fig. 1k; Methods).
Single-cell features of pathogenic mtDNA mutations
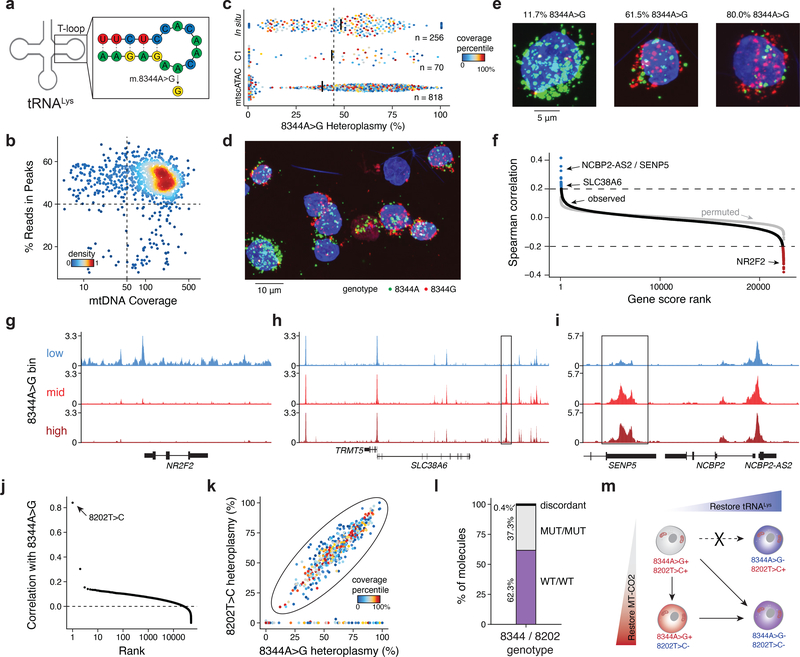
We used mtscATAC-seq to identify pathogenic mtDNA mutations, and gain insights into their impact. The GM11906 lymphoblastoid cells used in the mixing experiment (Fig. 1) were derived from a patient with myclonic epilepsy with red ragged fibers (MERFF), a mitochondrial disorder that in 80–90% of cases is caused by a 8344A>G mutation that alters tRNA function2 (Fig. 2a). Bulk ATAC-seq analyses of these cells estimated a population heteroplasmy of 44% for the 8344A>G allele, consistent with previous reports17. We retained 818 high-quality data GM11906 cells with at least 50x single-cell mtDNA coverage and 40% reads in peaks (Fig. 2b). Interestingly, we observed a broad range of heteroplasmy (0% to 100%) for the 8344A>G allele, with a median of 38%, consistent with the bulk ATAC-seq data (Fig. 2c) and previous family studies of this mutation18. We independently replicated the distribution of heteroplasmy levels using the Fluidigm scATAC-seq platform19 and in situ genotyping20 (Fig. 2c–e; Extended Data Fig. 2a; Supplementary Table 2).
Figure 2 -. Pathogenic mtDNA variability and clonal evolution in cells derived from a patient with MERRF.
(a) Schematic of the mitochondrial lysine tRNA secondary structure with sequence and the pathogenic single nucleotide variant (8344A>G). (b) Quality control filtering for GM11906 single cells based on mean mtDNA genome coverage and percentage of nuclear reads in chromatin accessibility peaks. (c) Quantification of 8344A>G heteroplasmy variability in single GM11906 cells across three technologies. Numbers (n) of cells plotted are shown. Color represents the within-assay coverage percentile. Black bars indicate the median heteroplasmy per technology; the dotted line presents the mean heteroplasmy as determined for bulk ATAC-seq. (d) Field of view for in situ genotyped GM11906 cells, highlighting (e) single cells with low, medium, and high heteroplasmy as indicated for the pathogenic allele. Representative image selected from one of seven fields of view for one experiment. (f) Per-gene score Spearman correlations with the 8344A>G allele heteroplasmy. The grey dots show values for a permutation. Pseudo bulk chromatin accessibility track plots are shown for the (g) NR2F2, (h) TRMT5, and (i) SENP5/ NCBP2-AS2 loci. Pseudo-bulk groups were binned based on 0–10% (low), 10–60% (mid), and 60–100% (high) 8344A>G heteroplasmy. (j) Per-mutation heteroplasmy correlation with 8344A>G allele. The 8202T>C mutation is highlighted as the most correlated mutation. (k) Single-cell heteroplasmy for two indicated mutations. The circled population represents a double-positive population for both mutations. (l) Abundances of each variant on single molecule sequencing reads in the double positive population. (m) Schematic of the co-evolution of two subclonal populations marked by indicated mutations detected based on single-cell genotyping data. Putative cell transitions are indicated with solid arrows that may be a result of selective pressure of the pathogenic variant or genetic drift.
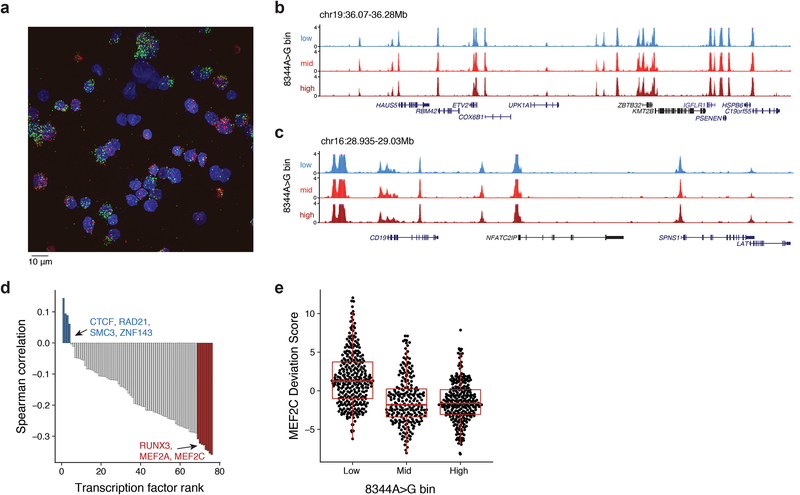
Analysis of matched chromatin profiles highlighted specific loci and transcription factor (TF) activities that are associated with different levels of the 8344A>G allele. First, promoter accessibility scores9,10 of 32 and 94 genes were positively or negatively correlated, respectively, with single-cell 8344A>G heteroplasmy, corresponding to a <1% false discovery rate (FDR) (Fig. 2f; Methods). Binning cells into high (>60%; n=273), intermediate (10–60%; n=228), and low (<10%; n=313) heteroplasmy for the pathogenic allele highlighted distinct chromatin features near the NR2F2, TRMT5, and SENP5/ NCBP2-AS2 loci (Fig. 2g–i). Notably, nearby genes have been broadly linked to mitochondria biology21–24. The accessibility profiles at other loci were virtually indistinguishable (Extended Data Fig. 2b,c), suggesting that the observed variations (Fig. 2g–i) may be a consequence of disease allele heteroplasmy. Furthermore, we identified TFs whose activity may be associated with the mutation by scoring TF binding sites (from ChIP-seq data; Methods). In particular, MEF2A and MEF2C were strongly anti-correlated with pathogenic heteroplasmy (Extended Data Fig. 2d,e). Notably, the transcription factor MEF2 is a target of mitochondrial apoptotic caspases, supporting a model where pathogenic allele heteroplasmy may regulate nuclear factor activity25. These analyses demonstrate the potential to study the altered cellular circuits resulting from pathogenic mtDNA variants in a heteroplasmy-dependent manner.
Notably, a second mutation, 8202T>C (bulk heteroplasmy 34%) was the most correlated mutation with the 8344A>G variant (Fig. 2j). Using MITOMAP26, we annotated the non-synonymous variant (phenylalanine to serine) as a “probably damaging” mutation in the cytochrome C oxidase II (MT-CO2) gene. 456 of 818 GM11906 cells were positive for both mutations (>5% heteroplasmy), whereas the remaining cells showed 0% heteroplasmy for either both mutations or 8202T>C alone, but not 8344A>G alone (Fig. 2k). Of the 5,230 reads that covered both variants, 99.6% exclusively contained either both mutated or wildtype alleles (Fig. 2l). The co-occurrence of both mutations on the same haplotype and the presence of 8344A>G+/8202T>C- cells suggests the evolution of at least two subclonal populations, each spanning the complete spectrum from low to very high 8344A>G heteroplasmy (Fig. 2k,m), demonstrating how mtscATAC-seq can enhance our understanding of clonal dynamics in the context of mitochondrial disease.
Inference of mutations for clonal lineage tracing
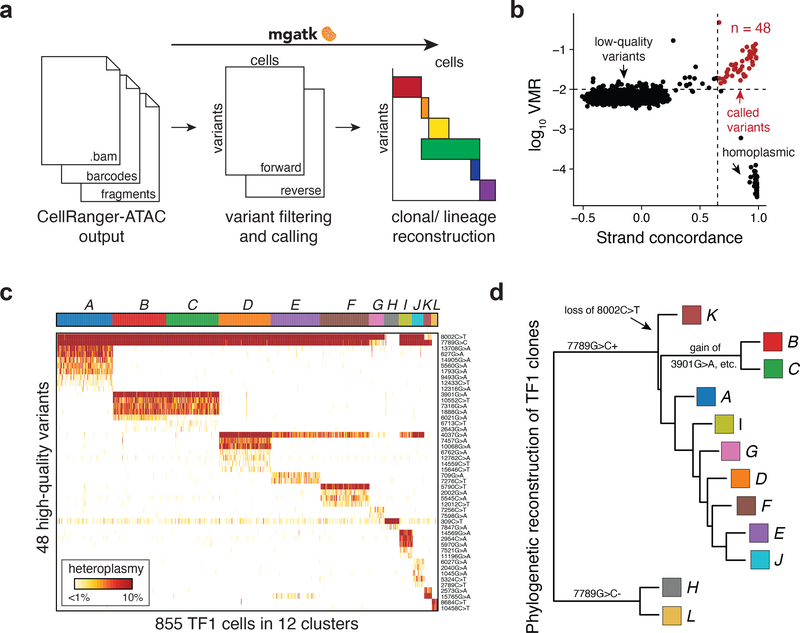
To facilitate clonal tracing of human cells based on reliable mtDNA variation, we developed the Mitochondrial Genome Analysis Toolkit (mgatk; Fig. 3a; Methods), a computational pipeline to identify clonal substructure in complex populations profiled using mtscATAC-seq. Here, we define clonal mutations as those with similar heteroplasmy that may genetically mark an individual cell and its immediate descendants to distinguish it from other more distantly related cells. Recent variant callers developed for single-cell genotyping were designed to separate amplicon error from true mutations27 or account for allelic dropout28, neither of which predominantly confound heteroplasmy estimates from mtscATAC-seq (Methods). Instead, mgatk focuses specifically on clonal mtDNA variant calling in single cells, by leveraging the deep per-cell coverage from mtscATAC-seq. Specifically, mgatk identifies high-confidence clonal mutations by aggregating signal across cells, leveraging between-cell variability (per mutation variance mean ratio; VMR) and strand bias (Pearson correlation of counts per strand; Fig. 3a; Methods). Thus, rather than calling variants in individual cells, mgatk leverages the high-throughput nature of our data to identify between-cell properties to distinguish signal from noise. The resulting mutations are then used as a feature set for downstream analyses, such as the inference of clonal families.
Figure 3 -. Identification of high-confidence variants and subclonal structure in TF1 cells.
(a) Schematic of mgatk workflow. (b) Identification of high-confidence variants from high strand concordance in paired-end sequencing data and high variance mean ratio (VMR). (c) Unsupervised clustering of TF1 cells using 48 high-quality variants into 12 population clusters. Each column is a cell. Rows show detected mutation. Heatmap color indicates percent heteroplasmy. (d) Phylogenetic reconstruction of clonal TF1 groups. The tree was constructed using neighbor joining; each tip represents a cell cluster from (c).
We validated mgatk by identifying anticipated clonal substructure in the 855 TF1 cells (>50x mitochondrial genome coverage) profiled in the mixture experiment (Fig. 1). Because these cells were expanded from 30 individually sorted TF1 cells, we expected to observe multiple sub-clones6. We identified 48 reliable mtDNA variants by bivariate filtering of variants with a relatively high VMR and concordant heteroplasmy from both strands (Fig. 3b; Methods). Using these 48 variants as features, we determined 12 clonal cell subsets using a shared nearest neighbor clustering approach (Fig. 3c; Methods). Variants called by other approaches lacked sensitivity or had substantial strand bias compared to mgatk (Extended Data Fig. 3a–c; Methods) The 48 high-confidence variants enabled us to reconstruct a putative phylogenetic tree for the identified TF1 subclones (Fig. 3d).
Though mgatk was optimized for mtscATAC-seq data, its unsupervised application performed comparably well to our previous supervised identification of multiple hematopoietic colony specific variants from 935 cells profiled by SMART-seq26 (Extended Data Fig. 3d–h; Methods). Furthermore, variants identified by mgatk substantially outperformed other unsupervised approaches in discerning cells that shared a clonal origin (Methods). However, as SMART-seq2 and other scRNA-seq methods detect a substantial number of false-positive variants, corroboration by mtDNA sequencing is highly recommended6; conversely, mtscATAC-seq captures DNA directly, minimizing potential artifacts. Simulations with empirically-derived parameters indicated that mtscATAC-seq has high sensitivity, high positive predictive value (PPV), and low dropout, particularly for sub-clonal variants of at least 5% heteroplasmy with at least ~50x coverage per cell (Extended Data Fig. 3i,j; Methods). Overall, the combination of mtscATAC-seq and mgatk provide a robust and high-throughput means to identify high-quality mtDNA variants associated with single cell states.
Clonal heterogeneity in human malignancies
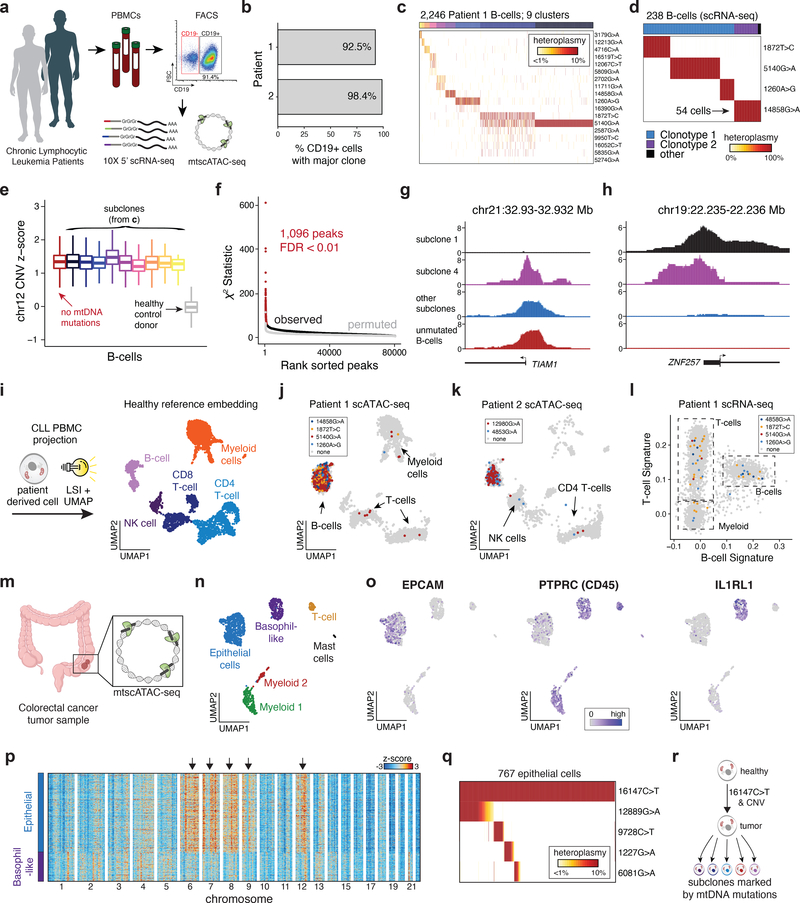
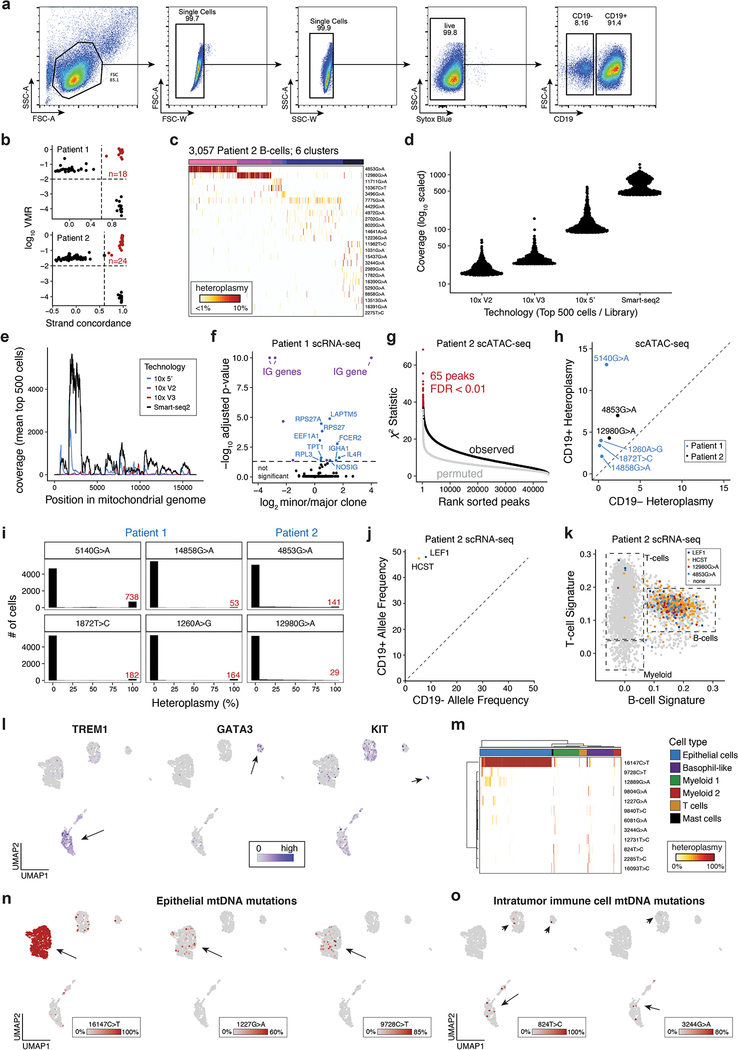
To evaluate mtscATAC-seq in vivo, we studied cells from patients with presumed clonal malignancies. We first profiled peripheral blood mononuclear cells (PBMCs) from two patients with chronic lymphocytic leukemia (CLL), which is conventionally characterized as a monoclonal B-cell malignancy (Fig. 4a; Extended Data Fig. 4a). Single-cell B-cell receptor sequencing by 5’ scRNA-seq confirmed a predominantly monoclonal population of leukemic cells in both patients (Fig. 4b; Methods). Based on our previous work, we hypothesized that somatic mtDNA mutations may arise during tumorigenesis, which mark and enable tracking of genetic subclones to aid in resolving intra-tumor heterogeneity6. We collected 23,467 high-quality mtscATAC-seq profiles (mean 55.5x mtDNA coverage; 11,423 unique nuclear fragments per cell and 70.8% in peaks), and applied mgatk to CD19+ leukemic cells to reveal 43 mutations and 15 putative subclones across the two patients (Fig. 4c; Extended Data Fig. 4b,c). This marked genetic diversity in a perceived highly clonal malignancy reinforces the effectiveness of our approach to identify rare subclonal structure, including a cluster marked by the 12067C>T mutation present in 0.4% of the leukemic population (Fig. 4c).
Figure 4 -. Clonal and functional heterogeneity in human malignancies resolved by somatic mtDNA mutations.
(a) Schematic of experimental design. Populations of peripheral blood mononuclear cells (PBMCs) from two CLL patients were separated by FACS or magnetic bead enrichment and profiled with mtscATAC-seq and 10× 5’ scRNA-seq. (b) Fraction of CD19+ cells with major B cell receptor (BCR) clonotype as determined from V(D)J receptor sequencing. (c) Inference of subclonal structure from somatic mtDNA mutations for patient 1. Cells (columns) are clustered based on mitochondrial genotypes (rows). Colors at the top of the heatmap represent clusters or putative subclones. Color bar, heteroplasmy (% allele frequency). (d) Clonotype receptors (columns) associated with somatic mtDNA mutations (rows) from patient 1. Colors at the top of the heatmap represent BCR clonotypes. Color bar, heteroplasmy (% allele frequency). (e) Estimated copy number of chromosome 12 across putative subclones for patient 1. Patient derived cells showed elevated DNA read counts of chromosome 12, consistent with a trisomy for this chromosome (Methods). Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. (f) Sub-clone associations with accessible chromatin. Red dots denote peaks associated at a false-discovery rate of <0.01. (g,h) Examples of subclone-associated differential accessibility peaks near the (g) TIAM1 and (h) ZNF257 promoters. (i) Schematic of scATAC projection framework using latent semantic indexing (LSI) and UMAP. A healthy PBMC reference embedding with indicated cell types is shown. (j) Projection of cells collected from Patient 1 and (k) Patient 2. Colors indicate cells positive for indicated somatic mtDNA mutations. Non-B-cells are highlighted. (l) Gene signature plots of PBMCs from single-cell RNA-seq for Patient 1 corroborating mtDNA mutations in non-B-cells. (m) Schematic showing mtscATAC-seq profiling of a colorectal cancer resection specimen. (n) Two dimensional embedding of all quality controlled tumor derived cells using UMAP showing the distribution of cells based on Louvain clustering and annotation based on marker gene scores as exemplified in panel (o) and Extended Data Fig. 4l. (o) Projection of marker gene scores for indicated genes EPCAM, PTPRC and IL1RL1. Color bar, gene score activity. (p) Inferred CNV profiles for indicated cell types (x axis) and chromosomes. Arrows indicate relative increase of copy numbers in the epithelial tumor cells. Cells from the basophil-like population are shown as a control group of cells. Color bar, z-score transformed fragment abundance. (q) Inference of subclonal structure from somatic mtDNA mutations in colorectal cancer. Epithelial cells (columns) are clustered based on mitochondrial genotypes (rows). Color bar, heteroplasmy (% allele frequency). (r) Putative model of clonal evolution of the profiled colorectal cancer specimen as suggested based on integrated analysis of nuclear CNV and somatic mtDNA mutation profiles.
Next, we related the mtDNA clones with both their chromatin profiles and receptor clonotypes, leveraging the mtDNA coverage from 5’ scRNA-seq (Extended Data Fig. 4d,e) to link to variants identified from mtscATAC-seq. Interestingly, leukemic cells with the 14858G>A mtDNA mutation did not carry the predominant BCR clonotype, presenting a distinct sub-clonal population showing various differentially-expressed genes (Fig. 4b,d; Extended Data Fig. 4f; Methods). Moreover, all cells in Patient 1 were positive for trisomy 12 (Methods), a common cytogenetic abnormality in CLL29, suggesting that the copy number alteration preceded the somatic mtDNA diversity detected (Fig. 4e). Performing a per-peak association with our putative subclones, we observed hundreds of loci associated with subclonal structure in these tumors (Fig. 4f; Extended Data Fig. 4g), including promoters of the ZNF257 and TIAM1 genes, the latter of which had been associated with chemoresistance in CLL and colorectal cancer30,31 (Fig. 4g,h). These results provide a broad basis for how mtscATAC-seq can resolve epigenetic differences in malignant sub-populations at single-cell resolution.
Among the identified variants from mgatk, six mutations (four in patients 1, two in patient 2) attained homoplasmy in a subset of cells and were markedly enriched in the CD19+ population (Extended Data Fig. 4h,i). Notably, the same variants were also identified in T lymphocytes, natural killer (NK), and myeloid cells (Fig. 4i–l; Extended Data Fig. 4j,k). These results point to the possible involvement of an early progenitor cell with residual multi-lineage capacity in the pathogenesis of CLL, as suggested by previous reports32–34. These results could further be corroborated in the scRNA-seq data of patient 2 upon integration of calling somatic mutations in nuclear genes (i.e. chr4:109,084,804A>C “LEF1” and chr19:36,394,730G>A “HCST”; identified by exome sequencing) (Extended Data Fig. 4j,k).
Next, we profiled a human colorectal cancer resection (Fig. 4m). Using variance in chromatin accessibility and marker gene scores, we identified six major cell populations, including tumor-derived epithelial cells and distinct immune cell populations (Fig. 4n,o; Extended Data Fig. 4l). Using integrated calling of somatic chromosomal copy number variants (CNV) (Fig. 4p; Methods) and mtDNA mutations (Fig. 4q), we suggest a model where copy number gains on chromosomes 6, 7, 8, 9, and 12 and a homoplasmic 16147C>T variant are shared across the dominant malignant cell population (Fig. 4p–r). Multiple additional mtDNA mutations then further resolve subclonal structure within the malignant cells, as well as in non-malignant immune cells (Extended Data Fig. 4m–o). Taken together, our results highlight the utility of mtscATAC-seq/ mgatk platform to enable the retrospective inference of cellular population dynamics in malignancies6.
Linking cell state to fate in hematopoietic differentiation
The multi-modal output of mtscATAC-seq simultaneously informs about cell state and clonal relationships, allowing us to study complex physiologic processes, where genetic barcoding is not possible. We focused on hematopoiesis, a process thought to be sustained by 10,000–100,000s distinct hematopoietic stem/progenitor cells (HSPCs) under steady state35,36, potentially requiring the sampling of large cell numbers to capture the full spectrum of clonal diversity.
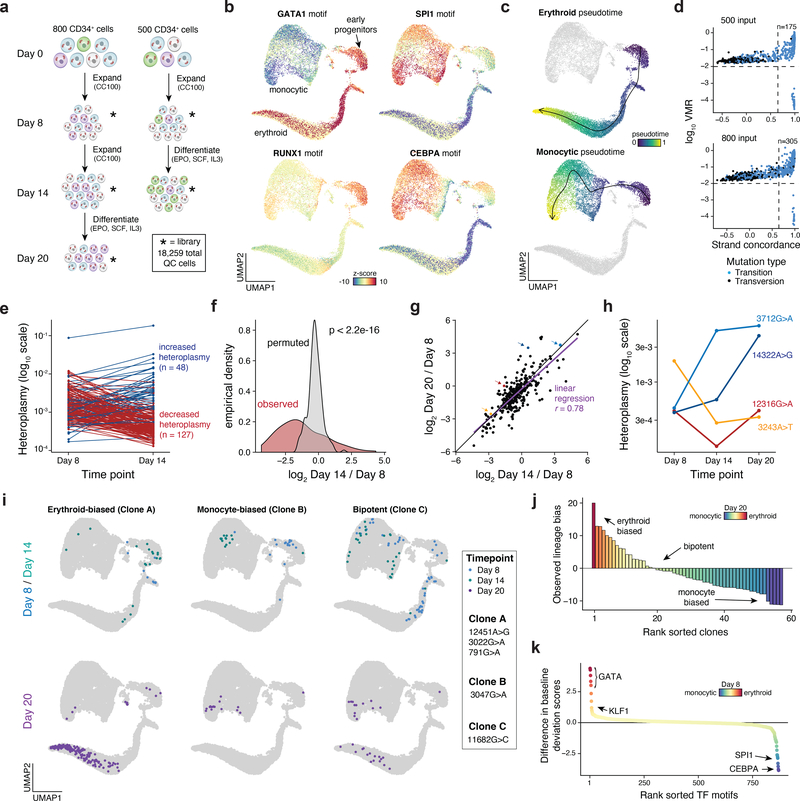
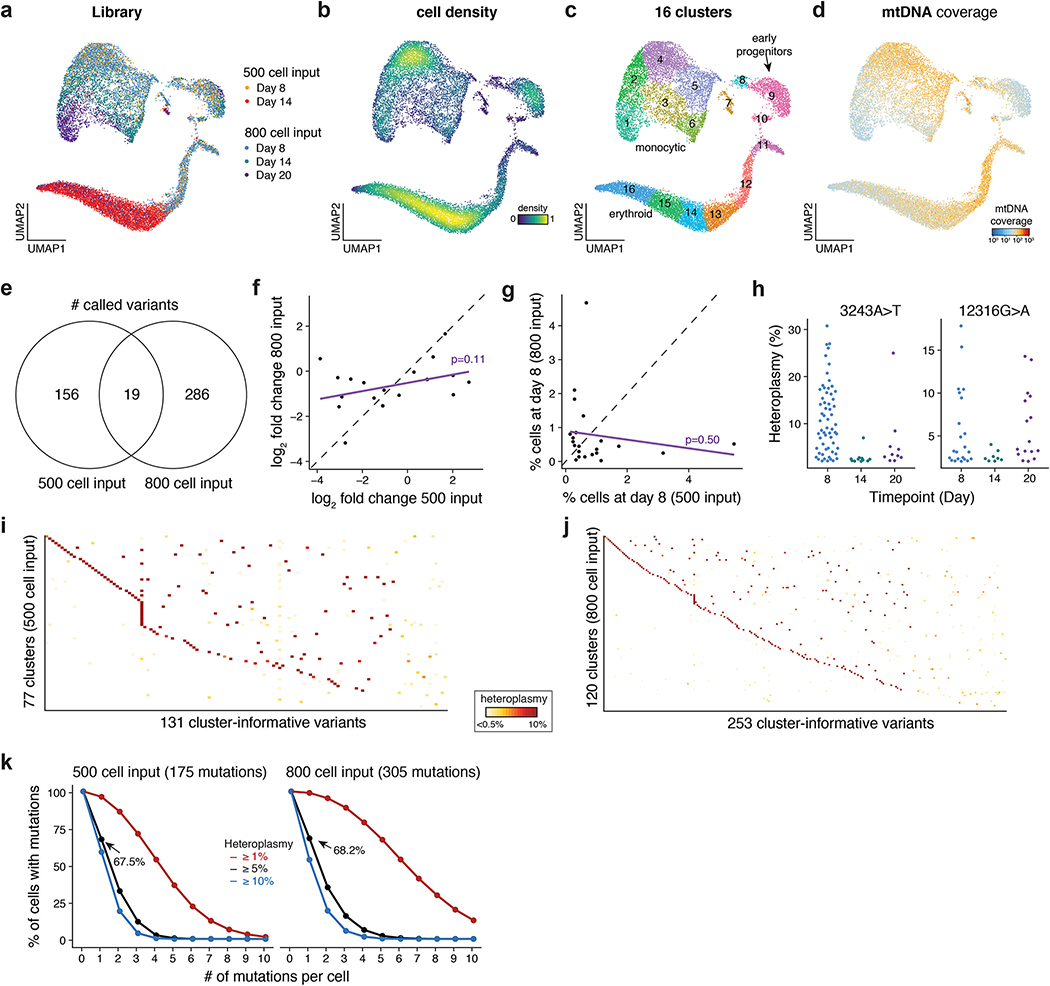
We first benchmarked mtscATAC-seq in an in vitro model of human hematopoiesis, where clonal contributions could be anticipated. We cultured ~500 or ~800 CD34+ HSPCs in progenitor expansion media, before induction of monocytic or erythroid differentiation. Over the course of 20 days we profiled cells from two independent cultures (two and three timepoints for the 500 and 800 cell input, respectively), yielding 18,259 high quality mtscATAC-seq cell profiles (Fig. 5a; Methods), with a mean of 24,944 unique nuclear fragments per cell, 49.1% of which were in accessibility peaks, and a mean 74.8x mtDNA coverage per cell. Dimensionality reduction37, TF motif scoring38, and inference of pseudotime trajectories highlighted differentiation continuums from HSPCs to either the erythroid or monocytic fates (Fig. 5b,c; Extended Data Fig. 5a–d; Methods). These findings verify that mtscATAC-seq can reconstruct cell state transitions comparable to previous scATAC-seq studies9,10,39–41.
Figure 5 -. Clonal lineage tracing across accessible chromatin landscapes and time in an in vitro model of hematopoiesis.
(a) Schematic of experimental design. Approximately 800 or 500 CD34+ HSPCs were derived from the same donor, expanded, and differentiated in two independent cultures over the course of 20 days as shown. Stars represent timepoints/ populations of cells that were profiled via mtscATAC-seq. (b) Two dimensional embedding of all quality controlled cells using UMAP. Single-cell TF motif deviation scores for indicated factors are shown in color for all cells. (c) Pseudotime trajectories for monocytic and erythroid trajectories are depicted. (d) Identification of high confidence variants derived from both cultures. The number of variants passing both thresholds (dotted lines) is indicated. (e) Changes in heteroplasmy for 175 variants identified from the 500 input culture from day 8 to day 14. Values represent the mean over all single-cells in the library. (f) Increased variability in heteroplasmy shifts for the 500 cell input culture. P-value is reported from a two-sided Kolmogorov–Smirnov test comparing the observed and permuted distributions log fold-changes of heteroplasmy. (g) Comparison of heteroplasmy shifts for the 800 cell input culture. Linear regression indicates that most of the variability in heteroplasmy changes at the late time point (day 20, y-axis) can be explained by the intermediate time point (day 14, x-axis). Colored dots are mutations highlighted in the next panel. (h) Heteroplasmy trajectories for four selected mutations from (g). Values represent the mean over all single-cells in the library for the indicated time point. (i) Three examples of clonal populations marked by indicated mutations identified in the 800 cell input culture that result in erythroid, monocytic, or bipotent lineage outcomes. (j) Systematic identification of clonal outcomes using the late time point (day 20). y-axis depicts the difference between z-score in erythroid and monocytic bias of a single clone. (k) Differences in transcription factor motif activity comparing erythroid-biased and monocytic biased clones at the earliest sampled time point (day 8).
Mgatk identified 175 and 305 high-confidence, heteroplasmic variants in the 500 cell and 800 cell input cultures, respectively, which were enriched for transitions (96.0 and 94.8%; Fig. 5d; Methods), consistent with previous findings6. In both cultures, there were substantial shifts in heteroplasmy, including significantly wider distribution of allele frequency fold changes than expected if the HSPCs underwent differentiation in a homogeneous manner (Fig. 5e,f; Kolmogorov–Smirnov p<2.2×10−16). Along with our sequential sampling experiment, the heteroplasmy change in the 800-cell input culture from the second sampling largely explained the third (Fig. 5g), suggesting that clonal contributions largely did not diverge further during continued differentiation. However, our sequential clonal tracing captures complexities in these temporal cell state transitions, including putative clone proliferation dynamics, such as cells that expanded earlier (3712G>A) or later (14322A>G) (Fig. 5h). Analysis of 19 shared mutations between the two cultures suggested that proliferation capacity was independent of the specific mutations as their heteroplasmy fold-changes were not correlated between the two experiments (Extended Data Fig. 5e–g).
Interestingly, we observed six “confirmed” pathogenic mutations between the two cultures, including 12316G>A and 3243A>T (Fig. 5h), both of which alter mitochondrial tRNA function26, possibly explaining their observed decreased population frequencies over the course of the culture. Each of these six mutations occurs at a maximum of 0.1% allele frequency in the bulk population, but exceed 30% heteroplasmy in some individual cells (Extended Data Fig. 5h).
Combining the mtDNA mutation and clonal status with the cells’ chromatin profiles, we inferred properties and possible fates of HSPCs, distinguishing bi-potent progenitors from those biased in favor of an erythroid vs. monocytic fate. We partitioned the cells from the two cultures to 197 clonal groups by mtDNA mutations with most cells carrying at least one high-quality somatic mtDNA mutation (Extended Data Fig. 5i–k; Methods). We then examined the states of the cells in each clone, to identify HSPCs from day 8 in clones with biased (enriched) membership of monocytic or erythroid cells on day 20 (Fig. 5i). Specifically, of the 57 clonal populations with at least 10 cells at day 20 we observed in the 800 input culture, 10 were erythroid-biased and 21 were monocytic-biased (z-score >5; Fig. 5j; Methods). Next, we examined the chromatin features of HSPCs in biased clones and in bi-potent ones. Indeed, well characterized erythroid (GATA1 and KLF1) or monocytic TF motifs (SPI1 and CEBPA) were more accessible in day 8 cell clones that preferentially gave rise to daughter cells of erythroid or monocytic lineage by day 20, respectively (Fig. 5k; Methods). However, when restricting this analysis towards day 8 cells within the early progenitor cluster (cluster 9; Extended Data Fig. 5c), this association diminishes, though our power to detect such lineage biasing features (if present and causal for such observations) may be limited given the number of cells profiled at this stage (n=257).
Clonal tracing in human hematopoiesis in vivo
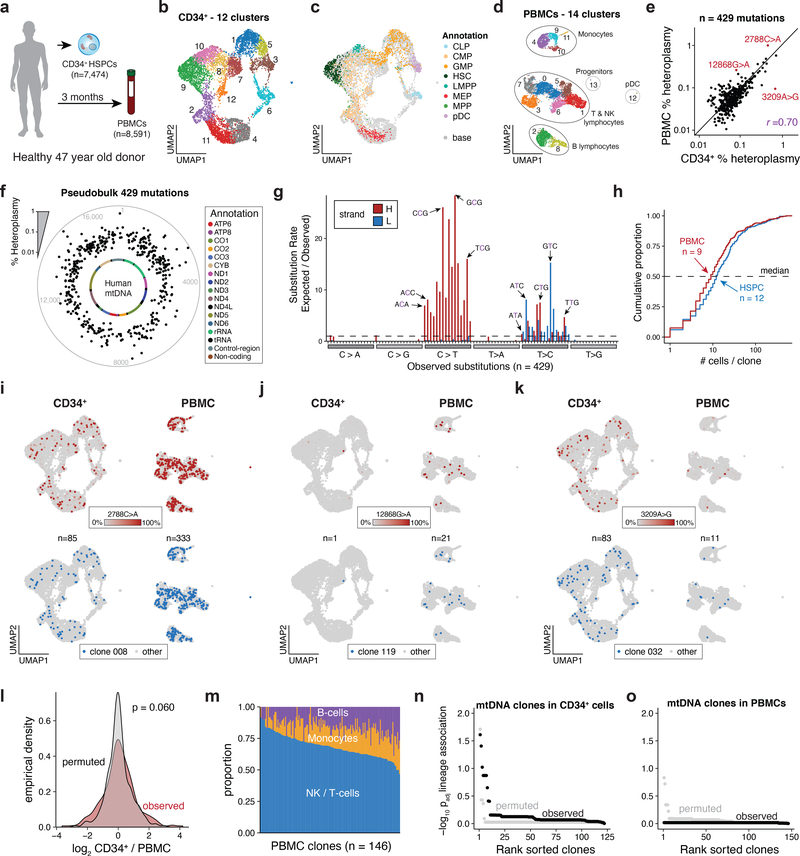
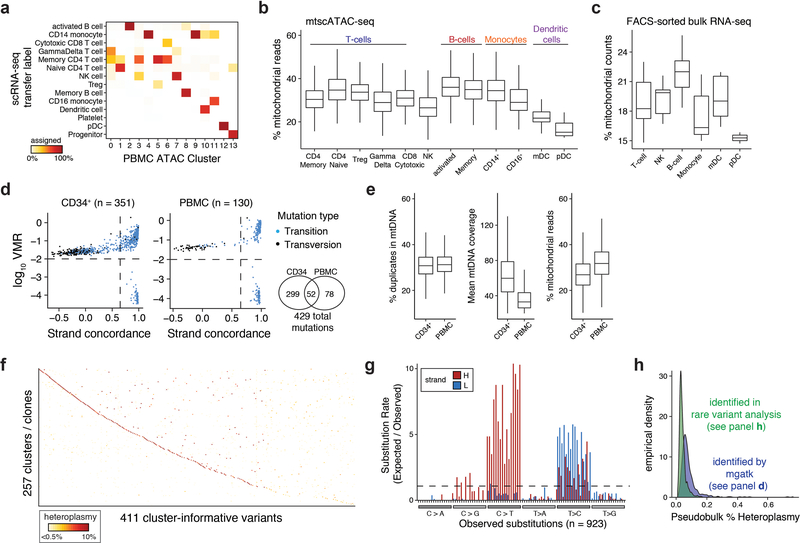
Finally, we utilized mtscATAC-seq to gain insights into the clonal architecture of hematopoiesis in vivo35,36. We profiled bone marrow-derived CD34+ HSPCs (n=7,474 quality-controlled cells) along with PBMCs (n=8,591) that were obtained after a three months interval from a 47 year old healthy donor (Fig. 6a). Using reference scATAC-seq39 and scRNA-seq data, we annotated cell states, revealing cellular heterogeneity and distinct hematopoietic lineages (Fig. 6b–d; Extended Data Fig. 6a). Our high-quality chromatin accessibility (mean of 23,551 and 9,874 unique nuclear fragments for CD34+ and PBMCs, respectively) and mtDNA data enabled detailed analysis of cell states, including the inference of relatively low mtDNA copy number in plasmacytoid dendritic cells (pDCs), further corroborated by analysis of bulk RNA-seq data42, and consistent with a previous report of mitophagy in DCs43 (Extended Data Fig. 6b,c).
Figure 6 -. Cellular population dynamics in native hematopoiesis in vivo resolved by mtDNA mutations.
(a) Schematic of experimental design. CD34+ HSPCs and PBMCs were derived from the same healthy donor at 0 and 3 months, respectively, and processed using mtscATAC-seq. (b,c) Two dimensional embedding of all quality controlled CD34+ cells using (b) UMAP colored by Louvain clustering or (c) by cell cluster annotation using previously published reference data. CLP = common lymphoid progenitor, CMP = common myeloid progenitor, GMP = granulocyte monocyte progenitor, HSC = hematopoietic stem cell, LMPP = lymphoid-primed multipotent progenitors, MEP = megakaryocyte-erythrocyte progenitors, MPP = multipotent progenitor, pDC = plasmacytoid dendritic cells. (d) Two dimensional embedding of all quality controlled PBMCs using UMAP colored by the distribution of cells based on Louvain clustering and annotation using scRNA-seq data derived label transfer (Extended Data Fig. 6a). (e) % heteroplasmy (log10 scale) of mgatk nominated variants and respective allele frequencies in pseudobulk CD34+ HSPC (x axis) and PBMC populations (y axis). Indicated select variants are further highlighted in panels (i-k). (f) Distribution of mgatk nominated mutations along the mitochondrial genome averaged over both populations (pseudobulk). Inner circle, mitochondrial genome; dots, % heteroplasmy of each mutation; outer gray circle, genome coordinates; annotation shows color coded mitochondrial genes. (g) Substitution rate (observed over expected) of mgatk identified heteroplasmic mutations (y-axis) in each class of mononucleotide and trinucleotide change resolved by the heavy (H) and light (L) strand of the mitochondrial genome. (h) Empirical cumulative distribution plots of the number of cells per clone for both HSPCs and PBMCs. The median number of cells per clone n is shown for each of the two populations. (i-k) Specific mutations (top) and cell clones to which they belong (bottom) marking CD34+ cells and PBMCs chromatin accessibility profiles (as in b-d). Number of cells n assigned to the respective clonal groups are shown for the CD34+ HSPC and PBMC cell populations. Color bar, heteroplasmy (% allele frequency). (l) Distribution of heteroplasmy shifts in the CD34+ HSPC over the PBMC cell population. P-value: two-sided Kolmogorov–Smirnov test comparing the observed and permuted distributions log2 fold-changes of clonal abundances. (m) Relative proportion of cells from indicated hematopoietic lineages (y axis) in each clone (x axis) identified in PBMCs. (n,o) Summary statistics of (n) CD34+ HSPC and (o) PBMC association between lineage (cell state) and clone. Adjusted p-values (of lineage association) represent the Benjamini-Hochberg adjusted Chi-squared goodness of fit per clone with at least 10 cells.
Within the HSPCs and PBMCs, mgatk called 351 and 130 high-confidence variants, respectively (HSPCs had greater mtDNA coverage than the PBMCs), 52 of which were shared among both compartments (Extended Data Fig. 6d,e). Although the 429 unique mutations were only present at low frequencies (<1%) in pseudobulk populations (Fig. 6e,f), allele frequencies in individual cells showed considerable homoplasmy (Extended Data Fig. 6f), and the mutational signatures of identified mtDNA variants were consistent with previous reports (Fig. 6g)6,44.
A community detection algorithm partitioned cells into 257 clonal groups with a median 9 and 12 cells per clone in the PBMC and HSPC compartments, respectively, noting that 92% of clones contained less than 1% of assayed cells (Fig. 6h; Extended Data Fig. 6g; Methods). Focusing on a select set of highly heteroplasmic and homoplasmic variants, we observed clonal patterns that may reflect physiologic waves of hematopoietic activity, both in terms of expansion in the HSPC compartment and in terms of contribution to the PMBC compartment (Fig. 6e,i–k). For instance, clone 008 (marked by 2788C>A) and clone 119 (12868G>A) are present in distinctive proportions in HSPCs with variable output 3 months later as reflected in their different abundance in the PBMC compartment (Fig. 6i,j). By contrast, clone 032 (3209A>G) had similar prevalence in HSPCs as clone 008 but reduced output in the following months based on decreased detection in PBMCs (Fig. 6k). Overall, our results suggest relative stable clonal output over the assessed time interval, with observed shifts in heteroplasmy in the HSPC and PBMC populations, either reflecting undersampling (Fig. 6l) or clonal succession45. These findings clearly support stable propagation of mutations present in stem and progenitor cells to the peripheral blood (Fig. 6e,i–k), and indicate that steady state hematopoiesis is fueled by a large pool of HSPCs where the contributions of individual clones to healthy blood cell production is low (<1%), consistent with previous reports35,36.
To further understand the clonal contributions to the major lineages of peripheral blood, we examined the association between clonal output and inferred cell state from the mtscATAC-seq data. While we observed variability in composition of inferred clones (Fig. 6m), such a distribution is statistically consistent with random sub-sampling of cell states (Fig. 6n,o). These results stand in contrast to the observations of biased clonal output (Fig. 5), which may reflect conditions in an in vitro system, where fate decisions may be restricted by limited cytokine availability. Moreover, these observations may further be confounded by distinct longevity of different cell types or the averaging of rare clones not detectable from the current sample size. In this regard, additional analysis designed to discover high-confidence mtDNA mutations present in no more than three HSPCs recovered an additional 923 distinct mtDNA mutations (Extended Data Fig. 6h; Methods). Though rare, these mutations showed concordant mutational spectra and significantly lower frequency in the pseudobulk population (Extended Data Fig. 6h,i) and may mark quiescent or low activity clones.
Taken together, our in vivo analysis demonstrates the potential, along with some of the challenges, to dissect complex physiologic systems. Our results highlight the ability of our framework to facilitate systematic studies aimed at investigating clonal population structures at single-cell resolution in vivo, which were previously limited to model organisms or gene therapy trials46–50.
Discussion
Here, we develop a high-throughput platform for measuring mtDNA mutation heteroplasmy along with accessible chromatin states in thousands of single-cells. We verify data standards (Fig. 1), chart the cis- and trans- effects of pathogenic mutations (Fig. 2), and infer subclonal population structure (Fig. 3), all from a single experiment. By leveraging somatic mtDNA variation in more complex settings, our results further indicate the potential of natural genetic mtDNA barcodes to resolve clonal heterogeneity within malignancies (Fig. 4), and assess clonal dynamics in hematopoiesis (Fig. 5 and 6), while also obtaining rich information on variation in cell state. Unlike conventional high-throughput scRNA-seq approaches that suffer from uneven coverage of mitochondrial RNA, a high false positive error rate6, or require a priori knowledge of specific variants51, our framework enables de novo discovery of variants to enable the inference of subclonal structure in complex settings, including tissue specimens directly obtained from patients. We expect that additional improvements in variant calling, clonal detection methods, and heteroplasmy-specific distance functions will aid to resolve cellular hierarchies in greater detail.
In addition to pathogenic mitochondrial variants, such as 8344A>G, our high-throughput platform should facilitate the examination of functional mtDNA mutations in relatively common disease settings1. Specifically, alterations in mtDNA have been associated with a variety of complex human diseases, including Alzheimer’s Disease52, Parkinson’s Disease53, cardiomyopathies54, pediatric cancers55 and more generally aging phenotypes1,56. As our approach facilitates rapid genotyping and concomitant chromatin profiles in thousands of cells, potential molecular consequences of mtDNA variants may now be dissected (Fig. 2), which is not otherwise possible using bulk approaches5.
Despite the relatively small size of the mitochondrial genome, the prevalence of somatic mutations, though not necessarily present in every cell, enabled inferences about cellular population dynamics in complex human tissues6,45 (Fig. 6). For future applications, we emphasize that care should be taken with respect to biological conclusions, which may require validation via orthogonal methodology across multiple donors. For example, our analyses in the context of malignancies (Fig. 4) provides a vignette of integrating nuclear point mutations, copy number alterations, immune receptor rearrangements, and mtDNA variation to further resolve clonal structure and functional heterogeneity. Though the hematopoietic system was the focus of our investigations (with the exception of the colorectal cancer sample), we expect our mtscATAC-seq framework to be compatible with most human tissues6,45. Overall, the advances presented here now enable new avenues to study the role of cellular dynamics in human health and disease.
ONLINE METHODS
Cell lines and cell culture
TF1 cells (ATCC) were maintained in Roswell Park Memorial Institute Medium (RPMI) 1640, 10% fetal bovine serum (FBS), 2 mM L-Glutamine and 2 ng/ml recombinant human Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) (Peprotech) and incubated at 37°C and 5% CO2. GM11906 cells (Corriell) were maintained in Roswell Park Memorial Institute Medium (RPMI) 1640, 15% fetal bovine serum (FBS) and 2 mM L-Glutamine and incubated at 37°C and 5% CO2.
Primary cells and cell culture
CD34+ hematopoietic stem and progenitor cells were obtained from the Fred Hutchinson Hematopoietic Cell Processing and Repository (Seattle, USA) or StemCell Technologies. The CD34+ samples were de-identified and approval for use of these samples for research purposes was provided by the Institutional Review Board and Biosafety Committees at Boston Children’s Hospital. Healthy donor peripheral blood mononuclear cells were obtained from StemCell Technologies. CD34+ cells were thawed and cultured in StemSpan II with 1x CC100 (StemCell Technologies, Inc.) at 37°C and 5% CO2. At indicated time points, these cells were seeded in media supporting the differentiation into monocytic and erythroid cells57,58. Briefly, cells were cultured at a density of 105 - 106 cells per milliliter (ml) in IMDM supplemented with 2% human AB plasma, 3% human AB serum, 1% penicillin/streptomycin, 3 IU/ml heparin, 10 mg/ml insulin, 200 mg/ml holo-transferrin, 1 IU erythropoietin (Epo), 10 ng/ml stem cell factor (SCF) and 1 ng/ml IL-3 and incubated at 37°C and 5% CO2. For mtscATAC-seq processing at indicated time points and when additional cells were to be maintained to enable sampling of cells at a later time, ⅓ of the cultured cells were maintained and ⅔ of the cells were forwarded to single cell sequencing as described below.
Chronic lymphocytic leukemia samples
Cryopreserved peripheral blood mononuclear cells from chronic lymphocytic leukemia (CLL) patients consented on institutional review board approved protocols were obtained from AllCells (Patient 1) or from Adrian Wiestner at the National Institute of Health (Patient 2). Cytogenetic analysis of Patient 1 CLL cells detected an extra copy of chromosome 12 (trisomy 12) as detected by fluorescence in situ hybridization (FISH). Cryopreserved cells were thawed by serial dilution in RPMI with 10% fetal bovine serum. B lymphocytes were isolated using the negative selection Mojosort Human Pan B Cell Isolation Kit (Biolegend, 480082) and CD19 negative immune cells were isolated from a separate aliquot using the positive selection Mojosort Human CD19 selection Kit (Biolegend, 480106).
Flow cytometry analysis and sorting
For flow cytometry analysis and sorting, cells were washed in FACS buffer (1% FBS in PBS) before antibody staining. For the CLL patient derived PBMC staining a FITC-conjugated CD19 antibody (HIB19, 302206, Biolegend) was used at 1:50 dilution. For live/ dead cell discrimination Sytox Blue was used according to the manufacturer’s instructions (Thermo Fisher, S34857). FACS analysis was conducted on a BD Bioscience Fortessa flow cytometer at the Whitehead Institute Flow Cytometry core. Data was analyzed using FlowJo software v10.4.2. Cell sorting was conducted using the Sony SH800 sorter with a 100 μm chip at the Broad Institute Flow Cytometry Facility. Sytox Blue (ThermoFisher) was used for live/ dead cell discrimination.
Colorectal cancer sample
A primary untreated colorectal tumor was surgically resected from an 84-year-old female patient with pathologically diagnosed colorectal adenocarcinoma at Massachusetts General Hospital. Written informed consent for tissue collection was provided in compliance with IRB regulations (IRB compliance protocol number 02–240; Broad Institute ORSP project number ORSP-1702). For mtscATAC-seq, fresh tissue was collected into RPMI 1640 medium supplemented with 2% human serum (Sigma), cut into 1 mm2 pieces, and enzymatically digested for 20 min at 37°C using the Human Tumor Dissociation Kit (Miltenyi Biotec). The cell suspension was passed through 70 μm cell strainers and centrifuged for 7 min at 450 g at 4°C. Supernatant was removed and cells were subject to ACK Lysis Buffer (Life Technologies) for 2 min on ice, centrifuged for 7 min at 450 g at 4°C, and resuspended in RPMI 1640 supplemented with 2% human serum (Sigma). The single cell suspension was stained with Zombie Violet in PBS (Invitrogen) for 10 min on ice, then stained for 15 min with antibodies (Biolegend) against human CD235a, CD326, CD45, CD66b, lineage cocktail (CD2, CD3, CD19, CD20, CD56), subsequently fixed with 1% formaldehyde, quenched in 0.125 M glycine, washed and sorted for Zombie Violet-negative, CD235a-negative, CD66b-negative cells into a 1.5 ml DNA LoBind tube (Eppendorf) prior to cell lysis and mtscATAC-seq processing as described below.
Single cell ATAC-seq (C1 Fluidigm)
The C1 Fluidigm platform using C1 single cell Auto Prep IFC for Open App and Open App Reagent Kit were used for the preparation of single cell ATAC-seq libraries as previously described19. Briefly, cells were washed and loaded at 350 cells/μl. Successful cell capture was monitored using a bright-field Nikon microscope and was >85%. Lysis and tagmentation reaction and 8 cycles of PCR were run on chip, followed by 13 cycles off chip using custom index primers and NEBNext High-Fidelity 2X PCR Master Mix (NEB). Individual libraries were pooled and purified using the MinElute PCR kit (QIAGEN) and quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent).
Single cell ATAC-seq and mtscATAC-seq
ScATAC-seq libraries were generated using the 10x Chromium Controller and the Chromium Single Cell ATAC Library & Gel Bead Kit (#1000111) according to the manufacturer’s instructions (CG000169-Rev C; CG000168-Rev B) or as detailed below with respect to the modifications enabling increased mtDNA yield and genome coverage. 1.5 ml or 2 ml DNA LoBind tubes (Eppendorf) were used to wash cells in PBS and downstream processing steps. After washing cells were fixed in 0.1 or 1% formaldehyde (FA; ThermoFisher #28906) in PBS for 10 min at RT, quenched with glycine solution to a final concentration of 0.125 M before washing cells twice in PBS via centrifugation at 400 g, 5 min, 4°C. Cells were subsequently treated with lysis buffer (10mM Tris-HCL pH 7.4, 10mM NaCl, 3mM MgCl2, 0.1% NP40, 1% BSA) for 3 min for primary cells and 5 min for cell lines on ice, followed by adding 1 ml of chilled wash buffer and inversion (10mM Tris-HCL pH 7.4, 10mM NaCl, 3mM MgCl2, 1% BSA) before centrifugation at 500 g, 5 min, 4°C. The supernatant was discarded and cells were diluted in 1x Diluted Nuclei buffer (10x Genomics) before counting using Trypan Blue and a Countess II FL Automated Cell Counter. If large cell clumps were observed a 40 μm Flowmi cell strainer was used prior to processing cells according to the Chromium Single Cell ATAC Solution user guide with no additional modifications. Briefly, after tagmentation, the cells were loaded on a Chromium controller Single-Cell Instrument to generate single-cell Gel Bead-In-Emulsions (GEMs) followed by linear PCR as described in the protocol using a C1000 Touch Thermal cycler with 96-Deep Well Reaction Module (BioRad). After breaking the GEMs, the barcoded tagmented DNA was purified and further amplified to enable sample indexing and enrichment of scATAC-seq libraries. The final libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent).
We further note the following related to mtscATAC-seq optimizations: Comparison of mtDNA cross-contamination between cell lines using data from Fig. 1b suggested higher levels at 0.1% formaldehyde (contamination 1.54%) compared to 1% formaldehyde fixation (contamination 1.14%). Therefore, cells were fixed in 1% formaldehyde for 10 min at RT. This has yielded excellent results and has been used throughout the manuscript unless indicated. Additional incubation (30 min to 12 h) at 60°C to further facilitate decrosslinking prior to the first 72°C elongation step did not improve results (data not shown) and we recommend using the PCR conditions specified in the 10x scATAC-seq protocol. Related to 10x Chromium microfluidic chip handling, cell loading, and recovery, we have followed the general recommendations from 10x Genomics and observe concordant results relative to their standard protocol. As hematopoietic cell suspensions were used for protocol optimizations, additional modifications may be required to obtain optimal results for other tissues of interest.
Single cell RNA-seq
ScRNA-seq libraries were generated using the 10x Chromium Controller and the Chromium Single Cell 5′ Library Construction Kit and human B cell and T cell V(D)J enrichment kit according to the manufacturer’s instructions. Briefly, the suspended cells were loaded on a Chromium controller Single-Cell Instrument to generate single-cell Gel Bead-In-Emulsions (GEMs) followed by reverse transcription and sample indexing using a C1000 Touch Thermal cycler with 96-Deep Well Reaction Module (BioRad). After breaking the GEMs, the barcoded cDNA was purified and amplified, followed by fragmenting, A-tailing and ligation with adaptors. Finally, PCR amplification was performed to enable sample indexing and enrichment of scRNA-Seq libraries. For T cell and B cell receptor sequencing, target enrichment from cDNA was conducted according to the manufacturer’s instructions. The final libraries were quantified using a Qubit dsDNA HS Assay kit (Invitrogen) and a High Sensitivity DNA chip run on a Bioanalyzer 2100 system (Agilent).
mtscATAC-seq sequencing and preprocessing
All libraries were sequenced using Nextseq High Output Cartridge kits and a Nextseq 550 sequencer (Illumina). 10x scATAC-seq libraries were sequenced paired end (2 × 72 cycles). 10× 5’ scRNA-seq libraries were sequenced as recommended by the manufacturer. Raw sequencing data was demultiplexed using CellRanger-ATAC mkfastq. Raw sequencing reads for all libraries were aligned to the regular and modified (for the mtDNA black list) hg19 reference genome using CellRanger-ATAC count version 1.0 (for cell-line mixing experiment) and version 1.2 (for all other samples).
With respect to mtscATAC-seq sequencing depth and cell numbers, we further note that for hematopoietic cells we have generally aimed to match the estimated overall library complexity of the sample, e.g. sequence 100 million reads for a library with an estimated complexity of 100 million unique fragments (estimated exclusively from the nuclear genome). Furthermore, we have aimed to obtain at least 20x mitochondrial genome coverage after removal of PCR duplicated reads to enable confident mtDNA mutation calling. Mitochondrial genome coverage may improve with deeper sequencing than used here. Moreover, because mtDNA content may vary from one cell type or state to another, the required sequencing depth may vary and higher coverage may be readily achieved in some cell types, which would in turn enable more confident detection of low frequency mutations.
We cannot currently specify general guidance for the number cells to be profiled, as this will inevitably depend on the specific context (i.e. tissue and question of interest). Generally, this will be a function of the “clonality” of each tissue and the diversity of cell types and states, the complexity of which we currently may not be able to accurately anticipate, given the relative lack of data in this area for many human tissues. All methods, when applied to a random sampling of cells, including genetic engineering approaches, are more likely to detect dominant clones, whereas the resolution of lower frequency clones ultimately improves with an increasing number of cells sequenced. Based on our experience with data in this manuscript, we suggest that profiles from as few as ~1,000 cells can highlight subclonal structures in malignant cell populations (Fig. 4). For steady state hematopoiesis ~10,000 cells have provided initial informative insights (Fig. 6), though deeper profiling may be desired depending on the question at hand.
Masked reference genome and NUMT comparison
To effectively assign putative multi-mapping reads to the mtDNA, we modified the existing CellRanger-ATAC reference genome by hard-masking nuclear mitochondrial DNA segments (NUMT). These regions were detected by simulating reads of length 20 from the reference mtDNA genome and encoding 1 base “errors” via the ART program59. Simulated reads were then aligned to the reference genome (with the mitochondrial chromosome excluded). As these reads were simulated to originate from the mtDNA genome but aligned to the nuclear genome, we hard masked these regions using bedtools60. Comparisons of data from Fig. 1 were performed by re-aligning the same datasets to the reference genome with and without masking. Complete documentation to reproduce the masking and modification of the CellRanger-ATAC reference genome are available as part of the mgatk wiki (https://github.com/caleblareau/mgatk/wiki).
To estimate the number of accessible NUMT fragments that would be assigned to mtDNA, we considered two different approaches. First, we used a public GM12878 dataset from 10x Genomics (https://www.10xgenomics.com/solutions/single-cell-atac/) that was aligned to the standard hg19 reference and counted the number of fragments per cell overlapping our NUMT blacklisted regions, which resulted in a mean 1.4 and median 1.0 fragments per cell. Second, we used a compendium of DNase accessible peaks from 164 distinct samples from the ENCODE15 and Roadmap14 consortia, and estimated that these samples contained a mean 22.6 peaks overlapping our NUMT blacklist. Next, using the GM12878 peakset and the same scATAC-seq dataset, we determined that a mean 4.1% of the GM12878 DNase peaks were detected over all cells. The product of these two numbers (22.6*0.041=0.93 fragments/cell) provides an alternative estimate for the number of accessible chromatin fragments overlapping NUMTs (~1 fragment) that were blacklisted. As our mtscATAC-seq assay generates ~5,000–10,000 mtDNA fragments, we conclude that our blacklist approach yields negligible NUMT contamination.
Comparison of experimental conditions
For all comparisons shown in the boxplots and violin plots, the top 1,000 cells/barcodes based on chromatin library complexity were plotted. The top 1,000 number was chosen to ensure the selection of real cells rather than barcode multiplets61 or other barcodes associated with low counts. For the overall coverage comparison (Fig. 1g), the top 2,000 cells based on nuclear complexity were averaged (to represent the expected 2,000 cell yield from the experiment).
Cells were assigned TF1, doublet, or GM11906 using the sum of alleles at homoplasmic mitochondrial SNP loci (Extended Data Fig. 1d) using a 99% threshold for assignment to either major cell-type for our final protocol. We assigned barcodes as cell doublets (Fig. 1d,e) when this 99% threshold was not met for the major celltype. For both mtDNA and chromatin complexity estimation (Extended Data Fig. 1e), we used the number of unique and duplicate fragments as part of the CellRanger-ATAC (chromatin) and mgatk (mitochondria) output as inputs into the Lander-Waterman equation62, which estimates the total number of unique molecules present given these two measurements. Complexity measures were computed per barcode passing the knee filter from the default CellRanger-ATAC execution.
To verify that cell type-specific accessible peaks were retained in mtscATAC-seq, we determined 77,704 peaks present in either the TF1 or GM11906 cell lines using the regular 10x scATAC-seq conditions. These were determined from assigning barcodes to either cell line using mtDNA SNPs and calling peaks on the aggregate bulk population as previously described9. We repeated this peak calling procedure with our mtscATAC-seq data, identifying 72,887 peaks that overlapped the 77,704 peaks (93.8%).
To model the residual variation in mtDNA coverage (Fig. 1g), we computed rolling averages of GC content and mean coverage after masked alignment in 50 bp bins with a 25 bp step size (Extended Data Fig. 1j).
Mitochondrial pathogenic variants
We queried MITOMAP26 version r102 and filtered for “Confirmed” pathogenic base-substitution variants. 46 variants were annotated to alter tRNA function whereas 42 were annotated to alter protein coding sequences in one or more protein-coding genes. Two additional variants were annotated to alter rRNA function.
In situ detection of mtDNA heteroplasmy
Sample preparation and imaging
All solutions below were prepared in 1x phosphate buffered saline (PBS), and incubations were carried out at RT unless otherwise specified. Two million GM11906 cells were fixed with 2 ml 1% paraformaldehyde for 10 min and quenched by adding 666 μl 1 M Tris-HCl pH 8 and incubation for 5 min. Cells were then permeabilized with 0.5% Triton-X 100 for 20 min and embedded in 4% acrylamide gels63. The mitochondrial target sequence (on the antisense strand) was made accessible for hybridization by enzymatic removal of the sense strand64,65: restriction digest with 0.5 U/μl XbaI at 37°C for 1 h, followed by adding 0.2 U/μl lambda exonuclease (both New England Biolabs) at 37°C for 30 min. The oligonucleotide probe sequences against the wildtype (/5PHOS/ACCAACACCTCTTTACtaataCAGCCAATCTCGGGAACGCTGAAGAcggcACGTACGTGTTAAAGATTAAGAGA) and mutant (/5PHOS/GCCAACACCTCTTTACtaataCTGTGAGTCTCGGGAACGCTGAAGAcggcTTCCTTCCGTTAAAGATTAAGAGA) alleles were pooled at 100 nM each in 2x SSC and 20% formamide, hybridized to the cell gels at 37°C overnight, and circularized with 6 U/μl T4 ligase (Enzymatics) for 2 h. Rolling circle amplification, crosslinking, and in situ sequencing were performed as previously described20. The cell gel was stained with DAPI (Thermo Fisher) and imaged on a Nikon Eclipse Ti microscope with a Yokogawa CSU-W1 confocal scanner unit and an Andor Zyla 4.2 Plus camera using a Nikon Plan Apo 60X/1.40 objective. Z-stack images spanning 24 μm at 0.4 μm intervals were acquired in the following channels: 405 nm excitation with a 452/45 emission filter; 488 nm excitation with a 525/50 emission filter; 561 nm excitation with a 579/34 emission filter.
Image processing and heteroplasmy quantification
Each image stack was transformed into 2D by taking the maximum intensity projection across z-planes. Individual nuclei boundaries were defined by performing watershed segmentation on the DAPI staining. Wild-type and mutant probes were detected using a local maxima finder and uniquely assigned to individual cells based on spatial proximity. Probes that could not be unambiguously assigned to a cell were excluded from heteroplasmy and coverage measurements.
Epigenomic correlates with pathogenic heteroplasmy
To identify chromatin accessibility features associated with pathogenic heteroplasmy in the GM11906 cell line, we considered two approaches that complemented our estimation of heteroplasmy at the single-cell level. First, to assess cis-associations, we computed single-cell gene scores as previously described9,10 and computed per-gene associations with heteroplasmy using Spearman correlation (Fig. 2f). To establish a background distribution, we permuted heteroplasmy per-cell and recomputed the per-gene association statistic. We reported the number of gene scores correlated with heteroplasmy if the magnitude of the Spearman correlation exceeded 0.2. However, we note that a 1% false positive rate from the permutation testing would be a threshold of 0.087, resulting in 752 positively and 1,992 negatively correlated gene scores. We reported the more conservative results after examination of the accessible chromatin tracks where loci exceeding a magnitude 0.2 correlation revealed more robust peak differences. Second, to assess trans-associations, we downloaded a compendium of 78 high-quality ChIP-seq peak sets from lymphoblastoid cell lines from the ENCODE project15. Per single-cell deviation scores were computed for these factors using chromVAR38.
Variant calling and evaluation
Overview
To best identify informative clonal mutations from our mtscATAC-seq assay, we first considered existing variant calling approaches. Notably, algorithms designed for genotyping typically utilize a Bayesian framework to determine the empirical probability of a certain non-reference allele being truly observed at a particular location. In this setting, the ploidy of the genome is often parameterized in the model, and the allele frequency directly influences the confidence of detecting the mutation. As mtDNA copy number per cell is variable and informative clonal mutations may occur at very low allele frequencies, we found these existing approaches to be unsuitable for our mtscATAC-seq assay. Therefore, we developed a variant calling framework to identify high-confidence heteroplasmic mutations in a manner that 1) is largely independent of the mean allele frequency; 2) is robust to variability in genome ploidy of a cell; and 3) utilizes the features intrinsic to the high-throughput single-cell mtDNA data, including near-uniform deep coverage, minimal dropout per-cell, and thousands of single-cells per experiment. Our resulting variant calling framework, mgatk, achieves these goals.
Analysis of mtscATAC-seq data from this manuscript revealed that certain positions with substantial heteroplasmy across biological diverse sources was primarily driven by sequencing error. These “recurrently-mutated” loci were due in part to several low-complexity stretches in the mitochondrial genome. However, by further evaluation of these variants, we determined that the erroneous heteroplasmy was primarily driven by one strand, reflective of a photobleaching effect from surrounding “G”s on successive cycles66.
Identification of subclonal variants with mgatk
The raw output of the CellRanger-ATAC count execution, specifically the barcodes passing knee and the position-sorted .bam file, serve as inputs into the command-line interface of mgatk. This execution produces intermediate plaintext sparse matrix files of PCR-deduplicated, per-cell, per-strand count of all alleles at all positions in the reference mitochondrial genome.
To determine high quality variants to infer clonal cell populations, mgatk then computes per variant summary statistics that are used to define high-quality variants. First, it computes a Pearson correlation coefficient between allele counts for all cells that have at least one count observed for the alternate allele (i.e. removing 0,0 points from the calculation). Intuitively, a high correlation captures the agreement of heteroplasmy between the strands and mitigates a widespread technical bias of sequencer photobleaching (Extended Data Fig. 3c). Explicitly, the Pearson correlation coefficient is the “strand concordance” value in Fig. 3b and 5d, and Extended Data Fig. 3d,e, 4b, and 6d. For all applications in this paper, we used a threshold of 0.65. Next, we compute a per-variant variance mean ratio (VMR; y-axis of the same figures) and subsequently filter out variants with a VMR < 0.01 (Fig. 3b, 5d and Extended Data Fig. 3d,e, 4b, and 6d). Default values for these two thresholds were based on performance in the hematopoietic clone data (Extended Data Fig. 3). Finally, mgatk reports the number of cells where the variant was confidently detected, defined by the mutation being detected in at least two fragments aligned to both strands. Here, we require the variant to be confidently detected in at least five cells for downstream analyses (which minimizes the inclusion of mutations that would not be associated with subclonal structure). While the workflow enables custom user-defined thresholds, we consistently applied these stated thresholds across the datasets in this study.
When visualizing variants in heatmaps, we have utilized different dynamic ranges (such as up to 10% or up to 100% heteroplasmy) to help display mutations in the relevant context of each figure. In general, we recommend visualizing variant x cell heatmaps at a variety of dynamic ranges to ensure best results. Specifically, the mutations displayed in Fig. 3c are of low frequency that mark smaller subclonal groups of cells. Conversely, variants shown in Fig. 4d are highly heteroplasmic or homoplasmic, which would not be conveyed when keeping an upper threshold of 10% heteroplasmy for visualization.
Finally, while our approach works for mtscATAC-seq and full-length scRNA-seq methods (e.g. SMART-seq2; Extended Data Fig. 3d–h), our approach is not appropriate for 3’ scRNA-seq methods (as data from such platforms are typically only derived from sequencing one strand).
Comparisons to other approaches
To compare our proposed variant calling approach to other tools, we analyzed the 855 TF1 single cells (Fig. 3) profiled in this manuscript. First, our execution of monovar28 failed as the genotype likelihood model is a function of a factorial of the max depth, which cannot be stored for the extremely deep coverage that results from our protocol. We then evaluated samtools/bcftools67 and FreeBayes68, treating each of the 855 cells as individual samples. To compare to mgatk (Extended Data Fig. 3a,b), the resulting .vcf files from each of these tools were filtered to remove clear homoplasmic variants and that had a variant quality ≥100. While our analyses indicated mgatk had greater sensitivity in resolving heteroplasmic variants informative for subclonal structure, relaxing this variant quality threshold did not improve detection of these informative variants and instead resulted in far more variants with strand discordance (Extended Data Fig. 3c). Finally, we acknowledge that other variant calling tools, such as GATK, utilize a Fisher’s exact test to flag variants with high strand discordance that can be removed in downstream processing. We found this approach to be unsuitable for this data due to the high copy-number, resulting in extremely-small p-values for all variants, including those that clearly correlated with subclonal structure.
Simulations
We estimated the sensitivity and positive predictive value (PPV) of mtscATAC-seq using a simulation where we varied mutation heteroplasmy and mutation coverage (Extended Data Fig. 3i). For each of 10,000 iterations per condition, we simulated data for 1,000 cells such that 100 cells contained the subclonal mutation (denoted by the set I). For heteroplasmy p (p ∈ {0.02,0.05,0.15,0.25,0.35,0.45}) and coverage n (n ∈ {20,50,100}), we simulated the variant allele frequency (AF) for cell i ∈ I as:
The simulated allele frequencies for the 900 cells that lacked the mutation (denoted by the set J), were computed in an analogous manner instead using a value q, corresponding to the contamination (or noise) of mtscATAC-seq. From our experiments in Fig. 1, we empirically derived q = 0.19. Thus, for cell j ∈ J,
For ‘detection’, we required the cell to have at least half of the simulated heteroplasmy (p / 2). Sensitivity and PPV were reported using I as the set of true positives, and J as the set of true negatives by the mean of the 10,000 iterations per condition.
To estimate the dropout rate of a mutation, defined by zero observations of the alternate allele, we simulated m = 10,000 observations for each value (indexed by k) of n and p and computed the ratio of draws of a binomial distribution that were identically zero to the total number of draws:
All code to reproduce all simulations is contained in the online resources.
Evaluation of mgatk with SMART-seq2 data
To further benchmark our variant calling algorithm, we reanalyzed 895 high-quality cells from poly-clonal hematopoietic cells carrying somatic mtDNA mutations identified from SMART-seq2 scRNA-seq data6. Previously aligned .bam files were re-processed with mgatk for each donor, and variant calling mirror the parameters established in the TF1 example (i.e. strand concordance ≥ 0.65; -log10(VMR) ≥ 2; see Extended Data Fig. 3g,h). From these samples, we had previously identified 78 variants showing subclonal structure using a supervised approach (i.e. the per-cell colony annotations were used in the identification of the variants). This set of 78 variants represents a “silver standard” as variants showed disproportionate heteroplasmy in a particular clone based on a Mann-Whitney U-test previously described6.
Overall, mgatk identified 103 variants across the two donors. This set replicated 64 of the 76 (84.2%) previously identified sub-clonal variants. The variants that were not replicated were rarer in the population of cells (p=0.00045; Wilcoxen Rank-Sum Test; Extended Data Fig. 3f). While we generally believe the mgatk variant calling approach to be sensitive to low-frequency variants, we note that this supervised variant calling procedure (when clonal annotations are known) is theoretically better-powered to detect low-frequency mutations. However, we note that one previously-identified variant, 4214T>C, had only non-zero heteroplasmy on one strand, strongly suggestive of an artifactual variant that was nonetheless identified by our previous supervised approach6.
To evaluate the efficacy of variant identification approaches for inferring clones, we tested their ability to correctly classify true-positive pairs of cells that were derived from the same clone6. We computed per cell pair mtDNA cosine similarity metric, using mutations identified by three unsupervised approaches (mgatk, bcftools, and FreeBayes), as well as our previous supervised approach for each donor. Area under the receiver operating curve (AUROC, Extended Data Fig. 3g,h) were computed and can be interpreted as the efficacy of classifying pairs of cells from the same clone based on sets of mtDNA variants.
TF1 analyses
To identify putative subclones, we used the square root of the heteroplasmy matrix as inputs into the FindNeighbors / FindClusters functions from Seurat69 with slight modifications for these functions (cosine distance metric, k.param = 10; resolution = 1.0). In principle, this approach identifies communities of cells whose overall mutations are similar (using a shared nearest neighbors approach), and subclones are identified using a modularity optimization. Finally, we performed tree reconstruction using neighbor-joining on the cosine distance between the average heteroplasmy of cells per clone using hierarchical clustering.
Chronic lymphocytic leukemia scATAC analyses
For each mtscATAC-seq library, cells were processed using CellRanger-ATAC with default settings, including the `--force-cells 6000` flag. Each library was further filtered such that cells had minimum 50% fragments in accessibility peaks, 1,000 unique nuclear fragments, and 20x mtDNA coverage. Somatic mtDNA mutations were identified using mgatk with the default parameters for the CD19 positive cells profiled with mtscATAC-seq (Extended Data Fig. 4b). Putative sub-clones were identified using the mutations for patient 1 (n=18) and patient 2 (n=24) separately using the FindNeighbors/ FindClusters functions from Seurat with a cosine distance function on the square root of the heteroplasmy matrix. We used parameters for patient 1 (k.param = 20; resolution = 0.2; Fig. 4c) and patient 2 (k.param = 30; resolution = 1.0; Extended Data Fig. 4c) to effectively identify subclones. For visualization of cell by mutation heatmaps, subsets of cells from Patient 1 (2,246/5,624; Fig. 4c) and Patient 2 (3,057/5,874; Extended Data Fig. 4c) were visualized as the remaining cells had largely 0% heteroplasmy at called mutations.
To determine copy number alterations (Fig. 4e), we first constructed overlapping 10Mb bins genome-wide using a step size of 2Mb. Next, we overlapped the .fragments.tsv file from the 10x CellRanger-ATAC output with these bins to compute a bin x cell matrix for both the CLL samples as well as a healthy control PBMC sample. Next, we computed a per-cell, per-bin z-score of the number of fragments after normalizing each cell to a consistent sequencing depth. The chromosome 12 z-score (Fig. 4e) represents the per-cell mean of the z-scores from the bins mapping to this chromosome. To interpret the z-score, we computed the percentage of unique autosomal reads mapping to chromosome 12 for the CLL (8.1%) and healthy PBMC samples (mean 5.3%). The 53% increase in reads mapping to chromosome 12 in CLL cells supported trisomy (rather than a higher copy number) as the chromosomal aberration.
To identify chromatin accessibility peaks associated with mtDNA mutation-derived subclones, we performed a series of χ2 association tests. After binarizing the chromatin accessibility count per-peak, per-cell, a contingency table of dimension n x 2 was assembled, where n is the number of subclones per tumor. The resulting chi-squared statistics were associated with p-values using n - 1 degrees of freedom, and correction for multiple testing was performed using the Benjamini–Hochberg procedure. To further visualize a null association statistics, we permuted the subclone annotations per peak to visualize a null distribution of the chi-square statistics (see gray from Fig. 4f; Extended Data Fig. 4g). The TIAM1 and ZNF257 loci were selected based on strong association (both in the top 10 most-associated peaks) and proximity to annotated transcription start sites.
To identify non-B-cells with mtDNA mutations, we first embedded a healthy PBMC 5k cell sample from the 10x Genomics public dataset using LSI and UMAP as previously described37. Using the LSI components the projection capability of UMAP, we projected CD19 negative cells from both CLL donors onto the reduced dimension space (Fig. 4j,k). Cells were annotated as positive for specific mtDNA mutations if the heteroplasmy exceeded 20% (corresponding to at least 4 unique molecules containing the alternate allele; Fig. 4j,k).
Colorectal cancer scATAC-seq analyses
The colorectal cancer sequencing library was processed with CellRanger-ATAC with default settings. Each cell was further filtered such that it had a minimum 40% of fragments overlapping a compendium of DNase hypersensitivity peaks (integrated in the CellRanger-ATAC workflow), 1,000 unique nuclear fragments, and 10x mtDNA coverage. Somatic mtDNA mutations were identified using mgatk using default parameters. Dimensionality reduction, clustering, and gene activity scores were determined using standard processing via Seurat and Signac69. Single-cell copy number inference was performed as described in the CLL scATAC analysis section, and the reported amplified chromosomes were corroborated by Whole-Exome Sequencing data (data not shown).
Exome sequencing
Enriched CLL cells and in vitro expanded CD3+ T lymphocytes to serve as a germline control were subjected to whole exome sequencing using the clinical somatic exome workflow through the Broad Institute Genomics Platform. The exome product targets 35.1 Mb with a total bait size of 38.9 Mb and are optimized to cover the following: 99% of ClinVar variants; complete Mitochondrial genome; full ACMG59 gene list; Online Mendelian Inheritance in Man (OMIM) putative gene sequences; Catalogue of Somatic Mutations in Cancer (COSMIC) variants; Internal ‘ONCO Panel’ and additional key promoters and other motifs that have been identified as potential cancer hot spots. Automated library preparation occurs as follows. Samples were plated at a concentration of 2 ng/μl and volume of 50 μl (total 100 ng input) into fresh matrix tubes allowing positive barcode tracking throughout the process.
Samples were sheared to yield ~180 bp size distribution. Kapa Hyperprep kits were used to construct libraries in a process optimized for somatic samples, involving end repair, adapter ligation with forked adaptors containing unique molecular indexes and addition of P5 and P7 sample barcodes via PCR. After SPRI purification libraries were quantified with Pico Green. Libraries were normalized and equimolar pooling was performed to prepare multiplexed sets for hybridization. Sample pools were then split and hybridized in up to 8 separate reaction wells to accommodate volumes. Automated capture was performed, followed by PCR of the enriched DNA and SPRI purification.
Multiplex pools were quantified with Pico Green and DNA fragment size was estimated using Bioanalyzer electrophoresis. Final libraries were quantitated by qPCR and loaded across the appropriate number of Illumina flow cell lanes to achieve the target coverage. Completed exomes contained >= 85% of target bases covered at >= 50x depth and ranged from 130–160x mean coverage of the targeted region. Both tumor and normal samples were processed and used for variant identification.
CLL scRNA-seq analyses
5’ scRNA-seq libraries, including VDJ sequencing, were processed using default parameters with CellRanger 3.1.0. Mitochondrial genotyping was conducted using mgatk with the “--umi-barcode” tag specifying the SAM tag from the CellRanger .bam output marking the error-corrected UMI barcode. Cell-type specific signatures (Fig. 4k; Extended Data Fig. 4k) were computed using Seurat’s AddModuleScore69 where gene bins were computed on a control set of healthy PBMCs. Cell-type specific genes were determined from the Immune Cell Atlas (available here: https://github.com/caleblareau/immune_cell_signature_genes). Two nuclear variants, chr4:109,084,804A>C (“LEF1”; p.S112A) and chr19:36,394,730G>A (“HSCT”; p.A56T), encoded missense mutations that were detected using whole-exome sequencing and somatic mutation calling. These mutations were covered by the 5’ scRNA-seq libraries, enabling single-cell examination (Extended Data Fig. 4k). Cells were annotated as positive for mtDNA mutations if at least two distinct UMIs supported the mutation (Fig. 4l; Extended Data Fig. 4k). Datasets used for the comparison of scRNA-seq technologies (Extended Data Fig. 4d,e) are detailed in Supplementary Table 4.
In vitro CD34+ cell culture analyses
For each mtscATAC-seq library, cells were processed using CellRanger-ATAC with default settings, including the `--force-cells 6000` flag. Each library was further filtered such that cells had minimum 25% fragments in accessibility peaks, 1,000 unique nuclear fragments, and 20x mtDNA coverage. Cutoffs were determined from examination of the density of each parameter. Somatic mtDNA mutations were identified using default thresholds from mgatk for each culture independently.
Clustering and embedding using Uniform Manifold Approximation and Projection70 (UMAP) were performed on the top 30 reduced dimensions from Latent Semantic Indexing (LSI) as previously described for the chromatin accessibility features37. Annotation of cell states were determined using transcription factor motif scoring via chromVAR38 with default parameters, noting that the background peak selection was performed using all libraries merged. Pseudotime trajectories were defined using a semi-supervised approach from LSI and embedding as previously described10.
To determine cell clones, we used the mutations by cells matrix as input to the FindNeighbors/ FindClusters functions from Seurat with hyperparameters k.param = 10, resolution = 1.5, and cosine distance function, which yielded good separation of the rare cell clones. Clone-specific mutations were shown for all mutations exceeding 0.5% mean heteroplasmy in cell clones (Extended Data Fig. 5i,j). We defined erythroid and monocytic cells in the day 20 library as those that exceeded a 0.5 pseudotime score along the specific axes (from Fig. 5c) and retained 57 clones from the 800 cell culture that had at least 10 total cells that were differentiated. To compute the lineage bias z-score (Fig. 5j), we computed the fraction of monocytic/erythroid labels in a cell clone and permuted these labels 100 times over the day 20 library. Finally, to infer putative lineage-priming chromatin accessibility, we identified 10 erythroid-biased and 21 monocytic-biased clones (z-score >5 from Fig. 5j) and computed the mean transcription factor deviation scores38 from the day 8 cells belonging to each clone. The difference in means between the erythroid and monocytic-biased clones represents the putative lineage bias score and is plotted in Fig. 5k.
In vivo hematopoiesis analyses
The four mtscATAC-seq libraries (2x PBMC; 2x CD34+ HSPC cells) were processed using CellRangerATAC-count with the `--force-cells 6000` flag. Each library was further filtered such that cells had minimum 25% (CD34+ HSPCs) or 60% (PBMCs) fragments in accessibility peaks, 1,000 unique nuclear fragments, and 20x mtDNA coverage. Cutoffs were determined from examination of the density of each parameter. Somatic mtDNA mutations were identified using default thresholds from mgatk for each sample separately.
To define cell states for the CD34+ HSPC dataset, clustering and embedding using Uniform Manifold Approximation and Projection70 (UMAP) were performed on the top 30 reduced dimensions from LSI as previously described37 for the chromatin accessibility features and utilized for the PBMC data. Here, we utilized the previously published peak set37 to facilitate projection of FACS-sorted progenitors (Fig. 6c). For the PBMC data, clustering, reduced dimensionality, and gene activity scores were determined using standard processing via Seurat and Signac69. This workflow was utilized to facilitate high resolution cell-type label transfer from an existing public 10x scRNA-seq v3 PBMC dataset (Extended Data Fig. 6a).
To determine cell clones, we used the mutations by cells matrix as input to the FindNeighbors/ FindClusters functions from Seurat with hyperparameters k.param = 10, resolution = 3.5, and cosine distance function, which produced cell clones, where one mtDNA variant often corresponded to one cluster (Extended Data Fig. 6g). To determine putative clonal lineage bias (Fig. 6m–o), we performed a Chi-squared goodness of fit for the observed per-clone proportions compared to the total proportions of cells. For the CD34+ HSPC data, we used the 12 chromatin clusters (Fig. 6c) and for the PBMC data the three main large clusters (T/NK cells; B-cells, monocytes; Fig. 6d). Here, clones were filtered such that at least 10 cells were present in the analyzed clones.
To identify the 923 additional rare variants (Extended Data Fig. 6h,i), we identified mutations that met the following criteria: a) “confidently detected” with at least 2 unique fragments aligning to both the top and bottom strand (minimum 4 total reads) in 1, 2, or 3 cells; b) present at no more than 5% heteroplasmy in no more than 5 cells (to further exclude the possibility of unaccounted bias). We emphasize that none of the additional 923 mutations overlapped with the 429 clonal variants identified using the standard mgatk processing.
DATA AVAILABILITY
Data associated with this work is available at GEO accession GSE142745.
CODE AVAILABILITY
Software and documentation for mitochondrial variant calling via mgatk is available at http://github.com/caleblareau/mgatk. Custom code to reproduce all analyses and figures is available at https://github.com/caleblareau/mtscATACpaper_reproducibility.
Extended Data
Extended Data Fig. 1: Additional validation of biotechnological and computational basis for single-cell mtDNA genotyping.
(a) Comparison of chromatin library complexity (estimated number of unique fragments) across screened lysis conditions as shown in Fig. 1. (b) The same variable lysis conditions showing the TSS rate per cell. (c) BioAnalyzer traces of mtscATAC-seq library fragment size distribution for regular conditions and mtDNA-enriched conditions. (d) Heteroplasmy heatmap of single cells (columns) for 43 private homoplasmic mutations (rows) in the TF1 or GM11906 cell lines with (left) and without (right) FA treatment. Color bar, heteroplasmy (% allele frequency). (e) Comparison of mtDNA fragment complexity and chromatin complexity between the original/ regular 10x scATAC protocol and modified lysis conditions with and without formaldehyde (FA) treatment. (f) Heteroplasmy of sum of single-cell ATAC-seq libraries with variable FA treatment. (g) Schematic, method, and results of improving mtDNA genome coverage via hard-masking the reference genome (Methods). (h) Comparison of % reads mapping to mtDNA and (i) chromatin complexity with (red) and without (blue) the hard masking. (j) Comparison of average coverage of mtscATAC-seq (y axis) and GC content (x axis) at each 50bp bin (dot) in the mtDNA genome. (k) Accessible chromatin landscapes aggregated from single cells near the ETV2 locus for both cell lines as assayed via regular scATAC-seq and mtscATAC-seq. For boxplots in (a,b,e,h,i), each condition represents the top 1,000 cells (based on chromatin complexity) for one experiment. Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range.
Extended Data Fig. 2: Further inferences in analysis of the GM11906 (MERRF) lymphoblastoid cell line.
(a) Alternative field of view for GM11906 in situ genotyping imaging experiment. Representative image selected from one of seven fields of view for one experiment. Pseudo bulk accessibility track plots are shown for the (b) ETV2 and (c) CD19 loci. Pseudo-bulk groups represent 0–10% (low), 10–60% (mid), and 60–100% (high) m.8344A>G heteroplasmy. (d) Spearman correlation of heteroplasmy against the ChIP-seq deviation scores computed via chromVAR. Each bar is a single transcription factor with selected factors highlighted. (e) Depiction of MEF2C deviation scores from chromVAR for m.8344A>G heteroplasmy bins, corresponding to 0–10% (Low), 10–60% (Mid), and 60–100% (High). Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. Bins contain single cells collected over one experiment where bins correspond to high (>60%; n=273), intermediate (10–60%; n=228), and low (<10%; n=313) heteroplasmy (see Fig. 2c).
Extended Data Fig. 3: Supporting information for somatic mtDNA mutation calling via mgatk.
(a) Venn diagrams depicting comparisons of heteroplasmic mutations identified by mgatk, samtools/ bcftools, and (b) FreeBayes. (c) Comparison of heteroplasmy estimated from reads aligned to either strand. The top row are three variants called specifically by mgatk; 3549C>A was identified only by FreeBayes. 7399C>G and 546A>C were called specifically by bcftools. (d) Identification of 67 and (e) 36 heteroplasmic variants from previously published SMART-seq2 hematopoietic colony data. Blue variants represent known RNA-editing events. (f) Comparison of population heteroplasmy values for variants replicated by mgatk from a previous supervised approach. Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. Statistical test: two-sided Mann-Whitney U Test. (g) Concordance between discerning cells sharing a clonal origin based on colony-specific mtDNA mutations and their unsupervised identification using indicated algorithms (mgatk, bcftools, FreeBayes) and previously described supervised approach6. Receiver operating characteristic (ROC) using the per cell pair mtDNA similarity metric to identify pairs of cells sharing a clonal origin based on sets of mtDNA variants. The number of variants in each set is also depicted. (h) Area under the ROC (AUROC) is denoted for each donor group and indicated variant caller as depicted in (g). Each bar represents the statistic from one evaluation per donor per tool. (i) Estimated sensitivity (y axis, left), positive predictive value (y axis, right), and (j) estimated % dropout (y axis) for mtscATAC-seq at different simulated levels of heteroplasmy (x axis; Methods). Vertical line: 5% heteroplasmy for a subclonal mutation. The in-graph numbers indicate the values from the curve at a single-cell heteroplasmy of 5% with colors corresponding to different per-cell coverage values in the simulation.
Extended Data Fig. 4: Supporting information for clonal and functional heterogeneity in malignant populations revealed by mtDNA mutations.
(a) Flow cytometry gating strategy of CLL patient derived PBMCs showing expansion of CD19+ cells. (b) Identification of high-confidence variants for Patient 1 (top) and Patient 2 (bottom). The number of variants n is indicated. (c) Inference of subclonal structure from somatic mtDNA mutations for patient 2. Cells (columns) are clustered based on mitochondrial genotypes (rows). Colors at the top of the heatmap represent clusters or putative subclones. Color bar, heteroplasmy (% allele frequency). (d) Dot plots showing the mitochondrial genome coverage (log10; y-axis) for the top 500 cells per technology for four indicated scRNA-seq technologies. (e) The mean per-position mitochondrial genome coverage for the same 500 cells as in (d). (f) Volcano plot showing differential gene expression analysis from major and minor clonotypes defined by BCR sequence. Immunoglobulin (IG) genes are shown in purple; all other genes with an FDR < 0.05 are shown in blue. (g) Results for per-peak chi-squared association with sub-clonal group. Each dot is a peak rank-sorted by the chi-squared statistic. (h) Heteroplasmy from the sum of single-cells in the CD19+ and CD19- mtscATAC-seq experiments for indicated mutations and patients. (i) Histograms showing the distribution of heteroplasmy across the profiled population of cells for six selected variants, four from Patient 1 (left) and two from Patient 2 (right). The number of variants in the top heteroplasmy bin (>90%) are shown in red. (j) Allele frequency from the sum of single cells from the 5’ CD19+ and CD19- scRNA-seq libraries for two indicated variants - chr4:109,084,804A>C (“LEF1”) and chr19:36,394,730G>A (“HSCT”). (k) Corroboration of T cells based on gene expression signatures and carrying indicated somatic nuclear and mtDNA mutations (Patient 2). (l) Gene activity scores supporting cell type annotations in Fig. 4n. Arrows: cluster enriched for respective gene score. (m) All mtDNA mutations (rows) by cells (columns) observed in the CRC tumor. Columns are colored by defined chromatin cell state defined as in Fig. 4n. (n,o) Chromatin-derived UMAP with cells marked by select mtDNA mutations enriched in (n) epithelial and (o) immune cells. Color bar: heteroplasmy (% allele frequency).
Extended Data Fig. 5: Supporting information for clonal lineage tracing across accessible chromatin landscapes and time in an in vitro model of hematopoiesis.
(a) Depiction of single-cell UMAP embedding showing the original distribution of cells for each library/ time point, (b) relative cell density, (c) Louvain cluster, and (d) mitochondrial DNA coverage per single cell. (e) Overlap of variants called for each of the two datasets. (f) Comparison of log2 fold change in heteroplasmy from day 14 to day 8 for 19 overlapping variants. The p-value shown is for the beta 1 coefficient of the depicted linear regression model. (g) Proportion of cells (%) at day 8 of the 500 cell (x axis) and 800 cell (y axis) input culture carrying shared mtDNA variants as derived from panel (e) suggests limited clonal overlap. (h) Known pathogenic mtDNA mutations detected from a healthy donor. Each dot is a cell separated by the sampled library. All cells with a heteroplasmy of at least 2% are shown. (i) Depiction of unsupervised clustering of groups of cells based on shared somatic mtDNA mutations (y-axis) with corresponding individual mtDNA mutations (x-axis) associated with each cluster for the 500 cell input and (j) 800 cell input culture. Color bar, heteroplasmy (% allele frequency). (k) Fraction of cells (y-axis) carrying number of somatic mtDNA variants (x-axis) above indicated thresholds (≥1%, ≥5%, ≥10% heteroplasmy; red, black, and blue lines, respectively) for indicated cultures.
Extended Data Fig. 6: Support information for cellular population dynamics in native hematopoiesis in vivo resolved by mtDNA based tracing.
(a) Assignment probabilities (%, colorbar) of scRNA-seq data derived transfer labels (rows) across mtscATAC-seq derived Louvian data clusters (columns) as identified in Fig. 6d. (b) Distribution of percent mitochondrial reads derived from mtscATAC-seq data (y axis) across PBMC populations (x axis). (c) Percent mitochondrial counts (y axis) in FACS sorted populations (x axis) from bulk RNA-seq data. (d) Identification of high confidence variants from CD34+ HSPC and PBMC cell populations. Number of variants passing both thresholds (dotted lines) is indicated. A Venn diagram depicts the overlap of shared mutations. (e) Percent duplicates of sequenced mtDNA fragments, mean mtDNA coverage and percent mitochondrial reads for CD34+ HSPC and PBMC cell populations as derived from mtscATAC-seq data. Boxplots: center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. (f) Distribution of maximum level of heteroplasmy of mgatk derived variants from (d) in individual cells. (g) Unsupervised clustering of groups of cells based on shared somatic mtDNA mutations (y-axis) with corresponding individual mtDNA mutations (x-axis) associated with each cluster/clone. (h) Fold-change (observed over expected) of identified rare mutations (y axis) in each class of mononucleotide and trinucleotide change from the CD34+ HSPC data. (i) Comparison of pseudobulk allele frequencies from mgatk identified variants (blue) and rare variants (green). Boxplots for (b,c,e): center line, median; box limits, first and third quartiles; whiskers, 1.5x interquartile range. Bounds are contained within the data range shown. Sample sizes exceed 100 single cells from one experiment.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Erik Bao, Jacob Ulirsch, Evgenij Fiskin, and members of the Sankaran and Regev labs for helpful discussion. We acknowledge support from the Broad Institute and the Whitehead Institute Flow Cytometry core facilities. This research was supported by National Institutes of Health grants F31 CA232670 (C.A.L.), R01 CA208756 (N.H.), P01 CA206978 (C.J.W. and G.G.), U10 CA180861 (C.J.W.), R01 DK103794 (V.G.S.), and R33 HL120791 (V.G.S.), a gift from Arthur, Sandra, and Sarah Irving (N.H.), a gift from the Lodish Family to Boston Children’s Hospital (V.G.S.), the New York Stem Cell Foundation (NYSCF, V.G.S.), and the Howard Hughes Medical Institute and Klarman Cell Observatory (A.R.). S.G. is supported by funding from the Kay Kendall Leukaemia Fund. K.P. is supported by a research fellowship of the German Research Foundation (DFG) and a Stand Up To Cancer Peggy Prescott Early Career Scientist Award in Colorectal Cancer Research. G.G. is supported by the Paul C. Zamecnick chair. C.J.W. is a scholar of the Leukemia and Lymphoma Society. F.C. and J.D.B were supported by the Allen Distinguished Investigator Program. V.G.S. is a NYSCF-Robertson Investigator. We are grateful to the patients who made this work possible.
COMPETING INTERESTS
The Broad Institute has filed for a patent related to lineage tracing using mtDNA mutations where C.A.L., L.S.L., C.M., J.D.B., A.R., and V.G.S are named inventors. J.D.B. holds patents related to ATAC-seq. N.H. and C.J.W are co-founders, equity holders, and SAB members of Neon Therapeutics, Inc, and receive research funding from Pharmacyclics. G.G. receives research funding from IBM and Pharmacyclics. A.R. is a founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics, and an SAB member of Syros Pharmaceuticals, Neogene Therapeutics, Asimov, and ThermoFisher Scientific.
REFERENCES
- 1.Stewart JB & Chinnery PF The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat. Rev. Genet 16, 530–542 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Shoffner JM & Wallace DC Mitochondrial genetics: principles and practice. Am. J. Hum. Genet 51, 1179–1186 (1992). [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott HR, Samuels DC, Eden JA, Relton CL & Chinnery PF Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet 83, 254–260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris J et al. Pervasive within-Mitochondrion Single-Nucleotide Variant Heteroplasmy as Revealed by Single-Mitochondrion Sequencing. Cell Rep. 21, 2706–2713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang E et al. Age-Related Accumulation of Somatic Mitochondrial DNA Mutations in Adult-Derived Human iPSCs. Cell Stem Cell 18, 625–636 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ludwig LS et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell 176, 1325–1339.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J et al. Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodato MA et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lareau CA et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol 37, 916–924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satpathy AT et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross MG et al. Characterizing and measuring bias in sequence data. Genome Biol. 14, R51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green B, Bouchier C, Fairhead C, Craig NL & Cormack BP Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob. DNA 3, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dames S et al. The development of next-generation sequencing assays for the mitochondrial genome and 108 nuclear genes associated with mitochondrial disorders. J. Mol. Diagn 15, 526–534 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Wallace DC & Chalkia D Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol 5, a021220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc 10, 442–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S-P et al. Increased COUP-TFII expression in adult hearts induces mitochondrial dysfunction resulting in heart failure. Nat. Commun 6, 8245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zunino R, Schauss A, Rippstein P, Andrade-Navarro M & McBride HM The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci 120, 1178–1188 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Powell CA et al. TRMT5 Mutations Cause a Defect in Post-transcriptional Modification of Mitochondrial tRNA Associated with Multiple Respiratory-Chain Deficiencies. Am. J. Hum. Genet 97, 319–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kugeratski FG et al. Hypoxic cancer–associated fibroblasts increase NCBP2-AS2/HIAR to promote endothelial sprouting through enhanced VEGF signaling. Sci. Signal 12, eaan8247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brusco J & Haas K Interactions between mitochondria and the transcription factor myocyte enhancer factor 2 (MEF2) regulate neuronal structural and functional plasticity and metaplasticity. J. Physiol 593, 3471–3481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lott MT et al. mtDNA Variation and Analysis Using Mitomap and Mitomaster. Curr. Protoc. Bioinformatics 44, 1.23.1–26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohrson CL et al. Linked-read analysis identifies mutations in single-cell DNA-sequencing data. Nature Genetics vol. 51 749–754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zafar H, Wang Y, Nakhleh L, Navin N & Chen K Monovar: single-nucleotide variant detection in single cells. Nat. Methods 13, 505–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roos-Weil D et al. Mutational and cytogenetic analyses of 188 CLL patients with trisomy 12: A retrospective study from the French Innovative Leukemia Organization (FILO) working group. Genes, Chromosomes and Cancer vol. 57 533–540 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Izumi D et al. TIAM1 promotes chemoresistance and tumor invasiveness in colorectal cancer. Cell Death & Disease vol. 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofbauer SW et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood vol. 123 2181–2188 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Damm F et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 4, 1088–1101 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Kikushige Y et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 20, 246–259 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Alizadeh AA & Majeti R Surprise! HSC are aberrant in chronic lymphocytic leukemia. Cancer cell vol. 20 135–136 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Lee-Six H et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osorio FG et al. Somatic Mutations Reveal Lineage Relationships and Age-Related Mutagenesis in Human Hematopoiesis. Cell Rep. 25, 2308–2316.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granja JM et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol 37, 1458–1465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schep AN, Wu B, Buenrostro JD & Greenleaf WJ chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buenrostro JD et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 173, 1535–1548.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cusanovich DA et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 174, 1309–1324.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pliner HA et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi J et al. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 47, D780–D785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jovanovic M et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science 347, 1259038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju YS et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lareau CA, Ludwig LS & Sankaran VG Longitudinal assessment of clonal mosaicism in human hematopoiesis via mitochondrial mutation tracking. Blood Adv 3, 4161–4165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature vol. 548 456–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biasco L et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell 19, 107–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scala S et al. Dynamics of genetically engineered hematopoietic stem and progenitor cells after autologous transplantation in humans. Nat. Med. 24, 1683–1690 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Nam AS et al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 571, 355–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corral-Debrinski M et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics 23, 471–476 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Bender A et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet 38, 515–517 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Lee SR & Han J Mitochondrial Mutations in Cardiac Disorders. Adv. Exp. Med. Biol 982, 81–111 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Triska P et al. Landscape of germline and somatic mitochondrial DNA mutations in pediatric malignancies. Cancer Res. (2019) doi: 10.1158/0008-5472.CAN-18-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun N, Youle RJ & Finkel T The Mitochondrial Basis of Aging. Mol. Cell 61, 654–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS ONLY REFERENCES
- 57.Hu J et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giani FC et al. Targeted Application of Human Genetic Variation Can Improve Red Blood Cell Production from Stem Cells. Cell Stem Cell 18, 73–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang W, Li L, Myers JR & Marth GT ART: a next-generation sequencing read simulator. Bioinformatics 28, 593–594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lareau CA, Ma S, Duarte FM & Buenrostro JD Inference and effects of barcode multiplets in droplet-based single-cell assays. Nat. Commun 11, 866 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lander ES & Waterman MS Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2, 231–239 (1988). [DOI] [PubMed] [Google Scholar]
- 63.Chen F, Tillberg PW & Boyden ES Optical imaging. Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Dekken H, Pinkel D, Mullikin J & Gray JW Enzymatic production of single-stranded DNA as a target for fluorescence in situ hybridization. Chromosoma 97, 1–5 (1988). [DOI] [PubMed] [Google Scholar]
- 65.Larsson C et al. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat. Methods 1, 227–232 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Schwartz S, Oren R & Ast G Detection and removal of biases in the analysis of next-generation sequencing reads. PLoS One 6, e16685 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garrison E & Marth G Haplotype-based variant detection from short-read sequencing. arXiv [q-bio.GN] (2012). [Google Scholar]
- 69.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol (2018) doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this work is available at GEO accession GSE142745.