Abstract
Introduction
Stromal cell derived factor-1a (SDF-1a) and its receptor CXCR4 modulate stem cell recruitment to neural injury sites. SDF-1a gradients originating from injury sites contribute to chemotactic cellular recruitment. To capitalize on this injury-induced cell recruitment, further investigation of SDF-1a/CXCR4 signaling dynamics are warranted. Here, we studied how exogenous SDF-1a delivery strategies impact spatiotemporal SDF-1a levels and the role autocrine/paracrine signaling plays.
Methods
We first assessed total SDF-1a and CXCR4 levels over the course of 7 days following intracortical injection of either bolus SDF-1a or SDF-1a loaded nanoparticles in CXCR4-EGFP mice. We then investigated cellular contributors to SDF-1a autocrine/paracrine signaling via time course in vitro measurements of SDF-1a and CXCR4 gene expression following exogenous SDF-1a application. Lastly, we created mathematical models that could recapitulate our in vivo observations.
Results
In vivo, we found sustained total SDF-1a levels beyond 3 days post injection, indicating endogenous SDF-1a production. We confirmed in vitro that microglia, astrocytes, and brain endothelial cells significantly change SDF-1a and CXCR4 expression after exposure. We found that diffusion-only based mathematical models were unable to capture in vivo SDF-1a spatial distribution. Adding autocrine/paracrine mechanisms to the model allowed for SDF-1a temporal trends to be modeled accurately, indicating it plays an essential role in SDF-1a sustainment.
Conclusions
We conclude that autocrine/paracrine dynamics play a role in endogenous SDF-1a levels in the brain following exogenous delivery. Implementation of these dynamics are necessary to improving SDF-1a delivery strategies. Further, mathematical models introduced here may be utilized in predicting future outcomes based upon new biomaterial designs.
Electronic supplementary material
The online version of this article (doi:10.1007/s12195-020-00643-y) contains supplementary material, which is available to authorized users.
Keywords: CXCL12, CXCR4, Chemokines, Modeling
Introduction
Stromal cell-derived factor-1a (SDF-1a)/chemokine receptor 4 (CXCR4) is a chemokine/receptor pair responsible for the migration of various cell types.8,25,33 SDF-1a induces migration by binding to its receptor CXCR4. Cells detect gradients of SDF-1a through spatial and temporal sensing of occupied receptors thereby guiding cells along the gradient.10 For example, neural stem cells (NSCs) migrate large distances towards locally secreted SDF-1a in vivo following multiple sclerosis inflammation, hypoxic-ischemic cerebral injury, and traumatic brain injury.5,11,12 However, injury-induced migration is transient where the number of immature cells present in the lesion area drops off around 14 days in a TBI model.38 We and others are interested in exploiting this signaling cascade via controlled release biomaterial devices to modulate/tune NSC recruitment for maximal regenerative capacity. The mechanism that the SDF-1a/CXCR4 cascade guides cells across large distances is not fully understood, particularly considering that the half-life of SDF-1a is 26 min.15 Therefore, understanding how SDF-1a chemotactic gradients are created in complex tissue microenvironments is critical for successful drug carrier designs.
One mechanism cells use to generate far-reaching chemotactic gradients is autocrine/paracrine signaling.3,9,14,23 This cell signaling loop stimulates cells to secrete additional signal locally and to pass that signal along to neighboring cells who in turn engage in the same process. SDF-1a/CXCR4 autocrine/paracrine signaling contributes to the long-distance migration of numerous cell types outside the central nervous system. For example, metastatic cancer cell migration of skin, breast, and ovarian cancers are dependent upon SDF-1a/CXCR4 autocrine/paracrine signaling.3,14,23 Further, studies are beginning to elucidate the role of SDF-1a autocrine/paracrine signaling in the peripheral nervous system. A recent study by Gao et al. showed that the SDF-1a/CXCR4 autocrine loop promotes Schwann cell migration in vitro.9 Specifically, SDF-1a administration led to significantly increased Schwann cell migration and upregulation of SDF-1a compared to non-treated controls.9 Therefore, SDF-1a/CXCR4 may act in a similar autocrine/paracrine fashion in the central nervous system to establish chemotactic gradients that contribute NSC migration.
We hypothesize that autocrine/paracrine signaling is a critical component of SDF-1a gradient formation in the brain. We used in vivo and in vitro signaling assays in combination with in silico modeling to elucidate and model the kinetics of SDF-1a propagation following exogenous SDF-1a delivery. In doing so, we may better exploit SDF-1a signal dynamics to control cell recruitment. We addressed our hypothesis in three ways: (1) by comparing different SDF-1a delivery strategies in vivo via bolus injection or sustained release nanoparticles (NPs) over 7 days, (2) by investigating SDF-1a autocrine/paracrine signaling in vitro through exogenous application of SDF-1a on neural cell cultures and measuring changes in gene expression, (3) by creating a mathematical model to study SDF-1a spatiotemporal dynamics using COMSOL Multiphysics®.
Materials and Methods
Intracortical Injection of AFSDF-1a and AFSDF-1a Loaded NPs
We fabricated poly(lactic-co-glycolic) acid NPs to deliver SDF-1a using a previously established water/oil/water emulsion technique.6 Recombinant SDF-1a conjugated with AlexaFluor647 (AFSDF-1a; Almac, Craigavon, UK) was used for exogenous SDF-1a delivery to distinguish between delivered SDF-1a and total SDF-1a levels. PLGA NPs were fabricated to release approximately 30 ng of AFSDF-1a in the first 24 h followed by 9 ng over the next 6 days. To best match this expected release, the bolus injections contained 30 ng of AFSDF-1a. All animal procedures were approved by Arizona State University’s Institute of Animal Use and Care Committee (IACUC) with previously published protocols.7 Briefly, 3 μL injections were performed centered over 1.5 mm anterior of bregma and 1.5 mm lateral of midline at a depth of 0.8 mm into the cortical tissue of adult CXCR4-EGFP transgenic mice, n = 4–5 animals (kindly donated by Dr. Richard Miller).7
Immunohistochemistry and Image Processing
Immunohistochemistry was used to detect total SDF-1a levels following 1, 3, and 7 days post-injection as previously described.7 Acute time points were selected based upon both the 26 min half-life of SDF-1a and the NP burst release of SDF-1a occurring within 24 h. Animals were euthanized and perfused following specified time points with cold phosphate buffer followed by 4% paraformaldehyde. Brains were extracted and fixed in 4% paraformaldehyde followed by incubation in 30% sucrose. Brains were embedded in OCT and stored at – 80 °C until cryo-sectioning was performed. 25 μm sections were blocked, permeabilized, and incubated with rabbit polyclonal anti-SDF-1a (Abcam, Cambridge, MA) overnight at 4°C followed by a 2 h incubation with goat anti-Rabbit IgG Alexa Fluor555 (Thermo Fisher Scientific, Waltham, MA) at room temperature. Stained sections were visualized using fluorescence microscopy (DMI6000B, Leica). Tile scans of each brain slice were prepared for further analysis with consistent acquisition settings. ImageJ (National Institutes of Health, Bethesda, MD) was used to quantify the percent immunopositive area of total SDF-1a (exogenous + endogenous) surrounding the injection. The region of interest quantified was 1200 μm on each side of the injection by 1200 μm deep. Each image was thresholded using the tissue’s contralateral signal, and threshold values were used to calculate the area fraction of SDF-1a positive stain. For each group/time point, 4–5 animals with 3–5 tissue sections per animal were quantified.
Primary Cell Cultures
Primary glial cell cultures were obtained based on established protocols and in accordance with Arizona State University’s IACUC approved protocol.22,27 Neonatal CXCR4-EGFP mice (aged P0–P2) were collected, anesthetized, and sacrificed via rapid decapitation. Tail clips were obtained for genotyping. Brains were dissected out in cold Hank’s Balanced Salt Solution (HBSS) using super fine forceps and micro dissecting knives. The meninges, cerebellum, and midbrain were removed and both hemispheres from each pup were placed in fresh HBSS at 4 °C for temporary storage. Briefly, each tube containing dissected brain hemispheres was triturated 3 times and centrifuged. Supernatant was aspirated and 0.25% Trypsin (0.1% EDTA) was added to each tube and incubated at 37 °C for 20 min. Equal amounts growth media (Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12) with 10% Fetal bovine serum (FBS)) were added to neutralize suspensions. Suspensions were then washed, DNAse was added, and tubes were vortexed briefly to break up any large tissue fragments. Cells were washed 2 more times and resuspended in fresh growth media. Suspensions were filtered through a 70 μm cell strainer and then plated at a concentration of 1 T75 flask per hemisphere. Mixed glial cultures were grown for 3 days before receiving a media change and then allowed to grow for 11 more days without a media change to enrich microglia in the cultures. On day 14, cultures were harvested with trypsin and magnetic bead separation was performed according to manufacturer’s instructions for either CD11b (microglia) or Glast (astrocytes) (Miltenyi Biotec, Bergisch Gladbach, Germany).
Reverse Transcription Quantitative Real Time Polymerase Chain Reaction (RT-qPCR)
Sorted primary microglia and astrocyte cultures as well as a bEnd.3 cell line (ATCC CRL-2299, kindly donated by Dr. Rachel Sirianni) were used to evaluate the effects of exogenous SDF-1a application on expression of SDF-1a and CXCR4. Each cell type was plated in a 6-well plate, n = 3–6 for both control and experimental samples for each time point. Exogenous murine recombinant SDF-1a (Peprotech, Rocky Hill, NJ) treated cultures were exposed at 400 ng/mL for 30 min intervals up to 120 min. Control cultures were not exposed to SDF-1a. At each time interval after SDF-1a application, media was removed and cells were washed before performing RNA isolation using the RNeasy Plus Kit (Qiagen, Hilden, Germany). Isolated RNA was tested for quality using the Agilent Bioanalyzer and RNA concentration was determined using the Thermo Fisher Scientific Nanodrop. RNA was converted to cDNA (Bio-rad S1000 thermo cycler) using the SuperScript™ IV VILO™ Master Mix (Invitrogen, Carlsbad, CA) and included a reverse transcriptase (RT) negative control. Primers for SDF-1a (forward: 5′-CGCCAGAGCCAACGTCAAGC-3′, reverse: 5′-TTCGGGTCAATGCACACTTG-3′), CXCR4 (forward: 5′-CGGTACCTCGCTATTGTCCA-3′, reverse: 5′-CTGTCATCCCCCTGACTGA-3′), and GAPDH (reference gene, forward: 5′-AATGTGTCCGTCGTGGATCTG-3′, reverse: 5′-CAACCTGGTCCTCAGTGTAGC-3′) were designed using the National Center for Biotechnology Information (NCBI) Primer Blast tool and purchased as custom oligonucleotides (IDT, Newark, NJ). The qPCR reaction was carried out using Luminaris Color HiGreen qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA) on the Analytik Jena Qtower 2.0. The thermal cycling protocol consisted of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s (40 cycles). Genomic DNA contamination was assessed by using a RT negative control, where acceptable samples had at least 10 quantification cycles (Cq) smaller than the RT negative sample. Primer efficiency was calculated by creating a standard curve. To account for variations in primer efficiencies, the Pfaffl method was used to quantify relative expression ratios between cultures ± SDF-1a.26 Specificity of the primers was confirmed by running PCR products against a ladder on gels and by confirming single products via the melting profile.
Mathematical Modeling
Two principle models were created using COMSOL Multiphysics software to reproduce the spatiotemporal patterns of total SDF-1a and CXCR4 from the immunohistochemistry experiments. All models contained three equations describing the mass transport of 3 dilute species: soluble SDF-1a, SDF-1a/CXCR4 complexes, and unbound CXCR4. This transport is governed by the following mass balance equation:
| 1 |
where ∂ci/t represents the accumulation of the species. The diffusive transport of the species is represented by where Di is the diffusion coefficient and ci is the concentration of the species. The last term needed is the reaction term, Ri, which is used to model the rate of production and removal of the species. All models used the same 2D geometry that represented a tissue section with a centered injection site. The first model was based upon diffusion only, which assumed that no autocrine/paracrine signaling was taking place. The second model incorporated production of SDF-1a by cells; the production rate was modeled using Michaelis-Menten kinetics so that SDF-1a production was linearly proportional to the concentration of SDF-1a/CXCR4 complexes when there are few complexes but there is a maximum SDF-1a production rate when there are many complexes. This method has been previously established for modeling autocrine signaling.29 The reaction term for soluble SDF-1a included this SDF-1a induced production and loss of soluble SDF-1a due to degradation. The reaction term of unbound CXCR4 included a constant basal production rate, an endocytosis rate, association with SDF-1a (as a loss term because then this becomes a complex rather than unbound), and an additional production rate modeled in Michaelis–Menten form based on the number of complexes. The reaction term of SDF-1a/CXCR4 complexes included association of soluble SDF-1a and unbound CXCR4 and loss of complex (to receptor-mediated endocytosis). A detailed outline of model equations and terms can be found in Appendix A. A corresponding parameter list was generated (Table 1) that includes transport properties, Michaelis–Menten constants, and baseline concentration values. For each of the diffusion and autocrine models, 2 sub-models were created: bolus injected SDF-1a and SDF-1a loaded NPs (Fig S2). The NP model was created by fitting SDF-1a release data to the Korsmeyer–Peppas equation and taking the derivative to provide the rate of change in SDF-1a over time (Fig S3).17 All models were computed over the course of 7 days to compare against the experimental data. Cross-section plots were created to visualize the amount of total soluble SDF-1a present over time as computed by each model. These plots were compared against the immunohistochemistry trends.
Table 1.
COMSOL parameters.
| Name | Value | Description |
|---|---|---|
| Vmax | 1e−9[mol/(s*m3)] | Max reaction rate, soluble SDF-1a |
| Km | 1e−10[mol/m3] | Substrate concentration for half-maximal SDF-1a reaction |
| Dc | 1e−11[m2/s]13 | Diffusion coefficient, SDF-1a |
| kdeg | 0.000406[1/s] | Degradation rate of SDF-1a |
| ka | 50[1/((mol/m3)*s)] | Association rate constant, SDF-1a with CXCR4 |
| ke | 0.00105[1/s]36 | Endocytosis rate constant of complex |
| Vr | 0.35e−12[mol/(s*m3)] | Max reaction rate, CXCR4 |
| Kr | 1e−10[mol/m3] | Substrate concentration for half-maximal CXCR4 Reaction |
| Vrc | 1e−16[mol/(s*m3)] | Baseline production rate of CXCR4 |
| crt | 1e−9[mol/(m3)] | Initial concentration of CXCR4 |
| blc | 1e−6[mol/(m3)] | Baseline concentration of soluble SDF-1a |
Sensitivity Analysis
To examine model sensitivity, we adjusted parameter values for all four models and visualized the output plots for SDF-1a (diffusion and autocrine models) and CXCR4 (autocrine models only). Increases/decreases were made to each parameter value, and results were plotted alongside the results using the original parameter value. Lines representing a 10% increase and 10% decrease in the baseline results were plotted to visualize if the tested parameter values were within a 10% tolerance of the original data. This process was repeated until the changed parameter resulted in plots that fell within 10% of the standard results. Exceptions were used when baseline values were less than 10−13 mol/m3. These values were considered zero and any plots where values fell below this due to parameter changes were also considered zero and acceptable. Amount of change in each parameter value was recorded for each model.
Statistical Analysis
All quantitative results were analyzed using GraphPad Prism software Version 8.4.3 (GraphPad Software, Inc., La Jolla, CA). Statistical analysis of differences between groups were performed by one-way analysis of variance with Tukey post-hoc testing where p < 0.05 was considered significant. Logarithmic transformation was performed on the SDF-1a immunopositivity and CXCR4+ cell density data prior to one-way analysis of variance with Tukey post-hoc testing (Fig. 3). All data were graphed as the mean ± standard deviation or standard error of the mean.
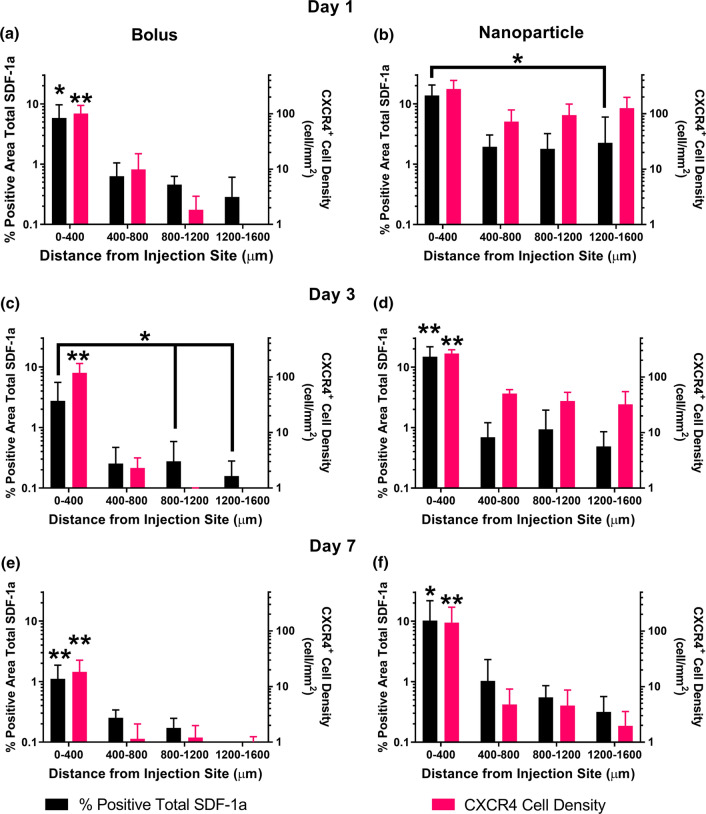
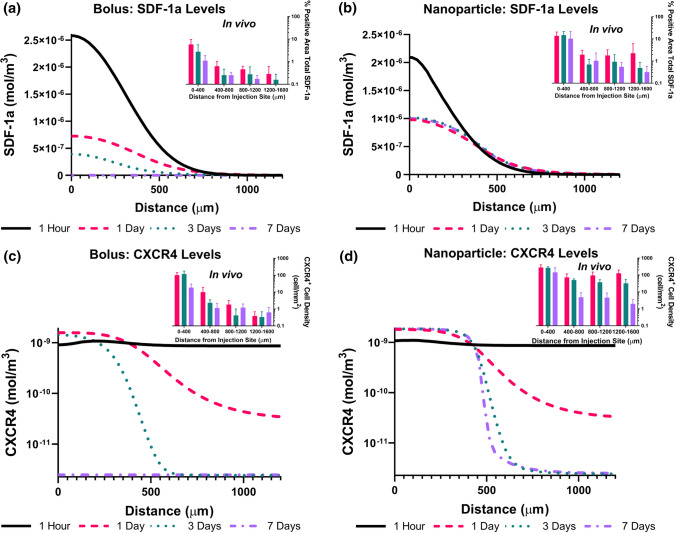
Figure 3.
Spatial trends of total SDF-1a (black) immunostaining and CXCR4 (pink) cell density 1, 3, and 7 days post injection. Day 1 and day 3 bolus (a, c) spatial distribution of SDF-1a and CXCR4 declines steadily across the tissue. Day 1 and day 3 NP (b, d) spatial distribution reveals sustained levels of SDF-1a and CXCR4 across 1600 μm of tissue, suggesting production of SDF-1a must be occurring endogenously. Spatial levels diminish on day 7 for the bolus group (e) and remain persistent immediate to the injection for the NP group (f). Mean ± SEM. n = 4–5 animals per group. Single asterisks above error bars indicate significance compared to all groups within data set where *p < 0.05, **p < 0.01.
Results
Sustained SDF-1 Immunostaining Suggests Autocrine/Paracrine Signaling
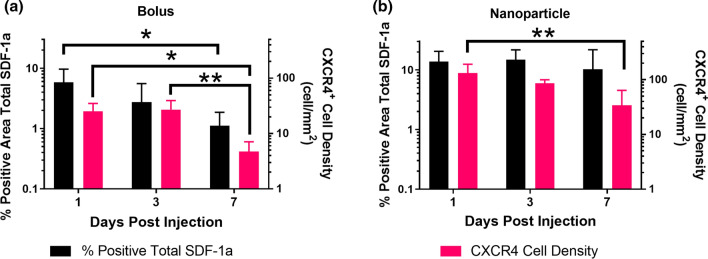
We first studied the impact of exogenous SDF-1a delivery on local SDF-1a and CXCR4 levels via bolus injection or injection of SDF-1a loaded NPs over 7 days, first reported in Dutta et al.7 We utilized fluorescent labeled exogenous SDF-1a and compared against total SDF-1a immunostaining to uncover differences that may be attributed to autocrine/paracrine signaling. Here, we focused on the temporal and spatial trends of total SDF-1a quantification and CXCR4+ cell density following exogenous delivery first identified in our prior publication.7 Visually, we reported higher levels of total SDF-1a immunostaining and CXCR4+ cell density in the NP group on day 1 than the bolus group (Fig. 1). Additionally, the pattern of immunostaining for the NP group on day 1 showed sustained levels across the tissue sections. Quantitation of both total SDF-1a and CXCR4+ surrounding the injection site showed distinct temporal trends between the bolus injected SDF-1a and NP delivered SDF-1a. Levels of total SDF-1a immunopositivity were highest at day 1 and significantly decreased between day 1 and day 7 following bolus SDF-1a delivery (Fig. 2a, p = 0.0469). In contrast, more total SDF-1a was detected in the NP group across all time points and did not exhibit any significant changes over 7 days (Fig. 2b). Likewise, CXCR4+ cell density was higher in the NP group than the bolus group, suggesting that the presentation of SDF-1a impacts cell response and potential recruitment (Fig. 2). Specifically, significant decreases in CXCR4+ cell density occurred between 1 and 7 days and 3 and 7 days in the bolus group (p = 0.0115 and p = 0.0067, respectively) and between 1 and 7 days in the NP group (p = 0.0052). We then analyzed the total SDF-1a immunopositivity and CXCR4+ density spatially across the 7 days. On day 1, the NP group exhibited near constant levels of both SDF-1a and CXCR4+ density out to 1600 μm from the injection site, where the only statistical difference found was between 0–400 and 1200–1600 μm for SDF-1a (Fig. 3b, p < 0.05). In contrast, the bolus group immunostaining and cell density declined across the tissue on day 1 with significant differences between 0–400 μm and all subsequent tissue sections (Fig. 3a, p < 0.05). On day 3, the trends remained the same for both groups where the NP group remained higher and more sustained in both SDF-1a immunostaining and CXCR4+ cell density (Figs. 3c and 3d). By day 7 the SDF-1a area and CXCR4+ cell density diminished for the bolus group (Fig. 3e). However, the NP group exhibited sustained levels to that of day 1 and day 3 immediate to the injection site (Fig. 3f). Both the bolus and NP group exhibited significant differences in immunostaining and cell density between the 0–400 μm region and all other regions (p < 0.05). In summary, the NP group showed spatially constant levels of SDF-1a and CXCR4+ cell density on days 1 and 3 whereas the bolus group exhibited a decline.
Figure 1.
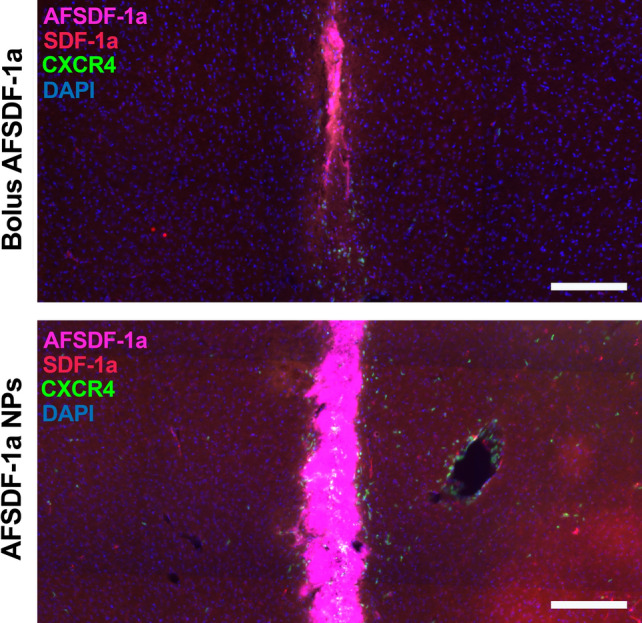
Spatial distribution of total SDF-1a 1 day after exogenous delivery. Representative cortical tissue sections from bolus delivered AFSDF-1a (top) and AFSDF-1a loaded NPs (bottom) stained for cell nuclei (DAPI, blue) and total SDF-1a (red), with EGFP-CXCR4 (green) and exogenous AFSDF-1a (pink). Increased levels of total SDF-1a immunostaining and CXCR4 are present visually across the cortex for the NP group. Immunostaining reveals steady levels of total SDF-1a across the cortex for the NP group in contrast to the spatially decreasing pattern for the bolus group. Consistent spatial levels of total SDF-1a suggest autocrine/paracrine signaling is occurring. Figure is adapted from Dutta et al. with copyright permissions.7 Scale bar = 200 μm.
Figure 2.
Temporal trends of total SDF-1a immunostaining and CXCR4 cell density 1, 3, and 7 days post injection. Total SDF-1a (black) and CXCR4 (pink) immunopositivity were persistent across 7 days for the NP group (b) and gradually declined over 7 days for the bolus group (a). Overall sustainment of total SDF-1a immunopositivity and CXCR4 activity across the 7 day time window suggests autocrine/paracrine signaling. Mean ± SEM. n = 4–5 animals per group. *p < 0.05, **p < 0.01.
Brain Microglia, Astrocytes and Endothelial Cells Respond Dynamically to Exogenous SDF-1a In Vitro
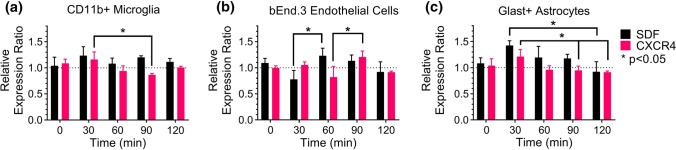
To determine whether autocrine/paracrine signaling is induced by SDF-1a in the brain we examined microglia, endothelial cells, and astrocytes response to exogenous SDF-1a. Each cell type responded in a unique, dynamic manner to exogenous SDF-1a stimulation. Microglia expressed higher levels of SDF-1a over the time course but never reached statistical significance perturbation (Fig. 4a). Microglia CXCR4 gene expression was significantly different between 30 and 90 min, suggesting its role in the autocrine/paracrine loop. In comparison to the upregulation of SDF-1a in microglia at 30 min, brain endothelial cell upregulation of SDF-1a occurred later at 60 min and was significantly higher than the 30 min level (Fig. 4b). Endothelial cells exhibited the largest reduction in expression of CXCR4 amongst all cell types at 60 min, which was significantly lower than expression levels at 90 min. This result suggests endothelial cells may be more sensitive to ligand-induced desensitization. Astrocytes exhibited the largest changes in gene expression amongst the three cell types (Fig. 4c). SDF-1a expression increased at 30 min compared to baseline and subsequently significantly decreased by 120 min. CXCR4 expression mirrored this trend with increased expression at 30 min then significantly declined by 90 and 120 min. Collectively, each of the cell types increased SDF-1a expression in response to exogenous SDF-1a, demonstrating participation in autocrine/paracrine signaling. Increases in SDF-1a expression coincided with changes in CXCR4 expression, confirming CXCR4 is part of the autocrine/paracrine loop. Suggestion of both ligand induced activation and delayed negative feedback of CXCR4 were observed across all cell types.
Figure 4.
Microglia (a), brain endothelial cells (b), and astrocytes (c) engage autocrine/paracrine signaling via dynamic response to exogenous SDF-1a. Significant changes in SDF-1a (black) and/or CXCR4 (pink) mRNA expression were found across all cell types. Changes in expression are dynamic and distinct over time for each cell type. n = 3–6 cultures per cell type. Mean ± SEM. *p < 0.05.
In Silico Model of Diffusion only Predicts Undetectable SDF-1a After 1 h
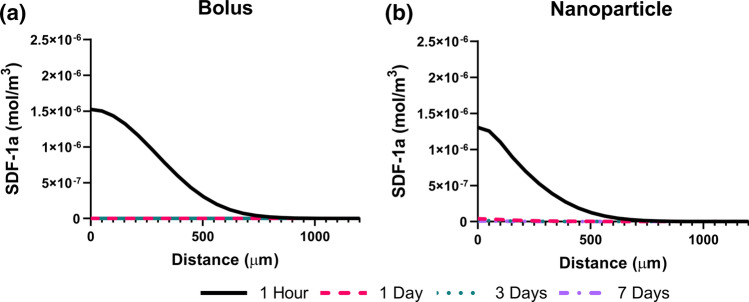
We next created mathematical models that were representative of the SDF-1a temporal trends observed in vivo. The first models included diffusion only kinetics and could not recapitulate sustained levels of SDF-1a over the period of days. Instead, both the bolus delivery and NP delivery model predicted undetectable levels of SDF-1a for after the 1-h time period (Figs. 5a and 5b). The SDF-1a concentration profiles depicted by the diffusion only models are consistent with the very short half-life of SDF-1a reported in the literature.
Figure 5.
Diffusion only model predicts undetectable SDF-1a after 1 hour. Diffusion-only model concentration plots of soluble SDF-1a for bolus (a) and NP (b) delivered SDF-1a across 1 hour, 1 day, 3 days, and 7 days. Arc length (X-axis) represents equidistant space from the injection. Separate colors/line styles used to distinguish between time points (1 hour, 1 day, 3 days, 7 days). Lack of concentration plot lines indicates undetectable soluble SDF-1a present at corresponding time points. Both bolus (a) and NP (b) trends are not consistent with in vivo data, suggesting another mechanism is required to sustain SDF-1a signal spatiotemporally.
In Silico Model with Autocrine/Paracrine Signaling Predicts Persistent SDF-1a Levels Over Time
Our in vitro and in vivo analyses indicated presence of autocrine/paracrine signaling. Therefore, we developed models that included autocrine/paracrine signaling kinetics. By fitting values for Michaelis–Menten reaction rates and constants, we created a model for bolus delivery of SDF-1a that was consistent with our in vivo results. The bolus delivered SDF-1a model with autocrine/paracrine kinetics showed SDF-1a levels sustained for 3 days like that of our immunohistochemistry results (Fig. 6a). In the same fashion, our autocrine/paracrine model of NP delivered SDF-1a resulted in SDF-1a levels sustained for 7 days (Fig. 6b). Further, the overall trends showed that bolus delivered SDF-1a gradually decreased over the 7 days whereas the NP delivered SDF-1a remained at the same level over the 7 days, like that of the in vivo trends. We also examined our models against the in vivo CXCR4 data. For the bolus CXCR4 data the model accurately represented the in vivo trends where CXCR4 activation occurs out to 500 μm at day 1 and decreases one-tenth by 1000 μm (Fig. 6c). Day 3 was also represented where CXCR4 levels drop off after 300 μm. Day 7 was not predicted by our model as the in vivo data show CXCR4 activity near the injection but our model predicts a lack of activity. For the NP group, the CXCR4 trends resulting from our model at day 1 appear like the bolus model, where CXCR4 declined around 500 μm (Fig. 6d). However, the in vivo data showed sustained levels further from the injection which was not predicted. The model also calculated that CXCR4 levels will drop off around 500 μm from the injection at both day 3 and day 7. This prediction matched day 7 in vivo data, but under predicted day 3. Overall our models predicted the in vivo observations for both SDF-1a and CXCR4 to a degree that discerned differences between the exogenous delivery methods.
Figure 6.
Autocrine/paracrine model replicates in vivo results. Autocrine/paracrine model concentration plots of soluble SDF-1a for bolus (a, c) and NP (b, d) delivered SDF-1a across 1 hour (black), 1 day (pink), 3 days (teal), and 7 days (purple). Arc length (X-axis) represents equidistant space from the injection. Addition of autocrine/paracrine kinetics flattens concentration curves of both SDF-1a and CXCR4, yielding spatiotemporal sustainment and matching most in vivo results.
Parameter Sensitivity
To test the robustness of each of our models, parameter sensitivity was performed. Parameter sensitivity analysis measured maximal changes in parameter values that did not affect the outcome significantly (± 10% of original results). As expected, the diffusion only models were not impacted by changes in parameters km, ka, ke, Vr, kr, Vrc, and crt since upstream signaling parameter Vmax was set to zero for these models (Table 2). Further, the diffusion only models allowed for larger changes in the diffusion coefficient, degradation rate, and delivered amount of SDF-1a than the autocrine models. This can be explained by the fine-tuned dynamics of autocrine systems.29 The autocrine models were the most sensitive to changes in soluble SDF-1a degradation and complex endocytosis, where rate adjustments of less than ± 0.005% would significantly impact the model predictions. Likewise, this is representative of the highly regulated nature of autocrine/paracrine biological systems. All other parameters could be adjusted between ± 0.005% and 5% without changing the resulting plots ± 10%.
Table 2.
Parameter sensitivity values.
| Parameter | Original | Bolus autocrine (%) | NP autocrine (%) | Bolus diffusion | NP diffusion |
|---|---|---|---|---|---|
| Vmax | 1.00E−09 | 0.004 | 0.004 | N/A* | N/A* |
| km | 1.00E−10 | 0.01 | 0.01 | No impact** | No impact** |
| Dc | 1.00E−11 | 0.10 | 0.10 | 10.00% | 10.00% |
| kdeg | 0.000406 | 0.002 | 0.002 | 5.00% | 5.00% |
| ka | 50 | 0.008 | 0.008 | No impact** | No impact** |
| ke | 0.00105 | 0.005 | 0.005 | No impact** | No impact** |
| Vr | 3.50E−13 | 0.006 | 0.006 | No impact** | No impact** |
| kr | 1.00E−10 | 0.009 | 0.009 | No impact** | No impact** |
| vrc | 1.00E−16 | 5.00 | 5.00 | No impact** | No impact** |
| crt | 1.00E−09 | 0.01 | 0.01 | No impact** | No impact** |
| blc | 1.00E−06 | 0.10 | 0.10 | 5.00% | 5.00% |
*Parameter is set to zero for model
**Parameter is multiplied by zero or dependent on unchanging parameter
Discussion
The results of our previously published in vivo study demonstrated that sustained exogenous SDF-1a therapy elicits an endogenous SDF-1a response.7 Considering the 26-min half-life of SDF-1a, the immunohistochemistry results analyzed here suggests autocrine/paracrine signaling prolongs SDF-1a presence following exogenous administration. Further, we showed that sustained release of exogenous SDF-1a from degradable NPs engages autocrine/paracrine signaling longer resulting in higher, constant levels of SDF-1a across the cortex compared to bolus delivery. The differences between these two delivery strategies indicated that the presentation of SDF-1a is critical to the duration of endogenous signaling and therapeutic potential. These results align with other studies where controlled release of SDF-1a prolonged SDF-1a gradients and subsequently stem cell recruitment.19
We demonstrated that autocrine/paracrine signaling contributes to establishing SDF-1a gradients. Microglia, astrocytes, and endothelial cells significantly modulate both endogenous SDF-1a and CXCR4 levels as a result of exogenous administration. These results were consistent to SDF-1a/CXCR4 signaling in cancer progression where SDF-1a autocrine loops contribute to cell migration and invasion.14,16,23 Our identification of cell types engaging SDF-1a/CXCR4 autocrine/paracrine signaling in the brain justify the need to further utilize this cascade as a tool for repair. SDF-1a is a known stimulator of astrocyte proliferation.1,4 A recent study by Mao et al. suggests that SDF-1a delivery increases radial glial cell proliferation following traumatic brain injury and that these cells may act as a cellular scaffold to aid in the migration of immature neurons.21 Here, we are the first, to our knowledge, to confirm that astrocytes participate in SDF-1a/CXCR4 autocrine/paracrine signaling that may be tuned for maximal endogenous repair. Future studies should investigate ways to best modify this signaling loop for increased cell recruitment. If we can further understand SDF-1a/CXCR4 gene regulation, we may design a better delivery strategy to increase spatiotemporal presence of SDF-1a for enhanced signaling and repair.
While our results show CXCR4 cell density and gene expression coinciding with changes in SDF-1a, we cannot neglect the potential implications of CXCR7. CXCR7 was first identified as a cognate receptor to SDF-1a in T lymphocytes 15 years ago.2 While relatively new, research on the role of CXCR7 in the brain is being established. Liu et al. found CXCR7 is a functional receptor for SDF-1a in brain endothelial cells and modulates their migration in vitro.20 Therefore, it is possible that the changes we found in SDF-1a expression were due in part to CXCR7. Moreover, CXCR7 can also be a negative regulator for SDF-1a where CXCR7 activation reduces levels of CXCR4 and downstream SDF-1a induced cellular events.24,35 The extent to which CXCR7 impacts spatiotemporal levels of SDF-1a in the brain and its role in SDF-1a autocrine/paracrine dynamics requires further investigation.
To validate our findings and create a representative model of SDF-1a levels following exogenous SDF-1a delivery, we evaluated two different mathematical model types. We found that the model that only incorporated diffusion was not able to reproduce the spatiotemporal trends observed in vivo. This result suggests that diffusion only is not responsible for sustained gradients of SDF-1a and there must be another mechanism involved. The lack of sustained gradients based on diffusion only is predictable based upon the short half-life of SDF-1a and transport via diffusion. As suggested by our gene expression results, inclusion of autocrine/paracrine kinetics in the models were required to achieve the same SDF-1a levels observed in vivo. Specifically, constant levels of SDF-1a across large sections of tissue were indicative of endogenous SDF-1a production. The only way constant levels could be achieved is through production of the molecule in conjunction with degradation and diffusion. Adding molecule production to our models allowed for SDF-1a curves to flatten (decreased slope), which would not be possible in a diffusion-only model where SDF-1a levels could only decline over time at a steady rate. SDF-1a constant levels were demonstrated by adding autocrine/paracrine kinetics to our models where flat curves of SDF-1a and CXCR4 were achieved.
We found that the parameters for soluble SDF-1a degradation rate constant and the SDF-1a/CXCR4 complex endocytosis rate constant were sensitive to less than ± 0.005% change. This behavior seems to be a product of the cellular machinery rather than model limitations as cells are known to tightly regulate chemokine signaling through endocytosis.31,32 In the same fashion, others have noted that epidermal growth factor receptor (EGFR) system signaling requires threshold amounts of ligand to trigger downstream signaling, yet, EGFR overstimulation shuts off signal propagation through endocytic control.30 If a similar phenomenon is at play for SDF-1a/CXCR4, one can postulate that if the rates of SDF-1a degradation or receptor endocytosis could be physically adjusted then autocrine/paracrine signals could be reduced or extended experimentally.
Current SDF-1a release systems focus primarily on extending the overall duration of exogenous SDF-1a activity. Sustained release platforms such as hydrogels, scaffolds, particles, and particle composite systems have been utilized to extend SDF-1a release for cell recruitment.18,28,34,37 Sustained release platforms are limited in their efficacy since they do not account for autocrine/paracrine engagement and desensitization. There are a lack of systems that work to alter the presentation of SDF-1a such that it is informed by the cellular machinery. Our model allows for experimentally testable predictions to be made and includes these dynamics. Our model results will inform next generation release systems by providing a platform to test SDF-1a release strategies for maximal SDF-1a/CXCR4 output and therefore cell recruitment.
Conclusion
The SDF-1a/CXCR4 signaling axis remains a promising tool for increasing cell recruitment following injury and disease. SDF-1a autocrine/paracrine kinetics and their impact on overall sustainment of SDF-1a was investigated here. We identified cell types that contribute to SDF-1a autocrine/paracrine signaling within the brain, the extent to which they impact SDF-1a levels and validated that this signaling is required to create sustained gradients for cellular migration. Knowing that autocrine/paracrine dynamics play a role in brain cell responses, the future of SDF-1a/CXCR4 based therapies should work to best control this signaling mechanism for specific outputs and improved therapies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Electronic supplementary material 1 (DOCX 2033 kb)
Acknowledgments
This work was supported by the NSF CBET 1454282 (SES) and NIH 1DP2HD084067 (SES). We thank Dr. Rachael Sirianni, Dr. Barbara Smith, Dr. Richard Miller, and Crystal Willingham for technical and materials support. We would like to acknowledge Brandon Neldner from the KE cores facilities at Arizona State University for assistance with technical setup and experimental design for the flow cytometry analysis. We also thank Scott Bingham from the Arizona State University DNA lab for assistance with RNA analysis.
Conflicts of interest
Kassondra N. Hickey declares that she has no conflict of interest. Shannon M. Grassi declares that she has no conflict of interest. Michael R. Caplan declares that he has no conflict of interest. Sarah E. Stabenfeldt declares that she has no conflict of interest. Sarah E. Stabenfeldt has received research grants NSF CBET 1454282 and NIH 1DP2HD084067.
Research Involving Animal Rights
All institutional and national guidelines for the care and use of laboratory animals were followed and approved by the appropriate institutional committees.
Research Involving Human Studies
No human subjects research was performed in this study.
Appendix A: Model Components
The tissue was modeled in 2D with a rectangular section of tissue. The injection sub-domain was a rectangle as tall as the tissue but only 200 μm wide and placed in the horizontal (x-direction) center of the tissue. For each transported species, the initial values and reaction terms varied depending upon the geometric location. Equations below include the terms and values for each location. Initial values and reaction terms for soluble SDF-1a transport varied upon delivery method while SDF-1a/CXCR4 complex and unbound CXCR4 transport remained constant between models. Therefore, separate values and terms for soluble SDF-1a transport are listed below for bolus delivery and NP delivery. To create the diffusion only models, maximum reaction velocity was set to zero for both soluble SDF-1a and unbound CXCR4 such that no downstream signaling could occur past SDF-1a and CXCR4 binding.
Soluble, Extracellular SDF-1a Transport for Bolus Delivery
| A1 |
| A2 |
| A3 |
Soluble, Extracellular SDF-1a Transport for Nanoparticle Delivery
| A4 |
| A5 |
| A6 |
SDF-1a/CXCR4 Complex Transport
| A7 |
| A8 |
Unbound CXCR4 Transport
| A9 |
| A10 |
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bajetto A, et al. Stromal cell-derived factor-1α induces astrocyte proliferation through the activation of extracellular signal-regulated kinases 1/2 pathway. J. Neurochem. 2001;77:1226–1236. doi: 10.1046/j.1471-4159.2001.00350.x. [DOI] [PubMed] [Google Scholar]
- 2.Balabanian K, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- 3.Bernat-Peguera A, et al. PDGFR-induced autocrine SDF-1 signaling in cancer cells promotes metastasis in advanced skin carcinoma. Oncogene. 2019;38:5021–5037. doi: 10.1038/s41388-019-0773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonavia R. Chemokines and their receptors in the CNS: expression of CXCL12/SDF-1 and CXCR4 and their role in astrocyte proliferation. Toxicol. Lett. 2003;139:181–189. doi: 10.1016/S0378-4274(02)00432-0. [DOI] [PubMed] [Google Scholar]
- 5.Carbajal KS, Schaumburg C, Strieter R, Kane J, Lane TE. Migration of engrafted neural stem cells is mediated by CXCL12 signaling through CXCR4 in a viral model of multiple sclerosis. PNAS. 2010;107:11068–11073. doi: 10.1073/pnas.1006375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dutta D, Fauer C, Mulleneux HL, Stabenfeldt SE. Tunable controlled release of bioactive SDF-1α via specific protein interactions within fibrin/nanoparticle composites. J. Mater. Chem. B. 2015;3:7963–7973. doi: 10.1039/C5TB00935A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta D, Hickey K, Salifu M, Fauer C, Willingham C, Stabenfeldt SE. Spatiotemporal presentation of exogenous SDF-1 with PLGA nanoparticles modulates SDF-1/CXCR4 signaling axis in the rodent cortex. Biomater. Sci. 2017;5:1640–1651. doi: 10.1039/C7BM00489C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dziembowska M, Tham TN, Lau P, Vitry S, Lazarini F, Dubois-Dalcq M. A role for CXCR4 signaling in survival and migration of neural and oligodendrocyte precursors. Glia. 2005;50:258–269. doi: 10.1002/glia.20170. [DOI] [PubMed] [Google Scholar]
- 9.Gao D, Sun H, Zhu J, Tang Y, Li S. CXCL12 induces migration of Schwann cells via p38 MAPK and autocrine of CXCL12 by the CXCR4 receptor. Int J Clin Exp Pathol. 2018;11:3119–3125. [PMC free article] [PubMed] [Google Scholar]
- 10.Iglesias, P. A. Dynamics of gradient sensing and chemotaxis. In: Encyclopedia of Cell Biology Elsevier, 2016, pp. 4–9. http://linkinghub.elsevier.com/retrieve/pii/B9780123944474400015. Accessed 24 Apr 2017.
- 11.Imitola J, et al. Directed migration of neural stem cells to sites of CNS injury by the stromal cell-derived factor 1α/CXC chemokine receptor 4 pathway. Proc. Natl. Acad. Sci. 2004;101:18117–18122. doi: 10.1073/pnas.0408258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh T, et al. The relationship between SDF-1/CXCR4 and neural stem cells appearing in damaged area after traumatic brain injury in rats. Neurol. Res. 2009;31:90–102. doi: 10.1179/174313208X332995. [DOI] [PubMed] [Google Scholar]
- 13.Jansma AL, Kirkpatrick JP, Hsu AR, Handel TM, Nietlispach D. NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27. J. Biol. Chem. 2010;285:14424–14437. doi: 10.1074/jbc.M109.091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang H, Mansel R, Jiang W. Genetic manipulation of stromal cell-derived factor-1 attests the pivotal role of the autocrine SDF-1-CXCR4 pathway in the aggressiveness of breast cancer cells. Int. J. Oncol. 2005 doi: 10.3892/ijo.26.5.1429. [DOI] [PubMed] [Google Scholar]
- 15.Kirkpatrick B, Nguyen L, Kondrikova G, Herberg S, Hill WD. Stability of human stromal-derived factor-1α (CXCL12α) after blood sampling. Ann. Clin. Lab. Sci. 2010;40:257–260. [PubMed] [Google Scholar]
- 16.Kojima Y, et al. Autocrine TGF-β and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor- promoting mammary stromal myofibroblasts. Proc. Natl. Acad. Sci. USA. 2010;107(46):20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korsmeyer RW, Gurny R, Doelker E, Buri P, Peppas NA. Mechanisms of solute release from porous hydrophilic polymers. Int. J. Pharm. 1983;15:25–35. doi: 10.1016/0378-5173(83)90064-9. [DOI] [PubMed] [Google Scholar]
- 18.Krieger JR, Ogle ME, McFaline-Figueroa J, Segar CE, Temenoff JS, Botchwey EA. Spatially localized recruitment of anti-inflammatory monocytes by SDF-1α-releasing hydrogels enhances microvascular network remodeling. Biomaterials. 2016;77:280–290. doi: 10.1016/j.biomaterials.2015.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K-W, Johnson NR, Gao J, Wang Y. Human progenitor cell recruitment via SDF-1α coacervate-laden PGS vascular grafts. Biomaterials. 2013;34:9877–9885. doi: 10.1016/j.biomaterials.2013.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Carson-Walter E, Walter KA. Chemokine receptor CXCR7 is a functional receptor for CXCL12 in brain endothelial cells. PLoS ONE. 2014;9:e103938. doi: 10.1371/journal.pone.0103938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao W, Yi X, Qin J, Tian M, Jin G. CXCL12 promotes proliferation of radial glia like cells after traumatic brain injury in rats. Cytokine. 2020;125:154771. doi: 10.1016/j.cyto.2019.154771. [DOI] [PubMed] [Google Scholar]
- 22.Marek R, Caruso M, Rostami A, Grinspan JB, Sarma JD. Magnetic cell sorting: a fast and effective method of concurrent isolation of high purity viable astrocytes and microglia from neonatal mouse brain tissue. J. Neurosci. Methods. 2008;175:108–118. doi: 10.1016/j.jneumeth.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Miyanishi N, Suzuki Y, Simizu S, Kuwabara Y, Banno K, Umezawa K. Involvement of autocrine CXCL12/CXCR4 system in the regulation of ovarian carcinoma cell invasion. Biochem. Biophys. Res. Commun. 2010;403:154–159. doi: 10.1016/j.bbrc.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Naumann U, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS ONE. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng H, et al. Stromal cell-derived factor 1-mediated CXCR4 signaling in rat and human cortical neural progenitor cells. J. Neurosci. Res. 2004;76:35–50. doi: 10.1002/jnr.20045. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saura J, Tusell JM, Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
- 28.Shen X, et al. Sequential and sustained release of SDF-1 and BMP-2 from silk fibroin-nanohydroxyapatite scaffold for the enhancement of bone regeneration. Biomaterials. 2016;106:205–216. doi: 10.1016/j.biomaterials.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 29.Shvartsman SY, Wiley HS, Deen WM, Lauffenburger DA. Spatial range of autocrine signaling: modeling and computational analysis. Biophys. J . 2001;81:1854–1867. doi: 10.1016/S0006-3495(01)75837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigismund S, et al. Threshold-controlled ubiquitination of the EGFR directs receptor fate. EMBO J. 2013;32:2140–2157. doi: 10.1038/emboj.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Signoret N, et al. Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol. 1997;139:651–664. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Signoret N, et al. Differential regulation of CXCR4 and CCR5 endocytosis. J. Cell Sci. 1998;111:2819–2830. doi: 10.1242/jcs.111.18.2819. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi H, et al. Intravenously transplanted human neural stem cells migrate to the injured spinal cord in adult mice in an SDF-1- and HGF-dependent manner. Neurosci. Lett. 2007;426:69–74. doi: 10.1016/j.neulet.2007.08.048. [DOI] [PubMed] [Google Scholar]
- 34.Thevenot PT, Nair AM, Shen J, Lotfi P, Ko C-Y, Tang L. The effect of incorporation of SDF-1α into PLGA scaffolds on stem cell recruitment and the inflammatory response. Biomaterials. 2010;31:3997–4008. doi: 10.1016/j.biomaterials.2010.01.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uto-Konomi A, et al. CXCR7 agonists inhibit the function of CXCL12 by down-regulation of CXCR4. Biochem. Biophys. Res. Commun. 2013;431:772–776. doi: 10.1016/j.bbrc.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesan S, Rose JJ, Lodge R, Murphy PM, Foley JF. Distinct mechanisms of agonist-induced endocytosis for human chemokine receptors CCR5 and CXCR4. Mol. Biol. Cell. 2003;14:3305–3324. doi: 10.1091/mbc.e02-11-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B, et al. Nanoparticle-modified chitosan-agarose-gelatin scaffold for sustained release of SDF-1 and BMP-2. Int. J. Nanomed. 2018;13:7395–7408. doi: 10.2147/IJN.S180859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yi X, et al. Cortical endogenic neural regeneration of adult rat after traumatic brain injury. PLoS ONE. 2013;8:e70306. doi: 10.1371/journal.pone.0070306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic supplementary material 1 (DOCX 2033 kb)