Abstract
Saraca asoca (Roxb.) Willd. (subfamily Detarioideae, family Fabaceae) is a perennial evergreen sacred medicinal tree classified under ‘vulnerable’ by the IUCN. The chloroplast (cp) genome/plastome which follows uniparental inheritance contains many useful genetic information because of its conservative rate of evolution. The assembled cp genome of S. asoca which maps as a conserved circular structure revealed extensive rearrangement in gene organization, comprising total length 160,003 bp including LSC, SSC, IRa, and IRb, and GC content was 35.26%. Herein a set of rbcL and matK gene were established using molecular phylogenetic analyses for molecular typing of S. asoca.
Keywords: Plastome, Saraca asoca, Detarioideae, Fabaceae, Molecular authentication, Simple sequence repeat, Genomic rearrangement
1. Introduction
Saraca asoca (Roxb.) de Wilde [family Fabaceae, subfamily Detarioideae (APG IV, 2016, LPWG, 2017)], commonly known as ‘asoca’ (Fig. 1A-B), indigenous to Assam, E. Pakistan, Upper Burma, Malaya, Ceylon and South India (Singh et al., 2015), is one of the most sacred tree of the Indian subcontinent (Murthy et al., 2008, Mollik et al., 2010). Apart from its various pharmacological significance e.g. antimicrobial (Seetharam et al., 2003, Shirolkar et al., 2013), anticancer (Cibin et al., 2012), anti-inflammatory (Cibin et al., 2012, Saha et al., 2012), antiarthritic (Preethi and Krishnakumar, 2011) activities, the barks, leaves, flowers, and seeds of ‘asoca’ have extensively been used against uterine infections and as astringent in the cases of the internal haemorrhoids in modern as well as in the Indian traditional systems of medicine (Nudrat et al., 2005, Singh et al., 2015).
Fig. 1.
Saraca asoca. A. The tree in the flowering stage. B. An enlarged view of inflorescence.
The continuous development in the next-generation sequencing (NGS) platforms (Shendure et al., 2017), and bioinformatics tools (Yang and Rannala, 2012) including cloud computing for genomic data analysis (Kwon et al., 2015, Langmead and Nellore, 2018) during last two decades have (a) greatly propelled to sequencing of the organellar genome e.g. mitochondria (Kozik et al., 2019), chloroplast (Daniell et al., 2016) and whole genome (Chen et al., 2018), (b) revolutionized the understanding of various biological disciplines (Ali et al., 2020) e.g. tree of life (Philippe et al., 2005, Rokas, 2006), evolution of plant genomes (Wendel et al., 2016), gene families and gene function (Leebens-Mack et al., 2019), conservation biology (Johnson and Koepfli, 2014, Wambugu et al., 2018), and (c) alleviate the enhancement of the agronomic traits (Rogalski et al., 2015, Daniell et al., 2016, Lima et al., 2016). The over-exploitation of S. asoca from the wild habitat due to increasing commercial demand of the bark of ‘asoca’ as crude drug material leads it to vulnerable (IUCN, 2019); hence, the characterization of plastome/whole chloroplast (cp) genome of ‘asoca’ and its genetic comparison will facilitate the development of DNA markers for diversity assessment, conservation, and in unraveling function of genes and gene families to produce its enhanced agronomic traits through genetic engineering.
2. Materials and methods
2.1. Leaf sampling and DNA sequencing
The green young leaves material of S. asoca was collected [voucher information: ‘MAA & TKPAN-116′ (BHAG, KSUH)] from the tree growing at conservatory of the botanical garden, Tilka Manjhi Bhagalpur University (TMBU), Bhagalpur, India, without harming the plant, were used for the extraction of DNA using # DNeasy Plant Mini Kit (QIAGEN). The de novo sequencing (as a single end run of 51 bp) was performed at Illumina platform, Illumina Pipeline 1.3.2 (Nie et al., 2012) was used for base calling.
2.2. Cp genome assembly and annotation
The raw reads were first filtered using fastqc. The high-quality reads were then assembled using spades (Bankevich et al., 2012), and annotated using the online tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) at Tamarindus indica L. (GenBank NC_026685.1) as reference (Hansen et al., 2007, Tillich et al., 2017). The repeat structure and small inversion (Timme et al., 2007, Yang et al., 2010, Maia et al., 1991, Doorduin et al., 2011, Castro et al., 2013, Beier et al., 2017) in cp genome were analyzed.
2.3. Comparison of cp genome and phylogenetic analysis
The cp genome of S. asoca was compared with the five other complete Detarioideae (Fabaceae) cp genomes including Crudia harmsiana Wild., (NC_036743.1), Daniellia pilosa (J. Léonard) Estrella, (NC_036744.1), Guibourtia leonensis J. Leonard, (NC_036742.1) and Tamarindus indica L. (NC_026685.1) by aligning using Kalign (Lassmann and Sonnhammer, 2005) and UPGMA analysis (Sneath and Sokal, 1973) employing MEGA X (Kumar et al., 2018) followed by the verification of the taxon proximity under UPGMA tree with MAUVE alignment.
The plant DNA barcoding genes i.e. rbcL and matK of adulterant species (a) Bauhinia variegata L. (GU135196, GU135033), (b) Mesua ferrea L. (KY654490, JN114759), (c) Polyalthia longifolia (Sonn.) Thwaites (JX856748, AY518786), (d) Shorea robusta Gaertn. (KY654498, KY973059) and (e) Trema orientalis (L.) Blume (KY654502, AB924756) were retrieved from the GenBank, and analyzed together with the sequences of the S. asoca (KY678341, KC592386). The sequences were aligned (Thompson et al., 1994), and the molecular phylogenetic analyses by Maximum Evolution method (Rzhetsky and Nei, 1992) rooted using outgroup Sarcandra glabra [Clade: Angiosperms, Order: Chloranthales, Family: Chloranthaceae (KP208901, JN407112) were performed using MEGA X (Kumar et al., 2018).
3. Results and discussion
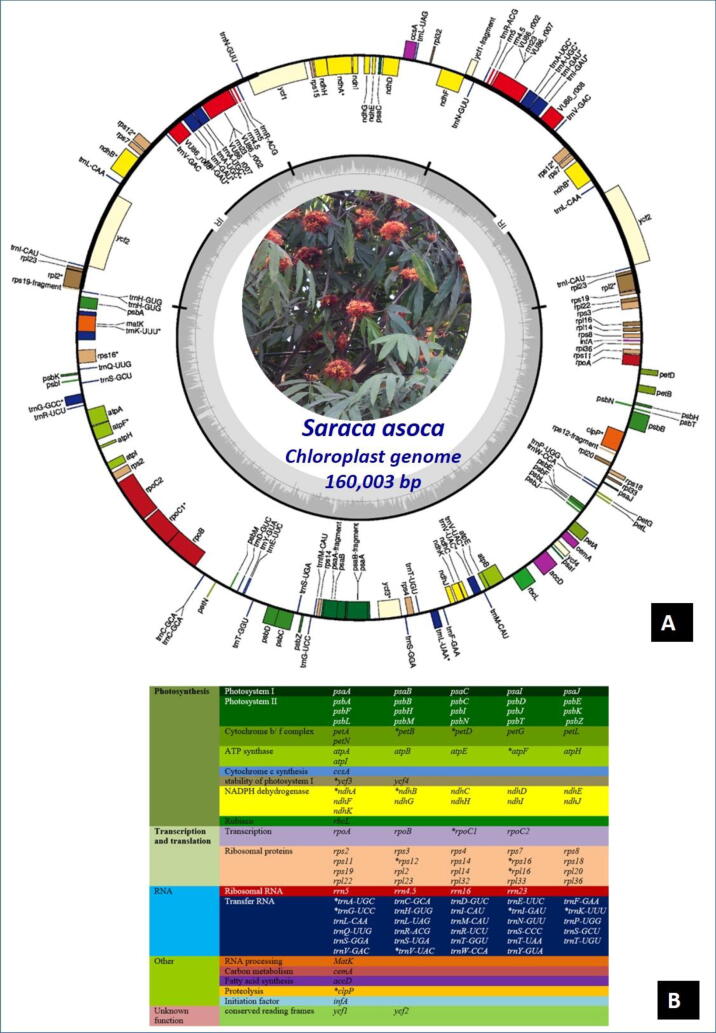
The assembled cp genome maps as a conserved circular structure (Fig. 2A), comprising total length 160,003 bp including LSC, SSC, IRa, and IRb, and GC content was 35.26% (NCBI GenBank accession number: MN866115) as similar to those of other angiosperms (Daniell et al., 2016). The cp genome possessed 111 genes (97 CDS, 29 tRNA, 4 rRNA genes) (Fig. 2B). Twelve of the CDS and eight of the tRNAs contain introns; 18 of these contain single intron, and two genes (ycf3 and clpP) possess 2 introns each (Fig. 2A).
Fig. 2.
A. The cp geneome map Saraca asoca, B. the genes of different groups are color-coded.
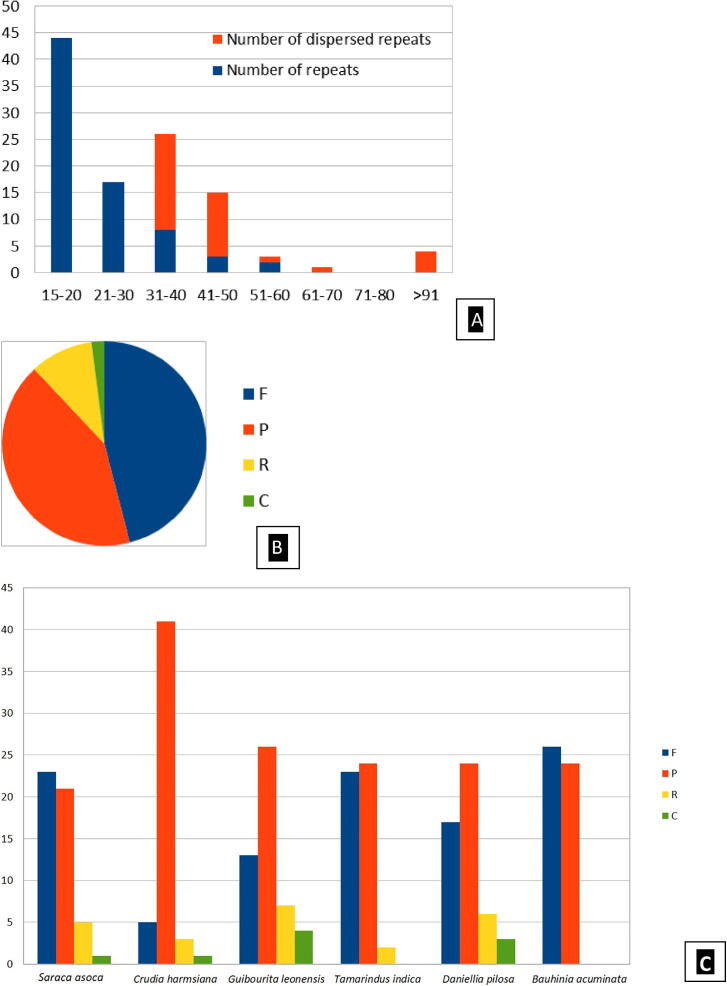
The tandem and dispersed repeats were analyzed for S. asoca cp genome. It is evident that the number of tandem and dispersed repeats were more in 15–20 bp and 31–40 bp category, respectively (Fig. 3A). The repeat structures of S. asoca and other five species of Fabaceae were analyzed by REPuter and were compared. The forward and palindrome repeats were common in these species (Fig. 3B-C). A total of 70 different SSR loci repeated more than 1 time (Table 1), contribute to the A–T richness of cp genome. The repeat regions play very significant roles in genome recombination (Yang et al., 2010). The SSRs are highly polymorphic due to higher mutation rate that affects the number of repeat units (Tsai et al., 2008).
Fig. 3.
The repeat structure analysis of the cp genome S. asoca. [A. The frequency of repeat by length; B. The repeat type; C. Comparison among six sequenced Fabaceae cp genomes (F: forward, P: palindrome, R: reverse, C: complement orientations)].
Table 1.
The SSR loci of S. asoca cp genome.
| S. | Type | SSR | Size | Starts | End |
|---|---|---|---|---|---|
| 1 | P1 | (T)10 | 10 | 2993 | 3002 |
| 2 | p2 | (TA)6 | 12 | 3933 | 3944 |
| 3 | p2 | (CT)6 | 12 | 9477 | 9488 |
| 4 | p2 | (TA)6 | 12 | 9799 | 9810 |
| 5 | p1 | (A)11 | 11 | 11,185 | 11,195 |
| 6 | p1 | (A)10 | 10 | 11,518 | 11,527 |
| 7 | p1 | (T)10 | 10 | 14,444 | 14,453 |
| 8 | p1 | (A)10 | 10 | 15,746 | 15,755 |
| 9 | c | (T)11seq (T)14 | 105 | 16,111 | 16,215 |
| 10 | c | (T)15 seq (A)10 | 109 | 17,236 | 17,344 |
| 11 | p1 | (T)12 | 12 | 18,454 | 18,465 |
| 12 | p1 | (T)13 | 13 | 18,906 | 18,918 |
| 13 | p1 | (A)11 | 11 | 19,421 | 19,431 |
| 14 | p1 | (A)10 | 10 | 48,187 | 48,196 |
| 15 | p1 | (A)14 | 14 | 50,369 | 50,382 |
| 16 | p1 | (T)14 | 14 | 51,455 | 51,468 |
| 17 | p2 | (TA)8 | 16 | 51,746 | 51,761 |
| 18 | p1 | 10(A) | 10 | 53,129 | 53,138 |
| 19 | p1 | (T)10 | 10 | 53,454 | 53,463 |
| 20 | p1 | (T)14 | 14 | 54,019 | 54,032 |
| 21 | c | (AT)7 seq (T)11 | 119 | 59,271 | 59,389 |
| 22 | p1 | (T)10 | 10 | 59,795 | 59,804 |
| 23 | c | (AT)6 seq (AT)6 seq (AT)7 | 163 | 60,249 | 60,411 |
| 24 | p3 | (TAT)5 | 15 | 60,634 | 60,648 |
| 25 | p1 | (A)10 | 10 | 61,434 | 61,443 |
| 26 | c | (T)11 g(A)10 | 22 | 63,236 | 63,257 |
| 27 | p1 | (T)12 | 12 | 65,351 | 65,362 |
| 28 | p1 | (T)10 | 10 | 65,958 | 65,967 |
| 29 | p4 | (TTAA)6 | 24 | 69,696 | 69,719 |
| 30 | p1 | (T)12 | 12 | 73,240 | 73,251 |
| 31 | p1 | (T)10 | 10 | 74,761 | 74,770 |
| 32 | c | (A)10 seq (A)9 | 89 | 76,392 | 76,480 |
| 33 | c | (A)10 seq (AT)6 | 58 | 77,232 | 77,289 |
| 34 | p1 | (T)11 | 11 | 77,940 | 77,950 |
| 35 | p1 | (A)11 | 11 | 79,224 | 79,234 |
| 36 | p1 | (T)10 | 10 | 80,731 | 80,740 |
| 37 | p1 | (G)10 | 10 | 82,977 | 82,986 |
| 38 | p1 | (A)15 | 15 | 84,348 | 84,362 |
| 39 | p1 | (C)11 | 11 | 84,762 | 84,772 |
| 40 | p2 | (AT)6 | 12 | 85,254 | 85,265 |
| 41 | p1 | (A)10 | 10 | 91,245 | 91,254 |
| 42 | p1 | (T)10 | 10 | 91,781 | 91,790 |
| 43 | p1 | (T)13 | 13 | 92,481 | 92,493 |
| 44 | p1 | (A)14 | 14 | 93,452 | 93,465 |
| 45 | p1 | (A)12 | 12 | 93,799 | 93,810 |
| 46 | p2 | (AT)10 | 20 | 94,894 | 94,913 |
| 47 | p2 | (TA)6 | 12 | 96,515 | 96,526 |
| 48 | p2 | (AT)6 | 12 | 96,648 | 96,659 |
| 49 | p1 | (A)12 | 12 | 101,516 | 101,527 |
| 50 | p1 | (T)10 | 10 | 103,439 | 103,448 |
| 51 | p2 | (TA)7 | 14 | 103,785 | 103,798 |
| 52 | p1 | (T)15 | 15 | 105,822 | 105,836 |
| 53 | p1 | (T)10 | 10 | 106,166 | 106,175 |
| 54 | p2 | (AT)6 | 12 | 106,328 | 106,339 |
| 55 | p1 | (T)12 | 12 | 108,073 | 108,084 |
| 56 | p1 | (T)10 | 10 | 109,129 | 109,138 |
| 57 | c | (A)11 seq (T)10 | 38 | 109,557 | 109,594 |
| 58 | p1 | (A)13 | 13 | 112,044 | 112,056 |
| 59 | p2 | (AT)6 | 12 | 112,200 | 112,211 |
| 60 | c | (AT)7 t(TA)7 | 29 | 112,852 | 112,880 |
| 61 | p1 | (A)10 | 10 | 114,163 | 114,172 |
| 62 | p1 | (A)13 | 13 | 115,814 | 115,826 |
| 63 | p1 | (T)10 | 10 | 116,265 | 116,274 |
| 64 | p1 | (T)10 | 10 | 117,561 | 117,570 |
| 65 | p1 | (T)10 | 10 | 120,635 | 120,644 |
| 66 | p1 | (A)10 | 10 | 121,197 | 121,206 |
| 67 | p1 | (T)10 | 10 | 126,128 | 126,137 |
| 68 | p1 | (T)13 | 13 | 130,202 | 130,214 |
| 69 | p1 | (A)11 | 11 | 131,334 | 131,344 |
| 70 | p1 | (T)10 | 10 | 133,319 | 133,328 |
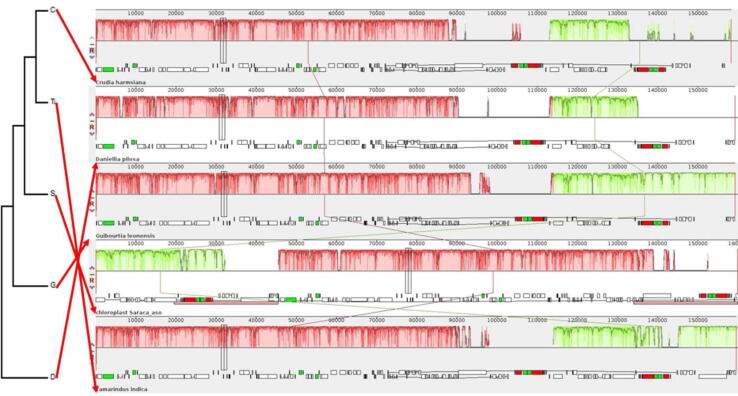
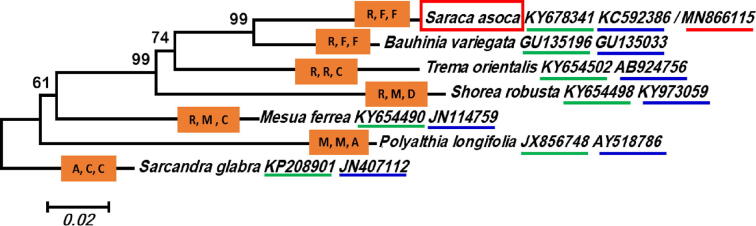
The comparison of cp genome of S. asoca with the five other complete Detarioideae (Fabaceae) cp genomes e.g. C. harmsiana, D. pilosa, G. leonensis, and T. indica revealed extensive rearrangement in gene organization (Fig. 4). Further, the ME tree from the set of the GenBank accession number [Bauhinia variegata [(Clade: Rosids, Order: Fabales, Family: Fabaceae; GenBank accession number: GU135196, GU135033)], Mesua ferrea [Clade: Rosids, Order: Malpighiales, Family: Calophyllaceae; GenBank accession number: KY654490, JN114759)], Polyalthia longifolia [Clade: Magnoliids, Order: Magnoliales, Family: Annonaceae; GenBank accession number: JX856748, AY518786)], Shorea robusta [Clade: Rosids, Order: Malvales, Family: Dipterocarpaceae; GenBank accession number: KY654498, KY973059] and Trema orientalis [Clade: Rosids, Order: Rosales, Family: Cannabaceae; GenBank accession number: KY654502, AB924756)] of rbcL and matK [- the cp genes used in the plant DNA barcoding (CBOL, 2009)] with the sequence of S. asoca (KY678341, KC592386/MN866115) revealed the optimal tree with the sum of branch length 0.57133802 (Fig. 5), and have potential to be used as molecular typing of S. acoca from its adulterants (Hegde et al., 2018) as NMR spectroscopy (Urumarudappa et al., 2016) and rbcL-ISSR based DNA barcodes (Hegde et al., 2018) are least user-friendly.
Fig. 4.
The MAUVE alignment of cp genomes of five different Detarioideae, showing genomic rearrangement.
Fig. 5.
The minimum evolution tree based on combined sequence of rbcL and matK gene representative species of Rosids. (R, F, F: Clade: Rosids, Order: Fabales, Family: Fabaceae; R, M, C: Clade: Rosids, Order: Malpighiales, Family: Calophyllaceae; M, M, A: Clade: Magnoliids, Order: Magnoliales, Family: Annonaceae; R, M, C: Clade: Rosids, Order: Malvales, Family: Dipterocarpaceae; R, R, C: Clade: Rosids, Order: Rosales, Family: Cannabaceae).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to extend their sincere appreciation to the Researchers Supporting Project number (RSP-2020/154), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mohammad Ajmal Ali, Email: ajmalpdc@gmail.cm.
Arun Bahadur Gurung, Email: arunbgurung@gmail.com.
References
- Ali M.A., Kim S.-Y., Pan T.K., Al-Hemaid F., Elshikh M.S., Elangbam M., Lee J., Farah M.A., Al-Anazi K.M. Complete chloroplast genome of vulnerable medicinal plant Saraca asoca (Fabaceae) Mitochondrial DNA Part B. 2020;5(1):754–755. doi: 10.1080/23802359.2020.1715300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APG, IV An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linnean Soc. 2016;181:1–20. [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A., Dvorkin M., Kulikov A.S., Lesin V., Nikolenko S., Pham S., Prjibelski A., Pyshkin A., Sirotkin A., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformation. 2017;33(16):2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro I., Pinto-Carnide O., Ortiz J.M., Martin J.P. Chloroplast genome diversity in Portuguese grapevine (Vitis vinifera L.) cultivars. Mol. Biotechnol. 2013;54(2):528–540. doi: 10.1007/s12033-012-9593-9. [DOI] [PubMed] [Google Scholar]
- CBOL Plant Wording Group A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Dong W., Zhang J., Guo X., Chen J., Wang Z., Lin Z., Tang H., Zhang L. The sequenced angiosperm genomes and genome databases. Front. Plant Sci. 2018;9:418. doi: 10.3389/fpls.2018.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibin T.R., Devi D.G., Abraham A. Chemoprevention of two-stage skin cancer in vivo by Saraca asoca. Integr. Cancer. Ther. 2012;11:279–286. doi: 10.1177/1534735411413264. [DOI] [PubMed] [Google Scholar]
- Daniell H., Lin C.S., Yu M., Chang W.J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:1–29. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin L., Gravendeel B., Lammers Y., Ariyurek Y., Chin A.W.T., Vrieling K. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. NA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D.R., Dastidar S.G., Cai Z., Penaflor C., Kuehl J.V., Boore J.L., Jansen R.K. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae) Mol. Phylogenet. Evol. 2007;45:547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Hegde S., Archana S., Harsha V.H., Sanjiva D.K., Subarna R. Molecular identification of Saraca asoca from its substituents and adulterants. 3 Biotech. 2018;8:161. doi: 10.1007/s13205-018-1175-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN, 2019. IUCN red list of threatened species. Version 2011.1. www.iucnredlist.org. Accessed 6th November 2019.
- Johnson W.E., Koepfli K. The role of genomics in conservation and reproductive sciences. In: Holt W., Brown J., Comizzoli P., editors. Reproductive sciences in animal conservation. Advances in experimental medicine and biology. Springer; New York, New York: 2014. [DOI] [PubMed] [Google Scholar]
- Kozik A., Rowan B.A., Lavelle D., Berke L., Schranz M.E., Michelmore R.W., Christensen A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019;15(8) doi: 10.1371/journal.pgen.1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T., Yoo W.G., Lee W.-J., Kim W., Kim D.-W. Next-generation sequencing data analysis on cloud computing. Genes Genom. 2015;37:489–501. [Google Scholar]
- Langmead B., Nellore A. Cloud computing for genomic data analysis and collaboration. Nat. Rev. Genet. 2018;19(4):208–219. doi: 10.1038/nrg.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E.L. Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinform. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J.H., Barker M.S., Carpenter E.J. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M.S., Woods L.C., Cartwright M.W., Smith D.R. The (in)complete organelle genome: exploring the use and non-use of available technologies for characterizing mitochondrial and plastid chromosomes. Mol. Ecol. Resour. 2016;16(6):1279–1286. doi: 10.1111/1755-0998.12585. [DOI] [PubMed] [Google Scholar]
- LPWG The Legume Phylogeny Working Group, 2017. Nasim, A., Babineau, M., Bailey, C.D., Banks, H., Barbosa, A.R., Pinto, R.B., Boatwright, J.S., Borges, L.M., Brown, G.K., Bruneau, A., Candido, E., Cardoso, D., Chung, K.-F., Clark, R.P., Conceição, A.S., Crisp, M., Cubas, P., Delgado-Salinas, A., Dexter, K.G., Doyle, J.J., Duminil, J., Egan, A.N., Estrella, M., Falcão, M.J., Filatov, D.A., Fortuna-Perez, A.P, Fortunato, R.H., Gagnon, E., Gasson, P., Rando, J.G., Tozzi, A.M.G.A., Gunn, B., Harris, D., Haston, E., Hawkins, J.A., Herendeen, P.S., Hughes, C.E., Iganci, J.R.V., Javadi, F., Kanu, S.A., Kazempour-Osaloo, S., Kite, G.C., Klitgaard, B.B., Kochanovski, F.J., Koenen, E.J.M., Kovar, L., Lavin, M., le Roux, M., Lewis, G.P., de Lima, H.C., López-Roberts, M.C., Mackinder, B., Maia, V.H., Malécot, V., Mansano, V.F., Marazzi, B., Mattapha, S., Miller, J.T., Mitsuyuki, C., Moura, T., Murphy, D.J., Nageswara-Rao, M., Nevado, B., Neves, D., Ojeda, D.I., Pennington, R.T., Prado, D.E., Prenner, G., Paganucci de Queiroz, L., Ramos, G., Filardi, F.L.R., Ribeiro, P.G., Rico-Arce, M.L., Sanderson, M.J., Santos-Silva, J., São-Mateus, W.M.B., Silva, M.J.S., Simon, M.F., Sinou, C., Snak, C., de Souza, É.R., Sprent, J., Steele, K.P., Steier, J.E., Steeves, R., Stirton, C.H., Tagane, S., Torke, B.M., Toyama, H., da Cruz, D.T., Vatanparast, M., Wieringa, J.J., Wink, M., Wojciechowski, M.F., Yahara, T., Yi, T., Zimmerman, E., 2017. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66 (1), 44–77.
- Maia G.L., Falcao-Silva Vdos S., Aquino P.G., de Araujo-Junior J.X., Tavares J.F., da Silva Nagano Y, Ishikawa H., Matsuno R., Sasaki Y. Nucleotide sequence and expression of the ribosomal protein L2 gene in pea chloroplasts. Plant Mol. Biol. 1991;17:541–545. doi: 10.1007/BF00040653. [DOI] [PubMed] [Google Scholar]
- Mollik M.A.H., Hossan M.S., Paul A.K., Taufiq-Ur-Rahman M., Jahan R., Rahmatullah M. A comparative analysis of medicinal plants used by folk medicinal healers in three districts of Bangladesh and inquiry as to mode of selection of medicinal plants. Ethnobot. Res. Appl. 2010;8:195–218. [Google Scholar]
- Murthy S.M., Mamatha B., Shivananda T.N. Saraca asoca – an endangered plant. Biomed. 2008;3:224–228. [Google Scholar]
- Nie X., Lv S., Zhang Y., Du X., Wang L., Biradar S.S., Tan X., Wan F., Weining S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora) PLoS One. 2012;7 doi: 10.1371/journal.pone.0036869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudrat S.Z., Usha M., Khan I.A., Khanum A. Ukaaz Publication; Hyderabad, India: 2005. Medicinal and aromatic plants of India, Part I. [Google Scholar]
- Philippe H., Delsuc F., Brinkmann H., Lartillot N. Phylogenomics. Annu. Rev. Ecol. Evol. Syst. 2005;36:541–562. [Google Scholar]
- Preethi F., Krishnakumar K. Anti-inflammatory activity of the barks of Saraca indica Linn. Pharmacol. Online. 2011;2:657–662. [Google Scholar]
- Rogalski M., do Nascimento Vieira L., Fraga H.P., Guerra M.P. Plastid genomics in horticultural species: Importance and applications for plant population genetics, evolution, and biotechnology. Front. Plant Sci. 2015;6:586. doi: 10.3389/fpls.2015.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A. Genomics and the tree of life. Science. 2006;313(5795):1897–1899. doi: 10.1126/science.1134490. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A., Nei M. A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 1992;9:945–967. [Google Scholar]
- Saha J., Mitra T., Gupta K., Mukherjee S. Phytoconstituents and HPTLC analysis in Saraca asoca (Roxb.) Wilde. Int. J. Pharm. Pharm. Sci. 2012;4:96–99. [Google Scholar]
- Seetharam N., Sujeeth H., Jyothishwaran G., Barad A., Sharanabasappa G., Shabana P. Antibacterial activity of Saraca asoca bark. Indian J. Plant Sci. 2003;65:658–659. [Google Scholar]
- Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H. DNA sequencing at 40: Past, present and future. Nature. 2017;550:345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- Shirolkar A., Gahlaut A., Chhillar A.K., Dabur R. Quantitative analysis of catechins in Saraca asoca and correlation with antimicrobial activity. J. Pharm. Anal. 2013;3:421–428. doi: 10.1016/j.jpha.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Anantha Krishna T.H., Subban K., Gini C.K., Jinu M.V., Chelliah J. Phytomedicinal importance of Saraca asoca (Ashoka): an exciting past, an emerging present and a promising future. Curr. Sci. 2015;109(10):1790–1801. [Google Scholar]
- Sneath P.H.A., Sokal R.R. Freeman; San Francisco: 1973. Numerical Taxonomy. [Google Scholar]
- Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq – versatile and accurate annotation of organelle genomes. Nuc. Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme R.E., Kuehl J.V., Boore J.L., Jansen R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- Tsai L.C., Wang J.C., Hsieh H.M., Liu K.L., Linacre A., Lee J.C. Bidens identification using the noncoding regions of chloroplast genome and nuclear ribosomal DNA. Forensic Sci. Int. Genet. 2008;2:35–40. doi: 10.1016/j.fsigen.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urumarudappa S.K., Gogna N., Newmaster S.G., Venkatarangaiah K., Subramanyam R., Saroja S.G., Gudasalamani R., Dorai K., Ramanan U.S. DNA barcoding and NMR spectroscopy-based assessment of species adulteration in the raw herbal trade of Saraca asoca (Roxb.) Willd, an important medicinal plant. Int. J. Legal. Med. 2016;130(6):1457–1470. doi: 10.1007/s00414-016-1436-y. [DOI] [PubMed] [Google Scholar]
- Wambugu P.W., Ndjiondjop M.-N., Henry R.J. Role of genomics in promoting the utilization of plantgenetic resources in genebanks. Functional Genom. 2018;17(3):198–206. doi: 10.1093/bfgp/ely014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F., Jackson S.A., Meyers B.C., Wing R.A. Evolution of plant genome architecture. Genome Biol. 2016;17:37. doi: 10.1186/s13059-016-0908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Zhang X., Liu G., Yin Y., Chen K., Yun Q., Zhao D., Al-Mssallem I.S., Yu J. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.) PLoS One. 2010;5 doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Rannala B. Molecular phylogenetics: principles and practice. Nat. Rev. Genet. 2012;13(5):303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]