Abstract
Background
Obesity is a reported risk factor for various health problems. Genome-wide association studies (GWASs) have identified numerous independent loci associated with body mass index (BMI). However, most of these have been focused on Europeans, and little evidence is available on the genetic effects across the life course of other ethnicities.
Methods
We conducted a cross-sectional study to examine the associations of 282 GWAS-identified single nucleotide polymorphisms with three BMI-related traits, current BMI, BMI at 20 years old (BMI at 20), and change in BMI (BMI change), among 11,586 Japanese individuals enrolled in the Japan Multi-Institutional Collaborative Cohort study. Associations were examined using multivariable linear regression models.
Results
We found a significant association (P < 0.05/282 = 1.77 × 10−4) between BMI and 11 polymorphisms in or near FTO, BDNF, TMEM18, HS6ST3, and BORCS7. The trend was similar between current BMI and BMI change, but differed from that of the BMI at 20. Among the significant variants, those on FTO were associated with all BMI traits, whereas those on TMEM18 and HS6SR3 were only associated with BMI at 20. The association of FTO loci with BMI remained, even after additional adjustment for dietary energy intake.
Conclusions
Previously reported BMI-associated loci discovered in Europeans were also identified in the Japanese population. Additionally, our results suggest that the effects of each loci on BMI may vary across the life course and that this variation may be caused by the differential effects of individual genes on BMI via different pathways.
Key words: obesity, body mass index, genome wide association study, polymorphisms
INTRODUCTION
Obesity is a known risk factor for various diseases1–3 and has increased globally in recent decades. Studies in twins and families suggest the existence of genetic factors for obesity.4–7 In recent years, genome-wide association studies (GWASs) have identified genes for common traits and enabled the identification of numerous obesity-related genetic variants.8–11 Among obesity-related traits, body mass index (BMI) is a well-established measure for evaluating obesity,12,13 and BMI-associated GWASs have reported a substantial number of related polymorphisms. Among these studies, the largest population meta-analysis study included 339,224 individuals and identified 97 BMI-associated loci.14
One recent GWAS conducted in a Japanese population using data from the BioBank Japan (BBJ) project reported 112 new BMI-associated loci15 and indicated genetic differences between European and East Asians. Apart from this study, however, most GWASs have been conducted in participants of European ancestry, and findings in non-Europeans are not currently sufficient.16,17 In addition, not only does BMI vary among ethnicities, but it also changes over the life course. For example, weight changes markedly in adulthood, and weight gain in early to middle adulthood is common.18,19 Numerous BMI-associated GWASs have been conducted using BMI determined at a single point in a person’s life, and it remains unclear at what age BMI is most likely to be affected by genetic variants or how these variants affect weight gain.
Therefore, the aims of this study were to identify BMI-associated loci in the Japanese population and to investigate how genetic factors affect BMI through the life course.
METHODS
Study population
The study was conducted using data from the Japan Multi-Institutional Collaborative Cohort (J-MICC) Study. Details of the J-MICC study have been described elsewhere.20 Briefly, the J-MICC study was launched in 2005 to investigate gene-environment interactions of lifestyle-related diseases. The subjects of the present study were volunteers aged 35 to 69 years who provided blood samples and information on their lifestyle via a questionnaire. The J-MICC study had recruited 102,145 participants by June 2018. Written informed consent was obtained from all participants. A total of 14,539 participants were randomly selected to be genotyped from a total of 47,163 participants from the 12 original participating sites: Aichi, Chiba, Fukuoka, Kagoshima, KOPS, Kyoto, Okazaki, Sakuragaoka, Saga, Shizuoka-Daiko, Takashima, and Tokushima. Genotyping was performed at the RIKEN Center for Integrative Medicine using a HumanOmniExpressExome-8 v1.2 BeadChip array (Illumina Inc., San Diego, CA, USA). Quality control (QC; described below) was conducted for the remaining 14,539 participants, and 422 participants whose genotyping data did not meet the QC filters were excluded. Among the remaining 14,091 participants, 32 were excluded due to a lack of questionnaire data.
In addition, participants were excluded from the study if they had missing or extreme values for self-reported weight at baseline (<30 kg), weight at age 20 (<30 kg), height (<130 cm), and current BMI (>60) and BMI at 20 (>60) (n = 413). Patients with cancer at baseline were also excluded from the analysis (n = 2,114). Fifty-four patients with both being outlier and cancer were included in these numbers, so a total of 2,473 participants were excluded. The remaining 11,586 subjects were analyzed (eFigure 1).
Quality control and genotype imputation
Participants with inconsistent baseline information on sex between the questionnaire and genotyping results were excluded (n = 26). The identity-by-descent method implemented in PLINK software21,22 identified 388 close relationship pairs (pi-hat >0.1875); one sample of each pair was excluded. According to principal component analysis with the 1000 Genomes reference panel (phase 3),23 34 subjects with non-Japanese ancestry were detected and excluded.24 The remaining samples met the sample-size genotype call rate criterion (≥0.99). Single nucleotide polymorphisms (SNPs) with genotype call rate <0.98, a Hardy-Weinberg equilibrium extract test P-value <1 × 10−6, a minor allele frequency of <0.01, or a departure from the allele frequency computed from the 1000 Genomes Project phase 3 EAS samples were removed. Non-autosomal SNPs were also removed. This QC filtering left 14,091 individuals. Genotype imputation was performed using SHAPIT225 and Minimac326 software based on the 1000 Genomes reference panel (phase 3). After genotype imputation, we excluded variants with an imputation quality score r2 < 0.3, resulting in 12 617 547 variants (J-MICC data set ver. 20180111).
Definition of BMI
BMI was calculated by dividing body weight in kilograms by the square of height in meters. We examined three BMI traits: (i) current BMI, defined as the BMI calculated based on the self-reported current weight and current height; (ii) BMI at 20, defined as the BMI calculated based on the self-reported weight at 20 years old and current height; and (iii) BMI change (per year), defined as (current BMI − BMI at 20)/(age − 20).
Measurement of lifestyle factors
Lifestyle factors, including smoking status and dietary intake, and medical information were evaluated using a self-administered questionnaire. Information on dietary intake of total energy was estimated using a food frequency questionnaire containing 47 food items27 based on the Standard Tables of Food Composition in Japan.28
SNP selection
We selected BMI-associated SNPs based on a previous Japanese study and GWAS catalog.29 First, we selected SNPs shown to be associated with BMI in a previous BBJ study that included Japanese-specific loci.15 We extracted loci in both trans-ethnic and Japanese specific, which are total of 261 significant loci reported by the BBJ study as the first group. Among these, SNPs on the X chromosome (n = 5) were excluded. Second, we selected 35 SNPs reported to be associated with “childhood body mass index” using the GWAS catalog29 as the second group. Third, we selected 45 SNPs reported to be associated with “longitudinal BMI measurement” in the GWAS catalog29 as the third group. Because 10 SNPs were duplicated in two or more groups, the total number of extracted SNPs was actually 326. We excluded SNPs with no available genotyping information in our J-MICC database (n = 10) and additionally excluded SNPs with a MAF less than 5% (n = 34), leaving 282 loci for analysis (eFigure 2, eTable 1, and eTable 2). Linkage disequilibrium (LD) was evaluated between SNPs on the same chromosome that showed a significant association with BMI by using both LDlink30 based on 1000 Genomes Project phase 3 (JPT) and our dataset. We defined two loci are in LD when R2 is above 0.80.
Statistical analysis
The relationship between each BMI-related phenotype and SNPs was analyzed using a crude and multivariable linear regression model to calculate the regression coefficient (β) and to evaluate the significance of the association (P-value). The adjusted model adjusted for age at baseline (continuous), age-squared (continuous), sex (men or women), birth year (continuous), and first five principal components (PCs) (continuous) in current BMI; sex, birth year, and five PCs in BMI at 20; age, age-squared, sex, birth year, first five PCs, and BMI at 20 in BMI change. At this level, we screened SNPs based on threshold P-values. Next, to evaluate the robustness of the association with the selected SNPs, we conducted a sex-stratified analysis and never-smokers only analysis. We also assessed heterogeneity among sex. To examine the potential effect of dietary energy intake on current BMI, we added total dietary energy intake to the adjusted model. Energy intake was defined as the individual total dietary energy intake divided by current weight.
All statistical analyses were conducted using Stata version 15.1 software (Stata Corp., College Station, TX, USA). P-values in both main analysis and stratified analyses were considered statistically significant at P < (0.05/282 = 1.77 × 10−4) following Bonferroni correction. We considered P-value <0.05 as significant only in assessing heterogeneity among sex.
Heritability analysis for each of three BMI phenotypes was performed with the use of genomic restricted maximum likelihood (GREML) method implemented in GCTA software.31 The analysis estimates the percentage of phenotypic variance explained by common SNPs. To estimate the heritability, we used the data set comprising the 11,586 subjects (5,169 males and 6,417 females) adopted for the association analysis as well as the 570,162 directly genotyped SNPs used for imputation. The phenotypic values for current BMI were adjusted for age at baseline, age-squared, birth year, sex, and first five principal components. The phenotypic values for BMI at 20 were adjusted for birth year, sex, and first five principal components. We applied two types of model to estimate the heritability of BMI change. The covariates in model 1 comprised age at baseline, age-squared, birth year, sex, and first five principal components. The covariates in model 2 included those of model 1 as well as BMI and BMI at 20. The estimation of the heritability was also performed using male and female subjects, separately. To estimate the genetic correlations among three BMI phenotypes, bivariate GREML32 was conducted using GCTA software. The estimate of heritability by GCTA is composed of two steps. At first, the genetic relationship matrix (GRM) between pairs of individuals was estimated from a set of SNPs using the GCTA option “--make-grm --thread-num 40”. This process is computationally intensive, so the process was performed by multi-thread computing with 40 CPU cores. Next, the heritability was estimated using the estimated GRM and the GCTA option “--reml”. We calculated the genetic correlations employing the same data sets, which were used to estimate the heritability. The genetic correlation was calculated using the same GRM and the GCTA option “--reml-bivar”. The phenotypic values for each of current BMI and BMI change were adjusted for age at baseline, age-squared, birth year, sex, and first five principal components. The phenotypic values for BMI at 20 were adjusted for birth year, sex, and first five principal components.
RESULTS
The baseline characteristics of the study subjects are shown in Table 1. All participants were Japanese and the 12 study sites were widely distributed throughout the western part of Japan. The study comprised 5,169 men (44.6%) and 6,421 women (55.4%), with an average age of 54.1 years. The averages of respective BMI phenotypes were as follows: 23.1 kg/m2 in current BMI, 20.9 kg/m2 in BMI at 20, and 0.069 kg/m2/year in BMI change.
Table 1. Baseline characteristics of study subjects.
| Number of subjects | 11,586 | |
| Age, years, mean (SD) | 54 | (9.4) |
| Sex, n (%) | ||
| Male | 5,169 | (44.6) |
| Female | 6,417 | (55.4) |
| Smoking status, n (%) | ||
| Never smoker | 6,865 | (59.3) |
| Ever smoker | ||
| Former smoker | 2,331 | (20.1) |
| Current smoker | 2,382 | (20.6) |
| Unknown | 8 | (0.1) |
| Area, n (%) | ||
| Chiba | 845 | (7.3) |
| Aichi Cancer Center | 695 | (6.0) |
| Okazaki | 956 | (8.3) |
| Shizuoka+Daiko | 1,883 | (16.3) |
| Takashima | 457 | (3.9) |
| Kyoto | 958 | (8.3) |
| Fukuoka | 715 | (6.2) |
| Saga | 1,753 | (15.1) |
| Kagoshima | 1,085 | (9.4) |
| Tokushima | 641 | (5.5) |
| KOPS | 1,112 | (9.6) |
| Shizuoka/Sakuragaoka | 490 | (4.2) |
| Birth year, mean (SD) | 1,953 | (10.1) |
| BMI phenotype, kg/m2, mean (SD) | ||
| Current BMI | 23.1 | (3.3) |
| BMI at 20 | 20.9 | (2.4) |
| BMI change | 0.069 | (0.098) |
BMI, body mass index; KOPS, Kyusyu Okinawa Population Study; SD, standard deviation.
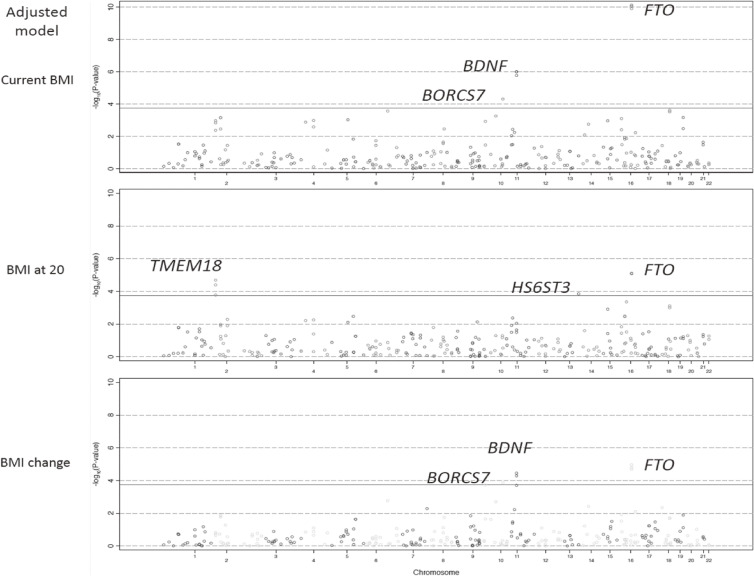
Among the 282 candidate loci, we identified a total of 11 SNPs with a statistically significant association with one or more BMI phenotypes in the adjusted analyses after applying the Bonferroni threshold. Table 2 shows the allele frequency of the significant SNPs and β estimates for the association with BMI. Among these loci, which were located near transmembrane protein 18 (TMEM18) (rs939584, rs13021737, and rs4854349), in brain-derived neurotrophic factor (BDNF) (rs11030100, rs6265, and rs11030104), and fat mass and obesity-associated gene (FTO) (rs1421085, rs11642015, rs1559302) showed LD with R2 ≥ 0.90 in both our dataset and LDlink (one locus on chromosome 16 (rs1559302) was not available in LDlink). In the non-stratified adjusted model, there was a significant association of 7 loci identified in the current BMI, 6 identified in the BMI at 20, and 6 identified in the BMI change. The allele frequency at each locus was consistent with that in the Japanese population in the 1000 Genome project (phase 3) database.33 Loci located in FTO were significantly associated with all BMI phenotypes, while loci located near TMEM18 and HS6ST3 were only associated with BMI at 20. Loci near TMEM18 and in BDNF were inversely associated, and loci in or near the remaining genes showed a positive association with each of the BMI phenotypes. Figure 1 shows the Manhattan plots for each BMI phenotype. We failed to identify any of the newly identified loci reported in a previous Japanese study among the significant SNPs in our adjusted models.15
Table 2. Eleven identified SNPs associated with any of the BMI phenotypes.
| SNP | Chr.a | Position (bp)a |
REF/ALT | Effect allele frequencyb |
Genotyped | Imputation quality score |
Candidate gene(s) | Current BMI | BMI at 20 | BMI change | Consistency for former reportsg |
||||||
| Adjustedd | Adjustede | Adjustedf | |||||||||||||||
| βb | SE | P-valuec | βb | SE | P-valuec | βb | SE | P-valuec | |||||||||
| rs939584 | 2 | 621558 | C/T | 0.898 | + | 1.000 | LOC105373352, TMEM18 | −0.220 | 0.067 | 1.07E-03 | −0.217 | 0.051 | 2.03E-05 | −0.003 | 0.002 | 1.52E-01 | − |
| rs13021737 | 2 | 632348 | A/G | 0.898 | − | 0.993 | LOC105373352, TMEM18 | −0.214 | 0.067 | 1.48E-03 | −0.208 | 0.051 | 4.02E-05 | −0.003 | 0.002 | 1.77E-01 | − |
| rs4854349 | 2 | 647861 | T/C | 0.895 | − | 0.996 | LOC105373352, TMEM18 | −0.191 | 0.067 | 4.26E-03 | −0.190 | 0.051 | 1.66E-04 | −0.002 | 0.002 | 2.30E-01 | − |
| rs4409766 | 10 | 104616663 | T/C | 0.295 | − | 0.996 | BORCS7-ASMT | 0.181 | 0.045 | 4.96E-05 | 0.036 | 0.034 | 2.93E-01 | 0.005 | 0.001 | 1.38E-04 | + |
| rs11030100 | 11 | 27677586 | G/T | 0.403 | − | 0.963 | BDNF-AS | −0.199 | 0.042 | 1.68E-06 | −0.082 | 0.031 | 8.95E-03 | −0.005 | −0.003 | 1.96E-04 | + |
| rs6265 | 11 | 27679916 | C/T | 0.403 | − | 0.996 | BDNF | −0.203 | 0.041 | 1.00E-06 | −0.071 | 0.031 | 2.26E-02 | −0.005 | 0.001 | 5.15E-05 | + |
| rs11030104 | 11 | 27684517 | A/G | 0.402 | − | 0.963 | BDNF-AS | −0.203 | 0.042 | 1.01E-06 | −0.067 | 0.031 | 3.18E-02 | −0.005 | 0.001 | 3.51E-05 | Not shownH |
| rs1927790 | 13 | 96922191 | T/C | 0.420 | + | 1.000 | HS6ST3 | 0.057 | 0.041 | 1.64E-01 | 0.119 | 0.031 | 1.39E-04 | 0.000 | 0.001 | 8.34E-01 | + |
| rs1421085 | 16 | 53800954 | T/C | 0.196 | − | 0.993 | FTO | 0.335 | 0.051 | 1.24E-10 | 0.173 | 0.039 | 8.02E-06 | 0.007 | 0.002 | 2.04E-05 | + |
| rs11642015 | 16 | 53802494 | C/T | 0.196 | − | 0.963 | FTO | 0.334 | 0.051 | 7.36E-11 | 0.174 | 0.039 | 8.41E-06 | 0.007 | 0.002 | 1.08E-05 | + |
| rs1558902 | 16 | 53803574 | T/A | 0.195 | − | 0.993 | FTO | 0.331 | 0.051 | 8.65E-11 | 0.174 | 0.039 | 7.69E-06 | 0.006 | 0.002 | 1.59E-05 | + |
ALT, alternative allele; BMI, body mass index; bp, base pair; Chr., chromosome; REF, reference allele; SE, standard error; SNP, single nucleotide polymorphism.
aPositions are based on Human Genome version 19 (hg19), build 37.
bAlternative alleles were treated as effect alleles.
cSignificant P-values (P ≤ 1.77E-04) are shown in bold.
dAdjusted model in current BMI were adjusted for age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), and the first five principal components (continuous).
eAdjusted model in BMI at 20 were adjusted for sex (male or female), birth year (continuous), and the first five principal components (continuous).
fAdjusted model in BMI change were adjusted for age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), the first five principal components (continuous), and BMI at 20 (continuous).
gIf the direction of effect allele in initially extracted BMI-related traits was consistent with that of former reports, “+”, if not, “−” is in the column.
hThe results of estimates were not shown, but only P-values were shown in former reports.
Figure 1. Manhattan plots of the association between candidate loci and each BMI phenotype in the adjusted model. The X-axis represents chromosomal position and the Y-axis represents −log10(P-value). The grey solid horizontal line indicates significance level (P = 1.77 × 10−4).
In sex-stratified analysis, several loci showed sex-specific association between BMI and genotype, but they did not show any heterogeneity in the 11 significant SNPs (eTable 3). In addition, to eliminate the disease-related weight change which derived from diabetes, we performed same analysis as original in current BMI with excluding participants with history of diabetes mellitus. The tendency of association between current BMI and significant SNPs were similar even after excluding those with diabetes (eTable 4). Given that smoking behavior has a strong effect on body weight,34–36 we conducted an analysis that focused on never smokers. In these subjects, the significant SNPs were similar to those in the adjusted models for the entire population (Table 3). We also examined the effect of food intake by adjusting for energy intake in the analysis of 7 significant loci (located in 3 genes) identified in the current BMI. After adjusting for energy intake, polymorphisms in FTO remained significant SNPs (Table 4).
Table 3. Eleven identified SNPs associated with any of the BMI phenotypes in never-smokers.
| SNP | Chr.a | Position (bp)a |
REF/ALT | Candidate gene(s) | Current BMI | BMI at 20 | BMI change | ||||||
| Adjustedd | Adjustede | Adjustedf | |||||||||||
| βb | SE | P-valuec | βb | SE | P-valuec | βb | SE | P-valuec | |||||
| rs939584 | 2 | 621558 | C/T | LOC105373352, TMEM18 | −0.287 | 0.088 | 1.03E-03 | −0.246 | 0.065 | 1.60E-04 | −0.006 | 0.003 | 3.12E-02 |
| rs13021737 | 2 | 632348 | A/G | LOC105373352, TMEM18 | −0.288 | 0.087 | 9.63E-04 | −0.254 | 0.065 | 9.05E-05 | −0.005 | 0.003 | 3.40E-02 |
| rs3800229 | 6 | 108996963 | G/T | FOXO3 | −0.254 | 0.063 | 5.96E-05 | −0.087 | 0.047 | 6.37E-02 | −0.006 | 0.002 | 1.25E-03 |
| rs143665886 | 7 | 115368366 | T/C | SNORA25B, TFEC | 0.037 | 0.053 | 4.85E-01 | 0.151 | 0.039 | 1.31E-04 | −0.001 | 0.002 | 5.27E-01 |
| rs4409766 | 10 | 104616663 | T/C | BORCS7-ASMT | 0.236 | 0.058 | 4.26E-05 | 0.075 | 0.043 | 8.11E-02 | 0.006 | 0.002 | 3.72E-04 |
| rs11030100 | 11 | 27677586 | G/T | BDNF-AS | −0.266 | 0.054 | 7.93E-07 | −0.117 | 0.040 | 3.50E-03 | −0.006 | 0.002 | 1.77E-04 |
| rs6265 | 11 | 27679916 | C/T | BDNF | −0.265 | 0.054 | 8.14E-07 | −0.097 | 0.040 | 1.49E-02 | −0.006 | 0.002 | 8.03E-05 |
| rs11030104 | 11 | 27684517 | A/G | BDNF-AS | −0.261 | 0.054 | 1.14E-06 | −0.090 | 0.040 | 2.37E-02 | −0.006 | 0.002 | 6.35E-05 |
| rs1421085 | 16 | 53800954 | T/C | FTO | 0.358 | 0.067 | 7.39E-08 | 0.164 | 0.050 | 9.61E-04 | 0.007 | 0.002 | 1.41E-04 |
| rs11642015 | 16 | 53802494 | C/T | FTO | 0.361 | 0.067 | 5.74E-08 | 0.163 | 0.050 | 1.00E-03 | 0.008 | 0.002 | 1.07E-04 |
| rs1558902 | 16 | 53803574 | T/A | FTO | 0.365 | 0.067 | 4.45E-08 | 0.164 | 0.050 | 9.24E-04 | 0.008 | 0.002 | 9.54E-05 |
ALT, alternative allele; BMI, body mass index; bp, base pair; Chr., chromosome; REF, reference allele; SE, standard error; SNP, single nucleotide polymorphism.
aPositions are based on Human Genome version 19 (hg19), build 37.
bAlternative alleles were treated as effect alleles.
cSignificant P-values (P ≤ 1.77E-04) are shown in bold.
dAdjusted model in current BMI were adjusted for age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), and the first five principal components (continuous).
eAdjusted model in BMI at 20 were adjusted for sex (male or female), birth year (continuous), and the first five principal components (continuous).
fAdjusted model in BMI change were adjusted for age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), the first five principal components (continuous), and BMI at 20 (continuous).
Table 4. Seven identified SNPs associated with the current BMI adjusted for total energy intake.
| SNP | Candidate gene(s) |
Current BMI | |||
| Adjusted modela | Adjusted model + total energyd/weighte | ||||
| βb | P-valuec | βb | P-valuec | ||
| rs4409766 | BORCS7-ASMT | 0.181 | 4.96E-05 | 0.036 | 2.93E-01 |
| rs11030100 | BDNF-AS | −0.199 | 1.68E-06 | −0.082 | 8.95E-03 |
| rs6265 | BDNF | −0.203 | 1.00E-06 | −0.071 | 2.26E-02 |
| rs11030104 | BDNF-AS | −0.203 | 1.01E-06 | −0.067 | 3.18E-02 |
| rs1421085 | FTO | 0.335 | 1.24E-10 | 0.174 | 8.02E-06 |
| rs11642015 | FTO | 0.334 | 7.36E-11 | 0.173 | 8.41E-06 |
| rs1558902 | FTO | 0.331 | 8.65E-11 | 0.174 | 7.69E-06 |
BMI, body mass index; SNP, single nucleotide polymorphism.
aAdjusted models were adjusted for age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), and the top 5 principal components (continuous).
bAlternative alleles were treated as effect alleles.
cSignificant P-values (P ≤ 1.77E-04) are shown in bold.
dTotal energy was estimated using a self-reported food frequency questionnaire.
eWeight indicates self-reported current weight.
To further examine potential heterogeneity of susceptibility loci across multiple BMI-related traits, we estimated the heritability of each BMI phenotype (Table 5). In all participants, the heritability was 27.2% in current BMI, and 21.6% in BMI at 20. Of note, the estimated heritability was quite different between sex especially in current BMI (male: 42.2% vs female: 25.9%), and the proportion of effects of genetic factor in male were clearly changed from BMI at 20 to current BMI, but more stable in females than males. In addition, the pair-wise genetic correlation analysis among three BMI-related traits showed that current BMI and BMI change had higher correlation (75.7%) in contrast to BMI at 20 and BMI change (13.0%) (Table 5).
Table 5. Heritability of each phenotype and genetic correlation between phenotypes.
| (i) Heritability | |||||||||
| All participants | Male | Female | |||||||
| (n = 11,586) | (n = 5,169) | (n = 6,417) | |||||||
| Heritability | SE | P-valuee | Heritability | SE | P-valuee | Heritability | SE | P-valuee | |
| Current BMIa | 0.272 | 0.030 | 0.00E+00 | 0.422 | 0.065 | 1.64E-11 | 0.259 | 0.053 | 4.99E-07 |
| BMI at 20b | 0.216 | 0.029 | 4.22E-15 | 0.254 | 0.064 | 2.35E-05 | 0.322 | 0.053 | 1.99E-10 |
| BMI changec | 0.158 | 0.030 | 3.34E-08 | 0.239 | 0.066 | 1.90E-04 | 0.166 | 0.052 | 1.05E-03 |
| BMI change (without adjustment for BMI at 20) | 0.116 | 0.029 | 4.64E-05 | 0.131 | 0.065 | 2.95E-02 | 0.163 | 0.052 | 1.19E-03 |
| (ii) Genetic correlation (n = 11,586) | ||||
| Genetic correlation (rG) |
SE |
P-valuee
(one-tailed, rG = 1) |
P-valuee
(one-tailed, rG = 0) |
|
| Current BMIa/BMI at 20b | 0.753 | 0.063 | 9.72E-05 | 3.33E-16 |
| Current BMIa/BMI changed | 0.757 | 0.063 | 3.07E-03 | 5.89E-08 |
| BMI at 20b/BMI changed | 0.130 | 0.148 | 5.84E-04 | 1.77E-01 |
BMI, body mass index; SE, standard error.
aThe covariates in current BMI were age (continuous), age-squared (continuous), birth year (continuous), sex (male or female), and first five principal components (continuous).
bThe covariates in BMI at 20 were adjusted for sex (male or female), birth year (continuous), and the first five principal components (continuous).
cThe covariates in BMI change were age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), the first five principal components (continuous), and BMI at 20 (continuous).
dThe covariates in BMI change were age (continuous), age-squared (continuous), sex (male or female), birth year (continuous), and the first five principal components (continuous).
eSignificant P-values were defined as P ≤ 0.05.
DISCUSSION
Our cross-sectional study within a prospective cohort study in a Japanese population showed that there was a significant association between BMI traits and polymorphisms in or near FTO, BDNF, TMEM18, HS6ST3, and BORCS7. All of these significant loci or genes have previously been reported in other ethnicities and are well-known BMI-associated polymorphisms.37–47 Our findings confirm the robustness of these associations in the Japanese population. Among the three BMI-related phenotypes examined in this study (current BMI, BMI at 20, and BMI change), the trend of association was similar in current BMI and BMI change, but differed from that in the BMI at 20. Interestingly, while FTO variants were associated with all BMI traits, TMEM18 and HS6ST3 were only associated with BMI at 20. Our results indicate that the effects of individual polymorphisms on BMI may vary across the life course.
A few studies have identified heterogeneities in genetic effects across age using both cross-sectional and longitudinal designs. Given that FTO loci reportedly explain the majority of the inter-individual variance among previously confirmed BMI-related loci,41 several studies have focused on FTO variants to explore the potential differential effects across the life course.48–51 One study revealed that the FTO variant identified by a GWAS in adults also showed a significant association in children and adolescents.48 This is consistent with our findings, suggesting that the FTO loci affect BMI across the life course. Another study showed that while there are consistent trends in the association of BMI with FTO loci, the effect size varies across life epoch.50 This study reported that among the four distinct phases of adulthood (18 to over 70 years), the strongest association between FTO loci and BMI was observed in young adults (18–25 years) relative to each life epoch.50 Similarly, another cohort study that conducted a longitudinal observation reported that the association between FTO polymorphisms and BMI strengthened with age, peaking at age 20 years, before weakening in later adult years.49 While the available evidence on TMEM18 loci is scarcer than that for FTO loci, several GWASs have reported an association between TMEM18 loci and BMI in both adults and children.14,38,43,52 Moreover, one study showed that there was no TMEM18 loci heterogeneity across the life course.50 In contrast, we found that TMEM18 variants were only associated with BMI at 20. However, the variants reported in previous life course studies differed from the loci examined in our study, which may explain the discordance in results. Importantly, our findings may suggest that the effect of some genes on BMI can differ across age while of other genes can be kept across age. In addition, each gene variant may exhibit different patterns of association with BMI, such that while one gene may have congenital effects, another may exhibit effects through an acquired pathway. Our findings in the current BMI and BMI change were similar, which supports the speculation that these associated significant genes affect BMI through an acquired pathway.
In particular, we think that dietary intake, for which habits are acquired after birth, may be the most important acquired pathway for obesity. Many obesity-related genes are expressed or are known to act in the central nervous system, especially in the hypothalamus, and the association between the expression of these genes and BMI may explain increases or decreases in appetite, such as via regulation of the feeding center in the hypothalamus.38,41 However, even after adjusting for total energy intake, FTO loci remained significantly associated with current BMI and BMI change. This may be because weight regulation by genetic factors is not simply dependent on feeding behavior, but also on other metabolic pathways and factors. For example, FTO loci are associated with an adipocyte thermogenesis regulation pathway involving IRX3 and IRX5,53 and BDNF is associated with physical activity.54,55 Given that the mechanism governing the effects of genes on obesity remains poorly understood, further studies are required to clarify the genetic associations with obesity.
Results of heritability and genetic correlation also supported our speculation that genes which affect current BMI and BMI at 20 are heterogeneous. The estimated heritability in current BMI was higher than in BMI at 20 especially in males, and genetic correlation between BMI change and BMI at 20 were only 13% in contrast to 75% of consistency in current BMI and BMI change. These results can make it reasonable to suppose that the degree of dependent on BMI by genetic factors are different by age and genetic effects vary across life course.
This study has limitations that warrant mention. First, the statistical power may have been limited because of the sample size and strict application of the Bonferroni threshold. This may explain the few significant variants obtained in this study compared to a previous Japanese study.15 However, the variants that we identified as being significant under these conditions are likely strongly associated with BMI in the Japanese population and may code potential target genes for the development of measures against obesity in the future. Second, because the analysis was restricted to individuals of Japanese ancestry, our results may not be generalizable to other ethnic populations. Third, although we assumed that BMI is changed monotonically through the life course, the actual change may vary depending on the time. As a result, genetic effects over a period of time may be underestimated, whereas those in another period overestimated.
In conclusion, previously reported BMI-associated genes discovered in European populations were also identified in a Japanese population. Additionally, some of the significant variants identified in this study were associated with BMI at 20, but not with current BMI or BMI change. This indicates that the genetic effects on BMI may vary across the life course via different pathways. Further studies are needed to identify changes in effect sizes across the life course and the mechanisms of genetic effects on obesity in various ethnicities.
ACKNOWLEDGMENTS
The authors thank all of the participants of the J-MICC study and the staff at each site for their cooperation. This study was supported by Grants-in-Aid for Scientific Research for Priority Areas of Cancer (No. 17015018) and Innovative Areas (No. 221S0001) and by a JSPS KAKENHI Grant (No. 16H06277) from the Japanese Ministry of Education, Culture, Sports, Science and Technology. This work was also supported in part by funding for the BioBank Japan Project from the Ministry of Education, Culture, Sports, Science and Technology from April 2003 to March 2015.
Conflicts of interest: None declared.
APPENDIX A. SUPPLEMENTARY DATA
The following is the supplementary data related to this article:
eTable 1. List of all SNPs examined
eTable 2. List of SNPs excluded from the initial candidate variants for analysis
eTable 3. Sex-stratified analysis of the 11 significant SNPs
eTable 4. Analysis of 11 significant SNPs excluding DM population in current BMI
eFigure 1. Consort diagram of the eligibility of subjects after merging questionnaire and genotype data. FFQ, food frequency questionnaire.
eFigure 2. Consort diagram of the selection of candidate loci. BBJ, BioBank Japan study15; BMI, body mass index; MAF, minor allele frequency; SNPs, single nucleotide polymorphisms.
REFERENCES
- 1.Haslam DW, James WPT. Obesity. Lancet. 2005;366(9492):1197–1209. 10.1016/S0140-6736(05)67483-1 [DOI] [PubMed] [Google Scholar]
- 2.Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371(9612):569–578. 10.1016/S0140-6736(08)60269-X [DOI] [PubMed] [Google Scholar]
- 3.Wilson PWF, D’Agostino RB, Sullivan L, Parise H, Kannel WB. Overweight and obesity as determinants of cardiovascular risk: the Framingham experience. Arch Intern Med. 2002;162(16):1867–1872. 10.1001/archinte.162.16.1867 [DOI] [PubMed] [Google Scholar]
- 4.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–351. 10.1023/A:1025635913927 [DOI] [PubMed] [Google Scholar]
- 5.Poulsen P, Vaag A, Kyvik K, Beck-Nielsen H. Genetic versus environmental aetiology of the metabolic syndrome among male and female twins. Diabetologia. 2001;44(5):537–543. 10.1007/s001250051659 [DOI] [PubMed] [Google Scholar]
- 6.Wardle J, Carnell S, Haworth CM, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87(2):398–404. 10.1093/ajcn/87.2.398 [DOI] [PubMed] [Google Scholar]
- 7.Zaitlen N, Kraft P, Patterson N, et al. . Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9(5):e1003520. 10.1371/journal.pgen.1003520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visscher PM, Brown MA, McCarthy MI, Yang J. Five Years of GWAS Discovery. Am J Hum Genet. 2012;90(1):7–24. 10.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loos RJF Genetic determinants of common obesity and their value in prediction. Best Pract Res Clin Endocrinol Metab. 2012;26(2):211–226. 10.1016/j.beem.2011.11.003 [DOI] [PubMed] [Google Scholar]
- 10.Fall T, Ingelsson E. Genome-wide association studies of obesity and metabolic syndrome. Mol Cell Endocrinol. 2014;382(1):740–757. 10.1016/j.mce.2012.08.018 [DOI] [PubMed] [Google Scholar]
- 11.Visscher PM, Wray NR, Zhang Q, et al. . 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 2017;101(1):5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher D, Visser M, Sepúlveda D, Pierson RN, Harris T, Heymsfield SB. How useful is body mass index for comparison of body fatness across age, sex, and ethnic groups? Am J Epidemiol. 1996;143(3):228–239. 10.1093/oxfordjournals.aje.a008733 [DOI] [PubMed] [Google Scholar]
- 13.Mei Z, Grummer-Strawn LM, Pietrobelli A, Goulding A, Goran MI, Dietz WH. Validity of body mass index compared with other body-composition screening indexes for the assessment of body fatness in children and adolescents. Am J Clin Nutr. 2002;75(6):978–985. 10.1093/ajcn/75.6.978 [DOI] [PubMed] [Google Scholar]
- 14.Locke AE, Kahali B, Berndt SI, et al. . Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama M, Okada Y, Kanai M, et al. . Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 2017;49(10):1458–1467. 10.1038/ng.3951 [DOI] [PubMed] [Google Scholar]
- 16.Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25(11):489–494. 10.1016/j.tig.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 17.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161–164. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NCD Risk Factor Collaboration (NCD-RisC) Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128·9 million children, adolescents, and adults. Lancet. 2017;390(10113):2627–2642. 10.1016/S0140-6736(17)32129-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushita Y, Takahashi Y, Mizoue T, Inoue M, Noda M, Tsugane S; JPHC Study Group . Overweight and obesity trends among Japanese adults: a 10-year follow-up of the JPHC Study. Int J Obes (Lond). 2008;32(12):1861–1867. 10.1038/ijo.2008.188 [DOI] [PubMed] [Google Scholar]
- 20.Hamajima N; J-MICC Study Group . The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for cancer. Asian Pac J Cancer Prev. 2007;8(2):317–323. [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, et al. . PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81(3):559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi-Kabata Y, Nakazono K, Takahashi A, et al. . Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83(4):445–456. 10.1016/j.ajhg.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. 10.1038/nmeth.2307 [DOI] [PubMed] [Google Scholar]
- 26.Das S, Forer L, Schönherr S, et al. . Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokudome S, Goto C, Imaeda N, Tokudome Y, Ikeda M, Maki S. Development of a data-based short food frequency questionnaire for assessing nutrient intake by middle-aged Japanese. Asian Pac J Cancer Prev. 2004;5(1):40–43. [PubMed] [Google Scholar]
- 28.Office for Resources Policy Division Science and Technology Policy Bureau, ed. Standard Tables of Food Composition in Japan -2015 - (Seventh Revised Version). http://www.mext.go.jp/en/policy/science_technology/policy/title01/detail01/1374030.htm.
- 29.The NHGRI-EBI Catalog of published genome-wide association studies. GWAS catalog. https://www.ebi.ac.uk/gwas/home. Accessed August 9, 2018. [DOI] [PMC free article] [PubMed]
- 30.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542. 10.1093/bioinformatics/bts474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.NCBI. 1000 Genome Browser. http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/.
- 34.Albanes D, Jones DY, Micozzi MS, Mattson ME. Associations between smoking and body weight in the US population: analysis of NHANES II. Am J Public Health. 1987;77(4):439–444. 10.2105/AJPH.77.4.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filozof C, Fernández Pinilla MC, Fernández-Cruz A. Smoking cessation and weight gain. Obes Rev. 2004;5(2):95–103. 10.1111/j.1467-789X.2004.00131.x [DOI] [PubMed] [Google Scholar]
- 36.Kim JH, Shim KW, Yoon YS, Lee SY, Kim SS, Oh SW. Cigarette smoking increases abdominal and visceral obesity but not overall fatness: an observational study. PLoS One. 2012;7(9):e45815. 10.1371/journal.pone.0045815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frayling TM, Timpson NJ, Weedon MN, et al. . A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Speliotes EK, Loos RJF, et al. . Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. 10.1038/ng.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyre D, Delplanque J, Chèvre JC, et al. . Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–159. 10.1038/ng.301 [DOI] [PubMed] [Google Scholar]
- 40.Scuteri A, Sanna S, Chen WM, et al. . Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. 10.1371/journal.pgen.0030115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speliotes EK, Willer CJ, Berndt SI, et al. . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. 10.1038/ng.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loos RJF, Lindgren CM, Li S, et al. . Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768–775. 10.1038/ng.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei YF, Zhang L, Liu Y, et al. . Meta-analysis of genome-wide association data identifies novel susceptibility loci for obesity. Hum Mol Genet. 2014;23(3):820–830. 10.1093/hmg/ddt464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Justice AE, Winkler TW, Feitosa MF, et al. . Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat Commun. 2017;8:14977. 10.1038/ncomms14977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. . Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. 10.1038/ng.274 [DOI] [PubMed] [Google Scholar]
- 46.Wen W, Zheng W, Okada Y, et al. . Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23(20):5492–5504. 10.1093/hmg/ddu248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warrington NM, Howe LD, Paternoster L, et al. . A genome-wide association study of body mass index across early life and childhood. Int J Epidemiol. 2015;44(2):700–712. 10.1093/ije/dyv077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.den Hoed M, Ekelund U, Brage S, et al. . Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59(11):2980–2988. 10.2337/db10-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardy R, Wills AK, Wong A, et al. . Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19(3):545–552. 10.1093/hmg/ddp504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graff M, Gordon-Larsen P, Lim U, et al. . The influence of obesity-related single nucleotide polymorphisms on BMI across the life course: the PAGE study. Diabetes. 2013;62(5):1763–1767. 10.2337/db12-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graff M, North KE, Richardson AS, et al. . BMI loci and longitudinal BMI from adolescence to young adulthood in an ethnically diverse cohort. Int J Obes (Lond). 2017;41(5):759–768. 10.1038/ijo.2016.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Felix JF, Bradfield JP, Monnereau C, et al. . Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet. 2016;25(2):389–403. 10.1093/hmg/ddv472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claussnitzer M, Dankel SN, Kim KH, et al. . FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neeper SA, Gómez-Pinilla F, Choi J, Cotman CW. Physical activity increases mRNA for brain-derived neurotrophic factor and nerve growth factor in rat brain. Brain Res. 1996;726(1–2):49–56. 10.1016/0006-8993(96)00273-9 [DOI] [PubMed] [Google Scholar]
- 55.Huang AM, Jen CJ, Chen HF, Yu L, Kuo YM, Chen HI. Compulsive exercise acutely upregulates rat hippocampal brain-derived neurotrophic factor. J Neural Transm (Vienna). 2006;113(7):803–811. 10.1007/s00702-005-0359-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.