Abstract
Parkinson’s disease (PD), a common neurodegenerative disorder, has a complex etiology where environmental and genetic factors intervene. While a number of genes and variants have been identified in recent decades as causative or protective agents of this condition, a limited number of studies have been conducted in mixed populations, such as Mexican Mestizos. The historical convergence of two founding groups and three ethnicities, and the increasing north-to-south gradient of Native American ancestry in Mexico resulted in a subpopulation structure with considerable genetic diversity. In this work, we investigate the influence of 21 known susceptibility variants for PD. Our case–control study, with a cohort of 311 Mexican Mestizo subjects, found a significant risk association for the variant rs1491942 in LRRK2. However, when stratification by ancestry was performed, a risk effect for MTHFR rs1801133 was observed only in the group with the highest percentage of European ancestry, and the PD risk effect for LRRK2 rs1491942 was significant in subjects with a higher ratio of Native American ancestry. Meta-analyses of these SNP revealed the effect of LRRK2 rs1491942 to be even more significant than previously described in populations of European descent. Although corroboration is necessary, our findings suggest that polymorphism rs1491942 may be useful as a risk marker of PD in Mexican Mestizos with greater Native American ancestry. The absence of associations with the remaining known risk factors is, in itself, a relevant finding and invites further research into the shared risk factors’ role in the pathophysiological mechanisms of this neurodegenerative disorder.
Subject terms: Parkinson's disease, Risk factors, Predictive markers
Introduction
Parkinson’s disease (PD), a common progressive and incurable neurodegenerative disorder, especially prevalent among the elderly, is estimated to affect >6 million people worldwide1–3. Clinical manifestations include motor symptoms, such as bradykinesia, resting tremor, rigidity, and deterioration of postural reflexes. In addition, non-motor alterations, e.g., sleep disorders, autonomic dysfunction, and cognitive impairment, adversely affect the quality of life, cause disability, or even mortality. Although the etiology of PD is complex, evidence suggests it is caused by the interaction of environmental and genetic factors1,3.
Studies conducted in recent decades have identified a number of genes and variants associated with PD4–12. It is estimated that 5–10% of all PD cases have a genetic etiology linked to forms with monogenic Mendelian inheritance patterns. These forms are attributed to various loci containing genes, such as SNCA, PRKN, PARK7, and LRRK2 (refs. 13–15). In the rest of the cases, called sporadic, genetic susceptibility factors have also been demonstrated. So far, >90 risk loci have been identified; the associated variants are mainly single-nucleotide polymorphisms (SNPs)5,8,10,12,14. Some of these SNPs are located within or very close to loci linked to the familial forms mentioned above, which indicates that changes in the sequence of these genes are likely to be implicated in the key biological processes of PD development14,16. The identification and functional characterization of these genetic changes have provided information on the cellular and subcellular mechanisms contributing to PD-related neurodegeneration15–21.
Despite advances in the typification of PD’s genetic susceptibility factors, interpretation of these findings is still controversial. This limitation is evidenced in a GWAS study by Foo et al.12, that investigated PD risk loci in an Asian cohort and then compared the results with those of European populations. Although they report substantial overlap in genetic risk factors, the similarities between the two groups are incomplete. In addition to these reported differences is the bias in information from ethnically diverse groups due to the scarcity of genomic data from populations other than Caucasians and Asians in current studies20,21. It is important to consider this bias as the differences in demographic histories and adaptation processes endured by different populations are likely to have influenced the genetic architecture of complex diseases such as PD in these groups.
The demographic history of a given population is one of the contributing factors to the impact genetic changes have on the incidence of PD in that population. When the number of individuals that gave rise to a population is limited, there is likely to be a representative bias of some of their alleles in the following generations. Migration processes, mutations, selective pressures, and genetic drift can contribute in a determinant way to the presence and frequency of allelic variations22–25. These changes are especially prevalent in mixed populations26 like Mexico, where the majority of individuals are Mestizo, i.e., of Native American, European, and African ancestral origins23,25,27.
According to the most accepted hypothesis, American natives originated from East Asian groups that crossed the Bering Strait ~16 thousand years ago. Once in America, they expanded from the northern to the southern continent, in different settlements from Alaska to Chile. As these original groups inhabited the new environments, they underwent adaptive processes, selective pressures, long migrations, and isolation, which resulted in a reduction of genetic diversity in the population (bottlenecks and founder effect). For the Native Americans in Mexico, a decisive second event occurred with the European conquest and colonization. The arrival of Spaniards accompanied by African slaves caused a decrease in the number of Native American settlers, due, among other factors, to their susceptibility to new European diseases and wars. Over time, the miscegenation of Europeans, surviving Native Americans, and Africans took place. These historical characteristics are all reflected in the heterogeneous structure of the current Mexican population, which shows significant genetic diversity compared with other populations24,28.
To date, 22 articles have been published that address the genetics of PD in Mexican Mestizos29–50. These studies analyzed alterations in 17 genes (SNCA, PINK1, PRKN, GBA, LRRK2, MTHFR, LRRK2, APOE, SYT11, DRD2, ANKK1, PARK7, MAPT, ALDH1, NR4A2, tRNAGln, and mtATP6). Their results identify eight SNPs as potential risk factors for PD in Mexican subjects (rs385705916, rs356220, rs356203, rs7684318, and rs2736990 in the SNCA gene, rs421016 in the GBA gene, rs35479735 in the NR4A2 gene, and rs1801133 in the MTHFR gene). When these findings were compared with the GWAS results from European and Asian populations, only one polymorphism (rs356203 in the SNCA gene) was found in common10. While this discrepancy may be due to insufficient statistical power, it could also be explained by genetic and environmental diversity among populations. In this work, we investigate the incidence of genetic variations that have previously been associated with PD in a Mexican Mestizo population. In addition, a novel panel of 32 Ancestry Informative Markers (AIMs)51 was used to estimate the gradient of European and Native American ancestry in our study subjects. This analysis of the subpopulation structure allowed us to assess PD risk association according to the percentage of Native American ancestry.
Results
Demographic and clinical characteristics
When comparing demographic and clinical characteristics between the groups, differences attributable to the place of recruitment are ruled out (Supplementary Table 1). The demographic and clinical characteristics of the 118 PD cases and 193 controls are summarized in Table 1. No differences are observed in age, sex, BMI, glucose levels, or cognitive deterioration; however, significant dissimilarities were found in total cholesterol and uric acid levels and frequency of depression. Total cholesterol levels were lower in PD cases at 175 mg/dl compared to controls at 195 mg/dl (p < 0.001). Similarly, uric acid levels were less in the cases (5.23 mg/dl) vs controls (6.035, p < 0.001). Also, the frequency of depression was higher in cases (72.32%) compared to controls (49.15%, p < 0.001). Case’s total Unified Parkinson’s Disease Rating Scale (UPDRS) scores were 72 ± 38, and UPDRS motor scores were 40 ± 23 with 2.5 ± 1 on the Hoehn and Yahr (HY) rating scale. The average age of PD onset was 64.08 ± 10.46 years; only 11 (9%) patients had an age of onset <50 years; in all cases, the subjects reported no family history of PD.
Table 1.
Clinical and demographic characteristic of the study population.
| Cases (n = 118) | Controls (n = 193) | p Value | |
|---|---|---|---|
| Males, n(%) | 60(50.8) | 97(50.2) | 0.999a |
| Age at enrollment, years | 69.92 ± 10.01 | 69.80 ± 8.63 | 0.914b |
| BMI, kg/m2 | 27.38 [19–38] | 27.19 [19–49] | 0.615c |
| Total cholesterol, mg/dl | 175 [100–270] | 195 [80–276] | 0.0001c |
| Glucose, mg/dl | 102 [67–217] | 110.60 [74–234] | 0.0181c |
| Uric acid, mg/dl | 5.23 ± 1.60 | 6.035 ± 1.38 | <0.0001b |
| Cognitive impairment, n(%) (by MMSE-test) | 37(32.74%) | 45(26.98%) | 0.298a |
| Depression, n(%) (by HAM-D test) | 81(72.32%) | 87(49.15%) | <0.001a |
| Age at onset, years | 64.08 ± 10.46 | ||
| Disease duration, years | 5.93 ± 4.93 | ||
| UPDRS total score | 72 ± 38 | ||
| UPDRS III score | 40 ± 23 | ||
| HY scale | 2.5 ± 1 |
Shows mean values (Standard deviation or interquartile range) and frequency (%). Skewness and kurtosis tests were performed for normality.
BMI body mass index, HAM-D Hamilton Depression Rating Scale, MMSE Mini-Mental State Exam, UPDRS Unified Parkinson’s Disease Rating Scale: total score and score for Part III—Motor Examination, HY Hoehn and Yahr scale.
Reported p values were determined with a aFisher’s exact test, bStudent’s t test, or cU Mann–Whitney.
Genotypic characteristics
We investigated 21 SNPs as associated factors in PD; however, the polymorphisms rs947211, rs356220, and rs2736990 were discarded from further analysis due to their pattern of linkage disequilibrium. The polymorphisms rs34778348 and rs33949390 in the LRRK2 gene were found to be monomorphic in our sample and were also discarded. Of the remaining 16 SNPs, a significant PD risk association was found for the allelic and genotype frequencies of the polymorphism rs1491942; all stated confidence intervals (CI) are 95%.
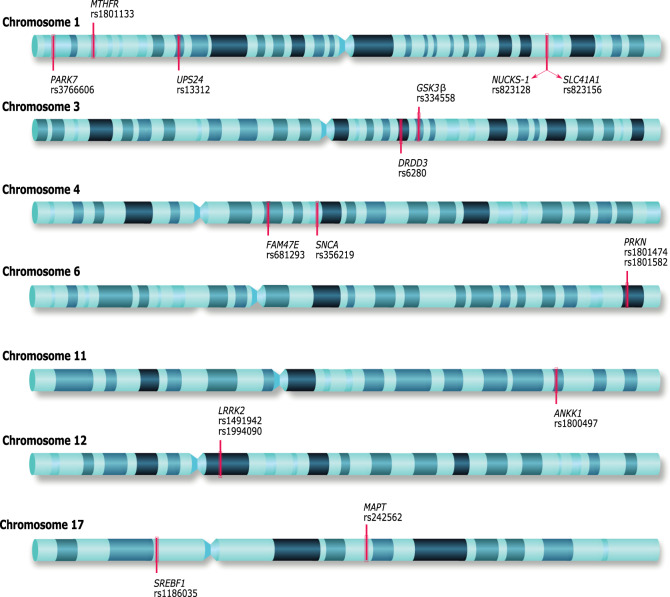
For SNP rs1491942 in the LRRK2 gene, the estimated risk association under an additive model was odds ratio (OR) 1.71 [1.22–2.40] p 0.002. The selected SNPs’ genomic localization is shown in Fig. 1; no deviation from Hardy–Weinberg equilibrium (HWE) is observed in the control group. The allelic, genotyping distribution, and OR estimation of the SNPs are shown in Table 2.
Fig. 1. Chromosomal location of the 16 single-nucleotide polymorphisms selected for this research.
Genes symbols and reference SNP numbers. Genes: PARK7 Parkinsonism associated deglycase, MTHFR Methylenetetrahydrofolate reductase; USP24 Ubiquitin specific peptidase 24, NUCKS1 Nuclear casein kinase and cyclin dependent kinase substrate SLC41A1 Solute carrier family 41 member 1, DRD3 Dopamine receptor D3, GSK3B Glycogen synthase kinase 3 beta, FAM47E family with sequence similarity 47 member E, SNCA Synuclein alpha, PRKN Parkin RBR E3 ubiquitin-protein ligase, ANKK1 Ankyrin repeat, and kinase domain containing 1, LRRK2 Leucine-rich repeat kinase 2, SREBF1 sterol regulatory element-binding transcription factor 1 MAPT Microtubule-associated protein tau.
Table 2.
Allele and genotype frequencies in PD patients and controls.
| Gene SNP | Group | MA n (freq) | pallelic | Genotype | pgenotype | ORa | pOR | ||
|---|---|---|---|---|---|---|---|---|---|
|
LRRK2 rs1491942 |
PD | 155 (0.67) | 0.003 | CC | CG | GG | 0.01 |
1.71 (1.22–2.40) |
0.002 |
| 14 (0.12) | 53 (0.45) | 51 (0.43) | |||||||
| CNT | 205 (0.53) | 44 (0.22) | 93 (0.48) | 56 (0.29) | |||||
|
MTHFR rs1801133 |
PD | 137 (0.58) | 0.01 | CC | CT | TT | 0.041 |
1.54 (1.11–2.15) |
0.01 |
| 23 (0.19) | 53 (0.45) | 42 (0.36) | |||||||
| CNT | 183 (0.47) | 55 (0.29) | 93 (0.48) | 45 (0.23) | |||||
|
USP24 rs13312 |
PD | 20 (0.09) | 0.99 | CC | CG | GG | 0.99 |
0.91 (0.49–1.71) |
0.79 |
| 96 (0.83) | 18 (0.16) | 1 (0.01) | |||||||
| CNT | 24 (0.10) | 104 (0.83) | 20 (0.16) | 2 (0.02) | |||||
|
PARK7 rs3766606 |
PD | 20 (0.03) | 0.67 | GG | GT | TT | 0.83 |
0.87 (0.49–1.54) |
0.64 |
| 99 (0.84) | 18 (0.15) | 1 (0.09) | |||||||
| CNT | 37 (0.06) | 157 (0.81) | 35 (0.18) | 1 (0.05) | |||||
|
NUCKS1 rs823128 |
PD | 32 (0.13) | 0.90 | AA | AG | GG | 0.56 |
0.94 (0.58–1.50) |
0.80 |
| 90 (0.76) | 24 (0.20) | 4 (0.04) | |||||||
| CNT | 55 (0.14) | 142 (0.74) | 47 (0.24) | 4 (0.02) | |||||
|
SLC41A1 rs823156 |
PD | 51 (0.22) | 0.29 | AA | AG | GG | 0.09 |
0.79 (0.54–1.17) |
0.25 |
| 76 (0.64) | 33 (0.28) | 9 (0.08) | |||||||
| CNT | 99 (0.26) | 105 (0.54) | 77 (0.40) | 11 (0.06) | |||||
|
GSK3B rs334558 |
PD | 85 (0.36) | 0.43 | AA | GA | GG | 0.44 |
1.14 (0.81–1.61) |
0.42 |
| 51 (0.43) | 49 (0.41) | 18 (0.15) | |||||||
| CNT | 127 (0.33) | 86 (0.45) | 87 (0.47) | 20 (0.10) | |||||
|
DRD3 rs6280 |
PD | 99 (0.42) | 0.16 | TT | TC | CC | 0.23 |
0.75 (0.54–1.05) |
0.10 |
| 42 (0.36) | 52 (0.44) | 24 (0.20) | |||||||
| CNT | 188 (0.49) | 51 (0.26) | 96 (0.49) | 46 (0.24) | |||||
|
FAM47E/ SCARB2 rs6812193 |
PD | 49 (0.21) | 0.4 | CC | CT | TT | 0.31 |
1.2 (0.80–1.82) |
0.37 |
| 78 (0.66) | 31 (0.27) | 9 (0.08) | |||||||
| CNT | 69 (0.18) | 131 (0.68) | 55 (0.26) | 7 (0.04) | |||||
|
SNCA rs356219 |
PD | 90 (0.38) | 0.21 | GG | AG | AA | 0.29 |
0.79 (0.57–1.11) |
0.18 |
| 47 (0.40) | 52 (0.44) | 19 (0.16) | |||||||
| CNT | 168 (0.44) | 60 (0.31) | 98 (0.51) | 35 (0.18) | |||||
|
PARK2 rs1801474 |
PD | 36 (0.15) | 0.48 | CC | CT | TT | 0.58 |
1.18 (0.74–1.87) |
0.48 |
| 85 (0.72) | 30 (0.25) | 3 (0.03) | |||||||
| CNT | 51 (0.13) | 148 (0.77) | 39 (0.20) | 6 (0.03) | |||||
|
PARK2 rs1801582 |
PD | 27 (0.11) | 0.73 | CC | CG | GG | 0.94 |
1.09 (0.65–1.83) |
0.77 |
| 93 (0.78) | 23 (0.19) | 2 (0.017) | |||||||
| CNT | 60 (0.16) | 155 (0.80) | 35 (0.18) | 3 (0.015) | |||||
|
ANKK1 rs1800497 |
PD | 103 (0.44) | 0.62 | CC | CT | TT | 0.63 |
0.91 (0.65–1.26) |
0.58 |
| 41 (0.35) | 51 (0.43) | 26 (0.22) | |||||||
| CNT | 177 (0.46) | 58 (0.30) | 93 (0.48) | 42 (0.22) | |||||
|
LRRK2 rs1994090 |
PD | 32 (0.14) | 0.62 | TT | TG | GG | 0.58 |
1.16 (0.71–1.88) |
0.55 |
| 87 (0.74) | 30 (0.25) | 1 (0.008) | |||||||
| CNT | 46 (0.12) | 150 (0.78) | 40 (0.21) | 3 (0.015) | |||||
|
MAPT rs242562 |
PD | 76 (0.32) | 0.12 | AA | AG | GG | 0.29 |
0.75 (0.53–1.06) |
0.11 |
| 51 (0.43) | 57 (0.48) | 10 (0.08) | |||||||
| CNT | 149 (0.39) | 68 (0.35) | 101 (0.52) | 24 (0.12) | |||||
|
RAIL/ SREBF1 rs11868035 |
PD | 112 (0.47) | 0.16 | AA | AG | GG | 0.39 |
1.29 (0.92–1.81) |
0.13 |
| 36 (0.31) | 52 (0.44) | 30 (0.25) | |||||||
| CNT | 160 (0.41) | 72 (0.37) | 82 (0.43) | 39 (0.20) | |||||
MA minor allele frequency, PD Parkinson disease patients, CNT controls.
pallelic p value allelic comparison, pgenotype p value genotype comparison, pOR p value OR
aOR (CI 95%) additive model adjusted by sex, age, and ancestry.
Possible differences due to the Mexican population’s heterogeneity were explored by subdividing the sample into quartiles according to their percentage of Native American ancestry (Supplementary Table 2). The first group included 78 individuals with the lowest percentage (ranges from 32–52%), groups two and three were each made up of 78 individuals with intermediate ranges (52.1–56.5% and 56.6–65%, respectively), while 77 individuals with the highest Native American percentage of the sample (≤66%) were in the fourth group. We found differences between these groups in the genotype frequencies and OR estimations of SNPs rs1801133 and rs1491942 (Table 3). When comparing cases and controls, the genotypic frequency for rs1801133 was significantly different (p = 0.03) in the group with the lowest Native American percentage (ranges from 32 to 52%; OR 2.02 [CI 95% 1.02–4.04] p 0.043) in an additive model. No statistical differences were observed in any of the other three groups.
Table 3.
Allele and genotype frequencies in PD patients and controls by percentage of Native American ancestry.
| ID Marker | Group 1 (32–52%) | Group 2 (52.1–58.5%) | Group 3 (56.6–65%) | Group 4 (≥66%) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAa n (freq) | Genotype | OR [95% CI] | MAa n (freq) | Genotype | OR [95% CI] | MAa n (freq) | Genotype | OR [95% CI] | MAa n (freq) | Genotype | OR [95% CI] | |||||||||
| rs1801133 | CC | CT | TT | 2.02 | CC | CT | TT | 1.54 | CC | CT | TT | 1.54 | CC | CT | TT | 1.11 | ||||
| PD |
31 (0.53) |
9 (0.31) |
9 (0.31) |
11 (0.38) |
[1.02–4.03] |
34 (0.57) |
5 (0.17) |
16 (0.53) |
9 (0.30) |
[0.77–2.26] |
40 (0.62) |
5 (0.17) |
14 (0.44) |
13 (0.40) |
[0.77–2.76] |
32 (0.60) |
4 (0.15) |
14 (0.52) |
9 (0.33) |
[1.56–2.11] |
| CNT |
38 (0.39) |
17 (0.35) |
26 (0.53) |
6 (0.12) |
40 (0.42) |
16 (0.33) |
24 (0.50) |
8 (0.17) |
48 (0.52) |
12 (0.26) |
20 (0.44) |
14 (0.30) |
57 (0.57) |
10 (0.20) |
23 (0.43) |
17 (0.34) |
||||
| p = 0.09 | p = 0.02 | p = 0.04 | p = 0.07 | p = 0.2 | p = 0.2 | p = 0.3 | p = 0.5 | p = 0.24 | p = 0.87 | p = 0.9 | p = 0.75 | |||||||||
| rs1491942 | CC | CG | GG | 1.0 | CC | CG | GG | 1.5 | CC | CG | GG | 2.26 | CC | CG | GG | 2.94 | ||||
| PD |
38 (0.66) |
4 (0.14) |
12 (0.41) |
13 (0.45) |
[0.50–1.99] |
37 (0.62) |
5 (0.17) |
27 (0.56) |
12 (0.40) |
[0.75–3.04] |
44 (0.68) |
2 (0.07) |
16 (0.50) |
14 (0.43) |
[1.04–4.91] |
36 (0.66) |
3 (0.12) |
12 (0.44) |
12 (0.44) |
[1.38–6.23] |
| CNT |
64 (0.65) |
6 (0.12) |
22 (0.45) |
21 (0.43) |
51 (0.53) |
9 (0.19) |
13 (0.43) |
1 (0.38) |
50 (0.54) |
9 (0.19) |
24 (0.52) |
13 (0.28) |
40 (0.40) |
20 (0.40) |
20 (0.40) |
10 (0.20) |
||||
| p = 0.99 | p = 0.9 | p = 0.99 | p = 0.32 | p = 0.4 | p = 0.2 | p = 0.1 | p = 0.1 | p = 0.04 | p = 0.002 | p = 0.01 | p = 0.01 | |||||||||
Group 1 n = 78 subjects; group 2 n = 78 subjects; group 3 n = 78 subjects; group 4 n = 77 subjects.
PD Parkinson disease group, CNT control group.
aMA minor allele, B additive model.
For rs1491942, significant allelic and genotypic frequency is observed in the subgroups with higher Native American ancestry. This polymorphism was estimated to be a PD risk factor in subjects with ≤56.5% Native American ancestry. In the subgroup with 56.6–65%, the estimation was (OR 2.26 [1.04–4.91] p 0.04), and in subjects with ≤66% Native American ancestry (OR 2.94 [1.38–6.23] p 0.01) in an additive model.
Meta-analyses were performed to clarify the PD risk association of SNPs rs1801133 and rs1491942; the flow chart is shown in Fig. 2. Briefly, 137 articles were retrieved in the database search. Of these, the following were eliminated; 26 were duplicates, 69 had irrelevant content, 12 articles contained insufficient genotype data, 2 lacked control groups, and 4 were meta-analyses. After analyzing the remaining 24 papers, two more studies were ruled out due to insufficient quality (Newcastle–Ottawa Scale System Studies <5). The present study’s findings were also included in the meta-analyses.
Fig. 2. PD risk association.

Flow diagram of the search and inclusion process of studies for the meta-analysis of the SNPs rs1801133 in the MTHFR gene and rs1491942 in the LRRK2 gene.
Meta-analysis: the association of MTHFR rs1801133 with PD risk
Nineteen studies were included in this meta-analysis (conducted on 11 European, 6 Asiatic, and 2 Mexican Mestizo populations); together, these studies comprised 2832 cases and 9074 controls. The summary characteristics of the selected studies are shown in Supplementary Table 3. No significant associations were observed for rs1801133 polymorphism and PD risk when considering an additive, dominant, or recessive model in the overall population (Table 4 and Supplementary Figs 1–3). However, in the subgroup analysis by ethnicity, there was a significant PD risk association in individuals of European ancestry under a dominant model with OR 1.17 [1.11–1.36] p 0.036 (Table 4 and Supplementary Figs 4–6). No significant association with PD was evident in Asian or Mexican Mestizo samples in any of the models considered (Tables 1 and 4, and Supplementary Figs 4–6). A Begg’s test detected no publication bias p < 0.005 (Table 4).
Table 4.
Summary of meta-analyses.
| SNP | Ethnicity | Studies | Genetic model | OR | 95% CI | p Value | Model | I2% | Begg’s test p Value |
|---|---|---|---|---|---|---|---|---|---|
| rs1801133 | Overall | 20 | T vs C | 1.12 | 0.98–1.28 | 0.094 | R | 64.3 | 0.347 |
| TT + TC vs CC | 1.14 | 0.96–1.36 | 0.111 | R | 53.5 | 0.417 | |||
| TT vs TC + CC | 1.15 | 0.89–1.48 | 0.271 | R | 53.9 | 0.721 | |||
| European | 12 | T vs C | 1.12 | 0.94–1.37 | 0.190 | R | 53.7 | 0.631 | |
| TT + TC vs CC | 1.17 | 1.11–1.36 | 0.036 | F | 13.8 | 0.537 | |||
| TT vs TC + CC | 1.12 | 0.78–1.61 | 0.407 | R | 56.9 | 0.537 | |||
| Asian | 6 | T vs C | 1.19 | 0.90–1.58 | 0.211 | R | 73.2 | 0.260 | |
| TT + TC vs CC | 1.22 | 0.87–1.72 | 0.240 | R | 70.4 | 0.260 | |||
| TT vs TC + CC | 0.95 | 0.72–1.27 | 0.750 | F | 38.7 | 0.060 | |||
| Mexican Mestizos | 2 | T vs C | 1.05 | 0.50–2.19 | 0.894 | R | 90.9 | 0.999 | |
| TT + TC vs CC | 0.95 | 0.32–2.72 | 0.926 | R | 87.5 | 0.999 | |||
| TT vs TC + CC | 1.14 | 0.46–2.83 | 0.769 | R | 84.7 | 0.999 | |||
| rs1491942 | Overall | 6 | C vs G | 1.25 | 1.10–1.14 | 0.012 | R | 81.7 | 0.548 |
| European | 4 | C vs G | 1.14 | 1.07–1.22 | <0.001 | F | 26.1 | 0.308 | |
| Asian | 2 | C vs G | 1.43 | 0.85–2.42 | 0.181 | R | 95.8 | 0.999 | |
| Mexican Mestizos | 1 | C vs G | 1.69 | 1.29–2.32 | 0.203 | R | ND | ND |
R random effects model, F fixed-effects model.
Meta-analysis: the association of LRRK2 rs1491942 with PD risk
Five studies were included in this meta-analysis (three considered only a European population, one only Asiatic, one European and Asiatic subjects, and one Mexican Mestizo). These studies included 13,117 cases and 10,154 controls. The summary characteristics of the selected studies are shown in Supplementary Table 4. The published data were only enough to evaluate the additive model of this polymorphism. A significant association was observed between LRRK2 rs1491942 and PD in the overall population and for the Caucasian group (Table 4, and Supplementary Figs 7 and 8). The p value of the Begg’s regression test revealed no publication bias p < 0.05 (Table 4).
Discussion
Detection of the genetic susceptibility factors of PD has been the aim of a growing number of investigations. However, as most of these studies focus on European and Asian populations, specific populations such as Mexicans are underrepresented in these findings. Our investigation in a Mexican Mestizo population of known susceptibility factors for PD identified an association for MTHFR rs1801133 and LRRK2 rs1491942 gene variants. These have previously been identified as risk variants for PD in European populations52–55.
The MTHFR gene located on chromosome 1p36.3 synthesizes the homodimeric cytoplasmic flavoprotein methylenetetrahydrofolate reductase. This gene is involved in the metabolism of the amino acids homocysteine and methionine, synthesis of nitrogen bases, methylation processes, and gene regulation56–60. While multiple polymorphisms have been described for MTHFR, the SNP rs1801133, also called C677T, is the most frequently investigated due to its functional impact. This polymorphism is associated with altered folate distribution, which decreases MTHFR enzyme activity in the catalytic region and may increase homocysteine levels57. In patients with PD, the C677T variant has been associated with increased homocysteine levels that precipitate damage mechanisms promoting neurodegeneration. Therefore, this variant is currently a target of PD research; however, the results have been contradictory42,53,58,61–74.
In our overall sample, the allelic and genotype distribution was not found to be associated with PD. However, when stratification by ancestry is performed, the risk association was observed in the group with the highest percentage of European ancestry. This information is consistent with the results of our meta-analysis and coincides with other works that describe a significant PD risk association of the rs1801133 variant in the European population52,53. Although verification is necessary, our findings suggest that heterogeneity in the structure of the subpopulations may explain the differences in findings for SNP rs1801133 in PD studies.
Contrary to our findings, a previous study in Mexican subjects reported the C677 allele as a PD risk factor42. However, the population of their study was limited to subjects from the northeastern and central regions of Mexico; therefore, geographical differences in the contribution of Native American and European ancestry in the Mexican population could explain this discrepancy, as Mexico has been shown to have an increasing north-to-south gradient of Native American ancestry27,28,75. However, as Garcia et al. did not report their sample’s subpopulation structure, this possibility cannot be assessed.
The LRRK2 gene has been widely associated with pathophysiological mechanisms of both familial and sporadic PD76. Based on this gene’s protein sequence, several domains have been identified, such as interaction with other proteins, dimerization, GTPase, and kinase activity. These domains suggest functions in different regulatory mechanisms; cell signaling, protein complex formation, synaptic vesicle trafficking, protein recycling via retrograde trafficking pathways, autophagy regulation, among others54,55,77. Although mutations in this gene are present in 1–13% of PD cases, the role these variants play in the disease is still a subject of research and debate.
Concordant with previous studies78–80, the minor G allele for SNP rs1491942 was identified as a PD risk factor in our cohort. The same effect was observed in the genotype under dominant and recessive models. Our meta-analysis showed a PD risk association for G allele in the overall population (OR 1.25 [1.10–1.41] p 0.012); however, when adjusted for ethnicity, the association was conserved in Caucasian (OR 1.14 [1.07–1.22] p < 0.001) and Mexican populations (OR 1.69 [1.20–2.32] p 0.02), but not for Asians (OR 1.43 [0.84–2.42] p 0.18). Interestingly, subdividing our Mexican Mestizo sample by their percentage of ethnicity revealed a risk association only for the groups with >56% of Native American ascendance (Table 3). The obtained evidence suggests a risk association for rs1491942 and PD in the Mexican population with an even greater effect than previously described in populations of European descent. Although this result will need corroboration, it suggests that polymorphism rs1491942 may be useful as a risk marker of PD in Mexican Mestizos, particularly for subjects with greater Native American ancestry.
The spectrum and frequency of individual variants differ among ethnic groups and geographical locations, making comparisons across populations difficult. Our results highlight the importance of factoring the subpopulation structure into the analysis of genetic factors of PD in ethnically diverse populations. Replication studies must consider these differences when identifying and comparing PD risk factors in distinct populations.
While the moderate number of samples and polymorphisms analyzed are limitations of this work, a significant PD risk association was found for polymorphism rs1491942 in our sample of Mexican Mestizos. However, these differences were dependent on the subject’s percentage of ethnic ancestry.
Other limitations of our work include the moderate number of samples analyzed and low statistical power; replication studies are needed to corroborate these results. To our knowledge, of all published data on PD risk variants in Mexican Mestizo individuals, this is the first study to consider ancestry and includes a greater number of SNPs. Nonetheless, this work’s scope is limited; large-scale genomic studies are needed to map loci and risk variants shared with other groups and identify additional population-specific genetic variations. However, technological, economic, and ethical issues make it difficult to collect sufficient data from underrepresented groups81. On the other hand, the discrepancies between the replication studies conducted in different populations may be attributed to genetic and environmental diversity. While genomic studies contribute greatly to our understanding of complex diseases, they rarely integrate relevant information such as environmental factors (exposure to toxins, lifestyle habits, and nutritional aspects). These factors vary significantly between populations and have been linked to the development of various disorders, including PD82–84. Although case–control association studies of candidate SNPs, such as this one, continue to be a viable option for poorly studied groups with high genetic diversity, the inclusion of environmental factors to the analysis of complex traits is necessary to validate or rectify the role attributed to risk loci identified in other populations.
In summary, our case–control study found PD risk association for the polymorphisms MTHFR rs1801133 and LRKK2 rs1491942 in the sample of Mexican Mestizo subjects. When relevant data from meta-analyses of these two SNPs and the proportion of ethnic ancestry were integrated into analysis, MTHFR rs1801133 was found to confer susceptibility to PD in subjects with a high percentage of European ancestry, and a more significant effect of LRKK2 rs1491942 was detected in Mexican Mestizo individuals with a high percentage of Native American ancestry. The authors consider that the association of these two SNPs and none of the other known PD-related markers derived from European and Asian cohorts merits further investigation into the functional consequences (e.g., changes in gene expression or alterations in protein levels or activity) of these shared risk factors. Identifying and studying the risk factors common to all populations will help elucidate the key biological processes of PD development.
The absence of the remaining 14 PD risk associations in our sample indicates the need for a GWAS of the Mexican population with subpopulation analysis to identify PD-associated variants that are rare in non-European populations and, therefore, not included as known genetic risk factors. Furthermore, identifying differences in LD structure around the causal variants within this population could lead to insights that shed light on the complex role of genetics in this neurological disorder.
Methods
Patients and controls
For the case–control study conducted between 2015 and 2017, 311 subjects were recruited from three hospitals; in the city of Durango, General Hospital 450, and Hospital Santiago Ramón y Cajal ISSSTE, and in Mexico City, General Hospital Dr. Manuel Gea González. To assure representation of Mexican Mestizos, only Spanish-speaking subjects, born in Mexico with Mexican ascendancy (at least parents and grandparents), were considered. The cohort included 118 patients (60 males and 58 females, mean age 69 ± 10 years) diagnosed with PD by an experienced neurologist according to the UK Parkinson’s Disease Society Brain Bank clinical diagnostic criteria, but with no familial history of the disease. The control group consisted of 193 unrelated individuals age- and sex-matched with the PD patients (96 males and 97 females mean age 69 ± 8 years), with no PD diagnosis or a personal or familial history of neurodegenerative diseases.
The subjects’ demographic characteristics, clinical data, and lifestyle were recorded. Their cognitive condition was evaluated with the Mini-Mental State Exam and depression with the Hamilton Depression Rating Scale. The UPDRS score and HY scale were used for determining PD severity.
The internal Ethics and Research Committees of the participating hospitals (no. 49-21-2015/no. Eel-56-2013) approved this study, and it was carried out per the Declaration of Helsinki’s ethical principles for medical research involving human subjects developed by the World Medical Association. All participants gave written informed consent.
Peripheral blood samples were collected from all the subjects. Genomic DNA was isolated from whole blood using a QIAamp DNA extraction Kit (Qiagen, Hilden, Germany). DNA purity and concentration were determined spectrophotometrically, and the samples were stored at −80 °C until use. For biochemical determinations, the blood samples were centrifuged at 3000 r.p.m. for 15 min. The serum and total cholesterol, uric acid, and glucose levels were then quantified using the Random Access Automatic Biochemical Analyzer for Clinical Chemistry and Turbidimetry A15 (BioSystems S.A.).
Selection of single-nucleotide polymorphisms and genotyping
For the selection of variants, a search was performed in the Pubmed database, considering articles published up to September 2014 using the keywords: “Parkinson’s disease,” “Polymorphism, Single Nucleotide,” or/and “Genetic Association Studies,” or/and “Genome-wide Association Study.”
For the final selection, preference was given to variants with a reported contribution to PD’s pathophysiological mechanisms or a PD association identified by GWAS studies (see Supplementary Table 5 for the selected SNPs’ characteristics). The 21 SNPs selected for genotyping were all associated with PD risk in more than one previous study.
Nine of these variants were linked to PD in at least two independent, unrelated cohort studies. They comprise: rs13312, a variant in the 3′-untranslated region of the ubiquitin-specific protease 24 gene (USP24), associated with (PARK10)85,86, a susceptibility locus for PD; rs3766606, an intronic modification in the deglycase gene (PARK7), linked with parkinsonism in Chinese and European populations87,88; rs1801474 and rs1801582, two missense variants in the parkin RBR E3 ubiquitin-protein ligase gene (PRKN), associated with PD risk89–91; rs1800497, often correlated with neurological disorders, including PD, is located in the coding region of the ankyrin repeat and kinase domain containing the gene (ANKK1), which controls dopamine synthesis in the brain50,92,93; rs1801133, in the methylenetetrahydrofolate reductase gene (MTHFR), possibly implicated in PD61,65; rs334558, a polymorphism in the glycogen synthase kinase 3 beta gene (GSK3B) potentially a protective factor for PD in Asian populations94,95; rs6280, in the dopamine receptor D3 gene (DRD3), implicated in both PD vulnerability and motor complications96,97; and rs242562, a polymorphism in the microtubule-associated protein tau gene (MAPT), associated with PD98.
The 12 additional variants were selected because of their PD association reported in complete genome studies (GWAS). These include: rs823128, rs823156, and rs947211, three variants vinculated with the susceptibility locus PARK16 in Asian and European populations99,100; rs2736990, rs356220, and rs356219, polymorphisms in the synuclein alpha gene (SNCA), proposed as PD risk factors in Asian and European populations100–103; rs1491942, rs33949390, and rs34778348, variants in the LRRK2 gene, frequently associated with PD worldwide104; and the variants, rs6812193 in FAM47E, associated with a significant risk of developing PD; and rs11868035 in SREBF1 (refs. 78,104).
Genotyping was carried out using predesigned TaqMan SNP Genotyping Assays (by Applied Biosystem CA) with the following assay ids, SNP id and gene name (C_7516392_10, rs1491942; C__63497592_10, rs33949390; C__63498855_10, rs34778348 C_1867882_10, rs1994090 LRRK2; C_1020193_10, rs356219; C_3208948_10 rs2736990; C_1020192_10, rs356220 SNCA, C_8701299_10, rs1801582; C_8947865_10, rs1801474 PRKN; C_2966873_10, rs3766606 PARK7; C_998739_10, rs13312 USP24; C_31139749_10, rs6812193 FAM47E; C_949770_10, rs6280, DRD3; C_3224431_10, rs1800497,ANKK1; C_3202957_10, rs242562, MAPT; C_375742_10, rs823156; C_8721272_10, rs947211, SLC41A1; C_1202883_20, rs1801133 MTHFR; C_31463202_10, rs11868035 SREBF1; C_11451241_10, rs823128 NUCKS1; and C_905680_10, rs334558 GSK3B).
The real-time polymerase chain reaction (PCR) with allelic discrimination analysis was performed according to the standard protocol. Briefly, 10 ng of genomic DNA mixed with 0.625 µL of Taqman SNP genotyping assay and 5 µL of Universal PCR Master Mix (Applied Biosystem CA) adjusted with nuclease-free water for a final volume of 20 µL per well. The mix was added to a 48-well plate and amplified 40 cycles in a StepOne machine (Applied Biosystems, Foster City, CA USA). All subjects were genotyped; 10% of the assays were randomly selected for replication, and these tests were all consistent with our initial results.
A validated panel of 32 AIMs designed for Mexican individuals was used for the stratification correction and estimation of global ancestry51. The SNP genotyping assays were generated with the OpenArray® platform by Quantstudio™ (Applied Biosystem, CA), per the manufacturer’s recommendations.
The comparative analysis was performed using ADMIXTURE software set at k = 2 to discriminate between European and Native American ancestries. A dataset of 95 non-related individuals from the European Utah population (CEU) plus 38 individuals of Mayan or Zapoteca origin was included to represent the parental populations51.
Statistical analysis
Since study participants were recruited from both Northeastern Mexico and Mexico City, differences in the demographic and clinical characteristics potentially attributable to the place of recruitment were compared using the Mann–Whitney test for continuous variables and Fisher’s exact test for the categorical variables. When comparing differences in demographics and clinical characteristics between PD cases and controls, either a Student’s t test or Mann–Whitney test was used depending on the distribution of the continuous variables. Chi-square and Fisher’s exact tests were used to assess differences between groups for categorical variables. For association analysis of the SNPs, we defined the ancestral alleles as the major allele, i.e., higher in frequency (according to the National Center for Biotechnology Information SNP database). The HWE of the control group was verified with a chi-square test. Linkage disequilibrium was examined using Haploview software (Broad Institute, Cambridge, MA, USA) and genotyping data from the 1000 Genomes project. Differences in genotyping and allelic frequency distribution between cases and controls were compared, using the Fisher exact test. Each SNP’s association with PD was evaluated using logistic regression models adjusted for age, sex, and percentage of Native American ancestry. OR and 95% CI were calculated for the associations, and p values reported. To counteract the problem of multiple comparisons, the Bonferroni correction was used to test the “universal null hypothesis”, i.e., that all tests are not significant. The threshold for statistical significance after this correction was p < 0.003.
To determine whether the subpopulation structure factored into the incidence of PD-associated SNPs, the differences were calculated considering each subject’s percentage of Native American ancestry. The sample was divided into quartiles. Group 1 had the lowest percentage of Native American ancestry ranging from 32 to 52%, group 2 contained 52.1–58.5%, group 3 from 56.6 to 65%, and group 4 had the highest percentage ≥66%. The association with PD was evaluated for these groups using stratified logistic regression models adjusted for age and sex; p values <0.05 were considered statistically significant.
Furthermore, meta-analyses were carried out for the SNPs shown to have a significant association with PD. The following search criteria were used to identify related studies in the PUBMED and ScienceDirect databases: papers published before July 2019 using the keywords: Parkinson’s disease (PD) and SNP (or polymorphism or mutation or variant) rs1801133 (or C677T or Ala222Val and MTHFR) and/or rs1491942 (and LRRK2). In addition, potentially relevant literature was identified from the reference section of related studies. The following selection criteria were used: (1) human case–control design, (2) evaluation of genetic susceptibility to PD, (3) OR reported with 95% confidence interval, or enough data to estimate the OR, and (4) English language publication. Exclusion criteria were: (1) duplicate studies, (2) animal studies, case reports, and conference abstracts, (3) only familial PD research, and (4) evaluation of the associations between SNPs and PD therapy response or prognosis. Also, the selected studies’ quality was assessed using the Newcastle–Ottawa Scale105. Studies were scored independently by two reviewers, and articles with scores <5 were discarded. The associations between polymorphism rs1491942 in the LRRK2 gene and PD susceptibility were estimated based on pooled ORs and 95% CI. The p value of Cochran’s Q statistic was evaluated to determine heterogeneity. If p < 0.10 or I² > 50, a random effects model was used; in the absence of heterogeneity, a fixed-effects model was used. The Z test was used to determine if the OR was significant, and a p value <0.05 was considered statistically significant. Publication bias was determined using a Begg’s linear regression test; a p value <0.05 was considered evidence of bias. A sensitivity analysis was also performed. The STATA software (version 13.0; STATA Corporation, USA) was used for all specified statistical analyzes.
Supplementary information
Acknowledgements
This study was supported by a grant awarded to Oscar Arias-Carrión by CONACyT (FOSISS 2015:262327). Elizabeth Romero-Gutiérrez is a doctoral student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and is the recepiente of CONACyT fellowship 254033. We thank Ma. Luisa Ordoñez-Sánchez for technical assistance.
Author contributions
E.R.-G.: research project execution and data acquisition; statistical analysis execution; first draft of the manuscript, and final approval. P.V.-C., H.M.-M., J.S.-P., and T.T.-L.: data acquisition; review and critique of the statistical analysis; review, critique and final approval of the manuscript. O.A.-C.: conception and organization of the research project; review and critique of the statistical analysis; writing, editing, critique, and final approval of the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-021-00157-y.
References
- 1.Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 2.Dorsey ER, Bloem BR. The Parkinson pandemic-A call to action. JAMA Neurol. 2018;75:9–10. doi: 10.1001/jamaneurol.2017.3299. [DOI] [PubMed] [Google Scholar]
- 3.Zesiewicz TA. Parkinson disease. Continuum. 2019;25:896–918. doi: 10.1212/CON.0000000000000764. [DOI] [PubMed] [Google Scholar]
- 4.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 5.Lill CM, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma M, et al. Large-scale replication and heterogeneity in Parkinson disease genetic loci. Neurology. 2012;79:659–667. doi: 10.1212/WNL.0b013e318264e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalls MA, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang D, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat. Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwaki H, et al. Genetic risk of Parkinson disease and progression:: an analysis of 13 longitudinal cohorts. Neurol. Genet. 2019;5:e348. doi: 10.1212/NXG.0000000000000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nalls MA, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grenn, F. P. et al. The Parkinson’s disease genome-wide association study locus browser. Mov. Disord.35, 2056–2067 (2020). [DOI] [PMC free article] [PubMed]
- 12.Foo, J. N. et al. Identification of risk loci for Parkinson disease in Asians and comparison of risk between Asians and Europeans: a genome-wide association study. JAMA Neurol.77, 746–754 (2020). [DOI] [PMC free article] [PubMed]
- 13.Marras C, et al. Nomenclature of genetic movement disorders: Recommendations of the international Parkinson and movement disorder society task force. Mov. Disord. Off. J. Mov. Disord. Soc. 2016;31:436–457. doi: 10.1002/mds.26527. [DOI] [PubMed] [Google Scholar]
- 14.Karimi-Moghadam A, Charsouei S, Bell B, Jabalameli MR. Parkinson disease from Mendelian forms to genetic susceptibility: new molecular insights into the neurodegeneration process. Cell. Mol. Neurobiol. 2018;38:1153–1178. doi: 10.1007/s10571-018-0587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumaran R, Cookson MR. Pathways to Parkinsonism Redux: convergent pathobiological mechanisms in genetics of Parkinson’s disease. Hum. Mol. Genet. 2015;24:R32–R44. doi: 10.1093/hmg/ddv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte C, Gasser T. Genetic basis of Parkinson’s disease: inheritance, penetrance, and expression. application Clin. Genet. 2011;4:67–80. doi: 10.2147/TACG.S11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labbe C, Lorenzo-Betancor O, Ross OA. Epigenetic regulation in Parkinson’s disease. Acta Neuropathol. 2016;132:515–530. doi: 10.1007/s00401-016-1590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari R, et al. Stratification of candidate genes for Parkinson’s disease using weighted protein-protein interaction network analysis. BMC Genomics. 2018;19:452. doi: 10.1186/s12864-018-4804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunati A, Lesage S, Brice A. The genetic landscape of Parkinson’s disease. Rev. Neurol. 2018;174:628–643. doi: 10.1016/j.neurol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:1080. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 21.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHugh C, Brown L, Thornton TA. Detecting heterogeneity in population structure across the genome in admixed populations. Genetics. 2016;204:43–56. doi: 10.1534/genetics.115.184184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet. 2019;20:520–535. doi: 10.1038/s41576-019-0144-0. [DOI] [PubMed] [Google Scholar]
- 26.Conomos MP, et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic community health study/study of Latinos. Am. J. Hum. Genet. 2016;98:165–184. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva-Zolezzi I, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc. Natl Acad. Sci. USA. 2009;106:8611–8616. doi: 10.1073/pnas.0903045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubi-Castellanos R, et al. Pre-Hispanic Mesoamerican demography approximates the present-day ancestry of Mestizos throughout the territory of Mexico. Am. J. Phys. Anthropol. 2009;139:284–294. doi: 10.1002/ajpa.20980. [DOI] [PubMed] [Google Scholar]
- 29.Sesar A, et al. Synaptotagmin XI in Parkinson’s disease: new evidence from an association study in Spain and Mexico. J. Neurol. Sci. 2016;362:321–325. doi: 10.1016/j.jns.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Garcia S, et al. The rs3857059 variant of the SNCA gene is associated with Parkinson’s disease in Mexican Mestizos. Arq. Neuropsiquiatr. 2016;74:445–449. doi: 10.1590/0004-282x20160061. [DOI] [PubMed] [Google Scholar]
- 31.Salas-Leal AC, et al. rs3764435 associated With Parkinson’s disease in Mexican Mestizos: case-control study reveals protective effects against disease development and cognitive impairment. Front Neurol. 2019;10:1066. doi: 10.3389/fneur.2019.01066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia S, et al. Low prevalence of most frequent pathogenic variants of six PARK genes in sporadic Parkinson’s disease. Folia Neuropathol. 2014;52:22–29. doi: 10.5114/fn.2014.41741. [DOI] [PubMed] [Google Scholar]
- 33.Yescas P, et al. Low frequency of common LRRK2 mutations in Mexican patients with Parkinson’s disease. Neurosci. Lett. 2010;485:79–82. doi: 10.1016/j.neulet.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Del Rincon Mde L, et al. The L444P GBA mutation is associated with early-onset Parkinson’s disease in Mexican Mestizos. Clin. Genet. 2013;84:386–387. doi: 10.1111/cge.12084. [DOI] [PubMed] [Google Scholar]
- 35.Guerrero Camacho JL, et al. High frequency of Parkin exon rearrangements in Mexican-mestizo patients with early-onset Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2012;27:1047–1051. doi: 10.1002/mds.25030. [DOI] [PubMed] [Google Scholar]
- 36.Miranda-Morales EG, et al. H1/H2 MAPT haplotype and Parkinson’s disease in Mexican mestizo population. Neurosci. Lett. 2019;690:210–213. doi: 10.1016/j.neulet.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 37.Monroy-Jaramillo N, et al. Genetic mutations in early-onset Parkinson’s disease Mexican patients: molecular testing implications. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2014;165B:235–244. doi: 10.1002/ajmg.b.32228. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez-Jirano LJ, et al. [Frequency of the IVS4+66A-G polymorphism in the alpha-synuclein gene in patients with Parkinson’s disease in north-western Mexico] Rev. Neurol. 2007;44:15–17. [PubMed] [Google Scholar]
- 39.Davila-Ortiz de Montellano DJ, Rodriguez-Violante M, Fresan A, Monroy-Jaramillo N, Yescas-Gomez P. [Frequency of single nucleotide polymorphisms and alpha-synuclein haplotypes associated with sporadic Parkinson’s disease in the Mexican population] Rev. Neurol. 2016;63:345–350. [PubMed] [Google Scholar]
- 40.Martinez Saucedo O, et al. Detencion de polimorfismos en el gen PARKIN como biomarcadores predictivos de la enfermedad de Parkinson. Rev. Mexi. Neuroci. 2004;5:7–12. [Google Scholar]
- 41.Davila-Ortiz DM, Monroy-Jaramillo N, Rodriguez-Violante M, Lopez-Lopez M, Yescas-Gomez Detección de mutaciones puntuales en el gen de alfa-sinucleína en pacientes mexicanos con enfermedad de Parkinson y herencia autosómica dominante, i. Arch. Neurosci. 2012;2012:17–21. [Google Scholar]
- 42.Garcia S, et al. Association of the rs1801133 variant in the MTHFR gene and sporadic Parkinson’s disease. Folia Neuropathol. 2015;53:24–28. doi: 10.5114/fn.2015.49971. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz-Sanchez E, et al. Association of polymorphisms and reduced expression levels of the NR4A2 gene with Parkinson’s disease in a Mexican population. J. Neurol. Sci. 2017;379:58–63. doi: 10.1016/j.jns.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 44.Garcia S, et al. Association of mitochondrial variants A4336G of the tRNAGln gene and 8701G/A of the MT-ATP6 gene in Mexicans Mestizos with Parkinson disease. Folia Neuropathol. 2019;57:335–339. doi: 10.5114/fn.2019.89859. [DOI] [PubMed] [Google Scholar]
- 45.Gallegos-Arreola MP, et al. Apolipoprotein E genotypes in Mexican patients with Parkinson’s disease. Dis. Markers. 2009;27:225–230. doi: 10.1155/2009/617863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez M, et al. Apolipoprotein E epsilon4 allele is associated with Parkinson disease risk in a Mexican Mestizo population. Mov. Disord. Off. J. Mov. Disord. Soc. 2007;22:417–420. doi: 10.1002/mds.21340. [DOI] [PubMed] [Google Scholar]
- 47.Cervantes-Arriaga A, et al. [ApoE polymorphisms and dopaminergic replacement therapy in Parkinson’s disease] Rev. Med. Inst. Mex. Seguro Soc. 2014;52:14–18. [PubMed] [Google Scholar]
- 48.Garcia S, et al. Analysis of the rs13306560 functional variant in the promoter region of the MTHFR gene in sporadic Parkinson s disease. Neuro Endocrinol. Lett. 2017;38:257–260. [PubMed] [Google Scholar]
- 49.Ramirez-Jirano LJ, et al. [116C-G polymorphism of the alpha-synuclein gene in patients with Parkinson disease] Salud Publica Mex. 2006;48:289–290. doi: 10.1590/S0036-36342006000400001. [DOI] [PubMed] [Google Scholar]
- 50.Cervantes-Arriaga Amin, R-VM, Davila Ortíz de Montellano David, Petra Yescas, Elisa. Alonso-Vilatela. Relación entre el polimorfismo DRD2/ANKK1 y el desarrollo de complicaciones motoras en enfermedad de Parkinson. Neurologia Argent. 2014;7:28–33. doi: 10.1016/j.neuarg.2014.09.001. [DOI] [Google Scholar]
- 51.Huerta-Chagoya A, et al. A panel of 32 AIMs suitable for population stratification correction and global ancestry estimation in Mexican mestizos. BMC Genet. 2019;20:5. doi: 10.1186/s12863-018-0707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y, Zhu RX, He ZY, Liu X, Liu HN. Association of MTHFR C677T with total homocysteine plasma levels and susceptibility to Parkinson’s disease: a meta-analysis. Neurol. Sci. 2015;36:945–951. doi: 10.1007/s10072-014-2052-6. [DOI] [PubMed] [Google Scholar]
- 53.Liu L, et al. MTHFR C677T and A1298C polymorphisms may contribute to the risk of Parkinson’s disease: a meta-analysis of 19 studies. Neurosci. Lett. 2018;662:339–345. doi: 10.1016/j.neulet.2017.10.060. [DOI] [PubMed] [Google Scholar]
- 54.Paisan-Ruiz C, Lewis PA, Singleton AB. LRRK2: cause, risk, and mechanism. J. Parkinsons Dis. 2013;3:85–103. doi: 10.3233/JPD-130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rideout HJ, Stefanis L. The neurobiology of LRRK2 and its role in the pathogenesis of Parkinson’s disease. Neurochem. Res. 2014;39:576–592. doi: 10.1007/s11064-013-1073-5. [DOI] [PubMed] [Google Scholar]
- 56.Goyette P, et al. Human methylenetetrahydrofolate reductase: isolation of cDNA, mapping and mutation identification. Nat. Genet. 1994;7:195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- 57.Frosst P, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 58.Yasui K, Kowa H, Nakaso K, Takeshima T, Nakashima K. Plasma homocysteine and MTHFR C677T genotype in levodopa-treated patients with PD. Neurology. 2000;55:437–440. doi: 10.1212/WNL.55.3.437. [DOI] [PubMed] [Google Scholar]
- 59.Liew SC, Gupta ED. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases. Eur. J. Med. Genet. 2015;58:1–10. doi: 10.1016/j.ejmg.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Miranda-Morales E, et al. Implications of DNA methylation in Parkinson’s disease. Front. Mol. Neurosci. 2017;10:225. doi: 10.3389/fnmol.2017.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan L, et al. Association of the MTHFR rs1801131 and rs1801133 variants in sporadic Parkinson’s disease patients. Neurosci. Lett. 2016;616:26–31. doi: 10.1016/j.neulet.2016.01.031. [DOI] [PubMed] [Google Scholar]
- 62.Wullner U, Kolsch H, Linnebank M. Methylenetetrahydrofolate reductase in Parkinson’s disease. Ann. Neurol. 2005;58:972–973. doi: 10.1002/ana.20696. [DOI] [PubMed] [Google Scholar]
- 63.Bialecka M, et al. Association of COMT, MTHFR, and SLC19A1(RFC-1) polymorphisms with homocysteine blood levels and cognitive impairment in Parkinson’s disease. Pharmacogenet. Genomics. 2012;22:716–724. doi: 10.1097/FPC.0b013e32835693f7. [DOI] [PubMed] [Google Scholar]
- 64.Caccamo D, et al. Effect of MTHFR polymorphisms on hyperhomocysteinemia in levodopa-treated Parkinsonian patients. Neuromolecular Med. 2007;9:249–254. doi: 10.1007/s12017-007-8006-x. [DOI] [PubMed] [Google Scholar]
- 65.de Lau LM, et al. Methylenetetrahydrofolate reductase C677T genotype and PD. Ann. Neurol. 2005;57:927–930. doi: 10.1002/ana.20509. [DOI] [PubMed] [Google Scholar]
- 66.Dorszewska J, et al. Oxidative DNA damage and level of thiols as related to polymorphisms of MTHFR, MTR, MTHFD1 in Alzheimer’s and Parkinson’s diseases. Acta Neurobiol. Exp. 2007;67:113–129. doi: 10.55782/ane-2007-1639. [DOI] [PubMed] [Google Scholar]
- 67.Fong CS, et al. Association of MTHFR, MTR, and MTRR polymorphisms with Parkinson’s disease among ethnic Chinese in Taiwan. Clin. Chim. Acta. 2011;412:332–338. doi: 10.1016/j.cca.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Gorgone G, et al. Coenzyme Q10, hyperhomocysteinemia and MTHFR C677T polymorphism in levodopa-treated Parkinson’s disease patients. Neuromolecular Med. 2012;14:84–90. doi: 10.1007/s12017-012-8174-1. [DOI] [PubMed] [Google Scholar]
- 69.Kumudini N, et al. Association of seven functional polymorphisms of one-carbon metabolic pathway with total plasma homocysteine levels and susceptibility to Parkinson’s disease among South Indians. Neurosci. Lett. 2014;568:1–5. doi: 10.1016/j.neulet.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 70.Religa D, et al. Hyperhomocysteinemia and methylenetetrahydrofolate reductase polymorphism in patients with Parkinson’s disease. Neurosci. Lett. 2006;404:56–60. doi: 10.1016/j.neulet.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez-Oroz MC, et al. Homocysteine and cognitive impairment in Parkinson’s disease: a biochemical, neuroimaging, and genetic study. Mov. Disord. Off. J. Mov. Disord. Soc. 2009;24:1437–1444. doi: 10.1002/mds.22522. [DOI] [PubMed] [Google Scholar]
- 72.Todorovic Z, et al. Homocysteine serum levels and MTHFR C677T genotype in patients with Parkinson’s disease, with and without levodopa therapy. J. Neurol. Sci. 2006;248:56–61. doi: 10.1016/j.jns.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 73.Yuan RY, et al. Methylenetetrahydrofolate reductase polymorphisms and plasma homocysteine in levodopa-treated and non-treated Parkinson’s disease patients. J. Neurol. Sci. 2009;287:64–68. doi: 10.1016/j.jns.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 74.Zahra C, et al. Genetic causes of Parkinson’s disease in the Maltese: a study of selected mutations in LRRK2, MTHFR, QDPR and SPR. BMC Med. Genet. 2016;17:65. doi: 10.1186/s12881-016-0327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romero-Hidalgo S, et al. Demographic history and biologically relevant genetic variation of Native Mexicans inferred from whole-genome sequencing. Nat. Commun. 2017;8:1005. doi: 10.1038/s41467-017-01194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tan EK, et al. Multiple LRRK2 variants modulate risk of Parkinson disease: a Chinese multicenter study. Hum. Mutat. 2010;31:561–568. doi: 10.1002/humu.21225. [DOI] [PubMed] [Google Scholar]
- 77.Redensek S, Trost M, Dolzan V. Genetic determinants of Parkinson’s disease: can they help to stratify the patients based on the underlying molecular defect? Front. Aging Neurosci. 2017;9:20. doi: 10.3389/fnagi.2017.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soto-Ortolaza AI, et al. GWAS risk factors in Parkinson’s disease: LRRK2 coding variation and genetic interaction with PARK16. Am. J. Neurodegener. Dis. 2013;2:287–299. [PMC free article] [PubMed] [Google Scholar]
- 79.Pihlstrøm L, et al. Supportive evidence for 11 loci from genome-wide association studies in Parkinson’s disease. Neurobiol. Aging. 2013;34:e1707–e1713. doi: 10.1016/j.neurobiolaging.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 80.Fang J, et al. Analysis of LRRK2, SNCA, and ITGA8 gene variants with sporadic Parkinson’s disease susceptibility in Chinese Han population. Parkinsons Dis. 2016;2016:3474751. doi: 10.1155/2016/3474751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McGuire, A. L. et al. The road ahead in genetics and genomics. Nat. Rev. Genet.21, 581–596 (2020). [DOI] [PMC free article] [PubMed]
- 82.Georgiou A, et al. Genetic and environmental factors contributing to Parkinson’s disease: a case-control study in the Cypriot population. Front. Neurol. 2019;10:1047. doi: 10.3389/fneur.2019.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsuboi Y. Environmental-genetic interactions in the pathogenesis of Parkinson’s disease. Exp. Neurobiol. 2012;21:123–128. doi: 10.5607/en.2012.21.3.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Priyadarshi A, Khuder SA, Schaub EA, Priyadarshi SS. Environmental risk factors and Parkinson’s disease: a metaanalysis. Environ. Res. 2001;86:122–127. doi: 10.1006/enrs.2001.4264. [DOI] [PubMed] [Google Scholar]
- 85.Li Y, et al. Genetic evidence for ubiquitin-specific proteases USP24 and USP40 as candidate genes for late-onset Parkinson disease. Hum. Mutat. 2006;27:1017–1023. doi: 10.1002/humu.20382. [DOI] [PubMed] [Google Scholar]
- 86.Haugarvoll K, et al. Fine-mapping and candidate gene investigation within the PARK10 locus. Eur. J. Hum. Genet. 2009;17:336–343. doi: 10.1038/ejhg.2008.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Marco EV, et al. DJ-1 is a Parkinson’s disease susceptibility gene in southern Italy. Clin. Genet. 2010;77:183–188. doi: 10.1111/j.1399-0004.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- 88.Chen, W. et al. [Association of the DJ-1 gene polymorphism with sporadic Parkinson’s disease in Sichuan province of China]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi25, 566–569 (2008). [PubMed]
- 89.Peng R, et al. Mutation screening and association analysis of the parkin gene in Parkinson’s disease patients from South-West China. Eur. Neurol. 2003;49:85–89. doi: 10.1159/000068505. [DOI] [PubMed] [Google Scholar]
- 90.Sanyal J, et al. Evaluation of PARKIN gene variants in West Bengal Parkinson’s disease patients. J. Hum. Genet. 2015;60:485–492. doi: 10.1038/jhg.2015.49. [DOI] [PubMed] [Google Scholar]
- 91.Biswas A, et al. Parkin polymorphisms: risk for Parkinson’s disease in Indian population. Clin. Genet. 2007;72:484–486. doi: 10.1111/j.1399-0004.2007.00878.x. [DOI] [PubMed] [Google Scholar]
- 92.McGuire V, et al. Association of DRD2 and DRD3 polymorphisms with Parkinson’s disease in a multiethnic consortium. J. Neurol. Sci. 2011;307:22–29. doi: 10.1016/j.jns.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dai D, et al. Polymorphisms of. Biomed. Rep. 2014;2:275–281. doi: 10.3892/br.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao DM, et al. GSK3β reduces risk of sporadic Parkinson’s disease in ethnic Chinese. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B:718–721. doi: 10.1002/ajmg.b.32075. [DOI] [PubMed] [Google Scholar]
- 95.Kwok JB, et al. GSK3B polymorphisms alter transcription and splicing in Parkinson’s disease. Ann. Neurol. 2005;58:829–839. doi: 10.1002/ana.20691. [DOI] [PubMed] [Google Scholar]
- 96.Hassan A, et al. Association of Parkinson disease age of onset with DRD2, DRD3 and GRIN2B polymorphisms. Parkinsonism Relat. Disord. 2016;22:102–105. doi: 10.1016/j.parkreldis.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajan R, Krishnan S, Sarma G, Sarma SP, Kishore A. Dopamine receptor D3 rs6280 is associated with aberrant decision-making in Parkinson’s disease. Mov. Disord. Clin. Pract. 2018;5:413–416. doi: 10.1002/mdc3.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu L, et al. MAPT rs242562 and GSK3B rs334558 are associated with Parkinson’s disease in central China. BMC Neurosci. 2014;15:54. doi: 10.1186/1471-2202-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Satake W, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 100.Simón-Sánchez J, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hill-Burns EM, et al. Identification of a novel Parkinson’s disease locus via stratified genome-wide association study. BMC Genomics. 2014;15:118. doi: 10.1186/1471-2164-15-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Edwards TL, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simón-Sánchez J, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur. J. Hum. Genet. 2011;19:655–661. doi: 10.1038/ejhg.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Do CB, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wells, G. S. B., O’Connell, D., Peterson, J., Welch, V. & Losos M. T. Ottawa: the Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Ottawa Hospital Research Institute http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.